Abstract
The major histocompatibility complex (MHC) plays an important role in immune response. Avian MHCs are not well characterized, only reporting highly compact Galliformes MHCs and extensively fragmented zebra finch MHC. We report the first genomic structure of an endangered Pelecaniformes (crested ibis) MHC containing 54 genes in three regions spanning ~500 kb. In contrast to the loose BG (26 loci within 265 kb) and Class I (11 within 150) genomic structures, the Core Region is condensed (17 within 85). Furthermore, this Region exhibits a COL11A2 gene, followed by four tandem MHC class II αβ dyads retaining two suites of anciently duplicated “αβ” lineages. Thus, the crested ibis MHC structure is entirely different from the known avian MHC architectures but similar to that of mammalian MHCs, suggesting that the fundamental structure of ancestral avian class II MHCs should be “COL11A2-IIαβ1-IIαβ2.” The gene structures, residue characteristics, and expression levels of the five class I genes reveal inter-locus functional divergence. However, phylogenetic analysis indicates that these five genes generate a well-supported intra-species clade, showing evidence for recent duplications. Our analyses suggest dramatic structural variation among avian MHC lineages, help elucidate avian MHC evolution, and provide a foundation for future conservation studies.
The major histocompatibility complex (MHC) is a cluster of immune and immunity-associated genes involved in infectious disease defense1. It is notable for the impressive polymorphism of class I and II genes2, which are responsible for presenting pathogen-derived peptides and triggering the adaptive immune response3. Class I molecules, assembled from an α chain and a connected β2-microglobulin, present intracellular antigens to CD8+ T cells4, while class II molecules, composed of an α chain and a β chain, present extracellular pathogens to CD4+ T cells5. The polymorphic peptide-binding regions (PBRs) are encoded by exons 2 and 3 in MHC-I genes or exon 2 in MHC-IIα and -IIβ loci. Expressed class I genes are generally divided into classical and non-classical genes, where the latter are mostly defunct or specialized, and weakly expressed or distributed tissue specifically6. In most birds, classical class I genes are further functionally categorized into major and minor loci7,8,9.
Ever since the mouse MHC was first revealed by tissue transplantation10, MHCs of many mammalian species have been characterized11,12,13. Generally, mammalian MHCs have a similar genomic structure with divergent class II “αβ” lineages (such as DRA-DRB, DQA-DQB, and DPA-DPB) and a separate class I region, which is divided by a complement cluster (the class III region). In contrast to mammals, the chicken MHC contains two genetically unlinked clusters: the B complex (MHC-B) and the Rfp-Y region14. The former, which contains class I (BF), class IIβ (BLB), and extended (BG) regions, has all the hallmarks of mammalian MHCs11,12,13 but lacks “αβ” units; however, MHC-IIα (BLA) is located roughly 5.6 cM away from the BF and BLB regions15. Furthermore, the chicken MHC-B is much smaller (about 0.2 Mb) than mammalian MHCs (several Mb) and consists of fewer genes with shorter introns but higher gene density. Therefore, the chicken is thought to have a “minimal essential” MHC14. Other Galliformes species, such as turkey16, pheasant17, and quail18, have MHCs quite similar to the chicken MHC, though the quail MHC is double-sized for a double-expanded number of duplicated genes. Thus, the characterized Galliformes B complexes are all relatively compact.
The zebra finch (Passerines) MHC is very different and comparatively more complex. In contrast to the streamlined chicken and human MHCs, the Passerines MHC contains substantive gene copies and exhibits regional fragmentation, dispersing the MHC across several different chromosomes and spanning a much more extensive genomic region than the chicken MHC19. Although the exact architecture is still unclear, remarkable differences in MHC organization among avian lineages are apparent.
Detailed information on MHC structures in other avian orders is limited because research efforts have focused on particular MHC genes such as the MHC-I and -IIβ genes7,20,21,22. These studies suggested that recent gene duplication may be the fundamental evolutionary dynamics for avian MHC class II genes21,22. However, Burri et al.23,24 identified two ancestral MHC-IIβ lineages (DAB1 and DAB2) in birds and accordingly speculated two ancestral MHC-IIα lineages in avian MHCs. Although partial DNA fragments (several kb) from the Green-rumped Parrotlet25 and White Pekin duck26 revealed that a IIα gene was adjacent to the IIβ gene(s), formal reports of these results from the perspective of MHC partial genomes are not available. Collectively, current avian MHC studies have demonstrated severe gene duplications; however, there is no evidence of divergent ancestral “αβ” lineages in avian genomes.
Frequent gene duplications and extensive repeats in the MHC region13,18 prevent the successful use of next-generation genome sequencing methods for assembling the MHC region, as revealed in birds19, non-avian reptiles27, and even structure-conserved mammals such as the giant panda (NCBI AilMel_1.0). As a result, resolution of whole MHC genomic structures depends on genomic DNA libraries.
The crested ibis (Nipponia nippon) is a medium-sized wading bird belonging to the Threskiornithidae family and the Pelecaniformes order and is classified as “endangered” by the IUCN28. This bird was historically distributed throughout northeast Asia29; however, its population declined dramatically during the early twentieth century30. This bird was believed to be extinct until seven birds (two pairs and three nestlings) were discovered in Yangxian, Shaanxi Province, China. Since then, the species has gradually recovered owing to the conservation efforts by China and other countries; however, all current individuals are descendants of these two pairs.
Previous molecular studies of the crested ibis mainly focused on microsatellites31 and mitochondrial DNA32. The only study of the crested ibis MHC examined genetic variation of a partial MHC-IIβ exon 2 in 36 samples and only isolated five cross-locus sequences33. Recently, our research group constructed a routine bacterial artificial chromosome (BAC) library and a non-gridded reverse-4D BAC library for the crested ibis, with genomic coverages of 7.8 and 35 folds respectively34, thereby providing an opportunity to resolve its MHC partial genome. In the present study, we aim to (1) determine the genomic structure and features of the crested ibis MHC by using the BAC libraries and (2) molecularly characterize Nipponia nippon MHC (Nini-MHC) class I and II genes. This work will provide a foundation for further conservation studies and provide new insights into the evolutionary history of avian MHCs.
