ABSTRACT
In this study, we characterized the molecular basis for binding of adenovirus (AdV) to the cytoplasmic face of the nuclear pore complex (NPC), a key step during delivery of the viral genome into the nucleus. We used RNA interference (RNAi) to deplete cells of either Nup214 or Nup358, the two major Phe-Gly (FG) repeat nucleoporins localized on the cytoplasmic side of the NPC, and evaluated the impact on hexon binding and AdV infection. The accumulation of purified hexon trimers or partially disassembled AdV at the nuclear envelope (NE) was observed in digitonin-permeabilized cells in the absence of cytosolic factors. Both in vitro hexon binding and in vivo nuclear import of the AdV genome were strongly reduced in Nup214-depleted cells but still occurred in Nup358-depleted cells, suggesting that Nup214 is a major binding site of AdV during infection. The expression of an NPC-targeted N-terminal domain of Nup214 in Nup214-depleted cells restored the binding of hexon at the NE and the nuclear import of protein VII (pVII), indicating that this region is sufficient to allow AdV binding. We further narrowed the binding site to a 137-amino-acid segment in the N-terminal domain of Nup214. Together, our results have identified a specific region within the N terminus of Nup214 that acts as a direct NPC binding site for AdV.
IMPORTANCE AdVs, which have the largest genome of nonenveloped DNA viruses, are being extensively explored for use in gene therapy, especially in alternative treatments for cancers that are refractory to traditional therapies. In this study, we characterized the molecular basis for binding of AdV to the cytoplasmic face of the NPC, a key step for delivery of the viral genome into the nucleus. Our data indicate that a 137-amino-acid region of the nucleoporin Nup214 is a binding site for the major AdV capsid protein, hexon, and that this interaction is required for viral DNA import. These findings provide additional insight on how AdV exploits the nuclear transport machinery for infection. The results could promote the development of new strategies for gene transfer and enhance understanding of the nuclear import of other viral DNA genomes, such as those of papillomavirus or hepatitis B virus that induce specific cancers.
INTRODUCTION
Adenoviruses (AdVs) are nonenveloped DNA viruses consisting of an icosahedral capsid of ∼90-nm diameter and an inner nucleoprotein core containing a linear double-stranded DNA genome of ∼36 kbp (1–3). The major structural component of the capsid is the hexon trimer that is present in 240 copies. On the outer surface of the capsid at each of the 12 vertices, fiber proteins are anchored to the penton base. A number of minor capsid proteins on the outer and inner surfaces of the virus particle help to stabilize the capsid (4). The DNA is directly associated with the core proteins, including protein X, the “terminal protein,” which is covalently linked to the 5′ DNA termini, protein VII, and protein V, which connects the core to the outer capsid.
AdV enters the cells by receptor-mediated endocytosis during which the virion becomes partially uncoated (3). Uncoating involves a series of events, culminating with endosomal membrane lysis by protein VI, which allows access of the particle to the cytosol (5). The partially disassembled capsid is then translocated along microtubules to the nucleus using the dynein/dynactin motor complex (6, 7). AdV then interacts with the nuclear envelope (NE) at nuclear pore complexes (NPCs) (8), and the viral genome is translocated into the nucleus by means of nuclear import receptors and/or histone H1 (8–10).
NPCs are evolutionarily conserved large protein complexes of ∼100 MDa spanning the NE that mediate trafficking into and out of the nucleus. Although small molecules passively diffuse through the NPC, macromolecules larger than ∼20 to 40 kDa are transported in an active manner. This pathway is mediated by cellular transport receptors, including the karyopherin beta family that facilitates the translocation of most proteins and certain RNAs (11, 12). NPCs are formed by ∼30 proteins, which are thought to be present in multiples of 8 copies (13). A third of all nucleoporins (Nups) contain intrinsically disordered regions enriched in Phe-Gly (FG) repeats. The FG repeat domains directly bind karyopherins (12) and play an essential role in trafficking of receptor-cargo complexes through the NPC (13). Many of the FG nucleoporins are localized to central regions of the NPC, but some occur at the NPC periphery. The peripheral FG nucleoporins include Nup214 and Nup358, which are located in fibrils that emanate from the cytoplasmic face of the NPC, and Nup153, which is concentrated in the “basket” that projects from the nuclear face of the NPC (14, 15).
Since the upper size limit for signal-mediated transport through the NPC is ∼40 nm (49), few viruses are sufficiently small to be imported into the nucleus without disassembly (16, 17). Accordingly, AdV genome import requires binding of the virus at the NPC and full capsid disassembly (17–19). A priori, AdV binding to the NPC is likely to be mediated by the hexon protein, which is the most abundant and outermost capsid protein, and by the cytoplasmic NPC fibrils, which are the most exposed structural components. AdV binding to the NPC was suggested to directly or indirectly involve the nuclear export receptor CRM-1 (20) and the cytoplasmic FG nucleoporins Nup214 (8) and Nup358 (21), which are known to be involved in binding of karyopherins during nuclear import/export (22, 23). The specific region involved in the docking has not been precisely evaluated.
To promote understanding of the mechanism of AdV interaction with the NPC, we used cells where Nup214 and Nup358 were depleted by RNA interference (RNAi) to study AdV and hexon binding to the NPC and import of the viral genome. Our studies revealed a specific role for Nup214 in hexon binding and genome import and mapped hexon binding to a 137-residue potentially unstructured region in the N terminus of Nup214.
MATERIALS AND METHODS
Cell lines and transfection.
Normal rat kidney (NRK) cells, human HEK293T cells, and human HeLa cells were maintained in complete Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. For transfection assays, 0.5 × 106 cells were seeded into each well of six-well dishes the day prior to transfection. For one well, 2 μg of DNA with 2 μl of Plus Reagent and 5 μl of Lipofectamine in Opti-MEM was used (Invitrogen). The next day, the cells were plated onto 10-well slides (ICN Biomedical) at 15,000 cells/well. The binding and AdV infection experiments were performed at 48 h posttransfection.
Plasmids.
All the plasmids were amplified by PCR using pBluescript containing human Nup214 as a template. The N-terminal (amino acids [aa] 1 to 1058) and central (amino acids 586 to 1058) domains of Nup214 were inserted in the pcDNA3.1D/V5-His-TOPO vector (Invitrogen). The N-terminal domains of Nup214 (aa 1 to 450, 1 to 586, and 450 to 586) were inserted in the pET101D/V5-His-TOPO vector (Invitrogen). These N-terminal domains and aa 587 to 723 were amplified by PCR using primers containing V5 (sequence found in the P and V proteins of the paramyxovirus, simian virus 5 [SV5]) and His tags and inserted in the pcDNA3.1 vector. The fragment of Nup214 consisting of aa 450 to 586 [Nup214(450–586)], which was amplified as a fusion with V5 and His tags by PCR, was inserted into pMalc2x vector (NEB) between HindIII and EcoRI sites. The pcDNA3.1D/LacZ-V5-His vector (Invitrogen) was used for a mock transfection control.
Expression of recombinant proteins.
V5-His fusion proteins were expressed in the BL21(DE3) Escherichia coli strain. Cells were lysed, and purification was performed on cleared lysate using Ni-nitrilotriacetic acid (NTA)-agarose beads (Qiagen). The proteins were eluted in transport buffer ([TB] 20 mM HEPES, pH 7.4, 110 mM potassium acetate, 2 mM magnesium acetate, 2 mM dithiothreitol [DTT], 1 mM EGTA, and protease inhibitors) with 200 mM imidazole. After imidazole removal, the proteins were concentrated and snap-frozen in liquid nitrogen. For the Nup214(450–586) fragment, the maltose binding protein (MBP) tag was removed by the addition of factor Xa for 3 h at room temperature (RT). The Nup214(450–586) fragment was eluted from beads, the buffer was exchanged, and the proteins were stored as described above.
Other recombinant protein fragments and proteins were expressed in E. coli and purified as described before: Nup358-1 and Nup358-4 fragments (24) and the C-terminal domain (aa 1861 to 2090) of Nup214 (25).
RNAi.
