Abstract
Dynamically regulated changes in chromatin states are vital for normal development and can produce disease when they go awry. Accordingly, much effort has been devoted to characterizing these states under normal and pathological conditions. Chromatin immunoprecipitation followed by sequencing (ChIP-seq) is the most widely used method to characterize where in the genome transcription factors, modified histones, modified nucleotides and chromatin binding proteins are found; bisulfite sequencing (BS-seq) and its variants are commonly used to characterize the locations of DNA modifications. Though very powerful, these methods are not without limitations. Notably, they are best at characterizing one chromatin feature at a time, yet chromatin features arise and function in combination. Investigators commonly superimpose separate ChIP-seq or BS-seq datasets, and then infer where chromatin features are found together. While these inferences might be correct, they can be misleading when the chromatin source has distinct cell types, or when a given cell type exhibits any cell to cell variation in chromatin state. These ambiguities can be eliminated by robust methods that directly characterize the existence and genomic locations of combinations of chromatin features in very small inputs of cells or ideally, single cells. Here we review single molecule epigenomic methods under development to overcome these limitations, the technical challenges associated with single molecule methods and their potential application to single cells.
Keywords: Single cell, single molecule, epigenomics, epigenetics, microfluidics, nanofluidics
1. Epigenetics and Epigenomics
The term “epigenetics” was originally coined by Conrad Waddington [1], and can be defined as the heritable changes in traits that occur without direct alteration of the underlying primary DNA sequence. At the molecular level, epigenetic phenomena are regulated by so-called epigenetic marks that include covalent changes to DNA and chromatin proteins, and remodeling of nucleosome position on the DNA. These are often controlled by non-coding RNAs and can respond to environmental influences. The epigenome refers to the constellation of epigenetic marks that exist in a cell and their chromosomal locations, which varies by cell type and developmental stage. Normal epigenomic states are important for cell-type specific gene expression and a variety of cellular and developmental processes. In humans, the aberrant placement of epigenetic marks has been linked to many diseases, including cancer (reviewed in [2]). The best studied DNA modification in mammals is 5-methylcytosine (5mC), which plays an important role in genomic imprinting, and the suppression of transposable elements [3]. Recently, it has been shown that 5mC can be converted to 5-hydroxymethylcytosine (5hmC) by the TET (Ten-Eleven-Translocation) family of proteins [4]. In addition to converting 5mC to 5hmC, TET proteins can catalyze further hydroxylation of 5hmC to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) in an enzymatic activity-dependent manner [5, 6]. Numerous histone modifications exist primarily on the protruding amino-terminal tails of histones [7]; these include methylation (mono-, di-, trimethylation), acylation, phosphorylation, ubiquitylation, sumoylation, ADP ribosylation, carboxylation, glutathionylation, O-GlcNAcylation, hydroxylation, formylation and citrullination. The covalent modifications are placed by “writer” activities (reviewed in [8–11]), removed by “eraser” activities (reviewed in [12–14]). They exert their cellular effects by altering chromatin structure and by recruiting “reader” factors (reviewed in [15, 16]), which in turn recruit additional factors necessary for epigenetically regulated processes.
2. Epigenomic methods
Characterization of epigenomes has been greatly enhanced by use of various high-throughput sequencing technologies, which have been used to construct detailed epigenomic landscapes in many systems. The currently used, highest resolution methods include bisulfite sequencing (BS-seq) to assess 5mC [17–20], variants of BS-seq to assess 5hmC [21, 22], and chromatin immunoprecipitation followed by sequencing (ChIP-seq) for the analysis of histone modifications and chromatin binding proteins [23–33]. Additional methods have been developed to assess epigenetically influenced chromatin states diagnostic for transcriptionally regulatory domains including DNAse-seq [34], FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) [35] and Sono-seq [36]. Among the technical challenges associated with these methods are biased amplification of bisulfite treated DNAs [37, 38] and the problem of antibody quality for ChIP studies. Antibody specificity can limit ChIP-seq accuracy and reproducibility. Standards have been established to validate antibody quality [32, 39] that include blotting approaches. Semi-synthetic DNA-barcoded nucleosome libraries may also prove useful [40] for antibody validation, and emerging microfluidic-based platforms [41] and the Nanostring nCounter platform [42] have the potential for streamlining this process. Synthetic affinity reagents [43] may ultimately replace antibodies that have batch-to-batch variation. Two issues that are more fundamental than these technical challenges are (1) that the cited epigenomic methods generally provide information about a single epigenetic feature at a time, rather than the combinations of epigenomic features that exist in the genome; and (2) that they work best with an abundance of materials, and not as well with inputs from few or even single cells. The remainder of the discussion focuses on why these are limitations, and approaches under development to overcome them.
3. Combinatorial Chromatin States
In describing the histone code hypothesis, Strahl and Allis suggested “that multiple histone modifications, acting in a combinatorial or sequential fashion on one or multiple histone tails, specify unique downstream functions” [44]. The strongest support for this at the time was provided by observations that the N-terminal tails of histones H3 and H4 contain multiple sites where covalent modifications may be placed. Since then, many lines of evidence lent support for the histone code hypothesis, and in particular, that combinations of chromatin modifications, and not just single modifications alone, are critical for genomic regulation. First, mass spectrometric analysis revealed that individual histones do in fact carry multiple modifications on their N-terminal tails [45, 46]. Second, plant homeodomain (PHD) and bromodomain (BRD) motifs within so-called “reader” proteins that bind to specific modified histone amino acids, bind their targets in a manner that is sensitive to the modification states of nearby amino acids [47]. The histone acetyltransferase p300 in particular was shown to display this property [40]. Third, the reader protein Bromodomain PHD-finger Transcription Factor (BPTF), which has a methyl lysine-binding PHD motif adjacent to an acetyl lysine-binding BRD motif, was shown to bind H3K4me2/3 containing nucleosomes preferentially when the nucleosome also carried H4K16ac [48]. The observation that modifications on two independent histone molecules together affected BPTF binding highlights the fact that biologically relevant combinatorial states are not limited to those in cis on a single histone molecule. Fourth, combinations of histone modifications influence the biochemical activities of factors that bind and further modify histones. For example, the demethylase KDM7A that targets methylated forms of H3K9 and H3K27 for demethylation [49] contains a PHD motif that binds H3K4me3, suggesting that KDM7A is directed to its H3K9me and H3K27me targets in chromatin by adjacent H3K4me3 [50].
The histone code hypothesis can be extended to include effects coordinated with DNA modifications, as the combined importance of DNA and histone modifications to gene expression has been documented. The NuRD complex contains methyl binding domain (MBD) proteins, which bind 5mC and 5hmC, histone deacetylases (HDAC) and chromatin remodeling activity [51]. Gene silencing by HDAC activity in these complexes is enabled by MBD recruitment of the complex to modified DNA [52]. Given the cross talk among chromatin modifications, it should come as no surprise that their effects are coordinated by mechanisms that sense the modifications in combination. As the number of known reader proteins [53] and chromatin modifications [6, 54] increases, so does the potential complexity of the histone code, or more broadly, the chromatin code. These trends elevate the importance of identifying and mapping the genomic locations of combinations of chromatin features in order to understand how those features regulate genomic information in normal and disease states.
4. Technologies that overcome some limitations of ChIP-seq and BS-seq
4.1. Re-ChIP and ChIP-BS-seq
The most widely used ChIP protocols query chromatin sources for chromatin features one at a time. Several sets of efforts have characterized where in the genome combinations of chromatin features can be found. One of these used sequential- or re-ChIP experiments, whereby chromatin immunoprecipitated with a first antibody was subjected to re-precipitation with a second antibody before analyzing the DNA [55–62]. In one application of re-ChIP, a bivalent state comprising H3K4me3 and H3K27me3 modifications at genes important for lineage specification was found in pluripotent stem cells [63]; in another application, histone variants H3.3 and H2AZ were found together on active promoters, enhancers and insulator regions [64]. Re-ChIP methods require large inputs of chromatin, given the inefficiencies with which each antibody precipitates the chromatin, and in some cases, the low abundance of the chromatin feature. There are few examples of whole genome re-ChIP studies. Studies with more than two sequential ChIP reactions will likely require antibodies or other affinity reagents with dissociation constants well below those of existing reagents in order to have high enough modification capture efficiencies; small reaction volumes that enable use of high concentrations of chromatin and capture reagents; and improvements in library preparation or sequencing methods that make most efficient use of the DNA isolated by ChIP.
In other efforts to define coincidence between 5mC and H3K27me3, DNA isolated by anti H3K27me3 ChIP was subjected to bisulfite sequencing [65]. In principle, this strategy can be applied to any DNAs isolated from a single or re-ChIP experiment, if sufficient amounts of DNA are recovered. Single molecule sequencing technologies have enabled the identification of DNA modifications, including 5mC and 5hmC without bisulfite and other chemistries (see below [66–68]). These provide an alternate means of characterizing the sequences and DNA modification states of chromatin isolated by ChIP.
