ABSTRACT
The clinical utility of the adeno-associated virus (AAV) gene delivery system has been validated by the regulatory approval of an AAV serotype 1 (AAV1) vector for the treatment of lipoprotein lipase deficiency. However, neutralization from preexisting antibodies is detrimental to AAV transduction efficiency. Hence, mapping of AAV antigenic sites and engineering of neutralization-escaping vectors are important for improving clinical efficacy. We report the structures of four AAV-monoclonal antibody fragment complexes, AAV1-ADK1a, AAV1-ADK1b, AAV5-ADK5a, and AAV5-ADK5b, determined by cryo-electron microscopy and image reconstruction to a resolution of ∼11 to 12 Å. Pseudoatomic modeling mapped the ADK1a epitope to the protrusions surrounding the icosahedral 3-fold axis and the ADK1b and ADK5a epitopes, which overlap, to the wall between depressions at the 2- and 5-fold axes (2/5-fold wall), and the ADK5b epitope spans both the 5-fold axis-facing wall of the 3-fold protrusion and portions of the 2/5-fold wall of the capsid. Combined with the six antigenic sites previously elucidated for different AAV serotypes through structural approaches, including AAV1 and AAV5, this study identified two common AAV epitopes: one on the 3-fold protrusions and one on the 2/5-fold wall. These epitopes coincide with regions with the highest sequence and structure diversity between AAV serotypes and correspond to regions determining receptor recognition and transduction phenotypes. Significantly, these locations overlap the two dominant epitopes reported for autonomous parvoviruses. Thus, rather than the amino acid sequence alone, the antigenic sites of parvoviruses appear to be dictated by structural features evolved to enable specific infectious functions.
IMPORTANCE The adeno-associated viruses (AAVs) are promising vectors for in vivo therapeutic gene delivery, with more than 20 years of intense research now realized in a number of successful human clinical trials that report therapeutic efficacy. However, a large percentage of the population has preexisting AAV capsid antibodies and therefore must be excluded from clinical trials or vector readministration. This report represents our continuing efforts to understand the antigenic structure of the AAVs, specifically, to obtain a picture of “polyclonal” reactivity as is the situation in humans. It describes the structures of four AAV-antibody complexes determined by cryo-electron microscopy and image reconstruction, increasing the number of mapped epitopes to four and three, respectively, for AAV1 and AAV5, two vectors currently in clinical trials. The results presented provide information essential for generating antigenic escape vectors to overcome a critical challenge remaining in the optimization of this highly promising vector delivery system.
INTRODUCTION
The adeno-associated viruses (AAVs), single-stranded DNA packaging viruses belonging to the Parvoviridae family, are promising vectors for gene delivery. There are over 100 AAV genomic isolates, and 13 human and nonhuman serotypes have been described (1). To date, no diseases have been associated with AAV infections. Recombinant AAV (rAAV) vectors can package foreign (nonviral) genes, transduce both dividing and nondividing cells, and induce long-term gene expression in nondividing cells (2). In addition, AAV serotypes have different transduction efficiencies for different tissues dictated by their capsid sequence (1, 3). These properties make AAVs desirable vectors for therapeutic gene delivery.
The AAV gene delivery system has been successfully utilized in several human clinical trials, including the treatment of hemophilia B with an rAAV8 vector expressing therapeutic levels of the factor IX protein (4) and the restoration of vision in Leber's congenital amaurosis patients with an rAAV2 vector encoding the retinal pigment epithelium-specific 65-kDa protein (5–7). AAV1, which displays better and quicker onset of transgene expression in skeletal muscle than AAV2 (8, 9), has also been used for several clinical trials, including the treatment of antitrypsin deficiency (10), lipoprotein lipase deficiency (11, 12), Pompe's disease (ClinicalTrials.gov registration no. NCT00976352), and muscular dystrophy (13). Significantly, in 2012, the European Commission approved an rAAV1 vector encoding lipoprotein lipase as a therapy treatment for patients with this enzyme deficiency (12). This approval represents the first realization of gene therapy as a viable clinical treatment.
However, despite the above successes, several obstacles must still be overcome to achieve full clinical efficacy in patient care and treatment for the AAV vector system. One of the most important of these is preexisting immunity. Serologic studies show that ∼40 to 70% of the human population has been exposed to AAVs (14–17). Neutralization by preexisting antibodies decreases AAV transduction efficiency, even at low antibody titers (18–20). For this reason, individuals with evidence of preexisting AAV antibodies were excluded from participation in a hemophilia B trial with the rAAV8 vector (4). Thus, improvement of the AAV vector system requires an understanding of how antibodies interact with the AAV capsid.
The T=1 icosahedral capsid of the AAVs is assembled from three overlapping viral proteins (VPs), VP1, VP2, and VP3, with a common C-terminal region, in a reported ratio of 1:1:10, respectively (21–26). Thus, the entire sequence of VP3 is contained within VP2, the entire sequence of which is, in turn, contained within VP1. The minor VP1 protein contains a unique N-terminal region (VP1u). The capsid structures of AAV1 to AAV9 have been determined by X-ray crystallography and/or cryo-electron microscopy (cryo-EM) and image reconstruction (cryo-reconstruction) (27–35; unpublished data). In all of these structures, only the VP3 common region (∼520 amino acids) is observed. The topology of this VP region consists of a conserved alpha helix (αA), a βA strand, and an eight-stranded antiparallel β-barrel (βB-βI) domain with large interstrand loops. The βBIDG sheet lines the interior of the capsid, and the interstrand loops between the β-barrel form the exterior capsid surface topology. Loops between the β-strand regions are named for their flanking strands; e.g., the largest of the surface loops is that between the βG and βH strands (GH loop), which consists of ∼230 amino acids. The surface features of the capsids include a depression at the 2-fold axes, three protrusions surrounding the 3-fold axes, a canyon surrounding a cylindrical channel at the 5-fold axes, and a wall/plateau between the depressions at the 2- and 5-fold axes (the 2/5-fold wall). Nine common AAV variable regions (VRs), VR-I to VR-IX, have been assigned at the apex of the large interstrand loops (28). These VRs contribute to local capsid surface topological differences between the AAV serotypes and contribute to their functional profiles, including receptor attachment, tissue tropism, transduction efficiency, and antigenic reactivity (9, 28, 30, 32, 34, 36–39).
The antigenically best-characterized AAV serotype is AAV2; it has been analyzed by directed evolution, peptide scanning, peptide insertion, and site-directed mutagenesis (40–43). Many of the antigenic residues were mapped to the VRs forming the 2/5-fold wall, i.e., VR-I, VR-III, and VR-IX, and the 3-fold protrusions, i.e., VR-IV, VR-V, and VR-VIII, although some residues were buried in the capsid interior (44). More recently, cryo-reconstruction has been used to visualize AAV2 antigenic epitopes for two neutralizing anticapsid monoclonal antibodies (MAbs), A20 (45) and C37-B (36), as well as for antibodies against AAV1, AAV5, AAV6, and AAV8 (36, 37). Significantly, these sites were also localized to the 2/5-fold wall and 3-fold protrusions despite a sequence diversity of ∼60 to 99% between the AAVs, suggesting a commonality in antibody recognition sites. However, to generate an antigenic profile that fully recapitulates the polyclonal response to these viruses that exists in vivo, several MAb epitopes must be mapped for all of the serotypes that are being considered for clinical application.
In this study, the epitopes for four MAbs, two that neutralize AAV1 (ADK1a and ADK1b), one that neutralizes AAV5 (ADK5b), and one that is nonneutralizing against AAV5 (ADK5a), were mapped by cryo-reconstruction. Their epitopes were localized to the 3-fold protrusion (ADK1a) and the 2/5-fold wall (ADK1b and ADK5a) and bridging the 3-fold protrusion and 2/5-fold wall (ADK5b). These regions are close to or overlap epitopes previously reported for antibodies against AAV2, AAV6, and AAV8, as well as two MAbs, 4E4 and 5H7, against AAV1, and MAb 3C5 against AAV5 (36, 37, 45). This study thus reaffirms the claim that the AAVs have antigenic regions in common, and the results begin to delineate the boundaries of the antigenic surface of the AAVs. These regions have the highest sequence and structural variability between the AAVs and are known to be receptor attachment and transduction determinants. Their overlap indicates that structure, rather than sequence alone, dictates host immune system reactivity. Significantly, equivalent regions of the capsid of autonomous parvoviruses serve as dominant antigenic epitopes and are also involved in similar capsid functions. These observations thus highlight a commonality in the development of the host humoral response against the parvovirus capsid, evolved to abrogate specific infectious functions. A combination of these visualized epitopes with information on functional regions of the capsid can help guide the genetic engineering of rAAV vectors that can evade antibody neutralization while retaining the parental tropism properties.
MATERIALS AND METHODS
Cell culture.
HEK293T (293T) and COS cells were maintained in Dulbecco modified Eagle medium supplement with 10% heat-inactivated fetal calf serum and a 1× antibiotic-antimycotic mixture (100 U/ml penicillin, 100 μg/ml streptomycin, and 250 ng/ml amphotericin B [Fungizone], diluted from 100× Antibiotic-Antimycotic [Gibco]) to prevent bacterial and fungal contamination. Cultures were incubated at 37°C in 5% CO2.
Vector production.
Purified rAAV1-green fluorescent protein (GFP) (UF11) vectors were kindly provided by Sanford L. Boye (Department of Ophthalmology, University of Florida College of Medicine, Gainesville, FL). The pTRUF11 gene was packaged into AAV1 capsids under the control of the synthetic chicken β-actin (CBA) promoter to express GFP as previously described (46). The rAAVl-GFP vectors were purified by iodixanol step gradients and a HiTrap Q column as previously described (47), and titers were determined by quantitative real-time PCR (qPCR) with primers specific for the CBA promoter.
rAAV5-GFP vectors expressing nucleus-localized GFP were produced as previously described (48). Briefly, 293T cells were triply transfected with three plasmids: pAAV5-NLS-GFP (under the control of the cytomegalovirus [CMV] promoter), pAAV5 RepCap, and the adenovirus (Ad) helper 449B (49). rAAV5-GFP vectors were purified by CsCl gradient centrifugation. DNase-resistant genome copy numbers were determined by qPCR with the TaqMan system (Applied Biosystems) and primer probes specific to the CMV promoter.
Neutralization assay.
For the AAV1 antibodies, 293T cells were seeded into 96-well plates at 1.0 × 104 to 3.0 × 104/well and incubated at 37°C for 24 h to achieve 70% confluence. rAAV1-GFP vectors, preincubated with different dilutions of hybridoma supernatants (1 to 1/1,000) of ADK1a, ADK1b, and ADK4 (negative control) at 37°C for 1 h, were then used to infect the cells at a ratio of 10,000 genome-containing vector particles per target cell (multiplicity of infection [MOI] = 10,000). Ad serotype 5, at an MOI of 1, was used as a helper during infection. Cells were harvested 48 h postinfection and resuspended in filtered 1× phosphate-buffered saline (PBS; 137 mM NaCl, 12 mM phosphate, 2.7 mM KCl, pH 7.4) for assessment of GFP expression by flow cytometry (FACSCalibur; BD Bioscience, San Jose, CA).
For the AAV5 antibodies, COS cells were seeded at a density of 5 × 103/well into 96-well plates 24 h prior to infection with 2 × 107 rAAV5-GFP vectors preincubated with serial dilutions (1/25 to 1/3,200) of ADK5a and ADK5b in medium for 1 h at room temperature. Cells were incubated at 37°C for 1 h and then washed with medium. At 24 h postinfection, cells were analyzed for GFP expression by flow cytometry (BD FACSArray bioanalyzer).
AAV VLP production and purification.
AAV1 and AAV5 virus-like particles (VLPs) were expressed by using a recombinant baculovirus-Sf9 expression system and purified as previously reported (50, 51). VLPs extracted from the 20 to 25% sucrose fraction were dialyzed overnight against 1× PBS with gentle stirring at 4°C. The VLP concentrations were determined by using readings of optical density at 280 nm and an extinction coefficient of 1.7 (for calculation of concentrations in mg/ml). The purity and integrity of purified VLPs were monitored by SDS-PAGE and negative-stain EM, respectively.
MAb purification and Fab fragment generation.