For gene silencing by RNAi, cells were transfected with the pSUPER vector (Oligoengine) expressing short hairpin RNAs (shRNAs) against Nup214 and Nup358. The oligonucleotides used for silencing of Nups were cloned in the pSUPER vector and had the following sequences: Nup214 sh214-1, 5′-GATCCCCTTGCCCAAGGAACGCTCGATTCAAGAGATCGAGCGTTCCTTGGGCAATTTTTA-3′; Nup214 sh214-2, 5′-GATCCCCCCCTAGCCAACAATCGGGAAATCATTTCAAGAGAATGATTTCCCGATTGTTGGCTAGGGTTTTTA-3′; Nup214 sh214-3, 5′-GATCCCCCGATAGCACAATGCTTGCCACGAAATTCAAGAGATTTCGTGGCAAGCATTGTGCTATCGTTTTTA-3′; Nup358 sh358, 5′-GATCCCCCGAGGTCAATGGCAAACTATTCAAGAGATAGTTTGCCATTGACCTCGTTTTTA-3′ (26). A peGFP-H1 vector without the inserted shRNA-encoding sequence containing two expression cassettes (one for enhanced green fluorescent protein [eGFP] and one for shRNA) was used as a control and for the reconstitution experiments (kindly provided by Claire Waterman).
Purification of virus and acid-treated virus.
An AdV5 vector (AdV) containing an eGFP expression cassette in the E1 region was propagated in the HEK293T cells. Virus was purified as previously described (27). AdV particles were partially disassembled according to the protocol of Wiethoff et al. (5). For this, the virus was dialyzed against the acetate buffer, pH 5.0, and incubated at 37°C for 10 min before being loaded onto a Nycodenz step density gradient. The partially disassembled virus was dialyzed against 5 mM HEPES, pH 7.4, 1 mM MgCl2, and 10% glycerol at 4°C. The fraction was analyzed for protein content by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining.
Purification of hexon.
Hexon was purified according to a protocol modified from Wodrich et al. (28). Infected cells were disrupted, and the lysate was cleared by two steps of centrifugation. The hexon-containing band was extracted and dialyzed against 10 mM bis-Tris propane, pH 7.0, using a 100-kDa cutoff Float-A-Lyser (Spectrum Lab) and then applied to a MonoQ fast protein liquid chromatography (FPLC) column (Pharmacia). Highly enriched hexon fractions were pooled and concentrated. The buffer was exchanged to TB. The purified hexon corresponds to trimeric hexon, as observed by negative-staining electron microscopy. The fraction was analyzed for protein content by SDS-PAGE after Coomassie staining and by immunoblotting using an affinity-purified rabbit anti-hexon antibody against peptides consisting of aa 230 to 243 and 430 to 445. No protein VI (pVI) was detected by immunoblotting using specific protein VI antibody (29).
Binding experiments in permeabilized cells.
The binding experiments in permeabilized cells were performed in NRK and HeLa cells. Cells were plated on 10-well slides the day before the experiment. Cells were permeabilized by treatment with 0.005% digitonin (Calbiochem) in TB for 5 min at RT. The cells were preincubated with TB alone for 15 min at 30°C. The binding reaction mixture was incubated for 30 min at 30°C or 4°C. The binding reaction mix contained 0.75 μM purified hexon, cytosol with or without ATP-regenerating system, and TB alone or TB with competitors (see below). Cytosol was isolated by digitonin lysis of HeLa cells as described by Kehlenbach et al. (30) and used at a final concentration of 4 mg/ml. Cells were fixed and processed for hexon and Nup detection as described below. All experiments were repeated at least three times.
Adenovirus infection following knockdown.
After initial transfection with pSUPER expressing different shRNAs and/or pcDNA3.1 expressing the central domain of Nup214 or the N-terminal domain of Nup214, HeLa cells were infected with AdV at 48 h posttransfection. Cells were infected using 10,000 virus particles/cell for fluorescence in situ hybridization (FISH) experiments or 1,000 virus particles/cell for pVII detection. For infection experiments, the virus was prebound to cells for 1 h on ice, followed by 2 h or 3 h of incubation at 37°C for FISH or 3 h at 37°C for pVII detection. Prior to detection, the cells were fixed with 3.7% formaldehyde as described above. The experiments were repeated at least three times with three independent transfections.
Immunofluorescence (IF).
The fixed cells were permeabilized with 0.2% Triton for 5 min, and detection of protein was performed by incubation with antibodies for 1 h. The primary antibodies used were 8C4 mouse anti-hexon antibody (Fitzgerald Industries), rabbit anti-Nup214 antibody (Bethyl Laboratories), affinity-purified guinea pig anti-Nup358 directed against the FG-rich domain (aa 996 to 1963) in the Nup358-1 clone provided by Nabeel Yaseen (31), and rabbit anti-pVII antibody (kindly provided by Daniel A. Engel). Fluorescein isothiocyanate (FITC)-, Texas Red (Jackson Laboratories)-, or Alexa-labeled antibodies (Invitrogen) were used as secondary antibodies. The Nup214 expressed domains were detected with FITC-labeled mouse monoclonal anti-V5 antibody (Invitrogen). Nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI; Sigma) or with To-Pro-3 (Molecular Probes). The cells were examined using a Bio-Rad 1024 or a Leica TCS SP5 laser scanning confocal microscope with a 63× oil immersion objective (Zeiss Plan Apo, 1.4 numerical aperture [NA], or Leica Plan Apo, 1.4 NA). Images were collected with Bio-Rad Lasersharp 2000 software or with Leica LAS AF software. Images were analyzed using Simple PCI software (Compix) or ImageJ software (NIH). The nuclear quantification or rim staining quantification was based on the nuclear space defined by the DAPI or TO-PRO-3 staining. In quantitative analysis, error bars indicate the standard deviations in three different experiments except for representative analysis, in which error bars indicate the standard deviation in three different fields. Statistical analysis was done using a two-tailed Student's t test. For the localization of the AdV genome and protein VII, a Leica SP8 laser scanning confocal microscope with a 63× oil immersion objective was used, and a representation of the staining was obtained using ImageJ software (NIH). Maximal projection of image stacks in the nucleus was performed; a Top Hat filter and StackReg plug-in (32) were applied.
FISH.
The detection of AdV DNA was performed following a modified protocol from Greber et al. (18). The probes were generated using random priming on purified AdV genome with a digoxigenin DNA labeling kit (Roche). The cells were stained, and IF images were acquired before the cells were further processed for FISH. The probe and the slide were predenatured for 10 min at 95°C in hybridization buffer, and then the mixture was additionally denatured for 5 min before hybridization overnight at 37°C. Mouse anti-digoxigenin antibody (Roche) and Alexa 488-labeled goat anti-mouse (Invitrogen) were used to detect AdV genome. The cells were examined as described above.
Pulldown and immunoblotting.
Total cell lysates from 106 cells were prepared and analyzed by Western blotting using standard procedures. Mouse monoclonal anti-V5 antibody (Invitrogen), rabbit anti-hexon (33), mouse RL1 (34), anti-hexon antibody (35), and horseradish peroxidase (HRP) secondary antibodies (Jackson ImmunoResearch) were used for immunoblotting. The pulldown was performed using a Dynabeads His tag isolation and pulldown kit according to the manufacturer's protocol (Invitrogen). Four micrograms of purified recombinant proteins or buffer alone and 2.5 μg of purified hexon were incubated together, and then 20 μl of beads was added. The unbound fraction was removed, and complexes were eluted with 300 mM imidazole. One-tenth of the input, one-third of the unbound fraction, and one-half of the bound fraction were loaded on SDS-PAGE gels for Western blotting.
RESULTS
Binding of partially disassembled AdV and purified hexon at the NE in digitonin-permeabilized cells.
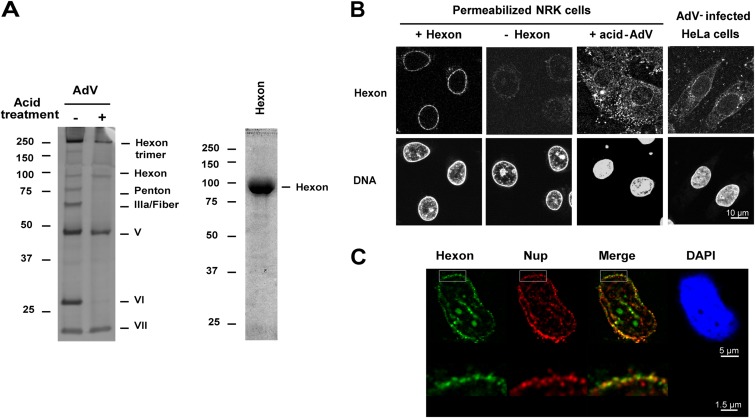
We used digitonin-permeabilized cells, which are functional for nuclear import (36), to analyze binding of AdV and purified hexon trimers to the NPC in vitro. Digitonin treatment selectively permeabilizes the plasma membrane while leaving the NE intact, thereby allowing virus entry into the cytosolic space. To mimic the exposure of virus to the low-pH environment of the endosome (3), the virus was pretreated with pH 5.0 buffer, followed by Nycodenz gradient purification. As described previously (5), this treatment induced dissociation of the penton base, fiber, protein IIIa, and protein VI (Fig. 1A, left). Removal of these proteins has been previously shown to be required for infection (37, 38). The low-pH treatment generates partially disassembled capsid, and the loss of capsid integrity was confirmed by transmission electron microscopy (data not shown). The purity of a trimeric hexon preparation was validated by SDS-PAGE (Fig. 1A, right) and by Western blotting using hexon and pVI-specific antibody (data not shown).