4.2. Statistical methods
With high quality, high coverage sequencing results in hand, the workflow for ChIP-seq data analysis consists of read mapping and peak calling, including peak modeling and identification (reviewed in [69]). Statistical strategies have been combined with single ChIP experiments to identify sites where combinations of chromatin features are likely to be found [42, 70–74]. These include use of hierarchical or k-means cluster analyses, heat map analysis, and Venn and Euler diagram analyses, (reviewed in [69]), however, direct observations using methods specifically designed to report combinations of chromatin features will be needed to validate these approaches. A distinct set of statistical considerations exists for single cell and single molecule analyses. These are discussed separately below.
4.3. Mass spectrometry
Mass spectrometry has been responsible for the dramatic expansion of the histone modifications known (reviewed in [75]). When applied to histones that were not previously fractionated, this can reliably identify modifications that arise together in pairwise and higher order combinations [45, 46]. This is true if the modifications are close enough on the peptide chain to remain on informative peptide fragments (reviewed in [76]). When amino acids of interest are localized on different histones, or far apart on a single histone, it is difficult or impossible to identify combinations of modifications. However, if nucleosomes are previously isolated by ChIP, quantitative mass spectrometry can reveal modifications on histones that associate with the modification recognized by the antibody [77]. A question still unaddressed by mass spectrometry is where on the DNA combinations of histone modifications reside, and if the histone modifications are associated with DNA modifications. In principle, it should be possible to perform ChIP and reserve the DNA for sequencing or bisulfite sequencing, and subject the isolated histones to mass spectrometry, but it is unlikely that this will enable unambiguous characterization of the range of histone modifications at a given locus.
4.4. Single molecule methods
Methods that interrogate individual molecules for a range of chromatin features may provide a means for determining which chromatin features are coincident, mutually exclusive, the frequency with which combinations actually appear in the genome and their location. Ensemble methods based on averaged measurements of typical single feature ChIP experiments cannot provide this information. Several distinct single molecule methods have been developed (reviewed in [78–82]), and can be applied to chromatin isolated from a population of cells, or from single cells (discussed below). Several of these methods rely on fluorescent antibodies to detect epigenetic marks. As is true with ChIP, data quality will depend on antibody binding specificity and affinity [39].
4.4.1. DNA curtains, ordered chromatin arrays, nanochannel squeezing and ChIP-string
DNA curtains are prepared by tethering the biotinylated ends of DNA to streptavidin localized in lipids anchored to a solid substrate in a flow cell (reviewed in [83]). When an aqueous medium is flowed through the cell, DNA fibers elongate (reviewed in [84]). These methods have been used to study variables influencing nucleosome deposition [85] and the effects of nucleosomes on DNA scanning by repair proteins [86]. Future embodiments of DNA curtains might make it possible to characterize where modifications exist on naturally occurring chromatin using fluorescent probes recognizing those modifications. This approach could identify patterns of coincidence and mutual exclusion of modifications on single chromatin fragments; however, further enhancements still would be required to reveal what DNA sequences are affected.
Ordered arrays of chromatin fragments stretched across devices fabricated with polydimethylsiloxane (PDMS) provide an alternative to molecular combing [87]. In this procedure the PDMS device contains micron scale pillars protruding from a surface, and spaced at micron scale intervals. Chromatin is placed between the device and a glass coverslip. Individual molecules bind to the pillars and are elongated across them by drawing the coverslip across the device. Once bound, histone H3 was detected using fluorescent antibodies and fluorescence microscopy. It should be possible to extend this method to report patterns of coincidence and mutual exclusion of chromatin features. Methods have not been developed to identify the underlying DNA sequences on ordered arrays.
DNA curtains and ordered arrays entail tethering chromatin in some manner prior to observation. In a complementary method, chromatin can be flowed through an elastic channel, which can be narrowed to a cross section of approximately 200nm × 200nm, thus confining linear fragments. Confinement can also be achieved in narrow rigid channels [88, 89]. When HeLa cell chromatin confined in elastic channels was stained with fluorescent antibodies detecting histone modifications, patterns of H4ac and H3K9me3 were observed [90]. Consistent with expectations, these modifications, that respectively report active and silent chromatin, were not coincident. This method was also use to characterize two histone modifications simultaneously in chromatin reconstituted from natural histones on T7 phage DNA [91]. Additionally, DNA methylation in experimentally methylated lambda DNA was detected using a fluorescently tagged MBD portion of MeCP2 [89].
The resolution of ordered arrays (at least 320 nm [87]) and elongation through confinement (6–10 kbp [88, 89]) correspond to as few as 25 nucleosomes, if chromatin was decompacted from the 30 nm fiber [92]. Further development of these methods is needed to determine the DNA sequences associated with fluorescent signals reporting chromatin features.
ChIP-string applied DNA isolated by ChIP to the NanoString nCounter platform [42]. This single molecule counting method reported the presence of chromatin features at 487 loci through use of fluorescent bar coded probes capable of uniquely identifying the loci. As originally developed, chromatin features were assayed individually. It is not clear if the method is sensitive enough for use with re-ChIP studies for dual mark assessment.
4.4.2. SMRT and nanopore sequencing
Recently developed and emerging single molecule sequencing technologies have led to the observation that modified bases can be detected during sequencing. These methods rely on two essential features of the sequencing platforms: First, sequenced DNA templates are those naturally existing in the cells of interest and not generated by amplification methods; and second, during sequencing, modified nucleotides can exhibit measurably distinct behaviors from unmodified nucleotides, thereby reporting their modification state. With the Pacific Biosciences SMRT platform, sequence data are determined by detecting incorporation of spectrally distinct fluorescent nucleotides during DNA synthesis guided by the naturally occurring template. The latency of incorporation of nucleotides is influenced by adjacent 5mC [66] and 5hmC [67] residues. With nanopore based methods, MBD1 protein bound to 5mC on a DNA template alters the current flowing through the sequencing nanopore [68]. MBD1 or similar probes might not be necessary to distinguish cytosines that are unmodified, methylated and hydroxymethylated on nanopore platforms [93–95]. In principle, pending further advancements with nanopore sequencing [96, 97], either of these single molecule sequencing approaches can enable detection of coincident chromatin modifications, if DNA isolated by ChIP is applied to the platforms.
4.4.3. SCAN
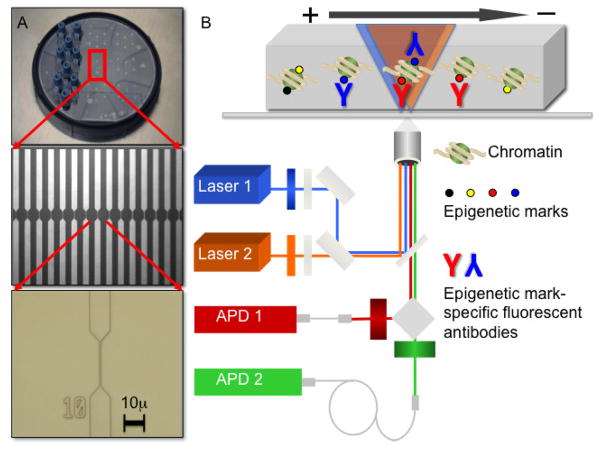
Single chromatin molecule analysis at the nanoscale (SCAN), is analogous in principle to flow cytometry (Figure 1). Flow cytometry entails binding cells to antibodies that recognize cell surface antigens and carry distinct fluorophores, then measuring the fluorescent properties of single cells as they flow through an inspection or focal volume. The results report the abundance of cells that carry different combinations of cell surface antigens. Similarly, chromatin fragments can be bound to fluorescent antibodies or other molecular probes that recognize different chromatin modifications, molecules can be driven by a voltage gradient through a fluidic channel, and defined combinations of chromatin modifications can be detected and their abundances measured [98, 99]. In this embodiment, SCAN is an analytical tool, capable of enumerating the abundance of chromatin modifications or their combinations. Further modifications enable SCAN to be used as a preparative tool to isolate chromatin molecules that carry those different states. This preparative mode is analogous to fluorescence activated cell sorting (FACS) and can provide molecules defined chromatin modifications for applications such as sequencing [100].
Figure 1. Experimental platform for single molecule epigenomics analyses.
A, SCAN device. Nanoscale channels were fabricated on 10 cm silica wafers and bonded to a second top wafer containing holes to access the channels (top). Cylinders attached over holes allow channels to be filled with chromatin or DNA solutions and fitted with electrodes to drive molecules through the channels electrokinetically. Red box outlined in top image contains 16 nanofluidic channels (middle), and wafers contain 27 such elements. Micrograph of one channel showing the 500 nm wide by 250 nm deep constriction where fluorescence detection occurred (bottom). B, Schematic of a wafer mounted on a confocal fluorescence microscope. Overlapping lasers (blue and orange cones) illuminate a 1.3 μm length of the channel forming a 160 aL (160×10−18 L) inspection volume where single molecule detection takes place. Chromatin is shown in a channel with black, yellow, red and blue circles depicting different epigenetic features. Antibodies recognizing those epigenetic features and carrying distinct fluorophores are bound to chromatin. After laser excitation, fluorescent emissions collected with a confocal aperture are detected with avalanche photodiodes (APDs) capable of single photon detection. Devices with this architecture enabled analyses of epigenomic marks individually and combinations [98, 99].
