Abstract
We report evidence for an unconventional type of allosteric regulation of a biomotor. We show that the genome-packaging motor of phage ϕ29 is regulated by a sensor that detects the density and conformation of the DNA packaged inside the viral capsid, and slows the motor by a mechanism distinct from the effect of a direct load force on the motor. Specifically, we show that motor-ATP interactions are regulated by a signal that is propagated allosterically from inside the viral shell to the motor mounted on the outside. This signal continuously regulates the motor speed and pausing in response to changes in either density or conformation of the packaged DNA, and slows the motor before the buildup of large forces resisting DNA confinement. Analysis of motor slipping reveals that the force resisting packaging remains low (<1 pN) until ∼70% and then rises sharply to ∼23 pN at high filling, which is a several-fold lower value than was previously estimated under the assumption that force alone slows the motor. These findings are consistent with recent studies of the stepping kinetics of the motor. The allosteric regulatory mechanism we report allows double-stranded DNA viruses to achieve rapid, high-density packing of their genomes by limiting the buildup of nonequilibrium load forces on the motor.
Introduction
A critical step in the assembly of many double-stranded DNA (dsDNA) viruses is packaging of the viral genome into preformed prohead shells (1–4). In phage ϕ29, a 19.3 kbp genome is packaged into a 42 × 54 nm prohead via an ∼4-nm-diameter portal channel (5). This channel is comprised of a ring of portal proteins (gene product 10 (gp10), also referred to as the head-tail connector). A ring of the packaging motor proteins (gp16) docks to the portal on the exterior of the prohead via an intervening ring of RNA molecules (prohead RNA (pRNA)) (Fig. 1). Gp16 converts chemical energy from ATP hydrolysis into mechanical work to translocate the DNA through the portal. This is a remarkable process because near-crystalline packing density of the DNA is achieved against large resistance forces that are thought to arise from DNA bending rigidity, electrostatic self-repulsion of DNA segments, and entropy loss (6–10).
Figure 1.
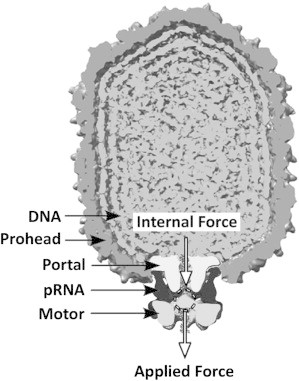
Components of the ϕ29 packaging complex. The packaging components are arranged based on superposition of the cryo-EM structure of the portal-pRNA-motor complex (after Morais et al. (48)) on that of the fully packaged virus (after Tang et al. (31)). The arrows schematically illustrate that both internal and applied forces exert loads on the motor at the site where the motor grips the DNA, opposite to the direction of translocation. Note: the dimensions of the prohead shell are 42 × 54 nm.
The dynamics of DNA translocation and forces generated by the molecular motor have been measured in the bacteriophage ϕ29, λ, and T4 systems via single DNA molecule manipulation with optical tweezers (11–22). A striking phenomenon observed in both ϕ29 and λ is that the motor velocity decreases sharply with increasing prohead filling and also decreases with increasing externally applied force (11,14,16).
Recently, we showed that the DNA inside phage ϕ29 undergoes nonequilibrium dynamics during packaging, which slows the motor, causes heterogeneity in the packaging rates of individual viruses, and causes frequent pauses in motor translocation (23). At high prohead filling, we showed that the DNA adopts a nonequilibrium conformation that relaxes on a timescale of >10 min, which is longer than the packaging reaction (∼6.6 min on average). The observed heterogeneity in packaging rates indicates that the DNA packaged in different individual viruses adopts different conformations and that the motor velocity depends not only on the length of DNA packaged but also on its conformation.
Here, we report that the amount and conformation of the DNA inside the prohead also indirectly influence motor function in a manner that is distinct from the influence of the forces that resist DNA confinement and directly load the motor. Much of the decrease in motor velocity with filling is attributable to this effect. Since only part of the slowing can be attributed to load forces, our findings show that previous analyses based on the assumption that load force is the only factor that slows the motor overestimated the resisting forces (11,13,16). The detailed measurements of motor slipping presented here suggest that the force resisting DNA confinement is quite low until ∼70% filling, but then builds rapidly during the final stages of packaging. The surprising implication of these findings is that besides being directly affected by the load force, the motor is also indirectly regulated by an allosteric interaction between the DNA packaged inside the prohead and the motor mounted on the outside. We further show that these findings are consistent with recent studies demonstrating highly coordinated burst-dwell stepping kinetics of the packaging motor (24).
Materials and Methods
The ϕ29 components were provided by Dr. Shelley Grimes and Dr. Paul Jardine and prepared as described previously (23). A 25 kbp dsDNA packaging substrate labeled at one end with biotin was prepared by PCR as described previously (23,25). The DNA was tethered to 2.1-μm-diameter streptavidin-coated microspheres, and prohead-motor complexes were preassembled and attached via anti-ϕ29 antibodies to 2.1 μm protein G-coated microspheres as described previously (13,16,23).
Packaging was initiated by bringing a microsphere carrying DNA into near contact with a microsphere carrying prohead-motor complexes in the presence of standard packaging buffer containing 25 mM Tris-HCl (pH 7.5), 50 mM NaCl, and 5 mM MgCl2 with 0.5 mM ATP, as described previously (13,16). Exchange between solutions containing ATP and γS-ATP was achieved using a custom-made microfluidic flow cell. Measurements were conducted in a channel filled with gently flowing ATP solution. The flow cell was attached to a translation stage, which allowed a tethered complex that was held fixed in the laboratory frame by the optical traps to be rapidly moved near the end of a capillary tube that delivered a 0.4 mM γS-ATP solution in the same background buffer, causing rapid (<1 s) solution exchange and stalling the motor. The motor was then restarted by moving the complex away from the capillary tube and back into the flowing ATP solutions, again achieving nearly instant solution exchange.