Results
BAC screening and sequencing
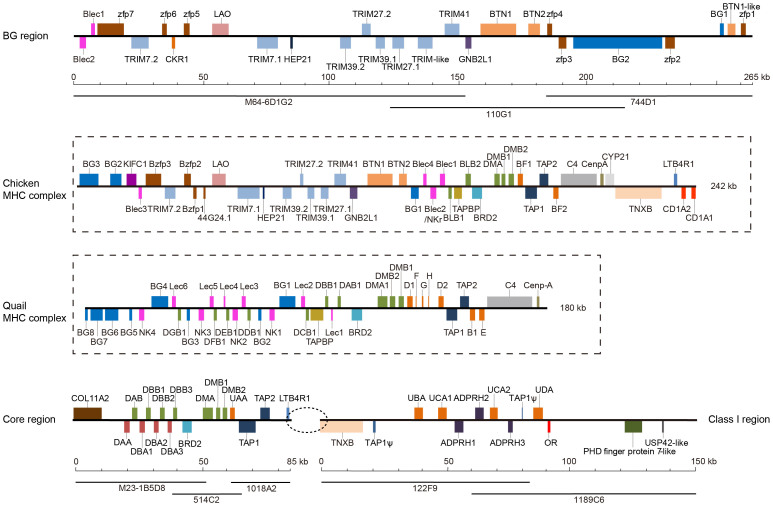
We obtained 25 MHC-positive BACs using 11 pairs of crested ibis-specific primers (Table S1) and sequenced eight clones (M64-6D1G2, 110G1, 744D1, M23-1B5D8, 514C2, 1018A2, 122F9, and 1189C6) depicting three minimum tiling paths (clusters) (Figure 1). From primer locus sources, we named the three clusters the BG (M64-6D1G2, 110G1, and 744D1), Core (M23-1B5D8, 514C2, and 1018A2), and Class I (122F9 and 1189C6) Regions with lengths of ~265 kb, ~85 kb, and ~150 kb, respectively, giving a total length of ~500 kb (Figure 1).
Figure 1. Genomic structure of the crested ibis MHC compared to the chicken and quail MHC-B complexes.
Colored boxes with different sizes show different gene loci of varied sizes, and upper/lower boxes indicate forward/reverse transcript orientations. The dotted oval between the Core and Class I Regions indicates the potential gap. The BACs involved in the minimum tiling path are depicted beneath the genomic maps.
Annotation of genes
Fifty-four genes were predicted in the three crested ibis MHC regions (Table S2). The BG Region ranges from the Blec genes to a zinc finger protein gene, member 1 (zfp1) (Figure 1 and Table S2), including a zfp gene family, a tripartite motif (TRIM) gene family, three butyrophilins (BTNs), two BGs, L-amino acid oxidase, Hep21, and guanidine nucleotide binding protein β-2-like 1.
The Core Region has a compact structure, with 17 predicted genes within an 85-kb segment (Figure 1) including collagen (COL11A2), four MHC-IIα genes (Nini-DAA and -DBAs 1–3), four MHC-IIβ genes (Nini-DAB and -DBBs 1–3), BRD2, DMA, two DMBs, one MHC-I gene (Nini-UAA), TAP1, TAP2, and LTB4R1 (Table S2). The Nini-IIα and -IIβ genes were arranged in tandem with opposite transcriptional orientations (Figure 1).
The Class I Region has a loose structure and shows 11 genes (TNXB, UBA, UCA1, ADPRH1, ADPRH2, UCA2, ADPRH3, UDA, OR, PHD finger protein 7, and USP42) and two pseudogenes (both TAP1) in a 150-kb range (Figure 1 and Table S2), presenting a sharp contrast to the 17 genes in the 85-kb class II Core Region (Figure 1). Four additional MHC-I loci were discovered in this region, giving five class I genes in the crested ibis. Unexpectedly, we found that both 122F9 and 1189C6 contain a pseudo TAP1 gene; the former presents just exons 7 and 8, while the latter exhibits only exon 11. The appearance of TAP1 adjacent to class I genes suggests a special relationship between the Class I and Core Regions.
Fluorescence in situ hybridization (FISH)
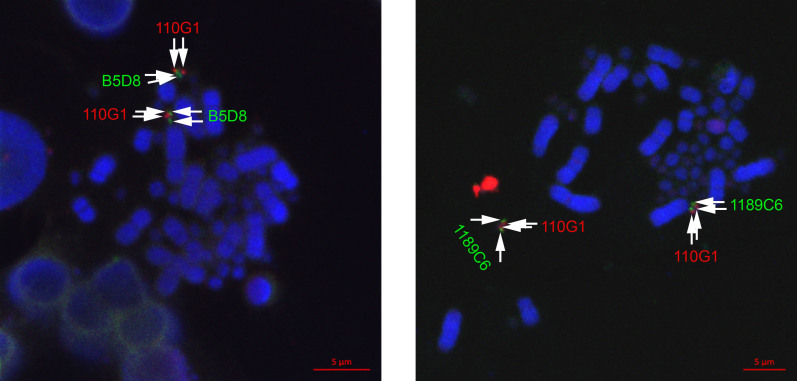
The crested ibis MHC shows a syntenic relationship with the chicken MHC in the BG and Core Regions, whereas the Class I Region is unique (Figure 1). We conducted two-color FISH using 110G1 (red), M23-1B5D8 (green), and 1189C6 (green) in the BG, Core, and Class I Regions, respectively, and localized these three clusters to the same chromosome (Figure 2). However, the crested ibis MHC is located in a microchromosome; hence, it is difficult to distinguish how the BG, Core, and Class I Regions correspond to the chicken MHC-located chromosome.
Figure 2. Two-color fluorescence in situ hybridization.
Probes were prepared from the BACs (110G1 in BG Region, M23-1B5D8 in Core Region, and 1189C6 in Class I Region) involved in minimum tiling paths (Figure 1). The BG, Core, and Class I Regions are linked and located in the same microchromosome.