4.4.4. Computational and statistical considerations for single molecule methods
There are at least three major computational and statistical challenges with single molecule methods. These include distinguishing signal from noise for each detection event; managing the large data sets that come from each experiment; difficulties performing data analysis in real time, which is preferable but less practical than performing analysis after data collection.
With the exception of nanopore-based approaches, each of the single molecule methods involves identification of fluorescence bursts from many molecules, and then extracting the characteristics from the data integrated over each burst. This is challenging, especially for platforms that assess molecular properties that change dynamically (SMRT sequencing and SCAN) vs. platforms that collect data from immobilized molecules whose properties do not change over the course of the experiment (DNA curtains, ordered chromatin arrays, nanochannel squeezing and ChIP-string; however, some embodiments of these methods could require dynamic analyses). The challenges arise because the event rate of particle detection is low and fluorescence background contributes significantly to the measured signal. Burst identification has been traditionally achieved by filtering and thresholding the detected photon flux based on the signal time-trace data through the use of statistical analysis and an iterative optimization of an intensity threshold [101–103]. However, development of more accurate statistical tools and faster data processing techniques are among the most important efforts in single molecule detection.
Fluorescent bursts are collected in one of two ways. With single-point detectors (photodiodes or photomultiplier tubes), the raw data from a photo-detector consists of digital pulses that can be time-tagged, binned or counted, processed to detect bursts and then different related quantities extracted. With two-dimensional detectors (CCD and CMOS arrays), data collection and analysis includes acquiring tens of thousands of images of dynamic samples, localizing and identifying individual molecules, and extracting the position of individual molecules. The computational cost of these tasks grows linearly with the number of detectors (or pixels in the arrays) and can easily exceed the capabilities of a typical desktop computer. For example, when a 512 × 512 pixel CCD is used for single molecule detection in high-throughput and multicolor imaging applications, one image generates about 0.5 Megabytes of data collected every 1 ms. Each hour of analysis entails nearly 30 Gigabytes of data.
Until now, single molecule experiments rely on processing data for analysis after collection instead of in real time, which can prove to be limiting. To overcome this limit will require development of new algorithms for molecule identification and tracking, and the use of high performance hardware such as graphics processor units and field programmable gate arrays. The latter has been incorporated into SCAN for single molecule sorting [100].
5. Analyzing Low Cell Inputs
Because single molecule methods provide a high level of sensitivity for chromatin analyses, they also hold promise for enabling analyses of very low cell inputs, including single cells. Such analyses are important for several kinds of studies, three examples of which are outlined below. First, embryonic development, from the single cell zygote to the morula and blastocyst stage when differentiation first begins, is known to involve dramatic changes in chromatin states that are vital to differentiation and development. However, little is known about how those states differ among cells in the preimplantation embryo, if they are affected by location in the embryo, cell-cell or cell-extracellular matrix contacts, and the sequence of changes that occur in the cells at different genomic loci. Mapping these states, spatially, temporally and genomically would provide an unprecedented view of molecular changes during development, which can only be achieved by single cell methods. Second, differentiation has been modeled using embryonic stem cells and culture conditions that drive differentiation into specific lineages. Chromatin states before and differentiation have been characterized in populations of cultured cells using ChIP [63] and bisulfite sequencing [104]. It is not known how individual cells responding to differentiation promoting signals behave, the course of those behaviors throughout differentiation in culture, and if they parallel what occurs in individual cells within the embryo. Third, beyond questions of differentiation and development, it is known that disease states can arise when individual cells exhibit deviations from normal behavior, including at the level of chromatin regulation. Ensemble analyses of tissues or groups of cells reveal population-averaged behaviors, but not the degree of variance among cells within the population, which can be revealed only by single cell analyses. Whether high variance in cellular behavior correlates with disease likelihood will require the application of single cell analytical methods. In addition to these three examples, single cell analyses can apply molecular criteria to assess the number of distinct cell types that exist in organisms and the heterogeneity that arises in clonally derived cells.
Single cell analytical methods have been developed to sequence bacterial genomes [105], mammalian genomes [106, 107], and transcriptomes by microarray [108, 109] and RNA sequencing [110–116] (reviewed in [117]). The C1 system from Fluidigm facilitates isolation of single cells, as well as on-device extraction and processing of RNA and DNA for such studies; however, this platform has not been adapted for chromatin isolation. In an effort to fill this gap, microscale devices have been developed capable of trapping low inputs of cells for standard ChIP [41, 118] and restriction enzyme based 5mC analysis [119], from single cells that enable the isolation and manipulation of single chromatin molecules using antibody conjugated microspheres and optical tweezers [120], or from single cells using a design that enables their integration with the SCAN platform [121]. The latter has been used for extracting chromatin from low cell inputs by sequentially flowing into the devices the reagents used for native chromatin extractions, followed by fluorescent antibodies needed for detecting chromatin features by SCAN (unpublished). Common to these devices is their nanoliter scale volume, which can ensure that reagent concentrations, including antibodies, are kept high, allowing efficient binding and high sensitivity of detection. Picoliter volumes are achievable with micron scale droplets, which hold promise for epigenomic analyses, either of single cells or single molecules. They are readily loaded with materials and reagents for analysis, can be subjected to high throughput fluorescent detection in flow channels [122] and droplets with defined properties can be selectively sorted for subsequent analyses [123].
Other efforts for single cell analyses of DNA and histone modification states have been developed that do not rely on custom-built microscale devices [124–126]. Hi-C methods were reported that characterize chromatin interactions in the genome of single cells [127]. Proximity ligation methods have detected coincidence of chromatin features by microscopy [128] in single cells. Other techniques have characterized nuclear lamina interactions with chromatin in single cells [129]. Analyses of 5mC have been reported for single cells using bisulfite methods [130, 131]. Further development of these and other single cell techniques will be necessary to address the kinds of questions raised above.
Aside from the technical issues associated with collecting epigenomic data from single molecules and single cells (discussed further below), are the statistical issues associated with extracting meaningful biological information from the data. ChIP-seq and BS-seq, when applied to materials isolated from tens of millions of cells, can reliably identify loci harboring a given epigenetic mark when sequencing is done at high enough coverage. However, when querying chromatin from few cells for combinations of epigenetic marks using single molecule approaches, coverage is fundamentally limited: Autosomal loci in a single cell can never be sequenced to more than 2X coverage even with completely efficient library preparation. Such limitations can be overcome by performing multiple single cell and single molecule experiments on a population of cells, with the sampling requirements dictated by the efficiency of each step in the workflow and the magnitudes of the effects being measured.
6. Technical challenges of single molecule SCAN and single cell chromatin analyses
Single molecule and single cell analytical methods bring new opportunities for discovery, and challenges for their implementation. Among the challenges of single molecule methods are achieving signal to noise ratios that enable reliable detection of chromatin features on individual molecules when using fluorescent detection reagents, and achieving high enough throughput rates in analytical and preparative modes to accomplish experimental objectives. Among the challenges of single cell analyses are availability of reagents with favorable binding constants to detect chromatin features with high efficiency, even at low analyte concentrations. The use of nanoliter volumes in microfluidic devices [118, 132, 133] or subnanoliter volumes in micrometer sized droplets [123, 134, 135] can help maximize those concentrations. Challenges common to single molecule and single cell analyses include development and use of high-efficiency library preparation and/or novel sequencing techniques that reliably gather sequence information from DNAs isolated using single molecule methods.
6.1. Signal-to-Noise ratio (SNR) for single-molecule detection (SMD)
Most of the single-molecule analytical methods for epigenomic measurements require measurement of emissions from fluorescent intercalators and antibodies bound to single chromatin molecules [87, 98–100]. The fluorescent signals from single molecules must be distinguished from hardware noise and background, which is complicated by the vast excess of solvent. The number of photons collected in detector (S) of an optical setup is
where Wabs is the molecular absorption rate, P (Watts) excitation beam power, σabs an absorption cross-section of the single fluorophore for excitation light, A (cm2) the cross-section of the excitation beam in the plane of the molecule, QY fluorescence quantum yield of a fluorophore, and Deff the global collection efficiency of the optics setup. The environment of the single molecule can add a background contribution modeled by a rate b per unit-volume and unit-excitation power. From these definitions, the signal-to-background ratio (SBR) can be expressed as
where t0 is the integration time of the detector and V is the intersection between the excitation and the detection volume [136]. SBR can be maximized by use of small focal volumes, large absorption cross sections (σabs) and fluorophores with high fluorescence quantum yields (QY). Accordingly, microfluidic, nanofluidic and microdroplet platforms are preferred for single molecule and single cell analyses.
Some detectors amplify the incoming signal (S × τ) before readout, introducing a gain (G) in the measured signal. In practice, the shot noise component of the incoming signal is itself multiplied by the gain G, and a correction factor F, called the excess noise factor, σamp = F × G × σin, where σin represents the standard deviation of the input signal and σamp that of the amplified signal. For a detector without gain, or for photon-counting detectors, G = 1 and F = 1. For negligible dark count rates and negligible readout noise component, the resulting SNR can thus be expressed generally as
SBR does not depend on the measurement duration, whereas SNR increases with the measurement duration and SBR. Achieving the required SNR requires close attention to maximizing signal while minimizing background. Higher excitation power produces higher SNR values, but the power cannot be increased arbitrarily because saturation accompanied by excess power can actually lead to reduced fluorescent emission due to photobleaching. Background can have different sources, including detector dark counts and residual laser scattered light or Raman scattering, some of which can in principle be minimized by sample or setup optimization.