Measurements were made at ∼23°C using a dual-trap optical tweezers system as described previously (13,16). Two measurement modes were used. In the force-clamp mode, the applied tension is held constant at a preset value by use of feedback control system that adjusts the separation between the two traps. In the fixed-trap position mode, the tension is allowed to build as packaging proceeds. The tweezers were calibrated as described previously (26,27). The tether length was computed from the measured force versus fractional extension relationship, and all velocities were calculated by linear fits to DNA tether length versus time in a 3 s sliding window. All error bars were determined by bootstrap analysis.
Pauses in which the motor temporarily stops translocating DNA for ∼1–10 s and slips in which segments of DNA rapidly exit the prohead are often observed (11). To score pausing events automatically, we employed a residence-time histogram method as described previously (18). Slips were scored for both fixed-force and fixed-position measurements as any increase in DNA tether length that occurred within ∼1 s and was larger than a threshold determined from an equal-sized ensemble of simulated packaging events that did not contain slips. Simulated packaging traces were generated according to the model described in Moffitt et al. (18), with added instrument noise as measured experimentally. The average frequency of pauses and slips as a function of the percentage of the length of the genome packaged for each ensemble was calculated in 5% genome length bins.
Results
We employed optical tweezers to directly measure the packaging of single DNA molecules into single phage ϕ29 proheads, using techniques modified from those we described previously (11,13,16,23). In brief, prohead-motor complexes were attached to one microsphere and DNA molecules were attached to a second microsphere. When a DNA molecule was brought into contact with the motor in the presence of ATP, packaging initiated and the DNA was translocated into the prohead (Fig. 2 A). We continuously tracked the length of the packaged DNA versus time by using a feedback control system to apply a constant force of 5 pN, which is small compared with the maximum force the motor can exert (>60 pN).
Figure 2.
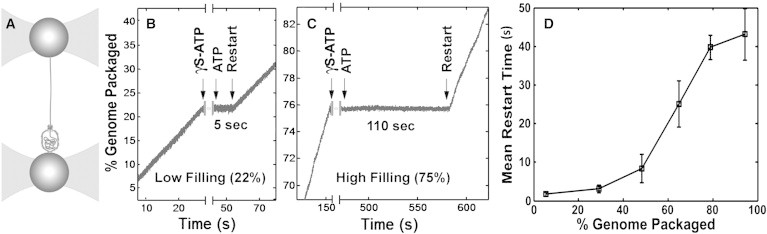
Nucleotide exchange experiments. (A) Schematic illustration of the experimental setup. A prohead-motor complex is attached to one microsphere and held in one optical trap (left), and a single DNA molecule is attached by one end to a second microsphere and its other end is packaged into the prohead. (B and C) Examples of nucleotide exchange experiments. The motor is stalled by addition of γS-ATP, ATP is reintroduced, and the motor is observed to restart after a delay (restart time). (B) Example of a measurement at low filling (22% genome packaged), showing a short restart time of 5 s. (C) Example of a measurement at high filling (75%), showing a very long restart time of 110 s. (D) Mean time to restart after nucleotide exchange. The dependence of the mean restart time after exchange from γS-ATP to ATP on prohead filling (n = 304 packaging events) is shown. Error bars indicate mean ± SE.
The most obvious way that DNA packaged inside the prohead can influence motor function is through forces that resist DNA translocation and directly load the motor, which we call internal force (11,12,18) (Fig. 1). Such forces are expected to arise because confinement of the DNA is an energetically unfavorable process (6–10). The internal force exerts a load on the part of the motor that grips and translocates the DNA through the portal channel, and slows the motor by reducing any force-dependent rate constants in the mechanochemical cycle. In standard theoretical analyses, the presence of a resisting load force Fload during a DNA translocation step of size Δx increases the mechanical work energy by an amount FloadΔx, which increases the reaction energy barriers (28,29). To probe the motor’s response, we can artificially apply a load force by applying tension to the unpackaged DNA tether (Fig. 1). The measured velocity v has been shown to decrease with increasing applied force Fload in accord with the formula v = 1/(a + b × exp(c × Fload)) predicted by Chemla et al. (12), where a, b, and c are constants related to the kinetic model parameters described in Chemla et al. (12). The motor also slows progressively as the prohead fills with DNA (11,13,16). Under the assumption that load force is the only factor that slows the motor, the above relationship was used to infer the magnitude of the internal force as the prohead filled (11,16). Surprisingly, however, we find that load force is not the only factor that influences motor function. We show that as the prohead fills, the building density of DNA has an additional effect on the motor that is distinct from the effect of load force and contributes significantly to the change in motor dynamics at high fillings.
Our first finding demonstrating this effect comes from measurements in which we stalled the motor with a nonhydrolyzable ATP analog (γS-ATP) and restarted it by reintroducing ATP. To accomplish that, we moved the packaging complex in front of a capillary dispensing γS-ATP and then moved it back into the main flow chamber containing ATP. In this manner, we achieved exchange back to ATP in <1 s. Strikingly, we find that the time it takes the motor to restart after this nucleotide exchange is strongly dependent on prohead filling. At low prohead filling, the motor restarts rapidly (within a few seconds; Fig. 2 B), but at high filling, the restart time can increase to longer than 1 min (Fig. 2 C). The average restart time measured for an ensemble of experiments increases continuously with increasing prohead filling (Fig. 2 D). This increase is not attributable to increasing load force because it was previously shown that the dissociation rate of γS-ATP measured at low filling is independent of the applied force (12). Therefore, our present finding implies an indirect effect of prohead filling on motor-ATP interactions.