Long and accurate PCR (La-PCR)
Unlike other previously investigated avian MHC regions, the Core Region contains several MHC-IIα and -IIβ gene copies, which are arranged repeatedly and alternatively (Figure 1). Therefore, we performed La-PCR to verify the manual assembly results. We divided the “COL11A2–BRD2” region into seven overlapping segments and designed beginning and end primers for the in-frame genes (Figure S1a). The La-PCR results (Figure S1b) agree with manual assembly in the Core Region (Figure 1). Furthermore, the La-PCR product sequences (P1–P7 segments) are identical to the assembly sequence of M23-1B5D8. Hence, La-PCR and sequencing rigorously confirm compact tandem MHC-II αβ dyads in the crested ibis genome.
Annotation of repeats
The overall GC content in the three regions is about 58.4%, which is higher than that in chicken (55.5%) and turkey (53.6%) MHC-B regions20. The Core Region has the highest GC content (67.1%), followed by the Class I (59.5%) and BG (55.0%) Regions. We identified 44 CpG islands, 39 tRNA elements, and 374 repetitive DNA elements (93 CR1/LTR repeats, 44 low complexity repeats, and 13 satellites) from the 500-kb MHC. The BG Region possesses all 39 tRNA elements, while the Class I Region has the highest density of repetitive DNA elements among these three regions, with 131 repeats within its 150-kb length, of which 61 are retroelements, and the long terminal repeats scatter with a frequency of 0.17 per kb.
Comparative genomic analysis
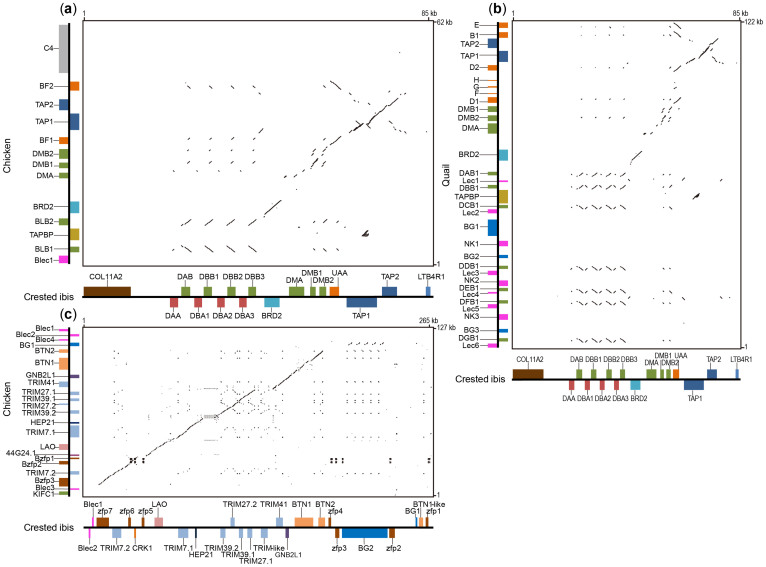
The identity plots of the Core Regions among crested ibis, chicken, and quail indicate some particular features of the crested ibis MHC: (1) presence of IIα genes in the class II region, (2) duplication of both IIα and IIβ genes to four each, (3) four tandem, tightly organized αβ dyads, and (4) occurrence of COL11A2 and loss of TAPBP (Figure 3a and 3b).
Figure 3. Identity plots of MHC regions.
Crested ibis Core Region vs. chicken (a) and quail (b), and crested ibis BG Region vs. chicken (c). Forward and reverse transcript orientations are respectively designated with upper and lower boxes on the horizontal axis or left and right boxes along the vertical axis. The reference sequence of quail MHC (AB078884) lacks the reported genes C4 and Cenp-A18,42, so we excluded them in the identity plot. For crested ibis vs. quail, we only compared the Core Region, since the extended TRIM-located region has not been reported in the quail MHC yet.
We plotted the identity matrix between the BG Regions of chicken and crested ibis (Figure 3c) and found that the initial (Blec2-Blec1) and final (zfp4-zfp1) fragments of the crested ibis have no synteny to the chicken MHC. However, the remaining region (zfp7-BTN2) in the crested ibis shows high structural consistency and sequence synteny with the related region in chicken. Notably, the TRIM genes in chicken and crested ibis are identically organized except for an additional predicted TRIM-like gene between TRIM41 and TRIM27.1 (Figure 3c).
Sequence and phylogenetic analyses
Class II genes
We obtained the full-length cDNA sequences of all Nini-IIα and -IIβ genes (Figure S2). These cDNA sequences perfectly match the class II haplotypes obtained by BAC sequencing, and cover three of the five partial IIβ exon 2 sequences previously reported33. The IIα and IIβ genes show four and six exons, respectively, and DBAs 1–3 encode the same amino acid sequences. We observed no frameshifts or in-frame premature-stop codons.
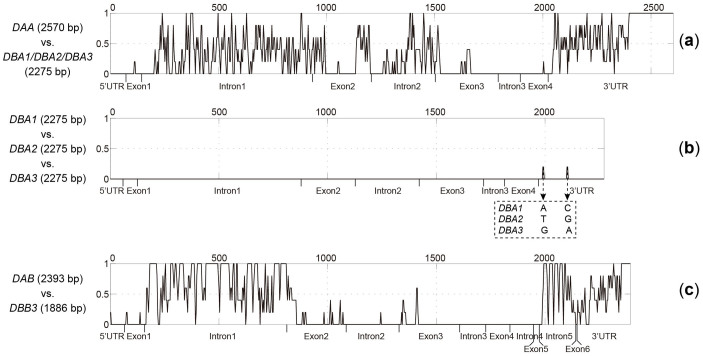
We aligned four full-length IIα sequences and found two obvious groups: DAA and DBA1/2/3 (Figure S2a). DAA introns 1 and 2, exons 2 and 3, and 3′UTR are highly divergent from DBA1/2/3 (Figure 4a). In particular, the 3′UTR of DAA is nearly double that of DBA1/2/3 (575 vs. 317 bp) (Figure 4a). DBAs 1–3 were nearly identical with only two single nucleotide polymorphisms (SNPs) in the 3′UTR (Figure 4b), which was verified in both individuals used for BAC library construction and RACE.
Figure 4. Variation distribution between aligned genomic sequences.