To estimate actual SBR and SNR values from a typical experiment, consider the single molecule fluorescence detection for Alexa Fluor dyes in water, using a single-photon avalanche photodiode detector (SPAD) to collect the signal. QY and σabs of Alexa Fluor 488 are 0.92 and 2.5 × 10−16 cm2 at 488-nm excitation (from the manufacturer), respectively. Using 100 μW of laser power, 1 ms integration time, 0.4 μm of channel width, and a 2 μm of length of laser spot along channel (Lfocal), we expect 7063 photons/ms emitted from a single AlexaFluor 488 molecule. With a high-performance optical microscope fitted with a high numerical aperture lens having 5% collection efficiency, there will be an estimated 353 photons/ms reaching the SPAD, leading to a SNR ~ 18.5 and SBR ~ 35.3. More favorable SNR and SBR values would be obtained by replacing organic dyes with quantum dots (QDs), because of their exceptional brightness and stability. QDs have the added benefit of excitation at low wavelengths and narrow emission spectra, which makes more efficient use of the spectrum for higher order multiplexing.
To improve SNR or SBR in SMD, several microscopic and spectroscopic techniques have been suggested and tested. These overcome a limitation of conventional widefield epi-fluorescence, which creates high background fluorescence from out-of-focus light. Each method modifies the excitation laser beam to further confine light on the nanoscale and improve the spatial resolution. This can be achieved by evanescent illumination using total internal reflection fluorescence microscopy [137], and by two photon excitation using two-photon fluorescence microscopy [138]. Confocal microscopy can also reduce background signals originating outside the focal plane using spatial filters [139]. Laser scanning and spinning disk confocal fluorescence microscopy have the added benefit of increasing spatial resolution in single molecule imaging [140]. Photo-activated localization microscopy (PALM) [141] and stochastic optical reconstruction microscopy [142] have been performed to obtain images with a resolution beyond the diffraction limit; these may be of particular value for ordered chromatin arrays or chromatin confinement methods where localization of chromatin features on large fragments is limiting. Other optical methods have been demonstrated to enhance single molecule detection techniques beyond the limits set by optical diffraction (reviewed in [143]).
6.2. Throughput
SCAN presents challenges distinct from flow cytometry and FACS. Micron scale fluidic channels suitable for single cell analyses are not suitable for single molecule analyses, because using such dimensions would mandate use of highly diluted samples to ensure single molecule analyses and result in low sample throughput. Therefore, submicron fluidic structures are used. When flowing molecules through a channel with a 500nm × 500nm cross section, and an inspection laser with a 1.3 μm beam diameter, the focal volume is 325 aL and Poisson statistics reveal the probability that the inspection volume will be occupied at a given concentration, by
where P(k) represents for the probability of finding exactly k number of molecules in the focal volume at a given time. λ is the average number of molecules in the focal volume. Thus, analyte concentrations must be kept low so that the probability of more than one molecule in the inspection volume P(≥ 2), is kept low. P(≥ 2) can be estimated by the expression (1 – probability of zero molecules in the focal volume – probability of one molecule in the focal volume). Requiring that two or more molecules are present in the detection volume no more than 1% of the time gives 0.01 ≥ P(≥ 2) = 1 − e−λ − λe−λ = 1 − e−λ(1 + λ), resulting in λ = 0.149. Then the maximum concentration of molecules (Cmax) that ensures this low likelihood of two molecules in the focal volume is given by
The throughput (NT) in number of molecules per unit time is then given by NT = λ/t0. Thus, the throughput of the single channel is estimated to 0.149 molecule/ms, using a 1ms value for t0 from the SNR discussion above. If the size of a human genome size is 3.2 × 106 kbp, and the size of prepared chromatin fragments is 500 bp, then the throughput in number of genomes per hour can be obtained as:
It will take about 12 hours to analyze one human genome equivalent in a single nanofluidic channel or just over 7 minutes on a 96 channel device like that shown in figure 2, if done with the same optical sensitivity that is possible with single channels. However, it is not likely that throughput will scale linearly with channel number for two reasons. First, SPADs that are ideal for single channels are not appropriate for detecting fluorescence from many channels, given their cost and engineering challenges to integrate 96 SPADs on a device; instead a 2D photon collection array is needed. These are available in the form of EM-CCD and sCMOS cameras; however, both camera formats have lower sensitivity than SPADs, requiring longer integration times. Parameters of high importance are fast frame rates, low dark current and readout noise, and high quantum efficiency and dynamic range. Pixel size and number should be appropriate to the geometry of the fluidic device, and data collection should be restricted to the informative pixels with relevant fluorescent emissions. Second, as channel number increases, so will the areas of excitation illumination and emission detection. These larger areas require lower magnification lenses with lower numerical apertures, which negatively affect photon collection efficiency [144]. Several hardware and data processing advances are needed for single molecule methods to achieve the genomic coverage currently enjoyed by ensemble methods like ChIP.
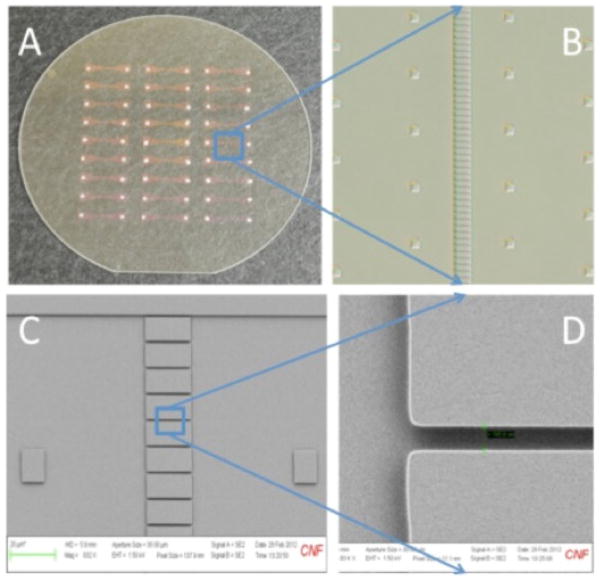
Figure 2. SCAN devices for increased throughput.
A, Optical micrograph of a 4-inch diameter fused silica wafer containing an array of 27 fluidic devices, each containing 96 parallel channels. B, Differential interference contrast optical micrograph of tens of the parallel fluidic channels in the center of a device. The square structures on each side of the central array are support pillars to prevent collapse when bonding to a cover wafer. C, Scanning electron micrograph showing approximately 8 parallel channels etched between microfluidic reservoirs on a single device. The channels are 20 microns in length (horizontal) and 500 nm deep. D, SEM close-up of an individual fluidic channel that is 750 nm in width. Devices with this parallel architecture can enable analyses like those previously reported [98, 99] with higher throughput, however, fluorescence detection will require replacing APDs with other detectors such as CMOS arrays, which will somewhat compromise sensitivity and hence throughput.
6.3. Reagents for detecting chromatin features and their dissociation constants (Kd)
Antibodies, DNA-binding proteins, and chromatin-binding proteins that bind specifically to their targets with high affinity are essential for single molecule chromatin analyses. Tests for specificity of antibodies recognizing histone modifications often use peptides corresponding to modified histone tails [145]. Kd measurements are rarely taken, but when reported, spanned four orders of magnitude, from 0.2nM to 2μM with high lot-to-lot variation [43, 146]. Kd values for proteins binding to DNA modifications ranged from 10nM to 50μM [147, 148]. Synthetic reagents to detect chromatin features hold promise for future studies [43].
Traditional ChIP-seq, which can use abundant sources of chromatin, does not require the highest affinity reagents as long as enough DNA can be recovered for high coverage sequence. However, single molecule and single cell analyses do rely on high affinity reagents or very small reaction volumes to enable high reagent concentrations. If we assume the Kd of an antibody or binding protein specific for a chromatin feature is 1 nM, and this concentration of detection reagent were used, then the concentration of the chromatin should be at least 10 nM for efficient binding. If a single cell were used for the chromatin source, and the chromatin were fragmented to 500 bp, then to achieve these concentrations requires a 2.2 nL reaction volume. For detection reagents with a 1μM Kd, we would want a chromatin concentration of 10μM, which for a single cell would require a reaction volume of 2.2 pL. These volumes can be achieved using microscale fluidic devices or micron scale droplets [123, 134, 135].