Our second finding demonstrating this effect comes from detailed studies of pausing in DNA translocation. In measurements with low load (5 pN), we observe short pauses (typically ∼1–10 s) in DNA translocation that occur with a frequency that increases strongly with increasing filling (Fig. 3 A), as also observed in earlier studies (11,23,24). Here, we present additional data showing that pausing measured at low prohead filling is not significantly induced by increasing load (Fig. 3 B). Thus, the observed increase in pausing with increasing filling cannot be attributed to a buildup of force resisting DNA translocation and instead must be attributed to an independent effect of prohead filling on motor function.
Figure 3.
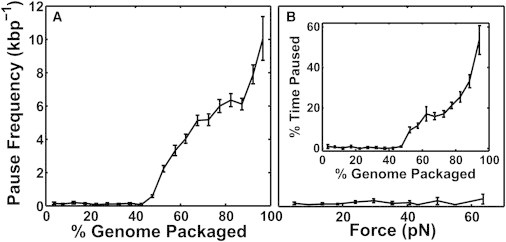
Analysis of pauses in motor translocation. (A) Mean frequency of pauses (number detected per packaged kilobasepair) versus prohead filling (n = 45). (B) Mean frequency of pauses versus applied force (n = 130). Inset shows the % time spent paused. All error bars indicate mean ± SE.
Our third finding demonstrating this effect comes from studies of motor slipping. Slips were defined as events in which a length of DNA (typically ∼30–150 bp) suddenly came backward out of the prohead. In sharp contrast to the trend observed with pausing, the frequency of slipping increased both with increasing filling (Fig. 4 A) and with increasing applied load (Fig. 4 B). We interpret these findings as indicating that slipping occurs when the motor loses its grip on the DNA, and that the probability of slipping increases with increasing load. In this case, the buildup of load force can be inferred by relating the frequency of slipping versus load force, measured at low filling, to the frequency of slipping versus filling (Fig. 4 C). The inferred internal force is very low (<1 pN) until ∼70% filling, where it then begins to build rapidly and reaches a maximum of ∼23 ± 7 pN at the end of packaging.
Figure 4.
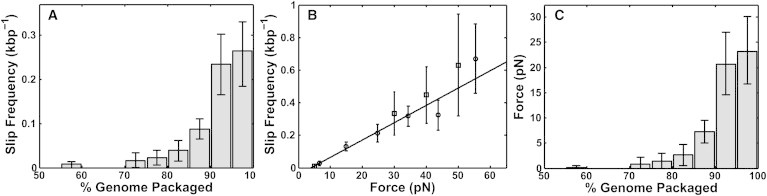
Analysis of motor slipping. (A) Mean frequency of slips (number detected per packaged kilobasepair) versus prohead filling measured with a 5 pN force clamp (n = 320 packaging events). (B) Mean frequency of slips versus applied force measured at low filling (<20%) in both force-clamp mode (where force is held constant at different preset values; circles, n = 190) and fixed-trap position mode (where force is allowed to build as packaging proceeds; squares, n = 130). The solid line is a linear fit to the data points. (C) Force resisting packaging versus prohead filling inferred from plots A and B. All error bars indicate mean ± SE.
In earlier analyses, we inferred internal force under the seemingly reasonable assumption that the decrease in motor velocity with filling is completely due to this force loading the motor (11,13,16). However, the magnitude of the internal force deduced under that assumption is several-fold higher than that inferred from our present measurements of slipping frequency (Fig. 4 B). Most notably, the motor velocity (i.e., the packaging rate not including pauses and slips) decreases by ∼50% going from 0% to 70% prohead filling (Fig. 5 A), but the measurements of slipping suggest that there is very little internal force (<1 pN) up to this point. Based on the measured velocity versus load force relationship (Fig. 5 B), 1 pN of internal force would only cause a ∼2% reduction in velocity, which cannot explain the measured ∼50% velocity decrease going from 0% to 70% filling. Thus, these findings further demonstrate that much of the slowing of the motor is not caused by load force and again must be attributed to an indirect effect of the packaged DNA on motor function.
Figure 5.
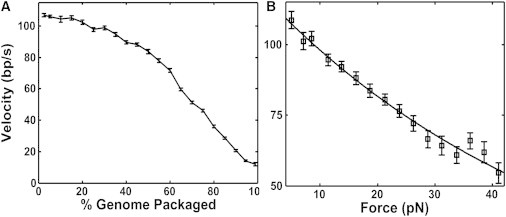
Motor velocity measurements. (A) Mean velocity versus prohead filling (n = 45). (B) Mean velocity versus applied force (n = 74). All error bars indicate mean ± SE.
Discussion
An unconventional type of allosteric regulation
We refer to the reported slowing of the motor with prohead filling due to modulation of motor-ATP interactions as allosteric regulation in analogy to the classical phenomenon wherein an enzyme’s activity at one site is regulated by the binding of an effector molecule at a second site. Here, the motor’s DNA translocase activity, which is coupled to the ATP hydrolysis cycle, is allosterically regulated by the presence of increasing amounts of DNA packaged inside the prohead. This allosteric signal negatively regulates the packaging rate by both reducing the motor’s translocation speed and increasing the frequency of pauses. This regulation likely helps to mitigate the formation of nonequilibrium DNA conformations that we have demonstrated can cause stalling and slipping of the motor (23,30).