Hybridoma cell supernatants for the ADK1a, ADK1b, ADK5a, and ADK5b MAbs were produced as previously reported (52). Intact IgGs were purified from the supernatants by Hi Trap protein G HP columns (GE Healthcare) with glycine-HCl, pH 2.7, as the elution buffer into 1 M Tris-HCl, pH 9.0, to achieve a neutral final pH. Purified IgGs were buffer exchanged into 20 mM sodium phosphate (pH 7.0)–10 mM EDTA. Fragment antigen-binding (Fab) fragments were generated by incubating the IgG samples with activated immobilized papain (Pierce) at the manufacturer-recommended enzyme/substrate ratio of 1:160 (vol/vol) at 37°C for 12 h. The digested samples were loaded onto a Hi Trap protein A column (GE Healthcare) to separate undigested IgG and the Fc (fragment, crystallizable) fragment from the Fab fragment, which is collected as the flowthrough. Purified Fab fragments were buffer exchanged into PBS and concentrated for virus capsid-Fab fragment complexing.
Virus capsid-Fab fragment complex preparation.
VLPs were mixed with Fab fragments at a ratio of one Fab fragment molecule per potential VP-binding site on the capsid (Fab fragment/VLP molar ratio of ∼60:1) for the ADK1a, ADK1b, and ADK5b complexes and at a ratio of two Fab fragment molecules per potential VP binding site for ADK5a. Complexes were incubated at 4°C for 1 h. Prior to sample vitrification for cryo-EM imaging, the complexes were examined by negative-stain EM to monitor capsid integrity and successful Fab fragment decoration.
Cryo-EM data collection.
Quantifoil (Quantifoil Micro Tools GmbH, Germany) or C-Flats (CF-2/2-4c-50; Protochips, Inc.) holey carbon grids were glow discharged in a Pelco easiGlow unit for 1 min prior to use. Three-microliter samples of the virus-Fab fragment complexes were loaded onto glow-discharged grids and vitrified by a manual plunge freezing device (AAV1-ADK1a) or a Vitrobot Mark IV (FEI) (AAV1-ADK1b, AAV5-ADK5a, and AAV5-ADK5b). The frozen grids were transferred into liquid nitrogen and then into FEI transmission EM cryo-sample holders. The data for the AAV1-ADK1a complex were collected with an FEI Sphera electron microscope operated at 200 kV at a magnification of ×50,000 with a 1.5- to 2.75-μm underfocus range under low-dose conditions (∼24 to 28 electrons/Å2) and on films. The films (42 in total) were scanned with a Nikon Super Coolscan 8000ED at a scan step size of 6.35 μm/pixel, giving a final image step size of 1.27 Å/pixel. The AAV1-ADK1b, AAV5-ADK5a, and AAV5-ADK5b complex data were collected with an FEI Tecnai TF20 electron microscope operated at 200 kV at a magnification of ×67,050 with a 1.5- to 3-μm defocus range and under low-dose conditions (∼20 electrons/Å2) and on a charge-coupled device (CCD). Forty-nine, 75, and 82 images, respectively, were recorded on a Gatan UltraScan 4000 CCD camera at a step size of 2.24 Å/pixel for these complexes.
Cryo-EM image reconstruction.
For the AAV1-ADK1a data set, the RobEM software package (http://cryoEM.ucsd.edu/programs.shtm) was used to extract individual particle images from the micrographs. Preprocessing of the selected images and estimation of the defocus levels were done as previously described (53). A random-model computation procedure (54) was used to generate an initial model at a resolution of ∼30-Å from 150 particle images. This map was then used to initiate full orientation and origin determination and refinement of the entire set of images by using the current version of Auto3DEM (55). Corrections to compensate for the effects of phase reversals in the contrast transfer functions of the images were performed as previously described (56, 57), but amplitude corrections were not applied. The resolution of the final reconstructed structure, which utilized 1,271 particle images, was estimated to be ∼11 Å on the basis of a conservative Fourier shell correlation (FSC) threshold of 0.5 (58). For the AAV1-ADK1b, AAV5-ADK5a, and AAV5-ADK5b data sets, the structures were determined as described for the AAV1-ADK1a complex by using AUTO3DEM and/or the EMAN2 software package (59) from particles reboxed from the same micrographs. This process was carried out to enable comparison of the structures from two independent software packages. The resolution estimate for the final reconstruction was comparable at ∼11 Å (FSC, 0.5) from 1,057 particle images for AAV1-ADK1b. For the AAV5 complexes, the values were ∼11 Å from 2,728 particle images for AAV5-ADK5a and ∼12 Å from 685 particle images for AAV5-ADK5b. The final density maps for the four complexes were compared to the AAV1 and AAV5 crystal structures (Research Collaboratory for Structural Bioinformatics Protein Data Bank [PDB] no. 3NG9 and 3NTT) to confirm their absolute handedness. Graphical representations of the reconstruction maps were generated by the Chimera software package (60).
Density map interpretation: pseudoatomic model fitting, difference map calculations, and prediction of antigenic epitopes.
The 60-mer VP capsid coordinates of the crystal structures of AAV1 and AAV5 (generated by icosahedral matrix multiplication using the Viperdb online server at http://viperdb.scripps.edu/) (61) were fitted by rigid-body rotation and translation into the respective cryo-reconstructed density maps of their AAV-Fab fragment complexes by using the “Fit in map” function in Chimera (60). Difference maps generated in Chimera, which subtracted the model map of the fitted AAV1 and AAV5 60-mer structures from the corresponding scaled AAV-Fab fragment cryo-reconstructed density map, were used to interpret the Fab fragment density. A generic Fab fragment structure (PDB no. 2FBJ) (62) was fitted into the positive difference maps, again by using the “Fit in map” function in Chimera as previous described (36). A generic Fab fragment structure was used because the sequence information for the four antibodies is not available. The correlation coefficients of a model map generated for the pseudoatomic coordinates of the AAV capsids fully decorated with Fab fragments (i.e., 60 copies) with the respective reconstructed density maps were 0.91 for AAV1-ADK1a, 0.96 for AAV1-ADK1b, 0.93 for AAV5-ADK5a, and 0.91 for AAV5-ADK5b. The coordinates of the fitted complexes were further examined in the COOT (63) and PyMol (http://www.pymol.org/; The PyMOL Molecular Graphics System, version 1.5.0.4; Schrödinger, LLC) programs to visualize the AAV-Fab fragment interfacing amino acid residues and capsid surface area occluded by the bound Fab fragment (the footprint). The Fab fragment-occluded residues were defined as the surface residues on the AAV capsid delineated by the Fab fragment density map. The area of this occluded region was determined on the basis of distances between “boundary atoms” measured in the COOT program and the shape of the footprint. These coordinates were also analyzed in the PDBePISA online software package (http://www.ebi.ac.uk/msd-srv/prot_int/) (64) to predict the interfacing (buried surface area [BSA]) and contact residues, defined here as the potential epitopes.
RESULTS
Neutralization.
The production and AAV1 and AAV5 capsid binding properties of the ADK1a, ADK1b, ADK5a, and ADK5b antibodies have been reported previously (52), but their abilities to neutralize infection by their respective serotypes were unknown. Thus, prior to mapping their footprints, we investigated their abilities to neutralize infection with rAAV-GFP vectors. For the AAV1 studies, the ADK4 MAb directed against the AAV4 capsid, which does not cross-react with AAV1 (52), was used as a negative control. Both the ADK1a and ADK1b antibodies were neutralizing, while in the presence of the nonspecific ADK4, the cellular GFP expression was relatively the same at all of the antibody ratios tested (Fig. 1A). For AAV5, the two antibodies displayed a difference in the ability to neutralize infection. ADK5b neutralized >50% of the transduction in COS cells by a dilution of 1/800, while ADK5a failed to neutralize 50% of the transduction at all of the dilutions tested (Fig. 1B).
FIG 1.
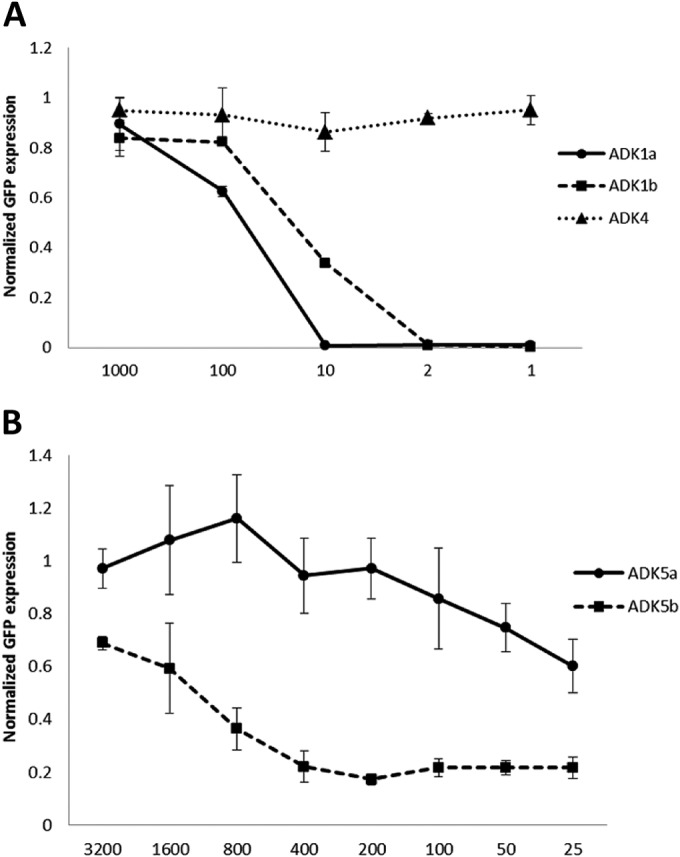
Neutralization by ADK antibodies. Shown are percentages of cellular GFP expression after rAAV1-GFP or rAAV5-GFP infection following preincubation with serial dilutions of ADK1a/1b and ADK4 (A) and ADK5a/5b (B). All of the data were normalized to the control, which was virus infection in the absence of antibody incubation. The data are average values of three replicates.
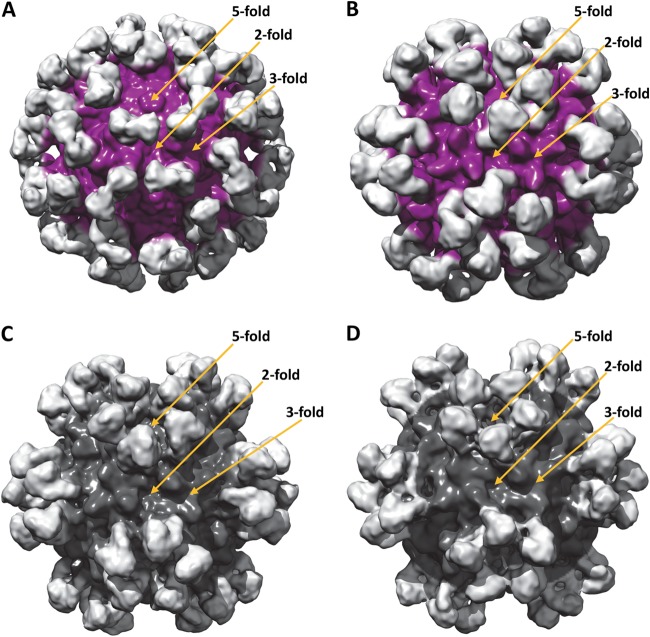
The three-dimensional (3D) cryo-reconstructed AAV1-Fab fragment complex structures.
The AAV1-ADK1a complex structure was determined to ∼11Å (Fig. 2A). Fab fragment densities were observed at the tip of each 3-fold protrusion bound at an estimated ∼45° angle relative to the capsid surface. For the AAV1-ADK1b complex, the structure was also determined to ∼11 Å by utilizing both EMAN2 and Auto3DEM (Fig. 2B). The resulting density maps from the two reconstruction programs were similar, consistent with their comparable resolution. The Fab fragment densities, projecting radially out from the capsid surface, were localized to the edge of the 2/5-wall, extending toward the 5-fold axes. Five Fab fragment molecules thus surround each 5-fold axis, with the 3-fold protrusion left exposed (Fig. 2B).
FIG 2.
AAV-Fab fragment complex structures determined by cryo-reconstruction. Shown are shaded surface representations of the AAV1-ADK1a (A), AAV1-ADK1b (B), AAV5-ADKa (C), and AAV5-ADK5b (D) complex structures, viewed along the 2-fold axis. The resolutions of the structures are 11, 11, 11, and 12 Å, respectively. The AAV capsid densities are purple (A, B) and dark gray (C, D), and the Fab fragment densities are light gray.
