Abstract
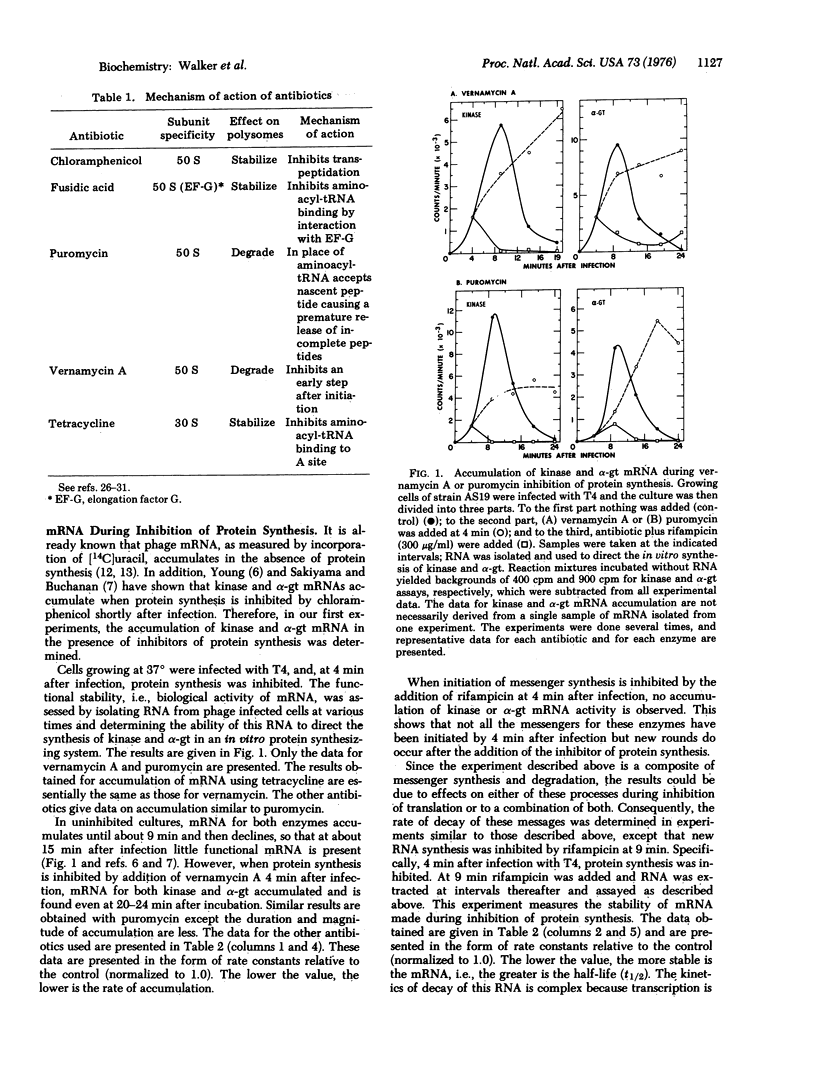
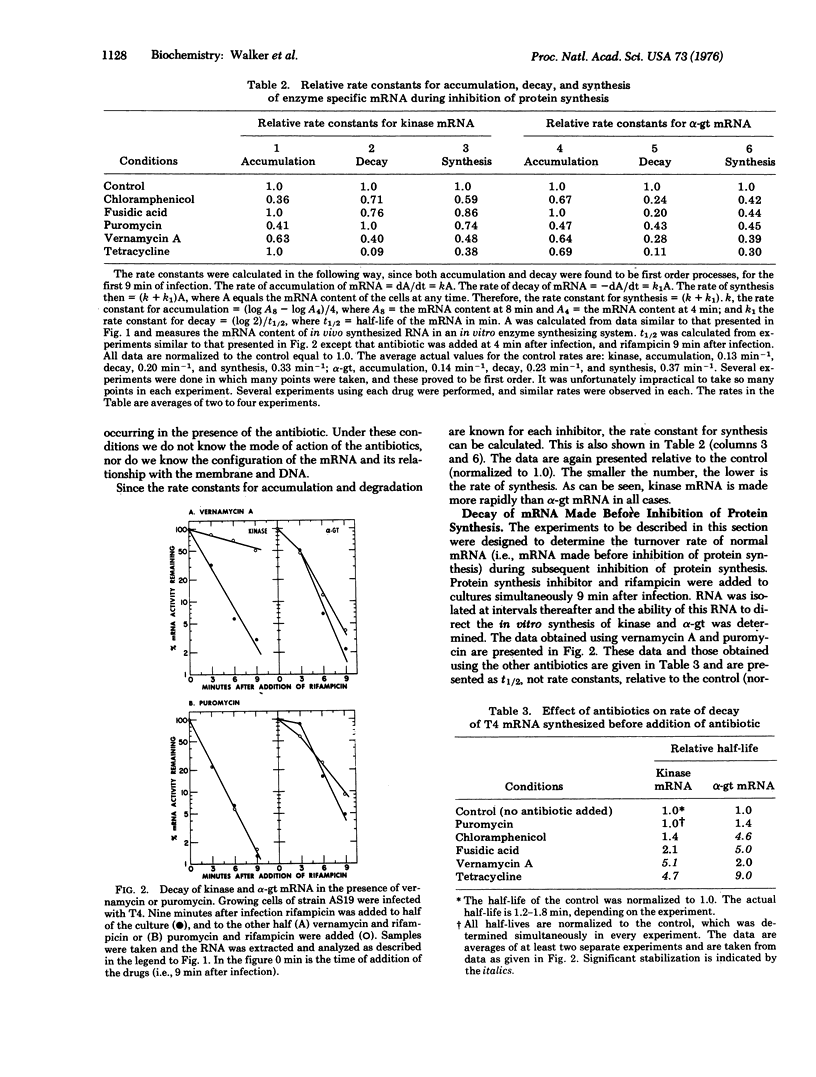
Antibiotics were used to inhibit protein synthesis at specific steps in the biosynthetic pathway. In this way, it was possible to study the coupling of protein synthesis to the accumulation of biologically active mRNA in T4-infected Escherichia coli. Functional mRNA for the phage enzymes deoxynucleotide kinase (EC 2.7.4.4; ATP: nucleoside monophosphate phosphotransferase or nucleosidemonophosphate kinase) and alpha-glucosyltransferase (EC 2.4.1.5; 1, 4-alpha-D-glucan: 1, 6-alpha-D-glucan 6-alpha-glucosyltransferase or dextrin dextranase) accumulated during inhibition of protein synthesis irrespective of the step in the synthesis of protein that was blocked. Under these conditions, however, the rate of mRNA synthesis for both enzymes was significantly inhibited. In contrast, the rate of degradation of these mRNAs was markedly dependent on the step in protein synthesis that was inhibited. That is, the site for mRNase action was different for each message. The most important step in protein synthesis required for the stability of deoxynucleotide kinase mRNA is the initiation step. A single ribosome bound to the 5' end of the deoxynucleotide kinase mRNA can stabilize the molecule. On the other hand, the initiation event does not seem to be important for stabilizing the alpha-glucosyltransferase mRNA. Instead, a high ribosome denisty on the alpha-glucosyltransferase messenger is required to achieve significant stability. Therefore, in studying messenger metabolism, it is important to focus on the functional stability of specific mRNAs instead of on total messenger since each mRNA can be metabolized differently.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ASTRACHAN L., VOLKIN E. Effects of chloramphenicol on ribonucleic acid metabolism in T2-infected Escherichia coli. Biochim Biophys Acta. 1959 Apr;32:449–456. doi: 10.1016/0006-3002(59)90618-3. [DOI] [PubMed] [Google Scholar]
- Cocito C., Voorma H. O., Bosch L. Interference of virginiamycin M with the initiation and the elongation of peptide chains in cell-free systems. Biochim Biophys Acta. 1974 Mar 27;340(3):285–298. doi: 10.1016/0005-2787(74)90274-3. [DOI] [PubMed] [Google Scholar]
