Abstract
Objective
To use high density genotyping to investigate the genetic associations of acute anterior uveitis (AAU) in patients both with and without ankylosing spondylitis (AS).
Method
We genotyped 1,711 patients with AAU (either primary or with AAU and AS), 2,339 AS patients without AAU, and 10,000 controls on the Illumina Immunochip Infinium microarray. We also used data on AS patients from previous genomewide association studies to investigate the AS risk locus ANTXR2 for its putative effect in AAU. ANTXR2 expression in mouse eyes was investigated by RT-PCR.
Results
Comparing all AAU cases with HC, strong association was seen over HLA-B corresponding to the HLA-B27 tag SNP rs116488202. Three non-MHC loci IL23R, the intergenic region 2p15 and ERAP1 were associated at genome-wide significance (P < 5×10−8). Five loci harboring the immune-related genes IL10-IL19, IL18R1-IL1R1, IL6R, the chromosome 1q32 locus harboring KIF21B, as well as the eye related gene EYS, were also associated at a suggestive level of significance (P < 5×10−6). A number of previously confirmed AS associations demonstrated significant differences in effect size between AS patients with AAU and AS patients without AAU. ANTXR2 expression was found to vary across eye compartments.
Conclusion
These findings, with both novel AAU specific associations, and associations shared with AS demonstrate overlapping but also distinct genetic susceptibility loci for AAU and AS. The associations in IL10 and IL18R1 are shared with inflammatory bowel disease, suggesting common etiologic pathways.
Keywords: Anterior uveitis, uveitis, ankylosing spondylitis, HLA-B27, ANTXR2
Acute anterior uveitis (AAU) has a cumulative incidence rate in the Caucasian general population of 0.2%, but in those who are HLA-B27 positive (8–10% of the Caucasian population) the cumulative incidence rate is 1%[1]. Recurrent AAU may lead to glaucoma, cataract development and significant visual loss. Uveitis is a major cause of eye disease, affecting an estimated 2 million Americans, and accounts for up to 10% of blindness[2, 3].
Evidence from both humans and animal models suggests a large genetic component to uveitis with strong familiality demonstrated[4]. The first-degree recurrence risk of AAU is 6% compared to a population prevalence of only 0.038–0.38%[5]. AAU occurs in 30–40% of individuals with ankylosing spondylitis (AS) suggesting a shared etiology[6]. It is strongly associated with HLA-B27 both in those with AS, and those without, with more than 50% of those with primary AAU being HLA-B27 positive[7]. There is evidence that genes other than HLA-B influence the risk of developing AAU. The prevalence of AAU in the HLA-B27 positive first-degree relatives of probands with AAU (13%) is much higher than in the normal HLA-B27 positive (Dutch) population (1%), indicating that other genetic factors besides HLA-B27 are involved[5]. In the same study, 11% of HLA-B27 positive first-degree relatives >45 years of age had AS compared with an expected frequency of AS in HLA-B27 carriers of ~1%, highlighting the strong co-familiality of AS and AAU. Other genetic associations described for AAU include HLA-A*02[8], HLA-DRB1*08:03[9], HLA-B*58[10], MICA[11], LMP2[12], CYP27B1[13], IL10[14], the complement components CFB, CFH and C2[15, 16], TNF[17, 18], the Killer Immunoglobulin Receptor (KIR) region[19], and suggestive linkage has been reported to the chromosome 9p region[20]. No findings have achieved genomewide significance (P < 5×10−8), and few associations have been replicated[6]. Few of these studies were adequetely powered to identify genes involved in AAU reliably so we sought to investigate its association in the largest data set assembled for this purpose to date.
PATIENTS AND METHODS
To identify AAU genetic associations, two main analyses were performed. AS patients with AAU (as cases) were first compared with AS patients without AAU (as controls). Whilst this analysis studies AAU genetic associations controlling for AS-comorbidity, potential issues such as delayed onset of uveitis and subclinical disease affect it. Therefore a second analysis compared all AAU patients with healthy controls (HC), with a subsequent heterogeneity test performed to assess whether associated SNPs had differing effect sizes in AS patients with and without uveitis. Genetic associations identified comparing AS patients with AAU to HC, that were not identified by the larger and better powered IGAS AS Immunochip study[21], are likely to be AAU-specific associations.
Immunochip Sample Collection and Phenotyping
AS cases (defined by the modified New York criteria[22]) of European descent either with (n=1,422) or without AAU (n=2,339) were recruited. Ophthalmologists also collected 289 patients with AAU (in whom the AS status was unknown). AS cases had either self-reported or ophthalmologist diagnosed AAU (Supplementary Table 1). Patients were positively selected on the basis of their phenotype, no exclusions were applied. Historical genotypes from 10,000 Caucasian controls from the 1958 British Birth Cohort and the UK National Blood Transfusion Service were used as common controls. All patients gave informed consent and ethical approval was obtained from all relevant institutional ethics committees.
After quality control (Supplementary Table 2) there remained 9,564 controls, 1,199 AS cases with AAU, 238 AAU cases with alone and 1,731 AS cases without AAU patients.
Samples were genotyped on the Illumina Immunochip microarray. Intensity data was processed and normalized in the Illumina GenomeStudio software and subsequently clustered with optiCall[23].
Quality Control
The thresholds used were a genotyping missingness rate of 0.03, between centre missingness threshold of 1×10−7, individual missingness rate of 0.03, a Hardy Weinberg Equilibrium (HWE) threshold in controls of 1×10−7. Heterozygosity versus missingness outliers beyond 3 SD were excluded. Identity by descent (IBDes) threshold of PI-HAT (Proportion(IBDes=2) + 0.5(IBDes=1)) 0.20 was used. Principal components were then computed using SHELLFISH (http://www.stats.ox.ac.uk/~davison/software/shellfish/shellfish.php) including the HAPMAP populations. Individuals identified as non-European, by model-based unsupervised clustering implemented in R by the MCLUST package, were excluded. Details of SNPs excluded are shown in Supplementary Table 3.
Association Analysis
Case-control analysis was performed with SNPTEST version 2.5 beta using the ‘expected’ method and including 10 eigenvectors, scree plots are shown in Supplementary Figure 1. Genomic inflation factor 1000 values for the Immunochip control SNPs were 1.058 for the AAU versus HC analysis and 1.035 for the AS with AAU versus AS without AAU analysis. The Q-Q plots for the studies are shown in Supplementary Figure 2. Results were deemed significant if they had P < 5 × 10−8 (genome-wide significance), and suggestive if they had P values > 5 × 10−8 but < 5 × 10−6. After conditional analyses, analyses with significance of P < 1 × 10−4 were reported.
Imputation
Data was phased with SHAPEIT version 2[24], and imputed with IMPUTE2[25] using the 1000G phase 1 integrated variant reference set. A post-imputation quality control threshold of 0.8 ‘info’ score from IMPUTE2 was used. Classical Major Histocompatibility Complex (MHC) alleles were imputed from the genotype data with SNP2HLA[26] against the supplied type 1 diabetes reference set. The carriage rate of HLA-B27 was calculated using imputed SNP2HLA doses and a dosage threshold of 0.6.
Tag SNP calculation
Imputed genotypes of each individual and their classical alleles imputed by SNP2HLA were used to calculate sensitivity and specificity of the SNPs for tagging classical alleles.
