Abstract
Genetically distinct strains of the plant bacterium Xylella fastidiosa (Xf) are responsible for a variety of plant diseases, accounting for severe economic damage throughout the world. Using as a reference the genome of Xf 9a5c strain, associated with citrus variegated chlorosis (CVC), we developed a microarray-based comparison involving 12 Xf isolates, providing a thorough assessment of the variation in genomic composition across the group. Our results demonstrate that Xf displays one of the largest flexible gene pools characterized to date, with several horizontally acquired elements, such as prophages, plasmids, and genomic islands (GIs), which contribute up to 18% of the final genome. Transcriptome analysis of bacteria grown under different conditions shows that most of these elements are transcriptionally active, and their expression can be influenced in a coordinated manner by environmental stimuli. Finally, evaluation of the genetic composition of these laterally transferred elements identified differences that may help to explain the adaptability of Xf strains to infect such a wide range of plant species.
[Supplemental material is available online at www.genome.org.]
The xylem-inhabiting bacterium Xylella fastidiosa (Xf; Hopkins and Mollenhauer 1973; Wells et al. 1987) has emerged as an important phytopathogen, implicated in the development of citrus variegated chlorosis (CVC) in orange trees, Pierce's disease (PD) in vineyards, phony peach disease (PP), periwinkle wilt and leaf scorch diseases in plum, elm, maple, oak, sycamore, and coffee (Hopkins 1989; Purcell and Hopkins 1996; Purcell et al. 1999; Hopkins and Purcell 2002). Due to the presence of economically important crops in this list, Xf has been the subject of increasing attention in recent years (Hartung et al. 1994; Chen et al. 1995; Pooler and Hartung 1995; Banks et al. 1999), leading to the complete genomic sequencing of a strain (9a5c) associated with CVC in Brazil, which turned out to be the first plant bacterium to have its complete genome elucidated. This work also involved an extensive in silico evaluation of the bacterium's presumed proteome, allowing the formulation of a virtual metabolome that provided a comprehensive view of the major biochemical processes that occur in this microorganism (Simpson et al. 2000).
The existence of different disease symptoms observed in a wide range of plant hosts (Pooler and Hartung 1995; Chen et al. 2000), possibly associated with genetically distinct Xf strains, has led researchers to hypothesize that total genome comparisons among such strains may uncover important information regarding genes involved in the interaction with specific hosts. These results could provide the basis for further epidemiological studies and more efficient control of these phytopathogens (Hendson et al. 2001). Thus, the genomes of two other strains isolated from oleander and almond trees were partially sequenced and annotated (http://www.jgi.doe.gov/jgi_microbial/html), and a fourth strain, isolated from grapevines and responsible for PD in California, has also been sequenced to completion (http://www.lbi.ic.unicamp.br/world/xf-grape). Total genome comparisons have been performed among these strains, indicating the presence of specific gene sets for each isolate (Bhattacharyya et al. 2002a,b; Costa de Oliveira et al. 2002). However, no attempt has been made to gather information about specific gene expression modulation under controlled experimental conditions. This problem becomes more relevant since the annotated genomes point to the existence of a high percentage of open reading frames (ORFs) whose functions are still unknown (over 50% of the genome from strain 9a5c), whereas genes assigned with putative functions may be associated with several, nonexclusive, possible pathogenicity mechanisms.
In this report, a spotted microarray carrying approximately 2200 ORFs from the genome of Xf 9a5c strain (Simpson et al. 2000) was hybridized with DNA from 11 Xf strains, obtained from different plant hosts and geographic origins. The results revealed the existence of a highly conserved core gene pool, containing all genes involved in the major biochemical pathways and cellular functions, which are likely to determine the adaptation of Xf to life in the plant xylem. In contrast, we found evidence that Xf displays a large flexible gene pool, characterized by the presence of several clusters containing foreign genes. Some of these clusters fulfilled the designation of genomic islands (GI), whereas others were considered putative GIs (Hacker and Kaper 2000; Hacker and Carniel 2001). The major GIs were consistently found in the citrus and coffee strains, but not in those obtained from other hosts. Moreover, transcriptome analysis revealed coordinated transcription of most genes within GIs and prophages, indicating that gene expression in these horizontally transferred elements can be controlled by growth conditions and, perhaps, other environmental stimuli. The number of genes contained in GIs, prophages, and plasmids indicates that Xf carries one of the largest flexible gene pools described to date, and dissection of its composition helps to explain the efficient adaptability of the group to infect such a wide range of plant species.
RESULTS
Genomic Comparison of Xf Strains
Evidence for the existence of laterally transferred genes in Xf was first reported by Simpson et al. (2000), who mapped the presence of two ORFs, originally found in Dichelobacter nodosus and Ralstonia eutropha, within cryptic prophages found in the genome of Xf strain 9a5c. Recent genome comparisons involving Xf strains obtained from citrus, almond, and oleander plants broadened this scenario, suggesting that a series of conjugation-related ORFs found in Xf strains could have been acquired from soil-inhabiting bacteria (Bhattacharyya et al. 2002a). Moreover, a large (∼67-kb) element present in the genome of Xf strain 9a5c was identified as a new prophage, probably derived from the Siphophage group of double-stranded bacteriophages (Bhattacharyya et al. 2002b).
However, these comparisons employed only in silico approaches and therefore, relied solely on information derived from completely or partially sequenced genomes, limiting the scope of the observations to a small number of strains. In the present work, a microarray hybridization approach allowed comparison of the genome from strain 9a5c with a significantly larger set of Xf strains, obtained from different hosts: two strains from citrus, plum, and grapevine and one strain from almond, coffee, elm, mulberry, and ragweed (Table 1). The results obtained allowed detection of missing and duplicated genes across the 12 Xf strains, generating the genomic profiles shown in Figure 1B (all genetic differences observed in the analyzed strains can be found in the supplemental material). Although the precise genomic composition seems to be unique for each strain, one can clearly visualize an overall pattern that differentiates the citrus/coffee group (group 1) from the isolates obtained from plum, ragweed, grape, mulberry, elm, and almond (group 2).
Table 1.
Xylella fastidiosaStrains Analyzed
| Strain | Host | Origin | Source |
| 9a5c (Citrus CVC) | Citrus sinensis cv. Valência | Macaubal, SP, Brazil | INRAa |
| SJ (Citrus SJ) | Citrus sinensis cv. Lima | Taquaritinga, SP, Brazil | UNAERPb |
| B14 (Citrus X1) | Citrus sinensis cv. Lima | Bebedouro, SP, Brazil | Centro APTAc |
| C13 (Coffee) | Coffea arabica | Cordeirópolis, SP, Brazil | Centro APTA |
| 9746 (Plum I) | Prunus salicina | Ponta Grossa, PR, Brazil | IAPARd |
| 35868 (Mulberry) | Morus rubra | Massachusetts, USA | ATCCe |
| 35870 (Almond) | Prunus amygdalus | California, USA | ATCC |
| 35871 (Plum II) | Prunussp. | Georgia, USA | ATCC |
| 35873 (Elm) | Ulmus americana | Washington, USA | ATCC |
| 35876 (Ragweed) | Ambrosia artemisiifolia | Florida, USA | ATCC |
| 35881 (Grape F1) | Vitis sp. | Florida, USA | ATCC |
| 700964 (Grape CA) | Vitis sp. | Temecula, CA, USA | ATCC |
INRA, Institut National de La Recherche Agronomique, Bordeaux, France.