6.4. Sorting single molecules
Whereas analytical SCAN can provide information about coincidence and mutual exclusion of chromatin features, and their relative abundance in samples [99], it does not report the genomic locations of those modifications. Methods that sort individual molecules have been developed [149], including one that is based on DNA modification status that was adapted to SCAN [100]. In proof of principle experiments, methylated DNA was reliably separated from unmethylated DNA by single molecule sorting. In those studies, DNAs were labeled with a fluorescent intercalator and MBD1 protein carrying a spectrally distinct fluorophore. In analytical SCAN, molecules are driven by voltage through a channel with a single input and output through a focal volume where fluorescence information is collected, but in preparative SCAN, the nanoscale fluidic device for sorting includes a bifurcation downstream of the focal volume (Figure 3). As single molecules are driven through a focal volume by the voltage potential, the fluorescent properties of the molecules are measured and the signals directed to a field programmable gate array (FPGA), which places the voltage potential along one outflow track or the other. In this manner, molecules with fluorescent properties diagnostic for the 5mC can be isolated. The false positive rate, whereby unmethylated molecules were inadvertently identified as methylated and isolated was under 2%; the false negative rate, whereby methylated molecules were inadvertently identified as unmethylated and excluded, was under 10% [100]. Ongoing efforts are focused on applying this approach to complex chromatin samples. The technical issues relevant to analytical SCAN that were described above also apply to preparative SCAN. Approaches using single molecules enclosed in droplets offer an alternative to SCAN sorting as droplets can be sorted using acoustic waves [122].
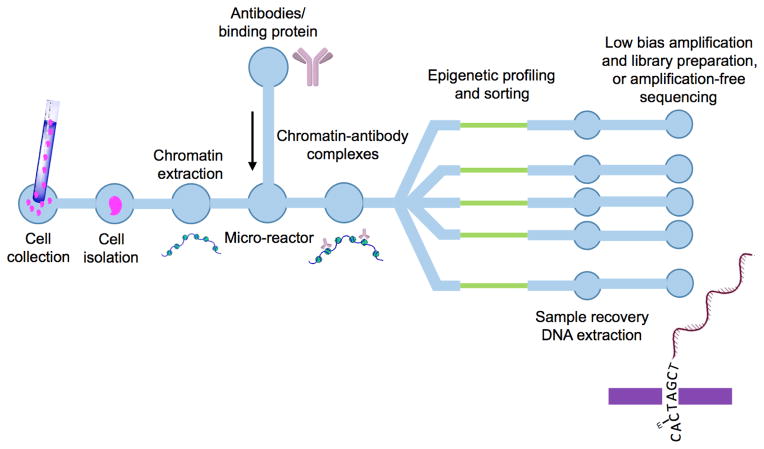
Figure 3. Potential integrated micro-/nano-device architecture for multiplexed epigenomic measurements and amplification-free sequencing from a single cell.
A schematic diagram depicting an integrated device for profiling combinations of epigenomic states from single cells using single molecule methods. Examples of each module of the integrated device were described in the text. From left, isolation of cells for analysis followed by extraction of chromatin from cells [41, 118, 121]; binding of antibodies or other reagents at high concentration in a low volume microreactor to detect epigenomic features [123, 133–135]; removal of unbound antibody can improve throughput of informative chromatin containing complexes; sorting of chromatin carrying various user defined combinations of epigenomic marks [100]; extraction of DNA from sorted chromatin; library preparation after low bias WGA [105, 155–157] followed by sequencing, or amplification free sequencing.
Once single molecules with user specified chromatin modifications are isolated, sequencing can reveal where combinations of modifications reside. DNA that is recovered will be present in picogram quantities or lower, which will require high-efficiency methods for sequencing. Library-free sequencing approaches will be ideal. Alternatively, highly efficient, low bias amplification and library preparation techniques are required.
6.4.1. Sequencing platforms
Each of the next generation sequencing technologies require distinct workflows of varying complexity to prepare templates for sequencing. DNA losses and inefficiencies at each stage present challenges for applications with very low DNA inputs. These can be minimized by performing library preparation reactions in nL volume scale devices [133], which in theory can be integrated with previously mentioned single molecule and single cell analytical platforms. Library preparation for the widely used Illumina HiSeq platforms requires repairing the DNA ends, single nucleotide A-tailing, adaptor ligation, and library amplification prior to final DNA purification and loading onto the sequencer.
Competing sequencers, notably those based on single molecule methods, can have simpler workflows. The formerly available Helicos sequencing platform required no prior ligation and amplification steps. Instead, single molecules, as they existed in cells, were sequenced directly after a 3′ tailing reaction and hybridization to tail-complementary sequences on the platform [150]. Accordingly, with this highly simplified workflow, the Helicos platform proved useful for ChIP-seq studies using as few as 50pg of input DNA [151]. While this platform was a success technically, it was short-lived as a commercial product.
The Pacific Biosciences SMRT system, as previously mentioned, sequences native single-molecule templates without any amplification, although templates must undergo repair, ligation, primer annealing, and DNA polymerase loading steps prior to deposition in zero mode wave guides, where single DNA molecules are optically monitored during DNA polymerase-directed incorporation of fluorescent nucleotides [152]. Given that library preparation is comparable in complexity to the Illumina system, it is not clear if this platform will be suitable for picogram and subpicogram quantities of DNA. As cited above, the Pacific Biosciences platform can report 5mC [66] and 5hmC [67] residues during sequencing based on the reaction kinetics of base incorporation – important features for combinatorial analysis.
Often called the fourth-generation of sequencing platforms, nanopore-based sensing devices provide another method for directly sequencing single DNA molecules (reviewed in [153]). The technology consists of a nanopore within a dielectric membrane that separates two salt solutions. Upon application of a current, the system drives ions through the nanopore where charged molecules, such as DNA, will create resistance, thereby partially blocking the pore as the DNA passes through. Using the translocation time and current blockade, each of the four nucleotides can be distinguished from one another and sequence data collected. A key advantage of this platform is that templates require no preparation beyond their purification, which will eliminate concerns of inefficiencies and losses associated with library preparation. However, it is not clear what will be the lower limit of template abundance required for sequencing on nanopore platforms, or their throughput. Published data reported the use of 1μM DNA solutions in a 60μL volume [97], but these values may drop as the technology matures. Like the SMRT-seq platform, 5mC and 5hmC can be distinguished from one another and unmodified cytosines in nanopores [68, 93, 95]. Nucleosome complexes can also be identified by nanopores [154], raising the possibility that histone modification states can be revealed as well.
6.4.2. Amplification
When single molecule methods are used to isolate chromatin for sequencing, the yields are likely to be lower than when ensemble methods like ChIP are used. Yields will be smaller still when low cell inputs are used. Whole-genome amplification is necessary to accrue enough material for sequencing. The most widely used methods for WGA using low DNA inputs include multiple displacement amplification (MDA) [105, 155], ligation-mediated PCR [156] and multiple annealing and looping-based amplification cycles (MALBAC) [157]. All methods suffer from some amplification bias, with PCR introducing more than MDA [155], and MDA potentially introducing more than MALBAC [157]. However, biases can be minimized with small reaction volumes [158]. These approaches have worked well in traditional single-cell analyses; however, they have yet to be tested thoroughly in epigenetic applications involving whole genome analysis using chromatin inputs from fewer than 5,000–10,000 cells [29, 159].
6.4.3. Tagmentation
Recently, an alternative to the standard methods of fragmentation and adapter ligation has been developed for preparing libraries for whole-genome sequencing. While traditional methods require multiple steps to generate adaptor-ligated fragments, the hyperactive derivative of the Tn5 transposase can be used to simultaneously fragment, end-repair, and tag the DNA with specific adaptors to both ends, all within a single-tube reaction. This method termed tagmentation, enables library preparation from subnanogram amounts of DNA [160]. Tagmentation has been adapted to bisulfite analysis of 5mC using low tens of nanograms of DNA [161, 162], and to analysis of open chromatin using as few as 500 cells [163].
7. Closing remarks
Many technical advances have been vital to modern molecular genetics research – DNA sequencing being prominent among them. It is clear that beyond the information carried by genomic sequences, combinations of chromatin features play vital roles in genomic control. Furthermore, variations in chromatin states that exist in different tissues and cells, and even within a given cell type, have great biological consequence. Accordingly, technical advances are required to characterize these chromatin states at high resolution. Just as the single nucleotide resolution provided by whole genome sequencing has been vital to understand the structure and function of genomes, we speculate that single molecule and single cell methods will be vital to understand phenotypes and mechanisms of inheritance not revealed by primary genomic sequences. Many independent methods that were described here hold promise for filling these needs. Ongoing development and integration of these methods are needed to fulfill this promise.
Table 1.