Interaction Analysis
This was completed using the formula:
where the uveitis phenotype ψ and the imputed SNP dosage of rs2032890(βSNPSNP, lead ERAP1 associated SNP) and imputed SNP2HLA dosage of the HLA-B27 (βB27B27) and ten eigenvectors were regressed with an interaction term between the SNP and HLA-B27 (βintSNP×B27). β0 is the intercept.
Comparison of SNP effects between AS with and without AAU
The HC were split into two by random sampling and allocated as controls to either the AS with AAU cases or the AS without AAU cases to ensure the independence of the two regressions. These two sets of cases and controls were then allocated a dummy variable of 1 or 0 coded as Z. The following regression model was then used:
where ψ is the uveitis phenotype (either 1=uveitis in the ‘AS with AAU’ cases, or 1=no uveitis in the ‘AS without AAU’ cases, controls in both sets were coded=0). βsnp is the regression co-efficient for the SNP dose, SNP is the dosage of the SNP genotype. are the regression coefficients of the ten principal components, PCi from 1–10 are the ten principal components. βZ is the regression co-efficient for the dummy variable Z. BZ2 is the regression co-efficient for the interaction term between the dummy variable and the SNP dosage: SNP×Z. β0 is the intercept. We also analysed the model with the HLA- B27 as a component:
In this model(βB27B27)is the SNP2HLA imputed dosage of HLA-B27.
Ankylosing spondylitis GWAS data for ANTXR2 analysis
The post quality control data from the Australo-Anglo-American Spondylitis Consortium (TASC) and Wellcome Trust Case Control Consortium-2 (WTCCC2) studies and an unpublished Canadian AS genome wide association study (GWAS) were taken through the same quality control process as the Immunochip data in this study. The Canadian study using the Illumina OmniExpress microarray and included 189 AS patients. Briefly, the WTCCC2-TASC study genotyped 3,023 cases and 8,779 controls[27]. The TASC study examined 2,951 AS cases and 6,658 HC, with 439 cases removed in QC[28].
ANTXR2 locus imputation was completed with SHAPEIT and IMPUTE2 identical to the Immunochip data. The MHC SNPs were used to impute classical alleles using SNP2HLA to determine HLA-B27 dosage (range 0–2).
Previously Reported AAU Genetic Associations
Where the exact SNP was not present on the Immunochip microarray, the SNP with the highest LD represented on the Immunochip was determined from 1000 genomes data.
Concordance of Effect Directions
To assess whether shared associations had the same or opposite directions of effect the in-phase alleles were calculated using HaploXT (http://genome.sph.umich.edu/wiki/Haploxt) and 1000 genomes reference data.
HLA-B27 Heterozygosity and Homozygosity Calculations
To calculate the odds of the disease for homozygosity and heterozygosity of HLA-B27 the SNP2HLA imputed doses of HLA-B27 in the AAU group versus HC were used. Contingency table analysis and the cross-products ratio was used.
ANTXR2 Expression RNA isolation
For each experiment, total RNA was isolated from a 25–50 mg piece of mouse lung or the dissected components pooled from 5 mouse eyes using the PureLink RNA kit (Ambion). Eyes were dissected to isolate the following components: (i) cornea, (ii) iris including ciliary body, (iii) choroid and sclera, (iv) retina. Briefly, tissues were collected, placed directly into lysis buffer containing β−mercaptoethanol and homogenised using a Tissue Lyzer II (Qiagen). The lysates was then put through PureLink RNA columns, treated with DNase I and eluted in RNase free water. The purity and quantity of RNA was assessed with a BioPhotometer (Eppendorf).
ANTXR2 Expression Real-time quantitative PCR
The relative expression of ANTXR1 and ANTXR2 mRNA was determined using a two-step RT-PCR assay by comparison to L32 mRNA. First, cDNA was generated from 2µg of total RNA using random primers and M-MLV Reverse Transcriptase (Promega). Second, cDNA samples were then used in the RT-PCR using the Bio-Rad SsoAdvanced Universal SYBR Green Supermix and run on a Bio-Rad CFX Connect system. The primer sequences used for ANTXR1 and ANTXR2 were from the Harvard Primer Bank (ANTXR1; ID# 32189436a1 and ANTXR2; ID# 13278124a1). The primer sequences for L32 were L32-F (5’-CATCGGTTATGGGAGCAAC-3’) and L32-R (5’-GCACACAAGCCATCTACTCAT-3’). Samples were run in triplicate and the assay repeated 3 times. The amounts of ANTXR1 and 2 mRNA in the lung and various eye compartments were normalized relative to L32 mRNA as lung has been shown to express both ANTXR1 and ANTXR2 genes[29].
RESULTS
To examine for the validity of the self-report diagnosis of AAU we calculated HLA-B27 carriage rates. The frequency of HLA-B27, as inferred from SNP2HLA imputation, was 81.8% (613/749) in the ophthalmologist diagnosed AAU group and 92.0% (633/688) in the self-report AAU group. Of note the self-report group were entirely AS patients whereas the ophthalmologist diagnosed AAU group included both AAU cases with and without AS.
Major Histocompatibility Complex Associations
AAU has been linked with classical MHC alleles previously so we examined the MHC for association. In the comparison of AS with AAU versus AS without AAU, single nucleotide polymorphisms (SNPs) in the MHC Class I region harboring HLA-B were strongly AAU-associated, see Supplementary Figure 3 (rs115879499, P=4.9×10−18, OR=1.4, 95% CI 1.2–1.5). Conditioning on this SNP showed association at rs9274411 in HLA-DQB1 (P=6.4×10−5, OR=1.2, 95% CI 1.1–1.3). Conditioning on these top 2 associations showed no further association (P > 1×10−4). When HLA-B27 negative subjects (as determined by SNP2HLA classical MHC allele imputation) were analysed separately there was moderate association at rs114199502 (P=2.6×10−5, OR=2.9 95% CI 1.3–6.6) between HLA-DRA and HLA-DRB5. Controlling for the effect of rs114199502 there was moderate association at the intronic variant rs71542449 in HLA-DQB2 (P=4.7×10−5, OR=0.57, 95% CI 0.39–0.84).