UNAERP, Unidade da Biotecnologia, Universidade de Ribeirão Preto, SP, Brazil.
Centro APTA Citros Sylvio Moreira, Instituto Agronômico de Campinas, Cordeirópolis, SP, Brazil.
IAPAR, Instituto Agronômico do Paraná, Paraná, Brazil.
ATCC, American Type Culture Collection, Manassas, VA, USA.
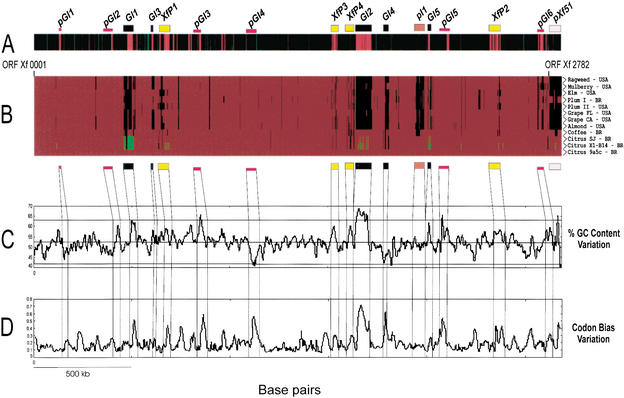
Figure 1.
A comparative and functional analysis of X. fastidiosa genomes. (A) The relative chromosomal position of genes that showed transcription upregulation in PW (red bars) and XDM (green bars) media. Most PW-overexpressed genes are located in clusters that coincide with prophages or GI regions. (B) The genomic profiles among 12 Xf isolates obtained from different hosts and disparate geographical origins. DNA from these isolates was labeled with Cy5-dCTP and submitted to competitive microarray hybridization with Cy3-labeled DNA from Xf strain 9a5c. ORFs were categorized as common to the two strains, exclusive to Xf strain 9a5c, or duplicated in the test strain, depending on the specific Cy5/Cy3 ratio. The figure shows the presence/absence of ∼2200 ORFs from Xf strain 9a5c across the genome of all tested strains. A red bar indicates the presence of a gene, a black bar, absence of the gene, and duplicated sequences are represented by a green bar. (C,D) The CG content and codon bias variations in strain 9a5c, respectively. The figure also indicates the relative positions of five genomic islands (GI1–GI5), six putative GIs (pGI1–pGI6), four integrated prophages (XfP1–XfP4), one plasmid integration region (pI1), and pXf51 plasmid, identified in the genome of Xf strain 9a5c.
Differences between the two groups were mainly associated with large sets of missing/duplicated ORFs, and some of these regions mapped within the laterally transferred genomic elements mentioned above. To verify whether other missing/duplicated regions could be the result of previously unnoticed prophages and GIs, we analyzed codon bias and GC content variation along the sequence of the Xf strain 9a5c chromosome (Fig. 1C,D). These analyses identified several regions where altered codon usage and variable GC content were simultaneously observed, and some of these regions were associated with chromosomal locations where missing/duplicated ORFs were mapped during our genomic comparisons. Four regions are related to integrated prophage sequences (XfP1, XfP2, XfP3, and XfP4) already described by Simpson et al. (2000), whereas others display several characteristics of GIs, as verified in other systems (Table 2; Hacker and Carniel 2001).
Table 2.
General Characteristics of Putative Horizontally Transferred Elements Found in the Genome of Xf Strain 9a5c
| Element | Integrase-like gene at the end | Next to tRNA | GC content variation | Codon bias | Overexpressed in PW medium | Relevant resident genes |
| GI1 XF0480-0536 | Transposases A and B | Asn-tRNA (5′) | Yes | Yes | Yes | 5 phage-related proteins 1 fimbrillin gene 1 virulence associated protein (VapE) |
| GI2 XF1718-1793 | Phage-related integrase | Gly-tRNA (5′) Thr-tRNA (3′) | Yes | Yes | Yes | 12 dehydrogenases/keto-reductases 1 fimbrillin gene 8 transcriptional regulators |
| GI3 XF0631-00641 | Phage-related integrase | Ser-tRNA (3′) | No | No | Yes | 1 DNA modification enzyme |
| GI4 XF1859-1885 | No | Lys-tRNA (5′) | Yes | Yes | No | 5 phage-related proteins from bacteriophage Pf3 |
| GI5 XF2108-2132 | Phage-related integrase | Cys-tRNA (5′) | Yes | Yes | Yes | 1 phage-related repressor protein 1 virulence-associated protein (VapE) |
| pGI1 XF0126-0140 | No | Ser-tRNA (5′) | No | Small | Yes | 2 copper resistance proteins 1 DNA polymerase III subunit |
| pGI2 XF0393-0437 | No | Gln-tRNA (5′) | Yes | Small | Partially at 3′ end | 4 toluene tolerance proteins 5 membrane transporters 3 recombination-associated nucleases |
| pGI3 XF855-0900 | No | Pro-tRNA (5′) | Yes | Yes | Partially at 3′ end | 7 membrane-related factors 1 stress-related factor (SurE) 1 hemagglutinin-like protein |
| pGI4 XF1119-1190 | No | Leu-tRNA (5′) Val-tRNA (3′) | Yes | Yes | No | Gyrase modulators (TedD/PmbA family) |
| pGI5 XF2156-2225 | No | Ser-tRNA (5′) | Yes | Yes | Partially at 3′ end | 2 DNA polymerase III subunits (from E. coli) 15 amino acid biosynthesis genes 1 hemagglutinin-like protein 1 aldehyde-resistance gene |
| pGI6 XF2715-2748 | No | tm-RNA (5′) | Yes | Yes | Yes | 10 restriction-modification proteins (in 2 operons) |
| PI1 XF2025-2087 | No | No | No | No | No | 15 conjugative genes (Tra, Trb operons) 4 efflux system proteins |
| XfP1 XF0678-0733 | Phage-related integrase | Val-tRNA (5′) | Yes | Small | Yes | 24 phage-related proteins, including replication, packaging and capside proteins 1 proteic killer and antidote (higA/higB) |
| XfP2 XF2478-2530 | Phage-related integrase | No | Yes | Yes | Yes | 24 phage-related proteins, including replication, packaging and capside proteins |
| XfP3 XF1564-1587 | No | No | Yes | Yes | Yes | Probably duplicated from XfP4 6 phage-related proteins |
| XfP4 XF1658-1711 | No | Arg-tRNA (3′) | Yes | Yes | No | 7 phage-related proteins |
The first region (named GI1) is an ∼37-kb-long segment of DNA encompassing ORFs XF0480 to XF0536. This region presents all the characteristics expected of a typical GI (Karlin 2001): (1) higher GC content, (2) altered codon bias, (3) insertion at the 3′ end of a tRNA gene (tRNA-N), and (4) it bears genes encoding an integrase at one end (ORFs XF0535 and XF0536). These ORFs display high similarity to a heterodimeric integrase found in association with an insertion element from Helicobacter pylori (Kersulyte et al. 2000). The above criteria have been used to characterize GIs from several microorganisms, and the ORF composition of GI1 is also in accordance with what has been verified in other systems, including 41 hypothetical proteins, six conserved hypothetical proteins, five phage-related factors (including a hemolysin-like enzyme), one lipid A biosynthesis enzyme, one fimbrillin, and the virulence-associated factor VapE from Dichelobacter nodosus (Bloomfield et al. 1997). These latter factors are typically present in pathogenicity islands (PAIs), a subset of GIs that transfer virulence-related genes among microorganisms and are currently viewed as key virulence determinants in many pathogenic bacteria, such as Vibrio cholerae, Shigella flexernii, Helicobacter pylori, enteropathogenic Escherichia coli (EPEC), and others (Bjorkholm et al. 2001; Dobrindt et al. 2002; Dziejman et al. 2002; Tauschek et al. 2002; Walker and Verma 2002). The changes associated with this region suggest that GI1 might be partly or entirely duplicated in the citrus strains, but absent in bacteria from the other hosts (except for coffee), indicating a remarkable level of transpositional activity during the evolution of the Xf group.
