SUMMARY
A pen infection-transmission experiment was conducted to elucidate the role of pathogen strain and environmental contamination in transmission of Escherichia coli O157:H7 (ECO157) in cattle. Five steers were inoculated with a three-strain mixture of ECO157 and joined with five susceptible steers in each of two experimental replicates. Faecal and environmental samples were monitored for ECO157 presence over 30 days. One ECO157 strain did not spread. Transmission rates for the other two strains were estimated using a generalized linear model developed based on a modified ‘Susceptible–Infectious–Susceptible’ mathematical model. Transmission rates estimated for the two strains (0·11 and 0·14) were similar. However, the rates significantly (P = 0·0006) increased 1·5 times for every 1-unit increase in the level of environmental contamination measured as log10 c.f.u. Depending on the level of environmental contamination, the estimated basic reproduction numbers varied from <1 to 8. The findings indicate the importance of on-farm measures to reduce environmental contamination for ECO157 control in cattle that should be validated under field conditions.
Key words: Cattle, E. coli O157, environmental contamination, transmission
INTRODUCTION
Escherichia coli O157:H7 (ECO157) usually causes uncomplicated diarrhoea, but occasionally, it is responsible for causing severe forms of illness including haemorrhagic colitis, and potentially lethal haemolytic uraemic syndrome (HUS) in humans [1, 2]. Cattle are the main hosts for ECO157 [3, 4]. Humans can become exposed to ECO157 through direct contact with an infected animal or a contaminated farm environment [5]. The most common mode of human exposure is through the consumption of contaminated foods including meat, milk, raw vegetables and fruits [6]. The majority of outbreaks or illnesses with ECO157 in humans, either foodborne or otherwise can be traced back to cattle [1, 7, 8].
A large body of information on the epidemiology of ECO157 in cattle has accumulated as awareness of human illness due to this pathogen has increased. ECO157 in cattle is known to transmit via the faecal–oral route [9]. Studies on the presence of ECO157 in cattle and their environments have revealed a seasonal pattern with increased prevalence of ECO157 faecal shedding during summer and early autumn [10–12]. Experimental inoculation studies have identified the sites of localization of the bacterium in the gastrointestinal tract of cattle, including the jejunum, ileum, caecum, colon and recto-anal junction [4]. The magnitude and duration of shedding and the associated risk factors have been studied under both experimental [13, 14] and observational [15, 16] settings. A few studies have quantified ECO157 transmission in cattle using mathematical modelling [12, 17, 18]. While these studies have been useful in describing ECO157 transmission dynamics and identified potential control strategies, the transmission rate [often partitioned into the direct and indirect (environmental) rate] used in those models and the contribution of the direct and environmental transmission routes to the overall infection spread, were largely assumed. The transmission rate, β, is defined as the product of contact rate (number of contacts per unit time) and the probability of transmission given the contact [19]. This is a very important quantity in modelling of infectious diseases because it determines the dynamics of the infection spread predicted by the model. Transmission rate may differ for different host–pathogen–environment settings that can be impacted by host population density and age-related susceptibility, pathogen strain colonization ability and environmental factors affecting any of these characteristics. Therefore, in mathematical modelling studies it is important yet challenging to choose a value that closely represents the true rate of transmission.
In 2004, Laegreid & Keen estimated the transmission of ECO157 in beef calves using seroconversion data [20]. More recently, another group used experimental transmission data to quantify ECO157 transmission in dairy calves [21]. However, there are two major problems with the transmission rates reported in these two studies: (i) they did not consider how the level of environmental contamination with the bacterium may influence transmission, which is important given that ECO157 is spread by the faecal–oral route and (ii) they did not consider the possibility of a difference in transmission among strains of ECO157. A recent, experimental inoculation study has highlighted the strain-specific differences in the probability of colonization post-inoculation and in the faecal shedding pattern of colonized cattle [22]. Given that different strains of ECO157 have different host colonization abilities and shedding patterns in cattle, it is likely that transmission dynamics and the transmission rate will also vary between strains of ECO157. Transmission experiments under controlled settings provide a valuable means to study the effect of a few selected factors (e.g. strain type) in which the confounding variation due to other factors is minimized [23].
The objective of this study was to estimate and compare the transmission rates for three different strains of ECO157 in steers under controlled experimental conditions that involved joining susceptible (non-inoculated) steers and steers inoculated with a three-strain mixture of ECO157. The hypotheses to be tested were: (i) that different strains of ECO157 have different rates of transmission from infectious to susceptible animals, and (ii) that the rate of transmission in cattle is affected by the level of ECO157 environmental contamination. In an effort to explain the mechanism by which environmental contamination influences ECO157 transmission in animals, an in vitro experiment was conducted to test survival of the three ECO157 strains in bovine faeces.
MATERIALS AND METHODS
Ethical statement
All procedures were approved by the University of Wisconsin–Madison School of Veterinary Medicine Institutional Animal Care and Use Committee (protocol no. A01388-0-03-09).
Transmission experiment
Twenty-two Holstein steers, aged 6–7 months, were selected for the transmission experiment. These steers had been used previously in a separate study involving single strain challenge experiments with one of three strains of ECO157 (but no infection transmission among animals) [24] and they were reused in the current experiment for ethical reasons (to minimize the number of animals to be sacrificed). Moreover, the experimental design partially controlled for the confounding effect of host factors which might have been introduced by enrolment of different animals. Following the termination of the single strain challenge experiments on 23 July 2010, the steers were cleared of residual infection with ECO157 by orally dosing them with 6 g neomycin (Neomycin Oral Solution 200 mg/ml; AgriLabs, USA), once daily for 5 days. After completing the course of the antibiotic treatment, the steers were confirmed to be clear of ECO157 by testing negative by culture and polymerase chain reaction (PCR) for at least three consecutive recto-anal mucosal swabs (RAMS) taken on alternate days. RAMS were obtained by inserting sterile cotton-tipped applicators 1–2 inches into the rectum and rubbing 3–4 times along the mucosal surface [25]. Out of the 22 steers, 12 were randomly selected and separated for inoculation as the infectious group and housed in two separate pens (six steers each) in one room. Pens within a room were separated by a corridor to prevent contact of animals between groups. The remaining 10 steers were housed in a separate room to serve as the susceptible contacts in two replicates of the transmission experiment. Pen environment was made free of ECO157 by disinfection and ECO157-free status was confirmed by taking environmental samples before housing the steers. The two replicates of the transmission experiment were conducted to capture possible variations between experiments, so that a reliable estimate of transmission could be measured. More than two replicates for the experiment were not feasible due to financial and ethical constraints. On day 0, the 12 steers in the infectious group were inoculated orally with a cocktail of three ECO157 strains, each at a dose of 105 colony-forming units (c.f.u.) [26]. The simultaneous inoculation of three strains was chosen in order to control for the confounding effect of host factors and under the assumption that the strains do not have any effect (synergistic or antagonistic) on each other. Each of the 12 steers was inoculated by mixing 1 ml inoculum with ~10 ml clean tap water in a drinking cup. A procedure described previously [24] was followed to ensure the inoculum was consumed. The infection status of the inoculated steers was determined 1 day post-inoculation (p.i.) by bacteriological culture of RAMS: the steers were considered infectious if they were shedding at least one of the three inoculated strains. The three inoculated strains were FRIK47, FRIK1641 and FRIK2533, which have been described in previous studies [22, 24]. Briefly, the three strains have different genotypes [27] (e.g. FRIK1641 and FRIK2533 lacked stx1 and hly933, respectively) and ecological histories (FRIK47 was involved in a ground beef-associated human outbreak [28], FRIK1641 was recovered from a raccoon and was selected as a representative of non-bovine adapted strains [29], and FRIK2533 was obtained from water on a cattle farm). For brevity, from here onwards we will refer to FRIK47, FRIK1641 and FRIK2533 strains as St1, St2 and St3, respectively. One day after confirmation of the infectious status (i.e. day 2 p.i.), five randomly selected animals each from the susceptible contact group were joined with five randomly selected infectious animals and each newly formed group of 10 animals was housed in one of the two separate rooms. The two groups of steers, each with five infectious and five susceptible steers, established the two replicates of the experiment. For brevity, we will refer to these two groups as G1 and G2. Both groups were followed for 30 days to monitor ECO157 colonization status and environmental contamination over time by testing RAMS and environmental samples, respectively. The length of 30 days was chosen based on the expectation that the duration of ECO157 infection will not exceed 30 days [30]. Shedding of ECO157 in cattle is intermittent in nature [22], and so we adopted a frequency of sampling every other day, which was considered frequent enough to capture the intermittent shedding behaviour and was logistically feasible.