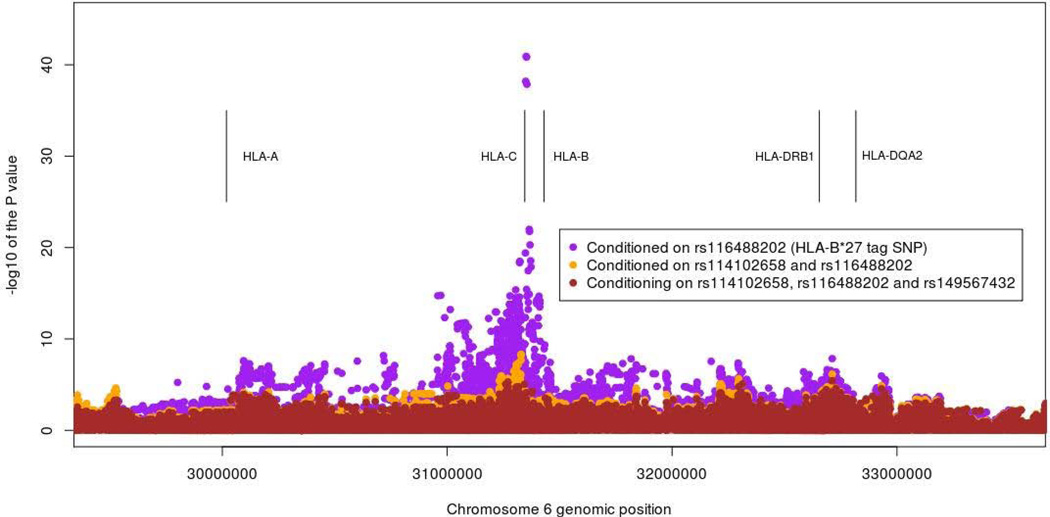
In the analysis of all AAU versus HC, strong association was also seen in the MHC, the lead SNP being the previously described[21] HLA-B27 tag SNP rs116488202 (P < 1×10−300, OR=16.8, 95% CI 15.0–18.7). On conditioning on this SNP the next associated SNP was rs114102658, between HLA-B and MICA, see Figure 1 (P=1.2×10−41, OR=17.4, 95% CI 16.7–19.4). Conditioning on both rs116488202 and rs114102658 lead association was seen with rs149567432, just centromeric to HLA-B (P=4.9×10−9, OR=9.0, 95% CI 8.2–9.9). None of these SNPs tag any classical class I allele accurately (all sensitivities and specificities < 70%). Conditioning on these top three signals, association was seen with rs114977878 in the MHC Class II locus in the gene HLA-DQA2 (P=3.8×10−6, OR=1.4, 95% CI 1.3–1.6). Conditioning on these 4 SNPs revealed association at rs115711695, an intronic variant in HLA-DRB5 (P=6.2×10−6, OR=1.7, 95% CI 1.5–1.9). Conditioning on these top 5 SNPs still left association in the HLA-B locus at rs114560492 (P=3.1×10−5, OR=1.9, 95% CI 1.7–2.0). Conditioning on these top 6 associations left no further residual signals (P > 1×10−4).
Figure 1.
Major histocompatibility complex association plot for all acute anterior uveitis versus healthy controls
Some AS patients do not carry HLA-B27 so to examine for MHC associations in this group we analysed HLA-B27 negative patients, as assessed by any imputed dose of the SNP2HLA imputed HLA-B27. In the all AAU versus HC analysis there was association at the HLA-B locus at rs115937001 (P=2.0×10−5, OR=1.8, 95% CI 1.5–2.3). This SNP tags HLA-B0801 with 76% specificity and 94% sensitivity. Conditioning on this SNP left no residual association (P>10−4).
Allelic diversity at classical HLA loci is extensive, and challenging to impute with single SNPs. Therefore imputation with multiple SNPs was performed with SNP2HLA, to provide a potentially more accurate assessment of classical MHC allele-disease associations[26]. In analysis of AS with AAU versus AS without AAU, the classical allele HLA-B27 was again strongly associated (P=1.4×10−16, OR=2.1, 95% CI 1.8–2.5). Controlling for the HLA-B27 effect showed residual association with HLA-DQB1:05 (P=2.1×10−5, OR=0.78, 95% CI 0.70–0.87). In the all AAU versus HC analysis, strong association with HLA-B27 was evident (P < 1×10−300 OR=59.7, 95% CI 51.4–69.5). Controlling for the HLA-B27 effect, HLA-DRB1:0103 was associated (P=2.0×10−5, OR=1.9, 95% CI 1.4–2.5).
The question of whether HLA-B27 exerts its influence through a dominant or additive genetic model was assessed. In the all AAU versus HC analysis heterozygosity for HLA-B27 (as determined by SNP2HLA imputation) confers OR for AAU of 66.8 (95% CI 66.7 – 67.0), homozygosity for HLA-B27 confers OR of 130.6 (95% CI 130.1 – 131.1), and the risk of two HLA-B27 alleles over one HLA-B27 allele confers an OR of 2.0 (95% CI 1.5 – 2.4). Thus homozygosity for HLA-B27 does confer additional risk of AAU over heterozygosity (see Supplementary Table 4).
The carriage rates for HLA-B27 (as determined by SNP2HLA imputation) were 91.2% in AS cases with AAU 80.6% in AS cases without AAU, and only 63.9% in the small cohort of AAU patients with unknown AS status (recruited by ophthalmologists).
Non-MHC Associations
A number of non-MHC associations have been described for AAU previously so we sought to examine non-MHC areas for association. In the comparison of AS cases with AAU versus AS cases without AAU, association was observed with variants within ERAP1 (rs2032890, P=9.0×10−6, OR=1.3, 95% CI 1.2–1.5), see Supplementary Table 5 and Supplementary Figure 4. In AS there is an interaction between ERAP1 and HLA-B27[27]. Controlling for HLA-B27 using SNP2HLA dosages[21] as a covariate in the analysis, association was essentially unchanged at rs2032890 (P=2.9×10−5, OR=1.3, 95% CI 1.2–1.5). Performing a regression analysis with an interaction term between HLA-B27 (as determined by SNP2HLA) and rs2032890 was negative (P=0.28). However, when subjects were split into HLA-B27 negative and HLA-B27 positive groups and the rs2032890 SNP was assessed with logistic regression in each group it was associated in the HLA-B27 positive group but not the negative group (P=1.6 × 10−5, OR=1.26, 95% CI 1.13 – 1.40 and P=0.76, OR=1.05, 95% CI 0.78 – 1.42 respectively).
In the all AAU cases vs HC analysis, genomewide significant associations were observed at ERAP1, the intergenic region chromosome 2p15, and IL23R. The observed associations were concordant with associations with AS, involving the same haplotypes with the same direction of effect (Table 1 and Supplementary Figure 5–12). In this study the logistic regression model examining the interaction between the ERAP1 SNP rs2032890 and HLA-B27 was strongly significant (P < 2×10−16). In HLA-B27 negative participants (as determined by SNP2HLA imputation) there was association over ERAP2 (lead SNP: rs4869314, P=8.8×10−5), consistent with that previously seen in HLA-B27 negative AS. After conditioning on this SNP no residual association was seen in ERAP2 (P > 10−4).
Table 1.
