ABSTRACT
Genetic robustness (tolerance of mutation) may be a naturally selected property in some viruses, because it should enhance adaptability. Robustness should be especially beneficial to viruses like HIV-1 that exhibit high mutation rates and exist in immunologically hostile environments. Surprisingly, however, the HIV-1 capsid protein (CA) exhibits extreme fragility. To determine whether fragility is a general property of HIV-1 proteins, we created a large library of random, single-amino-acid mutants in HIV-1 integrase (IN), covering >40% of amino acid positions. Despite similar degrees of sequence variation in naturally occurring IN and CA sequences, we found that HIV-1 IN was significantly more robust than CA, with random nonsilent IN mutations only half as likely to cause lethal defects. Interestingly, IN and CA were similar in that a subset of mutations with high in vitro fitness were rare in natural populations. IN mutations of this type were more likely to occur in the buried interior of the modeled HIV-1 intasome, suggesting that even very subtle fitness effects suppress variation in natural HIV-1 populations. Lethal mutations, in particular those that perturbed particle production, proteolytic processing, and particle-associated IN levels, were strikingly localized at specific IN subunit interfaces. This observation strongly suggests that binding interactions between particular IN subunits regulate proteolysis during HIV-1 virion morphogenesis. Overall, use of the IN mutant library in conjunction with structural models demonstrates the overall robustness of IN and highlights particular regions of vulnerability that may be targeted in therapeutic interventions.
IMPORTANCE The HIV-1 integrase (IN) protein is responsible for the integration of the viral genome into the host cell chromosome. To measure the capacity of IN to maintain function in the face of mutation, and to probe structure/function relationships, we created a library of random single-amino-acid IN mutations that could mimic the types of mutations that naturally occur during HIV-1 infection. Previously, we measured the robustness of HIV-1 capsid in this manner and determined that it is extremely intolerant of mutation. In contrast to CA, HIV-1 IN proved relatively robust, with far fewer mutations causing lethal defects. However, when we subsequently mapped the lethal mutations onto a model of the structure of the multisubunit IN-viral DNA complex, we found the lethal mutations that caused virus morphogenesis defects tended to be highly localized at subunit interfaces. This discovery of vulnerable regions of HIV-1 IN could inform development of novel therapeutics.
INTRODUCTION
Like many RNA viruses, HIV-1 derives its success from its evolvability. High mutation rates, large population sizes, and rapid replication all contribute to its ability to adapt to changing environmental pressures, such as immune responses, target cell availability, and therapy (1–3). Under these conditions, genetic robustness, which describes the capacity for biological entities to maintain function despite mutation, should be highly beneficial and offer a means of carrying a mutational load (3–5). Indeed, previous work with RNA viruses has provided evidence of natural selection for robustness and has suggested that under certain conditions, robustness can be favored over fitness (6–8).
These prior studies, and others which have explicitly investigated robustness in RNA viruses (9, 10), have done so primarily in the context of the full viral genome, which can mask potential variation in robustness among individual proteins or regions of the viral genome. To better understand how robustness might function at the level of individual viral proteins, we recently used a large library of random single-amino-acid mutants to investigate the robustness of the HIV-1 capsid (CA), both in vitro and in vivo. Strikingly, we found a remarkable degree of fragility (lack of robustness or sensitivity to mutation) in CA, with 70% of single-amino-acid changes resulting in replication-defective viruses in vitro. Requirements imposed during the assembly of mature virions were the predominant source of genetic fragility in CA (11). Investigation of the robustness of CA permitted examination of how competing selection pressures to maintain structure and function on the one hand and to diversify sequence in the face of immune pressure on the other can play out both in vitro and in vivo.
To determine whether the extreme genetic fragility exhibited by CA was unusual among HIV-1 proteins, we chose to evaluate the robustness of another critical HIV-1 protein that might also be expected to exhibit a high degree of fragility. HIV-1 integrase (IN), like HIV-1 CA, is an essential, highly conserved, multifunctional protein. The primary function of IN, as a component of the preintegration complex (PIC), is to mediate integration of the viral genome into host cell DNA through catalytic activities. Specifically, during 3′ processing, IN dimers bind at both of the long terminal repeats (LTRs) found at each end of the viral DNA (vDNA) and hydrolyze conserved GT dinucleotides to reveal recessed 3′ termini with exposed hydroxyl groups (12–15). In the subsequent DNA strand transfer reaction, these 3′ hydroxyls attack opposing strands of host chromosomal DNA, joining the vDNA ends to the host DNA's 5′ phosphates (16–18).
While the crystal structure for full-length HIV-1 IN has not yet been determined, much is known about the domain structures and functions of the 32-kDa, 288-amino-acid protein. An ∼49-amino-acid N-terminal domain (NTD) adopts a helix-turn-helix fold, contains an HH/CC zinc binding domain, and contributes to both IN multimerization and DNA binding (19). The central catalytic core domain (CCD), residues 50 to 212 (in some schemes, 59 to 202), is responsible for both 3′ processing and strand transfer reactions and also contributes to multimerization, vDNA binding, and host target DNA binding (20, 21). The CCD adopts an RNase H fold and contains a D,D(35)-E amino acid motif that binds divalent metal ions, specifically Mg2+ (20, 22). The C-terminal domain (CTD), which includes residues 213 to 288 (or in some schemes 223 to 270), adopts a beta barrel structure, resembling an Src homology 3 (SH3)-type fold, is involved in binding to host cell target DNA, and also contributes to multimerization (21, 23, 24). Interactions involving the three domains result in the formation of a tetramer comprised of a dimer of dimers, which, when bound to the vDNA, forms a complex known as the intasome (25). Determination of the crystal structure of the prototype foamy virus (PFV) intasome illuminated how IN domains contribute to protein multimerization, as well as to vDNA and host DNA binding (26, 27). By analogy to the PFV intasome crystal structure, new models of the HIV-1 intasome structure have been derived (28, 29).
Previous mutagenesis studies of IN have demonstrated that the various IN domains contribute to proviral integration and also to other steps in the viral life cycle, including virion assembly. Generally, IN mutants can be divided into two classes: class I mutants are those that are selectively defective for integration, while class II mutants are characterized by pleiotropic defects, including assembly and/or reverse transcription (30). Importantly, mutagenesis studies utilizing complementation have indicated the likelihood that IN forms a functional multimer (31–33). Other mutagenesis studies have also suggested a critical role for IN-binding cellular cofactors, for example, lens epithelium-derived growth factor (LEDGF) (34) and possibly transportin SR-2 (TRN-SR2, or TNPO3) (35) and integrase interactor (INI) 1/hSNF5 (36). Further work using scanning or site-directed mutagenesis has revealed individual IN amino acids that are likely to be involved in DNA binding or other critical IN functions (22, 37–44). Cumulatively, these studies have suggested that IN is comparatively sensitive to mutation. However, no previous study has analyzed a comprehensive panel of unbiased IN mutants, nor has there been any systematic measurement of IN robustness.
Here, we generated a large panel of random single-amino-acid mutants, which we used to measure the genetic robustness of HIV-1 IN. We also assessed the impact of fitness costs on IN variation in natural viral populations and reveal areas of mutational fragility using HIV-1 IN structural models. We find that IN is surprisingly robust in vitro compared to CA, with only 35% of single-amino-acid IN mutations resulting in nonviable viruses. Comparisons of in vitro fitness measurements with occurrence in natural subtype B HIV-1 populations reveal that IN mutations with fitness values of <40% of wild-type (WT) fitness rarely occur in vivo, a value that is remarkably similar to that obtained with the same analysis of CA mutations. Our data also show that certain IN subunit interfaces are particularly sensitive to mutation and are required for accurate particle assembly rather than integration. Comparison of random CA and IN mutant data sets demonstrates how differently two essential HIV-1 proteins can tolerate mutation in vitro but also reveals certain commonalities that arise in the two data sets. Understanding the tolerance of HIV-1 IN to mutation is particularly valuable given that IN is an increasingly important therapeutic target in HIV-1 infection (45).
MATERIALS AND METHODS
Plasmids and construction.
Creation of a randomly mutagenized IN library was done using approaches similar to those previously described for a random CA library (11). The proviral plasmid pNHGintBS is a derivative of the pNHGcapNM (GenBank accession number JQ686832) used for the CA library, with the following changes. The SpeI restriction enzyme site in pNHGcapNM was eliminated, and silent mutations were introduced so that the IN coding sequence was flanked by unique BstBI and SpeI sites at the 5′ and 3′ ends, respectively; this pNHGintBS construct is referred to as the parental or WT sequence. The GeneMorph II random mutagenesis kit (Agilent) was used in conjunction with the oligonucleotides designed to amplify IN coding sequences: 5′-GGA AAT GAA CAA GTA GAT GGG TTG GTC AGT GCT GGA ATT CGA AAA GTA CTA-3′ and 5′-GCT TTC CTT GAA ATA TAC ATA TGG TGT TTT ACT AGT CTT TTC CAT GTG TTA-3′. The mutagenized PCR product and the proviral plasmid pNHGintBS were digested with BstBI and SpeI and ligated. To ensure the appropriate mutagenesis frequency, approximately 285 mutagenized plasmid clones generated using GeneMorph II PCR conditions in which amounts of IN template varied, were sequenced. Subsequently, the mutant library was generated under conditions yielding the highest number of single nucleotide changes, and plasmid DNA was extracted from individual cultures derived from ∼768 colonies. Sequencing then revealed which single mutant clones were suitable for inclusion in the library. Final proviral plasmid DNA was then freshly isolated for library single mutants and subjected to analysis by restriction digest using BstBI and SpeI, at which point ∼2% of clones were removed for evidence of recombination.
Cell lines and transfection.
MT-4 suspension cells (NIH AIDS Reagent Repository) were maintained in RPMI with gentamicin and 10% fetal calf serum (FCS). Adherent 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with gentamicin and 10% FCS. For transfection experiments, 293T cells (American Type Culture Collection) were plated in 24-well plates at 1.8 × 105 cells per well and transfected the following day using polyethylenimine (Polysciences) and 500 ng of either NHGintBS (WT) or NHGintBS IN mutants described above. Approximately 12 h after transfection with the proviral plasmids, 293T cells were given fresh medium. At 40 h posttransfection, cell supernatants were harvested, filtered (0.22 μm), and arrayed in 96-well plates.
Viral replication and infectivity assays.
For single-cycle infectivity measurements, MT-4 cells were seeded in 96-well plates at 3 × 104 cells per well and inoculated with an amount of filtered supernatant corresponding to a multiplicity of infection (MOI) of ∼1 for the WT viral clone. To limit replication to a single cycle, 100 μM dextran sulfate was added 16 h later. At 48 h postinfection, cells were fixed in 4% paraformaldehyde (PFA). Alternatively, for spreading replication assays, MT-4 cells, in 96-well plates at 2 × 104 cells per well, were inoculated with a volume of filtered supernatant corresponding to an MOI of ∼0.01 for the WT viral clone. Cells were fixed in 4% PFA at ∼80 h postinfection. A Guava EasyCyte 5HT instrument was used for fluorescence-activated cell sorting (FACS) analysis for all infectivity and replication assays.
