Abstract
Chromosome replication is initiated by a universal mechanism in eukaryotic cells, involving the assembly and activation at replication origins of the CMG (Cdc45-MCM-GINS) DNA helicase, which is essential for the progression of replication forks. Disassembly of CMG is likely to be a key regulated step at the end of chromosome replication, but the mechanism was unknown until now. Here we show that the ubiquitin ligase known as SCFDia2 promotes ubiquitylation of CMG during the final stages of chromosome replication in Saccharomyces cerevisiae. The Cdc48/p97 segregase then associates with ubiquitylated CMG, leading rapidly to helicase disassembly. These findings indicate that the end of chromosome replication in eukaryotes is controlled in a similarly complex fashion to the much-better-characterized initiation step.
The initiation of chromosome replication involves a set of conserved and highly regulated factors, which recruit Cdc45 and the GINS complex to the Mcm2-7 helicase core at origins of replication, producing the active DNA helicase that contains 11 subunits and is known as CMG (Cdc45-MCM-GINS) (1, 2), around which the replisome is built (3–5). Helicase assembly represents the key regulated step during the initiation of chromosome replication (3), and the CMG helicase then gets a single opportunity to unwind the parental DNA duplex in each region of the genome (6), thus ensuring that every chromosome is copied just once. Whereas the Mcm2-7 helicase core can be loaded around DNA only during the G1 phase of the cell cycle (7–9), as an inactive double hexamer of two Mcm2-7 rings, the missing helicase subunits known as Cdc45 and GINS can be recruited only when cells enter S phase (4, 5). This produces active CMG complexes that each contains a single Mcm2-7 hexamer (1).
Once assembled, the CMG helicase stably encircles the parental DNA and must remain continuously associated with each DNA replication fork (6), whatever barriers are encountered during the elongation phase of DNA replication (10, 11), because CMG cannot normally be reloaded after initiation. Nevertheless, the convergence of two replication forks leads rapidly to disassembly of the CMG helicase, and thus of the entire replisome, by a mechanism that until now has remained enigmatic.
Results
CMG is ubiquitylated on its Mcm7 subunit
In native extracts of S-phase budding yeast cells, we discovered a modification of the Mcm7 protein (fig. S1), which was specifically detected in the small fraction of Mcm7 that associates with GINS and Cdc45 at DNA replication forks, as part of the CMG helicase (1). The modification of Mcm7 was more evident above neutral pH, and above of pH 8.5 we found that almost all of the Mcm7 protein that associated with GINS was present as a ladder of modified forms (fig. S1A). Several lines of evidence demonstrated that the modification corresponded to ubiquitylation of Mcm7 (Fig. 1A and fig. S1C), involving the addition of K48-linked ubiquitin chains (Fig. 1B and fig. S1D).
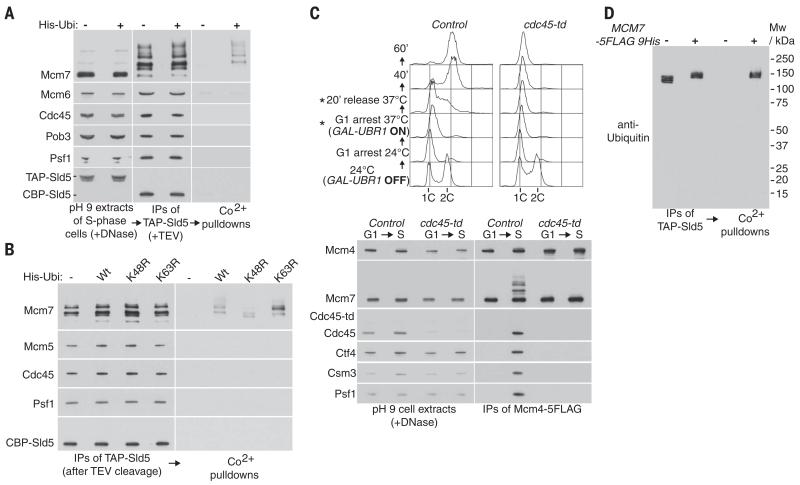
Fig. 1. The CMG DNA helicase is ubiquitylated on its Mcm7 subunit.
(A) Extracts of S-phase cells (YMM22 and YMM23), with or without a plasmid expressing His-tagged ubiquitin, were prepared at pH 9 before digestion of chromosomal DNA and isolation of the GINS component of the CMG helicase, via a TAP tag on the Sld5 subunit. The isolated complexes, representing a mixture of GINS and CMG helicase as part of the replisome progression complex (1), were then released from beads by cleavage with TEV protease before denaturation with 8 M urea (described in the materials and methods section), so that ubiquitylated factors could be isolated specifically on cobalt-coated magnetic beads. IPs, immunoprecipitates. (B) A similar experiment was performed with control cells (YMM22) or cells expressing wild-type (YMM23) ubiquitin, K48R ubiquitin (YMM89), or K63R ubiquitin (YMM90). (C) Control (YSS184) and cdc45-td (YMM74) cells were arrested in G1 phase at 24°C, before inactivation of Cdc45-td and release into S phase for 20 min, as described in the materials and methods. DNA content was measured by flow cytometry (asterisks denote samples that were used to prepare cell extracts). After digestion of chromosomal DNA in cell extracts, the Mcm4 subunit of the Mcm2-7 helicase core was then isolated by immunoprecipitation. This enriched specifically for two forms of Mcm2-7 complex, each of which are bound to chromatin (14): the inactive double hexamer at origins and the active CMG helicase at forks, the latter of which is the substrate for ubiquitylation on its Mcm7 subunit. (D) A similar experiment to that shown in (A) was performed with control cells (YASD375) or cells with tagged Mcm7 (YGDP483). The isolated material was resolved in a 4 to 12% gradient gel, before immunoblotting with anti-ubiquitin antibody (P4D1). Mw, molecular weight. Additional flow cytometry data for the experiments depicted in this figure can be found in fig. S14.
To confirm that Mcm7 was only ubiquitylated when it formed part of the CMG helicase, we isolated Mcm2-7 complexes that had been released from chromatin into yeast cell extracts by deoxyribonuclease (DNase) digestion. These comprised the inactive Mcm2-7 helicase core that is loaded at origins of replication before the initiation of chromosome replication (12, 13), and also the active CMG helicase that is present at DNA replication forks (1, 2)—these are the only two stable forms of the budding yeast Mcm2-7 complex under these extract conditions (14). Ubiquitylation of Mcm7 was only detected in isolated Mcm2-7 complexes and only upon entry into S phase (Fig. 1C, Control). Moreover, ubiquitylation was not detected if assembly of CMG was blocked by prior depletion of degron-tagged Cdc45 during G1 phase (Fig. 1C, cdc45-td). These findings define a ubiquitylation of the CMG helicase, which is specific for the Mcm7 subunit (Fig. 1D).
SCFDia2 is required for ubiquitylation of CMG in yeast extracts
Ubiquitylation of CMG was independent of the various E3 ubiquitin ligases and E2 ubiquitin conjugating enzymes that are known to regulate DNA replication and DNA repair in response to DNA damage [such as Rad18, Rad5, and Ubc13 that ubiquitylate PCNA (15, 16); the cullin Rtt101 and its partner Mms22 (17); and the SUMO-directed ubiquitin ligase Slx5-Slx8 (18, 19) (fig. S2)]. In contrast, ubiquitylation of CMG was completely blocked in the absence of the F-box protein known as Dia2 (fig. S2). The Dia2 protein maintains genome integrity and forms the substrate-targeting component of the SCFDia2 ubiquitin ligase (20–22), which associates with the “replisome progression complex” that assembles around the CMG helicase at replication forks (23).
In principle, the high efficiency of CMG ubiquitylation in these experiments could have had several very different explanations. One possibility was that almost all CMG complexes at DNA replication forks in vivo were ubiquitylated by SCFDia2. Alternatively, the yeast extract under these particular conditions might serve as an efficient and highly specific in vitro system, in which SCFDia2 drives ubiquitylation of the Mcm7 subunit of CMG helicase complexes that were derived from DNA replication forks in S-phase cells. In support of the latter idea, ubiquitylation of the helicase was blocked by an increased salt concentration in the extract (Fig. 2A, 700 mM salt), which does not affect the stability of CMG but prevents its association in vitro with dynamic partners (1, 24), including SCFDia2 (23). To prove that SCFDia2 was required for in vitro ubiquitylation of CMG, we developed an assay involving two yeast strains that were grown in parallel to each other. The first represented a recipient strain for the in vitro ubiquitylation assay and had a TAP tag on the Sld5 subunit of GINS, to allow the isolation of CMG helicase from the recipient yeast cell extracts. The second was a donor strain that provided the source of Dia2 for in vitro ubiquitylation of CMG. The recipient and donor strains were synchronized in S phase, before the two cultures were mixed in equal proportions and used to generate a common cell extract, from which recipient CMG was isolated by immunoprecipitation of TAP-Sld5. As controls, we showed that ubiquitylation of CMG was observed when both recipient and donor strains contained Dia2 (Fig. 2B, sample 1), but not if both strains lacked Dia2 (Fig. 2B, sample 2). Critically, we found that in vitro ubiquitylation of CMG complexes from dia2Δ recipient cells was partially restored when the donor strain contained wild-type levels of Dia2 (Fig. 2B, sample 3; note that the concentration of Dia2 was reduced twofold by the 1:1 mixture of DIA2 donor cells with dia2Δ recipient cells). Moreover, ubiquitylation was fully restored if the donor strain overexpressed DIA2 (Fig. 2B, sample 4). Finally, we showed that the addition of purified Dia2 was able to rescue the ubiquitylation of CMG in extracts of dia2Δ cells (Fig. 2C). These findings indicate that Dia2 promotes the ubiquitylation of CMG in yeast cell extracts.