Association results for the AS with AAU versus AS without AAU analysis
| rsid | Chr | Position* | P value | Risk Allele |
Protective Allele |
RAF (Cases/Controls) |
OR | OR Lower |
OR Upper |
Nearby Genes | Also AS Associated |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs2032890 | 5q15 | 96,121,152 | 2.11E-16 | A | C | 0.77/0.69 | 1.51 | 1.37 | 1.66 | ERAP1/ERAP2/LNPEP | Yes |
| rs4672507 | 2p15 | 62,570,573 | 2.05E-12 | T | A | 0.43/0.36 | 1.38 | 1.27 | 1.50 | intergenic | Yes |
| rs79755370 | 1p31 | 67,699,915 | 1.27E-08 | C | A | 0.96/0.94 | 1.80 | 1.45 | 2.23 | IL23R | Yes |
| rs12132349 | 1q32 | 200,875,242 | 1.57E-07 | T | A | 0.76/0.71 | 1.31 | 1.19 | 1.44 | KIF21B–C1orf106 | Yes |
| rs6690230 | 1q21 | 154,432,877 | 1.09E-06 | C | G | 0.41/0.36 | 1.23 | 1.13 | 1.34 | IL6R | Yes |
| rs17351243 | 1q32 | 206,959,527 | 1.46E-06 | A | G | 0.51/0.45 | 1.24 | 1.14 | 1.35 | IL10-IL19 | No |
| rs10197284 | 2q12 | 102,982,703 | 1.67E-06 | G | A | 0.26/0.22 | 1.25 | 1.14 | 1.38 | IL18R1-IL1R1 | No |
| rs665873 | 6q12 | 65,298,551 | 4.99E-06 | A | G | 0.98/0.97 | 2.03 | 1.48 | 2.79 | EYS | No |
Chr Chromosome,
Human genome build 19, RAF Risk allele frequency, OR Odds ratio, AS ankylosing spondylitis, AAU acute anterior uveitis
In the analysis of all AAU cases with HC, five loci had suggestive levels of association (P < 5×10−6). These loci included both loci known to be associated with AS (IL6R, chromosome 1q32) and novel loci not previously reported to be AS-associated including interleukin (IL)10 - IL19, the gene encoding the alpha chain of the IL-18 receptor (IL18R1)-IL1R1, and the EYS gene (Eyes shut Drosophila homolog, a gene associated with retinitis pigmentosa[30, 31]). The IL6R and chromosome 1q32 AAU associations are concordant with the AS associations (alleles are in phase and r2=0.98 and 1.0, respectively). On conditioning on the 1q32 association (rs12132349), a secondary association nearby at rs10920074 became apparent (P=1.7×10−7, OR=1.3 95% CI 1.2–1.4), LD between these SNPs is D’=0.87 and r2=0.48 (source: 1000 genomes). No further association was apparent after conditioning on both rs10920074 and rs12132349 (P > 1×10−4). At the IL18R1-IL1R1 association conditioning on rs10197284 shows a secondary signal at rs6750020 (P=1.2×10−4, OR=1.2, 95% CI 1.1–1.4), linkage disequilibirum (LD) between these SNPs is D’=0.92 and r2=0.85. Conditioning on rs6750020 in addition to rs10197284 removes all association (P>10−4). The IL18R1-IL1R1 AAU rs10197284 association is not in LD with the previously reported[21] suggestive AS association at rs4851529 (r2=0.00, D’=0.07). The other loci had no secondary signals on conditioning on the top SNP (P > 1×10−4).
The IL10 SNP rs17351243 has LD (D’=0.65, r2=0.52, source:1000 genomes) with the inflammatory bowel disease (IBD) associated SNP rs3024505[32], and the direction of association is the same. The IL18R1-IL1R1 SNP rs10197284 is in strong LD with previously reported celiac disease[33] and IBD[32] SNPs, and in lesser LD with asthma[34] and eosinophil count[35] SNPs (Supplementary Table 6). The IBD and celiac disease associations share the same direction of effect, but the asthma and eosinophil count SNPs have opposite directions of effect. The EYS SNP rs665873 has low frequency (HapMap CEU MAF=0.051) but is in tight LD with the common (MAF=0.43) statin-myopathy associated SNP rs3857532[36], (D’=0.91, r2=0.01, source:1000 genomes), and with six SNPs associated with retinitis pigmentosa[30] (D’=1, r2=0.001) (Supplementary Table 7). The AAU, statin-myopathy and retinitis pigmentosa SNPs all have the same direction of effect.
Odds Ratio Comparisons
We assessed whether the shared associations influence AS and AAU with differing magnitudes using a heterogeneity test. All SNPs associated with AAU (P<5×10−6) in the all AAU cases versus HC analysis and all SNPs associated with AS from the recently published IGAS Immunochip study[21] were assessed. We used two models, the first (model 1) has the SNP and principal components; the second (model 2) includes HLA-B27 dose as a model covariate, reflecting the previously demonstrated interaction between ERAP1 SNPs and HLA-B27 in AS.
The two AS-associated intergenic loci, chromosomes 2p15 and 21q22, both had significantly different effect sizes in AS patients with AAU versus HC compared to AS patients without AAU versus HC in model 2 but not model 1 (Table 2). At chromosome 2p15 both models showed a larger effect size in AS cases without AAU versus HC compared to AS with AAU versus HC, but this only reached statistical significance in model 2 (model 1: SNP rs4672507 P=0.17 and SNP rs6759298 P=0.13 and model 2: SNP rs4672507 P=0.06 and SNP rs6759298 P=0.05). The lead AAU associated ERAP1 SNP had a significantly stronger effect size in the AS with AAU versus HC compared to AS without AAU versus HC, even after taking into account HLA-B27 (model 2). The EYS SNP rs665873 had a much greater effect size (OR=0.92, 95% CI 0.88 – 0.97, P=6×10−5) in the AS with AAU compared to AS without AAU (OR=0.97, 95% CI 0.93 – 1.01, P=0.14).
Table 2.
Heterogeneity test results between AS with AAU versus healthy controls and AS without AAU versus healthy controls
| Model 1: SNP + Principle Components | Model 2: SNP + HLA-B27 + Principle Components | Interaction term | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP effects | SNP effects | ||||||||||
| SNP | Gene/Region | AS with AAU P value |
AS with AAU OR |
AS without AAU P value |
AS without AAU OR |
AS with AAU P value |
AS with AAU OR |
AS without AAU P value |
AS without AAU OR |
Model 1 P value |
Model 2 P value |
| rs2283790 | UBE2LE | 0.87 | 1.001 | 0.007 | 0.98 | 0.89 | 0.999 | 0.12 | 0.992 | 0.049 | 0.29 |
| rs2836883 | 21q22 | 0.0059 | 1.02 | 0.03 | 1.03 | 0.26 | 1.005 | 0.00017 | 1.019 | 0.19 | 0.04 |
| rs2032890 | ERAP1 | 1.80E-17 | 1.06 | 3.30E-04 | 1.03 | 0.02 | 1.02 | 0.02 | 1.01 | 0.001 | 0.08 |
| rs30187 | ERAP1 | 4.80E-19 | 1.06 | 1.04E-07 | 1.04 | 0.02 | 1.02 | 0.003 | 1.02 | 0.02 | 0.24 |
| rs10045403 | ERAP1 | 1.10E-14 | 1.05 | 5.30E-04 | 1.02 | 3.60E-04 | 1.02 | 0.03 | 0.01 | 0.005 | 0.5 |
| rs2910686 | ERAP2 | 0.18 | 1.01 | 0.03 | 0.99 | 0.71 | 1.00 | 0.14 | 0.99 | 0.01 | 0.43 |
| rs6759298 | 2p15 | 8.50E-16 | 1.06 | 1.70E-22 | 1.07 | 2.00E-06 | 1.02 | 9.70E-12 | 1.04 | 0.13 | 0.05 |
| rs4333130 | ANTXR2 | 0.048 | 0.98 | 3.00E-04 | 0.97 | 0.77 | 1.002 | 5.60E-03 | 0.98 | 0.28 | 0.04 |
| Suggestive | |||||||||||
| rs7282490 | ICOSLG | 0.98 | 1.00 | 0.007 | 1.02 | 0.41 | 1.004 | 0.01 | 1.013 | 0.054 | 0.17 |
| rs665873 | EYS | 6.10E-05 | 0.92 | 0.14 | 0.97 | 0.002 | 0.96 | 0.2 | 0.98 | 0.09 | 0.3 |
| rs4672507 | 2p15 | 4.00E-16 | 1.06 | 2.90E-22 | 1.07 | 2.60E-07 | 1.02 | 1.10E-12 | 1.04 | 0.17 | 0.06 |
SNP – single nucleotide polymorphism, AS ankylosing spondylitis, AAU acute anterior uveitis, OR odds ratio
ANTXR2 Odds Ratio Comparison
The AS associated ANTXR2 SNP rs4389526 showed a significant difference in effect size between AS with AAU and AS without AAU in model 2 (P=0.04). The rs4389526 SNP had an effect in the AS without AAU cohort (P=5.6×10−3, OR=0.98, 95% CI 0.97 – 0.99) but no effect in the AS with AAU cohort (P=0.77, OR=1.00, 95% CI 0.99 – 1.02).