Western blotting.
Virions, which were pelleted through 20% sucrose by centrifugation, and cell lysates were resuspended in SDS sample buffer, and proteins were separated by electrophoresis on NuPage 4 to 12% Bis-Tris gels (Novex). Proteins were then blotted onto nitrocellulose membranes and probed with either anti-HIV p24 capsid antibody (183-H12-5C), anti-HIV INT-4 antibody (46) (a gift from Michael Malim), or anti-clathrin heavy chain antibody (BD Biosciences; catalog no. 610499). Blots were then probed with fluorophore-conjugated goat anti-mouse secondary antibodies (Thermo Scientific) and scanned and quantified using a LI-COR Odyssey system.
Analysis of IN and CA variants.
All HIV-1 subtype B IN and MA sequences from 1980 to 2013 were selected from the Los Alamos HIV sequence database (www.hiv.lanl.gov/). Sequences with frame shifts, stop codons, or nucleotide ambiguities (N or IUPAC code) were excluded from the data set prior to random selection of either 1,000 sequences (for IN) or 4,174 sequences (for MA), from different individuals and locations. Sequences were aligned using MUSCLE (47), and Perl scripts were used to determine the number of mutations at each site in the alignment. The frequency of each amino acid and its polarity were determined for each variable site. The CA sequences referenced in this work were used previously (11).
Structural analysis.
Examination of the HIV-1 intasome was done with structure coordinates provided by Stephen Hughes (28) and Alan Engelman (29) using MacPyMOL. UCSF Chimera was utilized for analysis of solvent-accessible surface area. Statistical significance of differences in solvent-accessible surface area values between groups of IN mutants (viable versus nonviable mutants, frequent versus never-occurring fit mutants, no variability versus >10% variability residues, reduced IN incorporation versus no reduction mutants) was determined using unpaired, parametric t tests. Analysis of possible fitness differences between clones with either 0 or 1 silent mutation was also done using unpaired, parametric t tests.
RESULTS
Comparison of variation in natural populations of IN and CA sequences.
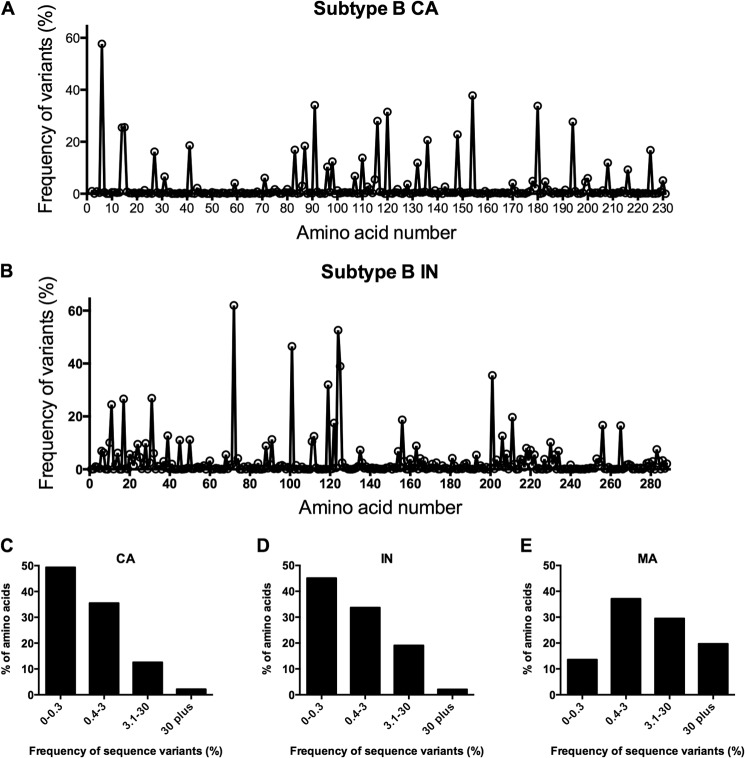
To determine whether the extreme fragility exhibited by the HIV-1 CA protein was unique to CA or shared by other HIV-1 proteins, we elected to examine a protein that might be expected to have similarly tight constraints on its sequence. We selected IN because it is a multifunctional protein with a size broadly similar to that of CA (32 kDa versus 24 kDa) and participates in several essential interactions and processes. First, we compared how frequently amino acid substitutions were present in cohorts of 1,000 naturally occurring HIV-1 subtype B CA and IN sequences. When depicted linearly, the variability among the sequences appeared similar for IN and CA (Fig. 1A and B). Comparable IN and CA variability was also evident when the frequencies of amino acids exhibiting different degrees of variability were plotted (Fig. 1C and D), and the similar degree to which IN and CA sequences are conserved was particularly apparent when contrasted with the variability found in naturally occurring subtype B matrix (MA) sequences (Fig. 1E). The similar levels of diversity in natural populations of CA and IN sequences suggested the possibility that CA and IN might be similarly tolerant of mutation.
FIG 1.
Naturally occurring variation in HIV-1 IN and CA. (A, B) Depictions of the frequency (%) of variants from WT that occur at each amino acid in 1,000 HIV-1 subtype B CA (A) or IN (B) sequences. Amino acid position is plotted on the x axis from left (N terminus) to right (C terminus). (C, D, E) A different representation of sequence variation wherein the y axis shows the percentage of amino acids within CA (C) or IN (D) or MA (E) with the indicated frequency (%) of variants on the x axis.
Construction of a library of random single-amino-acid-substitution mutants of HIV-1 IN.
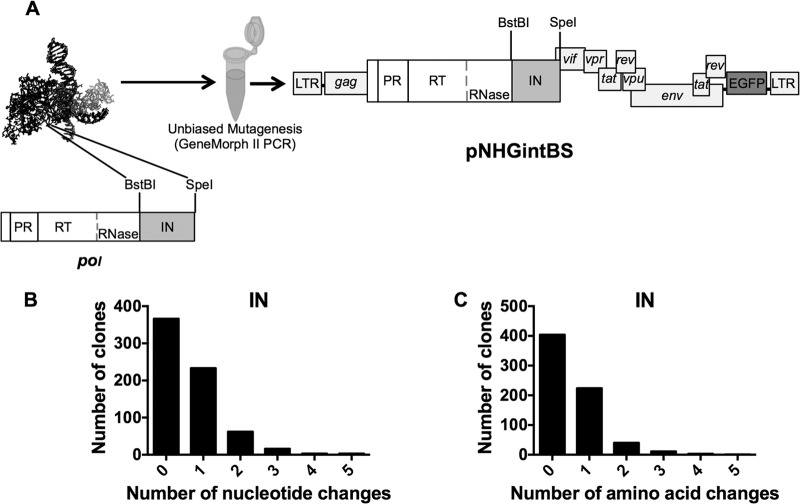
We previously constructed a library of random single-amino-acid CA mutants (11), and analogous procedures were used to design and construct a randomly mutagenized IN library (Fig. 2A). Briefly, a low-fidelity PCR method was used to generate a randomly mutagenized amplicon containing IN coding sequences (Fig. 2A). These mutagenized IN sequences were digested and inserted into a replication-competent proviral clone, pNHGintBS, here referred to as the WT, which was used as the parental clone in all experiments described herein. The pNHGintBS clone encodes enhanced green fluorescent protein (EGFP) in place of Nef, to facilitate measurements of virus replication. It is nearly identical to the pNHGcapNM (accession no. JQ686832) previously used for creation of a randomly mutagenized CA library (11) and differs only in that silent mutagenesis was used to create BstBI and SpeI sites flanking IN, into which the randomly mutagenized IN-encoding amplicon was inserted. During the IN mutant library construction, 770 single colonies were isolated, and proviral plasmid DNA was extracted and sequenced. After the removal of clones with failed sequencing reactions, chromatograms with more than one template, or evidence of recombination, 683 clones remained, and the distribution of their nucleotide changes (Fig. 2B) and amino acid substitutions (Fig. 2C) was determined. In pilot experiments, we optimized the mutagenesis rate; thus, few clones had multiple nucleotide changes, and the IN library therefore yielded a reasonable fraction of mutants with single nucleotide and single-amino-acid substitutions. From the resulting collection of clones that had single-amino-acid changes, clones with nonsense mutations, or duplicate mutations of clones already in the library, were purged. Ultimately, the random mutant IN library contained 156 single-amino-acid-substitution mutants, which covered 118 (41%) of the 288 IN residues. Mutated positions were fairly evenly distributed throughout the protein, although there was greater coverage of residues within the CCD, in part because of its size (Fig. 3C; see also Table 3). In total, 39% of the single-amino-acid IN substitutions were conservative with regard to change in polarity and hydrophobicity. In comparison, the random mutant CA library contained 135 single-amino-acid-substitution mutants, which covered 102 (44%) of the 231 CA residues and of which 46% were conservative with regard to change in polarity and hydrophobicity (11). Thus, the two mutant libraries are comparable, with a slight tendency to more conservative substitutions in the CA library.
FIG 2.
Production and characterization of the IN mutant library. (A) Schematic illustrating the PCR-based mutagenesis of the 864-bp HIV-1 IN coding sequence flanked by BstBI and SpeI restriction sites in the replication-competent proviral plasmid pNHGintBS. (B) Distribution of the number of nucleotide changes found in each clone in the IN library. Duplicate sequences, those that were represented more than once, are included. (C) Distribution of the number of amino acid changes found in each clone in the IN library. Clones containing duplicate mutations, nonsense mutations, or frameshift mutations are included.
FIG 3.