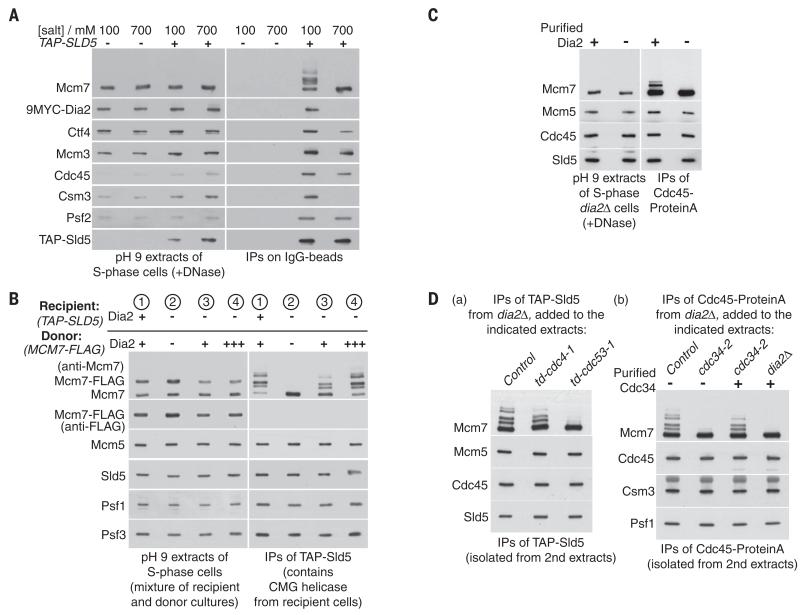
Fig. 2. SCFDia2 drives in vitro ubiquitylation of CMG in yeast cell extracts.
(A) Control (YHM117) and TAP-SLD5 cells (YHM132) were processed as above, except that the extracts were made in the presence of 700 mM salt where indicated. Note that the CMG helicase is still stable in the presence of 700 mM salt but does not associate with other replisome components such as Csm3 or Dia2. (B) To demonstrate that Dia2 promotes the in vitro ubiquitylation of CMG, we synchronized the indicated TAP-SLD5 recipient strains (1, YASD375; 2 to 4, YHM130) and MCM7-5FLAG donor strains (1 to 4, YTM305, YTM306, YTM305, and YTM312, respectively) in S phase at 30°C. Each of the indicated pairs of recipient and donor cultures were then mixed and used to prepare a single-cell extract at pH 9 (see materials and methods). After digestion of chromosomal DNA, the CMG helicase from recipient cells was isolated and monitored by immunoprecipitation of its TAP-Sld5 subunit. (C) dia2Δ CDC45-Protein A cells (YTM415) were synchronized in S phase, as above, and used to prepare a cell extract at pH 9. Purified Dia2 (from YTM532; see materials and methods) was added to the cell extracts as indicated, before immunoprecipitation of Cdc45-Protein A and detection of the associated CMG components by immunoblotting. (D) (a) Material containing CMG complexes was isolated from extracts of S-phase dia2Δ cells (YHM130) by immunoprecipitation of TAP-tagged Sld5 on IgG beads. The beads were then added to pH 9 extracts of control cells (YTM330), td-cdc4-1 cells (YTM445), or td-cdc53-1 cells (YTM444), which had been grown initially at 24°C, before shifting to 37°C for 60 min to inactivate Td-cdc4-1 and Td-cdc53-1 (see materials and methods for details). After reisolation of the beads, the indicated proteins were detected by immunoblotting. (b) Analogous experiments were performed with CMG-containing material that was isolated by immunoprecipitation of Cdc45-Protein from dia2Δ cells (YTM415). In this case, the beads were added to pH 9 extracts of control cells (YTM330), cdc34-2 cells (YTM376), or dia2Δ cells (YMM90), which were then shifted from 24° to 37°C for 60 min. As indicated, the extracts were supplemented with purified Cdc34, which was isolated on IgG beads from an extract of CDC34-TAP cells (YTM396) and then liberated with TEV protease.
The SCF represents a family of E3 ubiquitin ligases, of which each uses a different F-box protein to recruit substrates but all share a common scaffold comprising the cullin that in budding yeast is known as Cdc53. We showed that in vitro ubiquitylation of CMG was blocked by inactivation of Cdc53, but not by inactivation of the essential F-box protein Cdc4, which produces an equivalent arrest to cell cycle progression [Fig. 2D, (a)]. Similarly, we found that in vitro ubiquitylation of CMG was prevented by inactivation of the Cdc34 E2 enzyme, which ubiquitylates substrates of the SCF, and the defect could be rescued by addition of purified Cdc34 [Fig. 2D, (b)]. These findings demonstrate that SCFDia2 either ubiquitylates directly the Mcm7 subunit of the CMG helicase in yeast cell extracts or else is required for CMG to be ubiquitylated by another unknown ligase.
Dia2-dependent ubiquitylation links CMG to Cdc48 in vivo
To search for in vivo ubiquitylation of the CMG helicase at DNA replication forks, we isolated CMG complexes from high-salt extracts that blocked the in vitro ubiquitylation reaction (as shown in Fig. 2A). Ubiquitylated CMG helicase was not detected when cells entered S phase, even in the presence of DNA damage, or upon activation of the S-phase checkpoint by depletion of deoxynucleotide triphosphates (Fig. 3A). One possible explanation was that ubiquitylated CMG might exist only very transiently in vivo, either due to proteasomal degradation of ubiquitylated Mcm7, or else due to a disassembly reaction that could require the Cdc48/p97 segregase (25, 26). Inactivation of the Rpt6/Cim3 proteasome subunit blocks completion of the cell cycle (27), but the CMG helicase complex disappeared upon shifting an asynchronous culture of cim3-1 cells to 37°C (Fig. 3B) rather than accumulating in a ubiquitylated form. The disappearance of CMG was probably due to helicase disassembly at the end of S phase, because cim3-1 cells in S phase are able to finish DNA replication when the proteasome is inactivated at 37°C (fig. S3).
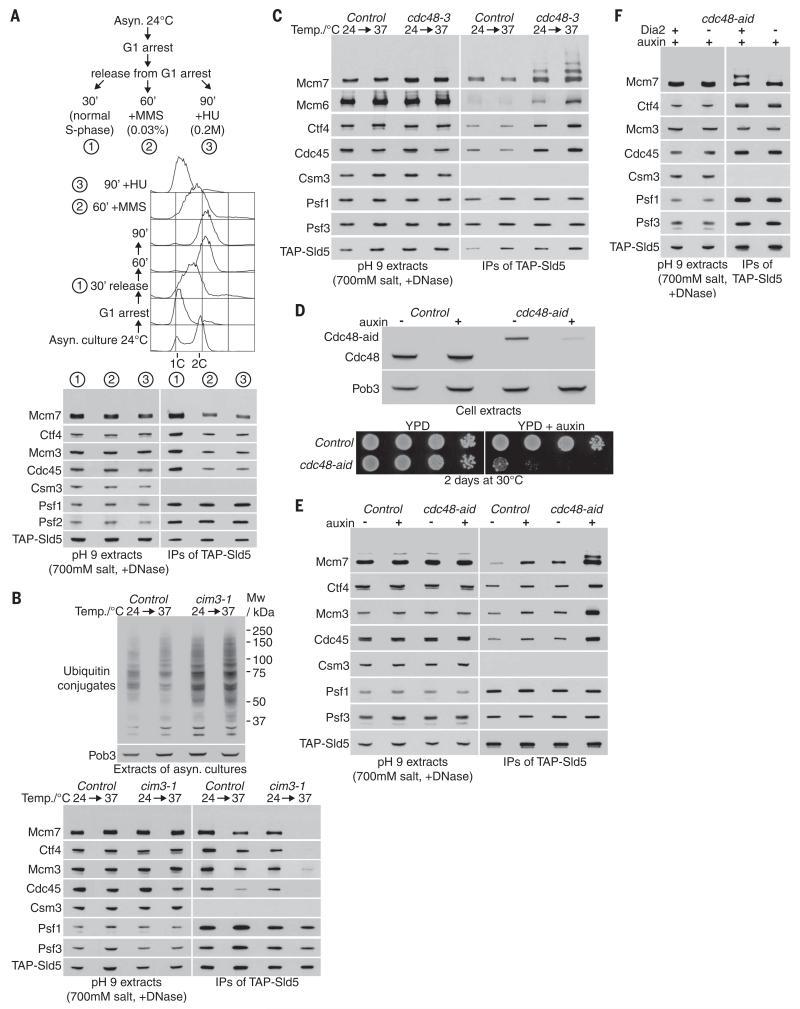
Fig. 3. Dia2-dependent ubiquitylation of CMG in vivo is revealed by inactivation of Cdc48.