The 3D cryo-reconstructed AAV5-Fab fragment complex structures.
The AAV5-ADK5a complex structure was determined to a resolution of ∼11 Å (Fig. 2C). The Fab fragment densities were located around the 5-fold axis, in the canyon close to the 2/5-fold wall with the 3-fold protrusions exposed. The AAV5-ADK5b complex structure was determined to a resolution of ∼12 Å (Fig. 2D). Interestingly, the ADK5b binding site was very similar to that of ADK5a with the Fab fragment density also located around the 5-fold axis, occluding residues on the 2/5-fold wall, but had a footprint that extended to the outside wall of the 3-fold protrusion facing the 5-fold axis (Fig. 2D). For both of the AAV5 Fab fragment structures, the antibody density projects radially outward from the capsid surface but adopts slightly different orientations (Fig. 2C and D). Neither Fab fragment structure made contact with amino acids on the side of the 3-fold protrusions facing the 3-fold axis (Fig. 2C and D).
Pseudoatomic models of the AAV-ADK1a/1b complexes predict potential neutralizing epitopes.
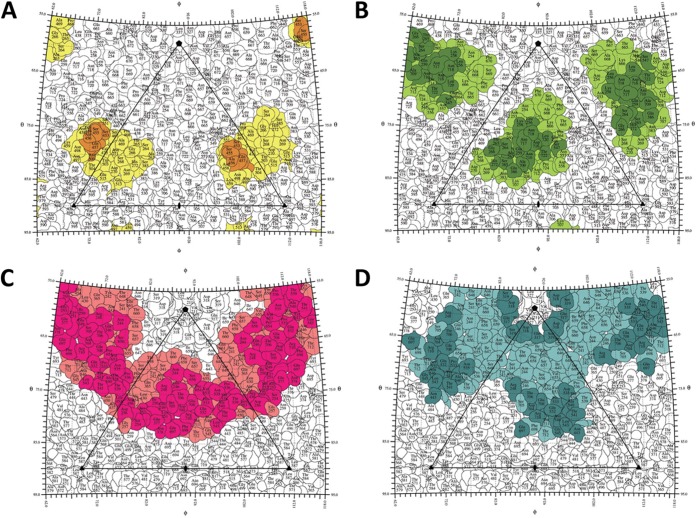
The pseudoatomic models of the AAV1-ADK1a and AAV-ADK1b complex structures were constructed by fitting the atomic models of AAV1 (residues 217 to 736 ordered in the crystal structure [50; unpublished data]) and a generic Fab fragment (62) into their respective cryo-reconstructed density maps (Fig. 3A to D). Analysis of the AAV1-Fab fragment interfaces, using the pseudoatomic models in PDBePISA, enabled the prediction of the potential epitopes for ADK1a and ADK1b on the AAV1 capsid surface (Table 1; Fig. 4A, orange, and B, dark green). In the AAV1-ADK1a complex, residues 448, 450, and 453 to 457 (AAV1 VP1 numbering) from the reference VP monomer and residue 500 from a 3-fold symmetry-related monomer were predicted to contact the Fab fragment molecule. Since the ADK1a Fab fragment binds on the tip of the 3-fold protrusion, the size of the contact area, which is the BSA between the interfacing atoms in the AAV1-ADK1a complex, was relatively small at ∼260 Å2. By visual inspection of the capsid-Fab fragment complex density map using the COOT and PyMol programs, the list of capsid surface residues occluded by the Fab fragment was more extensive, and the footprint increased to ∼450 Å2 (Table 1; Fig. 4A, orange and yellow, and 5). The Fab fragment binding site predicted for ADK1b was broader than that predicted for the ADK1a, with a contact area of ∼490 Å2. The predicted epitope for ADK1b contains residues 256, 258, 259, 261, 263 to 266, 272, 385, and 586 from the reference VP monomer and 547, 709, 710, 716 to 718, 720, and 722 from the 5-fold symmetry-related neighboring monomer (Table 1; Fig. 4B, dark green). Again, the Fab fragment footprint was more extensive, at ∼560 Å2. Residues within the footprint overlap for both Fab fragments (Table 1; Fig. 4A and B and 6A and B).
FIG 3.
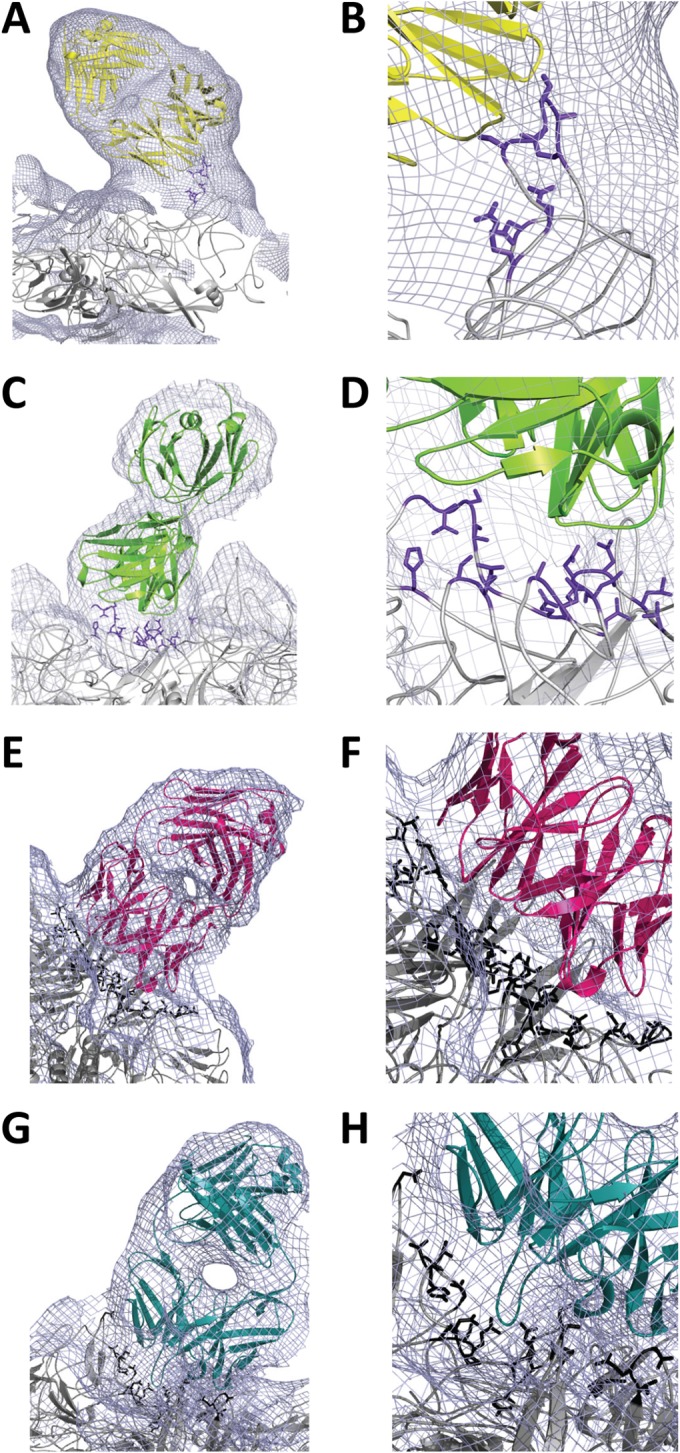
Pseudoatomic models of AAV-Fab fragment complex structures. Electron densities are shown as a gray mesh for the AAV1-ADK1a (A, B), AAV1-ADK1b (C, D), AAV5-ADKa (E, F), and AAV5-ADK5b (G, H) complex structures. The docked Fab fragment models are yellow, green, hot pink, and teal, respectively; the capsid contact residues (interface, AAV1 in purple blue and AAV5 in black) are shown in a stick representation. Panels B, D, F, and H are close-up views of the interface in panels A, C, E, and G, respectively.
TABLE 1.
Contact site and footprint residues for ADk1a/1b and ADK5a/5b on the AAV1 and AAV5 capsids, respectivelya
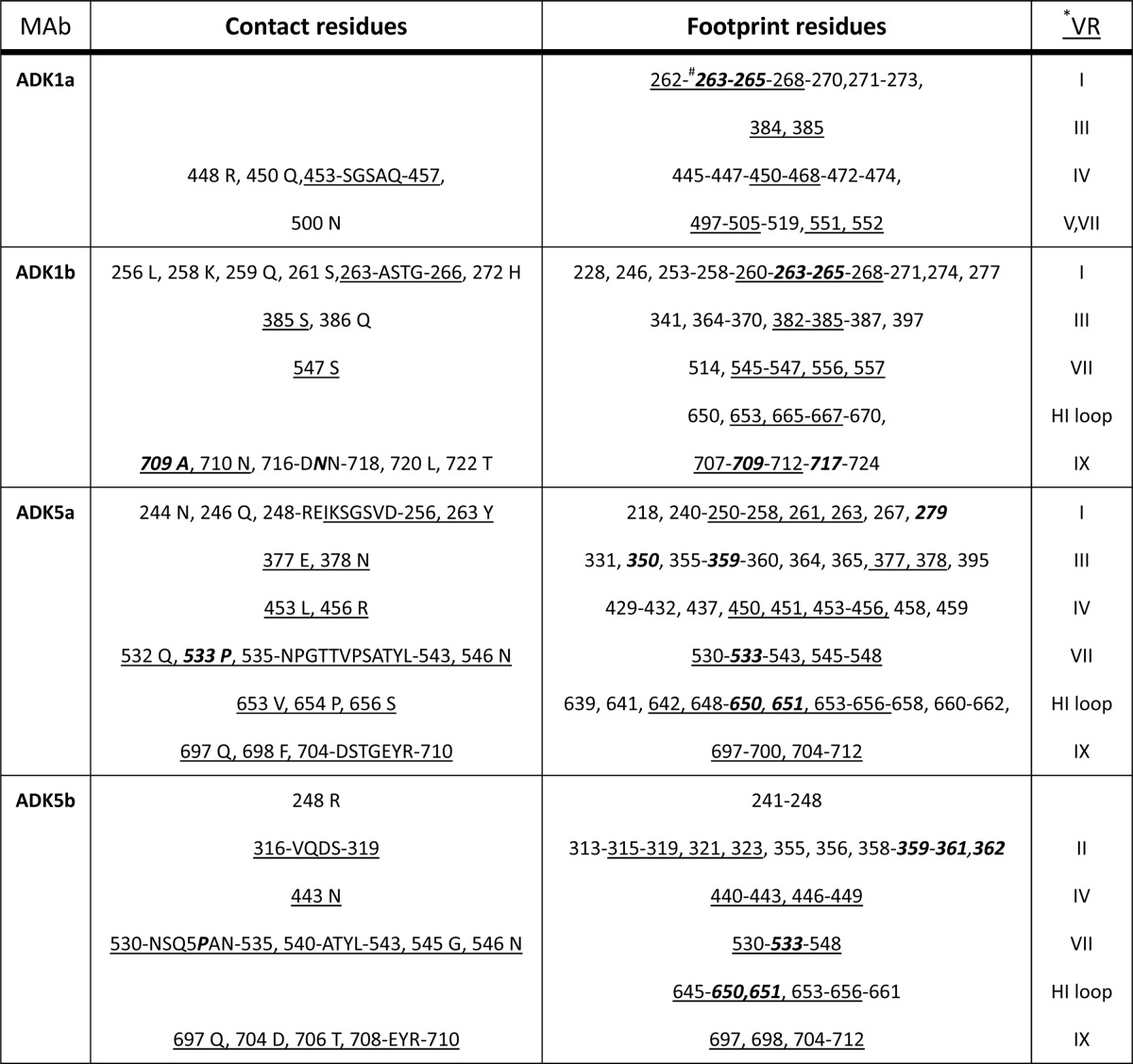
*, the residues in the VRs are underlined; #, those involved in AAV1 transduction or AAV5 receptor binding are bold and italicized.
FIG 4.