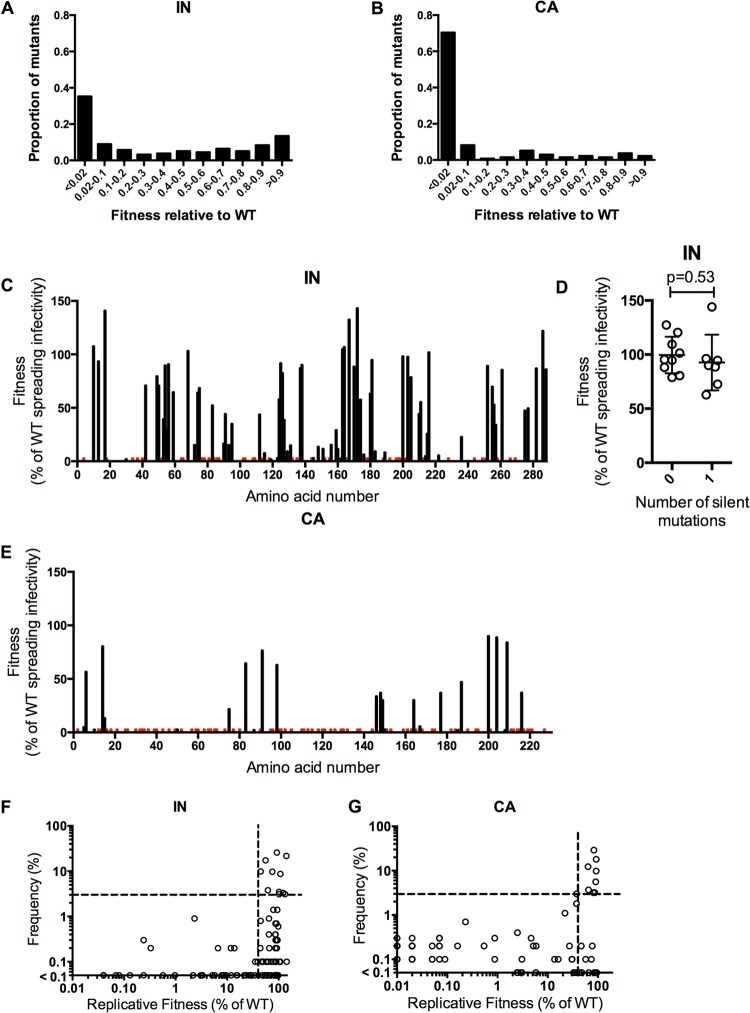
Fitness of randomly introduced and naturally occurring IN and CA mutations. (A, B) Distribution of mutational fitness effects (DMFE) for the single-amino-acid IN (A) and CA (B) mutant libraries. (C, E) Plots of fitness measurements for individual IN mutants (C) and CA mutants (E). Fitness, as a percentage of WT in a replicative fitness assay, is plotted against the position of mutation organized on the x axis from left (N-terminal amino acid) to right (C-terminal amino acid). For amino acids with more than one unique mutation, the smallest fitness value is plotted. Red bars indicate the locations of nonviable mutants (<2% of WT fitness), while gaps indicate residues for which no mutant was present in the library. For IN mutants in panel C, this is a graphical representation of the results found in Tables 1 and 2. (D) Fitness measurements for IN library clones containing either 0 (WT, NHGintBS) or 1 noncoding nucleotide change. (F, G) Plots of the frequencies with which IN (F) and CA (G) mutations that were present in the random mutant libraries occur in 1,000 HIV-1 subtype B CA and IN sequences (y axis) versus the measured replicative fitness of viruses carrying the same mutations (x axis). A horizontal dashed line indicates a frequency of 3%, above which mutations were considered to occur frequently. A vertical dashed line indicates 40% of WT replicative fitness, below which mutants occurred rarely in natural populations (frequency of <1%). Mutants with fitness below 0.01% of WT fitness, and that were not observed in nature, would appear on the origin of the graph and are not plotted.
TABLE 3.
IN mutant phenotypes by domain
| Region | No. of viable mutants | No. of nonviable mutants | Fragility (%)a | No. (%) of mutants resulting in reduced particle yieldb | No. (%) of mutants resulting in reduced IN levels in virionsc | No. (%) of mutants affecting clathrin particle incorporationd | No. (%) of nonviable mutants affected only by loss of infectiousnesse |
|---|---|---|---|---|---|---|---|
| NTD | 8 | 5 | 38 | 0 (0) | 6 (46) | 2 (15) | 0 (0) |
| CCD | 69 | 42 | 38 | 9 (8) | 25 (23) | 11 (10) | 25 (23) |
| CTD | 24 | 8 | 25 | 2 (6) | 3 (9) | 2 (6) | 5 (16) |
| IN total | 101 | 55 | 35 | 11 (7) | 34 (22) | 15 (10) | 30 (19) |
The percentage of mutants considered nonviable (<2% of WT replicative fitness).
Mutants with at least 2-fold reductions in particle production, as measured by levels of p24 in virions.
Mutants with at least 2-fold reductions in virion IN levels.
Mutants with at least 2-fold reductions in clathrin incorporation into virions.
All nonviable mutants with less than 2-fold reductions in both virion-associated IN and p24 levels.
Fitness measurements of IN mutants and comparison of the fitness effects of random CA and IN mutations.
To measure the genetic robustness of IN, we analyzed the fitness of the 156 random single-amino-acid mutants using methods identical to those previously used for the analysis of a random CA mutant library. Specifically, virus-containing supernatant harvested from transfected 293T cells was added to MT-4 cells (at a volume equivalent to a multiplicity of infection [MOI] of 1 for the WT virus), and single-cycle infectivity was measured. Alternatively, a volume equivalent to an MOI of 0.01 for the WT virus was added to MT-4 cells, and multiple rounds of replication were permitted until the WT virus had infected >50% of cells (∼80 h later). Fitness was determined using the number of infected (EGFP-positive) cells, measured by FACS and expressed as a percentage of the number of cells infected by the WT virus. As before in our analysis of a CA mutant library (11), an arbitrary cutoff was set at 2% of parental virus fitness; mutants with less than 2% of WT fitness in the spreading replication assay were considered nonviable.
Given the similarity in variability exhibited by natural populations of CA and IN sequences, it was surprising that only 35% of single-amino-acid IN substitutions resulted in nonviable virus, whereas twice as many, 70%, of single-amino-acid CA substitutions were lethal (11). The 101 viable IN mutations, with replicative fitness >2% of WT fitness, are listed in Table 1, while the 55 nonviable mutants are listed in Table 2. As was true for CA (11), comparison of Tables 1 and 2 demonstrates that different mutations at the same codon in IN could sometimes result in very different fitness outcomes, indicating that the type of amino acid change at some positions can be important. More frequently, however, different amino acid substitutions at the same position resulted in similar fitness outcomes (Tables 1 and 2). Analysis of fitness effects of the type of amino acid substitution with regard to specific charge, hydrophobicity, or molecular weight revealed no consistently significant trends with the exception that conservative substitutions (no polarity of hydrophobicity changes) typically resulted in higher fitness (mean fitness = 50% of WT fitness) than nonconservative mutations (mean fitness = 30% of WT fitness). Additionally, polar basic amino acids were typically more sensitive to hydrophobicity change than were other types of amino acids (see, for example, the effects of mutations at amino acids 34, 42, 78, 103, 166, 188, 228, 244, 258, 266, 269), but this was not always the case (amino acids 127, 156, 159, 160, 215, 236, 244) (Tables 1 and 2). The values in Tables 1 and 2 also provide an opportunity to make more general observations about the robustness of the three IN domains. Specifically, as is indicated in Table 3, the IN CTD was notably more tolerant of mutation than either the NTD or CCD.
TABLE 1.
Fitness of viable HIV-1 IN mutantsd
| IN mutation | Single cycle infectivity (% of WT)a | Replication (% of WT)b | Frequency (%) in subtype B isolatesc |
|---|---|---|---|
| E10D | 91.5 | 107.9 | 8.7 |
| E13V | 78.4 | 93.9 | 0 |
| S17N | 91.8 | 141.2 | 21.7 |
| P30H | 9.5 | 3.9 | 0 |
| P30S | 11.3 | 2.3 | 0.9 |
| K42I | 71.8 | 71.2 | 0 |
| K42R | 77.6 | 87.7 | 0.1 |
| A49V | 77.4 | 79.9 | 0 |
| M50I | 89.3 | 75.2 | 9.7 |
| M50L | 100.2 | 86.2 | 0.4 |
| M50V | 80.6 | 71.1 | 0.1 |
| Q53L | 63.9 | 39.7 | 0 |
| V54A | 87.0 | 89.9 | 0.1 |
| D55E | 45.4 | 30.3 | 0 |
| D55N | 79.9 | 65.7 | 0.2 |
| C56Y | 108.6 | 91.1 | 0.3 |
| G59E | 95.2 | 65.0 | 0.9 |
| C65S | 80.7 | 60.6 | 0 |
| L68S | 94.2 | 103.6 | 0.1 |
| V72D | 35.9 | 15.6 | 0 |
| L74 M | 73.6 | 64.5 | 0.4 |
| V75I | 99.7 | 73.4 | 0 |
| V75L | 90.1 | 68.8 | 0 |
| V77I | 82.5 | 56.8 | 0.1 |
| Y83H | 77.9 | 52.5 | 0 |
| P90L | 38.2 | 16.9 | 0 |
| P90Q | 73.8 | 66.4 | 0 |
| A91T | 73.3 | 44.6 | 9.9 |
| A91V | 81.6 | 46.5 | 0.1 |
| T93I | 19.5 | 15.5 | 0 |
| Q95L | 42.0 | 35.3 | 0 |
| L102F | 18.2 | 13.3 | 0 |
| T112I | 92.9 | 61.8 | 3.8 |
| T112R | 66.6 | 44.1 | 0.2 |
| V113L | 105.0 | 86.9 | 0.4 |
| T115I | 18.1 | 7.9 | 0 |
| S119N | 5.5 | 2.2 | 0 |
| S123N | 54.6 | 40.4 | 0 |
| S123T | 13.5 | 3.3 | 0 |
| T124I | 74.8 | 58.1 | 0.1 |
| T124S | 110.1 | 94.2 | 1.4 |
| T125A | 119.8 | 92.0 | 25.7 |
| V126F | 86.9 | 82.9 | 0 |
| K127T | 58.8 | 39.1 | 0.1 |
| A129S | 36.1 | 28.6 | 0 |
| A129T | 43.6 | 35.0 | 0.1 |
| A129V | 20.3 | 9.6 | 0 |
| W131C | 24.6 | 15.2 | 0 |
| Q137H | 97.9 | 87.4 | 0.3 |
| E138D | 103.7 | 90.4 | 0.7 |
| P145S | 19.6 | 3.3 | 0 |
| Q148R | 33.0 | 13.8 | 0.2 |
| I151T | 32.4 | 12.0 | 0 |
| K156T | 36.0 | 15.7 | 0 |
| K159 M | 44.7 | 29.5 | 0 |
| K160E | 26.5 | 11.8 | 0.2 |
| I162T | 6.0 | 3.1 | 0 |
| G163R | 117.1 | 105.1 | 0.1 |
| Q164H | 65.1 | 107.2 | 0.1 |
| D167E | 92.6 | 132.9 | 3.1 |
| E170K | 64.9 | 88.8 | 0.3 |
| L172F | 91.0 | 143.4 | 0.1 |
| T174I | 44.3 | 57.9 | 0 |
| V176I | 9.5 | 6.6 | 0.2 |
| V180I | 51.6 | 63.6 | 0 |
| F181Y | 68.3 | 95.1 | 0.3 |
| H183Q | 11.8 | 9.6 | 0 |
| G189E | 24.6 | 8.6 | 0 |
| G197W | 13.2 | 8.4 | 0 |
| E198Q | 89.8 | 91.5 | 0 |
| I200 M | 85.5 | 98.5 | 0.6 |
| I203L | 86.5 | 98.0 | 0 |
| I203 M | 101.5 | 101.5 | 3.5 |
| I204V | 82.6 | 87.0 | 0.7 |
| A205S | 84.3 | 79.0 | 1.4 |
| T210I | 45.3 | 44.6 | 0.8 |
| K211R | 63.0 | 55.7 | 17.4 |
| Q214E | 79.1 | 79.1 | 0.1 |
| Q214H | 79.0 | 54.8 | 0 |
| Q214L | 13.5 | 5.2 | 0 |
| K215R | 86.4 | 86.3 | 0.1 |
| K215T | 54.7 | 25.9 | 0 |
| Q216H | 83.6 | 102.1 | 3.1 |
| N222I | 14.8 | 5.8 | 0 |
| N222Y | 85.2 | 93.8 | 0 |
| K236I | 65.1 | 23.0 | 0 |
| K244N | 39.7 | 21.4 | 0 |
| I251V | 74.1 | 92.0 | 0.2 |
| Q252H | 77.4 | 89.6 | 0 |
| S255I | 87.1 | 70.1 | 0 |
| D256V | 98.5 | 53.3 | 0 |
| I257L | 77.0 | 65.3 | 0 |
| I257 M | 66.3 | 34.4 | 0 |
| I257V | 79.0 | 48.7 | 0.1 |
| K258R | 88.4 | 62.6 | 0.1 |
| P261S | 102.0 | 85.8 | 0 |
| M275L | 65.0 | 47.7 | 0 |
| G277S | 67.3 | 49.8 | 0.1 |
| A282T | 99.2 | 87.2 | 0 |
| D286N | 101.1 | 122.4 | 3.3 |
| D288G | 107.8 | 86.4 | 0.2 |
Fitness measurement in which MT-4 cells were inoculated, at MOI of 1 for the WT (NHGintBS) virus, with virus-containing supernatant from 293T cells transfected with single-residue IN mutant proviral plasmids. At 16 h postinfection, dextran sulfate was added to limit replication to a single cycle. Values shown are the percentages of infected cells (GFP+) compared to WT (NHGintBS).