FIG 1.
Binding of acid-treated AdV and purified hexon at the NE. (A) Intact AdV or low-pH-treated and purified AdV (1.5 μg) was analyzed by SDS-PAGE and silver staining (left). Purified hexon (2.5 μg) was analyzed by SDS-PAGE and Coomassie blue staining (right). The hexon trimer (left) is seen only with brief heating in SDS sample buffer. Values to the left of the blots are kilodaltons. (B) Digitonin-permeabilized NRK cells were incubated in transport buffer with low-pH-treated AdV or with purified hexon, washed, and analyzed by IF. HeLa cells were infected for 2 h by AdV. The hexon and nuclear (DNA) (DAPI/To-Pro-3) staining results are shown. (C) Permeabilized HeLa cells were incubated with purified hexon and analyzed as described for panel B. The nucleus stained with DAPI (in blue), hexon staining (in green), FG-repeat nucleoporin staining (in red), and a merged image of hexon and Nup staining are shown. Orthogonal views of NE from the boxed areas are shown below.
Purified trimeric hexon or low-pH-treated AdV was added to digitonin-permeabilized NRK cells in the absence or presence of cytosol, and the localization of hexon and Adv was determined by IF using anti-hexon antibody. In the presence of cytosol, purified hexon strongly accumulated in cytoplasmic areas around the nucleus and weakly associated with the nuclear rim; however, it was not transported into the nucleus (data not shown). In contrast, in the absence of cytosol, purified hexon was concentrated mainly in a perinuclear rim (Fig. 1B), and no staining could be observed in the absence of hexon indicating any cross-reactivity of the antibody for the NE (antibody control). The hexon localization largely overlapped that of Nups (Fig. 1C), indicating that hexon accumulates at the NPC with this assay. Nuclear rim accumulation also was seen for acidified AdV added to permeabilized cells in the absence of cytosol although binding to cytoplasmic areas occurred as well. This pattern resembled the distribution of hexon in HeLa cells 2 h after AdV infection (Fig. 1B). Because the cytosol-independent binding of hexon trimers to the NPC in permeabilized cells was very robust, this assay is used in the experiments below to dissect this interaction.
Loss of hexon binding at the NE in Nup214-depleted HeLa cells.
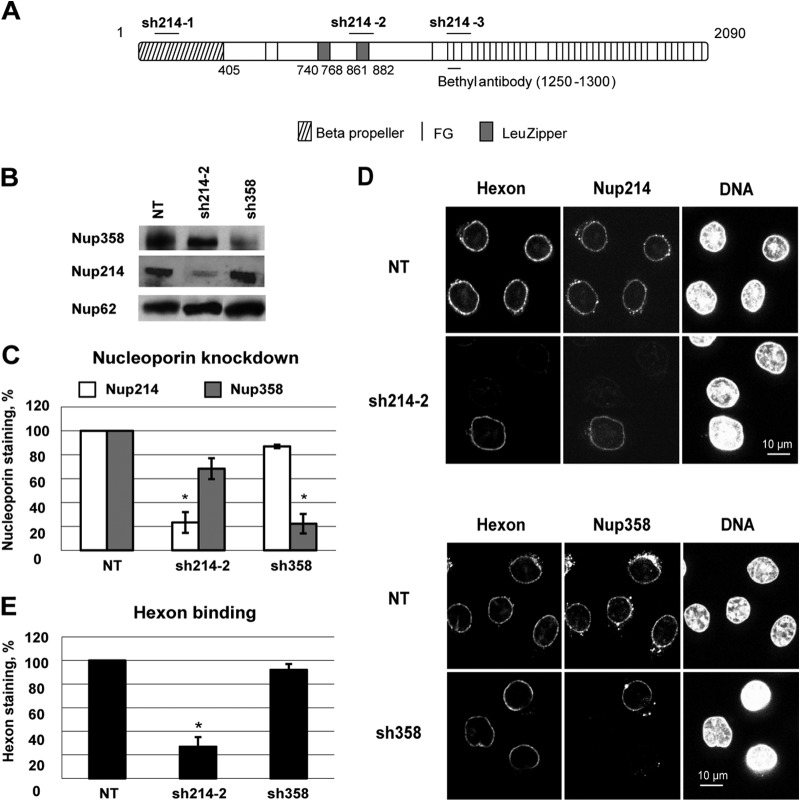
To determine whether Nup214 and Nup358 are required for binding of purified hexon to the NPC, we used RNAi to deplete HeLa cells of these Nups. Three separate shRNAs targeting different domains of the Nup214 RNA (Fig. 2A) were analyzed. In the majority of cells, most Nup214 was absent from the NE, as determined by IF, regardless of which shRNA was used (Fig. 2D and data not shown). Transfection with the sh214-2 plasmid reduced Nup214 expression by ∼80%, as determined by Western blotting (Fig. 2B and C), and also led to a smaller decrease (∼30%) in Nup358 expression. Transfection with the sh358 plasmid reduced the expression of Nup358 by 80% compared to levels in nontransfected cells but did not impact Nup214 expression (Fig. 2B and C).
FIG 2.
Loss of hexon binding at the NE in Nup214-depleted HeLa cells. (A) Schematic representation of targeting sites of shRNA directed against Nup214 (short horizontal bars). Full-length Nup214 (aa 1 to 2090) is represented as a large horizontal bar. A short horizontal bar indicates the epitope recognized by anti-Nup214 antibody (bethyl antibody). (B) Knockdown in HeLa cells 48 h after transfection. Cells were transfected with shRNA expression plasmids against Nup214 (sh214-2) or against Nup358 (sh358) or were not transfected (NT). Total lysates of cells were analyzed by SDS-PAGE and Western blotting using RL1 antibody. Under each condition, the expression of Nup358, Nup214, and Nup62 (as an internal control) is shown. (C) Quantification of the knockdown in HeLa cells 48 h after transfection. Cells were transfected as described for panel A and stained for Nup214 and Nup358. The mean fluorescence intensity of endogenous Nup staining was measured at the whole circumference of the nucleus (n = 31 to 63 cells for each condition of each experiment). The histogram shows the mean fluorescence intensity of endogenous Nup214 or Nup358 staining compared to NT cells, fixed at 100% (*, P < 0.05). (D) Representative images of hexon binding at the NE. Purified hexon was added 48 h after transfection to digitonin-permeabilized HeLa cells of expression plasmids encoding shRNA against Nup214 (sh214-2) or against Nup358 (sh358) or no plasmid (NT). The cells were analyzed by IF staining to detect the hexon, Nup214 (upper panels), or Nup358 (lower panels). The nucleus was stained with DAPI. (E) Quantitative analysis of hexon binding at the NE. HeLa cells were treated as described for panel D. The mean fluorescence intensity of hexon staining at the whole circumference of the nucleus was measured (n = 31 to 63 cells for each condition of each experiment). The histogram shows the mean fluorescence intensity of hexon staining in knockdown cells and compared to NT cells, set at 100 (*, P < 0.05).
Cells transfected with shRNA plasmids to deplete Nup214 or Nup358 were analyzed for the binding of purified hexon to the NE in the absence of cytosol after digitonin permeabilization. Hexon hardly bound to the NE of cells lacking Nup214 expression (Fig. 3D). In Nup214-depleted cell populations, hexon rim staining on average was reduced by 70% (Fig. 3E), which correlates well with the efficiency of Nup214 silencing. In contrast, hexon rim staining remained unchanged in Nup358-depleted cells (Fig. 3D, lower panels) compared to the level in nontransfected cells. In summary these data show that binding of hexon at the NE required Nup214 but not Nup358, suggesting that Nup214 is the primary binding partner for hexon exposed on the outer face of the NE.