(A) YASD375 was released into S phase either for 30 min (1), 60 min in the presence of 0.033% methyl methanesulfonate (MMS) (2), or 90 min in the presence of 0.2 M hydroxyurea (HU) (3) and then processed as in Fig. 1. DNA content was measured by flow cytometry. (B) Control (YASD375) and cim3-1 cells (YMM206) were grown at 24°C then incubated for a further hour at 24° or 37°C. The accumulation of ubiquitylated proteins was monitored by immunoblotting with an antibody specific for ubiquitin conjugates (FK2 antibody; upper panels). CMG was monitored as described above, by immunoprecipitation of TAP-tagged Sld5 (lower panels). (C) An analogous experiment to that in (B) was performed with control cells (YASD375) and cdc48-3 (YMM214). (D) The level of Cdc48 in control (YMM256) and cdc48-aid (YMM228) cells was monitored by immunoblotting before and after addition of auxin to the culture medium (upper panels). Serial dilutions of control (YJW15) and cdc48-aid (YMM203) cells were spotted onto the indicated media and incubated for 2 days at 30°C (lower panels). (E) Control (YMM256) and cdc48-aid cells (YMM228) were grown at 24°C, then incubated for 90 min in the presence or absence of auxin, as indicated, before processing as above. (F) Asynchronous cultures of cdc48-aid (YMM228) and cdc48-aid GALL-DIA2 (YMM283) were grown at 30°C in YPD media (see materials and methods for initial growth conditions of cdc48-aid GALL-DIA2), before addition of auxin for 2 hours.
In contrast, in vivo ubiquitylation of the Mcm7 subunit of CMG was detected, even at the permissive temperature of 24°C in cdc48-3 cells, and accumulated still further at the restrictive temperature of 37°C (Fig. 3C). Whereas similar amounts of GINS could be immunoprecipitated from control and cdc48-3 cells, more of the Cdc45 and Mcm2-7 proteins associated with GINS in the mutant, suggesting that the CMG helicase complex had accumulated in cells with defective Cdc48/p97 (Fig. 3C, also see below). These findings indicated that ubiquitylated CMG helicase exists in vivo but is normally a transient species that is rapidly processed in a reaction that requires the Cdc48 adenosine triphosphatase (ATPase).
To explore the regulation of CMG by ubiquitylation in vivo, we generated a degradable allele of Cdc48 (Fig. 3D) by fusion of the endogenous CDC48 gene to the auxin-inducible degron (28). Ubiquitylated CMG helicase was not detected in an asynchronously growing culture of cdc48-aid cells, but addition of auxin to the culture led to accumulation of CMG with ubiquitylated Mcm7 (Fig. 3E). Consistent with our in vitro data, in vivo ubiquitylation of the CMG helicase required Dia2 (Fig. 3F) but was independent of Rad5, Rad18, Ubc13, Slx5-Slx8, or Rtt101 (fig. S4). These data indicate that SCFDia2 drives CMG ubiquitylation in vivo, leading rapidly to Cdc48-dependent disassembly.
Dia2 is required for CMG disassembly
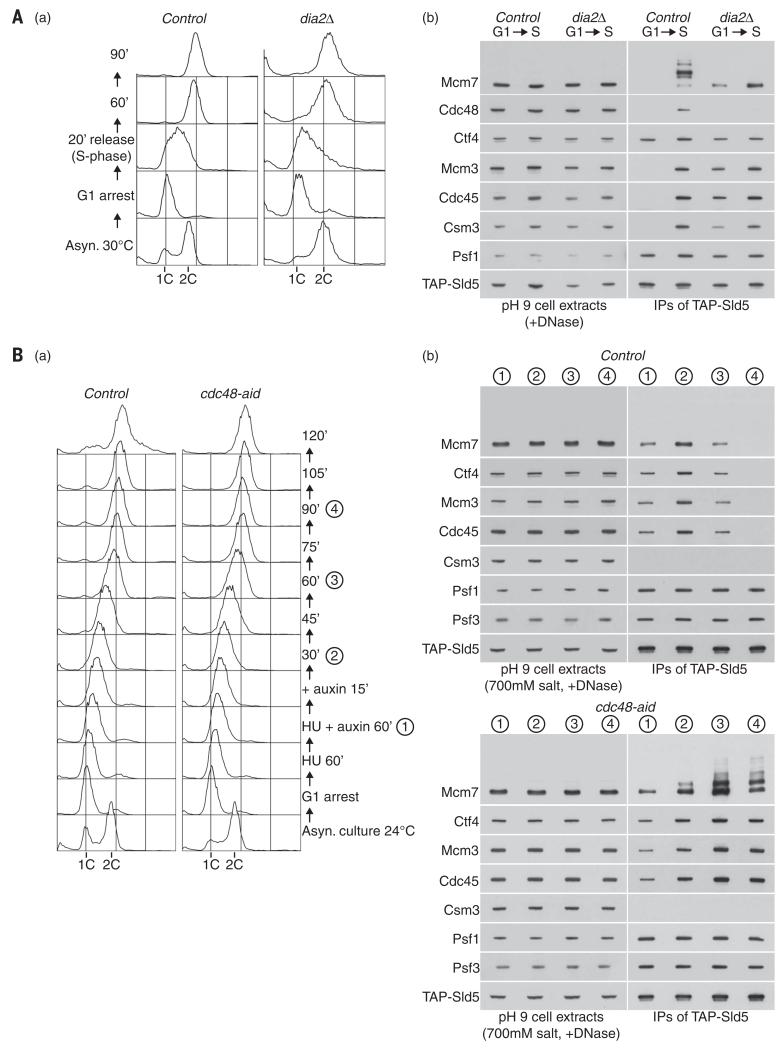
The CMG helicase is assembled during the initiation of chromosome replication and then remains associated with DNA throughout the elongation phase, until the convergence of two replication forks terminates DNA synthesis, and leads to helicase disassembly. Whereas the CMG helicase is not present before S phase in control cells (1), we found that dia2Δ cells contained stable CMG complexes even during G1 phase (Fig. 4 and fig. S5). This phenotype was not observed in the absence of other E3 ubiquitin ligases that regulate chromosome replication (fig. S6, A and B), or upon deletion of genes encoding other factors that preserve genome integrity at DNA replication forks, such as the Rrm3 helicase, the Blooms helicase Sgs1, Topoisomerase 3, or the recombination factors Rad51 and Rad52 (fig. S6, C and D).
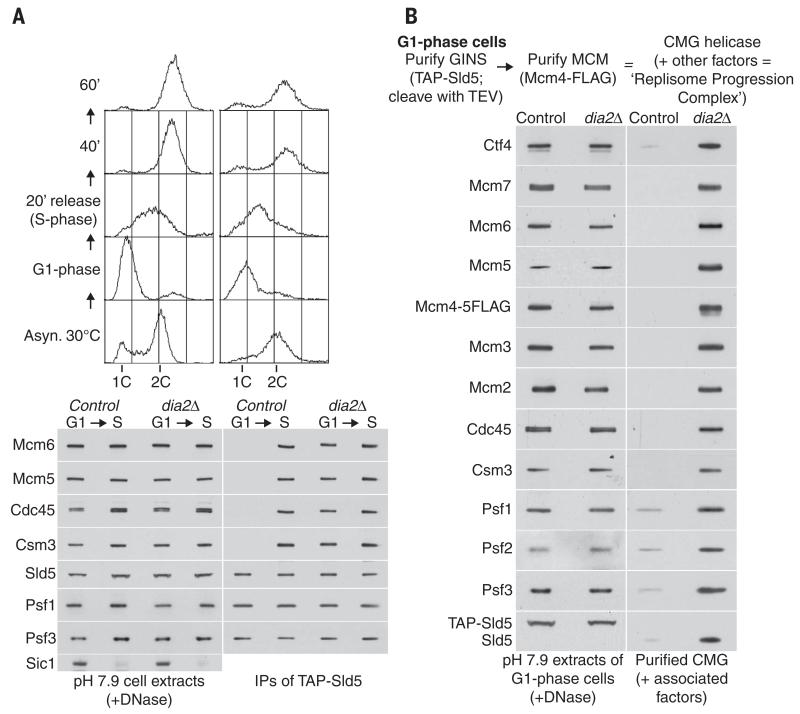
Fig. 4. CMG is present during the G1 phase of the cell cycle in the absence of Dia2.
(A) Control (YASD375) and dia2Δ (YHM130) cells were grown at 30°C, synchronized in G1 phase by addition of mating pheromone, and then released into S phase. Samples were taken at the indicated times (S phase, 20-min sample) and processed for flow cytometry (top panels) or else used to make cell extracts (containing 100 mM salt), before digestion of chromosomal DNA and immunoprecipitation of the TAP-tagged Sld5 subunit of GINS (lower panels). The association of GINS with Cdc45 and Mcm2-7, as well as with other components of the replisome progression complex such as Csm3 and Ctf4 (1), was monitored by immunoblotting of the indicated proteins. (B) Cell extracts were generated as above, from G1 phase control (YAG230-3) and dia2Δ (YTM687) cells. To screen for the presence of the CMG helicase, GINS was isolated as above, before release of the purified material from beads by cleavage with TEV protease and immunoprecipitation of Mcm4 (1). The final material was analyzed by immunoblotting, along with the cell extracts.