ADK antibody footprints on AAV capsids. Shown are capsid surface contact sites and footprint residues for ADK1a (A), ADK1b (B), ADK5a (C), and ADK5b (D) on their respective capsids on a stereographic roadmap projection viewed down a 2-fold axis (93). Residues are labeled. The surface area occluded by ADK1a is yellow (D) and includes the contact residues (predicted epitope) in orange. The surface area occluded by ADK1b is light green (B) and includes the contact residues (predicted epitope) in dark green. The surface area occluded by ADK5a is salmon (C) and includes the contact residues (predicted epitope) in hot pink. The surface area occluded by ADK5b is light teal (D) and includes the contact residues (predicted epitope) in teal. The boundary of each residue is black. The black triangle depicts the capsid asymmetric unit bounded by a 5-fold axis (filled pentagon) and two 3-fold axes (filled triangle) intercepted by a 2-fold axis (filled oval).
FIG 5.
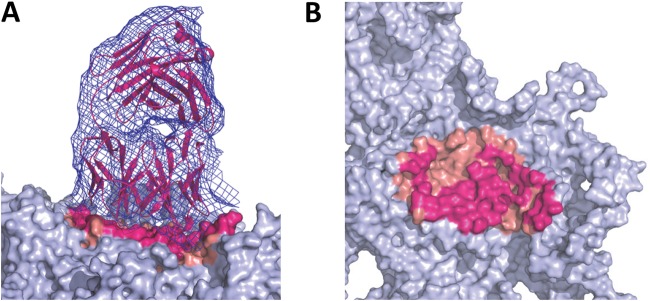
Identification of occluded residues. The AAV5-ADK5a complex structure is used to demonstrate the residue selection in the Fab fragment “occluded” footprint capsid surface area. (A) The density map of one Fab fragment (blue mesh) in the AAV5-ADK5a complex is shown on the left with the fitted Fab fragment structure (in magenta) on top of the AAV5 capsid (trimer in gray surface). The Fab fragment footprint is hot pink and salmon. The hot pink regions are the predicted contact sites (epitope), while the salmon regions are not in contact with the fitted model but form part of the Fab fragment footprint. (B) Side view of the image in panel A showing the ADK5a footprint. The Fab fragment density has been removed for clarity.
FIG 6.
A “polyclonal” footprint for AAV1. (A) The AAV1-Fab fragment cryo-reconstruction densities for the 4E4, 5H7, ADK1a, and ADK1b complexes with the capsid in light gray and the Fab fragment densities in red, blue, yellow, and green, respectively. (B) The occluded binding regions for the four anti-AAV1 antibodies 4E4, 5H7, ADK1a, and ADK1b are red, blue, yellow, and green, respectively. Regions of overlap between 4E4 and 5H7 are purple, those between 4E4 and ADK1a are orange, those between 4E4 and ADK1b are gray, and those between 5H7 and ADK1a are teal, and the regions shared by ADK1a and ADK1b are apple green. The occluded-area overlaps among 4E4, 5H7, and ADK1a are black. (C) Stereographic roadmap projection of the epitopes (contact residues) for 4E4, 5H7, ADK1a, and ADK1b, which are red, blue, yellow, and green, respectively, on the AAV1 capsid surface. The overlap regions are colored as in panel B. Residue labels and boundary definitions are as in Fig. 4.
The two predicted epitopes are within the VRs previously defined for the AAVs. For ADK1a, residues 450 and 453 to 457 are located in VR-IV (Table 1), which forms the apex of the 3-fold protrusion, and residue 500 is located in VR-V immediately adjacent to VR-IV and forms the middle of this protrusion (Table 1). The epitope for ADK1b contains residues 261 and 263 to 266 in VR-I, residue 385 in VR-III, residue 547 in VR-VII, and residues 709 and 710 in VR-IX (Table 1). These residues are located on the 2-/5-fold wall.
Pseudoatomic models of the AAV5-ADK5a/5b complexes predict potential epitopes with overlap between neutralizing and nonneutralizing interactions.
The PDB structure of AAV5 (27) and a generic Fab fragment structure (62) were fitted into the cryo-reconstructed density maps of AAV5-ADK5a and AAV5-ADK5b to build the pseudoatomic models of the complexes (Fig. 3E to H). Interface analysis in PDBePISA and interactive visual inspection in COOT and PyMol showed a large footprint for the ADK5a Fab fragment on the AAV5 capsid surface and a contact area of ∼1,000 Å2. The potential epitope for this Fab fragment includes residues 244, 246, 248, 249 to 256, 263, 377, 378, 536 to 539, 653, 654, and 656 from the reference VP monomer; 532, 533, 535, 538, 540 to 543, 546, 697, 698, and 704 to 710 from a 5-fold symmetry-related VP monomer; and 453 and 456 from a 3-fold symmetry-related VP monomer (Table 1; Fig. 4C, hot pink). There are numerous residues occluded by ADK5a binding, covering a footprint of ∼1,300 Å2 (Table 1; Fig. 4C, hot pink and salmon). The contact area for ADK5b is smaller than that for ADK5a, at ∼465 Å2, and also contained residues from three VP monomers: 319, 530 to 535, 540 to 546, 697, 704, 706, and 708 to 710 from the reference VP monomer; 248 and 316 to 318 from the 5-fold symmetry-related VP monomer; and residue 443 from the 3-fold symmetry-related VP monomer (Table 1; Fig. 4D, dark teal). The ADK5b footprint is ∼960 Å2 (Fig. 4D, dark teal and light-teal). Although ADK5b has a smaller footprint than ADK5a, the binding sites for the two Fab fragments overlap and share epitope residues, including 248, 532, 533, 535, 540, 541 to 543, 546, 697, 704, 706, 708, 709, and 710. Both Fab fragments occluded a large portion of the AAV5 capsid surface (Table 1; Fig. 4C and D and 7A and B), as observed for the previously reported 3C5 antibody (36).
FIG 7.
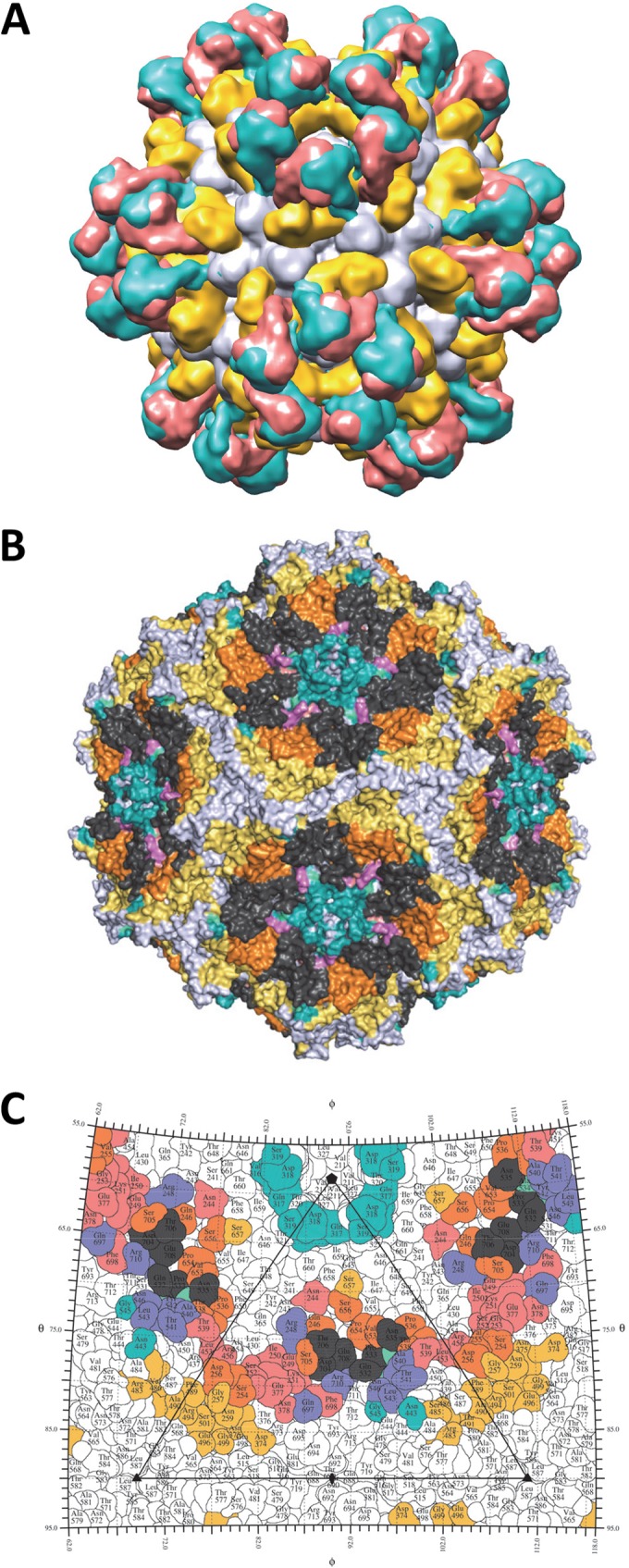
A “polyclonal” footprint for AAV5. (A) The AAV5-Fab fragment cryo-reconstruction densities for the 3C5, ADK5a, and ADK5b complexes with the capsid in light gray and the Fab fragment densities in yellow, salmon, and teal, respectively. (B) The occluded binding regions for three anti-AAV5 antibodies, 3C5, ADK5a, and ADK5b, which are yellow, salmon, and teal, respectively. Regions of overlap between 3C5 and ADK5a are orange, those between 3C5 and ADK5b are in cyan green, and those between ADK5a and ADK5b are purple. The occluded-area overlaps among 3C5, ADK5a, and ADK5a are dark gray. (C) Stereographic roadmap projection of the epitopes (contact residues) for 3C5, ADK5a, and ADK5b, which are yellow, salmon, and teal, respectively, on the AAV5 capsid surface. The overlap regions are colored as in panel B. Residue labels and boundary definitions are as in Fig. 4.
As for the AAV1-ADK1 complex structures, the predicted ADK5a and ADK5b epitopes contain residues in the AAV VRs (Table 1). For ADK5a, contact site residues 250 to 263 are in VR-I; 377 and 378 are in VR-III; 453 and 456 are in VR-IV; 532, 533, 535 to 543, and 546 are in VR-VII; and 697, 698, and 704 to 710 are in VR-IX. The epitope also includes residues 653, 654, and 656 located in the HI loop, which is on the canyon around the 5-fold axis. The epitope for ADK5b contains residues 316 to 319 in VR-II on the DE loop, which form the channel at the 5-fold axis; 443 in VR-IV; 530 to 535, 540 to 543, 545, and 546 in VR-VII; and 697, 704, 706, and 708 to 710 in VR-IX. Compared to the contact sites for ADK1a, which is on the peak of the 3-fold protrusion, both ADK5a and ADK5b bind in the 2/5-fold wall region, which is constituted by VR-I, VR-III, VR-IX, and part of VR-VII, similar to the binding site of ADK1b (Table 1).
DISCUSSION
An understanding of antigenic structure, especially information on capsid epitopes for both neutralizing and nonneutralizing antibodies, is important for the development of gene vector delivery systems. Toward this end, it is important that a complete “picture” of polyclonal reactivity be obtained since this represents the response of humans exposed to viruses. For the AAVs, previous studies have begun the task of accumulating this information by utilizing several approaches, including cryo-reconstruction, to map Fab fragment footprints onto capsid surfaces (44). However, only one or two capsid-Fab fragment structures are available for each AAV serotype studied. Here we expand this knowledge for AAV1 and AAV5, two promising therapeutic gene delivery vectors, with the goal of providing a more comprehensive view of their polyclonal antibody reactivity.
The mapped ADK1a and ADK1b epitopes, along with those for 4E4 and 5H7 (36), double the number of MAb footprints characterized for AAV1 to four (Fig. 6A to C). The mapped ADK1a epitope, at the 3-fold protrusion, overlaps the epitope for 4E4 at residues 456 and 457 (Table 2; Fig. 6C, orange). The 4E4 epitope overlaps residues 494 and 496 to 498 of the epitope for 5H7 (Table 2; Fig. 6C, purple). Thus, the epitopes for 4E4, 5H7, and ADK1a are tightly clustered (Fig. 6C, 4E4 [red], 5H7 [blue], ADK1a [yellow], common in 4E4 and ADK1a [orange], and common in 4E4 and 5H7 [purple]). The epitope for ADK1b, which binds on the 2/5-fold wall, does not overlap the predicted 4E4, 5H7, and ADK1a epitope residues; its occluded footprint overlaps 4E4 and ADK1a (Tables 1 and 3; Fig. 6B, dark gray and apple green, respectively).
TABLE 2.