Alignment gaps were considered. For each plot, the sequence length scale (bp) is given on the top, and gene regions are marked at the bottom. (a) Comparison between Nini-DAA and -DBA1/2/3 (nearly identical genes). (b) Comparison among Nini-DBAs 1, 2, and 3. The only two single nucleotide polymorphisms in the 3′UTR are listed below. (c) Comparison between the two most divergent IIβ sequences, Nini-DAB and -DBB3.
The IIβ alignments depict three sets of genes: DAB, DBB1, and DBB2/3 (Figure S2b). DBBs 2 and 3 are nearly identical except for three amino acid variations in exons 1, 4, and 5. DBB1 appears to be a hybrid, with its first half resembling DBB3 and the second half being identical to DAB. Therefore, we calculated pairwise sequence identity based on full-length genomic sequences of the IIβ genes. The results show that DBBs 1–3 share a relatively high identity (92.8–98.4%) but divergent from DAB (80.4–87.4%) (Table 1). The length of DBB1 (2018 bp) is also chimeric, falling between DAB (2393 bp) and DBB2/3 (1886 bp), suggesting that the DAB-like fragment in DBB1 may arise from recombination. Furthermore, the two most divergent loci are DAB and DBB3 (80.4%) (Table 1) with the greatest difference in intron 1 (Figure 4c). Hence, we further computed pairwise sequence identity for IIβ intron 1 and found two groups: DAB (660 bp) and DBB1/2/3 (285 bp) (Table 1).
Table 1. Pairwise sequence distances among Nini-IIβ genes.
| Identity (%) | ||||||
|---|---|---|---|---|---|---|
| DAB | DBB1 | DBB2 | DBB3 | |||
| Divergence (%) | Full-length | DAB (2393 bp) | - | 87.4 | 86.9 | 80.4 |
| DBB1 (2018 bp) | 13.8 | - | 98.4 | 92.8 | ||
| DBB2 (1886 bp) | 14.4 | 1.7 | - | 94 | ||
| DBB3 (1886 bp) | 22.7 | 7.5 | 6.3 | - | ||
| Intron1 | DAB (660 bp) | - | 49.8 | 49.8 | 49.8 | |
| DBB1 (285 bp) | 84.3 | - | 100.0 | 100.0 | ||
| DBB2 (285 bp) | 84.3 | 0.0 | - | 100.0 | ||
| DBB3 (285 bp) | 84.3 | 0.0 | 0.0 | - |
Notes: Percent identities were computed by comparing sequences directly, while the divergence values were calculated by comparing the branch lengths of sequence pairs with the total branch length of the phylogenetic tree.
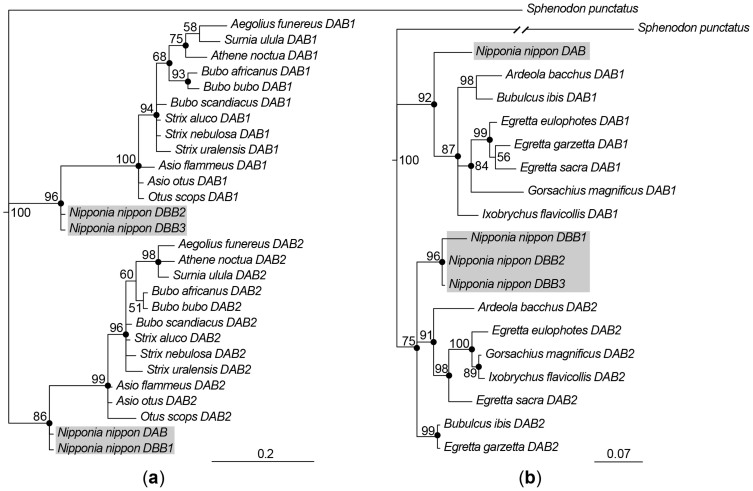
Considering the clear classification of Nini-IIβ genes into a separate DAB, a DBB2/DBB3 pair, and a DBB1 hybrid, we constructed exon 3-based phylogenetic trees in combination with owl IIβ genes, which contain an exon 3 fragment with two ancestral avian IIβ lineages23,24. The Bayesian and ML trees indicate that the four Nini-IIβ genes perfectly correspond to two owl IIβ clusters: Nini-DAB/DBB1 and -DBB2/DBB3 correspond to owl DAB2 and DAB1, respectively (Figure 5a). In view of the inter-Nini-IIβ variation hotspot in exon 2 (antigen presentation region) (Figure S2b), we built phylogenetic trees of exon 2 excluding antigen-binding sites (ABSs) for crested ibis and Ardeid birds, which split from the Threskiornithidae family 60 Mya35. The Bayesian and ML trees indicate that, similarly to two sets of IIα genes (Nini-DAA and -DBA1/2/3; Figure 4a and 4b), Nini-DAB clusters with Ardeid DAB1 loci whereas Nini-DBB1/2/3 are grouped with the Ardeid DAB2 genes (Figure 5b). Thus, the crested ibis MHC appears to retain two divergent ancestral class II “αβ” lineages with genetic recombination in Nini-DBB1.
Figure 5. Phylogenetic trees for IIβ exon 3 (a) and exon 2 (b) nucleotide sequences.
Numbers beside nodes indicate Bayesian support (%), and the filled circles on the nodes represent bootstrap values higher than 50% in maximum likelihood analysis. The crested ibis sequences are shaded. (a) The exon 3 tree includes crested ibis and owl sequences. Accession numbers of analyzed species are as follows: Aegolius funereus, EF641252, EF641253; Asio flammeus, EF641250, EF641251; Asio otus, EF641223, EF641224; Athene noctua, EF641247, EF641248; Bubo africanus, EF641244, EF641245; Bubo bubo, EF641236, EF641238; Bubo scandiacus, EF641233, EF641235; Otus scops, EF641257, EF641259; Strix aluco, EF641254, EF641256; Strix nebulosa, EF641240, EF641241; Strix uralensis, EF641242, EF641243; Surnia ulula, EF641226, EF641230; Sphenodon punctatus, DQ124232. (b) The exon 2 tree included the crested ibis and Ardeid sequences excluding the antigen-binding sites. Sequence sources are: Ardeola bacchus, HM991020, HM991044; Bubulcus ibis, HM991033, HM991054; Egretta eulophotes, HM991028, HM991052; Egretta garzetta, HM991035, HM991056; Egretta sacra, HM991037, HM991058; Gorsachius magnificus, HM991024, HM991048; Ixobrychus flavicollis, HM991026, HM991050; Sphenodon punctatus, DQ124231.