Measurement of infectivity in which MT-4 cells were inoculated, at MOI of 0.01 for the WT (NHGintBS) virus, with virus-containing supernatant from 293T cells transfected with single-residue IN mutant proviral plasmids. Multiple replication cycles were allowed over an 80-h period. Values shown are the percentages of infected cells (GFP+) compared to WT (NHGintBS).
Frequency with which the indicated mutant residue occurs in 1,000 HIV-1 subtype B IN sequences.
Viable, or infectious, mutants were those with at least 2% of WT infectivity in the spreading replication assay.
TABLE 2.
Nonviable IN mutants
| IN mutation | Frequency (%) in subtype B isolatesa |
|---|---|
| G4E | 0.3 |
| N18I | 0 |
| K34E | 0 |
| V37E | 0 |
| C40F | 0 |
| G52R | 0.2 |
| D64E | 0 |
| C65F | 0 |
| C65Y | 0 |
| A76S | 0 |
| A76T | 0 |
| V77F | 0 |
| H78L | 0 |
| S81N | 0 |
| I84V | 1.2 |
| A86T | 0 |
| A86V | 0 |
| E87G | 0 |
| L102S | 0 |
| K103E | 0 |
| W108R | 0 |
| P109Q | 0 |
| P109S | 0 |
| V113A | 0 |
| G118D | 0 |
| W132C | 0 |
| I135F | 0.1 |
| I135T | 0.1 |
| N144I | 0 |
| E152D | 0.1 |
| N155Y | 0.1 |
| V165L | 0 |
| R166I | 0 |
| M178V | 0.1 |
| A179V | 0 |
| I182 M | 0 |
| I182N | 0 |
| K188E | 0.1 |
| K188N | 0 |
| G192E | 0 |
| A196G | 0 |
| G197E | 0 |
| G197R | 0 |
| E198K | 0.2 |
| D202E | 0 |
| I204K | 0 |
| E212D | 0 |
| L213F | 0 |
| R228W | 0 |
| K244I | 0 |
| V249E | 0 |
| I251K | 0 |
| K258E | 0 |
| K266N | 0 |
| R269G | 0 |
Frequency with which the indicated mutant residue occurs in 1,000 HIV-1 subtype B IN sequences.
A comparison of the distributions of mutational fitness effects (DMFE) for IN and CA (Fig. 3A and B) illustrates that not only were there fewer mutants with lethal defects for IN, but there were also more mutants with intermediate, small, or no fitness reductions, reinforcing the notion that IN is considerably more robust than CA. The DMFE plot for IN appears quite similar to that reported for several RNA viruses (9, 10, 48), consistent with the notion that HIV-1 CA is unusual in exhibiting extreme genetic fragility. If the fitness of each IN and CA mutant is plotted (for codons with multiple mutations represented in the library, the smallest fitness value was selected) against its linear position in the protein, stark disparities in robustness are further highlighted (Fig. 3C and E). The IN mutant library obviously features significantly higher fitness values than the CA mutant library. IN also appears to display small, localized regions of relative fragility in a background of general mutational robustness, while the opposite appears to be true for CA. Notably, IN library clones with silent mutations had fitness values that were similar to the WT, suggesting that the dominant effect of the IN library mutations was via changes in protein rather than RNA function (Fig. 3D).
Comparison of randomly introduced and naturally occurring IN mutations.
Our previous analyses of the in vitro fitness of CA mutants and their occurrence in natural populations revealed an imperfect correlation. Mutations that conferred <40% of WT fitness were very rare in natural populations (11). However, our analysis revealed a population of so-called “fit but rare” mutants, i.e., those that have little effect on in vitro fitness but were rarely found in natural populations. This suggested the existence of some undetermined selective pressure, perhaps immunological, that acts on natural populations of CA sequences and was invisible in our in vitro fitness assays. To clarify the relationship between effects of mutations on in vitro fitness and their occurrence in natural populations, we examined the frequency with which the IN mutants in our library occurred in 1,000 HIV-1 subtype B IN sequences (Tables 1 and 2). Plots of mutant fitness against occurrence in natural populations revealed some key similarities in the behavior of IN and CA mutants (Fig. 3F and G; the CA plot is reproduced from reference 11). Specifically, as was the case for CA, it appeared that IN mutants were required to exhibit at least 40% of WT replicative fitness in vitro in order to occur at a frequency of >1% in natural populations. It was also notable that the IN mutant library, like the CA library, contained a significant number of mutations that had little impact on in vitro fitness but rarely occurred in natural populations (>40% of WT fitness, <0.3% frequency in natural populations). Table 4 lists 11 IN mutants that had >40% of WT fitness and occurred frequently in natural populations, as well as 22 IN mutants that had >40% of WT fitness yet were absent from the 1,000 naturally occurring IN sequences. Thus, the occurrence of mutations that lack major effects on in vitro fitness but are excluded from natural populations is even more evident for IN than for CA.
TABLE 4.
Opposing frequencies of fit IN mutants in vivo
| Fit IN mutation | Replication (% of WT)b | Frequency (%) in subtype B isolatesc |
|---|---|---|
| Occurring frequently in vivoa | ||
| E10D | 107.9 | 8.7 |
| S17N | 141.2 | 21.7 |
| M50I | 75.2 | 9.7 |
| A91T | 44.6 | 9.9 |
| T112I | 61.8 | 3.8 |
| T125A | 92.0 | 25.7 |
| D167E | 132.9 | 3.1 |
| I203 M | 101.5 | 3.5 |
| K211R | 55.7 | 17.4 |
| Q216H | 102.1 | 3.1 |
| D286N | 122.4 | 3.3 |
| Never occurring in vivod | ||
| E13V | 93.9 | |
| K42I | 71.2 | |
| A49V | 79.9 | |
| C65S | 60.6 | |
| V75I | 73.4 | |
| V75L | 68.8 | |
| Y83H | 52.5 | |
| P90Q | 66.4 | |
| S123N | 40.4 | |
| V126F | 82.9 | |
| T174I | 57.9 | |
| V180I | 63.6 | |
| E198Q | 91.5 | |
| I203L | 98.0 | |
| Q214H | 54.8 | |
| N222Y | 93.8 | |
| Q252H | 89.6 | |
| S2551 | 70.1 | |
| D256V | 53.3 | |
| I257L | 65.3 | |
| P261S | 85.8 | |
| M275L | 47.7 | |
| A282T | 87.2 |
Fit mutants that occur frequently are those single-amino-acid mutants with >40% of WT fitness that are found in >3% of 1,000 IN subtype B isolate sequences.
Replication assay is as described for Table 1.
Frequency in subtype B sequences is as described for Table 1.
Fit mutants that never occur are mutations that were not found in any of the 1,000 subtype B IN sequences.
Fitness effects and natural occurrence of IN mutations in the context of IN structural models.
Previously, our depiction of viable and nonviable capsid mutants on a hexameric CA structure revealed patterns in which viable mutants were preferentially located on exposed protein surfaces (11). While no crystal structure for the intact HIV-1 IN currently exists, as either part of the Gag-Pol precursor or part of the intasome or any other state, we used recently published models of the HIV-1 intasome to depict our IN mutations in the context of a plausible IN structure (Fig. 4A) (28, 29). The two published models are similar, and our analyses below do not depend on which model is actually used. However, our results are displayed using one model for simplicity, which was selected because it gave marginally easier visualization of the patterns. In this intasome model (Fig. 4A) (28), unique colors indicate the individual IN subunits, and gray designates DNA.
FIG 4.