Sampling, isolation and enumeration of ECO157
The protocol for collection of RAMS and environmental samples is described in Kulow et al. [24]. The RAMS from each individual steer and samples from selected environmental locations were collected the day after mixing (i.e. day 3 p.i.) and on alternate days thereafter for the follow-up period, for a total of 15 sampling occasions. Sampled environmental locations included: animal hides, feed troughs, drinking water, water-cup run-offs and pen floors. The details of the performed ECO157 isolation and enumeration and PCR confirmation were as described previously [24]. Briefly, RAMS were placed into a sterile tube containing 5 ml modified E. coli broth (mEC; Remel, USA) and novobiocin (0·02 mg/ml, mECnov; Remel). Drinking water (10 ml) was sampled directly from water cups. To collect samples of hides and feed troughs, we used sterile cellulose sponges (2 × 3·5 × 1 inch) moistened with mECnov broth; after sampling, a sponge was placed in a sterile Whirl-Pak (Nasco, USA) containing 10 ml mECnov broth. Samples from water-cup run-offs and pen floor were collected using sterile nylon ropes about 3-feet long and 8-mm thick (~52 g; Koch Industries, USA). Colonies suspected to be ECO157 were presumptively identified using the RIM E. coli O157:H7 latex test (Remel). For each sample containing colonies that were positive by the latex agglutination test, up to 10 randomly selected colonies were taken for strain differentiation (i.e. to identify St1, St2 and St3) using PCR. Enumeration results for RAMS and environmental samples were expressed as c.f.u./ml. The minimum detection limit was ~2 c.f.u./ml, i.e. 10 c.f.u./RAMS and 10 c.f.u./environmental sample.
Survival of ECO157 in bovine faeces
Bovine faeces collected from a local farm was processed as described previously [31] and the water content of the faeces was reduced to a final aw between 0·87 and 0·90 by heating small aliquots (~20–30 g wet feces) in a glass dish until an aw of 0·87–0·89 was achieved. This was done to prevent the growth of the strains during storage. The aw was determined using an Aqualab model 4TE aw meter (Decagon Devices, USA) according to the manufacturer's instructions. The faeces were then sterilized at 121°C, 15 psi, for 15 min, which did not significantly change the aw. The three bacterial strains (St1, St2, St3) were grown from a single colony in 5 ml Luria Bertani (LB) broth at 37°C for 22–24 h and diluted 1:10 in 1 × PBS to a concentration of ~108 c.f.u./ml. The diluted culture (650 μl) was mixed with 6·5 g dried faeces in sterile dishes and incubated at 30°C for 28 days. Samples were taken on days 0, 1, and 3 during the first week and twice a week thereafter. Samples (0·5 g faeces) were serially diluted in PBS and plated in duplicate on LB agar plates, incubated at 37°C for 22–24 h, and the number of c.f.u. enumerated. The detection limit was 20 c.f.u./g. Three independent experiments were performed.
Mathematical modeling and statistical analysis
To facilitate proper analysis of the data, the infectious status of the individual animals at each sampling day was determined and categorized into one of the three groups: Susceptible (S), Infectious (I) or Latent (L). In this study, Infectious (I) is used to indicate the shedding state of an animal that is capable of infecting (colonizing) susceptible animals. The latent (L) class is used to indicate an infected (colonized) animal during its transient non-shedding periods, when the animal is considered to be non-infectious. Thus, an infected animal is intermittently in either I or L class and a non-infected animal is in S class.
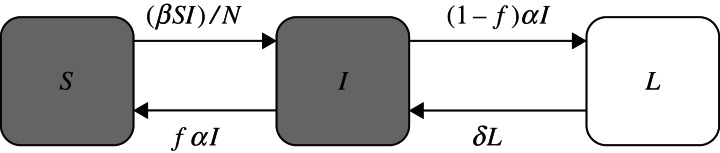
The infection status for each steer on each of the 15 sampling days was determined under the following assumptions that reflected the intermittent shedding pattern of ECO157 by the infected animals as reported in Gautam et al. [22]: (i) an animal testing positive was considered infectious (I), (ii) an animal testing negative could be classified either as latent (L) or susceptible (S) depending on the timing and the number of consecutive samples that were test-negative. If an animal was negative on ⩽4 consecutive samplings (i.e. ⩽8 days) following a positive sample, it was classified as latent (L), otherwise susceptible (S). A steer that was introduced into the experiment as a susceptible animal was considered susceptible until it tested positive for the first time. Similarly, an animal that was found negative on more than four consecutive sampling days (i.e. >8 days) after the last positive test was considered recovered from colonization, and thus susceptible again. If an animal tested negative towards the end of the study and it was not possible to apply the criterion mentioned above to classify the animal into one of the two possible non-shedding classes (L or S) because of too few negative samples, then that particular sampling observation on the individual animal was censored. Using the above information and considering that incidents of new infections (cases) at consecutive samplings are governed by a stochastic process, the transmission of strain-specific ECO157 can be described by a modified ‘Susceptible-Infectious-Susceptible’ (SISL) mathematical model where the Infectious state branches into a Latent state (Fig. 1). Although there were three strains in the transmission experiment, St3 was only detected a few times in only one inoculated steer and was not detected in any of the other inoculated and susceptible steers in the two groups. Since no new cases of cattle colonized with St3 were observed the transmission rate for this strain was considered zero (the statistical estimation of the transmission rate was not possible because no new case was observed).
Fig. 1.
Schematic representation of the SISL transmission model of ECO157 in cattle.