We note that this regulation is unlike typical allosteric regulation in two ways. First, the prohead filling sensor is nonspecific in the sense that it does not appear to involve binding of a specific DNA sequence to a complementary binding site. Studies using several different DNA substrates observed motor slowing that was independent of the sequence being packaged (11,13,16,23,30). Rather, the motor function is modulated by an increasing density of confined DNA, presumably through nonspecific interactions with the portal and/or prohead wall. Second, whereas binding of an effector molecule typically causes a discrete change in enzyme activity, here the velocity of the motor complex decreases continuously with increasing length of the packaged DNA. Since our experiments directly measure the rates of single-motor complexes, it is clear that this continuous change is due to the activity of individual motors and not simply to shifting subpopulations of complexes having two distinct activity levels (e.g., with effector bound versus unbound).
Structural basis for the allosteric effect
Structural studies based on cryo-electron microscopy (cryo-EM) have shown that the portal channel through which the DNA is translocated extends from the interior of the prohead to the exterior, where the motor protein is attached via the intervening pRNA (Fig. 1) (31–33). The force resisting DNA packaging is directly transmitted to the motor via its contacts with the section of DNA threaded through the channel (Fig. 6 A). However, it is quite clear that the section of DNA that is already packaged inside the prohead does not directly contact the motor protein and therefore cannot directly affect its operation. The packaged DNA only touches the inner wall of the prohead and a portion of the portal channel that extends into the prohead interior (31–33). Our results therefore imply that the packaged DNA must interact with the motor protein via a long-distance allosteric signal (Fig. 6 B). This signal must be transmitted ∼100 Å from the interior of the prohead to the motor on the exterior via the intervening prohead shell and/or portal protein and pRNA.
Figure 6.
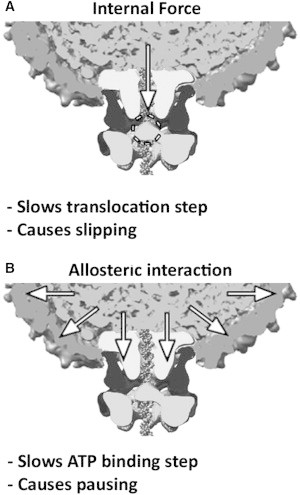
Two different mechanisms of regulation of the ϕ29 motor. Individual components of the packaging complex are labeled in Fig. 1. (A) Forces acting directly on the section of DNA that enters through the portal exert a direct load on the portion of the motor that grips and translocates the DNA (dashed circle). (B) DNA packaged inside the prohead interacts with the inner wall of the prohead and a portion of the portal protein that protrudes into the interior. This interaction allosterically regulates the motor protein attached to the exterior portion of the portal in a manner distinct from direct load. Images were constructed as described in Fig. 1.
Cryo-EM studies of several different phages, including ϕ29, have shown that the packaged DNA is in contact with the portion of the portal channel that is exposed to the interior (31,33–38). Two particularly striking observations are that 1), a ring of DNA appears to wrap around the portal, appearing to squeeze it; and 2), the portal adopts a different conformation when it is incorporated into the head than when it is isolated (36,37). Because the portal was only imaged at one filling level (fully packaged), it is unclear whether the alternate conformation represents one conformation of a two-state system or a single point in a continuum of conformational states. Our data support the later scenario for ϕ29 motor regulation, since we observe a continuous decrease in velocity for individual complexes and large heterogeneity among an ensemble of complexes (23). Mutant studies of phage P22 also indicated that residue changes in the portal protein can affect the length of packaged DNA (39). Unlike ϕ29, P22 packages by a headful mechanism in which the motor must excise a unit-length genome from a concatenated DNA substrate (3,4,36). The motor is triggered to terminate packaging and cleave the DNA after an appropriate length of the genome has been packaged. It was proposed that the conformational change observed in the P22 portal may be the signal that triggers the termination (36). Cryo-EM and molecular-dynamics studies of phage ε15 also revealed a well-resolved ring of averaged density located inside the narrow groove at the base of the portal (35,40). In simulations, this groove section often consisted of a variable arrangement of two or more independent DNA segments (23). Cryo-EM studies of ϕ29 virions also revealed a ring of DNA in contact with the portal, although it was touching the top of the portal rather than wrapping around it (33). Although ϕ29 does not package via a headful mechanism, a conformational change in its portal, induced by contact with the packaged DNA, could communicate the allosteric regulatory signal to the motor protein.
Distinction between the effects of load and allosteric regulation
Although we have distinguished motor slowing due to an internal force that directly loads the motor from that due to allosteric regulation, the latter could still be caused by conformational changes induced by the buildup of internal forces. As illustrated in the schematic diagram in Fig. 6, A and B, the force per unit area (pressure) exerted by the packaged DNA on the inner surface of the prohead wall and portal ring is presumably similar or equal, on average, to the force that resists translocation of DNA by the motor through the portal channel. However, our findings show that the onset of the slowing due to the allosteric mechanism occurs at a much lower filling than slowing due to force directly loading the motor. If the allosteric mechanism is indeed triggered by internal force, it must be much more sensitive to the magnitude of the force (i.e., triggered by lower forces than are needed to significantly slow the motor via load). Specifically, as discussed below, our analysis of motor slipping reveals that the allosteric effect begins to significantly slow the motor well before any changes in load-dependent behavior are detected (i.e., slipping). Although we could not detect any internal force below ∼70% filling, it is likely that a small internal force (e.g., <1 pN, which is too low for our slipping analysis to detect) does build up, as expected theoretically. Such a low force would not significantly slow the motor via load (Fig. 5 A), but could be more than sufficient to trigger motor slowing via the allosteric mechanism.