Comparison of methods
| Technique | Feature(s) measured | Information reported | Limitations | References |
|---|---|---|---|---|
| Ensemble methods | ||||
| ChIP-seq | Histone modifications Chromatin binding factors |
Genomic locations of individual chromatin features | Coincidence of multiple marks inferred; methods to improve inferences exist Requires high quality affinity reagents |
[42, 72, 73, 164] |
| re-ChIP or sequential ChIP | Histone modifications Chromatin binding factors |
Genomic locations of combinations of chromatin features | Full genome coverage not readily achieved Requires high quality affinity reagents |
[165] |
| BS-seq | 5mC or 5hmC | Genomic locations of either feature Can use DNA from ChIP for dual feature analysis |
Does not distinguish 5mC from 5hmC or report locations of 5fC or 5caC | [166] [65] |
| oxBS-seq | 5mC | Genomic locations of 5hmC Can be combined with BS-seq to distinguish locations of 5mC and 5hmC Can use DNA from ChIP for dual feature analysis |
Does not report locations of 5fC or 5caC | [22, 167] |
| TAB-seq | 5hmC | Genomic locations of 5hmC Can use DNA from ChIP for dual feature analysis |
Does not report locations of 5fC or 5caC | [21] |
| Mass spectrometry | Histone modifications | Abundance, identity and coincidence of histone modifications | Requires modifications reside on nearby amino acids on the same histone, unless combined with ChIP for prior purification of analyte Does not report genomic locations of histone modifications |
[46, 77, 168–171] |
| Single molecule methods | ||||
| DNA curtains | DNA binding proteins | Binding properties and dynamics of interactions between DNA binding proteins defined DNA substrates | Not yet adapted to naturally occurring chromatin | [83–86] |
| Ordered chromatin arrays | Histones | Presence of histones on chromatin; readily adaptable to detect histone modifications | Does not report sequence of DNA Requires high quality affinity reagents Limited resolution |
[87] |
| Chromatin elongated by confinement | Histone modifications and 5mC | Presence of histone modifications and 5mC | Does not report sequence of DNA Requires high quality affinity reagents Limited resolution |
[88, 89] |
| ChIP-string | Chromatin binding factors | Which of up to 800 genomic locations bind feature of interest | Does not provide full genome coverage Requires high quality affinity reagents |
[42] |
| SCAN | Histone modifications and 5mC, individually or in combination | Abundance of chromatin features Can be run in analytical or preparative modes |
Requires high quality affinity reagents Limited molecule per minute throughput in preparative mode |
[98–100] |
| SMRT sequencing | 5mC and 5hmC | Genomic locations of indicated features | Identification of features is based on latency of nucleotide incorporation, which is also influenced by sequence context | [66, 67] |
| Nanopore methods | 5mC and 5hmC | Presence of indicated features on a DNA template | May require high quality affinity reagents Further development needed for genome wide sequencing |
[68, 93–95] |
Acknowledgments
Funding support from NIH HG006850.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Waddington CH. The strategy of the genes; a discussion of some aspects of theoretical biology. London: Allen & Unwin; 1957. p. ix.p. 262. [Google Scholar]
- 2.Plass C, et al. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genet. 2013;14:765–80. doi: 10.1038/nrg3554. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 4.Tahiliani M, et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by the MLL Fusion Partner TET1. Science. 2009;324:930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–7. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–3. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–71. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 8.Shilatifard A. Chromatin Modifications by Methylation and Ubiquitination: Implications in the Regulation of Gene Expression. Annu Rev Biochem. 2006;75:243–69. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 9.Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn’t fit all. Nat Rev Mol Cell Biol. 2007;8:284–95. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 10.Cheng X, Blumenthal RM. Mammalian DNA methyltransferases: a structural perspective. Structure. 2008;16:341–50. doi: 10.1016/j.str.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartz YB, Pirrotta V. A new world of Polycombs: unexpected partnerships and emerging functions. Nat Rev Genet. 2013;14:853–64. doi: 10.1038/nrg3603. [DOI] [PubMed] [Google Scholar]
- 12.de Ruijter AJ, et al. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–49. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 14.Franchini DM, Schmitz KM, Petersen-Mahrt SK. 5-Methylcytosine DNA demethylation: more than losing a methyl group. Annu Rev Genet. 2012;46:419–41. doi: 10.1146/annurev-genet-110711-155451. [DOI] [PubMed] [Google Scholar]
- 15.Fatemi M, Wade PA. MBD family proteins: reading the epigenetic code. J Cell Sci. 2006;119:3033–7. doi: 10.1242/jcs.03099. [DOI] [PubMed] [Google Scholar]
- 16.Patel DJ, Wang Z. Readout of Epigenetic Modifications. Annu Rev Biochem. 2013;82:81–118. doi: 10.1146/annurev-biochem-072711-165700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu H, et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc. 2011;6:468–81. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- 18.Diep D, et al. Library-free methylation sequencing with bisulfite padlock probes. Nat Methods. 2012;9:270–2. doi: 10.1038/nmeth.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krueger F, et al. DNA methylome analysis using short bisulfite sequencing data. Nat Methods. 2012;9:145–51. doi: 10.1038/nmeth.1828. [DOI] [PubMed] [Google Scholar]
- 20.Miura F, et al. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 2012;40:e136. doi: 10.1093/nar/gks454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu M, et al. Base-Resolution Analysis of 5-Hydroxymethylcytosine in the Mammalian Genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Booth MJ, et al. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat Protocols. 2013;8:1841–1851. doi: 10.1038/nprot.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mardis ER. ChIP-seq: welcome to the new frontier. Nat Methods. 2007;4:613–4. doi: 10.1038/nmeth0807-613. [DOI] [PubMed] [Google Scholar]
- 24.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26:1351–9. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–80. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt D, et al. ChIP-seq: Using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009;48:248–248. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adli M, Zhu J, Bernstein BE. Genome-wide chromatin maps derived from limited numbers of hematopoietic progenitors. Nat Methods. 2010;7:615–8. doi: 10.1038/nmeth.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lefrancois P, Zheng W, Snyder M. ChIP-Seq using high-throughput DNA sequencing for genome-wide identification of transcription factor binding sites. Methods Enzymol. 2010;470:77–104. doi: 10.1016/S0076-6879(10)70004-5. [DOI] [PubMed] [Google Scholar]
- 29.Adli M, Bernstein BE. Whole-genome chromatin profiling from limited numbers of cells using nano-ChIP-seq. Nat Protoc. 2011;6:1656–68. doi: 10.1038/nprot.2011.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kidder BL, Hu G, Zhao K. ChIP-Seq: technical considerations for obtaining high-quality data. Nat Immunol. 2011;12:918–22. doi: 10.1038/ni.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z, Pugh BF. High-Resolution Genome-wide Mapping of the Primary Structure of Chromatin. Cell. 2011;144:175–186. doi: 10.1016/j.cell.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–31. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blecher-Gonen R, et al. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat Protoc. 2013;8:539–54. doi: 10.1038/nprot.2013.023. [DOI] [PubMed] [Google Scholar]
- 34.He HH, et al. Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat Methods. 2014;11:73–8. doi: 10.1038/nmeth.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giresi PG, Lieb JD. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements) Methods. 2009;48:233–9. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Auerbach RK, et al. Mapping accessible chromatin regions using Sono-Seq. Proc Natl Acad Sci U S A. 2009;106:14926–31. doi: 10.1073/pnas.0905443106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warnecke PM, et al. Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA. Nucleic Acids Res. 1997;25:4422–6. doi: 10.1093/nar/25.21.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voss KO, et al. Combating PCR Bias in Bisulfite-Based Cytosine Methylation Analysis. Betaine-Modified Cytosine Deamination PCR. Anal Chem. 1998;70:3818–3823. [Google Scholar]
- 39.Egelhofer TA, et al. An assessment of histone-modification antibody quality. Nat Struct Mol Biol. 2011;18:91–3. doi: 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen UT, et al. Accelerated chromatin biochemistry using DNA-barcoded nucleosome libraries. Nat Methods. 2014;11:834–40. doi: 10.1038/nmeth.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu AR, et al. High throughput automated chromatin immunoprecipitation as a platform for drug screening and antibody validation. Lab on a Chip. 2012;12:2190–2198. doi: 10.1039/c2lc21290k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ram O, et al. Combinatorial Patterning of Chromatin Regulators Uncovered by Genome-wide Location Analysis in Human Cells. Cell. 2011;147:1628–39. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hattori T, et al. Recombinant antibodies to histone post-translational modifications. Nat Methods. 2013;10:992–5. doi: 10.1038/nmeth.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 45.Johnson L, et al. Mass spectrometry analysis of Arabidopsis histone H3 reveals distinct combinations of post-translational modifications. Nucleic Acids Res. 2004;32:6511–8. doi: 10.1093/nar/gkh992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young NL, et al. High throughput characterization of combinatorial histone codes. Mol Cell Proteomics. 2009;8:2266–84. doi: 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garske AL, et al. Combinatorial profiling of chromatin binding modules reveals multisite discrimination. Nat Chem Biol. 2010;6:283–90. doi: 10.1038/nchembio.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruthenburg AJ, et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145:692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang C, et al. Dual-specificity histone demethylase KIAA1718 (KDM7A) regulates neural differentiation through FGF4. Cell Res. 2010;20:154–65. doi: 10.1038/cr.2010.5. [DOI] [PubMed] [Google Scholar]