A second large GI (named GI2) spans almost 67 kb, from ORF XF1718 to XF1792. It also displays typical GI characteristics, and ORF1718 is very similar to an integrase associated with a self-transmissible 105-kb element that carries the clcRABDE genes, which encode catechol-degrading enzymes in Pseudomonas putida (Ravatn et al. 1998). In Xf, this element is integrated at the 3′ end of tRNA-G and carries a series of 12 ORFs encoding different ketoreductases/dehydrogenases and eight transcription regulators. A few other ORFs have assigned functions associated with outer membrane structure (including one fimbrillin gene), DNA replication, phage-related metabolism, and others. However, the majority of ORFs found in this element also encode hypothetical and conserved hypothetical proteins (42 ORFs). As mentioned above, an independent study by Bhattacharyya et al. (2002b) identified this element as another cryptic prophage. However, we see no strong evidence to support this hypothesis, because two independent genome annotations were not capable of identifying a complete set of genes encoding prohead components, phage regulatory factors, and a full DNA replication machinery, as expected from a typical integrated prophage (Simpson et al. 2000; Battacharyya et al. 2002a,b). We shall, therefore, continue to refer to this element as a GI.
Three smaller GIs were identified, spanning from ORFs XF0631 to XF0641 (GI3), XF1859 to XF1885 (GI4), and XF2108 to XF2132 (GI5). Due to their reduced size (∼9, 15, and 16 kb, respectively), they should be referred to as genomic islets (Hacker and Carniel 2001). GI3 is the only GI identified that does not display significant variation in codon bias and GC content, but other than that, GI3 has all expected properties of a typical GI: It seems to be integrated at the 3′ end of tRNA-S and carries a phage-related integrase (ORF XF0631), along with nine hypothetical proteins and one DNA modifying enzyme. It is conserved across group 1 and in a few group 2 isolates (elm, plum I, and plum II), but missing in the remaining strains. The small variations in codon bias and GC content and the presence of GI3 in genomes other than citrus and coffee suggest that this islet might be more ancient than the others. GI4 is inserted next to tRNA-K and carries only 27 ORFs, five with a known function and all encoding phage-related proteins, probably derived from bacteriophage Pf3. This islet is likely missing in all group 2 isolates, with the exception of the mulberry strain, where most of its structure seems to be preserved. Similarly, this GI is conserved in the group 1 strains, although some degree of genomic instability could be found in the coffee strain and both citrus isolates. Surprisingly, no known integrase was found within GI4, and it is the only identified GI that does not display transcription upregulation when cells are grown in Periwinkle Wilt (PW) medium (see Methods). GI5, on the other hand, bears a phage-related integrase (ORF XF2132) and is inserted next to tRNA-C. It also carries a great number of ORFs with unknown function, and another copy of the D. nodosus vapE gene is present in its structure. This element displays a partial inverted duplication of the vapE-containing region of GI1 and seems to be conserved in the coffee strain, whereas it is duplicated in the two citrus isolates and partly missing in all group 2 representatives.
A series of other regions with altered codon bias and GC content were encountered throughout the genome of strain 9a5c, but because most of them did not seem to be associated with areas of genomic instability in the tested isolates, we decided to name them pGIs (for putative GIs). The general characteristics of these pGIs are listed in Table 2. They all resemble, to some extent, typical GIs, but the most striking observation is that most of them also carry PW-upregulated genes, either along their entire structure or associated with subsets of genes (see below).
Finally, a large 52-kb element located between ORFs XF2025 and XF2087 seems to be the result of plasmid-mediated gene integration. Although the majority of genes from this region are missing in the group 2 isolates, no alterations in GC content or codon bias were observed. Moreover, this element does not carry any integrase-like ORF and is not inserted in the vicinity of tRNA-like genes. A series of ORFs encoding bacterial conjugation factors (TraCDEF and TrbBCDEFGHIJLN) are present in this element. These tra/trb genes are typically found in widespread conjugationally transmitted plasmids, originally isolated from E. coli and other enterobacteria (Motallebi-Veshareh et al. 1992). This element also bears two plasmid stabilization proteins, ParD/E, reinforcing its possible plasmid-mediated origin. This element was named PI1 (for Plasmid Integration) and also carries a particularly noteworthy multidrug efflux system (ORFs XF2083 to XF2085), along with three transcription regulators (ORFs XF2038, XF2062, and XF2086) and a series of 34 hypothetical/conserved hypothetical protein-coding ORFs.
Transcriptome Analysis of Xf Strain 9a5c Grown in Different Media
Lateral gene transfer is currently viewed as an important mediator of bacterial evolution. However, acquisition of foreign genes present in GIs does not necessarily lead to the development of new phenotypes, because these newly acquired elements will only be expressed in the host cell if recognized by the transcriptional machinery and their expression control is efficiently coordinated with the rest of the genome (Hacker and Carniel 2001). Thus, the presence of so many horizontally transferred elements in Xf strain 9a5c raised questions about the functional role(s) played by their resident genes, because there is no evidence for their effective transcription in vivo. Moreover, because most GIs are lacking in some of the strains under analysis, it is possible that these elements are nonessential for bacterial survival.
Thus, to verify whether Xf can efficiently regulate the expression of genes present in any of the transposable elements mentioned above, microarray hybridizations were carried out with RNA samples obtained from bacteria grown in two very distinct media: Periwinkle Wilt (PW; Davis et al. 1981) and Xylella Defined Medium (XDM; Lemos and Alves 2001). PW contains amino acids, BSA, and peptone as sources of carbon, nitrogen, and other nutrients, whereas XDM contains only glycerol and glutamic acid. Due to the marked differences in the composition of these two media, they are able to induce variations in bacterial physiology and growth kinetics. Indeed, when cells were harvested for RNA extraction after several days of incubation, Xf grown in PW showed a twofold higher cellular density compared to cells grown in XDM, even though both cultures had reached early to mid-exponential growth phase at this point, as verified by OD600 measurements (data not shown).