Effect on motor-ATP interactions
Our finding that the motor restart time after the exchange from γS-ATP to ATP increases dramatically with prohead filling shows that the motor-ATP interaction is affected. In principle, either a decrease in the ATP binding rate or an increase in the ATP dissociation rate would slow the motor (12), but our measurements rule out the latter possibility. Fig. 2 D shows that the restart time at 70% filling is ∼30 s, but at this filling the motor velocity during packaging is ∼50 bp/s, which implies that one ATP is being hydrolyzed every 0.04 s (since ∼2 bp are packaged per ATP) (12). Thus, ATP binding, hydrolysis, DNA translocation, and ADP and phosphate release must all happen very rapidly in 0.04 s. Therefore, the 30 s restart time we measure must be attributed to slow dissociation of γS-ATP. Our measurements thus imply that the γS-ATP dissociation rate decreases with increasing prohead filling. Since γS-ATP has been shown to mimic ATP in its binding kinetics (12), this implies that if ATP had the opportunity to dissociate (as opposed to being rapidly hydrolyzed), its rate of dissociation would decrease with increasing prohead filling. A decreased dissociation rate, however, would increase motor velocity, and thus cannot explain the observed reduction with filling. Therefore, we attribute the reduction in motor velocity to a decrease in the ATP binding rate.
A decrease in the ATP binding rate could result from either weakening of the motor-ATP interaction or occlusion of the ATP binding pocket. Our evidence showing that the rate of ATP dissociation decreases with increasing filling suggests that it is due to occlusion. Such occlusion could be caused by a conformational change in the motor protein that affects the ATP binding pocket or by conformational changes in the prohead, portal, and/or pRNA components that result in steric hindrance of the entry of ATP into the binding pocket.
Contributions of load versus allosteric regulation to motor slowing
The magnitude of the internal forces implied by our measurements of motor slipping (Fig. 4 C) is significantly lower than those estimated previously under the assumption that internal force was the only factor slowing the motor (11,13,16). This discrepancy is explained by our present finding that prohead filling has an independent allosteric effect on motor function. Our measurements of motor slipping suggest that the internal force is quite low (<1 pN) until ∼70% filling, after which it rises sharply. Therefore, the ∼50% decrease in motor velocity going from 0% to 70% filling is almost entirely attributable to allosteric regulation.
In Fig. 7 A, we compare the measured dependence of motor velocity versus filling to that which would be expected due to the buildup of internal force loading the motor alone. We inferred the change in velocity by relating the internal force-filling relationship in Fig. 4 C to the measured force-velocity relationship in Fig. 5 B. This comparison shows that allosteric regulation begins to slow the motor significantly at ∼20% filling, whereas slowing due to the buildup of internal force starts much later, at ∼70% filling. We also plot for comparison the overall mean packaging rate, which includes the effect of motor pauses and slips (Fig. 7 A). At low filling (<50%), pauses and slips cause a negligible percent change, but pauses steadily increase in importance at high filling (see also Fig. 3 B, inset).
Figure 7.
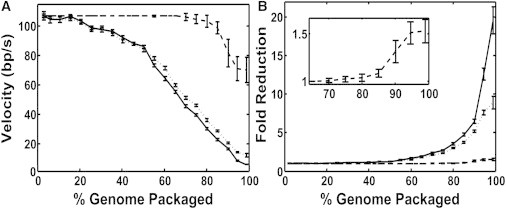
Factors that influence the reduction in motor velocity. (A) Measured mean packaging rate (solid line) and measured mean motor velocity (rate not including pauses and slips, dotted line) versus filling compared with the velocity that would be expected if the force plotted in Fig. 6B were the only factor slowing the motor (dashed line). (B) Fold decrease from the initial value of the measured packaging rate (solid line), measured motor velocity (dotted line), and expected velocity due to force alone (dashed line). The inset shows a zoomed-in plot of the dashed line. All error bars indicate mean ± SE.
Overall, the allosteric mechanism, which both slows the motor velocity and causes pausing, has a much larger effect on packaging kinetics than does internal force. Comparisons of the ratio of the initial velocity (Vmax) to the expected velocity if force alone slowed the motor and the ratio of Vmax to the measured velocity are plotted in Fig. 7 B. This shows that internal force alone is predicted to have only a minor impact on the overall velocity change, accounting for a ∼1.5-fold reduction from the initial value, whereas the actual reduction reaches ∼8-fold near the end of packaging. Thus, the allosteric interaction ultimately slows the motor velocity by an additional ∼5-fold. The reduction in the overall packaging rate, which includes the effect of pauses, reaches ∼20-fold near the end of packaging. Thus, allosterically induced pauses ultimately slow packaging by an additional ∼2.5-fold.
Force resisting DNA confinement and comparisons with theories
Our findings shed light on discrepancies between our earlier higher estimates of forces resisting packaging (internal force) and theoretical predictions (7–9,13,16,41–43). Our present measurements of slipping suggest very little buildup of force from 0% to 70% filling, followed by a rise to ∼23 pN near the end of packaging. This trend is in closer accord with theoretical predictions based on continuum mechanics models and measurements of forces that drive DNA ejection (44). Specifically, Tzlil et al. (7) calculated an initially low rise and maximum force of ∼25 pN based on a model assuming an inverse-spool conformation of DNA and interaction potential derived from measurements of DNA condensation by osmotic pressure. Caveats in this comparison are that they modeled phage λ, which has a different shape and genome length and slightly higher packing density, considered slightly higher ionic screening, and did not consider nonequilibrium effects (23,45). Using a similar model, but specifically considering the ϕ29 prohead size, shape, and packing density, Purohit et al. (8) also predicted a similar shape of the force versus filling curve, though they did not make absolute predictions about the forces independently of previously reported experimental estimates.