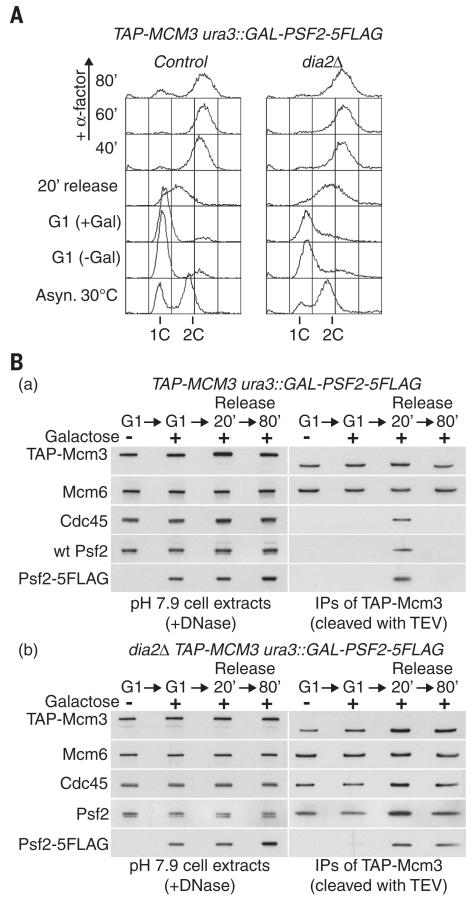
To monitor directly the assembly and disassembly of the CMG helicase during a single cell cycle, we arrested control and dia2Δ cells in G1 phase and then expressed a tagged version of the Psf2 subunit of GINS (in addition to the endogenous PSF2 gene) before allowing the cells to enter S phase and complete chromosome replication (Fig. 5A). When control cells entered S phase, tagged Psf2 was incorporated into CMG complexes that subsequently disappeared when replication was completed [Fig. 5B, (a)]. In contrast, newly formed CMG complexes were assembled on schedule when dia2Δ cells entered S phase, but CMG disassembly did not occur at the end of S phase [Fig. 5B, (b); see Psf2-5FLAG]. We observed the same result when a tagged version of Cdc45 was expressed from G1 phase onward in equivalent experiments (fig. S7), and it appears that the CMG persists into the next cell cycle in cells lacking Dia2, with much of the complex still associated with chromatin (fig. S8). Moreover, the persistence of CMG after S phase in dia2Δ cells did not reflect the unscheduled assembly of new CMG complexes (fig. S9). These findings demonstrate that Dia2 is essential for the disassembly of the CMG helicase, when budding yeast cells complete chromosome replication.
Fig. 5. Disassembly of the CMG helicase at the end of S phase requires Dia2.
(A) A plasmid containing the GAL-PSF2-5FLAG construct was integrated at the ura3 locus in control (YTM593) and dia2Δ (YTM592) cells, which were then synchronized at 30°C in G1 phase with mating pheromone, in medium lacking galactose. Cells were held in G1 phase for a further 60 min in the presence of galactose to induce expression of Psf2-5FLAG, before release for the indicated times in medium lacking mating pheromone. Alpha factor mating pheromone was added again from 40 min onward to prevent nuclei from entering into the next round of S phase, and DNA content was measured throughout the experiment by flow cytometry. (B) Cell extracts from the same experiment as depicted in (A) were treated with DNase before immunoprecipitation of TAP-tagged Mcm3. The material was released from beads with TEV protease before immunoblotting, to prevent TAP-Mcm3 from interfering with the similarly sized Mcm6 protein.
To begin to study how the role of Dia2 in CMG disassembly is regulated in vivo, we generated a yeast strain in which expression of the endogenous DIA2 gene was controlled by a weakened GALL version of the GAL1 promoter (29, 30). Whereas GALL-DIA2 behaved like control cells in medium containing galactose, repression of GALL-DIA2 led to the persistence of “old” CMG complexes in G1 phase (Fig. 6, A and B, stage 1), even upon prolonged incubation (Fig. 6, A and B, stage 2). Critically, however, reexpression of DIA2 in G1 phase led to efficient disassembly of these CMG complexes from previous cell cycles (Fig. 6, A and B, stage 3). Nevertheless, expression of GALL-DIA2 did not interfere with the assembly of new CMG complexes when cells entered the subsequent S phase (Fig. 6, A and B, stage 4). These data indicate that Dia2 plays a direct role in CMG disassembly but is only able to induce the disassembly of the terminated helicase complexes that are produced when cells complete chromosome replication.
Fig. 6. Dia2 induces disassembly of terminated CMG complexes but does not prevent assembly of active CMG complexes.
(A) (a) Cells in which the endogenous DIA2 gene had been placed under the control of the weak and inducible GALL promoter (YTM568) were grown at 30°C in medium lacking galactose (GALL-DIA2 OFF) and then synchronized in G1 phase by addition of mating pheromone (stage 1). The cells were then held for a further 60 min in G1 phase, either in the continued absence of galactose (GALL-DIA2 OFF, stage 2), or in the presence of galactose (GALL-DIA2 ON, stage 3) before subsequent release for 20 min into S phase (GALL-DIA2 ON, stage 4). (b) DNA content was monitored by flow cytometry. (B) Cell extracts were prepared, treated with DNase, and then used to isolate GINS as above, before monitoring of the associated factors by immunoblotting as indicated.
Dia2 drives CMG ubiquitylation during the completion of DNA replication
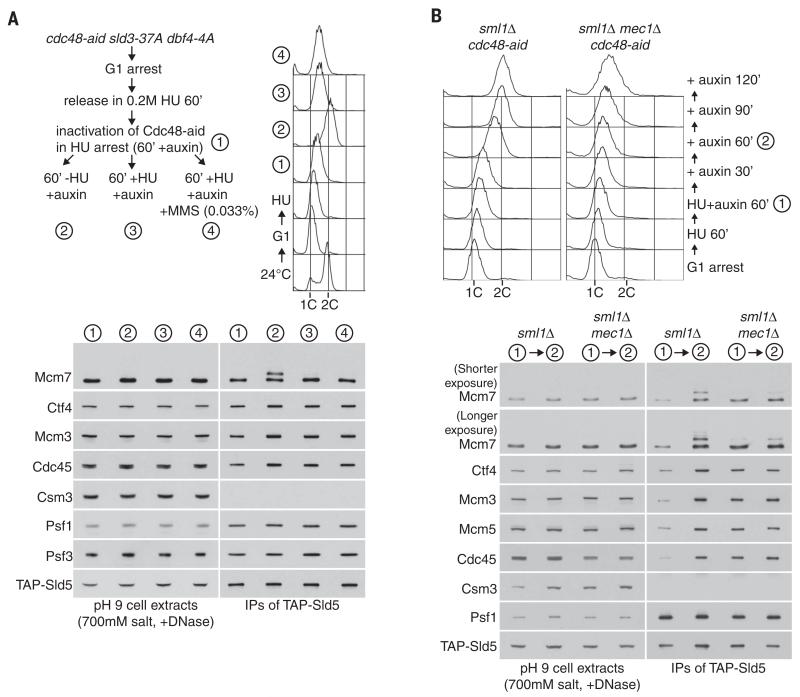
The action of SCFDia2 is likely to be a key regulated step in the dissolution of the CMG helicase, leading rapidly to Cdc48-dependent disassembly. To establish whether ubiquitylated CMG is a transient form of the helicase that is generated throughout S phase, or else is induced by specific signals, we examined the consequences of inactivating Cdc48-aid in early S phase. When the progression of DNA replication forks was inhibited by addition of hydroxyurea to the culture medium, inactivation of Cdc48-aid did not reveal ubiquitylation of CMG (Fig. 7A, stage 1), even after prolonged incubation (Fig. 7A, stage 3) or in combination with DNA damage (Fig. 7A, stage 4). In contrast, washing cells into fresh medium lacking hydroxyurea allowed them to complete DNA replication and led to the accumulation of ubiquitylated CMG (Fig. 7A, stage 2; also see fig. S10). This was not due to inhibition of ubiquitylation by the S phase checkpoint pathway (Fig. 7B), and instead these data indicate that ubiquitylation of CMG is linked to a late step in DNA replication. In similar experiments, we examined all 11 subunits of CMG in immunoprecipitates of GINS or Mcm2-7 and found that Mcm7 was the predominant target for ubiquitylation in vivo (fig. S11), mirroring the in vitro ubiquitylation reaction that occurs in yeast cell extracts (Fig. 1).
Fig. 7. The CMG helicase is ubiquitylated in vivo during a late step of DNA replication.
(A) cdc48-aid sld3-37A dbf4-4A (YMM309) was synchronized in G1 phase and then released for 60 min into S phase in the presence of 0.2 M hydroxyurea (HU) to allow early and late origins to fire (55). Auxin was then added to inactivate Cdc48-aid (1) before the culture was split and cells were released for 60 min in fresh medium lacking HU (2), held for a further 60 min in the presence of HU (3), or subjected to DNA damage (by addition of 0.033% MMS) in the continued presence of HU (4). The sld3-37A dbf4-4A background was used to allow late origins to fire in the HU arrest, thus ensuring that cells would have a similar number of CMG complexes both before and after release from HU arrest, making easier the comparison of the Mcm7 subunit of CMG in the two cases. DNA content was measured by flow cytometry. (B) A similar experiment was performed with control cells (YMM366, sml1Δ cdc48-aid ADH-TIR1) and mec1Δ (YMM368, mec1Δ sml1Δ cdc48-aid ADH-TIR1) strains. Inactivation of Cdc48-aid in hydroxyurea HU-arrested cells did not cause efficient accumulation of ubiquitylated CMG helicase in either control or mec1Δ cells, consistent with the notion that ubiquitylation only occurs during a late step of DNA replication. When cells were washed into fresh medium lacking hydroxyurea HU, completion of DNA replication in control cells led to the accumulation of ubiquitylated CMG helicase, but this was much less efficient in mec1Δ cells, in which the replisome is stable but is unable to resume remain functional after DNA replication stress (52).