Class I genes
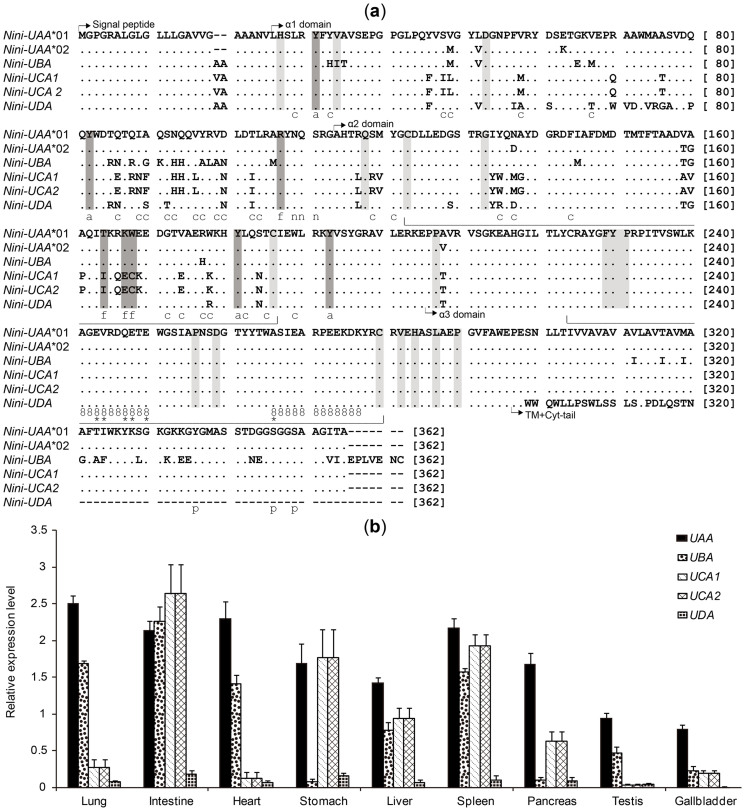
We successfully acquired the full-length cDNA sequences of all five class I genes including a new allele UAA*02 (Figure 6a), which is absent in target BACs above. UBA contains eight exons, as in most vertebrates, and the other four genes contain seven exons. UAA, UCA1, and UCA2 have lost intron 5. UDA is missing both intron 7 and exon 7, and carries frameshift mutation and premature termination in exon 5. UBA exon 8 is longer (seven residues) than the others. UCA1 and UCA2 encode the same amino acid sequence and are nearly identical in full-length nucleotide sequence with only a few SNPs.
Figure 6. Amino acid alignments (a) and expression levels (b) of Nini-MHC class I genes.
(a) Dots and dashes indicate identities and gaps within the first sequence. Conserved features along sequences are represented as follows: intra- and interdomain contacts (light grey shading), intradomain disulfide bridges (wide bracket), N-glycosylation site (“n”), CD8 binding sites (“8”), essential CD8 co-receptor sites (*), phosphorylated sites in cytoplasmic (cyt)-tail (“p”), antigen-peptide main chain-binding sites (i.e., antigen-binding sites anchoring two ends of peptides) (dark grey shading; “a” or “f” for A or F pocket, respectively)36,37,38,39,41, antigen-peptide non-main chain-binding sites (“c”) (i.e., antigen-binding sites binding the middle segment of peptides)37. (b) Expression levels of the five genes in nine different tissues were examined by qRT-PCR and normalized to the housekeeping gene GAPDH. The relative expression level of the Nini-I genes was calculated using the 2−ΔΔCT method54. The two nearly identical loci, UCA1 and UCA2, were simultaneously amplified using a single set of primers, and the average values were presented herein as the potential expression levels of each gene.
All but two functionally and structurally conserved Nini-I gene residues were highly conserved in crested ibis (Figure 6a). The first consists of eight residues (Figure 6a, dark grey) responsible for anchoring the N- (A pocket) and C- (F pocket) termini of the peptide main chains in α1 and α2 domains36,37,38. In the crested ibis, all class I genes except UCA1 and UCA2 maintain the consensus vertebrate “YYRTKWYY” sequence (“YYYTKWYY” in mammals)39. However, three substitutions in UCA1 and UCA2 are situated in the F pocket. The Y164I and K167E substitutions are regarded as neutral in extracellular proteins, whereas the W167C replacement is severely disfavored, as is any substitution for W16740. The second region comprises the 18 residues (Figure 6a, light grey) participating in intra- and inter-domain interactions of MHC-I molecules41 that are conserved in the five Nini-I genes, except for a favorable V34I substitution in UBA40.
The quantitative real-time PCR (qRT-PCR) results show that UAA was ubiquitously expressed in all tissues examined (Figure 6b). In contrast, UDA was weakly expressed in all tissues, with a bit higher levels in intestine and stomach. Relative to the dominantly expressed UAA, UBA was faintly present in stomach, pancreas, and gallbladder (<50% of UAA), whereas UCA1/2 were weakly expressed in lung, heart, testis, and gallbladder (<50% of UAA). Thus, the UBA and UCA1/2 exhibited variable expression levels among tissues and complementary roles in the lung, heart, stomach, pancreas, and testis. Generally, Nini-I genes were expressed at comparatively higher levels in the intestine and spleen, and lower in the testis and gallbladder.