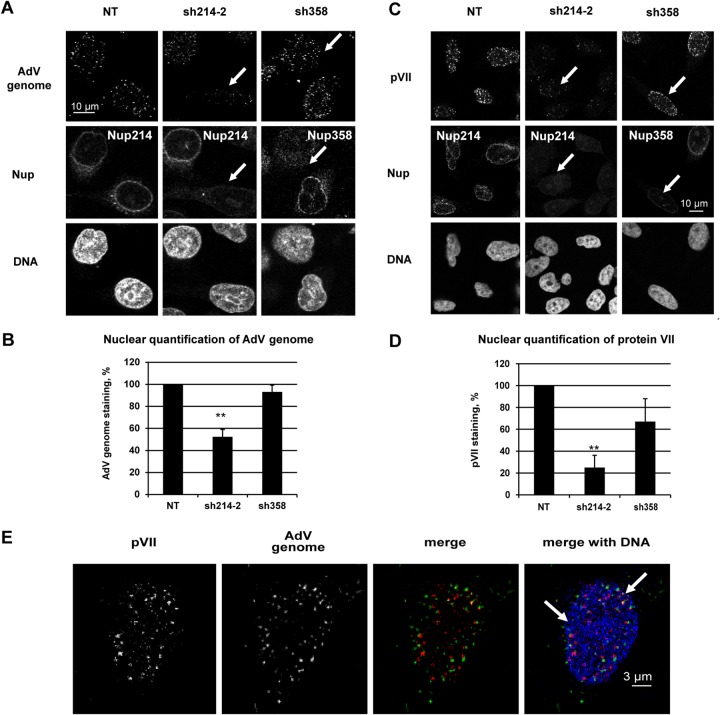
FIG 3.
Reduction of viral DNA import in Nup214-depleted HeLa cells. HeLa cells were transfected with shRNA expression plasmids against Nup214 (sh214-2) or against Nup358 (sh358) or were not transfected (NT). The cells were infected with AdV for 2 h (A and B) or 3 h (C, D, and E). (A) Reduction of AdV genomes in the nucleus of infected Nup214-depleted HeLa cells. Cells were immunostained to detect Nup214 or Nup358 (as indicated) and incubated with DAPI to detect the nucleus. The AdV genome was specifically detected by FISH using anti-digoxigenin antibody. The white arrows show the nucleus with decreased Nup214 and AdV genome staining and the nucleus with depleted Nup358 but with AdV genome staining present. (B) Quantification of AdV genomes in the nucleus of HeLa cells. The histogram shows the mean fluorescence intensity of AdV genome staining indicated as a percentage. The mean fluorescence intensity of AdV genome staining into the nucleus was measured (n = 17 to 36 cells for each condition of each experiment) and compared to the level in NT cells, set at 100 (**, P < 0.01). (C) Reduction of protein VII in the nucleus of infected Nup214-depleted HeLa cells. Cells were immunostained as described for panel A. Protein VII was detected using anti-pVII antibody. The white arrows indicate the nucleus with a loss of Nup214 and protein VII staining and the nucleus with a loss of Nup358 but with protein VII staining present. (D) Quantification of protein VII in the nucleus of HeLa cells. The histogram shows the mean fluorescence intensity of pVII staining indicated as a percentage of stained cells. The mean fluorescence intensity of pVII staining into the nucleus was measured (n = 64 to 114 cells for each condition of each experiment) and compared to the level in NT cells, set a 100 (**,P < 0.01; significant). (E) AdV genome and protein VII localization in the nucleus of HeLa cells. Cells were immunostained to detect protein VII using anti-pVII antibody (red) and DAPI (blue) to detect the nucleus. The AdV genome was detected by FISH using anti-digoxigenin antibody (green). Representations of maximal projections of image stacks and merged pictures are shown. The white arrows highlight overlapping staining of pVII and the AdV genome, shown as yellow dots.
Reduction of viral DNA import in Nup214-depleted HeLa cells.
The AdV genome is transported into the nucleus via the NPC in order to initiate virus replication and particle assembly. To assess the relationship of hexon association with Nup214 and the import of the genome, we infected HeLa cells with AdV 48 h after Nup214/Nup358 shRNA transfection. The infection was limited to 2 or 3 h of incubation of AdV in order to prevent interference by newly synthesized viral proteins. The AdV genome was detected by fluorescence in situ hybridization (FISH) under denaturing conditions. Fluorescent dots representing the AdV genome were observed in the nucleus and in the cytoplasm in nontransfected and in Nup358-depleted cells (Fig. 3A). In contrast, the presence of AdV DNA was substantially reduced in the nucleus in Nup214-depleted cells (Fig. 3A). Detection was specific as no fluorescent dots were observed in noninfected cells (data not shown).
Quantification of the fluorescence images revealed a 50% reduction for the AdV genome signal in the nucleus of Nup214-depleted cells compared to the signal in nontransfected cells (Fig. 3B), while no significant reduction was observed for Nup358-depleted cells. To confirm the FISH results, we also analyzed the localization of the genome-associated protein VII (pVII) by IF. Prior to nuclear import, protein VII is inaccessible to antibody staining since it is located inside the capsid (1–3). Similar to our observations of AdV DNA, pVII fluorescent dots accumulated in the nucleus of untreated and Nup358-depleted cells (Fig. 3C). In contrast, a >75% reduction of protein VII IF was observed when cells were depleted for Nup214 compared to levels in nontransfected cells (Fig. 3D), while no major reduction was observed for Nup358-depleted cells.
To extend these results, we analyzed the localization of protein VII and the AdV genome in the same samples by a protocol involving immunostaining combined with FISH. With this method, protein VII was detectable in the nucleus as dots very similar to the dot-like signal of AdV genome and partially overlapping the latter (Fig. 3E). The AdV genome but not protein VII was also seen in the cytoplasm in the vicinity of the nucleus. Complete overlap of pVII and the AdV genome in the nucleus was not apparent, probably due to the denaturation treatment required for FISH. For subsequent analyses of nuclear delivery of the AdV genome, pVII staining was used as a surrogate for the AdV genome.
Collectively, these data show that Nup214 but not Nup358 is required for efficient delivery of the AdV genome to the nucleus in infected cells, similar to the requirements for binding of hexon to the NPC in permeabilized cells. Our observation that protein VII was detectable in the nucleus only in the presence of Nup214 suggests that Nup214 is a prerequisite for capsid disassembly leading to genome release.
Identification of hexon binding region in Nup214.
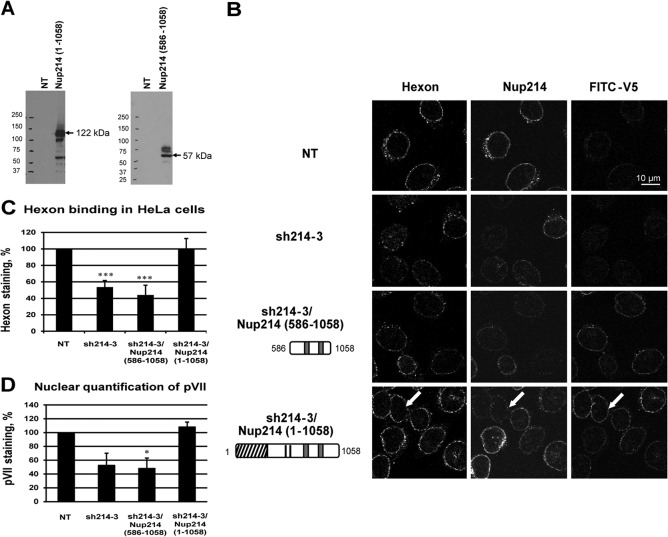
We expressed fragments of human Nup214 in Nup214-depleted HeLa cells to identify regions that can reconstitute hexon binding to the NE in permeabilized cells. Nup214 contains 2,090 aa and has an N-terminal domain (aa 1 to 586) containing a β-propeller structure (39), a coiled-coil-containing domain (aa 586 to 1058) with two leucine zippers, and a C-terminal domain (aa 1058 to 2090) enriched in FG repeats (Fig. 2A). Our initial analysis involved two amino-terminal fragments of Nup214 that contained the coiled-coil domain required to target the protein to the NPC (40): Nup214(586–1058) and Nup214(1–1058) (Fig. 4B). Knockdown was achieved with the sh214-3 plasmid, which does not target these exogenous Nup214 fragments. The Nup214 constructs were expressed as fusion proteins with a V5-His tag at the C terminus for discrimination from the endogenous Nup214. The antibody against endogenous Nup214 recognizes a domain of Nup214(1250–1300) that is not present on the ectopically expressed N-terminal Nup214 proteins (Fig. 2A and 4B). Western blotting verified expression of the recombinant proteins with some minor degradation products and two slower-migrating bands, presumably reflecting posttranslation modifications (Fig. 4A). IF staining validated the incorporation of these proteins into the NE (Fig. 4B, third column).
FIG 4.