Investigators have also modeled packaging using coarse-grained molecular dynamics simulations. Forrey and Muthukumar (9) calculated a maximum resisting force of ∼40 pN, which is higher than what we found here, although again they did not specifically model the ϕ29 prohead dimensions. Another caveat in this comparison is that they simulated DNA packaging at an initial rate ∼105× faster than the experimental rate due computational constraints, which could cause larger deviations from equilibrium and higher forces. Petrov and Harvey (43) also conducted molecular dynamics simulations using a DNA interaction potential derived from available osmotic pressure data for similar ionic conditions. They calculated resisting forces rising to ∼60 pN. Again, however, the simulated packaging was much faster than the natural speed (1 nm steps every 10 ns, although the authors pointed out that this timescale does not translate to experimentally measured time due the use of a coarse-grained model for DNA). Again, faster packaging could explain larger deviations from equilibrium and higher forces. Specifically, the finding of higher forces in these simulations may be due to an inability of the simulations to model the very slow DNA relaxation dynamics, which we have shown to occur on >10 min timescales (23).
Regulation responds to changes in both the length and conformation of packaged DNA
We have shown that packaging is not a quasi-static process, because at high filling the DNA forms nonequilibrium conformations that relax on a timescale longer than the packaging reaction (23). When packaging was stalled at ∼75% filling for ∼12 min (on average) to allow the DNA to relax, the average motor velocity increased by ∼23% after restarting. Notably, this change occurred with no change in prohead filling, indicating that this effect must be caused solely by a change in the conformation of the packaged DNA. At 75% filling, the internal force implied by our measurements of slipping is <2 pN. According to the measured force-velocity relationship (Fig. 5 B), at 75% filling the complete relaxation of 2 pN of force would only cause an ∼4% change in velocity, so the observed velocity increase after DNA relaxation cannot be attributed to the relaxation of internal force alone. Thus, this acceleration must be mostly attributed to modulation of motor activity via the allosteric regulatory mechanism.
We also found that the frequency and duration of pauses decreased dramatically after the DNA was allowed to relax (23). Since we have shown here that pausing is not significantly induced by the load force, the change in pausing dynamics after DNA relaxation must therefore be attributed to changes in the packaged DNA conformation that alter the interaction(s) responsible for the allosteric effect. Remarkably, these findings regarding both motor velocity and pausing imply that the allosteric sensor we have described responds not only to the prohead filling but also to the conformation of the packaged DNA.
Earlier studies by our lab (13) also showed that the decrease in velocity with filling is dependent on ionic conditions, which can change the packaged DNA conformation (46,47). In conditions where the DNA-DNA interaction is purely repulsive, higher screening (with Mg2+, cobalt hexamine3+, or spermidine3+) results in a later onset of motor slowing, consistent with decreased internal pressure causing decreased allosteric regulation. In addition, we recently found that spermidine at levels high enough to induce DNA condensation in solution (changing the DNA-DNA interaction to partly attractive) causes significant slowing of the motor as well as more frequent and longer pauses even at low filling levels (<50%) (30). Although this finding is contrary to theoretical studies that predicted reduced forces resisting DNA confinement (33), our interpretation is that attractive interactions exacerbate the formation of highly nonequilibrium DNA conformations that result in higher resistance forces and/or greater allosteric slowing.
Relationship with motor stepping kinetics
It was previously shown that packaging occurs in rapid bursts of four 2.5 bp steps separated by dwells (18). At low filling, the duration of the bursts is independent of ATP concentration but increases with increasing applied force, whereas the duration of the dwells depends on the ATP concentration but not the applied force. A study published very recently by Liu et al. (24) reported that the duration of the dwells also increases with increasing prohead filling. The conclusions we reach here are consistent with these findings. First, the reported increase in the duration of dwells with increasing prohead filling causes a load-independent decrease in the motor velocity, consistent with our findings. Second, our measurements of motor restart time after nucleotide exchange imply force-independent changes in motor-ATP interactions and suggest that motor slowing is due to slowed ATP binding. Previous measurements showed that motor velocity versus [ATP] follows Michaelis-Menten kinetics (12). Liu et al. (24) extended these measurements to show that the maximum velocity (Vmax) and Michaelis constant (KM), and the ratio of the two (Vmax/KM) decrease with increasing filling, suggesting that filling slows ATP binding, consistent with our conclusion. Third, our measurements of slipping frequency versus filling and versus force suggest that the internal force remains low (<1 pN) until ∼70% filling and then rises steeply to ∼23 pN near the end of packaging. Using a different method, Liu et al. (24) inferred internal force by relating the dependence of the duration of bursts of translocation steps on filling to their dependence on applied force, yielding a force that rises in a similar manner to a maximum value of ∼20 pN. The fact these two different methods of determining internal force yield very similar values provides strong support for the validity of these results.
Conclusions
We have presented three different findings that clearly indicate that prohead filling has a strong and indirect effect on the function of the motor distinct from the effect of a direct load force. First, motor-ATP interactions are strongly perturbed by filling, but not by load. Second, motor pausing increases sharply with increasing filling but not with load. Third, the maximum internal force resisting packaging inferred from measurements of motor slipping is several-fold lower than previous estimates based on motor velocity, which implies that much of the reduction in motor velocity is independent of load. The implication of these findings is that an allosteric signal acts to continuously reduce the packaging rate even before the buildup of significant internal force. This signal is propagated ∼100 Å from the interior of the prohead to the motor protein mounted on the exterior.