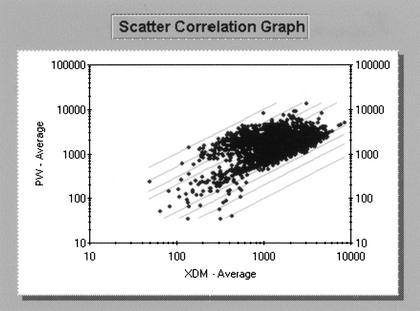
As expected, the two cultures revealed striking differences in their respective transcriptomes (see supplemental materials for a complete list of modulated genes). Surprisingly, only 55 genes were found to be overexpressed in XDM, and their transcription modulation seems to be much smaller compared to what is observed in PW-overexpressed genes (Fig. 2). Among these, 17 ORFs encode hypothetical and conserved hypothetical proteins. ORFs encoding stress-related factors, such as GroEL, GroES (ORFs XF0615 and XF0616), and SodA (ORF XF1921) were also found under these conditions, as were several ribosomal proteins. These findings are only partly consistent with the observations made by Paustian et al. (2002), who used microarray hybridizations to evaluate the transcriptional response of Pasteurella multocida to nutrient limitation, comparing bacteria grown in rich and minimal media. The Paustian studies showed that one-third of the bacterial genome appeared to have its transcription modulated over the course of the experiment. A greater number of genes, including those involved in energy metabolism, transport, protein synthesis, and binding were overexpressed in rich medium, whereas growth in minimal medium led to upregulation of a smaller number of genes, encoding enzymes associated with amino acid biosynthesis, membrane transport systems, and heat shock proteins. Some of the differences observed between the Paustian study and the findings reported herein might be due to the composition of XDM, which, although limited in nutrient availability compared to PW, cannot be considered minimal medium, which is yet to be developed for X. fastidiosa. Furthermore, it is important to note that most genes involved in major biosynthetic pathways were not modulated in any of the growth conditions. Although a few such genes were found to be upregulated in PW (see below), their expression could also be detected after hybridization with RNA obtained from cells grown in XDM, suggesting that X. fastidiosa might have most of these ORFs under the control of poorly induced and/or constitutive promoters, which may represent an adaptation to life in a naturally nutrient-poor environment such as the plant xylem.
Figure 2.
Results from competitive hybridizations between RNA samples obtained from cells grown in XDM medium (labeled with Cy3) and PW medium (labeled with Cy5). Each spot on the array is represented by a dot in the scatter plot shown above. The graph displays the specific, background-subtracted, average intensities for each gene on the array. Gray lines represent fold change values comparing the Cy3 and Cy5 intensities. Lines below the diagonal represent XDM/PW values of two-, three-, five- and 10-fold, respectively. Lines above the diagonal represent XDM/PW ratios of 1/2, 1/3, 1/5, and 1/10.
Growth in PW, on the other hand, leads to strong upregulation of 399 genes. Many of these genes are functionally related and seem to be organized in operons, such as the region encompassing ORFs XF0421 to XF0425—which appears to carry a recombination-related operon. Functional categorization of the PW-induced genes showed a great number of ORFs encoding hypothetical (166) and conserved hypothetical proteins (39), but representatives from all functional categories described by Simpson et al. (2000) seem to be upregulated in PW, including some pathogenicity-associated genes, such as the virulence proteins VapE (ORFs XF0506 and XF2121), AcvB (ORF XF0754), hemolysins (ORFs XF0668 and XF1011), and the hemagglutinin-like protein PspA (ORF XF0889). Interestingly, the neighboring ORFs XF0890, XF0891, XF0892, XF0894, XF0895, XF0896, and XF0897 seem to be submitted to the same transcriptional control as PspA, making it tempting to speculate that the hypothetical proteins encoded by these genes might be associated with PspA-related functions. Other ORFs are upregulated in PW, including genes associated with DNA replication and repair, cytochrome biogenesis, transport, amino acid biosynthesis, and other functions.
Surprisingly however, chromosomal mapping of PW-overexpressed genes showed that most of them were present within prophages, GIs, and pGIs detected in Xf strain 9a5c (Fig. 1A). Moreover, regulation of gene expression within these elements seems to occur in a coordinated manner, because large sets of contiguous genes were simultaneously upregulated. For example, in the large, 67-kb GI2, upregulation of nearly all genes could be observed. This coordinated control of transcription was also observed in the other GIs (with the exception of GI4, which bears no integrase gene), as well as in prophages XfP1, XfP2, and XfP3 (Fig. 1A). Although there is no evidence of phage particle release by Xf cells, the coordinated expression pattern observed across XfP1, XfP2, and XfP3 indicates that these elements might have conserved a relatively independent mechanism of transcription control. It was surprising, however, to see that the same phenomenon was associated with genes present in a genomic island, because there is no precedent for the existence of a similar control mechanism in GIs.
DISCUSSION
The recent advances in the field of comparative genomics resulted in the development of a new paradigm to explain bacterial evolution. It is now clear that prokaryotic genomes are composed of a core gene pool and a flexible gene pool (Hacker and Kaper 2000; Hacker and Carniel 2001). The majority of genes in the core gene pool encode proteins that play roles in basic cellular functions and exhibit a rather homogeneous GC content and codon usage preference. Elements from the flexible gene pool, on the other hand, often encode additional functions that are not essential for bacterial growth, but provide advantages under particular conditions, such as changes in the environment and adaptation to a new host. These genes are normally associated with phages, plasmids, integrons, and other transposable elements. Their contribution to the overall gene content has been estimated to vary among bacteria (Hacker and Carniel 2001). Some microorganisms, such as E. coli and Synechocystis sp., have larger flexible gene pools, corresponding to ∼18% and ∼16% of the total genome, respectively (Lawrence and Ochman 1998; Ochman et al. 2000). This percentage is around 10% in Methanobacterium thermoautrophicum and Mycoplasma pneumoniae, 5% in Helicobacter pylori and Deinococus radiodurans, and virtually 0% in Rickettsia prowasekii and Mycobacterium genitalium (Ochman et al. 2000). In Xf 9a5c, the number of genes carried by plasmid pXf51, prophages, pI1, and GIs add up to 510, which corresponds to ∼18% of the total number of ORFs annotated by Simpson et al. (2000). Thus, the flexible gene pool in this bacterium seems to be one of the largest described to date. Moreover, different Xf strains display variable genome sizes and carry strain-specific gene sets, which mainly include ORFs encoding conjugal transfer factors, restriction-modification systems, methyltransferases, hypothetical/conserved hypothetical proteins, and other genes typically found in horizontally transferred elements (Bhattacharyya et al. 2002a,b). Thus, strain-specific flexible gene pools are likely to be found within the Xf group, a theory that is reinforced by the presence of different plasmids and a putative plasmid-derived Pathogenicity Island in some North American isolates (Pooler et al. 1997; Bhattacharyya et al. 2002a,b).