In a closed population of size N = S+I+L = 10, the number of susceptible individuals that may become infectious per time interval, Δt, depends only on the transmission rate β (the rate at which a randomly chosen susceptible animal had successful infectious contacts in the time interval Δt), the number of susceptible (S) animals, the number of infectious (I) animals and the total population size N. If the number of new cases C at the end of each time interval (Δt) is considered a stochastic process based on a binomial distribution with S possible outcomes, then the transmission rate β can be estimated using a function of I, S, C, N and Δt [21] and can be represented for our SISL system as follows:
| (1) |
where S is the number of susceptible S(t) individuals at the start of the interval, I is the average number of infectious I(t) individuals during the interval, N is the population size (here N = 10) and Δt is the sampling interval. Taking log on both sides, equation (1) can be expressed as:
| (2) |
Here and onwards, log denotes the natural logarithm (base e). From equation (2), log(β) can be estimated from the transmission experiment data by using a generalized linear model (GLM) [32]. Equation (2) was extended into a covariate effects model to control for the effect of covariates on log(β) as follows:
| (3) |
The considered covariates were strain, St (St1 or St2; term μ1), experimental group, G (G1 or G2; term μ2), and the strain- and group-specific environmental contamination level on the preceding sampling day (E; term μ3). The level of environmental contamination during the immediately preceding sampling day was used because contamination level on the same day of sampling is unlikely to generate a new case on that very day. Lag times longer than 2 days (i.e. the preceding sampling date) were deemed biologically irrelevant because it has been estimated [22] that shedding occurs at the latest 2 days after successful inoculation. The overall level of environmental contamination for a given sampling day was determined by log10 transformation of the sum of the ECO157 counts from all positive environmental samples for a given strain and group on that day among the collected samples of animal hides, feed troughs, drinking water, water-cup run-offs and pen floor. The considered environmental contamination covariate was a proxy for some true, but impossible to measure, total environmental contamination on a particular day. However, because the measurements were consistently estimated for all strains, experimental groups and days, they were comparable.
The above model [equation (3)] was implemented in R version 2.13.1 (R Development Core Team, 2011) using GLM function with a complementary log-log link. The GLM version of the model equation is:
| (4) |
where C represents the number of new cases, S defines the number of trials of a binomial distribution, St is the strain of ECO157 (with St1 = 0 and St2 = 1), G is the experimental group (with G1 = 0 and G2 = 1), E is the pathogen load in the environment during the immediately previous sampling day (continuous on log10 scale) and offset = log[(I/N)*Δt].
In the offset term, note that if I = 0, this will produce an indeterminate value preventing the GLM from computing the transmission rate. On several sampling days, there were no infectious individuals (i.e. I = 0) for St2 in G1. For those days, in order to facilitate the computation, we added one infectious animal to the state I, and subtracted one from the state S (i.e. we used I+1 and S – 1). The implications of this assumption were assessed by comparing the model results from the full dataset with those produced for a subset of dataset after eliminating observations for St2 in G1.
In the model shown in equation (4) the coefficient λ0 can be interpreted as the transmission rate on the log scale (log(β)) for St1 in G1 when E = 0. The coefficient λ1 is the average difference in log(β) between the two strains of ECO157 after controlling for group and E, λ2 is the average difference in log(β) between the two groups after controlling for strain and E, and λ3 is the average increase in log(β) for a 1-unit increase in E after controlling for group and strain differences. For the specific combination of the group, strain and contamination load in E, log(β) was estimated, the exponentiation of which gives transmission rate, β. The intercept-only form of the model in equation (4) was run on the subsets of data for the individual strains to estimate the actual strain-specific transmission rates in the conducted experiment. It is important to note that β, as defined in this study, is frequency dependent and therefore, the term 1/N is absorbed into its parameterization [19].
The model in equation (4) was used to test the hypotheses that (i) transmission rates differ for two different ECO157 strains (St1, St2) and (ii) environmental contamination level influences the transmission rate of these two ECO157 strains in cattle. These hypotheses were tested by running several GLMs, including models with one covariate at a time (univariate analysis) and models with multiple covariates considered simultaneously (multivariable analysis). In these analyses we considered environmental contamination as a continuous variable (i.e. log10-transformed total ECO157 load) or dichotomized variable (representing whether the environment was contaminated or not with a particular strain on a particular day). When using environmental contamination as a continuous covariate, the validity of the linearity assumption was assessed graphically by plotting, and lowess smoothing, of complementary log-log probability of the outcome variable against the explanatory variable [33, 34]. Brown-Forsythe–Levene's test supported the validity of the assumption of homogeneity of variances between the strain-specific data (P = 0·08). The best-fitting model was selected using Akaike's Information Criterion (AIC = 77·26). Model fit was further evaluated by checking the over-dispersion parameter, which is the ratio of the deviance over the degrees of freedom. The over-dispersion parameter was slightly <1 for the final covariate model and 1 for the two strain-specific intercept models; it was therefore concluded that the final models did not have the over- or under-dispersion problem.
To compare survival of the three ECO157 strains in bovine faeces, a repeated-measures ANOVA was performed on the natural log (base e)-transformed counts of ECO157 for the three strains and the strain-specific decay rates were calculated using the standard exponential growth/decline equation:
where r = decay rate, N(t) = log(c.f.u. ECO157) at time t, N(0) = log(c.f.u. ECO157) at time zero, and t = time (in days). Time (t) was the length of time between the first and the last sampling day in this experiment (i.e. days 0 and 28).
The basic reproduction number (R0)
The SISL system in Figure 1 was solved analytically to obtain an expression for the basic reproduction number (R0). We used the next-generation matrix (NGM) method [35] to derive R0. The NGM can be derived from the transmission matrix F and transition matrix V:
The NGM, H, was obtained by taking the product of matrix F and the inverse of matrix V:
From the spectral radius of matrix H, R0 was determined to be β/αf. In the derived R0 expression 1/α represents the length of the infectious period before moving into the S compartment (indicating recovery) or into the L compartment (indicating temporary cessation of infectiousness). The term f can be interpreted as the weighting variable to account for the fact that only a fraction, f, of animals leaving state I enter into state S while the fraction (1−f) temporarily stops shedding and is thus considered latent (L). Because f reduces the recovery rate, it extends the total duration of infectious period (i.e. 1/αf). Previously, we estimated 1/α for St1 to be 4·3 [95% confidence interval (CI) 0·4–17·2] days and for St2 to be 5·3 (95% CI 0·9–12·8) days [22]. We also estimated f for St1 to be 0·25 and for St2 to be 0·5. These estimates indicate that compared to St2, St1 has on average larger α (i.e. shorter single episode of shedding) but smaller f (i.e. higher probability of extending infection through the mechanism of intermittent shedding) [22]. For an infected individual, the total duration of effective infectious period after discounting for the time spent in L (1/αf), was estimated as 17·2 (95% CI 1·6–68·8) days for St1 and 10·6 (95% CI 1·8–25·6) days for St2. The strain-specific R0 was then calculated using the derived expression R0 = β/αf.
RESULTS
Descriptive results
None of the inoculated and contact animals developed clinical symptoms during the experiments. All inoculated animals in G1 and four inoculated animals in G2 were shedding at least one of the three strains of ECO157 on day 1 p.i. In one of the inoculated steers, shedding was not detected until day 11 p.i. and this animal was considered susceptible until it was first detected as positive (when it was considered a new case). Four inoculated steers in G1 and three inoculated steers in G2 were excreting St1 on day 1 p.i., while only two inoculated steers were shedding St2 in both groups on day 1 p.i. Only one inoculated steer from G1 tested positive for St3 during the entire study period. That steer excreted St3 on days 1, 11, 13 and 15 p.i. at the average shedding level in positive RAMS of 0·95 log10 c.f.u./ml. None of the contact animals in either experimental group excreted St3 during the study period. Overall, average numbers of positive RAMS between the two experimental replicates for St1, St2 and St3 were 56·5, 18·5 and 2·5, respectively. The average (standard deviation, median) shedding level in positive RAMS for St1 was 2·12 (1·32, 1·91) log10 c.f.u./ml in G1 and 2·37 (1·56, 2·21) log10 c.f.u./ml in G2. The average (standard deviation, median) shedding level in positive RAMS for St2 was 1·77 (0·58, 1·64) log10 c.f.u./ml in G1 and 1·77 (1·12, 1·76) log10 c.f.u./ml in G2. The number of steers testing positive and negative for St1 and St2 in the two experimental groups for any given day is given in Table 1.