Analysis of ANTXR2 expression in the eye using real-time quantitative RT-PCR
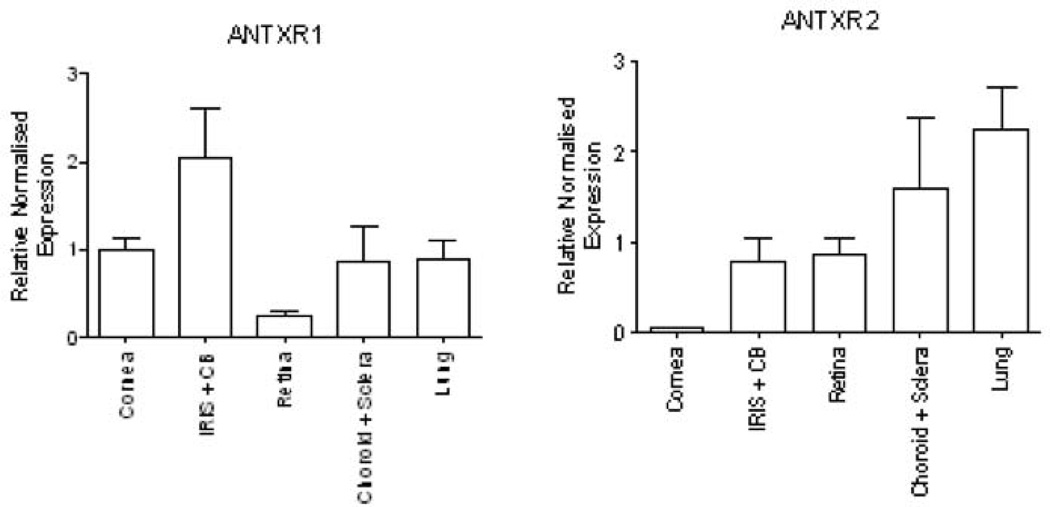
Given the negative association between ANTXR2 SNP rs4389526 and AS with AAU, we examined the expression of ANTXR2, and the related ANTXR1 (for comparison), in the eye using a murine model. ANTXR1 and ANTXR2 mRNA was detected in all the tested eye compartments, as well the lung (Figure 2). ANTXR1 expression was equivalent in the iris/ciliary body, cornea and choroid/sclera, and slightly lower in the retina (Figure 2A). By contrast, ANTXR2 was expressed most abundantly in the choroid/sclera, iris/ciliary body and retina, compared with the cornea, where expression of ANTXR2 was minimal (Figure 2B).
Figure 2.
Expression of ANTXR1 and ANTXR2 in eye compartments. Total RNA was isolated from different compartments of the eye, as well as the lung and real-time RT-PCR performed to determine the relative abundance of ANTRX1 and ANTRX2 mRNA by reference to L32 mRNA. The results represent mean and SEM from three independent experiments, each including triplicate samples
Previous AAU associations
A number of previous non-MHC associations have been reported for AAU. Therefore we sought to examine whether there was evidence to support their reported association. While not all previously reported AAU genetic associations were on the Immunochip microarray, we were able to investigate a number of regions previously reported to be associated with AAU. Of the previously reported AAU associations within the MHC, all were strongly associated in the AAU versus HC analysis (Table 3). After controlling for the lead MHC SNP (rs116488202), no residual association was seen with TNF SNPs previously reported to be AAU-associated, although nominal association was seen with TNF-238 (rs1800629, P=0.003). This association disappeared after conditioning on the next two associated HLA Class I SNPs rs114102658 and rs149567432 (P=0.08). Outside the MHC, nominally significant associations were observed with SNPs previously reported to be AAU-associated in IL10 and CYP27B1 (IL10, rs6703630, P=0.04 and CYP27B1, rs703842,P=0.003). As mentioned above, much stronger association was seen in the current study with the IL10 SNP rs17351243; this SNP is in moderately strong LD with the previously reported IL10 association rs6703630 (D’=0.87 and r2 =0.24 current dataset; D’=0.71 and r2=0.09 1000 Genomes)[14]. This suggests that they may tag the same association, as controlling for association at rs17351243 means rs6703630 becomes non-significant (P=0.7).
Table 3.
Previous AAU associations
| Chr | Position* | rsid | Gene | Uncond P value |
B27 Cond P value |
Top two Cond P value | Top Three Cond P value |
Reference |
|---|---|---|---|---|---|---|---|---|
| 1 | 206948639 | rs6703630 | IL10 | 0.0381204 | 14 | |||
| 1 | 206945381 | rs2222202 | IL10 | 0.550083 | 14 | |||
| 1 | 206945311 | rs3024490 | IL10 | 0.12393 | 14 | |||
| 6 | 31542482 | rs1799724 | TNF-857 | 7.04E-40 | 0.178664 | 0.01679 | 0.00718782 | 17,18 |
| 6 | 31543101 | rs361525 | TNF-308 | 2.74E-11 | 0.0680665 | 0.244505 | 0.361933 | 17,18 |
| 6 | 31543031 | rs1800629 | TNF-238 | 7.79E-32 | 0.00296247 | 0.0158237 | 0.0760507 | 17,18 |
| 6 | 31542308 | rs1799964 | TNF-103 | 2.67E-18 | 0.0645271 | 0.337868 | 0.303424 | 17 |
| 6 | 31542476 | rs1800630 | TNF-863 | 3.58E-09 | 0.244742 | 0.580368 | 0.440478 | 17 |
| 6 | 31914935 | rs1048709 | CFB | 4.11E-09 | 0.202212 | 0.749028 | 0.404371 | 16 |
| 12 | 58162739 | rs703842 | CYP27B1 | 0.00279726 | 13 |
Human genome build 19, Uncond: Unconditioned result. B27 Cond: Conditioned on HLA-B27 dose as imputed by SNP2HLA. Top two Cond: Result after conditioning on rs116488202 and rs114102658. Top Three Cond: Result after conditioning on rs116488202, rs114102658 and rs149567432
AS associations
Given the extensive co-morbidity between AS and AAU we examined all SNPs previously reported to be associated with AS in the AS with AAU versus AS without AAU, and the all AAU versus HC studies. The strength of association is significantly weaker than reported for AS, likely explained by small numbers of participants and hence greatly reduced power. The results are reported in Supplementary Table 8 and 9.