Several examples of GIs have been observed in different microorganisms associated with symbiotic and saprophytic behaviors, as well as other aspects of environmental adaptation and pathogenicity (Sullivan and Ronson 1998; Hacker and Kaper 2000; Hacker and Carniel 2001). Efforts have been made to understand the control of gene expression in GIs, which are believed to be coordinately regulated with the core genetic pool, allowing fully functional integration of GIs and other components of the flexible gene pool to the global regulatory network of the cell. For instance, the global regulator PhoP-PhoQ of Salmonella typhimurium is located in the ancestral chromosome (Deiwick et al. 1999), but can positively regulate expression of the mgtC genes located in a GI. Other examples are the ToxT and YbtA transcriptional activators of V. cholerae and Yersinia sp., respectively, which are located in GIs, but regulate the expression of genes located outside these regions (Carniel 2002; Sarkar et al. 2002). However, these analyses always addressed the study of gene expression in PAIs and other GIs through the individual evaluation of a few pathogenicity genes. Our array-based global analysis of gene expression in Xf demonstrated a situation where the entire length of several elements of the flexible gene pool have been subjected to upregulation. This phenomenon seems consistent with relatively recent acquisition of such horizontally transferred elements by Xf, with insufficient evolutionary time to guarantee proper gene expression coordination with the core genetic pool.
Interestingly, this upregulation does not seem to be related to differences in cell density or growth kinetics, because hybridizations comparing RNA samples obtained from cells harvested at early and late exponential growth phases did not show distinct expression profiles (data not shown). Thus, it is possible that these laterally acquired genes may be responding collectively to environmental stimuli, perhaps due to differences in media composition. This raises the possibility that GIs might not only be capable of transferring large gene sets among bacteria, but may also control their transcriptional activation in a rather independent manner. If that is the case, the ability of GIs to mediate the acquisition of new characteristics might have a much deeper impact on bacterial evolution than previously imagined, and a better assessment of the mechanisms that control gene expression in transposable elements might be of extreme importance to understand the processes that mediate pathogenicity and environmental adaptation of microorganisms.
In the case of Xf, these genes are likely to hold the key to explain the broad host range of this phytopathogen, because the overall genomic diversity observed among the strains is mainly due to elements from the flexible gene pool. As a result, most variations involve ORFs associated with phage metabolism, plasmid maintenance, bacterial conjugation, and enzymes associated with environmental response and adaptation, such as transcriptional regulators, drug-resistance enzymes, and virulence factors. Cell wall- and outer membrane-related structural components also accounted for significant differences among the tested strains. Particularly noteworthy is the observation of differences among fimbrillin genes, whose products may establish direct contacts with host cells and other bacteria (Park et al. 2001). One fimbrillin (ORF XF0539) was not found in the strains obtained from elm and plum, whereas the almond, grape, mulberry, and ragweed isolates lacked four of the fimbrillin genes from strain 9a5c (ORFs XF0487, XF0538, XF0539, and XF1791). These factors are adhesins of Type IV fimbriae found in numerous bacterial species that infect humans, animals, and plants and are important players in the process of host colonization and disease development (Kennan et al. 2001; van Doorn 2001). Another enzyme involved in modification of outer membrane carbohydrates (OafA–ORF XF0778) is missing from all North American strains except grape-FL. Virulence factors, such as bacteriocin, VapD, VapE and the virulence regulator XrvA (ORFs XF2007, XFa0052, XF0506, and XF0749, respectively) also displayed similar profiles, as did ORF XF1744, a hypothetical oxidoreductase, possibly associated with invasive capability, which is also missing in all group 2 strains (Gaillard et al. 1991). Although such differences among pathogenicity factors might help to explain the disparate pathogenies caused by Xf strains, recent evidence points to host-specific factors as key mediators of disease manifestation by these bacteria (Li et al. 2002).
The subdivision of Xf strains into at least two distinct groups is in accord with previous observations (Chen et al. 2000; Hendson et al. 2001; Mehta and Rosato 2001), but even inside each group, a considerable degree of variation can be observed. The citrus group bears duplicated regions not observed in the coffee strain. The latter isolate does not carry an ∼32-kb-long region of pI1—possibly excised by homologous recombination between two copies of the traC gene (ORFs XF2025 and XF2061). Within group 2, the almond/grape strains display a similar profile, whereas elm and plum II isolates are nearly identical to each other, contrasting with plum I. Independent studies found strain plum I to have a closer phylogenetic relationship with the citrus-coffee strains than with others (da Costa et al. 2000; Mehta and Rosato 2001). This finding is in accord with geographic distribution, as plum I was originally obtained from a diseased tree in the southern region of Brazil, being the first Xf isolate obtained in that country (French and Kitajima 1978). Interestingly, strain plum I was the only group 2 isolate to bear nearly all ORFs from pXf51, the large 51-kb plasmid found in CVC strain 9a5c. However, considering the overall genomic profile, it is likely that strain plum I derives directly from the North American isolates from group 2. It is tempting to speculate that this isolate might have been brought to Brazil as a graft or seedling contaminant and acquired pXF51 sequences through lateral gene transfer after coming into contact with other South American Xf strains, raising interesting possibilities concerning genetic interactions among Xf isolates in the wild and genomic plasticity within the group. In this sense, the observation that GI2 seems to be present only in group 1 suggests that it may have been acquired after geographical separation between North and South American strains. The remaining horizontally transferred elements, however, appear to display a more complex distribution pattern, hindering more conclusive evaluation of their evolutionary history. For instance, the mulberry strain seems to carry most sequences from GI4, which is lacking in other group 2 representatives. On the other hand, this very same strain does not carry sequences from GI3, which is present in elm, plum I, and plum II. Several sequences from prophage XfP1 are lacking in elm and plum II strains, but present in other group 2 isolates. These observations reinforce the idea that the flexible gene pool in Xf might display a complex, strain-specific evolutionary pattern.
Nonetheless, despite all of the differences categorized and discussed above, it must be noted that the overall similarity among Xf genomes was very high, with more than 90% ORF conservation between strain 9a5c and the group 2 representatives. The conservation of ORFs increased to ∼97% when strain 9a5c was compared to the coffee strain and to ∼99% when the comparison involved the other citrus isolates. Essentially, all genes involved in the major biosynthetic pathways and cellular functions are present in all analyzed Xf strains, suggesting that adaptation of the entire group to life in the plant xylem is likely to depend on genes from the highly conserved core gene pool, responsible for the major physiological characteristics already discussed by Simpson et al. (2000), which involve a mechanism for energy generation based on the utilization of carbohydrates, including enzymes for cellulose degradation, but with no alternative energy sources based on catabolism of fatty acids or amino acids. Fully equipped biochemical pathways guarantee biosynthesis of all amino acids, purines, pyrimidines, and other nutritional requirements, which are also met through the presence of an elaborated panel of membrane transporters for the efficient uptake of different ions and organic compounds. On the other hand, up to 18% of the genomic composition of strain 9a5c may be associated with elements from the flexible gene pool, which have been shown to carry important adaptation and putative pathogenicity factors. Moreover, we provide evidence that gene expression within these elements can be efficiently modulated by environmental stimuli, and that Xf strains are likely to exchange genetic material in the wild, contributing to the considerable genomic differences observed among strains from groups 1 and 2. Thus, an efficient mechanism for exchanging mobile elements in a large flexible gene pool may be of capital importance in mediating adaptation of Xf to new hosts.