Table 1.
Experimental infection transmission data for the two strains of ECO157 in each of the two experimental groups (G1 and G2). Inoculated steers were joined with the susceptible group on day 1 post-inoculation
| SDa | Db | G1 (n = 10) | G2 (n = 10) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ECO157 positive | Model input data | ECO157 positive | Model input data | ||||||||||||||||
| ICc | CCd | Ie | Sf | Cg | Lh | dti | Stj | Ek | ICc | CCd | Ie | Sf | Cg | Lh | dti | Stj | Ek | ||
| 0 | 1 | 4/5 | 0/5 | 4 | 6 | — | — | — | St1 | — | 3/5 | 0/5 | 3 | 7 | — | — | — | St1 | — |
| 2 | 3 | 3/5 | 0/5 | 4 | 6 | 0 | 0 | 1 | St1 | — | 3/5 | 0/5 | 3·5 | 7 | 1 | 0 | 1 | St1 | — |
| 4 | 5 | 3/5 | 0/5 | 3 | 7 | 0 | 0 | 2 | St1 | 2·0 | 2/5 | 0/5 | 3 | 6 | 0 | 1 | 2 | St1 | 0 |
| 6 | 7 | 3/5 | 0/5 | 3 | 7 | 0 | 0 | 2 | St1 | 3·2 | 2/5 | 2/5 | 3 | 6 | 2 | 2 | 2 | St1 | 6 |
| 8 | 9 | 3/5 | 1/5 | 4 | 7 | 2 | 0 | 2 | St1 | 2·6 | 4/5 | 3/5 | 4·5 | 4 | 1 | 2 | 2 | St1 | 4·8 |
| 10 | 11 | 2/5 | 1/5 | 4 | 6 | 0 | 0 | 2 | St1 | 3·5 | 2/5 | 0/5 | 7·5 | 3 | 1 | 0 | 2 | St1 | 2·8 |
| 12 | 13 | 2/5 | 1/5 | 3 | 6 | 0 | 1 | 2 | St1 | 0 | 4/5 | 0/5 | 2 | 6 | 0 | 2 | 2 | St1 | 2·9 |
| 14 | 15 | 2/5 | 2/5 | 4 | 6 | 1 | 1 | 2 | St1 | 3·2 | 3/5 | 0/5 | 4 | 6 | 0 | 0 | 2 | St1 | 0 |
| 16 | 17 | 3/5 | 1/5 | 4 | 5 | 0 | 2 | 2 | St1 | 0 | 3/5 | 0/5 | 3 | 7 | 0 | 0 | 2 | St1 | 0 |
| 18 | 19 | 3/5 | 1/5 | 4 | 5 | 0 | 1 | 2 | St1 | 0 | 3/5 | 0/5 | 3 | 7 | 0 | 0 | 2 | St1 | 0 |
| 20 | 21 | 4/5 | 0/5 | 4 | 5 | 0 | 1 | 2 | St1 | 1·9 | 1/5 | 2/5 | 4 | 7 | 2 | 0 | 2 | St1 | 0 |
| 22 | 23 | 4/5 | 1/5 | 4 | 5 | 0 | 1 | 2 | St1 | 0 | 1/5 | 3/5 | 3·5 | 6 | 1 | 1 | 2 | St1 | 3 |
| 24 | 25 | 3/5 | 0/5 | 5 | 5 | 0 | 0 | 2 | St1 | 0 | 1/5 | 2/5 | 4 | 5 | 0 | 1 | 2 | St1 | 0 |
| 26 | 27 | 3/5 | 1/5 | 3 | 6 | 0 | 1 | 2 | St1 | 0 | 2/5 | 3/5 | 3 | 5 | 0 | 2 | 2 | St1 | 0 |
| 28 | 29 | 1/5 | 1/5 | 4 | 6 | 0 | 0 | 2 | St1 | 0 | 1/5 | 3/5 | 5 | 5 | 0 | 0 | 2 | St1 | 0 |
| 30 | 31 | 3/5 | 0/5 | 3 | 6 | 1 | 2 | 2 | St1 | 0 | 2/5 | 3/5 | 4·5 | 5 | 1 | 1 | 2 | St1 | 1·8 |
| 0 | 1 | 2/5 | 0/5 | 2 | 8 | 0 | — | — | St2 | — | 2/5 | 0/5 | 2 | 8 | — | — | — | St2 | — |
| 2 | 3 | 0/5 | 0/5 | 2 | 8 | 0 | 0 | 1 | St2 | — | 1/5 | 1/5 | 2·5 | 8 | 1 | 0 | 1 | St2 | — |
| 4 | 5 | 0/5 | 0/5 | 0 | 10 | 0 | 0 | 2 | St2 | 0 | 1/5 | 2/5 | 2·5 | 8 | 1 | 0 | 2 | St2 | 0 |
| 6 | 7 | 0/5 | 0/5 | 0 | 10 | 0 | 0 | 2 | St2 | 0 | 1/5 | 0/5 | 3 | 7 | 0 | 0 | 2 | St2 | 0 |
| 8 | 9 | 0/5 | 0/5 | 0 | 10 | 0 | 0 | 2 | St2 | 0 | 1/5 | 0/5 | 1 | 8 | 0 | 1 | 2 | S2 | 3·7 |
| 10 | 11 | 0/5 | 0/5 | 0 | 10 | 0 | 0 | 2 | St2 | 0 | 2/5 | 3/5 | 2·5 | 8 | 3 | 1 | 2 | St2 | 4·1 |
| 12 | 13 | 0/5 | 0/5 | 0 | 10 | 0 | 0 | 2 | St2 | 0 | 2/5 | 0/5 | 5·5 | 5 | 1 | 0 | 2 | St2 | 2·7 |
| 14 | 15 | 0/5 | 0/5 | 0 | 10 | 0 | 0 | 2 | St2 | 0 | 2/5 | 1/5 | 2 | 6 | 0 | 2 | 2 | St2 | 0 |
| 16 | 17 | 1/5 | 0/5 | .5 | 10 | 1 | 0 | 2 | St2 | 0 | 2/5 | 0/5 | 3 | 5 | 0 | 2 | 2 | St2 | 0 |
| 18 | 19 | 0/5 | 0/5 | 1 | 9 | 0 | 0 | 2 | St2 | 0 | 2/5 | 2/5 | 2·5 | 6 | 1 | 2 | 2 | St2 | 0 |
| 20 | 21 | 0/5 | 0/5 | 0 | 10 | 0 | 0 | 2 | St2 | 0 | 1/5 | 0/5 | 4 | 5 | 0 | 1 | 2 | St2 | 0 |
| 22 | 23 | 0/5 | 0/5 | 0 | 10 | 0 | 0 | 2 | St2 | 0 | 2/5 | 0/5 | 1 | 8 | 0 | 1 | 2 | St2 | 0 |
| 24 | 25 | 1/5 | 0/5 | .5 | 10 | 1 | 0 | 2 | St2 | 0 | 1/5 | 0/5 | 2 | 8 | 0 | 0 | 2 | St2 | 0 |
| 26 | 27 | 0/5 | 0/5 | 1 | 9 | 0 | 0 | 2 | St2 | 0 | 0/5 | 1/5 | 1·5 | 9 | 1 | 0 | 2 | St2 | 0 |
| 28 | 29 | 0/5 | 0/5 | 0 | 10 | 0 | 0 | 2 | St2 | 0 | 1/5 | 1/5 | 1 | 8 | 0 | 1 | 2 | St2 | 0 |
| 30 | 31 | 0/5 | 0/5 | 0 | 10 | 0 | 0 | 2 | St2 | 0 | 1/5 | 0/5 | 2 | 7 | 0 | 1 | 2 | St2 | 1·2 |
a Sampling day; b days post-inoculation; c inoculated steers; d contact steers; e average number of infectious steers during the time interval before sampling. New cases were assumed to be infectious in between the time interval; f number of susceptible steers at the beginning of the time interval prior to sampling; g number of new cases per time interval; h number of intermittent non-shedding steers at the beginning of the time interval preceding sampling; i time interval between two samplings; j strain of ECO157; k number of ECO157 in the environmental sample during the preceding sampling time in log10 scale.