Cdc48-dependent CMG disassembly when replication terminates
In favor of Cdc48 playing a direct role in the disassembly of CMG, we found that Cdc48 copurified with ubiquitylated CMG helicase from extracts of control S-phase cells, but the association was lost in extracts of dia2Δ cells (Fig. 8A). To determine the consequences of ubiquitylating the CMG helicase during late S phase, we monitored the completion of S phase upon release from hydroxyurea arrest, either in control cells or after inactivation of Cdc48-aid. Both strains reached a 2C DNA content with similar kinetics [Fig. 8B, (a)], indicating that the progression of DNA replication forks was not prevented by depletion of Cdc48-aid. In control cells, CMG briefly accumulated upon release from hydroxyurea arrest and then disappeared completely as cells completed DNA replication [Fig. 8B, (b), Control], reflecting the normal process of helicase disassembly at the end of S phase. In marked contrast, helicase disassembly was completely blocked in the absence of Cdc48, and instead progression through DNA replication led to a progressive accumulation of the CMG helicase, coupled to ubiquitylation of its Mcm7 subunit [Fig. 8B, (b), cdc48-aid]. Isolation of the ubiquitylated CMG helicase from cell extracts was still dependent on digestion of the parental DNA (fig. S12), indicating that the ubiquitylation and disassembly of CMG normally occur when the helicase is still present on chromatin at the end of DNA replication. Together, these findings demonstrate that Cdc48 is essential for disassembly of the CMG helicase, when budding yeast cells complete chromosome replication. In addition to its recently identified roles in the cellular response to DNA damage and defective DNA synthesis (31-37), our results identify Cdc48 as a key player in the final stages of eukaryotic chromosome replication.
Fig. 8. Cdc48 is required for disassembly of CMG at the end of chromosome replication.
(A) Control (YASD375) and dia2Δ (YHM130) were grown as indicated, and DNA content was measured by flow cytometry (a). GINS was isolated from cell extracts, and the association of the indicated factors (including Cdc48) was monitored by immunoblotting (b). (B) Control (YMM256) and cdc48-aid cells (YMM228) were initially processed as described above for Fig. 7, but were then released from HU arrest for the indicated times in the presence of auxin.
Discussion
The encounter of two converged DNA replication forks leads locally to the termination of DNA synthesis in that particular region of the chromosome and to dissolution of the replisome at each fork. Our findings indicate that replisome dissolution is driven by the regulated disassembly of the stable CMG helicase, which uses its hexameric ring of Mcm2-7 proteins to encircle the parental DNA at replication forks. Our data identify two key features of helicase disassembly in budding yeast: First, there is an essential role for the F-box protein Dia2, which forms part of the E3 ligase SCFDia2 and drives ubiquitylation of the CMG helicase on its Mcm7 subunit. Second, the Cdc48 segregase is required to break ubiquitylated CMG into its three component parts. Once separated from GINS and Cdc45, the Mcm2-7 hexamer is much less stable (fig. S13), so that all three components of the CMG helicase are lost from the newly replicated DNA.
These data raise a series of questions regarding the final stages of chromosome replication in eukaryotes. The mechanisms by which CMG is first ubiquitylated at the end of chromosome replication and then disassembled in a Cdc48-dependent process remain to be elucidated in future studies. It will be interesting to determine how other factors, such as the MCM binding protein (MCM-BP) that is not found in budding yeasts (38, 39), might contribute to this process. Another candidate is the Pif1/Rrm3 family of helicases that is required for the timely resolution of converged DNA replication forks during termination (40, 41), although we found that budding yeast cells lacking Rrm3 are still able to disassemble the CMG helicase before the end of the cell cycle (fig. S6C). By analogy with the extensive studies over the past decade of CMG assembly during the initiation of chromosome replication, it will not only be necessary to define the list of factors that are required for helicase disassembly, but will also be important to develop experimental systems with which each step of the reaction can be recapitulated in vitro. Ultimately, the reconstitution of each step of CMG disassembly with purified components would then allow a more detailed exploration of the underlying mechanisms.
Much also remains to be learned about how cells ensure that the CMG helicase is never disassembled before replication terminates in each section of the genome, and yet is always disassembled when two DNA replication forks meet each other. Considering budding yeast, our data indicate that Dia2 specifically induces the disassembly of terminated CMG complexes in vivo (Fig. 6), but in yeast cell extracts is able to ubiquitylate CMG complexes that are derived from active forks (e.g., Fig. 1 and fig. S1) under in vitro conditions that presumably override some of the normal controls that act at DNA replication forks.
It seems likely that the ubiquitylation of CMG only occurs in vivo when two replication forks converge, although this remains to be demonstrated directly. A major challenge for future studies will be to identify which features of replication termination provide the signal for CMG ubiquitylation. It is possible that the CMG helicase undergoes a structural change or is otherwise modified during the latter stages of replication, in a manner that controls recruitment of the ubiquitin ligase and/or Cdc48. It is also conceivable that the latter factors are regulated independently of CMG during the termination of replication, although our data show that both Dia2 and Cdc48 can induce the disassembly of terminated CMG complexes, even during G1 phase (Fig. 6), suggesting that the cell cycle stage is not a key determinant of the disassembly reaction. Further outstanding questions include how the ubiquitylation of CMG is restricted to its Mcm7 subunit, and how ubiquitylation of Mcm7 is limited to the small fraction of this protein that is present at replication forks in the CMG complex. We note that our data do not address whether SCFDia2 or Cdc48 play any role in the removal of inactive Mcm2-7 complexes from the path of progressing replication forks.
Just as the main features of helicase assembly have been conserved across evolution from yeasts to humans, we envisage that disassembly of the CMG helicase will also prove to involve a universal mechanism in eukaryotic cells. In this case, a major challenge will be to identify the ubiquitin ligase(s) that ubiquitylate CMG in other species. The fission yeast Pof3 protein appears to be orthologous to budding yeast Dia2 (42), although it remains to be determined whether SCFPof3 controls disassembly of CMG. It is unclear whether homologs of Dia2 are also present in higher eukaryotes, and it is possible that other ligases might control disassembly of the CMG helicase at the end of chromosome replication.
We anticipate that the role of Cdc48/p97 in CMG disassembly will be conserved in all eukaryotes. Consistent with this view, inhibition of p97/Cdc48 in early embryos of Caenorhabditis elegans, or in extracts of Xenopus laevis eggs, was found to cause the persistence of GINS on mitotic chromatin (43). Although it was thought that this phenomenon was due to a failure to degrade the Cdt1 loading factor of the Mcm2-7 helicase core (43), our results suggest that these data might instead reflect a conserved role for p97 in the disassembly of ubiquitylated CMG helicase at the end of chromosome replication. Accordingly, Mcm7 is ubiquitylated on chromatin during chromosome replication in extracts of Xenopus eggs, and the removal of CMG subunits from chromatin is dependent on p97/Cdc48 (44). It will be important to determine which adaptors of Cdc48/p97 are required for helicase disassembly in different eukaryotic species. A better understanding of the CMG disassembly pathway in human cells might help to explain the potential efficacy in antitumor therapies of small-molecule inhibitors of the p97/Cdc48 ATPase (45).
Materials and methods
Yeast strains and plasmids
The budding yeast Saccharomyces cerevisiae strains that were used in this study are listed in table S1. A standard one-step polymerase chain reaction (PCR) approach was applied to generate yeast strains harboring individual gene deletions or with epitope tags at the C or N terminus of the target protein (46). All the modifications were initially made in the diploid W303-1, before derivation of haploids by tetrad analysis. The cdc48-aid allele was constructed by transformation of cells with a PCR product containing the auxin inducible degron tag (28) and the hphNT marker (29). The resultant cdc48-aid haploid cells were then crossed to cells expressing the rice Tir1 E3 ligase from the ura3 locus (a generous gift from K. Nishimura and M. Kanemaki). Deletions of the RAD5, RAD18, and UBC13 genes were obtained by transformation of a PCR cassette containing the hphNT marker (29). A similar approach was used to make the GALL-DIA2 strain (29), which allows very low expression of DIA2 in medium lacking galactose, due to point mutations in the TATA box of the GAL1 promoter (30). To generate alleles of CDC53 and CDC4 that would allow very efficient inactivation of protein function at 37°C, we tagged existing temperaturesensitive alleles with the heat-inducible degron cassette, as described previously (47). In this way, we inserted the degron cassette (48) into the corresponding genomic locus in cdc53-1 and cdc4-1 cells, producing the alleles td-cdc53-1 and td-cdc4-1. DNA fragments expressing Psf2-5FLAG, Cdc45-5FLAG9His, 5FLAG-Dia2, and ProteinA-3TEV-sites-Dia2 were cloned downstream of the GAL1 promoter in the pRS series of plasmids (49). The resultant vectors were then linearized within the marker sequence and integrated into the ura3 or leu2 locus of the corresponding yeast strains. The plasmid pRS423-CUP-His7-Ubi was a gift from H. Ulrich and was used to make the derivatives pRS423-CUP-His7-UbiK48R and pRS423-CUP-His7-UbiK63R by site-directed mutagenesis involving three-step PCR reactions.
Growth of yeast cells
Yeast cells were grown at 24°, 30°, or 37°C in YP medium supplemented with sugar. The composition of YP medium is 1% (w/v) yeast extract (Becton Dickinson, 212750) and 2% (w/v) bacteriological peptone (Oxoid, LP0037B). The medium was then supplemented with 2% glucose (YPD), 2% raffinose (YPRaff), or 2% galactose (YPGal). All experiments with dia2Δ cells were performed at 30°C because the cells are sensitive to cold (23). To synchronize the cells in G1 phase, we added α-factor mating pheromone (Pepceuticals Limited) to log-phase cell cultures (cell density: 7 × 106 cells/ml) to a final concentration of 7.5 μg/ml. When cells were grown at 24°C, additional aliquots of 2.5 μg/ml (for cdc48-aid cells, 5 μg/ml) were added after an hour and a half and then every 30min until more than 90% of cells were unbudded and had formed “shmoos.” When cells were grown at 30°C, additional aliquots of 2.5 μg/ml (for dia2Δ cells, 5 μg/ml) of α-factor were added every 15 min after 1 hour until ~90% cells were unbudded.