METHODS
Origin of the Isolates and Growth Conditions
Xf strains from different hosts and geographic origins (Table 1) were grown in either Periwinkle Wilt (PW) medium (Davis et al. 1981), which contained 4.0 g phytone peptone, 1.0 g tryptcase peptone, 1.2 g K2HPO4, 1.0 g KH2PO4, 0.4 g MgSO4, 0.1% hemin chloride, and 10 mL phenol red per L, or in Xylella Defined Medium (XDM), which contained 0.23 g K2HPO4, 0.10 g MgSO4.7H2O, 1.1 g glutamic acid, 4 mL glycerol, and 0.5 g ferric pyrophosphate per L (Lemos and Alves 2001). The bacteria were initially grown on solid medium and then transferred consecutively to increasing volumes of liquid medium (5, 20, 50, and 100 mL). In each step, the cells were incubated for 4 d in a rotatory shaker at 28°C. Because the bacteria grew adhering to the flask walls, the cellular mass was disrupted by pipetting and vortexing before proceeding to the next transfer. After the last step, the cellular mass was transferred to 200 mL of PW or XDM liquid medium and incubated for another 4 d. Bacterial growth in both PW and XDM media was evaluated through measurement of optical density (OD600). Aliquots (1.0 mL) were sampled at 24-h intervals over a 10-d period. The samples were centrifuged, and the resulting pellet was disrupted in 1.0 mL of saline (0.9% NaCl) and OD600 immediately read in a spectrophotometer. Three independent samples were analyzed for each time point.
Microarray Fabrication
Specific primer pairs for amplification of 2838 ORFs found in the genome of Xf strain 9a5c were designed employing the software Primer3 (kindly provided by Dr. S. Rozen at http://www.genome.wi.mit.edu/genome_software/other/primer3.html). Approximately 2500 primer pairs, representing 2205 different ORFs from the main chromosomal DNA and from the pXf51 plasmid, yielded specific PCR amplification products. Most of the amplified fragments ranged in size between 350 and 1000 base pairs. The PCR products obtained were purified with the QiaQuick 96 PCR purification system (QIAGEN) and spotted onto CMT-GAPS silane-coated slides (Corning), using an Affymetrix 417 Arrayer, according to the manufacturer's instructions. The printed microarrays were used for competitive hybridization experiments using DNA samples obtained from Xf strains. This approach allowed a global comparison of approximately 2200 ORFs from Xf strain 9a5c with the genome of the other 11 Xf isolates. Although the strain 9a5c genome is composed of more than 2800 ORFs, many of them were, actually, paralogous duplications scattered throughout the chromosome and/or small ORFs (<250 bp) encoding hypothetical proteins. Many of these sequences could not be efficiently and/or specifically amplified, due to their reduced size and chromosomal location (closely flanked by larger ORFs). We estimate, however, that these analyses cover more than 90% of the most relevant ORFs present in the genome from Xf strain 9a5c. The microarrays were also used to monitor the global transcription profile of Xf strain 9a5c under two different growth conditions.
Nucleic Acid Labeling and Hybridization Conditions
DNA samples were extracted according to the protocol described by Dungan (1989). Labeling reactions and purification were performed as described by Behr et al. (1999). Total RNA was isolated from Xf cells using the RNeasy RNA extraction kit (QIAGEN) and labeled by reverse transcription. Briefly, 30 μg of total bacterial RNA was mixed with 16 μg of random hexamers (Invitrogen) in a 30 μL final volume. Annealing was accomplished by incubation for 2 min at 75°C, 2 min at 55°C, 2 min at 45°C, 2 min at 37°C, and 2 min at 22°C, followed by addition of 6 μL SuperScript II reaction buffer (Invitrogen), 3 μL of 0.1 M dithiothreitol, 1 μL dNTP mix (10 mM dATP, 10 mM dGTP, 10 mM dTTP, 5 mM dCTP), 2 μL of 1.0mM Cy3- or Cy5-labeled dCTP (Amersham Biosciences), and 2 μL of SuperScript II reverse transcriptase (200 U/μL; Invitrogen) into the reaction. The cDNA synthesis was carried out at 42°C for 2 h. After labeling, the RNA was hydrolyzed with 0.1 N NaOH treatment and neutralized by the addition of 0.1 N HCl. The labeled cDNA was diluted to 500 μL with TE and concentrated using a Microcon-30 (Amicon) to 10 μL.
Arrays were hybridized overnight (42°C) in a GeneTac Hybridization Station (Genomic Solutions), in 6 × SSC, 5 × Denhardt's solution, 0.25 mg/mL sheared salmon sperm DNA, 0.5% SDS, and 2 μg of each labeled DNA or cDNA sample. After hybridization, slides were washed twice (42°C) in 0.5 × SSC, 0.01% SDS, followed by two washes in 0.06 × SSC, 0.01% SDS and two final washes in 0.06 × SSC. All washing steps consisted of 1 min of flow, followed by 5 min of incubation. Slides were then dried and submitted to fluorescence detection.
Image Acquisition and Analysis
Hybridized arrays were scanned in an Affymetrix 418 Array Scanner, and images were analyzed with Affymetrix Jaguar v 2.0. Quality control of the hybridized spots is automatically performed by the software, based on spot morphology and local signal-to-background ratio, using the Easy Threshold and Variable Circle Size Algorithm. In all experiments, reliable hybridization signals could be obtained for more than 90% of the arrayed probes (see Costa de Oliveira et al. 2002). For DNA–DNA hybridizations, normalization between the intensities in the two channels was achieved with the Jaguar Control Spots option, using a list of 30 control ORFs that had 100% sequence identity in the genomes of strains 9a5c and Grape CA. For each pair of strains, hybridizations were performed in triplicate. These data were consolidated into a GATC database with Affymetrix MicroDB v 2.0, and the averages from all six readings were submitted to scatter plot visualization with Affymetrix Data Mining Tool v 2.0. Statistical validation of fold change variations was performed with the aid of the Significance Analysis of Microarrays (SAM) method proposed by Tusher et al. (2001), using the software developed by B. Narasimham and available at http://www-stat.stanford.edu/∼tibs/SAM/index.html. Spots that showed a Reference/Test ratio <1:2 were considered to be present in greater copy number in the test over the reference strain, as proposed by Smoot et al. (2002), whereas spots that showed an average Reference/Test ratio >5:1 were considered to be missing in the test strain. The application of these criteria in a direct sequence comparison between strains 9a5c and Grape CA, which have been completely sequenced, provided an estimated error rate below 0.3% (Costa de Oliveira et al. 2002).
For cDNA–cDNA hybridizations, three independent experiments were performed with different RNA preparations. Hybridizations were performed in triplicate, generating nine intensity readings for each gene. Normalization was achieved through the sum of all intensities in each channel, using the Jaguar All Spots option. Normalized data were consolidated into a GATC database with Affymetrix MicroDB v 2.0, and genes that displayed statistically significant variations in gene expression were identified with the aid of the SAM method. A fold change cutoff of at least 2.0 was used in these analyses.