The environmental samples were collected on 15 sampling days from each of the two groups (G1 and G2) of animals following the mixing of infectious and susceptible steers (Table 2). In G1, St1 and St3 were recovered, respectively, on six and two out of 15 sampling occasions and St2 was never detected in any of the environmental samples. In G2, environmental samples were positive on seven, five and zero sampling occasions for St1, St2 and St3, respectively. For St1, the average load (standard deviation, median) of ECO157 in positive environmental samples was 2·74 (0·67, 2·93) log10 c.f.u./ml in G1 and 3·34 (1·52, 2·92) log10 c.f.u./ml in G2; for St2, the average load (standard deviation, median) was 2·38 (1·10, 2·68) log10 c.f.u./ml in G2 (no samples were positive in G1); and for St3, the average load (standard deviation, median) was 2·73 (0·30, 2·73) log10 c.f.u./ml in G1 (no samples were positive in G2).
Table 2.
ECO157 load (expressed as log10-transformed c.f.u./ml) in the environment during the infection transmission experiment for the two strains (St1 and St2) in each of the two experimental groups by sampling day post-inoculation
| Group | Env. type | Strain | Days post-inoculation | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 5 | 7 | 9 | 11 | 13 | 15 | 17 | 19 | 21 | 23 | 25 | 27 | 29 | 31 | |||
| 1 | Hide | St1 | 1·97 | 0·00 | 1·65 | 0·00 | 0·00 | 2·84 | 0·00 | 0·00 | 1·82 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 |
| 1 | Feeda | St1 | 0·00 | 0·00 | 1·86 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 |
| 1 | Water | St1 | 0·00 | 2·42 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 |
| 1 | Penb | St1 | 0·00 | 3·17 | 2·46 | 3·48 | 0·00 | 2·88 | 0·00 | 0·00 | 0·70 | 0·00 | 0·00 | 0·00 | n.a. | 0·00 | 0·00 |
| 1 | W.cupc | St1 | 0·90 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·90 | 0·00 | 0·00 | 0·00 | n.a. | 0·00 | 0·00 |
| 2 | Hide | St1 | 0·00 | 2·87 | 4·68 | 1·81 | 2·91 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 1·18 | 0·00 |
| 2 | Feeda | St1 | 0·00 | 0·00 | 2·51 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 |
| 2 | Water | St1 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 |
| 2 | Penb | St1 | 0·00 | 6·00 | 4·17 | 2·73 | 0·00 | 0·00 | 0·00 | n.a. | 0·00 | 3·00 | 0·00 | n.a. | 0·00 | n.a. | 1·59 |
| 2 | W.cupc | St1 | 0·00 | 0·00 | 3·61 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | n.a. | 0·00 | 1·71 | 1·34 |
| 1 | Hide | St2 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 |
| 1 | Feeda | St2 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 |
| 1 | Water | St2 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 |
| 1 | Penb | St2 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | n.a. | 0·00 | 0·00 |
| 1 | W.cupc | St2 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | n.a. | 0·00 | 0·00 |
| 2 | Hide | St2 | 0·00 | 0·00 | 0·00 | 4·12 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 1·18 | 0·00 |
| 2 | Feeda | St2 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 |
| 2 | Water | St2 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 |
| 2 | Penb | St2 | 0·00 | 0·00 | 3·72 | 2·73 | 2·68 | 0·00 | 0·00 | n.a. | 0·00 | 0·00 | 0·00 | n.a. | 0·00 | n.a. | 0·00 |
| 2 | W.cupc | St2 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | 0·00 | n.a. | 0·00 | 0·00 | 0·85 |
a Feed trough sample; b pen floor sample; c water-cup run-off sample.
n.a. Sample not collected because the rope was eaten by the steers; St3 was only detected from the environment in group 1 on one occasion from the hide (day 7 post-inoculation) and on two occasions from the pen floor surface (days 3 and 7 post-inoculation) and is not shown in the table.
Results of mathematical and statistical modeling
In the univariate analysis, the effect of environmental contamination level significantly influenced the transmission rate (P = 0·0002), the group effect was marginally significant (P = 0·052) and the strain effect was not significant. All three covariates were used in the multivariable GLM to assess the effect of strain on transmission rate. The average transmission rate per animal per day for St1 in experimental group G1 in the absence of bacterial contamination of the environment was 0·038 (Table 3). The transmission rate for St2 was on average 1·69 times greater than that for St1, albeit not statistically significantly different after controlling for the experimental group and environmental contamination.
Table 3.
Transmission rate (β) with 95% confidence interval (CI) estimated using the generalized linear model with covariates (strain, group, environment)
| Variables | β | 95% CI | P value |
|---|---|---|---|
| Baseline* | 0·038 | 0·0133–0·09 | |
| Strain | |||
| St2 | 1·69 | 0·65–4·37 | 0·27 |
| St1 | — | — | |
| Group | |||
| G2 | 1·56 | 0·58–4·61 | 0·4 |
| G1 | — | — | |
| E† | 1·5 | 1·19–1·9 | 0·0006 |
Corresponds to estimate for St1 in group 1 at log10 (env) = 0.
ECO157 load in the environment during the preceding sampling time (i.e. 2 days prior) in log10 scale.
After controlling for the strain and group factors, contamination of the environment during the immediately preceding sampling day significantly increased the transmission rate. On average, the transmission rate increased 1·5 times for every 1-unit (on log10 scale) increase in the environmental contamination. Table 4 shows the increase in the mean transmission rate β and the corresponding R0 for St1 and St2 for increasing levels of environmental contamination as predicted by the model. Table 5 shows the transmission rate and the corresponding R0 for the two strains describing the overall dynamics in the conducted experiments that accounts for the strain-specific levels of environmental contamination over time implicitly. The transmission rate estimated for St2 after eliminating observations from G1 slightly increased the estimated value, but did not alter the result significantly. This confirmed that adding an infectious and subtracting a susceptible animal for St2 on days when there were no detected infectious animals in G1 did not affect the conclusions of the study.
Table 4.
Transmission rate (β) and basic reproduction number (R0) with 95% confidence intervals (CI) for different levels of environmental contamination (E) represented in log10 scale
| Strain | Group | E = 0 | E = 1 | E = 3 | E = 5 | ||||
|---|---|---|---|---|---|---|---|---|---|
| β (95% CI) | R0 (95% CI) | β | R0 | β | R0 | β | R0 | ||
| St1 | G1 | 0·038 (0·013–0·09) | 0·65 (0·22–1·55) | 0·057 | 0·98 | 0·171 | 2·94 | 0·285 | 4·90 |
| G2 | 0·060 (0·008–0·42) | 1·03 (0·14–7·22) | 0·09 | 1·55 | 0·27 | 4·64 | 0·45 | 7·74 | |
| All | 0·045 (0·018–0·097) | 0·77 (0·31–1·67) | 0·068 | 1·17 | 0·203 | 3·49 | 0·338 | 5·81 | |
| St2 | G1 | 0·064 (0·009–0·39) | 0·68 (0·10–4·13) | 0·096 | 1·02 | 0·288 | 2·42 | 0·48 | 5·10 |
| G2 | 0·096 (0·005–1·82) | 1·02 (0·05–19·29) | 0·144 | 1·21 | 0·432 | 4·58 | 0·72 | 7·63 | |
| All | 0·086 (0·013–0·46) | 0·91 (0·14–4·88) | 0·129 | 1·37 | 0·387 | 4·10 | 0·645 | 6·84 | |
Table 5.