Commonality in anti-AAV antibody epitopesa
| Amino acid | AAV1 |
AAV2 |
AAV5 |
AAV8 ADK8 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 4E4 | 5H7 | ADK1a | ADK1b | A20 | C37B | 3C5 | ADK5a | ADK5b | ||
| 458 | N | T | ||||||||
| 493 | K | S | ||||||||
| 494 | T | T | A | |||||||
| 495 | D | D | ||||||||
| 496 | N | N | N | |||||||
| 497 | N | N | N | R | ||||||
| 498 | N | N | N | A | ||||||
| 499 | S | S | V | |||||||
| 500 | N | S | ||||||||
| 588 | S | S | N | |||||||
| 589 | T | R | T | |||||||
| 254 | N | N | ||||||||
| 256 | L | Q | Q | |||||||
| 258 | K | K | R | R | ||||||
| 259 | Q | E | ||||||||
| 261 | S | S | K | |||||||
| 262 | S | S | ||||||||
| 263 | A | G | ||||||||
| 264 | S | S | ||||||||
| 268 | G | G | ||||||||
| 385 | S | S | E | |||||||
| 386 | Q | Q | N | |||||||
| 546 | Q | Q | ||||||||
| 547 | S | P | P | P | ||||||
| 548 | N | N | N | |||||||
| 549 | E | G | G | G | ||||||
| 550 | G | T | T | |||||||
| 551 | T | T | ||||||||
| 552 | T | T | ||||||||
| 553 | Y | Y | ||||||||
| 554 | L | L | ||||||||
| 557 | K | N | N | |||||||
| 665 | V | V | ||||||||
| 666 | P | P | ||||||||
| 709 | A | V | Q | Q | Q | |||||
| 710 | N | F | ||||||||
| 716 | D | D | D | D | ||||||
| 717 | N | S | S | |||||||
| 718 | N | N | T | T | T | |||||
| 719 | G | G | ||||||||
| 720 | L | E | ||||||||
| 721 | Y | Y | ||||||||
| 722 | T | R | R | |||||||
Epitope residue numbers are based on AAV1 VP1; serotype-specific epitope residue types are shown.
TABLE 3.
Contact site and footprint residues for 4E4 and 5H7 on the AAV1 capsid and 3C5 on the AAV5 capsida
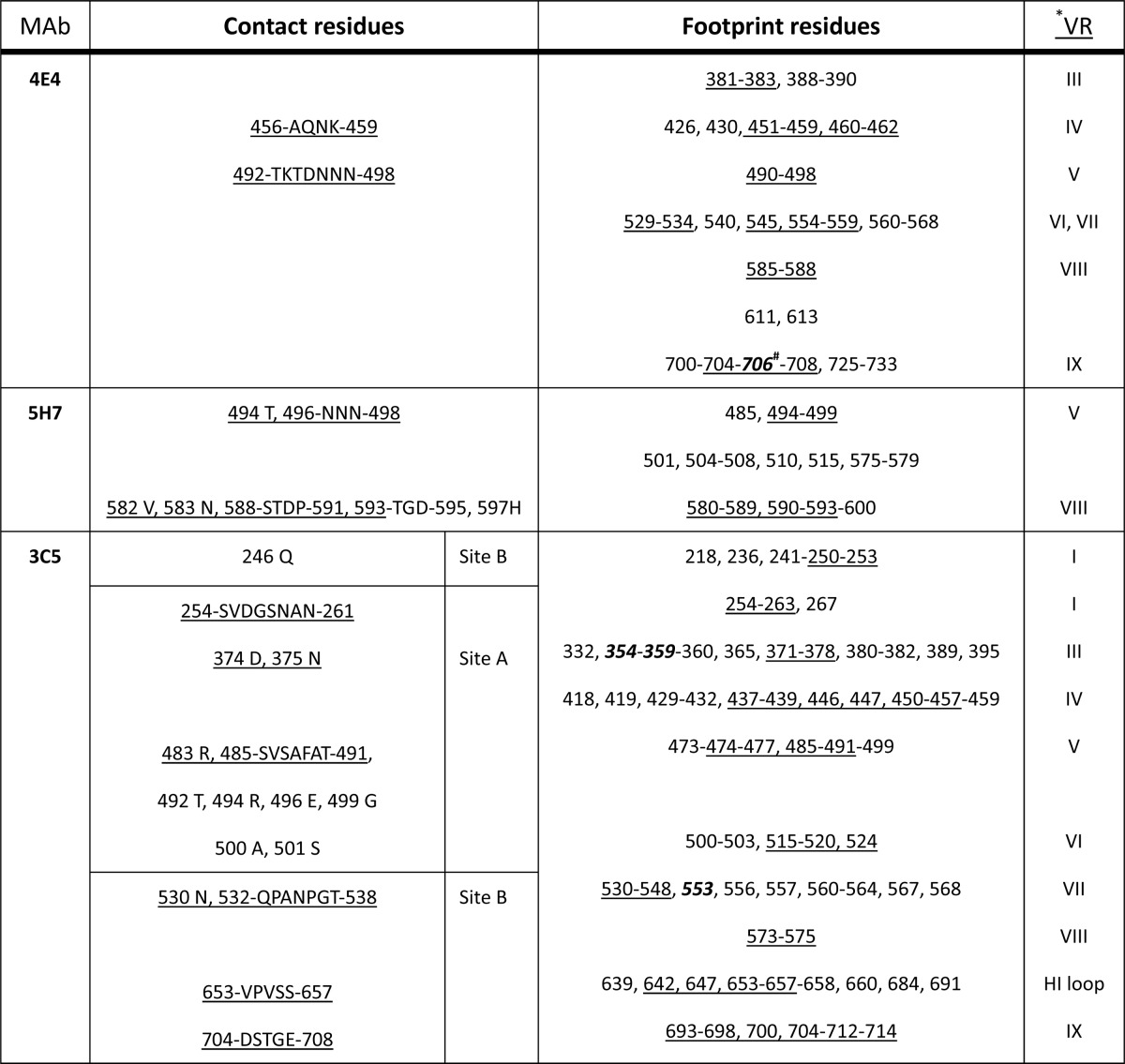
*, the residues in the VRs are underlined; #, bold and italicized numbers are residues involved in AAV1 transduction or AAV5 receptor binding.
The ADK1a and ADK1b antibodies both neutralize AAV1 infection in vitro. However, the neutralization mechanisms for these antibodies are not known. The overlap of the ADK1a epitope with those for 4E4 and 5H7 (Tables 2 and 3) also suggests that it may act by blocking cellular receptor attachment/cell surface binding, as previously reported for these two MAbs (65). Competition for receptor attachment has also been reported as the mechanism of neutralization for other anti-AAV antibodies. For example, C37-B, a MAb against AAV2, which binds to the top of the 3-fold protrusions (36, 42), has a footprint that overlaps residues important for AAV2's recognition of its heparan sulfate proteoglycan receptor (66–70). However, a similar attachment site does not equate to a similar neutralization mechanism. For example, the ADK8 MAb against AAV8, which binds in the capsid region equivalent to C37-B (Table 2), does not neutralize receptor attachment but rather acts at a step after cellular/prenuclear entry (37).
The ADK1b binding site overlaps that of the A20 MAb, which specifically recognizes and neutralizes AAV2 at a step after nuclear entry without inhibiting cellular attachment (42). Although the exact capsid function being abrogated by A20 is unknown, overlap of its footprint with a transduction “dead zone” described for AAV2 and the negative effect of mutations of its epitope residues on tissue transduction efficiency suggest a block of an essential interaction(s) (38, 71–73). While the AAV1 infection step neutralized by ADK1b remains to be elucidated, its contact site includes residues 263, 265, 709, and 717, which have been reported to affect muscle tropism and transduction efficiency (71). Significantly, residues 263 and 265 are also included in the occluded ADK1a footprint (Table 1; Fig. 4A, yellow). These observations support a neutralization mechanism that inhibits transduction for both antibodies, although ADK1a may be acting at an entry step while ADK1b has a postentry effect.
The ADK5a and ADK5b complex structures, along with that of AAV5-3C5 (36), increased the number of mapped antigenic footprints to three for AAV5. The 3C5 Fab fragment was reported to have two sites of contact with the AAV5 capsid surface, site A close to the 3-fold protrusions and site B on the 2/5-fold wall (36). The latter site was proposed to involve the complementarity determining region (CDR), and hence the “true” footprint, while site A was proposed to be mediated by the constant region but still indicates a Fab fragment contact site. The footprints of all three antibodies are similar and occlude a significant portion of the capsid surface but do not contact residues at the 3-fold axis, i.e., the center of the 3-fold protrusions (Fig. 7A to C). All three epitopes contain overlapping residues 532, 533, 535, 704, 706, and 708 on the 2/5-fold wall (Fig. 7C, dark gray). Interestingly, in a pairwise comparison of the footprints, the 3C5 epitope shares more residues with ADK5a (246, 254, 255, 256, 536, 537, 538, 653, 654, 705, and 707) than with ADK5b (530 and 534) (Table 2; Fig. 7C, orange and purple, respectively). The residues shared in the occluded footprint by these AAV5 antibodies are more extensive (Fig. 7B; Tables 1 and 3).
All three AAV5 MAbs have overlapping epitopes, yet only ADK5b is neutralizing. It has been shown that AAV5 can utilize sialic acid (SIA) as a receptor to infect cells (74–76), and two SIA binding sites, A and B, have been described (77). The A site is at the center of the 3-fold axis and includes residues 569, 570, 571, and 583 to 587; the B site is located under the HI loop close to the 5-fold axis and includes residues 277, 279, 280, 350 to 354, 359, 361 to 363, 533, and 650 to 652. There is no overlap between the AAV5 antigenic epitopes, which have no residues at the center of the 3-fold axis, and the A site. However, AAV5 residues in the B site affecting transduction in an SIA-independent manner overlap residues occluded by the ADK5a, ADK5b, and 3C5 footprints. All three footprints occlude residues 359 and 533, and the ADK5a and ADK5b footprints occlude residues 650 and 651, while ADK5a alone occludes 279 and 350 and ADK5b alone occludes residues 361 and 362. Since ADK5a/3C5 and ADK5b are nonneutralizing and neutralizing, respectively, the indication is that the SIA interactions mediated by residues 361 and 362 are prevented by ADK5b in its neutralization mechanism. Since this is not the primary SIA binding interaction for AAV5, it is possible that interaction with a sialylated coreceptor, such as the platelet-derived growth factor receptor (78, 79), is competed for by ADK5b binding. However, it is also possible that neutralization by ADK5b is due to the occlusion of residues on the wall of the 3-fold protrusion facing the 5-fold axis and/or the DE loop forming the 5-fold cylinder, which are unique to the epitope for this MAb. Significantly, the SIA A site has been implicated in SIA-dependent cellular transduction (77, 80). It is possible that the binding of ADK5b to the outer wall of protrusions has an effect on this function.
The mapped AAV5 antibody epitopes show that binding footprint commonality does not equate to having a neutralizing phenotype. The predicted epitopes for nonneutralizing MAbs 3C5 (site B) and ADK5a overlap the footprints for neutralizing MAbs A20 against AAV2 and ADK1b against AAV1. Intriguingly, as already stated above, the footprints for the AAV5 antibodies overlap but also have differences. Thus, the ADK5a and 3C5 footprints define regions of the AAV5 capsid that are not essential for transduction, while the nonoverlapping residues within the ADK5b footprint delineate a capsid surface that is important for infection.