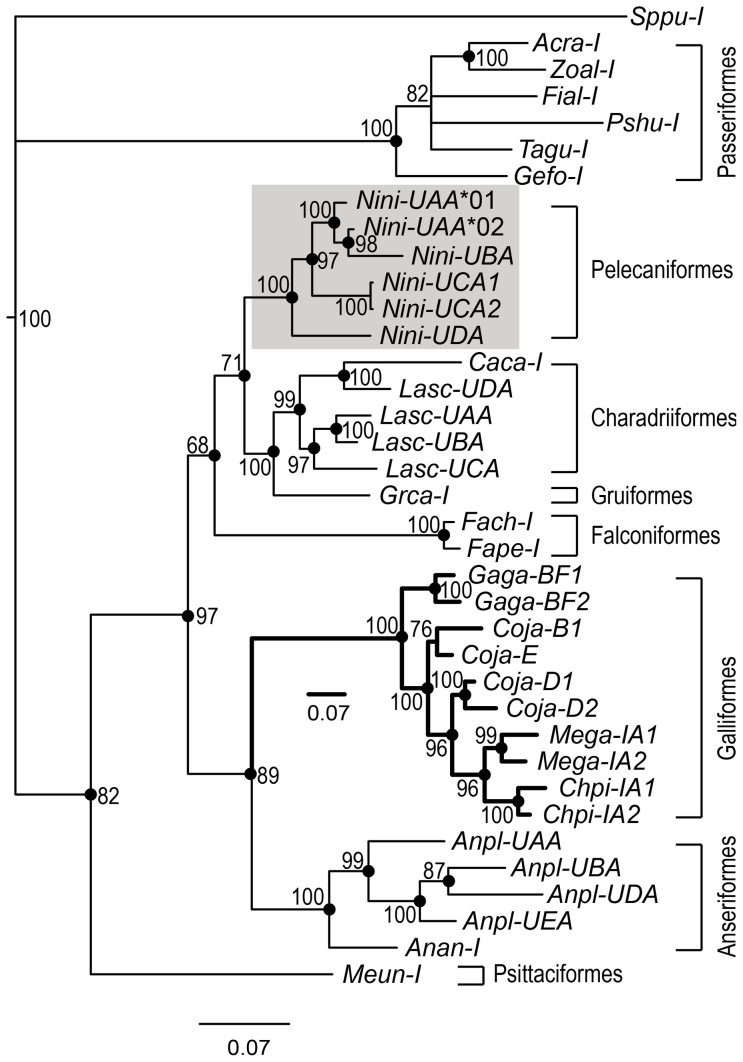
We constructed Bayesian and ML phylogenetic trees from the available PBR nucleotide sequences (exons 2 and 3) of avian MHC-I genes after removing ABSs. As revealed in mammalian class I genes6, the bird phylogeny has no obvious orthologous relationships (Figure 7), and most sequences cluster within species or orders. The phylogenetic trees exhibit four clusters for avian class I genes (Figure 7). The Passerines form a distinct cluster on the top, and the Psittaciformes branch from the remaining species as a second cluster at the bottom. The crested ibis displays a close relationship in the third clade with other wading and predatory birds, including Charadriiformes, Gruiformes, and Falconiformes. Finally, Galliformes and Anseriformes are grouped together as the Galloanserae. For the crested ibis, UDA is the basal branch of the five-gene-containing cluster (Figure 7) and is the most divergent Nini-I gene.
Figure 7. Phylogenetic tree of Nini-MHC class I genes.
The Bayesian tree was constructed based on the nucleotide sequences of exons 2 and 3 (excluding the antigen-binding sites). Numbers beside nodes indicate Bayesian support (%). The filled circles on the nodes represent bootstrap values higher than 50% in maximum likelihood analysis. The crested ibis sequences are shaded. Species information and sequence sources are: Gallus gallus (Gaga), AL023516; Coturnix japonica (Coja), AB078884; Meleagris gallopavo (Mega), DQ993255; Chrysolophus pictus (Chpi), JQ440366; Anas platyrhynchos (Anpl), AY885227; Anser anser (Anan), AY387648; Falco cherrug (Fach), XM005433165; Falco peregrines (Fape), XM005237695; Acrocephalus arundinaceus (Acar), AJ005507; Ficedula albicollis (Fial), XM005062204; Geospiza fortis (Gefo), XM005431466; Pseudopodoces humilis (Pshu), XM005534528; Taeniopygia guttata (Tagu), XM002186531; Zonotrichia albicollis (Zoal), XM005497294; Calidris canutus (Caca), KC205141; Larus scopulinus (Lasc), HM025953, HM015821, HM015820, HM008716; Grus canadensis (Grca), AF033106; Melopsittacus undulates (Meun), XM005139400; Sphenodon punctatus (Sppu), DQ145788.
Discussion
We characterize herein the first Pelecaniformes MHC. The compact Core Region of the crested ibis MHC, containing 17 genes within an 85-kb range, has a gene density similar to that of the chicken (19 loci within 92 kb)14 and quail (41 loci within 180 kb)18 core MHCs. Although the Passerine MHC is fragmented into several segments and has a much lower gene density19, our results suggest that the compact MHC structure is not limited to Galliformes.
The most dramatic structural feature in the crested ibis MHC is the compact organization of IIα and IIβ genes into four connected elementary “αβ” units within a 20-kb class II region, each consisting of one α and one β with opposite orientations (Figure 1), which differs from that observed in the Galliformes species, where the IIα gene exists as a single copy physically and genetically distant from IIβ genes. These four tandem αβ dyads also diverge from the parrot and duck MHCs, which preliminarily depicted a IIα gene adjacent to IIβ gene(s)25,26. In addition, although the quail also contains numerous duplicated IIβ genes, they were particularly mixed with Lec, NK, and BG genes (Figure 1), probably due to the repeated segmental duplication of IIβ, Lec, BG, and NK genes18. Furthermore, eight class II loci and nine MHC-related genes are condensed into an 85-kb Nini-MHC Core Region, sharply contrasting the loose distribution of class I genes. UAA is located in the Core Region, but the other four Nini-I loci (UBA, UCA1, UCA2, and UDA), two pseudo TAP1s, and 10 other genes are packed in a 150-kb Class I Region (Figure 1).