DISCUSSION
This study reports genetic associations specific to AAU, as well as shared associations with AS that have both similar and significantly different effect sizes and directions of association in the two related diseases.
We found evidence to suggest that MHC class I or class II alleles besides HLA-B27 contribute to AAU susceptibility. HLA-B08, which we found to be tagged by rs115937001 in HLA-B27 negative patients, has been associated previously with AAU and in addition with sarcoidosis, a disease in which AAU also occurs[37–39]. In conditional analyses HLA-DQB1:05 and HLA-DRB1:0103 have been implicated, and SNPs in or around HLA-DRA, HLA-DRB5 and HLA-DQA2 have also shown association with AAU. This suggests that additional non-HLA-B27 MHC factors affect the etiology of AAU. In our study HLA-B27 does show association even comparing AAU cases with and without AS, confirming the long held view that HLA-B27 is an AAU risk gene regardless of whether AS is present or not.
We observed a number of genetic associations shared with IBD[40, 41]. The IL10 gene is associated with IBD, and IL-10 is important in an animal model of experimentally-induced uveitis, as treatment with IL-10 abrogates disease in this model [42]. It has also been shown recently that IL-10 is important in IL-35 induced B regulatory cell suppression of EAU[43]. In humans with AAU, peripheral blood mononuclear cells (PBMCs) show up-regulation of both IL-10 and IL-19[44]. Patients with AS who also have IBD have much lower rates of HLA-B27, offering relative protection from AAU. In patients with IBD up to 3.8% develop uveitis, a rate substantially lower than in AS[45].
The suggestive association of the IL-18 receptor highlights the importance of the innate immune recognition of foreign microorganisms and the triggering of an appropriate adaptive immune response. Research has linked infection with organisms such as chlamydia with AAU, and the mechanism of this association may be related to inappropriate or abnormal activation of a cell-mediated immune response by IL-18[46, 47]. In sarcoidosis, a condition in which AAU occurs, enhanced expression of IL-18R alpha chain has been noted in CD4 T cells[48].
In considering the shared associations between AS and AAU, the intergenic region chromosome 21q22 and ANTXR2 have an effect in only AS patients without AAU, and not in AS patients with AAU. Further, the chromosome 2p15 intergenic region shows significantly greater association in AS without AAU. These findings suggest that there may be genetic subgroups in AS with heterogeneity in the genetic profiles of AS patients, and that genetic factors influence which AS patients develop AAU and which do not. A suggestive association between SNPs 17kb upstream of ANTXR2 and myopia has been described[49]. But with no effect in the AS with AAU cohort it seems ANTXR2 is potentially an AS risk locus alone. However, the difference in ANTXR2 expression across the different components of the eye is of interest in view of the positive genetic associations. The mechanisms for disease association have not been identified at either chromosomes 2p15 or 21q22; long non-coding mRNA transcripts have been identified, but their function is currently unknown[50].
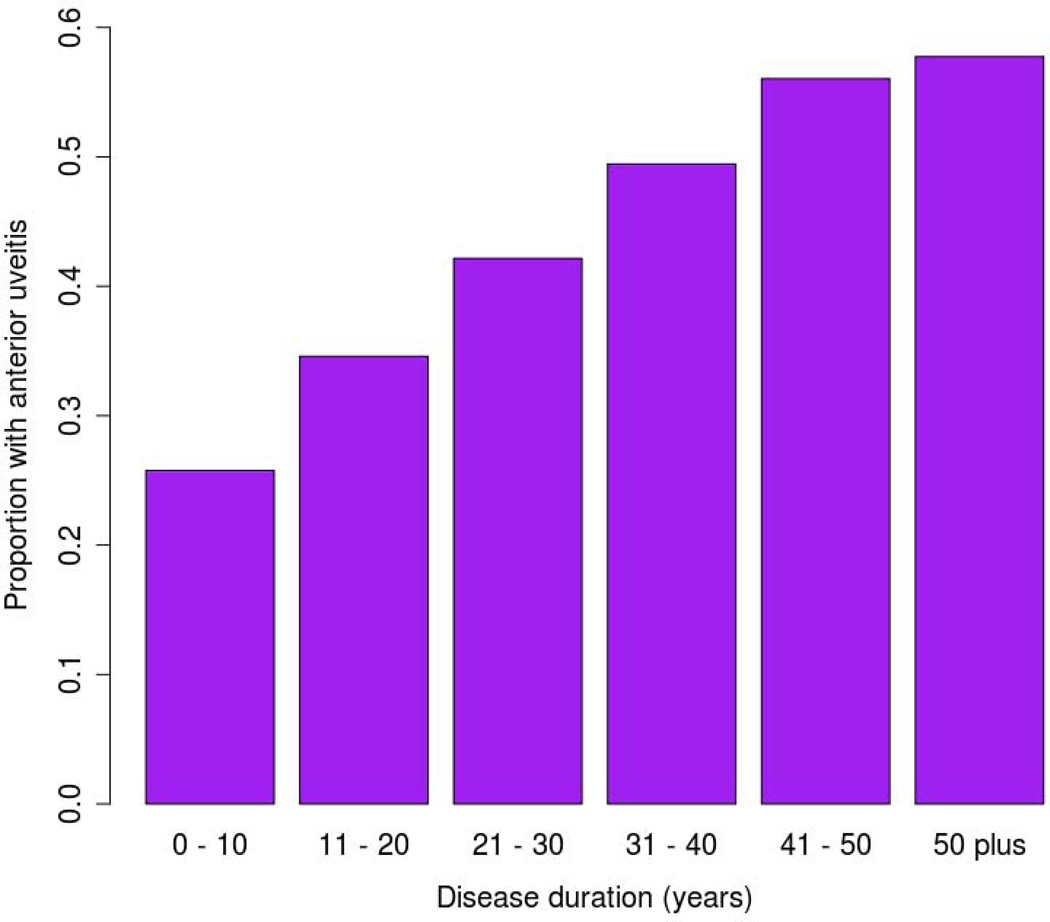
These results should be interpreted in light of several limitations; the study is underpowered and additional cases are likely to result in new associations. In addition, the suggestive associations require replication to be considered robustly associated. The use of self-report AAU is a potential limitation, although self-report has been shown to differ little from ophthalmologist diagnosis in SpA patients previously[51]. The high incidence of AAU in AS, and the progressive increase in AAU penetrance with disease duration, means that many AS cases currently classified as AAU-negative may ultimately develop AAU (Figure 3). There may also have been issues with under diagnosis of AAU in subclinical cases, AS treatment suppressing AAU manifesting or where the diagnosis was missed even if clinically apparent. Accurately quantifying the power of the study is not possible because the size of these effects is completely unknown. Finally there are environmental factors in the etiology of AAU, and therefore there is the potential issue of incomplete penetrance of AAU-susceptibility loci.
Figure 3.