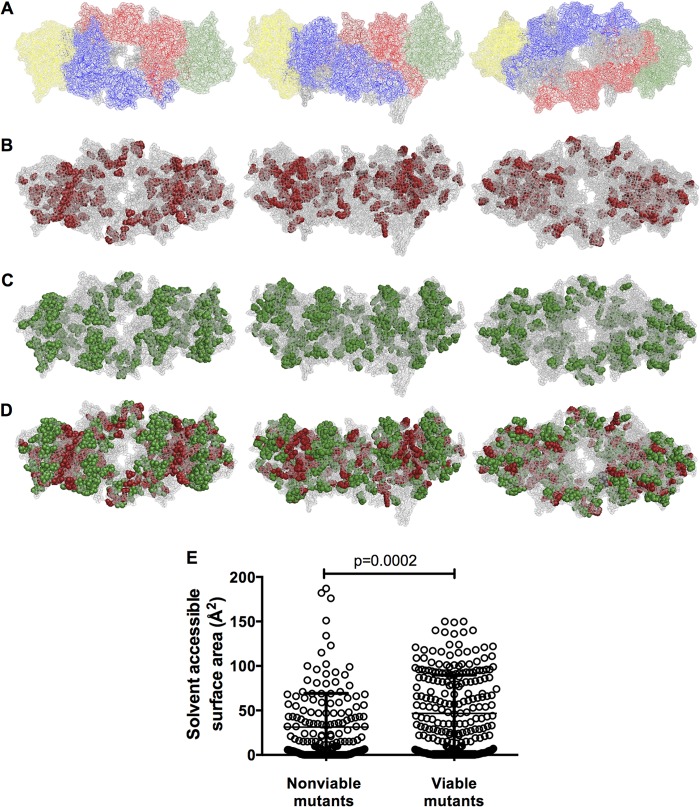
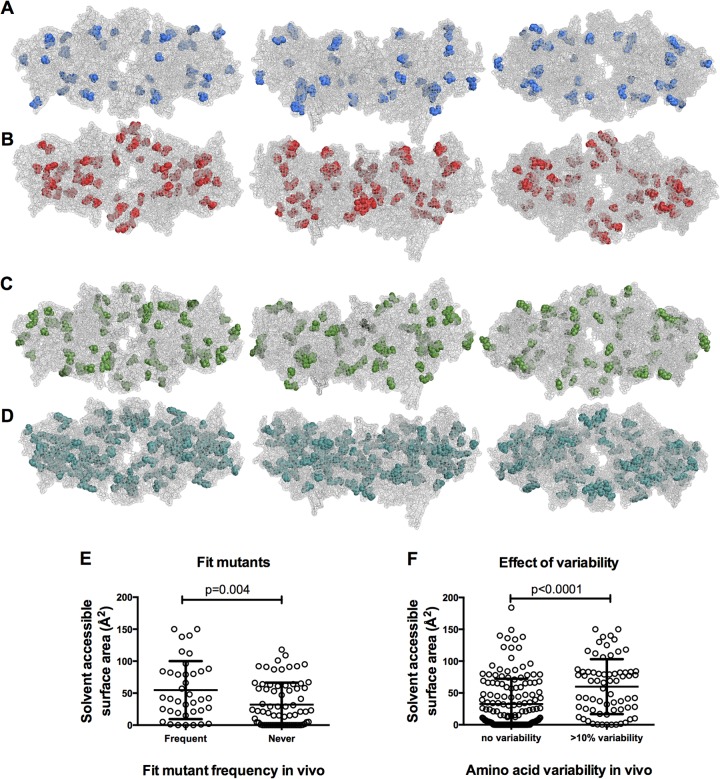
Viable and nonviable HIV-1 IN mutants displayed in the context of an intasome model. (A) Images of an HIV-1 intasome model (28), in which the individual IN subunits of the tetramer are shown in different colors. Inner subunits are shown in blue and red, while outer subunits are in green and yellow. Outer subunits include the CCD only, while inner subunits include full-length IN. DNA is shown in gray, and magnesium and zinc binding pockets are not visible. The image on the left displays the intasome from the (arbitrarily defined) top, the center image displays a side profile image, and the image on the right displays a view of the intasome from below. This model, and image arrangements, is used for all subsequent displays of the intasome. (B) Locations of nonviable (<2% WT replicative fitness) IN mutants displayed in red on the intasome model. Image views are as described for panel A. (C) Locations of viable (>2% WT replicative fitness) IN mutants displayed in green on the intasome model. (D) Locations of both nonviable (red) and viable (green) IN mutants on the same intasome model. (E) Correlation of mutant viability with amino acid surface exposure as measured by solvent-accessible surface area (in Å2). Mutated IN residues were designated either viable or nonviable based on the 2% replicative fitness cutoff, and solvent-accessible surface area values for each mutated residue in each IN subunit were plotted, in order to represent the various degrees of surface exposure for the same residue in different subunits (as available).
Upon displaying sites at which the nonviable IN mutants occur in the intasome model, two trends were observed. First, a subset of lethal mutations occurred at the interfaces between IN subunits, in particular the interface between the inner and outer subunits, while a second subset of lethal mutations appeared buried within the intasome structure (Fig. 4A and B). Conversely, amino acids that tolerate mutation appeared to occur preferentially in surface locations and away from subunit interfaces (Fig. 4C). These trends are especially apparent when viable and nonviable mutants are mapped onto the same intasome structure (Fig. 4D). Moreover, when the solvent-accessible surface area was measured for each IN amino acid, in each of the four subunits (as available), sites of lethal mutations exhibited significantly lower solvent-accessible surface area values than sites of nonlethal mutations (Fig. 4E; P = 0.0002).
We also compared the location of mutants that occurred frequently in natural IN sequences with the “fit but absent” subset of mutants (>40% WT replicative fitness in vitro but absent from natural sequences). Sites that were frequently mutated in naturally occurring IN sequences appeared to be scattered primarily in surface-exposed locations on the intasome (Fig. 5A). In contrast, the fit but absent mutants were often located in more buried positions (Fig. 5B). The solvent-accessible surface area values for these two categories of mutants confirmed that the fit but absent mutants are significantly more buried (P = 0.004) (Fig. 5E). While the degree of surface exposure does not entirely explain why some fit IN mutants seem to not occur in vivo, it appears to be a significant contributing factor. The different locations of the fit and frequently occurring versus the fit but absent categories of mutants are most easily explained by the notion that the fit but absent mutant class has fitness defects that are too subtle to be easily measured in vitro but are sufficient to suppress their occurrence in natural populations. Indeed, while the fit but absent IN mutants all (by definition) exhibited >40% fitness, the mean fitness of the fit and frequently occurring IN mutants (94.3% of WT fitness) was marginally higher than the fitness of the fit but absent mutants (71.7% of WT fitness), a small but significant difference (P = 0.01, unpaired parametric t test). This finding suggests that our in vitro fitness assay, in which replication over 3 to 4 days was measured, is insensitive in discerning modest fitness defects and that IN mutations conferring even very subtle fitness deficits are excluded from natural populations.
FIG 5.
Naturally occurring IN variability displayed on the intasome model. (A) IN mutants that were fit (>40% of WT replicative fitness) and occurred frequently (>3% of 1,000 HIV-1 subtype B IN sequences) are displayed in blue on an intasome model. (B) IN mutants that were fit (>40% of WT replicative fitness) but never occurred in 1,000 HIV-1 subtype B IN sequences are displayed in red on an intasome model. (C) IN amino acids that were variable from WT in >10% of 1,000 HIV-1 subtype B IN sequences are displayed in green on an intasome model. (D) IN amino acids for which there was no variation in 1,000 HIV-1 subtype B IN sequences are displayed in teal on an intasome model. (E) Comparison of solvent-accessible surface area values for fit (>40% of WT) IN mutants that occurred frequently (>3%) or never occurred in 1,000 subtype B IN sequences. All available subunit solvent-accessible surface area values were plotted for each amino acid. (F) Comparison of solvent-accessible surface area for IN amino acids that were variable in >10%, or invariant, in 1,000 HIV-1 subtype B IN sequences. All available subunit solvent-accessible surface area values were plotted for each amino acid.
Because the degree of surface exposure correlated with the frequency with which mutations in our IN library occurred in natural populations, we investigated whether the degree of surface exposure might more generally correlate with variability in vivo, irrespective of whether the fitness of a variant had been measured. In fact, IN amino acids that exhibited variation in >10% of naturally occurring sequences preferentially occurred in surface-exposed locations (Fig. 5C), while amino acids for which there was no variability in our natural population sample appeared generally more buried (Fig. 5D). Indeed, amino acids that were invariant had significantly lower solvent-accessible surface area values than those that showed variation (P < 0.0001) (Fig. 5F). Overall, therefore, the degree of surface exposure of a particular amino acid seems to be a good indicator of both how fit a mutant is and how likely it is to occur in natural populations.
Phenotypic characterization of the mutant library indicates that IN subunit interfaces play a key role during virion assembly.
Measurements of the replicative fitness of the IN mutant library could determine the robustness of IN but not the stage in the viral life cycle where deleterious mutations exert their effect. IN mutants have previously been shown to affect integration (class I mutants) or have pleiotropic effects (class II mutants), and either class of mutations could impact IN robustness. Therefore, we employed a simple biochemical analysis, namely, Western blotting, that could feasibly be applied to all 156 IN mutants to quantify the impact of each mutation on IN expression, particle production, and IN incorporation into particles. IN mutants affecting these phenotypes have previously been reported (39, 49). Since we have also previously shown that certain IN mutations could also affect clathrin incorporation into virions (50), we assessed this parameter, too.
Importantly, cell-associated Gag expression (p24 CA and Gag precursor Pr55) was similar to that of the WT for all IN mutants (Fig. 6). Moreover, consistent with the fitness measurements, most IN mutants did not exhibit any deficiency in IN expression, particle production, and IN incorporation into particles (Fig. 6; summarized in Table 3). Mutant N18I was excluded from further analyses, as the complete absence of signal on Western blots probed with anti-IN suggested that this mutation might have ablated the epitope for the antibody used. Thirty-four (22%) of the IN mutants had at least a 2-fold reduction in virion-associated IN levels, and 13 of these (8%) had a >5-fold reduction. A larger fraction, 46%, of the NTD mutants appeared to have reduced virion-associated IN levels compared to those of the CCD (23%) or CTD (9%) mutants. Moreover, mutants with reductions in virion-associated IN levels frequently occurred in clusters in the CCD (e.g., residues 78 to 83, 102 to 109, 129 to 132). Mutants with reduced virion-associated IN protein also exhibited decreased cell-associated IN, suggesting that these mutations either caused defects in proteolytic processing of the Gag-Pol precursor or instability of the mature processed integrase protein. Notably, virtually all of the IN mutants that exhibited reductions in cell- and virion-associated IN also displayed increases in the cell-associated p41 Gag product, indicating that these mutations induce defects in the extent or timing of proteolytic processing. Additionally, there was a large degree of concordance in the degree to which mutations affected IN expression/virion incorporation and the degree to which they affected clathrin incorporation. This was true for the intact clathrin heavy chain (HC) but was more prominent for a clathrin HC fragment whose occurrence is likely the result of cleavage by the HIV-1 protease in virions (Fig. 6, below the HC band). A subset of the mutants that exhibited reduced IN levels in cells and virions also exhibited reduced overall particle production. Specifically, 11 mutants (7%) generated at least 2-fold-decreased levels of virion-associated p24.
FIG 6.
Phenotypic characterization of IN mutants. Western blot analyses, using anti-CA, anti-IN, and anti-clathrin heavy chain antibodies, of virions and cell lysates generated using 293T cells transfected with the IN mutant proviral plasmids. The numbers shown below lanes indicate the fluorescence intensities (LiCOR) associated with the CA, IN, or clathrin HC protein that was pelleted from virion-containing supernatant. The blots shown below the anti-clathrin heavy chain (HC) blots are a secondary clathrin band appearing on the same blot that is assumed to be a product of digestion by HIV-1 protease in virions. Mutants were considered phenotypically different to WT if CA or IN protein levels were decreased by at least 2-fold.