Reconstitution of hexon binding by expression of the N-terminal domain of Nup214. (A) Expression of Nup214 domains in HeLa cells. HeLa cells were transfected with expression constructs corresponding to the N-terminal domain of Nup214 (aa 1 to 1058) or the coiled-coil domain of Nup214, Nup214(586–1058), or were not transfected (NT). Total lysates were analyzed by immunoblotting using V5 antibody. The predicted sizes of the domains are indicated by the arrows. (B) Representative images of hexon binding at the NE. HeLa cells were transfected with shRNA expression plasmid against Nup214 (sh214-3) alone or were cotransfected with the Nup214 coiled-coil domain expression plasmid [sh214-3/Nup214(586–1058)] or with the Nup214 N-terminal domain expression plasmid [sh214-3/Nup214(1–1058)] or were not transfected (NT). Purified hexon was added to digitonin-permeabilized HeLa cells. The cells were analyzed by IF staining using FITC anti-V5 antibody to detect the overexpressed Nup214 fragments or with the anti-hexon antibody and anti-Nup214 antibody. The white arrows indicate hexon staining around the nucleus of Nup214-depleted cells after overexpression of the N-terminal domain of Nup214(1–1058). Symbols in the schematic representations at left are as identified on Fig. 2A. (C) Quantitative analysis of hexon binding at the NE. The histogram presents the mean fluorescence intensity of hexon staining indicated as a percentage. The mean fluorescence intensity of hexon staining around the nucleus was measured (n = 43 to 91 cells for each condition of each experiment) and compared to that in NT cells, set at 100 (***, P < 0.001). (D) Reconstitution of the nuclear import of protein VII by expression of the N-terminal domain of Nup214 in Nup214-depleted cells. HeLa cells were transfected with expression plasmid eGFP-shRNA against Nup214 (eGFP-sh214-3) alone or were cotransfected with the Nup214 coiled-coil domain expression plasmid [eGFP-sh214-3/Nup214(586–1058)] or with the Nup214 N-terminal domain expression plasmid [eGFP-sh214-3/Nup214(1–1058)] or were not transfected (NT). Cells were infected with AdV for 3 h. The cells were analyzed by IF staining using anti-pVII antibody. The histogram presents the mean fluorescence intensity of pVII staining in the nucleus, indicated as a percentage. The mean fluorescence intensity of pVII staining around the nucleus was measured (n = 40 to 114 cells for each condition of each experiment) and compared to that in NT cells, set at 100 (*, P < 0.05).
In the permeabilized cell assay for hexon binding to the NE, no increase in nuclear rim staining of hexon was observed in cells expressing the coiled-coil domain of Nup214 (Fig. 4B, 3rd row, and C) compared to the sh214 control. In contrast, hexon staining was restored to the levels of the nontransfected (NT)control when the larger N-terminal region of Nup214 was expressed (Fig. 4B, 4th row, and C). This finding indicates that the coiled-coil domain of Nup214 by itself is not sufficient for the binding of hexon to the NE, whereas hexon binding is fully supported by the entire N-terminal region (aa 1 to 1058).
Reconstitution of the nuclear import of protein VII by complementation with recombinant Nup214 proteins.
Since the N-terminal region of Nup214 could restore nuclear binding of hexon, we asked if this domain could also restore nuclear delivery of the AdV genome in Nup214-depleted HeLa cells. Cells were first cotransfected with plasmids encoding eGFP and sh214, on the one hand, and exogenous fragments of human Nup214, on the other hand; cells were then infected with AdV and analyzed for the localization of protein VII. eGFP expression was used to track transfected cells. In Nup214-depleted HeLa cells, we observed a strong reduction of specific pVII staining in the nucleus. Reduced nuclear accumulation of pVII was also observed after expression of the coiled-coil domain of Nup214 (Fig. 4D). In contrast, strong nuclear staining with pVII was observed after complementation with the N-terminal region of Nup214, as observed for NT cells (Fig. 4D). This suggests that hexon binding to the N-terminal region of Nup214 is required for nuclear import of pVII and, by inference, AdV DNA.
Competition of Nup214 proteins for hexon binding at the NE.
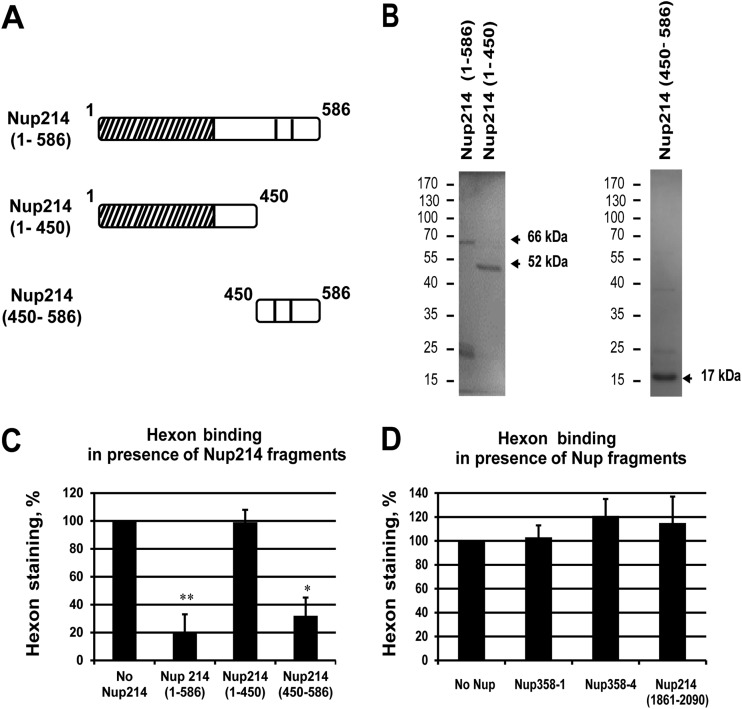
The previous experiments showed that the coiled-coil domain of Nup214 was not sufficient for the binding of hexon to the NE in permeabilized cells but that the first N-terminal 586 residues were also needed. To test if a specific domain of this N-terminal extension is involved in hexon binding, we expressed several soluble N-terminal Nup214 domains in bacteria and used the purified molecules in competition experiments. These included Nup214(1–586), Nup214(1–450), and Nup214(450–586) (Fig. 5A and B). All proteins appeared as prominent bands at the predicted molecular masses by SDS-PAGE and Coomassie blue staining although Nup214(1–586) also produced smaller additional bands around 25 kDa that contained the V5 epitope in addition to the full-length protein (66 kDa) (data not shown).
FIG 5.
Competition of hexon binding at the NE. Permeabilized HeLa cells (A, B, C, and D) or NRK cells (E) were incubated with purified hexon in transport buffer for 30 min in the presence or absence of soluble Nup fragments used as competitors and analyzed by IF staining using anti-hexon antibody. (A) Schematic representation of soluble Nup214 proteins used for hexon binding competition in HeLa cells as shown on Fig. 2A. (B) Expression of purified recombinant N-terminal proteins analyzed by SDS-PAGE and Coomassie staining. The expected size of each protein is indicated by the arrows. The migration and sizes of standard markers are shown on the left. (C) Quantitative analysis of hexon binding at the NE in HeLa cells in the presence of 2 μM concentrations of the soluble Nup214 proteins. The histogram shows the mean fluorescence intensity of hexon staining, indicated as a percentage, in the presence of Nup214(1–586), Nup214(1–450), or Nup214(450–586). The mean fluorescence intensity of hexon staining around the nucleus was measured (n = 36 to 57 cells for each condition of each experiment) and compared to the no-Nup214 condition, fixed at 100 (*, P < 0.05). (D) Representative analysis of hexon binding at the NE in HeLa cells in the presence of 2.5 μM concentrations of the soluble Nup proteins. The histogram shows the mean fluorescence intensity of hexon staining, indicated as a percentage, in the presence of Nup358-1 (aa 996 to 1963), Nup358-4 (aa 2500 to 3224), or Nup214 (aa 1861 to 2090). The mean fluorescence intensity of hexon staining around the nucleus was measured (n = 52 to 70 cells for each condition) and compared to the no-Nup condition, fixed at 100 (*, P < 0.05).
We analyzed the localization of exogenously added hexon in digitonin-permeabilized HeLa cells when cells were incubated with various Nup214 fragments at excess molar concentrations. Competition with the longer Nup214 domain (aa 1 to 586) strongly inhibited hexon staining around the nucleus, whereas the shorter Nup214 domain (aa 1 to 450) had little effect on hexon binding to the NE (Fig. 5C). The addition of the Nup214(450–586) domain also strongly reduced hexon staining around the nucleus. Quantification of hexon binding in the presence of the three domains indicated 80% reduction for Nup214(1–586) and Nup214(450–586) and only a 20% reduction for Nup214(1–450) in these competition studies. These data indicate that the most N-terminal part of the Nup214 region did not compete for hexon binding while the region between aa 450 and 586 of Nup214 reduced the binding significantly, thus further delineating the hexon binding site within the Nup214 region of aa 450 to 586 (Fig. 5C). This domain contains only two FG repeats, unlike the C-terminal domain of Nup214, which contains many repeats.