Transmission rate (β) and basic reproduction number (R0) with 95% confidence intervals (CI) for the two strains of ECO157 estimated using the intercept-only generalized linear model
| Strain | Mean β (95% CI) | Mean R0 (95% CI) |
|---|---|---|
| St1 | 0·11 (0·06–0·18) | 1·89 (1·03–3·10) |
| St2 | 0·14 (0·07–0·24) | 1·48 (0·74–2·54) |
ECO157 survival in bovine feces
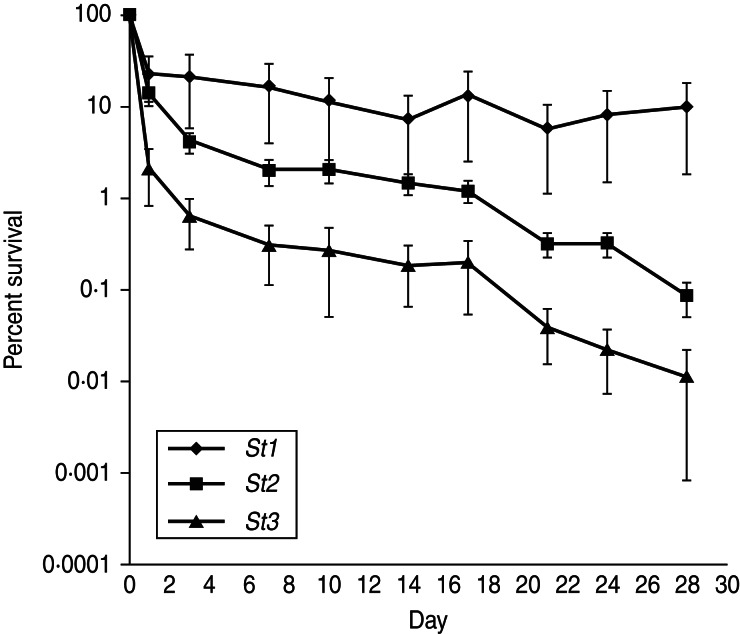
The average number of c.f.u.s of St1, St2 and St3 in faeces over a 28-day storage period is shown in Table 6 and the percent survival of each strain over time is presented in Figure 2. The growth of the three strains in LB was similar which resulted in essentially equal starting numbers of c.f.u.s (viable counts) at day 0, suggesting that the three strains have similar growth rates in the conditions tested. During the first 24 h post-inoculation in faeces, all three strains displayed around a 1-log reduction in viable counts with an average percent survival of 22%, 13% and 2%, respectively for St1, St2 and St3. After this initial decrease, the viable numbers of St1 remained relatively constant over the 28 days of incubation (2·54 × 106 c.f.u./g, 10% survivors). By contrast, St3 had the initial 1-log reduction followed by an additional 3-log reduction in viable counts during the 28 days of incubation (7·43 × 103 c.f.u./g, 0·01% survivors). The survival curve of St2 was in between those of St1 (best survival) and St3 (poorest survival) and had a 3-log reduction during the 28 days of incubation (2·79 × 104 c.f.u./g, 0·08% survivors). The mean decay rates estimated for St1, St2 and St3 were 0·21, 0·26, 0·39, respectively. Nevertheless, the results of the repeated-measures ANOVA and comparison of decay rates did not confirm existence of a significantly different survival in the three strains.
Table 6.
Average c.f.u. count with the standard deviation (s.d.), expressed per million (106), for three ECO157 strains, St1, St2 and St3, in bovine faeces during the survivability assay
| Day | St1 | St2 | St3 | |||
|---|---|---|---|---|---|---|
| Mean | s.d. | Mean | s.d. | Mean | s.d. | |
| 0 | 38·40 | 17·40 | 35·10 | 9·31 | 39·50 | 24·10 |
| 1 | 6·61 | 4·24 | 4·55 | 0·92 | 1·17 | 1·58 |
| 3 | 5·78 | 6·53 | 1·33 | 0·32 | 0·33 | 0·42 |
| 7 | 4·44 | 5·46 | 0·64 | 0·25 | 0·18 | 0·24 |
| 10 | 3·05 | 3·92 | 0·68 | 0·24 | 0·16 | 0·26 |
| 14 | 1·90 | 2·50 | 0·47 | 0·12 | 0·10 | 0·15 |
| 17 | 3·42 | 4·76 | 0·40 | 0·12 | 0·12 | 0·17 |
| 21 | 1·51 | 2·11 | 0·10 | 0·03 | 0·02 | 0·03 |
| 24 | 2·09 | 2·94 | 0·10 | 0·04 | 0·01 | 0·02 |
| 28 | 2·54 | 3·59 | 0·03 | 0·02 | 0·01 | 0·01 |
Fig. 2.
Comparison of the percent survival of the three ECO157 strains (St1, St2, St3) in sterilized bovine faeces over time.
DISCUSSION
The aim of this study was to quantify and compare the transmission rates of different strains of ECO157 in steers through a combination of an infection transmission experiment and mathematical modelling. The main finding of the study strongly supports the hypothesis that the transmission rate significantly increases with increasing levels of environmental contamination. Two of the three tested strains had similar transmission rates, but the third did not transmit at all, supporting the hypothesis that ECO157 strains are characterized by different transmission rates in the cattle population. In the following sections, we will describe these findings in the context of existing information and discuss important implications, strengths and limitations.
Previously, Laegreid & Keen [20] estimated R0 for ECO157 in beef calves in a cow-calf operation to be 5·25 (95% CI 3·87–6·64) based on time to seroconversion after birth, using final size of the infected population. This means that one calf could contribute to at most one case when it seroconverted for ECO157 and could not account for re-infection. In our study, we allowed the animals to contribute to more than one case by permitting them to join the susceptible group immediately after recovery from infection. Later, another group [21] estimated R0 of ECO157 in dairy calves to be 7·3, based on the transmission experiment data under field conditions (i.e. the calves were raised in outside pasture) with a sampling interval of 1 day, and in dairy cattle to be 0·7, based on the shedding prevalence with a sampling interval of 1 month. The sampling interval of 1 month that they used in dairy cattle is longer than the average duration of infectiousness for two strains of ECO157 we estimated in a previous study [22] (17·2 days for St1 and 10·6 days for St2), which may have resulted in the R0 estimate of <1 in dairy cattle. Furthermore, those studies only considered the infectious animals in the transmission model and did not consider the influence of ECO157 load in the environment; neither did they consider the intermittent shedding pattern explicitly in the model. In the current study we have included the effect of environmental contamination in the estimation of transmission rate (and showed how ECO157 transmission in cattle is affected by the level of bacterial contamination in the barn environment) and explicitly represented the intermittent shedding pattern in the model. If the duration of infectiousness is not corrected for the intermittent shedding pattern (i.e. by discounting the period of non-shedding), it will lead to overestimation of R0. The differences between our and their estimated values of R0 could be partly due to the differences in the level of environmental contamination with the bacterium, due to strain differences and/or the experimental setting. Regarding intermittent shedding as modelled in our study, it is possible that the recorded intermittent shedding actually represented shedding that alternated below and above the detection level. However, the detection level in our study was relatively low (2 c.f.u./ml). Therefore, even if an animal recorded as negative was actually shedding at a level of <2 c.f.u./ml, we believe that such an animal was less infectious than the animals that tested positive and that it is appropriate to consider such an animal as non-infectious.