- 50.Horton JR, et al. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol. 2010;17:38–43. doi: 10.1038/nsmb.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, et al. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–89. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 52.Nan X, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex [see comments] Nature. 1998;393:386–9. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 53.Musselman CA, et al. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol. 2012;19:1218–27. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan M, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–28. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chaya D, et al. Transcription factor FoxA (HNF3) on a nucleosome at an enhancer complex in liver chromatin. J Biol Chem. 2001;276:44385–9. doi: 10.1074/jbc.M108214200. [DOI] [PubMed] [Google Scholar]
- 56.Soutoglou E, Talianidis I. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science. 2002;295:1901–4. doi: 10.1126/science.1068356. [DOI] [PubMed] [Google Scholar]
- 57.Proft M, Struhl K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell. 2002;9:1307–17. doi: 10.1016/s1097-2765(02)00557-9. [DOI] [PubMed] [Google Scholar]
- 58.Henry KW, et al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–63. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Metivier R, et al. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–63. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 60.IJpenberg A, et al. In vivo activation of PPAR target genes by RXR homodimers. EMBO J. 2004;23:2083–91. doi: 10.1038/sj.emboj.7600209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Geisberg JV, Struhl K. Analysis of protein co-occupancy by quantitative sequential chromatin immunoprecipitation. Curr Protoc Mol Biol Chapter. 2005;21(Unit 21):8. doi: 10.1002/0471142727.mb2108s70. [DOI] [PubMed] [Google Scholar]
- 62.de Medeiros RB. Sequential chromatin immunoprecipitation assay and analysis. Methods Mol Biol. 2011;791:225–37. doi: 10.1007/978-1-61779-316-5_17. [DOI] [PubMed] [Google Scholar]
- 63.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 64.Jin C, et al. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009;41:941–5. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brinkman AB, et al. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res. 2012;22:1128–38. doi: 10.1101/gr.133728.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Flusberg BA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7:461–5. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Song CX, et al. Sensitive and specific single-molecule sequencing of 5-hydroxymethylcytosine. Nat Methods. 2012;9:75–7. doi: 10.1038/nmeth.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shim J, et al. Detection and quantification of methylation in DNA using solid-state nanopores. Sci Rep. 2013;3:1389. doi: 10.1038/srep01389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shin H, et al. Computational methodology for ChIP-seq analysis. Quantitative Biology. 2013;1:54–70. doi: 10.1007/s40484-013-0006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kharchenko PV, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2010;471:480–5. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu T, et al. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 2011;21:227–36. doi: 10.1101/gr.115519.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–9. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Giannopoulou EG, Elemento O. Inferring chromatin-bound protein complexes from genome-wide binding assays. Genome Res. 2013;23:1295–306. doi: 10.1101/gr.149419.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maehara K, et al. A co-localization model of paired ChIP-seq data using a large ENCODE data set enables comparison of multiple samples. Nucleic Acids Res. 2013;41:54–62. doi: 10.1093/nar/gks1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arnaudo AM, Garcia BA. Proteomic characterization of novel histone post-translational modifications. Epigenetics Chromatin. 2013;6:24. doi: 10.1186/1756-8935-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karch KR, et al. Identification and interrogation of combinatorial histone modifications. Front Genet. 2013;4:264. doi: 10.3389/fgene.2013.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Voigt P, et al. Asymmetrically Modified Nucleosomes. Cell. 2012;151:181–193. doi: 10.1016/j.cell.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marie R, Kristensen A. Nanofluidic devices towards single DNA molecule sequence mapping. Journal of biophotonics. 2012;5:673–686. doi: 10.1002/jbio.201200050. [DOI] [PubMed] [Google Scholar]
- 79.Zillner K, Németh A. Single-molecule, genome-scale analyses of DNA modifications: exposing the epigenome with next-generation technologies. Epigenomics. 2012;4:403–414. doi: 10.2217/epi.12.30. [DOI] [PubMed] [Google Scholar]
- 80.Matsuoka T, et al. Micro- and nanofluidic technologies for epigenetic profiling. Biomicrofluidics. 2013;7:041301. doi: 10.1063/1.4816835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aguilar CA, Craighead HG. Micro- and nanoscale devices for the investigation of epigenetics and chromatin dynamics. Nat Nanotechnol. 2013;8:709–18. doi: 10.1038/nnano.2013.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Levy-Sakin M, et al. Toward single-molecule optical mapping of the epigenome. ACS Nano. 2014;8:14–26. doi: 10.1021/nn4050694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Collins BE, et al. DNA curtains: Novel tools for imaging protein-nucleic acid interactions at the single-molecule level. Methods Cell Biol. 2014;123:217–34. doi: 10.1016/B978-0-12-420138-5.00012-4. [DOI] [PubMed] [Google Scholar]
- 84.Greene EC, et al. DNA curtains for high-throughput single-molecule optical imaging. Methods Enzymol. 2010;472:293–315. doi: 10.1016/S0076-6879(10)72006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Visnapuu ML, Greene EC. Single-molecule imaging of DNA curtains reveals intrinsic energy landscapes for nucleosome deposition. Nat Struct Mol Biol. 2009;16:1056–62. doi: 10.1038/nsmb.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gorman J, et al. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat Struct Mol Biol. 2010;17:932–8. doi: 10.1038/nsmb.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cerf A, Tian HC, Craighead HG. Ordered Arrays of Native Chromatin Molecules for High-Resolution Imaging and Analysis. ACS Nano. 2012;6:7928–7934. doi: 10.1021/nn3023624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Streng DE, et al. Stretching chromatin through confinement. Lab Chip. 2009;9:2772–4. doi: 10.1039/b909217j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lim SF, et al. DNA methylation profiling in nanochannels. Biomicrofluidics. 2011;5:34106–341068. doi: 10.1063/1.3613671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Matsuoka T, et al. Nanoscale squeezing in elastomeric nanochannels for single chromatin linearization. Nano letters. 2012;12:6480–6484. doi: 10.1021/nl304063f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lim SF, et al. Chromatin modification mapping in nanochannels. Biomicrofluidics. 2013;7:64105. doi: 10.1063/1.4833257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Olins AL, Olins DE. Spheroid chromatin units (v bodies) Science. 1974;183:330–2. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- 93.Wallace EV, et al. Identification of epigenetic DNA modifications with a protein nanopore. Chem Commun (Camb) 2010;46:8195–7. doi: 10.1039/c0cc02864a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wanunu M, et al. Discrimination of methylcytosine from hydroxymethylcytosine in DNA molecules. J Am Chem Soc. 2011;133:486–92. doi: 10.1021/ja107836t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Laszlo AH, et al. Detection and mapping of 5-methylcytosine and 5-hydroxymethylcytosine with nanopore MspA. Proc Natl Acad Sci U S A. 2013;110:18904–9. doi: 10.1073/pnas.1310240110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cherf GM, et al. Automated forward and reverse ratcheting of DNA in a nanopore at 5-A precision. Nat Biotechnol. 2012;30:344–8. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Manrao EA, et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat Biotechnol. 2012;30:349–53. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cipriany BR, et al. Single Molecule Epigenetic Analysis in a Nanofluidic Channel. Anal Chem. 2010;82:2480–2487. doi: 10.1021/ac9028642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Murphy PJ, et al. Single-molecule analysis of combinatorial epigenomic states in normal and tumor cells. Proc Natl Acad Sci U S A. 2013;110:7772–7. doi: 10.1073/pnas.1218495110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cipriany BR, et al. Real-time analysis and selection of methylated DNA by fluorescence-activated single molecule sorting in a nanofluidic channel. Proc Natl Acad Sci U S A. 2012;109:8477–82. doi: 10.1073/pnas.1117549109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Enderlein J, et al. Molecular Shot Noise, Burst Size Distribution, and Single-Molecule Detection in Fluid Flow: Effects of Multiple Occupancy. The Journal of Physical Chemistry A. 1998;102:6089–6094. [Google Scholar]
- 102.Grange W, et al. Detection of transient events in the presence of background noise. J Phys Chem B. 2008;112:7140–4. doi: 10.1021/jp7114862. [DOI] [PubMed] [Google Scholar]
- 103.Johnson J, Chen Y, Mueller JD. Characterization of brightness and stoichiometry of bright particles by flow-fluorescence fluctuation spectroscopy. Biophys J. 2010;99:3084–92. doi: 10.1016/j.bpj.2010.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Raghunathan A, et al. Genomic DNA amplification from a single bacterium. Appl Environ Microbiol. 2005;71:3342–7. doi: 10.1128/AEM.71.6.3342-3347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hou Y, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–85. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 107.Wang J, et al. Genome-wide Single-Cell Analysis of Recombination Activity and De Novo Mutation Rates in Human Sperm. Cell. 2012;150:402–12. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tietjen I, et al. Single-cell transcriptional analysis of neuronal progenitors. Neuron. 2003;38:161–75. doi: 10.1016/s0896-6273(03)00229-0. [DOI] [PubMed] [Google Scholar]
- 109.Kurimoto K, et al. Global single-cell cDNA amplification to provide a template for representative high-density oligonucleotide microarray analysis. Nat Protoc. 2007;2:739–52. doi: 10.1038/nprot.2007.79. [DOI] [PubMed] [Google Scholar]