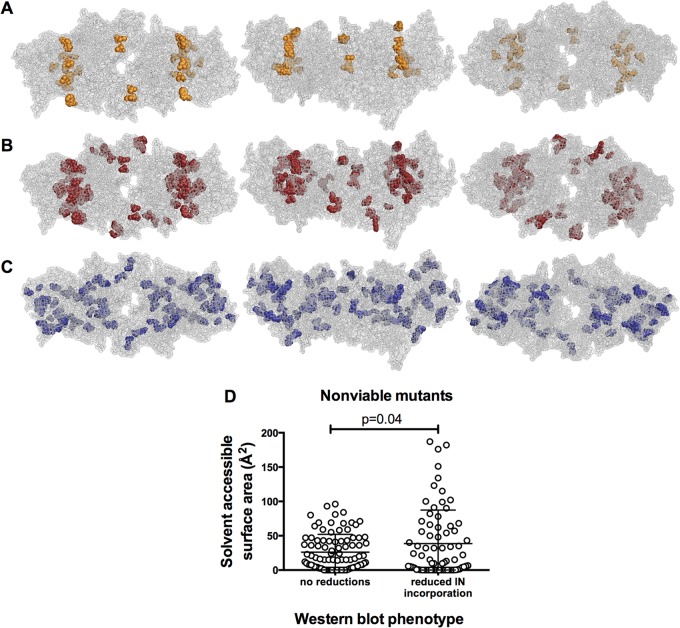
Notably, a display of the nonviable mutants with particle production deficits in the context of the intasome structure revealed a highly specific pattern of localization in which nearly all mutants displaying this phenotype targeted the interface between the inner and outer IN subunits (Fig. 7A). Moreover, the nonviable mutants that exhibited reduced levels of virion-associated IN and clathrin included an even greater number of mutants at the same interface between the inner and outer IN subunits (Fig. 7B).
FIG 7.
HIV-1 IN mutants affecting particle production and IN levels displayed in the context of an intasome model. (A) Nonviable IN mutants conferring at least a 2-fold reduction in particle production, displayed on an intasome model; (B) nonviable IN mutants conferring at least a 2-fold reduction in virion-associated IN levels, displayed on an intasome model; (C) nonviable IN mutants conferring no significant reductions in either particle production or virion-associated IN levels, displayed on an intasome model. These mutants exhibited a loss of virion infectiousness only. (D) Solvent-accessible surface area (in Å2) for IN mutants that either did or did not cause a 2-fold reduction in virion-associated IN protein levels. All available subunit solvent-accessible surface area values were plotted for each amino acid.
A different phenotype was exhibited by 24 (44%) nonviable mutants. These mutants had normal levels of IN protein in virions or cells and normal levels of particle production but exhibited reduced particle infectiousness. These mutants displayed a different pattern of localization and were scattered throughout the intasome, mostly in solvent-inaccessible locations (Fig. 7C). The degree to which these nonviable mutants were buried was significant compared to that of nonviable mutants with reduced virion-associated IN levels (P = 0.04) (Fig. 7D). Overall, a consideration of the effects of IN mutations on particle yield, composition, and infectiousness, in the context of IN structural models, highlights the role that certain portions of the IN protein, particularly subunit interfaces, play during virion assembly and other steps in the viral life cycle.
DISCUSSION
In this study, we first sought to compare the genetic robustness of HIV-1 IN with HIV-1 CA. Our results reveal a significant difference in the robustness of IN and CA, with just 35% of single IN amino acid substitutions resulting in nonviable virus (<2% of WT fitness), whereas twice as many mutants in CA (70%) gave this outcome. This finding is surprising, because enzymes are expected to be particularly genetically fragile, given the requirement that they adopt and change their tertiary structures in subtle, flexible ways (51). The relatively highly conserved nature of IN should also suggest intolerance to mutation (52). However, our finding that IN is relatively robust compared to CA is similar to previous findings with HIV-1 protease (53) and other viral proteins, such as the bacteriophage f1 gene V protein and varicella-zoster virus thymidine kinase, in which 35 to 40% of mutations are lethal (54, 55). Moreover, IN appears to exhibit a degree of robustness similar to that of several whole viral genomes, including those of tobacco etch virus, vesicular stomatitis virus, and bacteriophage Qβ, in which 41%, 40%, and 29% of random mutations are lethal (9, 10, 48). Furthermore, IN exhibits a level of robustness that is similar to that of nonviral enzymes, such as 3-methyladenine DNA glycosylase (AAG) and Campylobacter jejuni oligosaccharyltransferase PglB, in which 34% and 33% of mutations are lethal, respectively (56, 57). Thus, the relative robustness of HIV-1 IN compared to CA validates the notion that the extreme fragility of CA is exceptional.
The reasons for the greater robustness of HIV-1 IN than HIV-1 CA are not obvious. Both IN and CA are essential for viral replication and have relatively similar degrees of sequence variation in natural populations (Fig. 1A to D), suggesting that they should be similarly intolerant of mutation. Difference in function presumably underlies the observed difference in robustness. CA must maintain a stable structure that can adopt a range of CA-CA interactions that are finely tuned and able to adjust as virions assemble and disassemble into immature and mature configurations (58–61). Moreover, CA must also form interactions with a wide variety of host proteins (62–64). While IN forms critical interactions with LEDGF (65, 66), as well as interactions with TRN-SR2(TNPO3) (35, 67) and INI-1/hSNF5 (36, 68), and also undergoes multimerization processes (21, 69–71), the larger number of specialized CA-CA interactions that mediate accurate virion assembly may make CA exceptionally fragile (Fig. 3A to E) (11).
It is also conceivable that the disparities in the fragility of IN and CA could, at least partially, arise from differences in the underlying RNA structure. While it has previously been suggested that HIV-1 RNA structure might constrain variability and evolution (72–74), silent mutations introduced into some of the most highly conserved RNA hairpins in HIV-1 result in no significant virus replication defects (75). Similarly, introduction of synonymous mutations into other regions of the HIV-1 genome with apparently strong purifying selection against synonymous substitutions does not reduce viral fitness in vitro (76). Our own results indicate that random introduction of single silent mutations in both IN (Fig. 3D) and CA (11) did not frequently cause reduced fitness. Furthermore, the remarkable clustering of nonviable IN mutants at specific protein interfaces argues against the notion that underlying RNA structure is a major determinant of fitness in our mutant libraries. Others have also concluded that specific protein structure, including the degree of surface exposure, can significantly constrain HIV-1's capacity to explore sequence space and evolve (77).
The comprehensive nature of our mutagenized IN library enabled a more thorough understanding of the role of the individual IN domains in HIV-1 replication. Previous IN mutagenesis studies have elucidated the importance of either specific residues or domain functions (32, 33, 38–40, 42, 78) and are consistent with our results. However, the large-scale nature of our IN library permitted the observation of the most striking features of our analyses, including (i) the localization of many nonviable IN mutants to particular IN subunit interfaces in the intasome model (Fig. 4B to D) and (ii) the specific functional deficits exhibited by these mutants, namely, reduced particle production and reduced levels of IN in cells and virions. In clear contrast, nonviable mutants that generated normal levels of virions with normal protein content were more likely to occur in buried positions in the intasome model (Fig. 7A to C). While the structural models may not precisely represent structures of the full-length IN protein in the context of the Gag-Pol precursor or the intasome, the strong and defined patterns suggest that the HIV-1 intasome structural models we have utilized are likely quite accurate. Most IN mutants that conferred reduced particle production or virion-associated IN levels target the interface between the inner (blue and red, Fig. 4A) and outer (yellow and green, Fig. 4A) IN subunits within the clusters of contiguous amino acids 76 to 87, 102 to 113, 132 to 135, and 178 to 213. These amino acids form much of the CCD-CCD dimerization interface (21). Interestingly, while the function of the outer subunits of the intasome has not been clear (21), the considerable fragility of the subunit interfaces (Fig. 4B and D), and the fact that the IN mutants that display reductions in IN expression or incorporation also exhibit defective proteolytic processing (Fig. 6), suggests that IN multimerization is an important regulator of protease activity during virion assembly. While previous reports have hinted that individual amino acids at the inner-outer subunit, CCD-CCD interface could be important for IN multimerization (79–81), none has realized the full extent of the fragility of this IN domain and the importance it plays during HIV-1 assembly.
This CCD-CCD interface is known to be adjacent to, and include, some critical amino acids that participate in the interaction between HIV-1 IN and LEDGF (82). A new class of antiretroviral drugs, the allosteric integrase inhibitors (ALLINIs), engage IN at, or close to, the LEDGF binding sites and the CCD-CCD interface (for a review of ALLINIs and their mechanisms of action, see reference 83). Surprisingly, these drugs exert their effect during HIV-1 production rather than during integration (where LEDGF acts), suggesting that they affect IN subunit interactions rather than LEDGF binding (84). Together, these data suggest that the nonviable IN mutants described herein, like ALLINIs, affect replication by perturbing (either promoting or inhibiting) multimerization or binding steps at the CCD-CCD interface between the inner and outer subunits that are critical during the assembly of virions.
The finding that a major source of fragility in IN is defined by subunit multimerization during virion assembly is, in a sense, analogous to the finding that the major source of HIV-1 CA fragility is the requirement that it participate in precisely configured multimerization reactions during assembly (11). Overall, the data from HIV-1 IN and CA suggest the possibility that, in general, (i) specific structural interactions during virus particle assembly processes and (ii) the fraction of protein sequence that remains surface exposed or buried might be key determinants of overall mutational robustness and fragility in viral proteins. Accordingly, in both IN and CA, mutations that were compatible with viability were more likely to occur in solvent-accessible locations (Fig. 4E) (11). This is an extension of a long-appreciated concept, based on data from the 1960s, which indicated that hemoglobin residues at the surface were less sensitive to replacement than more internal residues (85). Consistent with this idea is the observation that surface-exposed IN amino acids were also more variable in natural populations (Fig. 5C, D, and F).
Our fitness measurements suggested that 40% of WT replicative fitness represented a cutoff, and mutants with fitness values less than this were unlikely to occur frequently in natural populations (Fig. 3F and G). The fact that the same minimum fitness value was obtained for frequently occurring CA and IN mutants suggests that this cutoff represents an accurate estimate of in vivo fitness restrictions. However, while there were many fit (>40%) mutants that occurred frequently in vivo, a significant number of fit (>40%) IN and CA mutants never or rarely occurred in our sample of 1,000 naturally occurring sequences (Table 4) (11). Despite their similar fitness values in vitro, frequently and rarely occurring IN mutants exhibited a clear difference in the location in IN structural models: IN mutations that rarely occurred in vivo typically represented buried amino acids, while those mutations that frequently occurred generally represented surface-exposed amino acids (Fig. 5A, B, and E). The simplest explanation of this anomaly is that fitness defects that are too subtle to be measured in an in vitro fitness assay are sufficient for exclusion in a more exacting in vivo environment.
Why do CA and IN exhibit similar degrees of variation in natural populations when CA is significantly more genetically fragile than IN? One consideration is that CA would be predicted to be under greater immunological pressure than IN because Gag is expressed at 10- to 20-fold-higher levels than Pol in infected cells. While both IN and CA have been documented to be under selective pressure generated by host immune CD8+ CTL responses, there is more evidence of CTL-imposed reductions in fitness in Gag/CA than in Pol/IN, suggesting the fragility exhibited by CA makes it particularly vulnerable to this type of pressure (86–91). Thus, the similar degrees of natural sequence variation exhibited by IN and CA could simply reflect a similar balance of opposing forces, namely, immunological selective pressure (driving sequence diversification) and fragility (driving sequence conservation). Moreover, the lower robustness of CA versus IN could help explain why individuals with immune responses to Gag during HIV-1 infection fare better than those with immune responses to other viral proteins (87).