Remarkably, the allosteric sensor responds to not only the quantity of the packaged DNA but also its conformation. We propose that this effect mitigates the formation of highly nonequilibrium DNA conformations, which we have shown can slow and stall the motor, and cause the DNA to slip out (23,30). Specifically, when the DNA is given less time to relax toward equilibrium, this mechanism slows the motor and induces pauses. Conversely, the motor speeds up and pauses less when the DNA is given more time to relax (23). Thus, the motor speed appears to be tuned by this regulatory mechanism to achieve sustainably fast packaging rates depending on the filling level and DNA conformation.
Acknowledgments
We thank Dr. Shelley Grimes and Dr. Paul Jardine for supplying the phi29 packaging components and invaluable advice on in vitro packaging reactions, Damian delToro and Mariam Ordyan for assistance with instrumentation and DNA constructs, and Michael Feiss and Stephen Harvey for comments on the manuscript.
This research was supported by National Science Foundation awards PHY-0848905 and MCB-1158328.
Footnotes
Zachary T. Berndsen and Nicholas Keller contributed equally to this work.
References
- 1.Smith D.E. Single-molecule studies of viral DNA packaging. Curr. Opin. Virol. 2011;1:134–141. doi: 10.1016/j.coviro.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chemla Y.R., Smith D.E. Single-molecule studies of viral DNA packaging. In: Rao V., Rossmann M.G., editors. Viral Molecular Machines. Springer; New York: 2012. pp. 549–584. [Google Scholar]
- 3.Casjens S.R. The DNA-packaging nanomotor of tailed bacteriophages. Nat. Rev. Microbiol. 2011;9:647–657. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- 4.Feiss M., Rao V.B. The bacteriophage DNA packaging machine. In: Rao V., Rossmann M.G., editors. Viral Molecular Machines. Springer; New York: 2012. pp. 489–509. [Google Scholar]
- 5.Morais M.C. The dsDNA packaging motor in bacteriophage ø29. In: Rao V., Rossmann M.G., editors. Viral Molecular Machines. Springer; New York: 2012. pp. 511–547. [DOI] [PubMed] [Google Scholar]
- 6.Riemer S.C., Bloomfield V.A. Packaging of DNA in bacteriophage heads: some considerations on energetics. Biopolymers. 1978;17:785–794. doi: 10.1002/bip.1978.360170317. [DOI] [PubMed] [Google Scholar]
- 7.Tzlil S., Kindt J.T., Ben-Shaul A. Forces and pressures in DNA packaging and release from viral capsids. Biophys. J. 2003;84:1616–1627. doi: 10.1016/S0006-3495(03)74971-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purohit P.K., Inamdar M.M., Phillips R. Forces during bacteriophage DNA packaging and ejection. Biophys. J. 2005;88:851–866. doi: 10.1529/biophysj.104.047134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forrey C., Muthukumar M. Langevin dynamics simulations of genome packing in bacteriophage. Biophys. J. 2006;91:25–41. doi: 10.1529/biophysj.105.073429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harvey S.C., Petrov A.S., Boz M.B. Viral assembly: a molecular modeling perspective. Phys. Chem. Chem. Phys. 2009;11:10553–10564. doi: 10.1039/b912884k. [DOI] [PubMed] [Google Scholar]
- 11.Smith D.E., Tans S.J., Bustamante C. The bacteriophage ϕ29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 12.Chemla Y.R., Aathavan K., Bustamante C. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122:683–692. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 13.Fuller D.N., Rickgauer J.P., Smith D.E. Ionic effects on viral DNA packaging and portal motor function in bacteriophage ϕ 29. Proc. Natl. Acad. Sci. USA. 2007;104:11245–11250. doi: 10.1073/pnas.0701323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuller D.N., Raymer D.M., Smith D.E. Measurements of single DNA molecule packaging dynamics in bacteriophage λ reveal high forces, high motor processivity, and capsid transformations. J. Mol. Biol. 2007;373:1113–1122. doi: 10.1016/j.jmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuller D.N., Raymer D.M., Smith D.E. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc. Natl. Acad. Sci. USA. 2007;104:16868–16873. doi: 10.1073/pnas.0704008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rickgauer J.P., Fuller D.N., Smith D.E. Portal motor velocity and internal force resisting viral DNA packaging in bacteriophage ϕ29. Biophys. J. 2008;94:159–167. doi: 10.1529/biophysj.107.104612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsay J.M., Sippy J., Smith D.E. The Q motif of a viral packaging motor governs its force generation and communicates ATP recognition to DNA interaction. Proc. Natl. Acad. Sci. USA. 2009;106:14355–14360. doi: 10.1073/pnas.0904364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moffitt J.R., Chemla Y.R., Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsay J.M., Sippy J., Smith D.E. Mutations altering a structurally conserved loop-helix-loop region of a viral packaging motor change DNA translocation velocity and processivity. J. Biol. Chem. 2010;285:24282–24289. doi: 10.1074/jbc.M110.129395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chistol G., Liu S., Bustamante C. High degree of coordination and division of labor among subunits in a homomeric ring ATPase. Cell. 2012;151:1017–1028. doi: 10.1016/j.cell.2012.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kottadiel V.I., Rao V.B., Chemla Y.R. The dynamic pause-unpackaging state, an off-translocation recovery state of a DNA packaging motor from bacteriophage T4. Proc. Natl. Acad. Sci. USA. 2012;109:20000–20005. doi: 10.1073/pnas.1209214109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Migliori A.D., Keller N., Smith D.E. Evidence for an electrostatic mechanism of force generation by the bacteriophage T4 DNA packaging motor. Nat. Commun. 