Although our data argue that the N-terminal part of Nup214 is the only critical region for hexon binding, we sought to determine whether the FG repeat-rich C-terminal region of Nup214(1861–2090) and other known FG repeat-rich regions of Nup358 (Nup358-1 and Nup358-4) also could compete for hexon binding (Fig. 5D). However, hexon staining was not diminished significantly after the addition of a 2-fold molar excess of these regions. Since other FG repeats of different Nups did not interfere with hexon binding to the NE, we concluded that FG repeats do not play a role in the hexon binding to Nup214.
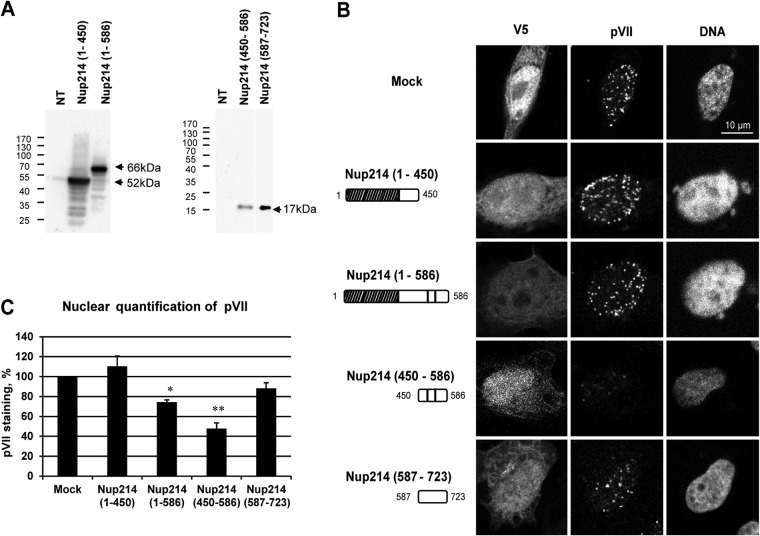
Reduction of protein VII nuclear localization following overexpression of soluble Nup214 fragments.
To complement the in vitro experiments involving competition of hexon binding to the NPC by Nup214 fragments (Fig. 6), we overexpressed N-terminal Nup214 fragments in cells and infected the cells with AdV. We analyzed pVII subcellular localization by IF at 3 h after infection. The Nup214 proteins were expressed at the expected molecular masses, as shown by Western blotting (Fig. 6A), and were localized throughout the cell (Fig. 6B). Nuclear accumulation of pVII was observed after overexpression of Nup214(1–450) as well as in mock-transfected cells, showing that this region of Nup214 does not prevent pVII nuclear import. A small decrease in nuclear accumulation of pVII was observed after overexpression of Nup214(1–586), and pVII accumulation in the nucleus was strongly reduced after overexpression of the Nup214(450–586) fragment (Fig. 6B and C). Although the Nup214(1–586) fragment also contains aa 450 to 586, these results suggest that full accessibility of the domain may not occur when it is not anchored in the NPC. The absence of pVII staining in the cytoplasm after overexpression of the Nup214(450–586) fragment indicates that the fragment was sufficient to prevent AdV binding to the NPC and the release of the genome but not sufficient to allow the capsid disassembly in the cytoplasm. The sequence of Nup214(450–586) is rich in proline, alanine, and serine (18%, 17%, and 24%, respectively). To test the specificity of this fragment for inhibition of pVII nuclear import, we also overexpressed a fragment of the same length enriched in similar amino acids just downstream of position 586, Nup214(587–723) (proline, alanine, and serine at 16%, 12%, and 20%, respectively). This fragment does not contain a coiled-coil domain, so it does not dimerize. Nuclear accumulation of pVII was observed after overexpression of Nup214(587–723) fragments, showing that this fragment did not compete for binding and pVII nuclear import. These data further indicate that nuclear binding is required for nuclear transport of the genome and that a specific small region (aa 450 to 586) in the N-terminal part of Nup214 binding is required for nuclear import of AdV DNA.
FIG 6.
Reduction of nuclear localization of protein VII in HeLa cells overexpressing soluble Nup214 proteins. HeLa cells were transfected with expression plasmids encoding soluble Nup214 proteins, Nup214(1–450), Nup214(1–586), Nup214(450–586), and Nup214(587–723), or with an empty plasmid expressing a V5-His tag (Mock) or were not transfected (NT). Cells were infected with AdV for 3 h. The cells were analyzed by IF staining using anti-V5 to detect the Nup214 proteins and anti-pVII antibody. (A) Expression analysis of soluble Nup214 proteins at 48 h posttransfection in HeLa cells. Cell lysates of NT cells or cells transfected with the different expression constructs were analyzed by Western blotting using anti-V5 antibody. The expected sizes of the Nup214 domains are indicated by the arrows. The migration and size standards are shown on the left side of each Western blot. (B) Representative images of protein VII localization in the nucleus of HeLa cells. Nuclei were stained with DAPI. Cells transfected by empty plasmid expressing a V5-His tag are shown (Mock). Symbols in the schematic representations at left are as identified on Fig. 2A. (C) Quantitative analysis of pVII in the nucleus of HeLa cells transfected with soluble Nup214 proteins. The histogram shows the mean fluorescence intensity of pVII staining, indicated as a percentage. The mean fluorescence intensity of pVII staining around the nucleus was measured (n = 36 to 76 cells for each condition of each experiment) and compared to that in mock cells, set at 100 (*, P < 0.05; **, P < 0.01).
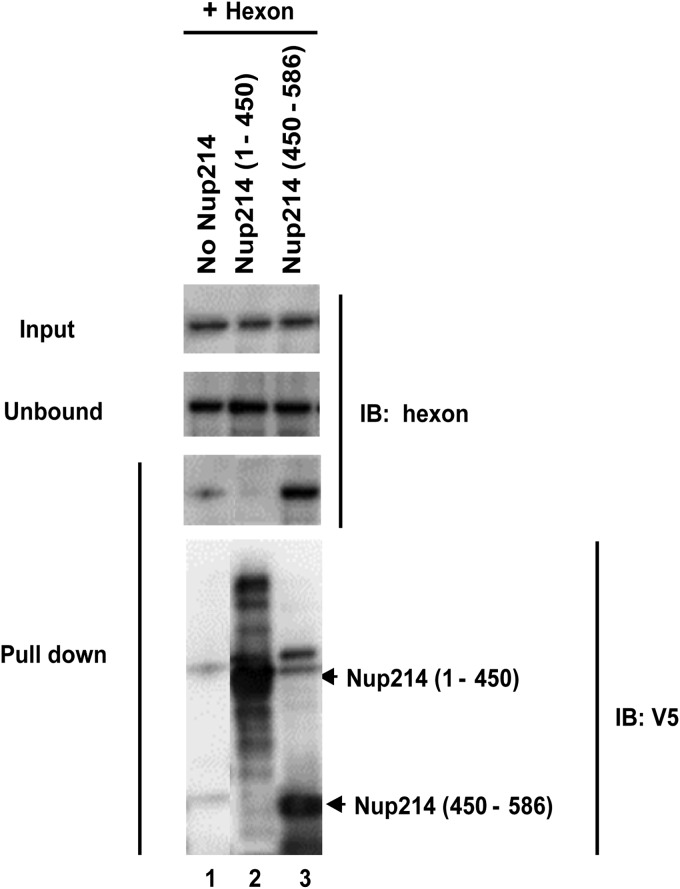
Direct interaction of hexon with the N-terminal region (aa 450 to 586) of Nup214.
Having obtained evidence for a direct, transport receptor-independent binding of hexon to Nup214 and localized the binding site to aa 450 to 586 of Nup214, we next confirmed the interaction biochemically. We incubated the soluble recombinant His-tagged Nup214 N-terminal proteins with purified hexon, Nup214(1–450), and Nup214(450–586) and captured the formed complexes on Ni-agarose beads (Fig. 7). The complexes bound were eluted and analyzed by SDS-PAGE. Western blotting revealed that purified hexon showed substantial interaction with Nup214(450–586) (Fig. 7, lane 3) but not Nup214(1–450) (Fig. 7, lane 2). A weak unspecific interaction of purified hexon for the beads alone was observed. Our finding that purified hexon showed a direct interaction with aa 450 to 586 of Nup214 is consistent with the ability of this fragment to inhibit AdV genome nuclear import.