Genome comparisons and transcription profiles were viewed using the programs Cluster and TreeView, available at http://www.microarrays.org/software (Eisen et al. 1998). For the visualization of comparative profiles from the 12 analyzed strains, we applied the method proposed by Smoot et al. (2002), where ORFs shared by the reference and each test strain was labeled 1, whereas ORFs exclusive to the reference strain, or present in greater copy number in the test over the reference strain were labeled 0 and −1, respectively.
WEB SITE REFERENCES
http://www.jgi.doe.gov/jgi_microbial/html; Accession to the partial genome sequences from Xf strains isolated from Almond and Oleander trees.
http://www.lbi.ic.unicamp.br/world/xf-grape; Accession to the complete genome sequence from Xf strains isolated from grapevines.
http://www-genome.wi.mit.edu/genome_software/other/primer3.html; Accession to software PRIMER3.
http://www-stat.stanford.edu/∼tibs/SAM/index.html; Accession to software SAM.
http://www.microarrays.org/software; Accession to softwares CLUSTER and TREEVIEW.
Acknowledgments
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), under the X. fastidiosa Functional Genome Program.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL Lnunes@umc.br; FAX 55-11-4798-7104.
E-MAIL reginaco@umc.br; FAX 55-11-4798-7106.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.930803.
REFERENCES
- 1.Banks D., Albibi, R., Chen, J., Lamikanra, O., Jarret, R.L., and Smith, B.J. 1999. Specific detection of Xylella fastidiosa Pierce's disease strains. Curr. Microbiol. 39: 85-88. [DOI] [PubMed] [Google Scholar]
- 2.Behr M.A., Wilson, M.A., Gill, W.P., Salamon, H., Schoolnik, G.K., Rane, S., and Small, P.M. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284: 1520-1523. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharyya A., Stilwagen, S., Ivanova, N., D'Souza, M., Bernal, A., Lykidis, A., Kapatral, V., Anderson, I., Larsen, N., Los, T., et al. 2002a. Whole-genome comparative analysis of three phytopathogenic Xylella fastidiosa strains. Proc. Natl. Acad. Sci. 99: 12403-12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhattacharyya A., Stilwagen, S., Reznik, G., Feil, H., Feil, W.S., Anderson, I., Bernal, A., D'Souza, M., Ivanova, N., Kapatral, V., et al. 2002b. Draft sequencing and comparative genomics of Xylella fastidiosa strains reveal novel biological insights. Genome Res. 12: 3707-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjorkholm B., Lundin, A., Sillen, A., Guillemin, K., Salama, N., Rubio, C., Gordon, J.I., Falk, P., and Engstrand, L. 2001. Comparison of genetic divergence and fitness between two subclones of Helicobacter pylori. Infect. Immun. 69: 7832-7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloomfield G.A., Whittle, G., McDonagh, M.B., Katz, M.E., and Cheetham, B.F. 1997. Analysis of sequences flanking the vap regions of Dichelobacter nodosus: Evidence for multiple integration events, a killer system, and a new genetic element. Microbiology 143: 553-562. [DOI] [PubMed] [Google Scholar]
- 7.Carniel E. 2002. Plasmids and pathogenicity islands of Yersinia. Curr. Top. Microbiol. Immunol. 264: 89-108. [PubMed] [Google Scholar]
- 8.Chen J., Lamikanra, O., Chang, C.J., and Hopkins, D.L. 1995. Randomly amplified polymorphic DNA analysis of Xylella fastidiosa Pierce's disease and oak leaf scorch pathotypes. Appl. Environ. Microbiol. 61: 1688-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J., Jarret, R.L., Qin, X., Hartung, J.S., Banks, D., Chang, C.J., and Hopkins, D.L. 2000. 16S rDNA sequence analysis of Xylella fastidiosa strains. Syst. Appl. Microbiol. 23: 349-354. [DOI] [PubMed] [Google Scholar]
- 10.Costa de Oliveira R., Yanai, G.M., Muto, N.H., Leite, D.B., de Souza, A.A., Coletta-Filho, H.D., Machado, M.A., and Nunes, L.R. 2002. Competitive hybridization on spotted microarrays as a tool to conduct comparative genomic analyses of Xylella fastidiosa strains. FEMS Microbiol. Lett. 216: 15-21. [DOI] [PubMed] [Google Scholar]
- 11.da Costa P.I., Franco, C.F., Miranda, V.S., Teixeira, D.C., and Hartung, J.S. 2000. Strains of Xylella fastidiosa rapidly distinguished by arbitrarily primed-PCR. Curr. Microbiol. 40: 279-282. [DOI] [PubMed] [Google Scholar]
- 12.Davis M.J., French, W.J., and Schaad, N.W. 1981. Axenic culture of the bacteria associated with phony disease of peach and plum leaf scald. Curr. Microbiol. 6: 309-314. [Google Scholar]
- 13.Deiwick J., Nikolaus, T., Erdogan, S., and Hensel, M. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31: 1759-1773. [DOI] [PubMed] [Google Scholar]
- 14.Dobrindt U., Hentschel, U., Kaper, J.B., and Hacker, J. 2002. Genome plasticity in pathogenic and nonpathogenic enterobacteria. Curr. Top. Microbiol. Immunol. 264: 157-175. [PubMed] [Google Scholar]
- 15.Dungan D., et al. 1989. Current protocols in molecular biology (ed. F.M. Ausubel), pp. 241–242. John Wiley and Sons, Inc., New York, NY.