To release from G1-phase arrest, cells were washed twice with fresh medium lacking α-factor. To arrest cells in early S phase upon release from mating pheromone arrest, cells were washed twice in YPD medium containing 0.2 Mhydroxyurea (H8627, Sigma-Aldrich, and 10872383, Molekula). For the experiment shown in fig. S3, an asynchronous culture of cim3-1 cells was arrested in early S phase by incubation for one generation time in medium containing 0.2 M hydroxyurea (cim3-1 cells are resistant to arrest in G1 phase with mating pheromone).
To arrest cells in G2-M phase, the medium was supplemented with nocodazole (Sigma-Aldrich, M1404) at 5 μg/ml. The cells were then incubated until ~90% had large buds. To induce DNA damage during S phase, cells were released into S phase in the presence of 0.033% methyl methanesulfonate [(MMS), M4016, Sigma-Aldrich]. To induce degradation of proteins fused to the auxin-inducible degron, we added 3-indolacetic acid (I3750, Sigma-Aldrich) to the culture medium at a final concentration of 0.5 mM.
For the experiment shown in Fig. 1C, involving depletion of Cdc45-td, cells were first grown at 24°C in YPRaff medium containing 0.1 mM CuSO4, before induction of GAL-UBR1 for 45 min in YPGal medium at 24°C (6). For the experiment in Fig. 2D, where Cdc53 and Cdc4 were inactivated, cells were grown at 24°C in YPRaff medium with 0.1 mM CuSO4, before induction of GAL-UBR1 for 35 min. For the experiment shown in Fig. 3F, GALL-DIA2 cdc48-aid cells were grown overnight at 30°C in YP medium supplemented with 2% sucrose and 0.1% galactose, to allow cells to grow well but also to minimize the initial expression level of Dia2. The culture was then switched to YPD medium for 8 hours at 30°C to repress GALL-DIA2 (we confirmed that cells would still have been able to grow for one more generation time), before addition of auxin to inactivate Cdc48-aid.
Analyzing cell growth by serial dilution on solid medium
Fresh colonies of yeast cells were grown on YPD plates, and serial 10-fold dilutions of cells were then made in phosphate-buffered saline, before plating cells in spots ranging from 5 × 104 to 5 × 10 cells. Cells were incubated at the indicated temperature, and the plates were scanned each day for 3 days.
Flow cytometry
Fixed cells for the measurement of DNA content were prepared as described previously (50). Samples were then analyzed using a FACSCalibur flow cytometer and FACSCanto II flow cytometer (Becton Dickinson), and the profiles were analyzed by CellQuest software (Becton Dickinson) or FlowJo software (TreeStar).
Preparation of denatured cell extracts containing trichloroacetic acid
For the experiments shown in Fig. 3, B (upper panel) and D, ~108 cells were used to generate total protein extracts in the presence of trichloroacetic acid to inactivate proteases, as described previously (51).
Immunoprecipitation of protein complexes from yeast cell extracts
Standard cell extracts (pH 7.9)
The preparation of cell extracts from frozen yeast cells, as well as the immunoprecipitation of protein complexes from extracts, was based on previously described methods (1, 23, 52). Briefly, 250 ml of a cell culture was pelleted at 200 × g for 3 min. Cells were washed once in 50 ml of 20 mM HEPES-KOH pH 7.9 buffer and once in 10 ml of lysis buffer (100 mM HEPES-KOH pH 7.9, 50 mM potassium acetate, 10 mM magnesium acetate, 2 mM EDTA) and then resuspended in three volumes of lysis buffer (1 g of cell pellet was taken as being equivalent to 1 ml of buffer) supplemented with 2 mM sodium fluoride, 2 mM sodium β-glycerophosphate pentahydrate, 1 mM dithiothreitol (DTT), 1% Protease Inhibitor Cocktail (P8215, Sigma-Aldrich), and 1× Complete Protease Inhibitor Cocktail (05056489001, Roche; one tablet dissolved in 1 ml water makes a 25× stock solution) before freezing dropwise in liquid nitrogen. We ground ~2.5 g of frozen yeast cells in a SPEX SamplePrep 6780 Freezer/Mill. After thawing, we measured the volume of the extract and then added one-quarter of the volume of glycerol mix buffer, comprising lysis buffer plus 50% glycerol, 300 mM potassium acetate, 0.5% detergent IGEPAL CA-630 (18896, Sigma-Aldrich), inhibitors, and DTT at the concentrations mentioned above, such that the extract contained 10% glycerol, 100 mM potassium acetate, and 0.1% IGEPAL CA-630. Chromosomal DNA was then digested by addition of 800 U/ml Benzonase Nuclease (71206, Merck) or 400 U/ml Pierce Universal Nuclease (123991963, Fisher) and incubation for 30 min at 4°C. Insoluble cell debris was pelleted in two high-speed centrifugation steps (at 25000 × g for 30 min and then at 100000 × g for 1 hour). To analyze cell extracts subsequently, 100 μl of concentrated Laemmli buffer (1.5 x) was added to 50 μl of cell extract [final concentration of Laemmli buffer corresponds to 6.66% (v/v) glycerol, 715 mM β-mercaptoethanol, 3% (w/v) sodium dodecyl sulfate, 62.5 mM Tris-HCl pH 6.9, and 0.00416% (w/v) Bromophenol Blue]. Cell extracts with added Laemmli buffer were then heated at 95°C for 5 min and stored at −80°C.
The remaining fraction of each cell extract (~2 ml) was then split into two aliquots, each of which was incubated for 2 hours at 4°C with 1.7 × 109 magnetic beads (Dynabeads M-270 Epoxy; 14302D, Life Technologies) that had been coupled to rabbit immunoglobulin G (IgG) (S1265, Sigma-Aldrich) or M2 anti-FLAG monoclonal antibody (F3165, Sigma-Aldrich). After the incubation, protein complexes bound on antibody-coupled magnetic beads were washed four times with 1 ml of wash buffer [100 mM HEPES-KOH pH 7.9, 100 mM potassium acetate, 10 mM magnesium acetate, 2 mM EDTA, 0.1% IGEPAL CA-630, 2 mM sodium fluoride, 2 mM sodium β-glycerophosphate pentahydrate, 1% Protease Inhibitor Cocktail (Sigma-Aldrich), and 1× Complete Protease Inhibitor Cocktail (Roche)]. The bound proteins were then eluted by heating at 95°C for 5 min in 50 μl Laemmli buffer and stored at −80°C.
pH 9 cell extracts and high-salt cell extracts
For the experiment in fig. S1A, all respective buffers for immunoprecipitation were made as indicated, with either HEPES-KOH (at pH 7.0, pH 7.9, and pH 8.5) or Tris-acetate (at pH 9). Buffers at pH 7.9 were used in experiments shown in Figs. 4 to 6 and figs. S5 to S9, S12, and S13. For all the other experiments, we used Tris-acetate buffers at pH 9. For the experiments in Figs. 1, 2, 4 to 6, and 8A, as well as figs. S1, S2, S6 to S9, and S13, the lysis buffer and wash buffers contained a final concentration of 100 mM potassium acetate. In other experiments, the buffers all contained 700 mM potassium acetate. In Fig. 2A and fig. S10, both 100 and 700 mM potassium acetate buffers were used, respectively.
More concentrated cell extracts from 1-liter cultures
Some experiments required more concentrated cell extracts, to improve visualization of ubiquitylated forms of Mcm7. In these experiments (Figs. 1, A, B, and D; and 3, C, E, and F; and figs. S1, C and D; S4; and S10), cell extracts were made from 1-liter cell cultures. After washing, the cells were finally resuspended in one-quarter of a volume of lysis buffer supplemented with 8 mM sodium fluoride, 8 mM sodium β-glycerophosphate pentahydrate, 4 mM DTT, 4% Protease Inhibitor Cocktail (Sigma-Aldrich), and 4× Complete Protease Inhibitor Cocktail (Roche). After grinding of the cells, we added 1 ml of lysis buffer (with the appropriate pH and salt concentration) supplemented with 2 mM sodium fluoride, 2 mM sodium β-glycerophosphate pentahydrate, 1 mM dithiothreitol, 1% Protease Inhibitor Cocktail (Sigma-Aldrich), and 1× Complete Protease Inhibitor Cocktail (Roche) to the thawing extracts. The extracts were then adjusted by addition of one-quarter of the volume of the appropriate glycerol mix as above, before digestion of chromosomal DNA with 3200 U/ml of Benzonase Nuclease or 1600 U/ml of Pierce Universal Nuclease.