Robustness should correlate with evolvability and perhaps capacity to evolve resistance to therapeutics and immune responses (5, 92). Understanding the robustness and fragility of individual proteins like HIV-1 IN and CA could help predict constraints on sequence, indicate vulnerable regions within protein structures, and aid the development of better antiviral strategies.
ACKNOWLEDGMENTS
We thank Michael Malim for generously providing murine monoclonal antibody INT-4 and Steven Smith, Barry Johnson, Stephen Hughes, Kellie Jurado, and Alan Engelman for the HIV-1 intasome model coordinates.
This work was supported by NIH grants R01AI50111 and P50GM103297.
REFERENCES
- 1.Holland J, Spindler K, Horodyski F, Grabau E, Nichol S, VandePol S. 1982. Rapid evolution of RNA genomes. Science 215:1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- 2.Sanjuan R, Nebot MR, Chirico N, Mansky LM, Belshaw R. 2010. Viral mutation rates. J Virol 84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elena SF. 2012. RNA virus genetic robustness: possible causes and some consequences. Curr Opin Virol 2:525–530. doi: 10.1016/j.coviro.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 4.de Visser JA, Hermisson J, Wagner GP, Ancel Meyers L, Bagheri-Chaichian H, Blanchard JL, Chao L, Cheverud JM, Elena SF, Fontana W, Gibson G, Hansen TF, Krakauer D, Lewontin RC, Ofria C, Rice SH, von Dassow G, Wagner A, Whitlock MC. 2003. Perspective: evolution and detection of genetic robustness. Evolution 57:1959–1972. doi: 10.1111/j.0014-3820.2003.tb00377.x. [DOI] [PubMed] [Google Scholar]
- 5.Lauring AS, Frydman J, Andino R. 2013. The role of mutational robustness in RNA virus evolution. Nat Rev Microbiol 11:327–336. doi: 10.1038/nrmicro3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanjuan R, Cuevas JM, Furio V, Holmes EC, Moya A. 2007. Selection for robustness in mutagenized RNA viruses. PLoS Genet 3:e93. doi: 10.1371/journal.pgen.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Codoner FM, Daros JA, Sole RV, Elena SF. 2006. The fittest versus the flattest: experimental confirmation of the quasispecies effect with subviral pathogens. PLoS Pathog 2:e136. doi: 10.1371/journal.ppat.0020136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montville R, Froissart R, Remold SK, Tenaillon O, Turner PE. 2005. Evolution of mutational robustness in an RNA virus. PLoS Biol 3:e381. doi: 10.1371/journal.pbio.0030381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanjuan R, Moya A, Elena SF. 2004. The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proc Natl Acad Sci U S A 101:8396–8401. doi: 10.1073/pnas.0400146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrasco P, de la Iglesia F, Elena SF. 2007. Distribution of fitness and virulence effects caused by single-nucleotide substitutions in tobacco etch virus. J Virol 81:12979–12984. doi: 10.1128/JVI.00524-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rihn SJ, Wilson SJ, Loman NJ, Alim M, Bakker SE, Bhella D, Gifford RJ, Rixon FJ, Bieniasz PD. 2013. Extreme genetic fragility of the HIV-1 capsid. PLoS Pathog 9:e1003461. doi: 10.1371/journal.ppat.1003461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller MD, Farnet CM, Bushman FD. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol 71:5382–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen H, Engelman A. 2001. Asymmetric processing of human immunodeficiency virus type 1 cDNA in vivo: implications for functional end coupling during the chemical steps of DNA transposition. Mol Cell Biol 21:6758–6767. doi: 10.1128/MCB.21.20.6758-6767.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei SQ, Mizuuchi K, Craigie R. 1997. A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J 16:7511–7520. doi: 10.1093/emboj/16.24.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherman PA, Fyfe JA. 1990. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci U S A 87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engelman A, Mizuuchi K, Craigie R. 1991. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell 67:1211–1221. doi: 10.1016/0092-8674(91)90297-C. [DOI] [PubMed] [Google Scholar]
- 17.Bushman FD, Fujiwara T, Craigie R. 1990. Retroviral DNA integration directed by HIV integration protein in vitro. Science 249:1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]
- 18.Bushman FD, Craigie R. 1991. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc Natl Acad Sci U S A 88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng R, Jenkins TM, Craigie R. 1996. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc Natl Acad Sci U S A 93:13659–13664. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulkosky J, Jones KS, Katz RA, Mack JP, Skalka AM. 1992. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol 12:2331–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Krishnan L, Cherepanov P, Engelman A. 2011. Structural biology of retroviral DNA integration. Virology 411:194–205. doi: 10.1016/j.virol.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR. 1994. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 23.Woerner AM, Klutch M, Levin JG, Marcus-Sekura CJ. 1992. Localization of DNA binding activity of HIV-1 integrase to the C-terminal half of the protein. AIDS Res Hum Retroviruses 8:297–304. doi: 10.1089/aid.1992.8.297. [DOI] [PubMed] [Google Scholar]
- 24.Eijkelenboom AP, Lutzke RA, Boelens R, Plasterk RH, Kaptein R, Hard K. 1995. The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat Struct Biol 2:807–810. doi: 10.1038/nsb0995-807. [DOI] [PubMed] [Google Scholar]
- 25.Guiot E, Carayon K, Delelis O, Simon F, Tauc P, Zubin E, Gottikh M, Mouscadet JF, Brochon JC, Deprez E. 2006. Relationship between the oligomeric status of HIV-1 integrase on DNA and enzymatic activity. J Biol Chem 281:22707–22719. doi: 10.1074/jbc.M602198200. [DOI] [PubMed] [Google Scholar]
- 26.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. 2010. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464:232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maertens GN, Hare S, Cherepanov P. 2010. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature 468:326–329. doi: 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson BC, Metifiot M, Ferris A, Pommier Y, Hughes SH. 2013. A homology model of HIV-1 integrase and analysis of mutations designed to test the model. J Mol Biol 425:2133–2146. doi: 10.1016/j.jmb.2013.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krishnan L, Li X, Naraharisetty HL, Hare S, Cherepanov P, Engelman A. 2010. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc Natl Acad Sci U S A 107:15910–15915. doi: 10.1073/pnas.1002346107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engelman A. 1999. In vivo analysis of retroviral integrase structure and function. Adv Virus Res 52:411–426. doi: 10.1016/S0065-3527(08)60309-7. [DOI] [PubMed] [Google Scholar]
- 31.Bushman FD, Engelman A, Palmer I, Wingfield P, Craigie R. 1993. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci U S A 90:3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drelich M, Wilhelm R, Mous J. 1992. Identification of amino acid residues critical for endonuclease and integration activities of HIV-1 IN protein in vitro. Virology 188:459–468. doi: 10.1016/0042-6822(92)90499-F. [DOI] [PubMed] [Google Scholar]
- 33.Vink C, Oude Groeneger AM, Plasterk RH. 1993. Identification of the catalytic and DNA-binding region of the human immunodeficiency virus type I integrase protein. Nucleic Acids Res 21:1419–1425. doi: 10.1093/nar/21.6.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng Y, Ao Z, Jayappa KD, Yao X. 2010. Characterization of the HIV-1 integrase chromatin- and LEDGF/p75-binding abilities by mutagenic analysis within the catalytic core domain of integrase. Virol J 7:68. doi: 10.1186/1743-422X-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Houwer S, Demeulemeester J, Thys W, Taltynov O, Zmajkovicova K, Christ F, Debyser Z. 2012. Identification of residues in the C-terminal domain of HIV-1 integrase that mediate binding to the transportin-SR2 protein. J Biol Chem 287:34059–34068. doi: 10.1074/jbc.M112.387944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yung E, Sorin M, Wang EJ, Perumal S, Ott D, Kalpana GV. 2004. Specificity of interaction of INI1/hSNF5 with retroviral integrases and its functional significance. J Virol 78:2222–2231. doi: 10.1128/JVI.78.5.2222-2231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiskerchen M, Muesing MA. 1995. Identification and characterization of a temperature-sensitive mutant of human immunodeficiency virus type 1 by alanine scanning mutagenesis of the integrase gene. J Virol 69:597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Engelman A, Craigie R. 1992. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol 66:6361–6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engelman A, Englund G, Orenstein JM, Martin MA, Craigie R. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol 69:2729–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engelman A, Liu Y, Chen H, Farzan M, Dyda F. 1997. Structure-based mutagenesis of the catalytic domain of human immunodeficiency virus type 1 integrase. J Virol 71:3507–3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jenkins TM, Esposito D, Engelman A, Craigie R. 1997. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J 16:6849–6859. doi: 10.1093/emboj/16.22.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen JC, Krucinski J, Miercke LJ, Finer-Moore JS, Tang AH, Leavitt AD, Stroud RM. 2000. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc Natl Acad Sci U S A 97:8233–8238. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai M, Zheng R, Caffrey M, Craigie R, Clore GM, Gronenborn AM. 1997. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat Struct Biol 4:567–577. doi: 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- 44.Johnson AA, Santos W, Pais GC, Marchand C, Amin R, Burke TR Jr, Verdine G, Pommier Y. 2006. Integration requires a specific interaction of the donor DNA terminal 5′-cytosine with glutamine 148 of the HIV-1 integrase flexible loop. J Biol Chem 281:461–467. doi: 10.1074/jbc.M511348200. [DOI] [PubMed] [Google Scholar]
- 45.Cahn P, Pozniak AL, Mingrone H, Shuldyakov A, Brites C, Andrade-Villanueva JF, Richmond G, Buendia CB, Fourie J, Ramgopal M, Hagins D, Felizarta F, Madruga J, Reuter T, Newman T, Small CB, Lombaard J, Grinsztejn B, Dorey D, Underwood M, Griffith S, Min S. 2013. Dolutegravir versus raltegravir in antiretroviral-experienced, integrase-inhibitor-naive adults with HIV: week 48 results from the randomised, double-blind, noninferiority SAILING study. Lancet 382:700–708. doi: 10.1016/S0140-6736(13)61221-0. [DOI] [PubMed] [Google Scholar]
- 46.Bouyac-Bertoia M, Dvorin JD, Fouchier RA, Jenkins Y, Meyer BE, Wu LI, Emerman M, Malim MH. 2001. HIV-1 infection requires a functional integrase NLS. Mol Cell 7:1025–1035. doi: 10.1016/S1097-2765(01)00240-4. [DOI] [PubMed] [Google Scholar]