FIG 7.
Direct binding of hexon to the Nup214(450–586) fragment. Soluble recombinant N-terminal fragments of Nup214 containing His tags were expressed and purified and coupled to nickel beads. The beads were incubated with purified hexon alone without Nup214 (lane 1), with purified hexon and Nup214(1–450) (lane 2), or with purified hexon and Nup214(450–586) (lane 3). The hexon was detected using anti-hexon antibody in the input (1/10) and unbound fractions and in the pulldown fractions. The presence of Nup214 fragments on the beads was detected using anti-V5 antibody (lower panel). IB, immunoblot.
DISCUSSION
Because the partially uncoated AdV particle is suggested to bind to the NPC via hexon, we used a permeabilized cell assay to map the Nup binding site for hexon binding at the NPC. The information from this assay was then used to validate the relevance of this interaction for AdV genome import into the nucleus. For initial insight on the NPC binding site for hexon, we used an shRNA knockdown approach to deplete either Nup214 or Nup358, the two major Nups localized at the cytoplasmic side of the NPC previously suggested to be involved in AdV nuclear import (8, 21). The depletion of each Nup was achieved without markedly affecting the expression of the other, as reported previously (23). The in vitro binding of hexon to the NE was specifically lost in Nup214-depleted cells and not in Nup358-depleted cells, consistent with work of Strunze et al. (21). Correspondingly, we observed loss of AdV genome import in Nup214-depleted cells but not in Nup358-depleted cells. These results strongly support an essential role of Nup214 in AdV docking to the NPC and genome import. In other studies, it has been suggested that kinesin-1 is involved in an active process engaging Nup214 and Nup358 to disrupt the permeability of the NPC and to uncoat the AdV DNA (21). Although a requirement for Nup358 was not detected under our conditions, the discrepancy between the two studies might be due to different multiplicities of infection (MOIs) and infection times after the Nup knockdown or to differences in silencing efficiencies.
Here, we have showed a direct interaction of hexon with an N-terminal region of Nup214 by biochemical assays and in the context of the NPC in digitonin-permeabilized cells. We found that highly purified hexon as well as low-pH-treated AdV particles efficiently bound to the NE of permeabilized cells in the absence of added cytosolic factors, suggesting that this interaction does not involve nuclear transport receptors. Consistent with the notion that this binding represents a docking step for AdV, we did not observe hexon nuclear import when energy and cytosol were added to the binding reaction mixture. These results agree with the previous report that hexon import is mediated by protein VI (28). In an effort to map the binding site on Nup214 for AdV, we performed reconstitution with Nup214 N-terminal fragments that retained information for targeting to the NPC and competition experiments. The expression of the N terminus of Nup214(1–1058) in Nup214-depleted cells, but not a fragment containing aa 586 to 1058, was sufficient to fully restore the binding of hexon to the NE. Consistently, a recombinant N-terminal fragment (Nup214 aa 1 to 586) strongly reduced the binding of hexon to the NE in competition experiments. These results showed that the coiled-coil domain (aa 586 to 1058), the minimal region for NPC binding of Nup214, was not involved in hexon binding, excluding a potential role of Nup88 or other associated Nups. We also showed direct binding in in vitro binding assays using purified hexon and a recombinant fragment, which supports the observation that hexon binds to aa 450 to 586 of Nup214. A multitude of in vivo competition assays indicate that binding to this region is important for AdV DNA import, both competition involving expression of NPC-targeted regions of Nup214 and competition with fragments of Nup214 that are not targeted to NPC.
Here, we identified a new binding site at the N terminus of Nup214 while a previous study reported that the C terminus of Nup214 (aa 1549 to 2090) is associated with AdV in vitro using cross-linking approaches (8). Nevertheless, the total restoration of binding and protein VII accumulation after expression of the N-terminal domain of Nup214 in Nup214-depleted cells does not exclude the role of other regions of Nup214 in AdV import. The N-terminal region of Nup214 contains a β-propeller structure (39) and two FG repeats. The β-propeller structure of Nup214 does not seem to be part of the binding site because the soluble N-terminal fragment (Nup214 aa 1 to 450) encompassing the β-propeller structure does not compete for hexon binding. This restricts the putative binding site to a 137-amino-acid fragment of highly disordered structure featuring two FG repeats. A role of the FG repeats in the binding is not supported by our experiments showing that several FG-enriched fragments were not able to compete for hexon binding in digitonin-permeabilized cells. Several tools for protein disorder prediction are in agreement to identify this fragment as a disordered structure: DisEMBL identified aa 375 to 582 as disordered by loops and aa 487 to 517 and 527 to 543 as disordered by hot loops, and GlobPlot software identified aa 454 to 584 as disordered (38, 41). Intrinsically disordered proteins are involved in cell signaling and can bind to a multitude of interacting proteins (42). A recent study used this disorder property to explain the specificity of interaction between Nup153 and transport receptors (43). Further analysis will be required to understand how hexon binds to this restricted, highly disordered fragment.
Nups including Nup214 and Nup358 are likely involved in the binding and nuclear transport of several other nuclear replicating viruses; however, each virus seems to have developed its own nuclear association strategy. The nuclear entry of the human immunodeficiency virus type 1 (HIV-1) (44) was reduced in Nup358-depleted cells. A hypothetical model for herpes simplex virus 1 (HSV-1) binding and uncoating at the NPC involved the binding of the capsid to the cytoplasmic filaments mediated by Nup358 (45), then Nup214, resulting in capsid destabilization (46). Indeed, the knockdown of Nup214 also delayed the onset of DNA replication in the nucleus of HSV-1-infected cells (47). In contrast to AdV, the NPC binding of HSV-1 capsid was importin β dependent (48). AdV could also bind in a nonspecific manner to the FG-enriched domain of Nup214 or other FG-enriched Nups of cytoplasmic fibrils, thereby allowing capsid dissociation from microtubules in the vicinity of a specific binding site in the N terminus of Nup214. Further delineation of the complex steps involved in virus binding and nuclear penetration will require concerted efforts in both structural and molecular cell biology. The identification of the specific domain on hexon interacting with Nup214 could promote the development of new strategies for gene transfer to target efficiently the nucleus.
ACKNOWLEDGMENTS
This work was supported by NIH grants AI55729 to L.G. and HL054352 to G.R.N. A.C. was supported by a grant from the FRM SPE20041102385. A.C, H.W., and M.K. were supported by a grant from the FRM DEQ20110421299. H.W. is an INSERM fellow.
We thank D. A. Engel and C. Waterman for sharing reagents with us. We acknowledge the technical support of William B. Kiosses (TSRI Microscopy Core), N. Dugot-Senant, C. Poujol, P. Legros, S Marais, M. Mondin, and F. Cordelieres (Bordeaux Imaging Center) for confocal microscopy.