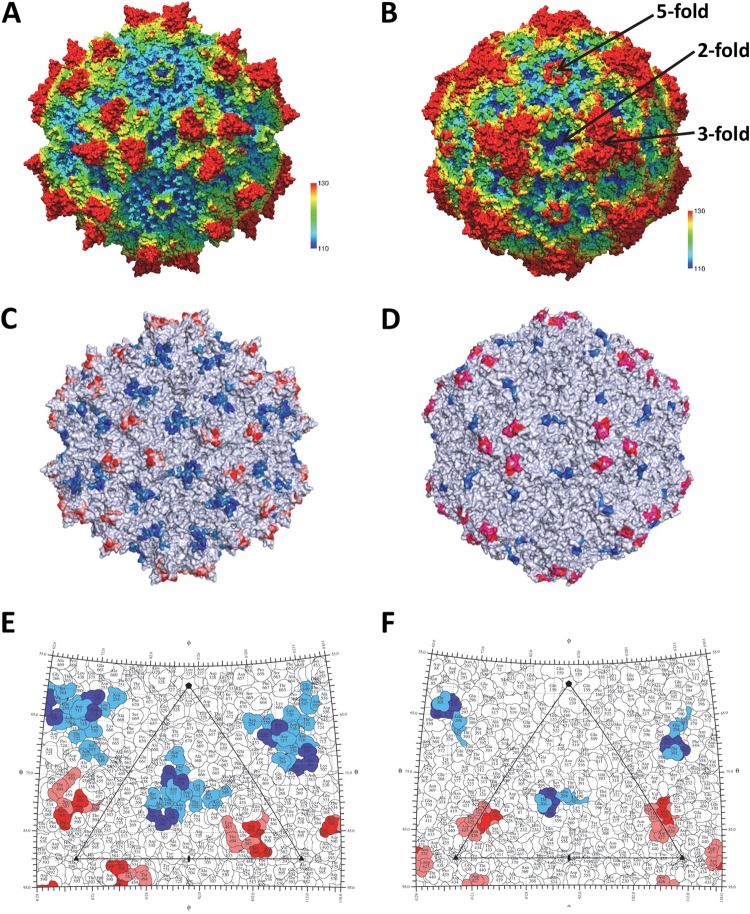
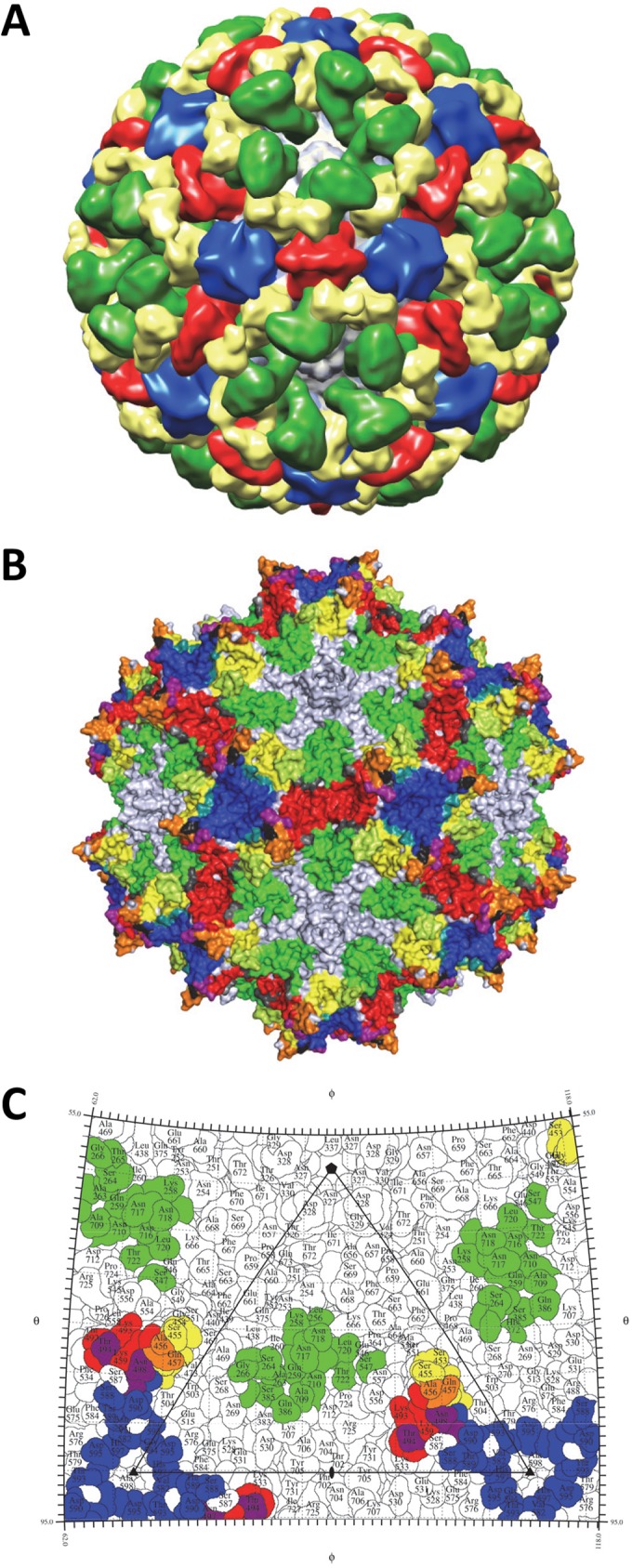
The wealth of antigenic structures now available for the AAVs, 10 (AAV1-4E4, AAV1/6-5H7, AAV1-ADK1a, AAV1-ADK1b, AAV2-A20, AAV2-C37B, AAV5-3C5, AAV5-ADK5a, AAV5-ADK5b, and AAV8-ADK8) (36, 37, 45) including the 4 reported here, facilitates the identification of the specific epitope residue positions that are most common for these viruses. The AAV-Fab fragment complex structures have been determined for viruses that span the spectrum of AAV sequence identity (∼60 to 99%) and structural similarity (∼80 to 90%) for the 13 human and nonhuman primate serotypes representing the more than 150 genomic sequences available. Thus, information on specific common epitope residues should identify the antigenic regions common to all AAVs. A superposition of the VP3 structures for the respective AAV serotypes identified the capsid residues shared by 3-fold protrusion and the 2-/5-fold wall binding MAbs (Table 2). Each epitope residue overlapped at least one from another antibody directed at the same AAV or a different serotype. This residue level comparison identified two common antigenic regions on the AAV capsid surface: one on the 3-fold protrusions and the other on the 2/5-fold wall (Fig. 8A, C, and E). In the previous epitope mapping studies that used cryo-reconstruction, 5/6 epitopes identified were localized to the 3-fold protrusions (4E4, 5H7, C37-B, 3C5 [site A], and ADK8) and only two were localized to the 2/5-fold wall (A20 and 3C5 (site B). The present study identified three additional 2/5-fold wall epitopes (ADK1a, ADK5a, and ADK5b), which enabled the confident delineation of the two common antigenic regions of the AAV capsid. This study thus defines AAV capsid surface “hot spots” for the engineering of antibody neutralization escape variants.
FIG 8.
The common parvovirus epitopes. (A and B) The AAV1 and CPV structures, respectively. The images are radially depth cued (colored from 110 to 130 Å) to visualize the capsid surface features, including the 5-fold channels, 3-fold protrusions, 2/5-fold wall, and 2-fold depressions. Panels C and E and panels D and F are capsid surface images and stereographic roadmap projections for AAV1 and CPV, respectively, showing the equivalent epitopes for the AAVs and CPV/FPV. In panels C and E, the epitope positions shared by three or four AAV serotypes are red and blue and those shared by two serotypes are pink and marine. In panels D and F, CPV/FPV epitopes identified by mutagenesis are red/blue (81) and those identified by cryo-reconstruction are red/blue and pink/marine (83).
Significantly, with the exception of the AAV1 ADK1 MAbs, which cross-react with closely related AAV6 (52), MAb 3C5 (against AAV5), which is weakly cross-reactive with AAV1 (65), and MAb A20, which cross-reacts with AAV3 (42), the AAV MAbs display mostly serotype-specific reactivity yet bind to equivalent capsid regions. In addition, the MAbs binding to similar sites abrogate different cellular functions for different serotypes, for example, receptor attachment for C37-B against AAV2 and an event that occurs after cellular/prenuclear entry for ADK8 against AAV8. These observations indicate that while commonalities in the antigenic structure of the AAVs exist, the capsid functions inhibited are serotype specific. This may be a reflection of serotype-specific interactions that differ between their multifunctional capsids.
The two common antigenic regions described for the AAVs are reminiscent of the two dominant epitopes proposed to occur on the surface of canine parvovirus (CPV) and feline panleukopenia virus (FPV), pathogenic members of the family Parvoviridae infecting dogs and cats, respectively. For these two viruses, mutagenesis, cell-based assays, and cryo-reconstruction for nine capsid-Fab fragment complexes defined two regions, the mounds at the 3-fold axis (site A) and the 2/5-fold wall (site B) (Fig. 8B, D, and F), as being antigenically dominant (81–85). These regions also play a role in host range evolution and receptor attachment. Significantly, the binding sites for different MAbs against other pathogenic autonomous parvoviruses, including Aleutian mink disease parvovirus (ADV), human parvovirus B19, and minute virus of mice (MVM), determined either by mutagenesis or cryo-reconstruction, also localize to these two capsid regions (86–89). The capsid sequences of the AAVs and these pathogenic parvovirus family members are highly divergent. For example, AAV1 shares only 20, 29, 20, 20, and 21% sequence identity, respectively, with ADV, B19, CPV, FPV, and MVM. Yet the above observations highlight a commonality in antigenic structure. The VP structures of these viruses share high similarity in topology, with 62, 50, 60, 67, and 71%, respectively, of their VP Cα positions superposable with the AAV1 VP Cα positions. The capsid surfaces of these viruses also have features in common, including depressions at the 2-fold axis and surrounding a cylindrical channel at the 5-fold axis. A difference is observed at the 2-/5-fold wall and the 3-fold axis. The AAVs, ADV, and B19 have three separate protrusions surrounding the 3-fold axis, while CPV, FPV, and MVM have a single protrusion (24, 25). As was previously stated for the AAVs, the amino acids located at the 2-/5-fold wall and the 3-fold axis are also the most divergent among these viruses. Thus, the observation of similar antigenic “hot spots” on the capsid surfaces of the AAVs and the pathogenic viruses implies that the driving force for antigenic reactivity to members of the family Parvoviridae is the structural features on the capsid surfaces and not the characteristics of epitope amino acids.
In summary, the available epitope information for different AAV serotypes has begun to delineate the boundary of their common antigenic structure. Capsid residues controlling receptor attachment and transduction efficiency are also known for several AAV serotypes (90–92). In the four antigenic structures reported, several of the predicted epitope and occluded residues (Table 1) are close to or overlap residues controlling these functions. This is consistent with the ability of these MAbs to neutralize infection. Thus, a combination of information on AAV antigenic structures and functional regions can help create a platform for the rational engineering of second-generation rAAV vectors designed to evade the preexisting immune response directed against the capsid while retaining required infectious functions.
ACKNOWLEDGMENTS
This project was funded by NIH R01 GM082946 (to M.A.-M., R.M., and N.M), R37 GM33050 (to T.S.B.), and 1S10 RR020016 (to T.S.B.), by NIH Intramural funds (to J.A.C.), by support from the University of Florida (UF) Division of Sponsored Research and the College of Medicine to establish cryo-EM facilities, the UF ICBR Electron microscopy and Hybridoma cores, and by support from the University of California—San Diego (UCSD) and the Agouron Foundation (to T.S.B.) to establish cryo-EM facilities at UCSD.