Proportion of ankylosing spondylitis patients with acute anterior uveitis by ankylosing spondylitis disease duration. Source: Ankylosing spondylitis GWAS patient cohort
Because of these problems we took advantage of the availability of a large study of AS-susceptibility loci, reasoning that any novel loci found with the AAU patient sets compared to HC in the small sample sizes used were highly likely to be AAU associated loci. From this analysis we have been able to identify a number of novel immune related loci associated with AAU. The two shared associations with IBD are of particular interest because these are not associations shared with AS. The pathways identified will help identify novel treatment strategies for this common and important disease and highlights further pathways shared between AS, IBD and AAU.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank all the subjects for contributing to the study. PR is funded by the National Health & Medical Research Council (NHMRC) of Australia and the University of Queensland Diamantina Institute. TAMC was supported by a fellowship from Saal van Zwanenbergstichting, The Netherlands. TMM was funded by the National Eye Institute (R01-EY013139). MAB is a Senior Principal Research Fellow of the NHMRC. The Australo-Anglo-American Spondylitis Consortium (TASC) study was funded by National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) grants P01-052915 and R01-AR046208. The study was supported by NIH/National Center for Advancing Translational Science (NCATS) UCLA CTSI Grant Number UL1TR000124, (Prime: University of California, Los Angeles), 3/1/13 to 2/28/14 and NIH/National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant Number PO1 AR052915, Program Project Grant – Genetics and Ankylosing Spondylitis (AS) Pathogenesis (PI: Reveille), 7/01/2013-6/30/2014. The Spondyloarthritis Research Consortium of Canada (SPARCC) was funded by The Arthritis Society. The Casey Eye Institute is supported by the Research to Prevent Blindness, New York, NY, USA. Funding support from Arthritis Research UK (grants 19536 & 18797) and support of the National Ankylosing Spondylitis Society and NIHR Thames Valley CLRN in the recruitment of cases.
Footnotes
Membership of consortia listed as authors is detailed in a Supplementary note.
No author has a potential conflict of interest in relation to this work.
References
- 1.Linssen A, Rothova A, Valkenburg HA, Dekker-Saeys AJ, Luyendijk L, Kijlstra A, et al. The lifetime cumulative incidence of acute anterior uveitis in a normal population and its relation to ankylosing spondylitis and histocompatibility antigen HLA-B27. Investigative ophthalmology & visual science. 1991;32:2568–2578. [PubMed] [Google Scholar]
- 2.Caspi RR. Understanding Autoimmune Uveitis through Animal Models The Friedenwald Lecture. Investigative ophthalmology & visual science. 2011;52:1873–1879. doi: 10.1167/iovs.10-6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gritz DC, Wong IG. Incidence and prevalence of uveitis in Northern California - The Northern California Epidemiology of Uveitis Study. Ophthalmology. 2004;111:491–500. doi: 10.1016/j.ophtha.2003.06.014. [DOI] [PubMed] [Google Scholar]
- 4.Pennesi G, Caspi RR. Genetic control of susceptibility in clinical and experimental uveitis. International reviews of immunology. 2002;21:67–88. doi: 10.1080/08830180212059. [DOI] [PubMed] [Google Scholar]
- 5.Derhaag PJ, Linssen A, Broekema N, de Waal LP, Feltkamp TE. A familial study of the inheritance of HLA-B27-positive acute anterior uveitis. American journal of ophthalmology. 1988;105:603–606. doi: 10.1016/0002-9394(88)90051-7. [DOI] [PubMed] [Google Scholar]
- 6.Robinson PC, Brown MA. Genetics of ankylosing spondylitis. Mol Immunol. 2014;57:2–11. doi: 10.1016/j.molimm.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Martin TM, Rosenbaum JT. An update on the genetics of HLA B27-associated acute anterior uveitis. Ocul Immunol Inflamm. 2011;19:108–114. doi: 10.3109/09273948.2011.559302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keino H, Sakai J, Usui M. Association between HLA-A2 in Japanese psoriasis arthritis and susceptibility to uveitis. Graefe's archive for clinical and experimental ophthalmology = Albrecht von Graefes Archiv fur klinische und experimentelle Ophthalmologie. 2003;241:777–778. doi: 10.1007/s00417-003-0706-9. [DOI] [PubMed] [Google Scholar]
- 9.Monowarul Islam SM, Numaga J, Fujino Y, Masuda K, Ohda H, Hirata R, et al. HLA-DR8 and acute anterior uveitis in ankylosing spondylitis. Arthritis Rheum. 1995;38:547–550. doi: 10.1002/art.1780380414. [DOI] [PubMed] [Google Scholar]
- 10.Orchard TR, Chua CN, Ahmad T, Cheng H, Welsh KI, Jewell DP. Uveitis and erythema nodosum in inflammatory bowel disease: clinical features and the role of HLA genes. Gastroenterology. 2002;123:714–718. doi: 10.1053/gast.2002.35396. [DOI] [PubMed] [Google Scholar]
- 11.Goto K, Ota M, Maksymowych WP, Mizuki N, Yabuki K, Katsuyama Y, et al. Association between MICA gene A4 allele and acute anterior uveitis in white patients with and without HLA-B27. American journal of ophthalmology. 1998;126:436–441. doi: 10.1016/s0002-9394(98)00100-7. [DOI] [PubMed] [Google Scholar]
- 12.Maksymowych WP, Jhangri GS, Gorodezky C, Luong M, Wong C, Burgos-Vargas R, et al. The LMP2 polymorphism is associated with susceptibility to acute anterior uveitis in HLA-B27 positive juvenile and adult Mexican subjects with ankylosing spondylitis. Ann Rheum Dis. 1997;56:488–492. doi: 10.1136/ard.56.8.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steinwender G, Lindner E, Weger M, Plainer S, Renner W, Ardjomand N, et al. Association between polymorphism of the vitamin D metabolism gene CYP27B1 and HLA-B27-associated uveitis. Is a state of relative immunodeficiency pathogenic in HLA B27-positive uveitis? PLoS One. 2013;8:e62244. doi: 10.1371/journal.pone.0062244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atan D, Fraser-Bell S, Plskova J, Kuffova L, Hogan A, Tufail A, et al. Cytokine polymorphism in noninfectious uveitis. Investigative ophthalmology & visual science. 2010;51:4133–4142. doi: 10.1167/iovs.09-4583. [DOI] [PubMed] [Google Scholar]
- 15.Yang MM, Lai TY, Tam PO, Chiang SW, Chan CK, Luk FO, et al. CFH 184G as a genetic risk marker for anterior uveitis in Chinese females. Mol Vis. 2011;17:2655–2664. [PMC free article] [PubMed] [Google Scholar]
- 16.Yang MM, Lai TY, Tam PO, Chiang SW, Ng TK, Liu K, et al. Association of C2 and CFB polymorphisms with anterior uveitis. Investigative ophthalmology & visual science. 2012;53:4969–4974. doi: 10.1167/iovs.12-9478. [DOI] [PubMed] [Google Scholar]
- 17.Kuo NW, Lympany PA, Menezo V, Lagan AL, John S, Yeo TK, et al. TNF-857T, a genetic risk marker for acute anterior uveitis. Investigative ophthalmology & visual science. 2005;46:1565–1571. doi: 10.1167/iovs.04-0932. [DOI] [PubMed] [Google Scholar]