We submit that the crested ibis Class I Region is close to the Core Region because a BAC (988E8) extending upstream of the Class I Region contains a DBB2-like exon 2 and a IIβ pseudogene fragment, and overlaps the TNXB segment in 122F9. However, this 50-kb BAC was too fragmented (20 contigs) to provide useful information. Furthermore, 1018A2 downstream of the Core Region (Figure 1) also produced several small, extensively fragmented contigs (unknown orientations) in addition to the LTB4R1-containing contig. Considering these points and the lack of further BACs to extend upstream and downstream from the ~40-fold BAC genomic library34, we presume that the Class I Region is tightly linked with the Core Region and that the current gap could be attributed to complex sequences and/or secondary structures. This speculation is strongly supported by the high content of long terminal repeats in the Class I Region, which shows a frequency of 0.17 per kb and is even higher than that reported in the MHC class II region of Passerines (0.14 per kb)19, previously the most fragmented avian MHC. Hence, we propose that the Nini-MHC Class I Region is an extended region of the core Nini-MHC-B complex and that its divergence from the conserved BRD2–TAP2 frame, as shown in Figure 3a and 3b, relaxed selection pressure, resulting in an uncondensed genomic structure. Likewise, although the quail class I region contains numerous duplicated genes, it radically differs from the Nini-MHC Class I region and is virtually an “expanded” (rather than “extended”) pattern of the chicken class I region (Figure 1); the Coja-D1, -F, -G, -H, and -D2 loci correspond to Gaga-BF1, and the Coja-B1 and -E correspond to Gaga-BF218,42.
The crested ibis MHC includes COL11A2, which is absent in the Galliformes MHC but located upstream of the Nini-MHC class II region (Figure 1). In the western clawed frog, COL11A2 is adjacent to one pair of IIαβ genes43, and the mammalian COL11A2 is always located at the boundary of the classical and extended class II regions44. Furthermore, the mammalian MHC generally exhibits multiple α and β genes in the classical class II region, which is divided into DR, DQ, and DP subregions. Thus, the genomic structure of the crested ibis MHC class II region resembles the mammalian MHC, and the “COL11A2-IIα-IIβ” structure represents an ancestral feature of tetrapod MHCs. This predicts that the αβ dyad structure should be common in avian MHCs, and the separation of IIα from IIβ in chicken and the extensively fragmented MHC in Passerines should be group-specific structural variations.
The chicken BG (extended region) begins with Blec genes (Figure 1), rather like COL11A2 in mammalian MHC extended regions. The crested ibis BG Region and its COL11A2 gene are divided into two clusters (Figure 1) with an unknown gap size but have confirmed locations in the same microchromosome (Figure 2). However, the chicken and crested ibis MHCs both display organized TRIM clusters in their BG regions (Figure 1). Thus, we speculate that either the chicken MHC lost a COL11A2-located segment, which induces the compact MHC-B complex (42 genes in a 242-kb range) (Figure 1), or the BG Region was transposed outside of the Core Region in the crested ibis MHC, leading to the uncondensed BG genomic structure (26 genes in a 265-kb region) (Figure 1).
C4, Cenp-A, and CYP21 genes are commonly seen in Galliformes birds but were neither identified in the crested ibis class II segment nor in Passerines MHC19. Meantime, LTB4R1 and TNXB, which are at the downstream of these three genes in chicken, are respectively located at the boundaries of Core Region and Class I Region in crested ibis (Figure 1), showing a reverse order compared to that in chicken. Thus, we propose that C4, Cenp-A, and CYP21 genes are likely situated in the presumed gap between the Core and Class I Regions, and translocation of LTB4R1 occurred once in chicken or crested ibis.
Burri et al.23,24 identified two distinct IIβ clusters from phylogenetic trees of owl IIβ exon 3, which are interpreted as a pair of ancestral avian IIβ lineages that emerged ~100 Mya. During avian MHC evolution, each lineage lost once, resulting in only a single IIβ lineage in some avian orders. Herein, the four Nini-IIβ genes show two sets of exon 3 sequences that are highly similar to the two owl IIβ lineages (Figure 5a), suggesting that the Nini-IIβ genes retained both ancestral IIβ lineages. Although Burri et al.23 failed to detect two divergent lineages in owl exon 2 due to the homogenizing effect of gene conversion, we discovered two sets of IIβ exon 2 groupings between the crested ibis and Ardeidae (Figure 5b), further supporting the retention of two ancestral avian IIβ lineages.
Burri et al.24 also predicted that the birds developed two sets of ancestral IIα genes to pair with two divergent IIβ lineages. This hypothesis is first verified in our study, which reveals two suites of divergent Nini-IIα paralogs (Figure 4a and 4b, and Figure S2a). Furthermore, we noticed that the two sets of Nini-IIα paralogs (DAA and DBA1/2/3) were perfectly coincident with the two Nini-IIβ paralogs (DAB and DBB1/2/3) identified by PBR exon 2 (Figure 5b) and intron 1 (Table 1). Thus, the four tandem αβ dyads in the Nini-MHC suggest that the DAA/DAB pair may represent one ancestral “αβ” unit, while the other three αβ pairs (DBA1/DBB1, DBA2/DBB2, and DBA3/DBB3) may be derived from another ancestral αβ dyad through two recent duplication events. DBA3/DBB3 might be the most primitive unit because DBB3 is most divergent from DAB at the whole sequence level (Table 1). By combining these observations with our identification of the DBB1 hybrid between DAB and DBB3 (Figure 5 and Figure S2b), we propose that (1) the DAA/DAB and DBA3/DBB3 pairs represent the two sets of ancestral avian αβ dimers, (2) DBA1/DBB1 and DBA2/DBB2 are duplicated from DBA3/DBB3, and (3) recombination occurred in the β gene of the DBA1/DBB1 unit, shaping a new DBA1/DBB1' dimer.
The physical proximity of the IIα and IIβ genes provides a condition for coevolution. The Nini-IIα and Nini-IIβ variation distributions largely overlap (Figure 4a and 4c), implying that some overlapped variable sites may encode interacting residues between the α and β chains. The mammalian DRα/β45 and chicken BLA/BLB15 typically contain a non-polymorphic IIα gene and one or more highly polymorphic IIβ loci, which may arise from separate recombination of the α and β genes8,45, where the non-polymorphic α gene evolves to best fit the polymorphic β gene partners. However, the crested ibis displays tightly linked IIα and IIβ genes with two suites of dramatically divergent paralogs in a pattern similar to the mammalian DQα/β45, where both DQA and DQB genes vary. However, the loyal DBA1 is linked with a recombined DBB1', possibly arising from intra-locus (rather than inter-locus) recombination between the original DBB1 sequence and other undetermined DBB1 alleles.