Although not significantly different, the mean transmission rate for St1 was smaller than that estimated for St2. However, due to a substantially longer duration of infectiousness of St1, the mean R0 values computed for this strain for several different fixed values of environmental contamination were similar to those estimated for St2 (Table 4). Furthermore, the overall R0 estimated for St1 was higher than that estimated for St2 (Table 5), which may be explained by a higher average load of St1 in positive environmental samples (2·74 log10 c.f.u./ml in G1 and 3·34 log10 c.f.u./ml in G2) compared to St2 (no positives in G1 and 2·38 log10 c.f.u. in G2). According to Velthuis et al. [36], our study had a very low power to detect a difference between St1 and St2 even if such a difference exists, but would have had a reasonable power (up to 80%) to detect a difference between St3 and either of the strains St1 or St2 should the data for St3 have allowed statistical comparison. That said, detecting a small difference in strains, such as that between St1 and St2, would have a limited practical value from the control point of view and would not be of interest.
The estimate of the transmission rate for the two strains when the environment is completely clean may represent the contribution of direct transmission from an infectious to susceptible animal, or somewhat less likely although not impossible, indirect transmission through environmental locations not represented in our sampling plan. When E = 0, the corresponding R0 values for the strains were <1 suggesting that infection would die out if there is only direct transmission present. With an increase in the contamination level, R0 quickly exceeds 1, indicating the critical role of bacterial concentration in the environment in causing an outbreak and sustaining transmission. Provided that our results are generalizable to field conditions, the identified influence of environmental contamination in the spread of ECO157 indicates the importance of control strategies which either aim to decrease the frequency or rate of ECO157 faecal shedding, such as vaccination [31] or promote elimination of the pathogen from the environment such as through barn cleaning [37]. Moreover, the results indicate a major role of environmental transmission and only a minimal role of direct transmission in the epidemiology of ECO157 in cattle. These results have implications for the current practice of mathematical modelling in epidemiology of ECO157 in cattle. Assuming that the results obtained for the ECO157 strains and experimental conditions evaluated in this study hold for all ECO157 strains and in field conditions, the results suggest that the infection spread should be modelled with a predominant rate of environmental transmission (i.e. occurring through the contaminated environment) while the rate of direct transmission should be negligible. This is important because mathematical models of ECO157 in cattle have largely used assumed arbitrary values for the rate of direct and environmental transmission [12, 18, 38].
From previous studies [22, 24] we know that host-to-host variation plays an important role in ECO157 colonization and shedding. Accordingly, it was important to control for the confounding effect of host factors in the infection transmission experiment. However, it is also important to understand whether there could be any between-strain interactions (e.g. competition) that would mask the true colonization or transmission characteristics of the strains represented in this study. While we did not have the means to verify whether transmission could have been different if the steers were infected with a single strain, we did have the data from a separate single strain inoculation experiment [22, 24] to compare colonization and shedding patterns of these strains on the same experimental animals. The shedding and colonization pattern for the three strains in steers inoculated with a mixture of strains compared closely with the shedding and colonization pattern when steers were inoculated with a single strain. For example, St3 was not shed by the inoculated steers during the single strain inoculation experiment [24] and it was not shed by the steers when inoculated with a mixture of strains. St1 showed better colonization ability with more episodes of intermittent shedding than St2 in the single strain experiment [22] and a similar pattern was observed in the steers inoculated with the mixture of strains in the transmission experiment. Based on these similarities, we feel comfortable assuming that transmission characteristics would have been similar had the transmission experiment been conducted separately for St1, St2 and St3 on these same animals (which would have been flawed because of the effect of age of the steers on ECO157 colonization and shedding patterns [39, 40]).
We used a cut-off of four sampling days to determine whether a steer was latent or recovered, very similar to what has been described by previous studies [21, 41]. It is possible, but not likely, that the cut-off choice was incorrect to the extent that it would significantly alter the results. All animals were rigorously tested to ensure they were free of ECO157 at the beginning of the experiment. Considering that the test used could detect ECO157 at a concentration of ⩾10 c.f.u./RAMS, it is highly unlikely that any steer would continue to shed below this level for three consecutive sampling days (6 days) without clearing from infection. False-negative results, if any, would therefore be minimal and unlikely to have affected the findings of the experiment.
In an attempt to explain the results of the infection transmission experiment described here we conducted assays of survivability of the three ECO157 strains in bovine faeces. The observed trend suggested that St1 survived better than St2 and even more so than St3 (Fig. 2), which would explain why St1 was isolated more often than St2 (and substantially more often than St3) from the environmental samples during the transmission experiment. However, despite the trends, no statistically significant difference was found in the strains which may have been due to the low number of replicates or because there truly is no difference.
In summary, the results indicate that one of the three tested strains did not transmit at all in cattle while the other two strains had considerable, but indistinguishable, transmission rates. For the two transmitted strains, the main finding is that the level of environmental contamination strongly, positively influences the rate of infection transmission in animals. One of the two strains (St1) had on average higher R0 values compared to the other strain (St2), which was attributed to its longer duration of infectiousness and the ability of this strain to maintain a higher load of the pathogen in the environment. This implies that the on-farm measures to reduce shedding duration and decrease the pathogen load in the environment are important in the transmission and control of ECO157 in cattle. It also has implications on the practice of mathematical modelling of ECO157 spread and the underlying understanding of the contribution of the direct and environmental transmission to the overall infection spread.
ACKNOWLEDGEMENTS
We thank Amanda Yang who performed the in vitro competition experiments of the ECO157 strains at the University of Wisconsin (UW). We also thank Kelly Anklam, and Kelly M. Pertzborn for their assistance with the in vivo ECO157 transmission experiment at the UW and the personnel from the Livestock Laboratory at the UW–Madison for their assistance in handling and disposal of animals. The authors thank two anonymous reviewers for their excellent suggestions on how to improve the manuscript.