- 47.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Domingo-Calap P, Cuevas JM, Sanjuan R. 2009. The fitness effects of random mutations in single-stranded DNA and RNA bacteriophages. PLoS Genet 5:e1000742. doi: 10.1371/journal.pgen.1000742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cannon PM, Wilson W, Byles E, Kingsman SM, Kingsman AJ. 1994. Human immunodeficiency virus type 1 integrase: effect on viral replication of mutations at highly conserved residues. J Virol 68:4768–4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang F, Zang T, Wilson SJ, Johnson MC, Bieniasz PD. 2011. Clathrin facilitates the morphogenesis of retrovirus particles. PLoS Pathog 7:e1002119. doi: 10.1371/journal.ppat.1002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner A. 2005. Robustness and evolvability in living systems. Princeton University Press, Princeton, NJ. [Google Scholar]
- 52.Ceccherini-Silberstein F, Malet I, D'Arrigo R, Antinori A, Marcelin AG, Perno CF. 2009. Characterization and structural analysis of HIV-1 integrase conservation. AIDS Rev 11:17–29. http://www.aidsreviews.com/files/2009_11_1_017-029.pdf. [PubMed] [Google Scholar]
- 53.Loeb DD, Swanstrom R, Everitt L, Manchester M, Stamper SE, Hutchison CA III. 1989. Complete mutagenesis of the HIV-1 protease. Nature 340:397–400. doi: 10.1038/340397a0. [DOI] [PubMed] [Google Scholar]
- 54.Terwilliger TC, Zabin HB, Horvath MP, Sandberg WS, Schlunk PM. 1994. In vivo characterization of mutants of the bacteriophage f1 gene V protein isolated by saturation mutagenesis. J Mol Biol 236:556–571. doi: 10.1006/jmbi.1994.1165. [DOI] [PubMed] [Google Scholar]
- 55.Suzutani T, Lacey SF, Powell KL, Purifoy DJ, Honess RW. 1992. Random mutagenesis of the thymidine kinase gene of varicella-zoster virus. J Virol 66:2118–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo HH, Choe J, Loeb LA. 2004. Protein tolerance to random amino acid change. Proc Natl Acad Sci U S A 101:9205–9210. doi: 10.1073/pnas.0403255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ihssen J, Kowarik M, Wiesli L, Reiss R, Wacker M, Thony-Meyer L. 2012. Structural insights from random mutagenesis of Campylobacter jejuni oligosaccharyltransferase PglB. BMC Biotechnol 12:67. doi: 10.1186/1472-6750-12-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua Y, Whitby FG, Stout CD, Sundquist WI, Hill CP, Yeager M. 2009. X-ray structures of the hexameric building block of the HIV capsid. Cell 137:1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Krausslich HG. 2009. Structure and assembly of immature HIV. Proc Natl Acad Sci U S A 106:11090–11095. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Forshey BM, von Schwedler U, Sundquist WI, Aiken C. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol 76:5667–5677. doi: 10.1128/JVI.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jouvenet N, Bieniasz PD, Simon SM. 2008. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature 454:236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schaller T, Ocwieja KE, Rasaiyaah J, Price AJ, Brady TL, Roth SL, Hue S, Fletcher AJ, Lee K, KewalRamani VN, Noursadeghi M, Jenner RG, James LC, Bushman FD, Towers GJ. 2011. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog 7:e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luban J, Bossolt KL, Franke EK, Kalpana GV, Goff SP. 1993. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73:1067–1078. [DOI] [PubMed] [Google Scholar]
- 64.Lee K, Mulky A, Yuen W, Martin TD, Meyerson NR, Choi L, Yu H, Sawyer SL, Kewalramani VN. 2012. HIV-1 capsid-targeting domain of cleavage and polyadenylation specificity factor 6. J Virol 86:3851–3860. doi: 10.1128/JVI.06607-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. 2003. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem 278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 66.Maertens G, Cherepanov P, Pluymers W, Busschots K, De Clercq E, Debyser Z, Engelborghs Y. 2003. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J Biol Chem 278:33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- 67.Christ F, Thys W, De Rijck J, Gijsbers R, Albanese A, Arosio D, Emiliani S, Rain JC, Benarous R, Cereseto A, Debyser Z. 2008. Transportin-SR2 imports HIV into the nucleus. Curr Biol 18:1192–1202. doi: 10.1016/j.cub.2008.07.079. [DOI] [PubMed] [Google Scholar]
- 68.Morozov A, Yung E, Kalpana GV. 1998. Structure-function analysis of integrase interactor 1/hSNF5L1 reveals differential properties of two repeat motifs present in the highly conserved region. Proc Natl Acad Sci U S A 95:1120–1125. doi: 10.1073/pnas.95.3.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kalpana GV, Goff SP. 1993. Genetic analysis of homomeric interactions of human immunodeficiency virus type 1 integrase using the yeast two-hybrid system. Proc Natl Acad Sci U S A 90:10593–10597. doi: 10.1073/pnas.90.22.10593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jenkins TM, Engelman A, Ghirlando R, Craigie R. 1996. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J Biol Chem 271:7712–7718. doi: 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- 71.Hare S, Di Nunzio F, Labeja A, Wang J, Engelman A, Cherepanov P. 2009. Structural basis for functional tetramerization of lentiviral integrase. PLoS Pathog 5:e1000515. doi: 10.1371/journal.ppat.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sanjuan R, Borderia AV. 2011. Interplay between RNA structure and protein evolution in HIV-1. Mol Biol Evol 28:1333–1338. doi: 10.1093/molbev/msq329. [DOI] [PubMed] [Google Scholar]
- 73.Zanini F, Neher RA. 2013. Quantifying selection against synonymous mutations in HIV-1 env evolution. J Virol 87:11843–11850. doi: 10.1128/JVI.01529-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.ter Brake O, von Eije KJ, Berkhout B. 2008. Probing the sequence space available for HIV-1 evolution. AIDS 22:1875–1877. doi: 10.1097/QAD.0b013e328309efe3. [DOI] [PubMed] [Google Scholar]
- 75.Knoepfel SA, Berkhout B. 2013. On the role of four small hairpins in the HIV-1 RNA genome. RNA Biol 10:540–552. doi: 10.4161/rna.24133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mayrose I, Stern A, Burdelova EO, Sabo Y, Laham-Karam N, Zamostiano R, Bacharach E, Pupko T. 2013. Synonymous site conservation in the HIV-1 genome. BMC Evol Biol 13:164. doi: 10.1186/1471-2148-13-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Woo J, Robertson DL, Lovell SC. 2010. Constraints on HIV-1 diversity from protein structure. J Virol 84:12995–13003. doi: 10.1128/JVI.00702-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Limon A, Devroe E, Lu R, Ghory HZ, Silver PA, Engelman A. 2002. Nuclear localization of human immunodeficiency virus type 1 preintegration complexes (PICs): V165A and R166A are pleiotropic integrase mutants primarily defective for integration, not PIC nuclear import. J Virol 76:10598–10607. doi: 10.1128/JVI.76.21.10598-10607.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Berthoux L, Sebastian S, Muesing MA, Luban J. 2007. The role of lysine 186 in HIV-1 integrase multimerization. Virology 364:227–236. doi: 10.1016/j.virol.2007.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Al-Mawsawi LQ, Hombrouck A, Dayam R, Debyser Z, Neamati N. 2008. Four-tiered pi interaction at the dimeric interface of HIV-1 integrase critical for DNA integration and viral infectivity. Virology 377:355–363. doi: 10.1016/j.virol.2008.04.030. [DOI] [PubMed] [Google Scholar]
- 81.Serrao E, Thys W, Demeulemeester J, Al-Mawsawi LQ, Christ F, Debyser Z, Neamati N. 2012. A symmetric region of the HIV-1 integrase dimerization interface is essential for viral replication. PLoS One 7:e45177. doi: 10.1371/journal.pone.0045177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A. 2005. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc Natl Acad Sci U S A 102:17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Engelman A, Kessl JJ, Kvaratskhelia M. 2013. Allosteric inhibition of HIV-1 integrase activity. Curr Opin Chem Biol 17:339–345. doi: 10.1016/j.cbpa.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sharma A, Slaughter A, Jena N, Feng L, Kessl JJ, Fadel HJ, Malani N, Male F, Wu L, Poeschla E, Bushman FD, Fuchs JR, Kvaratskhelia M. 2014. A new class of multimerization selective inhibitors of HIV-1 integrase. PLoS Pathog 10:e1004171. doi: 10.1371/journal.ppat.1004171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Perutz MF, Lehmann H. 1968. Molecular pathology of human haemoglobin. Nature 219:902–909. doi: 10.1038/219902a0. [DOI] [PubMed] [Google Scholar]
- 86.Rodriguez WR, Addo MM, Rathod A, Fitzpatrick CA, Yu XG, Perkins B, Rosenberg ES, Altfeld M, Walker BD. 2004. CD8+ T lymphocyte responses target functionally important regions of protease and integrase in HIV-1 infected subjects. J Transl Med 2:15. doi: 10.1186/1479-5876-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N, Bishop K, van der Stok M, Nair K, Khan N, Crawford H, Payne R, Leslie A, Prado J, Prendergast A, Frater J, McCarthy N, Brander C, Learn GH, Nickle D, Rousseau C, Coovadia H, Mullins JI, Heckerman D, Walker BD, Goulder P. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med 13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 88.Brockman MA, Chopera DR, Olvera A, Brumme CJ, Sela J, Markle TJ, Martin E, Carlson JM, Le AQ, McGovern R, Cheung PK, Kelleher AD, Jessen H, Markowitz M, Rosenberg E, Frahm N, Sanchez J, Mallal S, John M, Harrigan PR, Heckerman D, Brander C, Walker BD, Brumme ZL. 2012. Uncommon pathways of immune escape attenuate HIV-1 integrase replication capacity. J Virol 86:6913–6923. doi: 10.1128/JVI.07133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Crawford H, Matthews PC, Schaefer M, Carlson JM, Leslie A, Kilembe W, Allen S, Ndung'u T, Heckerman D, Hunter E, Goulder PJ. 2011. The hypervariable HIV-1 capsid protein residues comprise HLA-driven CD8+ T-cell escape mutations and covarying HLA-independent polymorphisms. J Virol 85:1384–1390. doi: 10.1128/JVI.01879-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brockman MA, Brumme ZL, Brumme CJ, Miura T, Sela J, Rosato PC, Kadie CM, Carlson JM, Markle TJ, Streeck H, Kelleher AD, Markowitz M, Jessen H, Rosenberg E, Altfeld M, Harrigan PR, Heckerman D, Walker BD, Allen TM. 2010. Early selection in Gag by protective HLA alleles contributes to reduced HIV-1 replication capacity that may be largely compensated for in chronic infection. J Virol 84:11937–11949. doi: 10.1128/JVI.01086-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martinez-Picado J, Prado JG, Fry EE, Pfafferott K, Leslie A, Chetty S, Thobakgale C, Honeyborne I, Crawford H, Matthews P, Pillay T, Rousseau C, Mullins JI, Brander C, Walker BD, Stuart DI, Kiepiela P, Goulder P. 2006. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J Virol 80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McBride RC, Ogbunugafor CB, Turner PE. 2008. Robustness promotes evolvability of thermotolerance in an RNA virus. BMC Evol Biol 8:231. doi: 10.1186/1471-2148-8-231. [DOI] [PMC free article] [PubMed] [Google Scholar]