- Cohen P. S., Ennis H. L. The requirement for potassium for bacteriophage T4 protein and deoxyribonucleic acid synthesis. Virology. 1965 Nov;27(3):282–289. doi: 10.1016/0042-6822(65)90107-8. [DOI] [PubMed] [Google Scholar]
- Cohen P. S., Natale P. J., Buchanan J. M. Transcriptional regulation of T4 bacteriophage-specific enzymes synthesized in vitro. J Virol. 1974 Aug;14(2):292–299. doi: 10.1128/jvi.14.2.292-299.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cundliffe E. The mode of action of fusidic acid. Biochem Biophys Res Commun. 1972 Mar 10;46(5):1794–1801. doi: 10.1016/0006-291x(72)90053-8. [DOI] [PubMed] [Google Scholar]
- Dahlberg A. E., Lund E., Kjeldgaard N. O. Some effects of antibiotics on bacterial polyribosomes as studied by gel electrophoresis. J Mol Biol. 1973 Aug 25;78(4):627–636. doi: 10.1016/0022-2836(73)90284-2. [DOI] [PubMed] [Google Scholar]
- Ennis H. L., Cohen P. S. Control of phage and host ribonucleic acid synthesis in phage T4 infected Escherichia coli. Virology. 1968 Oct;36(2):193–200. doi: 10.1016/0042-6822(68)90136-0. [DOI] [PubMed] [Google Scholar]
- Gurgo C., Apirion D., Schlessinger D. Polyribosome metabolism in Escherichia coli treated with chloramphenicol, neomycin, spectinomycin or tetracycline. J Mol Biol. 1969 Oct 28;45(2):205–220. doi: 10.1016/0022-2836(69)90100-4. [DOI] [PubMed] [Google Scholar]
- Imamoto F. Diversity of regulation of genetic transcription. I. Effect of antibiotics which inhibit the process of translation on RNA metabolism in Escherichia coli. J Mol Biol. 1973 Feb 25;74(2):113–136. doi: 10.1016/0022-2836(73)90102-2. [DOI] [PubMed] [Google Scholar]
- Natale P. J., Buchanan J. M. DNA-directed synthesis in vitro of T4 phage-specific enzymes. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2513–2517. doi: 10.1073/pnas.69.9.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale P. J., Buchanan J. M. DNA-directed synthesis in vitro of T4 phage-specific enzymes. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2513–2517. doi: 10.1073/pnas.69.9.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale P. J., Ireland C., Buchanan J. M. Sizing of two bacteriophage T4 specific messenger ribonucletic acids formed in vitro and in vivo. Biochem Biophys Res Commun. 1975 Oct 27;66(4):1287–1293. doi: 10.1016/0006-291x(75)90499-4. [DOI] [PubMed] [Google Scholar]