This work was supported by the National Science Foundation grants NSF-EF-0913042 to D.D. and NSF-EF-0913367 to R.I. funded under the American Recovery and Reinvestment Act of 2009.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Valcour JE, et al. Associations between indicators of livestock farming intensity and incidence of human Shiga toxin-producing Escherichia coli infection. Emerging Infectious Diseases 2002; 8: 252–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tesh VL, Obrien AD. The pathogenic mechanisms of Shiga toxin and the Shiga-like toxins. Molecular Microbiology 1991; 5: 1817–1822. [DOI] [PubMed] [Google Scholar]
- 3.Chapman PA, et al. Cattle as a possible source of verocytotoxin-producing Escherichia-coli-O157 infections in man. Epidemiology & Infection. 1993; 111: 439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baines D, Lee B, McAllister T. Heterogeneity in enterohemorrhagic Escherichia coli O157:H7 fecal shedding in cattle is related to Escherichia coli O157:H7 colonization of the small and large intestine. Canadian Journal of Microbiology 2008; 54: 984–995. [DOI] [PubMed] [Google Scholar]
- 5.Crump JA, et al. An outbreak of Escherichia coli O157: H7 infections among visitors to a dairy farm. New England Journal of Medicine 2002; 347: 555–560. [DOI] [PubMed] [Google Scholar]
- 6.Scallan E, et al. 2011 Foodborne illness acquired in the United States – major pathogens. Emerging Infectious Diseases 2011; 17: 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michel P, et al. Temporal and geographical distributions of reported cases of Escherichia coli O157 : H7 infection in Ontario. Epidemiology & Infection 1999; 122: 193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffin PM, Tauxe RV. The epidemiology of infections caused by Escherichia-coli O157-H7, other enterohemorrhagic Escherichia-coli, and the associated hemolytic uremic syndrome. Epidemiologic Reviews 1991; 13: 60–98. [DOI] [PubMed] [Google Scholar]
- 9.Zhang XS, Woolhouse ME. Escherichia coli O157 infection on Scottish cattle farms: dynamics and control. Journal of Royal Society Interface. 2010; 8: 1051–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barkocy-Gallagher GA, et al. Seasonal prevalence of Shiga toxin-producing Escherichia coli, including O157:H7 and non-O157 serotypes, and Salmonella in commercial beef processing plants. Journal of Food Protection 2003; 66: 1978–1986. [DOI] [PubMed] [Google Scholar]
- 11.Hancock DD, et al. A longitudinal study of Escherichia coli O157 in fourteen cattle herds. Epidemiology & Infection 1997; 118: 193–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gautam R, et al. Modeling the effect of seasonal variation in ambient temperature on the transmission dynamics of a pathogen with a free-living stage: example of Escherichia coli O157:H7 in a dairy herd. Preventive Veterinary Medicine 2011; 102: 10–21. [DOI] [PubMed] [Google Scholar]
- 13.Brown C, et al. Experimental Escherichia coli O157:H7 carriage in calves. Applied Environmental Microbiology 1997; 63: 27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cray W Jr, Moon H. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Applied Environmental Microbiology 1995; 61: 1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chase-Topping ME, et al. Risk factors for the presence of high-level shedders of Escherichia coli O157 on Scottish farms. Journal of Clinical Microbiology 2007; 45: 1594–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn JR, Thompson RA. Prevalence of Shiga-toxigenic Escherichia coli O157:H7 in adult dairy cattle. Journal of American Veterinary Medical Association 2004; 224: 1151–1158. [DOI] [PubMed] [Google Scholar]
- 17.Matthews L, et al. Heterogeneous shedding of Escherichia coli O157 in cattle and its implications for control. Proceedings of the National Academy of Sciences USA 2006; 103: 547–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner J, et al. A model appropriate to the transmission of a human food-borne pathogen in a multigroup managed herd. Preventive Veterinary Medicine 2003; 57: 175–198. [DOI] [PubMed] [Google Scholar]
- 19.Keeling MJ, Rohani P. Modeling Infectious Diseases in Humans and Animals. New Jersey: Princeton University Press, 2008, pp. 17. [Google Scholar]
- 20.Laegreid WW, Keen JE. Estimation of the basic reproduction ratio (R0) for Shiga toxin-producing Escherichia coli O157:H7 (STEC O157) in beef calves. Epidemiology & Infection 2004; 132: 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schouten JM, et al. Transmission and quantification of verocytotoxin-producing Escherichia coli O157 in dairy cattle and calves. Epidemiology & Infection 2009; 137: 114–123. [DOI] [PubMed] [Google Scholar]
- 22.Gautam R, et al. The strain-specific dynamics of Escherichia coli O157:H7 faecal shedding in cattle post inoculation. Journal of Biological Dynamics 2012; 6: 1052–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Jong MC, Kimman TG. Experimental quantification of vaccine-induced reduction in virus transmission. Vaccine. 1994; 12: 761–766. [DOI] [PubMed] [Google Scholar]
- 24.Kulow MJ, et al. Differences in colonization and shedding patterns after oral challenge of cattle with three Escherichia coli O157:H7 strains. Applied Environmental Microbiology 2012; 78: 8045–8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rice DH, et al. Rectoanal mucosal swab culture is more sensitive than fecal culture and distinguishes Escherichia coli O157:H7-colonized cattle and those transiently shedding the same organism. Journal of Clinical Microbiology 2003; 41: 4924–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Besser TE, et al. Escherichia coli O157:H7 infection of calves: infectious dose and direct contact transmission. Epidemiology & Infection 2001; 127: 555–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzales TK, et al. A high-throughput open-array qPCR gene panel to identify, virulotype, and subtype O157 and non-O157 enterohemorrhagic Escherichia coli. Molecular and Cellular Probes 2011; 25: 222–230. [DOI] [PubMed] [Google Scholar]
- 28.Perna NT, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 2001; 409: 529–533. [DOI] [PubMed] [Google Scholar]
- 29.Shere JA, Bartlett KJ, Kaspar CW. Longitudinal study of Escherichia coli O157:H7 dissemination on four dairy farms in Wisconsin. Applied Environmental Microbiology 1998; 64: 1390–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shere JA, et al. Shedding of Escherichia coli O157:H7 in dairy cattle housed in a confined environment following waterborne inoculation. Applied Environmental Microbiology 2002; 68: 1947–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stasic AJ, Lee Wong AC, Kaspar CW. Osmotic and desiccation tolerance in Escherichia coli O157:H7 requires rpoS (sigma(38)). Current Microbiology 2012; 65: 660–665. [DOI] [PubMed] [Google Scholar]
- 32.Becker NG. Analysis of Infectious Disease Data. London, New York: Chapman and Hall, 1989, pp. 1–223. [Google Scholar]
- 33.Dohoo IR, Martin W, Stryhn H. Veterinary Epidemiologic Research, 2nd edn. Prince Edward Island, Canada: VER Incorporated, 2010. [Google Scholar]
- 34.Park S, et al. Generic Escherichia coli contamination of spinach at the preharvest stage: effects of farm management and environmental factors. Applied Environmental Microbiology 2013; 79: 4347–4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van den Driessche P, Watmough J. Reproduction numbers and sub-threshold endemic equilibria for compartmental models of disease transmission. Mathematical Biosciences 2002; 180: 29–48. [DOI] [PubMed] [Google Scholar]
- 36.Velthuis AGJ, et al. Design and analysis of small-scale transmission experiments with animals. Epidemiology & Infection 2007; 135: 202–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, et al. A stochastic model for transmission, extinction and outbreak of Escherichia coli O157:H7 in cattle as affected by ambient temperature and cleaning practices. Journal of Mathematical Biology. Published online: 18July2013. doi: 10.1007/s00285-013-0707-1 [DOI] [PubMed] [Google Scholar]
- 38.Turner J, et al. A network model of E. coli O157 transmission within a typical UK dairy herd: the effect of heterogeneity and clustering on the prevalence of infection. Journal of Theoretical Biology 2008; 254: 45–54. [DOI] [PubMed] [Google Scholar]
- 39.Cho S, et al. Cattle-level risk factors associated with fecal shedding of Shiga toxin-encoding bacteria on dairy farms, Minnesota, USA. Canadian Journal of Veterinary Research 2009; 73: 151–156. [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen EM, et al. Influence of age, sex and herd characteristics on the occurrence of Verocytotoxin-producing Escherichia coli O157 in Danish dairy farms. Veterinary Microbiology 2002; 88: 245–257. [DOI] [PubMed] [Google Scholar]
- 41.Jeong KC, et al. Reduction of Escherichia coli O157:H7 shedding in cattle by addition of chitosan microparticles to feed. Applied & Environmental Microbiology 2011; 77: 2611–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]