Isolation of His-tagged ubiquitylated Mcm7
For the experiments in Fig. 1, A, B, and D; and fig. S1D, material containing CMG was isolated as described above, by immunoprecipitation of TAP-Sld5 at pH 9 and with 100 mM potassium acetate from 1-liter cell cultures. Both aliquots of magnetic beads with bound complexes were then washed three times with 1 ml of pH 9 wash buffer (100 mM Tris-acetate pH 9, 100 mM potassium acetate, 10 mM magnesium acetate, 0.1% IGEPAL CA-630, 2 mM sodium fluoride, 2 mM sodium β-glycerophosphate pentahydrate) and once with 1 ml of pH 7.9 wash buffer (100 mM HEPES-KOH pH 7.9, 100 mM potassium acetate, 10 mM magnesium acetate, 0.1% IGEPAL CA-630, 2 mM sodium fluoride, 2 mM sodium β-glycerophosphate pentahydrate) before resuspending in 50 μl of TEV cleavage buffer (100 mM HEPES-KOH pH 7.9, 100 mM potassium acetate, 10 mM magnesium acetate, 0.1% IGEPAL CA-630). Protein complexes were then released from the magnetic beads by the addition of 1 μl (10 U) of AcTEV protease (12575015, Life Technologies) to each aliquot and incubation at 24°C for 2 hours with agitation. Released protein complexes were then denatured by the addition of 450 μl of denaturation buffer per 50 μl [8.88 M urea, 555 mM sodium chloride, 0.11 mM sodium dihydrogen phosphate, 2.22% (v/v) Triton X-100, 11.1 mM Tris-HCl pH 8.0] and incubated with Dynabeads His-tag Isolation and Pulldown (10103D, Life Technologies) for 2 hours at 4°C (we used 100 μl of magnetic bead slurry per isolation). The His-tagged proteins isolated via cobalt-affinity chromatography were then eluted from the magnetic beads by boiling for 5 min at 95°C in Laemmli buffer containing 200 mM imidazole, 5% (w/v) sodium dodecylsulfate, 150 mM Tris-HCl pH 6.7, 30% glycerol, 715 mM β-mercaptoethanol, and 0.0125% Bromophenol Blue.
Lambda protein phosphatase assay
Immunoprecipitates of TAP-Sld5 were treated with lambda phosphatase (Lambda PP, P0753, New England Biolabs) as follows: Magnetic beads with bound complexes were washed twice with buffer comprising 100 mM Tris-acetate pH 9.0, 100 mM potassium acetate, 10 mM magnesium acetate, 0.1% IGEPAL CA-630, and 1× Complete Protease Inhibitor Cocktail (Roche). The beads were then washed a further two times with ice-cold phosphatase buffer (New England Biolabs; 1× buffer: 50 mM HEPES, 10 mM sodium chloride, 2 mM dithiothreitol, 0.01% Brij 35, pH 7.5 at 25°C) supplemented with 1× Complete Protease Inhibitor Cocktail (Roche). The beads with bound complexes were then resuspended in 47 μl of phosphatase buffer with 1 mM manganese chloride and 1× Complete Protease Inhibitor Cocktail (Roche). In the sample where phosphatase inhibitors were added, the reaction buffer was supplemented with 50 mM sodium fluoride and 10 mM sodium orthovanadate. We then added 1200 units of lambda phosphatase per sample (3 μl in a total reaction volume of 50 μl) and incubated the reactions for 30 min at 30°C. Magnetic beads with bound complexes were then washed twice with wash buffer [100 mM Tris-acetate pH 9, 100 mM potassium acetate, 10 mM magnesium acetate, 2 mM EDTA, 0.1% IGEPAL CA-630, 2 mM sodium fluoride, 2 mM sodium β-glycerophosphate pentahydrate, 1% Protease Inhibitor Cocktail (Sigma-Aldrich) and 1× Complete Protease Inhibitor Cocktail (Roche)], and the proteins were eluted from the magnetic beads by the addition of Laemmli buffer and boiling.
Purification of Cdc34 or Dia2 for in vitro complementation experiments
Cdc34-TAP or TAP-Dia2 was isolated from 2.5 g of frozen yeast as described above, using magnetic beads coupled to rabbit IgG. After a two-hour incubation with yeast extracts at 4°C, the IgG-coated beads were washed twice with 1 ml of pH 9 wash buffer supplemented with phosphatase and protease inhibitors, once with 1 ml of pH 7.9 wash buffer and once with 1 ml of TEV cleavage buffer. The magnetic beads were then incubated with agitation for 1 hour at 24°C in 80 μl of TEV cleavage buffer supplemented with 4 μl (40 U) of AcTEV protease. After elution, the supernatant was removed, and 40 μl of supernatant was added to each milliliter of the recipient cell extract (see below for details).
Purification of CMG material from dia2Δ cells for in vitro assays
For the experiment in Fig. 2D, (a), TAP-Sld5 was isolated with associated factors from 2.5 g of S phase dia2Δ cells, as described above. After immunoprecipitation, 1.7 × 109 of beads per sample were washed four times with 1 ml of pH 9 wash buffer supplemented with phosphatase and protease inhibitors. The beads were then added to 1 ml of a second cell extract (after the removal of insoluble cell debris) from control, cdc34-2, or dia2Δ cells and were incubated for 2 hours at 4°C before washing and analysis as above. A similar approach was taken for the experiment in Fig. 2D, (b), except that CMG-containing material was isolated from extracts of S-phase dia2Δ cells by immunoprecipitation of Cdc45-ProteinA.
Immunoblotting
Protein samples were resolved on SDS–polyacrylamide gel electrophoresis (PAGE) gels (various concentrations of polyacrylamide used from 5 to 12%) or NuPAGE Novex 4 to 12% Midi Bis-Tris gels (NP0301, Life Technologies) with NuPAGE MOPS SDS buffer (NP000102, Life Technologies). Molecular weight markers used were SDS-PAGE Molecular Weight Standards, Broad Range (161–0317, Bio-Rad) and Precision Plus Protein Kaleidoscope Standards (161-0375, Bio-Rad). Resolved proteins were transferred onto either nitrocellulose membrane Hybond-ECL (Amersham, GE Healthcare) or polyvinylidene difluoride membrane Hybond-P (Amersham, GE Healthcare) with a Trans-Blot SD Semi-Dry Transfer Cell (Bio-Rad) or to a nitrocellulose iBlot membrane (Life Technologies) with the iBlot Dry Transfer System (Life Technologies).
Polyclonal antibodies against replisome components have been described previously (1, 53, 54). Tagged proteins were detected with polyclonal anti-FLAG antibody (F-7425, Sigma-Aldrich), monoclonal anti-MYC (9E10; Cancer Research UK), or peroxidase-anti peroxidase complex (P1291, Sigma-Aldrich). Ubiquitin conjugates were detected with either the monoclonal P4D1 anti-ubiquitin antibody (3936, Cell Signaling) or the monoclonal FK2 antibody (BML-PW8810, Enzo Life Sciences) that preferentially recognizes polyubiquitylated conjugates. In addition, specific chain linkages of ubiquitin were detected with monoclonal antibodies raised against di-ubiquitin molecules with K48 linkage (Apu2; 05-1307, Millipore) or K63 linkage (Apu3; 05-1308, Millipore). A polyclonal antibody specific to the yeast SUMO protein Smt3 was kindly donated by H. Ulrich. A polyclonal antibody against Cdc48 was raised in sheep against the N-terminal 25-kD portion of the protein, which was expressed as a recombinant His-tagged protein in Escherichia coli. Conjugates to horse-radish peroxidase of anti-sheep IgG from donkey (Sigma, A3415), anti-rabbit IgG from rabbit (Fisher, GZNA93401ML), or anti-mouse IgG from horse (PI-2000 Vector Labs) were used as secondary antibodies before the detection of chemoluminescent signals on Hyperfilm ECL (Amersham, GE Healthcare) using ECL Western Blotting Detection Reagent (GE Healthcare).
Supplementary Material
ACKNOWLEDGMENTS
We thank Cancer Research UK, the Medical Research Council, and the Wellcome Trust (references 097945/B/11/Z for flow cytometry and 102943/Z/13/Z for an award to K.L.) for funding our work. We are grateful to M. Foltman for assistance in the early stages of the project; H. Ulrich, M. Foltman, and S. Sengupta for plasmids; R. Deshaies, M. Kanemaki, H. Morohashi, K. Nasmyth, K. Nishimura, P. Zegerman, and S. Sengupta for providing strains; A. Gambus for helpful discussions; I. Hagan and C. Wilkinson for their support during the latter stages of this work; and all members of our group for helpful discussions.