- 110.Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–82. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 111.Tang F, et al. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010;6:468–78. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hashimshony T, et al. CEL-Seq: Single-Cell RNA-Seq by Multiplexed Linear Amplification. Cell Reports. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 113.Ramskold D, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–82. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Picelli S, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 115.Yan L, et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 2013;20:1131–9. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 116.Deng Q, et al. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343:193–6. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- 117.Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. 2013;14:618–30. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 118.Wu AR, et al. Automated microfluidic chromatin immunoprecipitation from 2,000 cells. Lab Chip. 2009;9:1365–70. doi: 10.1039/b819648f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lorthongpanich C, et al. Single-cell DNA-methylation analysis reveals epigenetic chimerism in preimplantation embryos. Science. 2013;341:1110–2. doi: 10.1126/science.1240617. [DOI] [PubMed] [Google Scholar]
- 120.Oana H, et al. Non-destructive handling of individual chromatin fibers isolated from single cells in a microfluidic device utilizing an optically driven microtool. Lab Chip. 2013;14:696–704. doi: 10.1039/c3lc51111a. [DOI] [PubMed] [Google Scholar]
- 121.Benitez JJ, et al. Microfluidic extraction, stretching and analysis of human chromosomal DNA from single cells. Lab Chip. 2012;12:4848–54. doi: 10.1039/c2lc40955k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schmid L, Weitz DA, Franke T. Sorting drops and cells with acoustics: acoustic microfluidic fluorescence-activated cell sorter. Lab Chip. 2014 doi: 10.1039/c4lc00588k. [DOI] [PubMed] [Google Scholar]
- 123.Abate AR, et al. DNA sequence analysis with droplet-based microfluidics. Lab Chip. 2013;13:4864–9. doi: 10.1039/c3lc50905b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kantlehner M, et al. A high-throughput DNA methylation analysis of a single cell. Nucleic Acids Res. 2011;39:e44. doi: 10.1093/nar/gkq1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Market-Velker BA, Fernandes AD, Mann MR. Side-by-side comparison of five commercial media systems in a mouse model: suboptimal in vitro culture interferes with imprint maintenance. Biol Reprod. 2010;83:938–50. doi: 10.1095/biolreprod.110.085480. [DOI] [PubMed] [Google Scholar]
- 126.Gomez D, et al. Detection of histone modifications at specific gene loci in single cells in histological sections. Nat Methods. 2013;10:171–7. doi: 10.1038/nmeth.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nagano T, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hattori N, et al. Visualization of multivalent histone modification in a single cell reveals highly concerted epigenetic changes on differentiation of embryonic stem cells. Nucleic Acids Res. 2013;41:7231–9. doi: 10.1093/nar/gkt528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kind J, et al. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–92. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 130.Guo H, et al. Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res. 2013;23:2126–35. doi: 10.1101/gr.161679.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Smallwood SA, et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Meth. 2014;11:817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Geng T, et al. Histone modification analysis by chromatin immunoprecipitation from a low number of cells on a microfluidic platform. Lab Chip. 2011;11:2842–8. doi: 10.1039/c1lc20253g. [DOI] [PubMed] [Google Scholar]
- 133.Tan SJ, et al. A microfluidic device for preparing next generation DNA sequencing libraries and for automating other laboratory protocols that require one or more column chromatography steps. PLoS ONE. 2013;8:e64084. doi: 10.1371/journal.pone.0064084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kemna EW, et al. High-yield cell ordering and deterministic cell-in-droplet encapsulation using Dean flow in a curved microchannel. Lab Chip. 2012;12:2881–7. doi: 10.1039/c2lc00013j. [DOI] [PubMed] [Google Scholar]
- 135.Guo MT, et al. Droplet microfluidics for high-throughput biological assays. Lab Chip. 2012;12:2146–55. doi: 10.1039/c2lc21147e. [DOI] [PubMed] [Google Scholar]
- 136.Michalet X, et al. Detectors for single-molecule fluorescence imaging and spectroscopy. J Mod Opt. 2007;54:239. doi: 10.1080/09500340600769067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Paige MF, Bjerneld EJ, Moerner WE. A Comparison of Through-the-Objective Total Internal Reflection Microscopy and Epifluorescence Microscopy for Single-Molecule Fluorescence Imaging. Single Molecules. 2001;2:191–201. [Google Scholar]
- 138.So PT, et al. Two-photon excitation fluorescence microscopy. Annu Rev Biomed Eng. 2000;2:399–429. doi: 10.1146/annurev.bioeng.2.1.399. [DOI] [PubMed] [Google Scholar]
- 139.Rigler R, et al. Fluorescence correlation spectroscopy with high count rate and low background: analysis of translational diffusion. European Biophysics Journal. 1993;22:169–175. [Google Scholar]
- 140.Weber MA, Stracke F, Meixner AJ. Dynamics of single dye molecules observed by confocal imaging and spectroscopy. Cytometry. 1999;36:217–23. [PubMed] [Google Scholar]
- 141.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–5. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 142.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–5. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wenger J, Rigneault H. Photonic methods to enhance fluorescence correlation spectroscopy and single molecule fluorescence detection. Int J Mol Sci. 2010;11:206–21. doi: 10.3390/ijms11010206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Emory JM, Soper SA. Charge-coupled device operated in a time-delayed integration mode as an approach to high-throughput flow-based single molecule analysis. Anal Chem. 2008;80:3897–903. doi: 10.1021/ac800447x. [DOI] [PubMed] [Google Scholar]
- 145.Goens G, et al. Characterization and quality control of antibodies used in ChIP assays. Methods Mol Biol. 2009;567:27–43. doi: 10.1007/978-1-60327-414-2_2. [DOI] [PubMed] [Google Scholar]
- 146.Nishikori S, et al. Broad ranges of affinity and specificity of anti-histone antibodies revealed by a quantitative peptide immunoprecipitation assay. J Mol Biol. 2012;424:391–9. doi: 10.1016/j.jmb.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jorgensen HF, et al. Engineering a high-affinity methyl-CpG-binding protein. Nucleic Acids Res. 2006;34:e96. doi: 10.1093/nar/gkl527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Iurlaro M, et al. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol. 2013;14:R119. doi: 10.1186/gb-2013-14-10-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yamamoto T, Fujii T. Nanofluidic single-molecule sorting of DNA: a new concept in separation and analysis of biomolecules towards ultimate level performance. Nanotechnology. 2010;21:395502. doi: 10.1088/0957-4484/21/39/395502. [DOI] [PubMed] [Google Scholar]
- 150.Harris TD, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320:106–9. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 151.Goren A, et al. Chromatin profiling by directly sequencing small quantities of immunoprecipitated DNA. Nat Methods. 2010;7:47–9. doi: 10.1038/nmeth.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Levene MJ, et al. Zero-mode waveguides for single-molecule analysis at high concentrations. Science. 2003;299:682–6. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 153.Venkatesan BM, Bashir R. Nanopore sensors for nucleic acid analysis. Nat Nanotechnol. 2011;6:615–24. doi: 10.1038/nnano.2011.129. [DOI] [PubMed] [Google Scholar]
- 154.Soni GV, Dekker C. Detection of Nucleosomal Substructures using Solid State Nanopores. Nano Lett. 2012;12:3180–6. doi: 10.1021/nl301163m. [DOI] [PubMed] [Google Scholar]
- 155.Dean FB, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci U S A. 2002;99:5261–6. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Klein CA, et al. Comparative genomic hybridization, loss of heterozygosity, and DNA sequence analysis of single cells. Proc Natl Acad Sci U S A. 1999;96:4494–9. doi: 10.1073/pnas.96.8.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zong C, et al. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–6. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Marcy Y, et al. Nanoliter reactors improve multiple displacement amplification of genomes from single cells. PLoS Genet. 2007;3:1702–8. doi: 10.1371/journal.pgen.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shankaranarayanan P, et al. Single-tube linear DNA amplification (LinDA) for robust ChIP-seq. Nat Methods. 2011;8:565–7. doi: 10.1038/nmeth.1626. [DOI] [PubMed] [Google Scholar]
- 160.Adey A, et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Adey A, Shendure J. Ultra-low-input, tagmentation-based whole-genome bisulfite sequencing. Genome Res. 2012;22:1139–43. doi: 10.1101/gr.136242.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wang Q, et al. Tagmentation-based whole-genome bisulfite sequencing. Nat Protoc. 2013;8:2022–32. doi: 10.1038/nprot.2013.118. [DOI] [PubMed] [Google Scholar]
- 163.Buenrostro JD, et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–8. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 165.Furlan-Magaril M, Rincon-Arano H, Recillas-Targa F. Sequential chromatin immunoprecipitation protocol: ChIP-reChIP. Methods Mol Biol. 2009;543:253–66. doi: 10.1007/978-1-60327-015-1_17. [DOI] [PubMed] [Google Scholar]
- 166.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–36. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Booth MJ, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–7. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 168.Garcia BA, et al. Pervasive combinatorial modification of histone H3 in human cells. Nat Methods. 2007;4:487–9. doi: 10.1038/nmeth1052. [DOI] [PubMed] [Google Scholar]
- 169.Plazas-Mayorca MD, et al. One-pot shotgun quantitative mass spectrometry characterization of histones. J Proteome Res. 2009;8:5367–74. doi: 10.1021/pr900777e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Smits AH, et al. Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res. 2013;41:e28. doi: 10.1093/nar/gks941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jung HR, et al. Precision mapping of co-existing modifications in histone H3 tails from embryonic stem cells by ETD-MS/MS. Anal Chem. 2013;85:8232–9. doi: 10.1021/ac401299w. [DOI] [PubMed] [Google Scholar]