REFERENCES
- 1.Gao G, Vandenberghe LH, Alvira MR, Lu Y, Calcedo R, Zhou X, Wilson JM. 2004. Clades of adeno-associated viruses are widely disseminated in human tissues. J Virol 78:6381–6388. doi: 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark KR, Sferra TJ, Johnson PR. 1997. Recombinant adeno-associated viral vectors mediate long-term transgene expression in muscle. Hum Gene Ther 8:659–669. doi: 10.1089/hum.1997.8.6-659. [DOI] [PubMed] [Google Scholar]
- 3.Zincarelli C, Soltys S, Rengo G, Koch WJ, Rabinowitz JE. 2010. Comparative cardiac gene delivery of adeno-associated virus serotypes 1-9 reveals that AAV6 mediates the most efficient transduction in mouse heart. Clin Transl Sci 3:81–89. doi: 10.1111/j.1752-8062.2010.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nathwani AC, Tuddenham EG, Rangarajan S, Rosales C, McIntosh J, Linch DC, Chowdary P, Riddell A, Pie AJ, Harrington C, O'Beirne J, Smith K, Pasi J, Glader B, Rustagi P, Ng CY, Kay MA, Zhou J, Spence Y, Morton CL, Allay J, Coleman J, Sleep S, Cunningham JM, Srivastava D, Basner-Tschakarjan E, Mingozzi F, High KA, Gray JT, Reiss UM, Nienhuis AW, Davidoff AM. 2011. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med 365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bainbridge JW, Smith AJ, Barker SS, Robbie S, Henderson R, Balaggan K, Viswanathan A, Holder GE, Stockman A, Tyler N, Petersen-Jones S, Bhattacharya SS, Thrasher AJ, Fitzke FW, Carter BJ, Rubin GS, Moore AT, Ali RR. 2008. Effect of gene therapy on visual function in Leber's congenital amaurosis. N Engl J Med 358:2231–2239. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- 6.Maguire AM, Simonelli F, Pierce EA, Pugh EN Jr, Mingozzi F, Bennicelli J, Banfi S, Marshall KA, Testa F, Surace EM, Rossi S, Lyubarsky A, Arruda VR, Konkle B, Stone E, Sun J, Jacobs J, Dell'Osso L, Hertle R, Ma JX, Redmond TM, Zhu X, Hauck B, Zelenaia O, Shindler KS, Maguire MG, Wright JF, Volpe NJ, McDonnell JW, Auricchio A, High KA, Bennett J. 2008. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med 358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maguire AM, High KA, Auricchio A, Wright JF, Pierce EA, Testa F, Mingozzi F, Bennicelli JL, Ying GS, Rossi S, Fulton A, Marshall KA, Banfi S, Chung DC, Morgan JI, Hauck B, Zelenaia O, Zhu X, Raffini L, Coppieters F, De Baere E, Shindler KS, Volpe NJ, Surace EM, Acerra C, Lyubarsky A, Redmond TM, Stone E, Sun J, McDonnell JW, Leroy BP, Simonelli F, Bennett J. 2009. Age-dependent effects of RPE65 gene therapy for Leber's congenital amaurosis: a phase 1 dose-escalation trial. Lancet 374:1597–1605. doi: 10.1016/S0140-6736(09)61836-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. 2008. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol Ther 16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 9.Wu Z, Asokan A, Samulski RJ. 2006. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol Ther 14:316–327. doi: 10.1016/j.ymthe.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Brantly ML, Chulay JD, Wang L, Mueller C, Humphries M, Spencer LT, Rouhani F, Conlon TJ, Calcedo R, Betts MR, Spencer C, Byrne BJ, Wilson JM, Flotte TR. 2009. Sustained transgene expression despite T lymphocyte responses in a clinical trial of rAAV1-AAT gene therapy. Proc Natl Acad Sci U S A 106:16363–16368. doi: 10.1073/pnas.0904514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mingozzi F, Meulenberg JJ, Hui DJ, Basner-Tschakarjan E, Hasbrouck NC, Edmonson SA, Hutnick NA, Betts MR, Kastelein JJ, Stroes ES, High KA. 2009. AAV-1-mediated gene transfer to skeletal muscle in humans results in dose-dependent activation of capsid-specific T cells. Blood 114:2077–2086. doi: 10.1182/blood-2008-07-167510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pollack A. 20July2012. European agency backs approval of a gene therapy. New York Times, New York, NY. [Google Scholar]
- 13.Mendell JR, Rodino-Klapac LR, Rosales-Quintero X, Kota J, Coley BD, Galloway G, Craenen JM, Lewis S, Malik V, Shilling C, Byrne BJ, Conlon T, Campbell KJ, Bremer WG, Viollet L, Walker CM, Sahenk Z, Clark KR. 2009. Limb-girdle muscular dystrophy type 2D gene therapy restores alpha-sarcoglycan and associated proteins. Ann Neurol 66:290–297. doi: 10.1002/ana.21732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li C, Narkbunnam N, Samulski RJ, Asokan A, Hu G, Jacobson LJ, Manco-Johnson MJ, Monahan PE, Joint Outcome Study Investigators . 2012. Neutralizing antibodies against adeno-associated virus examined prospectively in pediatric patients with hemophilia. Gene Ther 19:288–294. doi: 10.1038/gt.2011.90. [DOI] [PubMed] [Google Scholar]
- 15.Calcedo R, Morizono H, Wang L, McCarter R, He J, Jones D, Batshaw ML, Wilson JM. 2011. Adeno-associated virus antibody profiles in newborns, children, and adolescents. Clin Vaccine Immunol 18:1586–1588. doi: 10.1128/CVI.05107-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boutin S, Monteilhet V, Veron P, Leborgne C, Benveniste O, Montus MF, Masurier C. 2010. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum Gene Ther 21:704–712. doi: 10.1089/hum.2009.182. [DOI] [PubMed] [Google Scholar]
- 17.Calcedo R, Vandenberghe LH, Gao G, Lin J, Wilson JM. 2009. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J Infect Dis 199:381–390. doi: 10.1086/595830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manno CS, Pierce GF, Arruda VR, Glader B, Ragni M, Rasko JJ, Ozelo MC, Hoots K, Blatt P, Konkle B, Dake M, Kaye R, Razavi M, Zajko A, Zehnder J, Rustagi PK, Nakai H, Chew A, Leonard D, Wright JF, Lessard RR, Sommer JM, Tigges M, Sabatino D, Luk A, Jiang H, Mingozzi F, Couto L, Ertl HC, High KA, Kay MA. 2006. Successful transduction of liver in hemophilia by AAV-factor IX and limitations imposed by the host immune response. Nat Med 12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 19.Hurlbut GD, Ziegler RJ, Nietupski JB, Foley JW, Woodworth LA, Meyers E, Bercury SD, Pande NN, Souza DW, Bree MP, Lukason MJ, Marshall J, Cheng SH, Scheule RK. 2010. Preexisting immunity and low expression in primates highlight translational challenges for liver-directed AAV8-mediated gene therapy. Mol Ther 18:1983–1994. doi: 10.1038/mt.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scallan CD, Jiang H, Liu T, Patarroyo-White S, Sommer JM, Zhou S, Couto LB, Pierce GF. 2006. Human immunoglobulin inhibits liver transduction by AAV vectors at low AAV2 neutralizing titers in SCID mice. Blood 107:1810–1817. doi: 10.1182/blood-2005-08-3229. [DOI] [PubMed] [Google Scholar]
- 21.Buller RM, Rose JA. 1978. Characterization of adenovirus-associated virus-induced polypeptides in KB cells. J Virol 25:331–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rose JA, Maizel JV Jr, Inman JK, Shatkin AJ. 1971. Structural proteins of adenovirus-associated viruses. J Virol 8:766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson FB, Ozer HL, Hoggan MD. 1971. Structural proteins of adenovirus-associated virus type 3. J Virol 8:860–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halder S, Ng R, Agbandje-McKenna M. 2012. Parvoviruses: structure and infection. Future Virol 7:253–278. doi: 10.2217/fvl.12.12. [DOI] [Google Scholar]
- 25.Chapman MS, Agbandje-McKenna M. 2006. Atomic structure of viral particles, p 107–123 InKerr JR, Cotmore SF, Bloom ME, Linden RM, Parrish CR (ed), Parvoviruses Edward Arnold Ltd., New York, NY. [Google Scholar]
- 26.Snijder J, van de Waterbeemd M, Damoc E, Denisov E, Grinfeld D, Bennett A, Agbandje-McKenna M, Makarov A, Heck AJ. 2014. Defining the stoichiometry and cargo load of viral and bacterial nanoparticles by Orbitrap mass spectrometry. J Am Chem Soc 136:7295–7299. doi: 10.1021/ja502616y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Govindasamy L, Dimattia MA, Gurda BL, Halder S, McKenna R, Chiorini JA, Muzyczka N, Zolotukhin S, Agbandje-McKenna M. 2013. Structural insights into adeno-associated virus serotype 5. J Virol 87:11187–11199. doi: 10.1128/JVI.00867-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Govindasamy L, Padron E, McKenna R, Muzyczka N, Kaludov N, Chiorini JA, Agbandje-McKenna M. 2006. Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J Virol 80:11556–11570. doi: 10.1128/JVI.01536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kronenberg S, Kleinschmidt JA, Bottcher B. 2001. Electron cryo-microscopy and image reconstruction of adeno-associated virus type 2 empty capsids. EMBO Rep 2:997–1002. doi: 10.1093/embo-reports/kve234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lerch TF, Xie Q, Chapman MS. 2010. The structure of adeno-associated virus serotype 3B (AAV-3B): insights into receptor binding and immune evasion. Virology 403:26–36. doi: 10.1016/j.virol.2010.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nam HJ, Lane MD, Padron E, Gurda B, McKenna R, Kohlbrenner E, Aslanidi G, Byrne B, Muzyczka N, Zolotukhin S, Agbandje-McKenna M. 2007. Structure of adeno-associated virus serotype 8, a gene therapy vector. J Virol 81:12260–12271. doi: 10.1128/JVI.01304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng R, Govindasamy L, Gurda BL, McKenna R, Kozyreva OG, Samulski RJ, Parent KN, Baker TS, Agbandje-McKenna M. 2010. Structural characterization of the dual glycan binding adeno-associated virus serotype 6. J Virol 84:12945–12957. doi: 10.1128/JVI.01235-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie Q, Bu W, Bhatia S, Hare J, Somasundaram T, Azzi A, Chapman MS. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc Natl Acad Sci U S A 99:10405–10410. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DiMattia MA, Nam HJ, Van Vliet K, Mitchell M, Bennett A, Gurda BL, McKenna R, Olson NH, Sinkovits RS, Potter M, Byrne BJ, Aslanidi G, Zolotukhin S, Muzyczka N, Baker TS, Agbandje-McKenna M. 2012. Structural insight into the unique properties of adeno-associated virus serotype 9. J Virol 86:6947–6958. doi: 10.1128/JVI.07232-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walters RW, Agbandje-McKenna M, Bowman VD, Moninger TO, Olson NH, Seiler M, Chiorini JA, Baker TS, Zabner J. 2004. Structure of adeno-associated virus serotype 5. J Virol 78:3361–3371. doi: 10.1128/JVI.78.7.3361-3371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gurda BL, Dimattia MA, Miller EB, Bennett A, McKenna R, Weichert WS, Nelson CD, Chen WJ, Muzyczka N, Olson NH, Sinkovits RS, Chiorini JA, Zolotutkhin S, Kozyreva OG, Samulski RJ, Baker TS, Parrish CR, Agbandje-McKenna M. 2013. Capsid antibodies to different adeno-associated virus serotypes bind common regions. J Virol 87:9111–9124. doi: 10.1128/JVI.00622-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gurda BL, Raupp C, Popa-Wagner R, Naumer M, Olson NH, Ng R, McKenna R, Baker TS, Kleinschmidt JA, Agbandje-McKenna M. 2012. Mapping a neutralizing epitope onto the capsid of adeno-associated virus serotype 8. J Virol 86:7739–7751. doi: 10.1128/JVI.00218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lochrie MA, Tatsuno GP, Christie B, McDonnell JW, Zhou S, Surosky R, Pierce GF, Colosi P. 2006. Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J Virol 80:821–834. doi: 10.1128/JVI.80.2.821-834.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raupp C, Naumer M, Muller OJ, Gurda BL, Agbandje-McKenna M, Kleinschmidt JA. 2012. The threefold protrusions of adeno-associated virus type 8 are involved in cell surface targeting as well as postattachment processing. J Virol 86:9396–9408. doi: 10.1128/JVI.00209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maheshri N, Koerber JT, Kaspar BK, Schaffer DV. 2006. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat Biotechnol 24:198–204. doi: 10.1038/nbt1182. [DOI] [PubMed] [Google Scholar]
- 41.Perabo L, Endell J, King S, Lux K, Goldnau D, Hallek M, Buning H. 2006. Combinatorial engineering of a gene therapy vector: directed evolution of adeno-associated virus. J Gene Med 8:155–162. doi: 10.1002/jgm.849. [DOI] [PubMed] [Google Scholar]
- 42.Wobus CE, Hugle-Dorr B, Girod A, Petersen G, Hallek M, Kleinschmidt JA. 2000. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J Virol 74:9281–9293. doi: 10.1128/JVI.74.19.9281-9293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huttner NA, Girod A, Perabo L, Edbauer D, Kleinschmidt JA, Buning H, Hallek M. 2003. Genetic modifications of the adeno-associated virus type 2 capsid reduce the affinity and the neutralizing effects of human serum antibodies. Gene Ther 10:2139–2147. doi: 10.1038/sj.gt.3302123. [DOI] [PubMed] [Google Scholar]