Footnotes
www.sciencemag.org/content/346/6208/1253596/suppl/DC1
Supplementary Text
Figs. S1 to S14
Table S1
References
REFERENCES AND NOTES
- 1.Gambus A, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. doi:10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 2.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. U.S.A. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. doi:10.1073/ pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, Araki H. Loading and activation of DNA replicative helicases: The key step of initiation of DNA replication. Genes Cells. 2013;18:266–277. doi: 10.1111/gtc.12040. doi:10.1111/gtc.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boos D, Frigola J, Diffley JF. Activation of the replicative DNA helicase: Breaking up is hard to do. Curr. Opin. Cell Biol. 2012;24:423–430. doi: 10.1016/j.ceb.2012.01.011. doi:10.1016/j.ceb.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 5.Labib K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 2010;24:1208–1219. doi: 10.1101/gad.1933010. doi:10.1101/gad.1933010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Labib K, Tercero JA, Diffley JFX. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. doi:10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- 7.Samson RY, Bell SD. MCM loading—an open-and-shut case? Mol. Cell. 2013;50:457–458. doi: 10.1016/j.molcel.2013.05.008. doi:10.1016/j.molcel.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 8.Takara TJ, Bell SP. Putting two heads together to unwind DNA. Cell. 2009;139:652–654. doi: 10.1016/j.cell.2009.10.037. doi: 10.1016/j.cell.2009.10.037. [DOI] [PubMed] [Google Scholar]
- 9.Yardimci H, Walter JC. Prereplication-complex formation: A molecular double take? Nat. Struct. Mol. Biol. 2014;21:20–25. doi: 10.1038/nsmb.2738. doi: 10.1038/nsmb.2738. [DOI] [PubMed] [Google Scholar]
- 10.Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 2005;19:1905–1919. doi: 10.1101/gad.337205. doi: 10.1101/gad.337205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell. 2006;21:581–587. doi: 10.1016/j.molcel.2006.01.030. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 12.Evrin C, et al. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl. Acad. Sci. U.S.A. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Remus D, et al. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Deursen F, Sengupta S, De Piccoli G, Sanchez-Diaz A, Labib K. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. EMBO J. 2012;31:2195–2206. doi: 10.1038/emboj.2012.69. doi: 10.1038/emboj.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- 16.Ulrich HD, Walden H. Ubiquitin signalling in DNA replication and repair. Nat. Rev. Mol. Cell Biol. 2010;11:479–489. doi: 10.1038/nrm2921. doi: 10.1038/nrm2921. [DOI] [PubMed] [Google Scholar]
- 17.Zaidi IW, et al. Rtt101 and Mms1 in budding yeast form a CUL4(DDB1)-like ubiquitin ligase that promotes replication through damaged DNA. EMBO Rep. 2008;9:1034–1040. doi: 10.1038/embor.2008.155. doi: 10.1038/embor.2008.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geoffroy MC, Hay RT. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat. Rev. Mol. Cell Biol. 2009;10:564–568. doi: 10.1038/nrm2707. doi: 10.1038/nrm2707. [DOI] [PubMed] [Google Scholar]
- 19.Sriramachandran AM, Dohmen RJ. SUMO-targeted ubiquitin ligases. Biochim. Biophys. Acta. 2014;1843:75–85. doi: 10.1016/j.bbamcr.2013.08.022. doi: 10.1016/j.bbamcr.2013.08.022. [DOI] [PubMed] [Google Scholar]
- 20.Blake D, et al. The F-box protein Dia2 overcomes replication impedance to promote genome stability in Saccharomyces cerevisiae. Genetics. 2006;174:1709–1727. doi: 10.1534/genetics.106.057836. doi: 10.1534/ genetics.106.057836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koepp DM, Kile AC, Swaminathan S, Rodriguez-Rivera V. The F-box protein Dia2 regulates DNA replication. Mol. Biol. Cell. 2006;17:1540–1548. doi: 10.1091/mbc.E05-09-0884. doi: 10.1091/mbc.E05-09-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan X, et al. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 23.Morohashi H, Maculins T, Labib K. The amino-terminal TPR domain of Dia2 tethers SCF(Dia2) to the replisome progression complex. Curr. Biol. 2009;19:1943–1949. doi: 10.1016/j.cub.2009.09.062. doi: 10.1016/j.cub.2009.09.062. [DOI] [PubMed] [Google Scholar]
- 24.Sengupta S, van Deursen F, de Piccoli G, Labib K. Dpb2 integrates the leading-strand DNA polymerase into the eukaryotic replisome. Curr. Biol. 2013;23:543–552. doi: 10.1016/j.cub.2013.02.011. doi: 10.1016/j.cub.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 25.Finley D, Ulrich HD, Sommer T, Kaiser P. The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics. 2012;192:319–360. doi: 10.1534/genetics.112.140467. doi: 10.1534/genetics.112.140467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 2012;14:117–123. doi: 10.1038/ncb2407. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- 27.Ghislain M, Udvardy A, Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993;366:358–362. doi: 10.1038/366358a0. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- 28.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 29.Janke C, et al. A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 30.Mumberg D, Müller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: Comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. doi: 10.1093/nar/ 22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Acs K, et al. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat. Struct. Mol. Biol. 2011;18:1345–1350. doi: 10.1038/nsmb.2188. doi: 10.1038/nsmb.2188. [DOI] [PubMed] [Google Scholar]
- 32.Bergink S, et al. Role of Cdc48/p97 as a SUMO-targeted segregase curbing Rad51-Rad52 interaction. Nat. Cell Biol. 2013;15:526–532. doi: 10.1038/ncb2729. doi: 10.1038/ncb2729. [DOI] [PubMed] [Google Scholar]
- 33.Davis EJ, et al. DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nat. Struct. Mol. Biol. 2012;19:1093–1100. doi: 10.1038/nsmb.2394. doi: 10.1038/nsmb.2394. [DOI] [PubMed] [Google Scholar]
- 34.Meerang M, et al. The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double-strand breaks. Nat. Cell Biol. 2011;13:1376–1382. doi: 10.1038/ncb2367. doi: 10.1038/ncb2367. [DOI] [PubMed] [Google Scholar]
- 35.Mosbech A, et al. DVC1 (C1orf124) is a DNA damage-targeting p97 adaptor that promotes ubiquitin-dependent responses to replication blocks. Nat. Struct. Mol. Biol. 2012;19:1084–1092. doi: 10.1038/nsmb.2395. doi: 10.1038/nsmb.2395. [DOI] [PubMed] [Google Scholar]
- 36.Mouysset J, et al. Cell cycle progression requires the CDC-48UFD-1/NPL-4 complex for efficient DNA replication. Proc. Natl. Acad. Sci. U.S.A. 2008;105:12879–12884. doi: 10.1073/pnas.0805944105. doi: 10.1073/pnas.0805944105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raman M, Havens CG, Walter JC, Harper JW. A genome-wide screen identifies p97 as an essential regulator of DNA damage-dependent CDT1 destruction. Mol. Cell. 2011;44:72–84. doi: 10.1016/j.molcel.2011.06.036. doi: 10.1016/j.molcel.2011.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jagannathan M, et al. A role for USP7 in DNA replication. Mol. Cell. Biol. 2014;34:132–145. doi: 10.1128/MCB.00639-13. doi: 10.1128/MCB.00639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishiyama A, Frappier L, Méchali M. MCM-BP regulates unloading of the MCM2-7 helicase in late S phase. Genes Dev. 2011;25:165–175. doi: 10.1101/gad.614411. doi: 10.1101/gad.614411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fachinetti D, et al. Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol. Cell. 2010;39:595–605. doi: 10.1016/j.molcel.2010.07.024. doi: 10.1016/j.molcel.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steinacher R, Osman F, Dalgaard JZ, Lorenz A, Whitby MC. The DNA helicase Pfh1 promotes fork merging at replication termination sites to ensure genome stability. Genes Dev. 2012;26:594–602. doi: 10.1101/gad.184663.111. doi: 10.1101/gad.184663.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katayama S, Kitamura K, Lehmann A, Nikaido O, Toda T. Fission yeast F-box protein Pof3 is required for genome integrity and telomere function. Mol. Biol. Cell. 2002;13:211–224. doi: 10.1091/mbc.01-07-0333. doi: 10.1091/mbc.01-07-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franz A, et al. CDC-48/p97 coordinates CDT-1 degradation with GINS chromatin dissociation to ensure faithful DNA replication. Mol. Cell. 2011;44:85–96. doi: 10.1016/j.molcel.2011.08.028. doi: 10.1016/j. molcel.2011.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moreno S. Priego, Bailey R, Campion N, Herron S, Gambus A. Polyubiquitylation drives replisome disassembly at the termination of DNA replication. Science. 2014;346:477–481. doi: 10.1126/science.1253585. [DOI] [PubMed] [Google Scholar]
- 45.Magnaghi P, et al. Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nat. Chem. Biol. 2013;9:548–556. doi: 10.1038/nchembio.1313. doi: 10.1038/nchembio.1313. [DOI] [PubMed] [Google Scholar]
- 46.Knop M, et al. Epitope tagging of yeast genes using a PCR-based strategy: More tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. doi: 10.1002/(SICI)1097-0061 (199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 47.Kanemaki M, Labib K. Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J. 2006;25:1753–1763. doi: 10.1038/sj.emboj.7601063. doi: 10.1038/sj. emboj.7601063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanchez-Diaz A, Kanemaki M, Marchesi V, Labib K. Rapid depletion of budding yeast proteins by fusion to a heat-inducible degron. Sci. STKE. 2004;2004:pl8. doi: 10.1126/stke.2232004pl8. [DOI] [PubMed] [Google Scholar]
- 49.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Labib K, Diffley JFX, Kearsey SE. G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat. Cell Biol. 1999;1:415–422. doi: 10.1038/15649. doi: 10.1038/15649. [DOI] [PubMed] [Google Scholar]
- 51.Foiani M, Marini F, Gamba D, Lucchini G, Plevani P. The B subunit of the DNA polymerase α-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol. Cell. Biol. 1994;14:923–933. doi: 10.1128/mcb.14.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Piccoli G, et al. Replisome stability at defective DNA replication forks is independent of S phase checkpoint kinases. Mol. Cell. 2012;45:696–704. doi: 10.1016/j.molcel.2012.01.007. doi: 10.1016/j. molcel.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 53.Gambus A, et al. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase α within the eukaryotic replisome. EMBO J. 2009;28:2992–3004. doi: 10.1038/emboj.2009.226. doi: 10.1038/ emboj.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Foltman M, et al. Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell Reports. 2013;3:892–904. doi: 10.1016/j.celrep.2013.02.028. doi: 10.1016/j.celrep.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 55.Zegerman P, Diffley JF. Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature. 2010;467:474–478. doi: 10.1038/nature09373. doi: 10.1038/nature09373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.