- 18.El-Shabrawi Y, Wegscheider BJ, Weger M, Renner W, Posch U, Ulrich S, et al. Polymorphisms within the tumor necrosis factor-alpha promoter region in patients with HLA-B27-associated uveitis: association with susceptibility and clinical manifestations. Ophthalmology. 2006;113:695–700. doi: 10.1016/j.ophtha.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Levinson RD, Martin TM, Luo L, Ashouri E, Rosenbaum JT, Smith JR, et al. Killer cell immunoglobulin-like receptors in HLA-B27-associated acute anterior uveitis, with and without axial spondyloarthropathy. Investigative ophthalmology & visual science. 2010;51:1505–1510. doi: 10.1167/iovs.09-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin TM, Zhang G, Luo J, Jin L, Doyle TM, Rajska BM, et al. A locus on chromosome 9p predisposes to a specific disease manifestation, acute anterior uveitis, in ankylosing spondylitis, a genetically complex, multisystem, inflammatory disease. Arthritis Rheum. 2005;52:269–274. doi: 10.1002/art.20777. [DOI] [PubMed] [Google Scholar]
- 21.Cortes A, Hadler J, Pointon JP, Robinson PC, Karaderi T, Leo P, et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat Genet. 2013;45:730–738. doi: 10.1038/ng.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Linden S, Valkenburg HA, Cats A. Evaluation of diagnostic criteria for ankylosing spondylitis. A proposal for modification of the New York criteria. Arthritis Rheum. 1984;27:361–368. doi: 10.1002/art.1780270401. [DOI] [PubMed] [Google Scholar]
- 23.Shah TS, Liu JZ, Floyd JA, Morris JA, Wirth N, Barrett JC, et al. optiCall: A robust genotype-calling algorithm for rare, low frequency and common variants. Bioinformatics. 2012;28:1598–1603. doi: 10.1093/bioinformatics/bts180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 25.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia X, Han B, Onengut-Gumuscu S, Chen WM, Concannon PJ, Rich SS, et al. Imputing amino Acid polymorphisms in human leukocyte antigens. PLoS One. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans DM, Spencer CC, Pointon JJ, Su Z, Harvey D, Kochan G, et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet. 2011;43:761–767. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reveille JD, Sims AM, Danoy P, Evans DM, Leo P, Pointon JJ, et al. Genome-wide association study of ankylosing spondylitis identifies non-MHC susceptibility loci. Nat Genet. 2010;42:123–127. doi: 10.1038/ng.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Q, Hesek ED, Zeng M. Transcriptional stimulation of anthrax toxin receptors by anthrax edema toxin and Bacillus anthracis Sterne spore. Microbial pathogenesis. 2007;43:37–45. doi: 10.1016/j.micpath.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abd El-Aziz MM, O'Driscoll CA, Kaye RS, Barragan I, El-Ashry MF, Borrego S, et al. Identification of novel mutations in the ortholog of Drosophila eyes shut gene (EYS) causing autosomal recessive retinitis pigmentosa. Investigative ophthalmology & visual science. 2010;51:4266–4272. doi: 10.1167/iovs.09-5109. [DOI] [PubMed] [Google Scholar]
- 31.Abd El-Aziz MM, Barragan I, O'Driscoll CA, Goodstadt L, Prigmore E, Borrego S, et al. EYS, encoding an ortholog of Drosophila spacemaker, is mutated in autosomal recessive retinitis pigmentosa. Nat Genet. 2008;40:1285–1287. doi: 10.1038/ng.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 36.Isackson PJ, Ochs-Balcom HM, Ma C, Harley JB, Peltier W, Tarnopolsky M, et al. Association of common variants in the human eyes shut ortholog (EYS) with statin-induced myopathy: evidence for additional functions of EYS. Muscle & nerve. 2011;44:531–538. doi: 10.1002/mus.22115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nussenblatt RB, Mittal KK. Iridocyclitis in black Americans: association with HLA B8 suggests an autoimmune aetiology. Br J Ophthalmol. 1981;65:329–332. doi: 10.1136/bjo.65.5.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith MJ, Turton CW, Mitchell DN, Turner-Warwick M, Morris LM, Lawler SD. Association of HLA B8 with spontaneous resolution in sarcoidosis. Thorax. 1981;36:296–298. doi: 10.1136/thx.36.4.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hedfors E, Lindstrom F, HLA-B8/DR3 in sarcoidosis. Correlation to acute onset disease with arthritis. Tissue Antigens. 1983;22:200–203. doi: 10.1111/j.1399-0039.1983.tb01192.x. [DOI] [PubMed] [Google Scholar]
- 40.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 2013;14:661–673. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 41.Costello M-E, Elewaut D, Kenna T, Brown MA. Microbes, the Gut and Ankylosing Spondylitis. Arthritis Research & Therapy. 2013;15:214. doi: 10.1186/ar4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayashi S, Guex-Crosier Y, Delvaux A, Velu T, Roberge FG. Interleukin 10 inhibits inflammatory cells infiltration in endotoxin-induced uveitis. Graefe’s archive for clinical and experimental ophthalmology = Albrecht von Graefes Archiv fur klinische und experimentelle Ophthalmologie. 1996;234:633–636. doi: 10.1007/BF00185297. [DOI] [PubMed] [Google Scholar]
- 43.Wang RX, Yu CR, Dambuza IM, Mahdi RM, Dolinska M, Sergeey YV, et al. Interleukin-35 induces regulatory B cells that suppress autoimmune disease. Nature medicine. 2014 doi: 10.1038/nm.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Z, Liu B, Maminishkis A, Mahesh SP, Yeh S, Lew J, et al. Gene expression profiling in autoimmune noninfectious uveitis disease. J Immunol. 2008;181:5147–5157. doi: 10.4049/jimmunol.181.7.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernstein CN, Blanchard JF, Rawsthorne P, Yu N. The prevalence of extraintestinal diseases in inflammatory bowel disease: a population-based study. The American journal of gastroenterology. 2001;96:1116–1122. doi: 10.1111/j.1572-0241.2001.03756.x. [DOI] [PubMed] [Google Scholar]
- 46.Wakefield D, Penny R. Cell-mediated immune response to chlamydia in anterior uveitis: role of HLA B27. Clin Exp Immunol. 1983;51:191–196. [PMC free article] [PubMed] [Google Scholar]
- 47.Huhtinen M, Puolakkainen M, Laasila K, Sarvas M, Karma A, Leirisalo-Repo M. Chlamydial antibodies in patients with previous acute anterior uveitis. Investigative ophthalmology & visual science. 2001;42:1816–1819. [PubMed] [Google Scholar]
- 48.Zhou Y, Yamaguchi E, Fukui Y, Konno S, Maeda Y, Kimata K, et al. Enhanced expression of interleukin-18 receptor alpha chain by CD4+ T cells in sarcoidosis. Chest. 2005;128:2497–2503. doi: 10.1378/chest.128.4.2497. [DOI] [PubMed] [Google Scholar]
- 49.Kiefer AK, Tung JY, Do CB, Hinds DA, Mountain JL, Francke U, et al. Genome-wide analysis points to roles for extracellular matrix remodeling, the visual cycle, and neuronal development in myopia. PLoS genetics. 2013;9:e1003299. doi: 10.1371/journal.pgen.1003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson PC, Brown MA. The genetics of ankylosing spondylitis and axial spondyloarthritis. Rheum Dis Clin North Am. 2012;38:539–553. doi: 10.1016/j.rdc.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 51.Zeboulon N, Dougados M, Gossec L. Prevalence and characteristics of uveitis in the spondyloarthropathies: a systematic literature review. Ann Rheum Dis. 2008;67:955–959. doi: 10.1136/ard.2007.075754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.