2014;5:4173. doi: 10.1038/ncomms5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berndsen Z.T., Keller N., Smith D.E. Nonequilibrium dynamics and ultraslow relaxation of confined DNA during viral packaging. Proc. Natl. Acad. Sci. USA. 2014;111:8345–8350. doi: 10.1073/pnas.1405109111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu S., Chistol G., Bustamante C. A viral packaging motor varies its DNA rotation and step size to preserve subunit coordination as the capsid fills. Cell. 2014;157:702–713. doi: 10.1016/j.cell.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuller D.N., Gemmen G.J., Smith D.E. A general method for manipulating DNA sequences from any organism with optical tweezers. Nucleic Acids Res. 2006;34:e15. doi: 10.1093/nar/gnj016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rickgauer J.P., Fuller D.N., Smith D.E. DNA as a metrology standard for length and force measurements with optical tweezers. Biophys. J. 2006;91:4253–4257. doi: 10.1529/biophysj.106.089524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.delToro D., Smith D.E. Accurate measurement of force and displacement with optical tweezers using DNA molecules as metrology standards. Appl. Phys. Lett. 2014;104:143701. doi: 10.1063/1.4871005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang M.D., Schnitzer M.J., Block S.M. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 29.Keller D., Bustamante C. The mechanochemistry of molecular motors. Biophys. J. 2000;78:541–556. doi: 10.1016/S0006-3495(00)76615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keller N., delToro D., Smith D.E. Repulsive DNA-DNA interactions accelerate viral DNA packaging in phage ϕ29. Phys. Rev. Lett. 2014;112:248101. doi: 10.1103/PhysRevLett.112.248101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang J., Olson N., Baker T.S. DNA poised for release in bacteriophage ϕ29. Structure. 2008;16:935–943. doi: 10.1016/j.str.2008.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tao Y., Olson N.H., Baker T.S. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell. 1998;95:431–437. doi: 10.1016/s0092-8674(00)81773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiang Y., Morais M.C., Rossmann M.G. Structural changes of bacteriophage ϕ29 upon DNA packaging and release. EMBO J. 2006;25:5229–5239. doi: 10.1038/sj.emboj.7601386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fokine A., Chipman P.R., Rossmann M.G. Molecular architecture of the prolate head of bacteriophage T4. Proc. Natl. Acad. Sci. USA. 2004;101:6003–6008. doi: 10.1073/pnas.0400444101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang W., Chang J., Chiu W. Structure of ε15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lander G.C., Tang L., Johnson J.E. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 37.Tang J., Lander G.C., Johnson J.E. Peering down the barrel of a bacteriophage portal: the genome packaging and release valve in p22. Structure. 2011;19:496–502. doi: 10.1016/j.str.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olia A.S., Prevelige P.E., Jr., Cingolani G. Three-dimensional structure of a viral genome-delivery portal vertex. Nat. Struct. Mol. Biol. 2011;18:597–603. doi: 10.1038/nsmb.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casjens S., Wyckoff E., Serwer P. Bacteriophage P22 portal protein is part of the gauge that regulates packing density of intravirion DNA. J. Mol. Biol. 1992;224:1055–1074. doi: 10.1016/0022-2836(92)90469-z. [DOI] [PubMed] [Google Scholar]
- 40.Petrov A.S., Lim-Hing K., Harvey S.C. Packaging of DNA by bacteriophage ε15: structure, forces, and thermodynamics. Structure. 2007;15:807–812. doi: 10.1016/j.str.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 41.Kindt J., Tzlil S., Gelbart W.M. DNA packaging and ejection forces in bacteriophage. Proc. Natl. Acad. Sci. USA. 2001;98:13671–13674. doi: 10.1073/pnas.241486298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purohit P.K., Kondev J., Phillips R. Mechanics of DNA packaging in viruses. Proc. Natl. Acad. Sci. USA. 2003;100:3173–3178. doi: 10.1073/pnas.0737893100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petrov A.S., Harvey S.C. Structural and thermodynamic principles of viral packaging. Structure. 2007;15:21–27. doi: 10.1016/j.str.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 44.Evilevitch A., Castelnovo M., Gelbart W.M. Measuring the force ejecting DNA from phage. J. Phys. Chem. B. 2004;108:6838–6843. [Google Scholar]
- 45.De Frutos M., Leforestier A., Livolant F. Relationship between the genome packaging in the bacteriophage capsid and the kinetics of DNA ejection. Biophys. Rev. Lett. 2014;9:81–104. [Google Scholar]
- 46.Qiu X., Rau D.C., Gelbart W.M. Salt-dependent DNA-DNA spacings in intact bacteriophage λ reflect relative importance of DNA self-repulsion and bending energies. Phys. Rev. Lett. 2011;106:028102. doi: 10.1103/PhysRevLett.106.028102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lander G.C., Johnson J.E., Evilevitch A. DNA bending-induced phase transition of encapsidated genome in phage λ. Nucleic Acids Res. 2013;41:4518–4524. doi: 10.1093/nar/gkt137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morais M.C., Koti J.S., Rossmann M.G. Defining molecular and domain boundaries in the bacteriophage ϕ29 DNA packaging motor. Structure. 2008;16:1267–1274. doi: 10.1016/j.str.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]