- 16.Dziejman M., Balon, E., Boyd, D., Fraser, C., Heidelberg, J., and Mekalanos, J. 2002. Comparative genomic analysis of Vibrio cholerae: Genes that correlate with cholera endemic and pandemic disease. Proc. Natl. Acad. Sci. 99: 1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisen M.B., Spellman, P.T., Brown, P.O., and Botstein, D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 95: 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.French W.J. and Kitajima, E.W. 1978. Occurrence of plum leaf scald in Brazil and Paraguay. Plant Dis. Reptr. 62: 1035-1038. [Google Scholar]
- 19.Gaillard J.L., Berche, P., Frehel, C., Gouin, E., and Cossart, P. 1991. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65: 1127-1141. [DOI] [PubMed] [Google Scholar]
- 20.Hacker J. and Carniel, E. 2001. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2: 376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hacker J. and Kaper, J.B. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54: 641-679. [DOI] [PubMed] [Google Scholar]
- 22.Hartung J.S., Beretta, J., Brlansky, R.H., Spisso, J., and Lee, R.F. 1994. Citrus variegated chlorosis bacterium: Axenic culture, pathogenicity, and serological relationships with other strains of Xylella fastidiosa. Phytopathology 84: 591-597. [Google Scholar]
- 23.Hendson M., Purcell, A.H., Chen, D., Smart, C., Guilhabert, M., and Kirkpatrick, B. 2001. Genetic diversity of Pierce's disease strains and other pathotypes of Xylella fastidiosa. Appl. Environ. Microbiol. 67: 895-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hopkins D.L. 1989. Xylella fastidiosa: Xylem-limited bacterial pathogen of plants. Annu. Rev. Phytopathology 27: 271-290. [Google Scholar]
- 25.Hopkins D.L. and Mollenhauer, H.H. 1973. Rickettsia-like bacterium associated with Pierce's disease of grapes. Science 179: 298-300. [DOI] [PubMed] [Google Scholar]
- 26.Hopkins D.L. and Purcell, A.H. 2002. Xylella fastidiosa: Cause of Pierce's disease of grapevine and other emergent diseases. Plant Dis. 86: 1056-1066. [DOI] [PubMed] [Google Scholar]
- 27.Karlin S. 2001. Detecting anomalous gene clusters and pathogenicity islands in diverse bacterial genomes. Trends Microbiol. 9: 335-343. [DOI] [PubMed] [Google Scholar]
- 28.Kennan R.M., Dhungyel, O.P., Whittington, R.J., Egerton, J.R., and Rood, J.I. 2001. The type IV fimbrial subunit gene (fimA) of Dichelobacter nodosus is essential for virulence, protease secretion, and natural competence. J. Bacteriol. 183: 4451-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kersulyte D., Mukhopadhyay, A.K., Shirai, M, Nakazawa, T., and Berg, D.E. 2000. Functional organization and insertion specificity of IS607, a chimeric element of Helicobacter pylori. J. Bacteriol. 182: 5300-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawrence J.G. and Ochman, H. 1998. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. 95: 9413-9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemos E.G.M. and Alves, L.M.C., 2001. A defined medium that allows the growth of X. fastidiosa CVC, using the information from annotation sequenced genes. In Annals from the 1st Xylella fastidiosa Functional Genome Symposium, Serra Negra, SP., pp. 24 Ed. FAPESP, São Paulo, Brazil.
- 32.Li W.-B., Zhou, C.-H., Pria, W.D., Jr., Teixeira, D.C., Miranda, V.S., Pereira, E.O., Ayres, A.J., He, C.-X., Costa, P.I., and Hartung, J.S. 2002. Citrus and coffee strains of Xylella fastidiosa induce Pierce's Disease in grapevine. Plant Dis. 86: 1206-1210. [DOI] [PubMed] [Google Scholar]
- 33.Mehta A. and Rosato, Y.B. 2001. Phylogenetic relationships of Xylella fastidiosa strains from different hosts based on 16S rDNA and 16S-23S intergenic spacer sequences. Int. J. Syst. Evol. Microbiol. 51: 311-318. [DOI] [PubMed] [Google Scholar]
- 34.Motallebi-Veshareh M., Balzer, D., Lanka, E., Jagura-Burdzy, G., and Thomas, C.M. 1992. Conjugative transfer functions of broad-host-range plasmid RK2 are coregulated with vegetative replication. Mol. Microbiol. 6: 907-920. [DOI] [PubMed] [Google Scholar]
- 35.Ochman H., Lawrence, J.G., and Groisman, E.A. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405: 299-304. [DOI] [PubMed] [Google Scholar]
- 36.Park H.S., Wolfgang, M., van Putten, J.P., Dorward, D., Hayes, S.F., and Koomey, M. 2001. Structural alterations in a type IV pilus subunit protein result in concurrent defects in multicellular behaviour and adherence to host tissue. Mol. Microbiol. 42: 293-307. [DOI] [PubMed] [Google Scholar]
- 37.Paustian M.L., May, B.J., and Kapur, V. 2002. Transcriptional response of Pasteurella multocida to nutrient limitation. J. Bacteriol. 184: 3734-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pooler M.R. and Hartung, J.S. 1995. Genetic relationships among strains of Xylella fastidiosa from RAPD-PCR data. Curr. Microbiol. 31: 134-137. [DOI] [PubMed] [Google Scholar]
- 39.Pooler M.R., Hartung, J.S., and Fenton, R.G. 1997. Sequence analysis of a 1296-nucleotide plasmid from Xylella fastidiosa. FEMS Microbiol. Lett. 155: 217-222. [DOI] [PubMed] [Google Scholar]
- 40.Purcell A.H. and Hopkins, D.L. 1996. Fastidious xylem-limited bacterial plant pathogens. Annu. Rev. Phytopathol. 34: 131-151. [DOI] [PubMed] [Google Scholar]
- 41.Purcell A.H., Saunders, S.R., Hendson, M., Grebus, M.E., and Henry, M.J. 1999. Causal role of Xylella fastidiosa in oleander leaf scorch. Phytopatology 89: 53-58. [DOI] [PubMed] [Google Scholar]
- 42.Ravatn R., Studer, S., Zehnder, A.J., and van der Meer, J.R. 1998. Int-B13, an unusual site-specific recombinase of the bacteriophage P4 integrase family, is responsible for chromosomal insertion of the 105-kilobase clc element of Pseudomonas sp. Strain B13. J. Bacteriol. 180: 5505-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarkar A., Nandy, R.K., Nair, G.B., and Ghose, A.C. 2002. Vibrio pathogenicity island and cholera toxin genetic element-associated virulence genes and their expression in non-O1 non-O139 strains of Vibrio cholerae. Infect. Immun. 70: 4735-4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simpson A.J., Reinach, F.C., Arruda, P., Abreu, F.A., Acencio, M., Alvarenga, R., Alves, L.M., Araya, J.E., Baia, G.S., Baptista, C.S., et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406: 151-157. [DOI] [PubMed] [Google Scholar]
- 45.Smoot J.C., Barbian, K.D., Van Gompel, J.J., Smoot, L.M., Chaussee, M.S., Sylva, G.L., Sturdevant, D.E., Ricklefs, S.M., Porcella, S.F., Parkins, L.D., et al. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. 99: 4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sullivan J.T. and Ronson, C.W. 1998. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl. Acad. Sci. 95: 5145-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tauschek M., Strugnell, R.A., and Robins-Browne, R.M. 2002. Characterization and evidence of mobilization of the LEE pathogenicity island of rabbit-specific strains of enteropathogenic Escherichia coli. Mol. Microbiol. 44: 1533-1550. [DOI] [PubMed] [Google Scholar]
- 48.Tusher V.G., Tibshirani, R., and Chu, G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. 98: 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Doorn J., Hollinger, T.C., and Oudega, B. 2001. Analysis of the type IV fimbrial-subunit gene fimA of Xanthomonas hyacinthi: Application in PCR-mediated detection of yellow disease in Hyacinths. Appl. Environ. Microbiol. 67: 598-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker J. and Verma, N. 2002. Identification of a putative pathogenicity island in Shigella flexneri using subtractive hybridization of the S. flexneri and Escherichia coli genomes. FEMS Microbiol. Lett. 213: 257-264. [DOI] [PubMed] [Google Scholar]
- 51.Wells J.M., Raju, B.C., Hung, H.Y., Weisburg, W.G., Mandelco-Paul, L., and Brenner, D.J. 1987. Xylella fastidiosa gen. nov. sp.nov.: Gram-negative, xylem-limited, fastidious plant bacteria related to Xanthomonas spp. Int. J. Syst. Bacteriol. 37: 136-143. [Google Scholar]