- Pestka S. Inhibitors of ribosome functions. Annu Rev Microbiol. 1971;25:487–562. doi: 10.1146/annurev.mi.25.100171.002415. [DOI] [PubMed] [Google Scholar]
- Sakiyama S., Buchanan J. M. Control of the synthesis of T4 phage deoxynucleotide kinase messenger ribonucleic acid in vivo. J Biol Chem. 1972 Dec 10;247(23):7806–7814. [PubMed] [Google Scholar]
- Sakiyama S., Buchanan J. M. In vitro synthesis of deoxynucleotide kinase programmed by bacteriophage "T4-RNA. Proc Natl Acad Sci U S A. 1971 Jun;68(6):1376–1380. doi: 10.1073/pnas.68.6.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakiyama S., Buchanan J. M. Relationship between molecular weight of T4 phag-induced deoxynucleotide kinase and the size of its messenger ribonucleic acid. J Biol Chem. 1973 May 10;248(9):3150–3154. [PubMed] [Google Scholar]
- Salser W., Gesteland R. F., Bolle A. In vitro synthesis of bacteriophage lysozyme. Nature. 1967 Aug 5;215(5101):588–591. doi: 10.1038/215588a0. [DOI] [PubMed] [Google Scholar]
- Sekiguchi M., Iida S. Mutants of Escherichia coli permeable to actinomycin. Proc Natl Acad Sci U S A. 1967 Dec;58(6):2315–2320. doi: 10.1073/pnas.58.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz J. A. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature. 1969 Dec 6;224(5223):957–964. doi: 10.1038/224957a0. [DOI] [PubMed] [Google Scholar]
- Trimble R. B., Maley F. In vitro synthesis of deoxynucleotide kinase, dihydrofolate reductase and deosycytidylate hydroxymethylase from RNA transcripts of T2 phage DNA. Biochem Biophys Res Commun. 1973 Jun 8;52(3):1026–1033. doi: 10.1016/0006-291x(73)91040-1. [DOI] [PubMed] [Google Scholar]
- Varmus H. E., Perlman R. L., Pastan I. Regulation of lac transcription in antibiotic-treated E. coli. Nat New Biol. 1971 Mar 10;230(10):41–44. doi: 10.1038/newbio230041a0. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Imamoto F. Differential stability of trp messenger RNA synthesized originating at the trp promoter and pL promoter of lambda trp phage. J Mol Biol. 1975 Feb 25;92(2):289–304. doi: 10.1016/0022-2836(75)90228-4. [DOI] [PubMed] [Google Scholar]
- Young E. T., 2nd Cell-free synthesis of bacteriophage T4 glucosyl transferase. J Mol Biol. 1970 Aug;51(3):591–604. doi: 10.1016/0022-2836(70)90010-0. [DOI] [PubMed] [Google Scholar]