- 44.Tseng YS, Agbandje-McKenna M. 2014. Mapping the AAV capsid host antibody response toward the development of second generation gene delivery vectors. Front Immunol 5:9. doi: 10.3389/fimmu.2014.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCraw DM, O'Donnell JK, Taylor KA, Stagg SM, Chapman MS. 2012. Structure of adeno-associated virus-2 in complex with neutralizing monoclonal antibody A20. Virology 431:40–49. doi: 10.1016/j.virol.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burger C, Gorbatyuk OS, Velardo MJ, Peden CS, Williams P, Zolotukhin S, Reier PJ, Mandel RJ, Muzyczka N. 2004. Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Mol Ther 10:302–317. doi: 10.1016/j.ymthe.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 47.Zolotukhin S, Potter M, Zolotukhin I, Sakai Y, Loiler S, Fraites TJ Jr, Chiodo VA, Phillipsberg T, Muzyczka N, Hauswirth WW, Flotte TR, Byrne BJ, Snyder RO. 2002. Production and purification of serotype 1, 2, and 5 recombinant adeno-associated viral vectors. Methods 28:158–167. doi: 10.1016/S1046-2023(02)00220-7. [DOI] [PubMed] [Google Scholar]
- 48.Kaludov N, Handelman B, Chiorini JA. 2002. Scalable purification of adeno-associated virus type 2, 4, or 5 using ion-exchange chromatography. Hum Gene Ther 13:1235–1243. doi: 10.1089/104303402320139014. [DOI] [PubMed] [Google Scholar]
- 49.Smith RH, Afione SA, Kotin RM. 2002. Transposase-mediated construction of an integrated adeno-associated virus type 5 helper plasmid. Biotechniques 33:204–206, 208, 210–211. [DOI] [PubMed] [Google Scholar]
- 50.Miller EB, Gurda-Whitaker B, Govindasamy L, McKenna R, Zolotukhin S, Muzyczka N, Agbandje-McKenna M. 2006. Production, purification and preliminary X-ray crystallographic studies of adeno-associated virus serotype 1. Acta Crystallogr Sect F Struct Biol Cryst Commun 62(Pt 12):1271–1274. doi: 10.1107/S1744309106048184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DiMattia M, Govindasamy L, Levy HC, Gurda-Whitaker B, Kalina A, Kohlbrenner E, Chiorini JA, McKenna R, Muzyczka N, Zolotukhin S, Agbandje-McKenna M. 2005. Production, purification, crystallization and preliminary X-ray structural studies of adeno-associated virus serotype 5. Acta Crystallogr Sect F Struct Biol Cryst Commun 61(Pt 10):917–921. doi: 10.1107/S1744309105028514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuck D, Kern A, Kleinschmidt JA. 2007. Development of AAV serotype-specific ELISAs using novel monoclonal antibodies. J Virol Methods 140:17–24. doi: 10.1016/j.jviromet.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 53.Baker TS, Olson NH, Fuller SD. 1999. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol Mol Biol Rev 63:862–922, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan X, Dryden KA, Tang J, Baker TS. 2007. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J Struct Biol 157:211–225. doi: 10.1016/j.jsb.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yan X, Sinkovits RS, Baker TS. 2007. AUTO3DEM—an automated and high throughput program for image reconstruction of icosahedral particles. J Struct Biol 157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bowman VD, Chase ES, Franz AW, Chipman PR, Zhang X, Perry KL, Baker TS, Smith TJ. 2002. An antibody to the putative aphid recognition site on cucumber mosaic virus recognizes pentons but not hexons. J Virol 76:12250–12258. doi: 10.1128/JVI.76.23.12250-12258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang X, Walker SB, Chipman PR, Nibert ML, Baker TS. 2003. Reovirus polymerase lambda 3 localized by cryo-electron microscopy of virions at a resolution of 7.6 A. Nat Struct Biol 10:1011–1018. doi: 10.1038/nsb1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Heel M, Schatz M. 2005. Fourier shell correlation threshold criteria. J Struct Biol 151:250–262. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 59.Rees I, Langley E, Chiu W, Ludtke SJ. 2013. EMEN2: an object oriented database and electronic lab notebook. Microsc Microanal 19:1–10. doi: 10.1017/S1431927612014043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 61.Carrillo-Tripp M, Shepherd CM, Borelli IA, Venkataraman S, Lander G, Natarajan P, Johnson JE, Brooks CL III, Reddy VS. 2009. VIPERdb2: an enhanced and web API enabled relational database for structural virology. Nucleic Acids Res 37:D436–D442. doi: 10.1093/nar/gkn840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suh SW, Bhat TN, Navia MA, Cohen GH, Rao DN, Rudikoff S, Davies DR. 1986. The galactan-binding immunoglobulin Fab J539: an X-ray diffraction study at 2.6-A resolution. Proteins 1:74–80. doi: 10.1002/prot.340010112. [DOI] [PubMed] [Google Scholar]
- 63.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 65.Harbison CE, Weichert WS, Gurda BL, Chiorini JA, Agbandje-McKenna M, Parrish CR. 2012. Examining the cross-reactivity and neutralization mechanisms of a panel of mAbs against adeno-associated virus serotypes 1 and 5. J Gen Virol 93:347–355. doi: 10.1099/vir.0.035113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kern A, Schmidt K, Leder C, Muller OJ, Wobus CE, Bettinger K, Von der Lieth CW, King JA, Kleinschmidt JA. 2003. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J Virol 77:11072–11081. doi: 10.1128/JVI.77.20.11072-11081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Levy HC, Bowman VD, Govindasamy L, McKenna R, Nash K, Warrington K, Chen W, Muzyczka N, Yan X, Baker TS, Agbandje-McKenna M. 2009. Heparin binding induces conformational changes in adeno-associated virus serotype 2. J Struct Biol 165:146–156. doi: 10.1016/j.jsb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O'Donnell J, Taylor KA, Chapman MS. 2009. Adeno-associated virus-2 and its primary cellular receptor—Cryo-EM structure of a heparin complex. Virology 385:434–443. doi: 10.1016/j.virol.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Opie SR, Warrington KH Jr, Agbandje-McKenna M, Zolotukhin S, Muzyczka N. 2003. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J Virol 77:6995–7006. doi: 10.1128/JVI.77.12.6995-7006.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Summerford C, Samulski RJ. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol 72:1438–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bowles DE, McPhee SW, Li C, Gray SJ, Samulski JJ, Camp AS, Li J, Wang B, Monahan PE, Rabinowitz JE, Grieger JC, Govindasamy L, Agbandje-McKenna M, Xiao X, Samulski RJ. 2012. Phase 1 gene therapy for Duchenne muscular dystrophy using a translational optimized AAV vector. Mol Ther 20:443–455. doi: 10.1038/mt.2011.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grimm D, Lee JS, Wang L, Desai T, Akache B, Storm TA, Kay MA. 2008. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J Virol 82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salganik M, Aydemir F, Nam HJ, McKenna R, Agbandje-McKenna M, Muzyczka N. 2014. Adeno-associated virus capsid proteins may play a role in transcription and second-strand synthesis of recombinant genomes. J Virol 88:1071–1079. doi: 10.1128/JVI.02093-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kaludov N, Brown KE, Walters RW, Zabner J, Chiorini JA. 2001. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J Virol 75:6884–6893. doi: 10.1128/JVI.75.15.6884-6893.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Walters RW, Yi SM, Keshavjee S, Brown KE, Welsh MJ, Chiorini JA, Zabner J. 2001. Binding of adeno-associated virus type 5 to 2,3-linked sialic acid is required for gene transfer. J Biol Chem 276:20610–20616. doi: 10.1074/jbc.M101559200. [DOI] [PubMed] [Google Scholar]
- 76.Seiler MP, Miller AD, Zabner J, Halbert CL. 2006. Adeno-associated virus types 5 and 6 use distinct receptors for cell entry. Hum Gene Ther 17:10–19. doi: 10.1089/hum.2006.17.10. [DOI] [PubMed] [Google Scholar]
- 77.Afione S, DiMattia M, Halder S, Di Pasquale G, Agbandje-McKenna M, Chiorini JA. 19November2014. Identification and mutagenesis of the AAV5 sialic acid binding region. J Virol doi: 10.1128/JVI.02503-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Di Pasquale G, Davidson BL, Stein CS, Martins I, Scudiero D, Monks A, Chiorini JA. 2003. Identification of PDGFR as a receptor for AAV-5 transduction. Nat Med 9:1306–1312. doi: 10.1038/nm929. [DOI] [PubMed] [Google Scholar]
- 79.Pilz IH, Di Pasquale G, Rzadzinska A, Leppla SH, Chiorini JA. 2012. Mutation in the platelet-derived growth factor receptor alpha inhibits adeno-associated virus type 5 transduction. Virology 428:58–63. doi: 10.1016/j.virol.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Excoffon KJ, Koerber JT, Dickey DD, Murtha M, Keshavjee S, Kaspar BK, Zabner J, Schaffer DV. 2009. Directed evolution of adeno-associated virus to an infectious respiratory virus. Proc Natl Acad Sci U S A 106:3865–3870. doi: 10.1073/pnas.0813365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Strassheim ML, Gruenberg A, Veijalainen P, Sgro JY, Parrish CR. 1994. Two dominant neutralizing antigenic determinants of canine parvovirus are found on the threefold spike of the virus capsid. Virology 198:175–184. doi: 10.1006/viro.1994.1020. [DOI] [PubMed] [Google Scholar]
- 82.Wikoff WR, Wang G, Parrish CR, Cheng RH, Strassheim ML, Baker TS, Rossmann MG. 1994. The structure of a neutralized virus: canine parvovirus complexed with neutralizing antibody fragment. Structure 2:595–607. doi: 10.1016/S0969-2126(00)00062-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hafenstein S, Bowman VD, Sun T, Nelson CD, Palermo LM, Chipman PR, Battisti AJ, Parrish CR, Rossmann MG. 2009. Structural comparison of different antibodies interacting with parvovirus capsids. J Virol 83:5556–5566. doi: 10.1128/JVI.02532-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hoelzer K, Shackelton LA, Parrish CR, Holmes EC. 2008. Phylogenetic analysis reveals the emergence, evolution and dispersal of carnivore parvoviruses. J Gen Virol 89:2280–2289. doi: 10.1099/vir.0.2008/002055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ohshima T, Mochizuki M. 2009. Evidence for recombination between feline panleukopenia virus and canine parvovirus type 2. J Vet Med Sci 71:403–408. doi: 10.1292/jvms.71.403. [DOI] [PubMed] [Google Scholar]
- 86.Kaufmann B, Lopez-Bueno A, Mateu MG, Chipman PR, Nelson CD, Parrish CR, Almendral JM, Rossmann MG. 2007. Minute virus of mice, a parvovirus, in complex with the Fab fragment of a neutralizing monoclonal antibody. J Virol 81:9851–9858. doi: 10.1128/JVI.00775-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chipman PR, Agbandje-McKenna M, Kajigaya S, Brown KE, Young NS, Baker TS, Rossmann MG. 1996. Cryo-electron microscopy studies of empty capsids of human parvovirus B19 complexed with its cellular receptor. Proc Natl Acad Sci U S A 93:7502–7506. doi: 10.1073/pnas.93.15.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bloom ME, Best SM, Hayes SF, Wells RD, Wolfinbarger JB, McKenna R, Agbandje-McKenna M. 2001. Identification of Aleutian mink disease parvovirus capsid sequences mediating antibody-dependent enhancement of infection, virus neutralization, and immune complex formation. J Virol 75:11116–11127. doi: 10.1128/JVI.75.22.11116-11127.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McKenna R, Olson NH, Chipman PR, Baker TS, Booth TF, Christensen J, Aasted B, Fox JM, Bloom ME, Wolfinbarger JB, Agbandje-McKenna M. 1999. Three-dimensional structure of Aleutian mink disease parvovirus: implications for disease pathogenicity. J Virol 73:6882–6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang LY, Halder S, Agbandje-McKenna M. 2014. Parvovirus glycan interactions. Curr Opin Virol 7:108–118. doi: 10.1016/j.coviro.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Halder S, Nam HJ, Govindasamy L, Vogel M, Dinsart C, Salome N, McKenna R, Agbandje-McKenna M. 2012. Production, purification, crystallization and structure determination of H-1 parvovirus. Acta Crystallogr Sect F Struct Biol Cryst Commun 68(Pt 12):1571–1576. doi: 10.1107/S1744309112045563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Drouin LM, Agbandje-McKenna M. 2013. Adeno-associated virus structural biology as a tool in vector development. Future Virol 8:1183–1199. doi: 10.2217/fvl.13.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xiao C, Rossmann MG. 2007. Interpretation of electron density with stereographic roadmap projections. J Struct Biol 158:182–187. doi: 10.1016/j.jsb.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]