REFERENCES
- 1.San Martin C. 2012. Latest insights on adenovirus structure and assembly. Viruses 4:847–877. doi: 10.3390/v4050847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nemerow GR, Stewart PL, Reddy VS. 2012. Structure of human adenovirus. Curr Opin Virol 2:115–121. doi: 10.1016/j.coviro.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith JG, Wiethoff CM, Stewart PL, Nemerow GR. 2010. Adenovirus. Curr Top Microbiol Immunol 343:195–224. doi: 10.1007/82_2010_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reddy VS, Nemerow GR. 2014. Structures and organization of adenovirus cement proteins provide insights into the role of capsid maturation in virus entry and infection. Proc Natl Acad Sci U S A 111:11715–11720. doi: 10.1073/pnas.1408462111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiethoff CM, Wodrich H, Gerace L, Nemerow GR. 2005. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J Virol 79:1992–2000. doi: 10.1128/JVI.79.4.1992-2000.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dodding MP, Way M. 2011. Coupling viruses to dynein and kinesin-1. EMBO J 30:3527–3539. doi: 10.1038/emboj.2011.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scherer J, Vallee RB. 2011. Adenovirus recruits dynein by an evolutionary novel mechanism involving direct binding to pH-primed hexon. Viruses 3:1417–1431. doi: 10.3390/v3081417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trotman LC, Mosberger N, Fornerod M, Stidwill RP, Greber UF. 2001. Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat Cell Biol 3:1092–1100. doi: 10.1038/ncb1201-1092. [DOI] [PubMed] [Google Scholar]
- 9.Hindley CE, Lawrence FJ, Matthews DA. 2007. A role for transportin in the nuclear import of adenovirus core proteins and DNA. Traffic 8:1313–1322. doi: 10.1111/j.1600-0854.2007.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wodrich H, Cassany A, D'Angelo MA, Guan T, Nemerow G, Gerace L. 2006. Adenovirus core protein pVII is translocated into the nucleus by multiple import receptor pathways. J Virol 80:9608–9618. doi: 10.1128/JVI.00850-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chook YM, Süel KE. 2011. Nuclear import by karyopherin-βs: recognition and inhibition. Biochim Biophys Acta 1813:1593–1606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tetenbaum-Novatt J, Rout MP. 2010. The mechanism of nucleocytoplasmic transport through the nuclear pore complex. Cold Spring Harb Symp Quant Biol 75:567–584. doi: 10.1101/sqb.2010.75.033. [DOI] [PubMed] [Google Scholar]
- 13.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. 2002. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol 158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoelz A, Debler EW, Blobel G. 2011. The structure of the nuclear pore complex. Annu Rev Biochem 80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 15.D'Angelo MA, Hetzer MW. 2008. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol 18:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whittaker GR, Kann M, Helenius A. 2000. Viral entry into the nucleus. Annu Rev Cell Dev Biol 16:627–651. doi: 10.1146/annurev.cellbio.16.1.627. [DOI] [PubMed] [Google Scholar]
- 17.Greber UF, Willetts M, Webster P, Helenius A. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75:477–486. doi: 10.1016/0092-8674(93)90382-Z. [DOI] [PubMed] [Google Scholar]
- 18.Greber UF, Suomalainen M, Stidwill RP, Boucke K, Ebersold MW, Helenius A. 1997. The role of the nuclear pore complex in adenovirus DNA entry. EMBO J 16:5998–6007. doi: 10.1093/emboj/16.19.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin-Fernandez M, Longshaw SV, Kirby I, Santis G, Tobin MJ, Clarke DT, Jones GR. 2004. Adenovirus type-5 entry and disassembly followed in living cells by FRET, fluorescence anisotropy, and FLIM. Biophys J 87:1316–1327. doi: 10.1529/biophysj.103.035444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strunze S, Trotman LC, Boucke K, Greber UF. 2005. Nuclear targeting of Adenovirus type 2 requires CRM1-mediated nuclear export. Mol Biol Cell 16:2999–3009. doi: 10.1091/mbc.E05-02-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strunze S, Engelke MF, Wang I-H, Puntener D, Boucke K, Schleich S, Way M, Schoenenberger P, Burckhardt CJ, Greber UF. 2011. Kinesin-1-mediated capsid disassembly and disruption of the nuclear pore complex promote virus infection. Cell Host Microbe 10:210–223. doi: 10.1016/j.chom.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Moroianu J, Hijikata M, Blobel G, Radu A. 1995. Mammalian karyopherin alpha 1 beta and alpha 2 beta heterodimers: alpha 1 or alpha 2 subunit binds nuclear localization signal and beta subunit interacts with peptide repeat-containing nucleoporins. Proc Natl Acad Sci U S A 92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutten S, Kehlenbach RH. 2006. Nup214 is required for CRM1-dependent nuclear protein export in vivo. Mol Cell Biol 26:6772–6785. doi: 10.1128/MCB.00342-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yaseen NR, Blobel G. 1999. Two distinct classes of Ran-binding sites on the nucleoporin Nup-358. Proc Natl Acad Sci U S A 96:5516–5521. doi: 10.1073/pnas.96.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitt I, Gerace L. 2001. In vitro analysis of nuclear transport mediated by the C-terminal shuttle domain of Tap. J Biol Chem 276:42355–42363. doi: 10.1074/jbc.M103916200. [DOI] [PubMed] [Google Scholar]
- 26.Bernad R, van der Velde H, Fornerod M, Pickersgill H. 2004. Nup358/RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/CAN and plays a supporting role in CRM1-mediated nuclear protein export. Mol Cell Biol 24:2373–2384. doi: 10.1128/MCB.24.6.2373-2384.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu E, Trauger SA, Pache L, Mullen T-M, Seggern DJV, Siuzdak G, Nemerow GR. 2004. Membrane cofactor protein is a receptor for adenoviruses associated with epidemic keratoconjunctivitis. J Virol 78:3897–3905. doi: 10.1128/JVI.78.8.3897-3905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wodrich H, Guan T, Cingolani G, Von Seggern D, Nemerow G, Gerace L. 2003. Switch from capsid protein import to adenovirus assembly by cleavage of nuclear transport signals. EMBO J 22:6245–6255. doi: 10.1093/emboj/cdg614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wodrich H, Henaff D, Jammart B, Segura-Morales C, Seelmeir S, Coux O, Ruzsics Z, Wiethoff CM, Kremer EJ. 2010. A capsid-encoded PPxY-motif facilitates adenovirus entry. PLoS Pathog 6:e1000808. doi: 10.1371/journal.ppat.1000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kehlenbach RH, Dickmanns A, Gerace L. 1998. Nucleocytoplasmic shuttling factors including Ran and CRM1 mediate nuclear export of NFAT in vitro. J Cell Biol 141:863–874. doi: 10.1083/jcb.141.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yaseen NR, Blobel G. 1999. GTP hydrolysis links initiation and termination of nuclear import on the nucleoporin Nup358. J Biol Chem 274:26493–26502. doi: 10.1074/jbc.274.37.26493. [DOI] [PubMed] [Google Scholar]
- 32.Thevenaz P, Ruttimann UE, Unser M. 1998. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process 7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 33.Saphire ACS, Guan T, Schirmer EC, Nemerow GR, Gerace L. 2000. Nuclear import of adenovirus DNA in vitro involves the nuclear protein import pathway and hsc70. J Biol Chem 275:4298–4304. doi: 10.1074/jbc.275.6.4298. [DOI] [PubMed] [Google Scholar]
- 34.Featherstone C, Darby MK, Gerace L. 1988. A monoclonal antibody against the nuclear pore complex inhibits nucleocytoplasmic transport of protein and RNA in vivo. J Cell Biol 107:1289–1297. doi: 10.1083/jcb.107.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snow CM, Senior A, Gerace L. 1987. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J Cell Biol 104:1143–1156. doi: 10.1083/jcb.104.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adam SA, Marr RS, Gerace L. 1990. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol 111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villén J, Li J, Cohn MA, Cantley LC, Gygi SP. 2004. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A 101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. 2003. Protein disorder prediction: implications for structural proteomics. Structure 11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Napetschnig J, Blobel G, Hoelz A. 2007. Crystal structure of the N-terminal domain of the human protooncogene Nup214/CAN. Proc Natl Acad Sci U S A 104:1783–1788. doi: 10.1073/pnas.0610828104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernad R, Engelsma D, Sanderson H, Pickersgill H, Fornerod M. 2006. Nup214-Nup88 nucleoporin subcomplex is required for CRM1-mediated 60 S preribosomal nuclear export. J Biol Chem 281:19378–19386. doi: 10.1074/jbc.M512585200. [DOI] [PubMed] [Google Scholar]
- 41.Linding R, Russell RB, Neduva V, Gibson TJ. 2003. GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res 31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. 2002. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol 323:573–584. doi: 10.1016/S0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 43.Milles S, Lemke EA. 2014. Mapping multivalency and differential affinities within large intrinsically disordered protein complexes with segmental motion analysis. Angew Chem Int Ed Engl 53:7364–7367. doi: 10.1002/anie.201403694. [DOI] [PubMed] [Google Scholar]
- 44.Zhang R, Mehla R, Chauhan A. 2010. Perturbation of host nuclear membrane component RanBP2 impairs the nuclear import of human immunodeficiency virus-1 preintegration complex (DNA). PLoS One 5:e15620. doi: 10.1371/journal.pone.0015620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Copeland AM, Newcomb WW, Brown JC. 2009. Herpes simplex virus replication: roles of viral proteins and nucleoporins in capsid-nucleus attachment. J Virol 83:1660–1668. doi: 10.1128/JVI.01139-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liashkovich I, Hafezi W, Kühn JM, Oberleithner H, Shahin V. 2011. Nuclear delivery mechanism of herpes simplex virus type 1 genome. J Mol Recognit 24:414–421. doi: 10.1002/jmr.1120. [DOI] [PubMed] [Google Scholar]
- 47.Pasdeloup D, Blondel D, Isidro AL, Rixon FJ. 2009. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J Virol 83:6610–6623. doi: 10.1128/JVI.02655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ojala PM, Sodeik B, Ebersold MW, Kutay U, Helenius A. 2000. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol Cell Biol 20:4922–4931. doi: 10.1128/MCB.20.13.4922-4931.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Panté N, Kann M. 2002. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell 13:425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]