Currently, the exact numbers and functional categorizations of MHC-I genes have been ascertained in only a few bird species. For example, the chicken B complex includes one major (BF1) and one minor (BF2) class I gene, while the Rfp-Y region contains one non-classical class I gene and one pseudogene9,14. Among the five MHC-I genes in duck, only Anpl-UAA is a dominantly expressed classical gene, and the others are a non-classical (Anpl-UDA) gene and unexpressed pseudogenes (Anpl-UBA, -UCA, and -UEA)7. The criteria for functional categorization in these studies generally involved comparison of sequence features (e.g., in-frame premature-stop codon, promoter sequence mutation, variation of conserved sites), expression (e.g., expression level, tissue specificity), and polymorphism.
Based on these criteria and several lines of evidence (detailed below), we classified the five Nini-MHC class I genes: one major classical gene (UAA); three minor classical loci (UBA, UCA1, and UCA2); and one non-classical locus (UDA). UDA carries a frameshift mutation and a resultant premature-stop codon in exon 5, leading to continuous variation in its transmembrane domain and lack of a cytoplasmic tail. UBA, UCA1, and UCA2 all contain variations on functionally and structurally conserved sites, which are more critical in UCA1 and UCA2 because the variable sites are responsible for main chain binding (Figure 6a), and the W167C substitution could be lethal. Furthermore, only UAA was ubiquitously expressed in all tissues examined. UBA and UCA1/UCA2 were expressed at different levels among the nine tissues and showed complementary patterns in some tissues (Figure 6b), probably indicating complementary roles in disease resistance. As UDA was faintly expressed in all tissues, but retained all conserved residues (Figure 6a), we propose that it might have become defunct only recently.
In most birds such as chicken14, and duck7, and even in amphibians46 and fish47, a single major class I gene is usually in close proximity to TAP genes which are responsible for translocation of endogenous pathogens to class I molecules48. In this study, we provide further support for this phenomenon by identifying a major classical locus, UAA, tightly linked to TAP1 and TAP2 in the Core Region (Figure 1). The other four class I loci, however, are located in the Class I Region (Figure 1), which is separated from the TAPs by several functional genes. Kaufman8 proposed that the evolution of a single predominant class I gene is probably due to its close proximity to TAP genes, providing an opportunity for the coevolution of both genes with associated roles in presenting antigen peptides. Thus, we predict that Nini-UAA in the Core Region may exhibit higher polymorphism than the other four Nini-I genes in the Class I Region. RACE identified two UAA alleles with six amino acid substitutions in PBR exons 2 and 3 (Figure 6a), thereby providing preliminary evidence to support this hypothesis.
We did not find any evidence of orthologous relationships with other birds and only showed that UDA is at the base of the Nini-MHC clade (Figure 7). These results suggest that the five Nini-I genes are all descendants from one ancestral copy. The coevolution between UAA and TAPs did not cause UAA to diverge further from the others; instead, UAA*02 clusters with UBA (Figure 7). UCA1 and UCA2 show nearly identical amino acid sequences (Figure 6a) and are therefore undoubtedly grouped together (Figure 7). As a consequence, the crested ibis MHC produces five similar class I genes, which are arranged at intervals in a range >115 kb (UAA–LTB4R1…TNXB–UDA; Figure 1).
Our study thus reveals that the crested ibis MHC develops tandem divergent αβ dyads in the condensed class II region but similar class I genes in the extended region. Furthermore, we predict that the fundamental structure of ancestral avian class II MHCs should be “COL11A2-IIαβ1-IIαβ2”, where two “αβ” units represent different dyad lineages. In light of the dramatic diversity in avian MHC architectures, there is a clear requirement for further investigation of MHC genomic landscapes in taxonomically diverse avian species to understand avian MHC evolution completely.
The MHC plays an important role in immune response. The crested ibis has undergone a severe bottleneck and shows a low level of genetic variation31,33, suggesting pathogen susceptibility. Thus, the Nini-MHC sequence reported in this study provides a foundation for examining genetic variation and adaptive evolution. Based on these information, the scientists are able to formulate effective strategies to improve survival rate, and optimize the population structure of founders for newly created populations (such as reintroduction) to ensure long-term persistence of this endangered bird.
Methods
Sample collection
This study involves two kinds of samples from three crested ibises; blood of individual 01 was collected in a previous study for BAC genomic library construction34, blood of individual 02 was used for FISH, tissues of individual 03 were subjected to mRNA extraction. The samples were obtained from the Louguantai (individual 01) and Deqing (02 and 03) Crested Ibis Breeding Centers of Shaanxi and Zhejiang Provinces, respectively. The individual 03 was immediately dissected after its death by accident, and tissues of lung, intestine, heart, stomach, liver, spleen, pancreas, testis, and gallbladder were collected and strictly stored in liquid nitrogen. All experiments were approved by the ethics committees of these two Centers and were carried out in accordance with the approved guidelines.
Primer design, BAC screening, and sequencing
Based on the reported exon 2 sequences of Nini-MHC-IIβ genes33, we designed a pair of primers for a 161-bp product with high PCR efficiency. In addition, by using the available sequences from chicken and other related species, we also designed degenerate primers for the IIα, MHC-I, DMA, TAP2, and BG genes. We ascertained the reliability of the PCR-generated product sequences by the Basic Local Alignment Search Tool (BLAST)49, and then developed a collection of species-specific primers for the Nini-MHC genes based on the determined sequences (Table S1).
BAC clones containing MHC loci were screened from both routine and reverse-4D libraries34 using the Nini-MHC gene-specific primers. The PCR-based BAC screening processes and identification of real positive clones were performed as previously described17,50. Once a positive BAC clone was screened out, we subjected it to BAC-end sequencing. The end sequences were repeat-masked with RepeatMasker (http://www.repeatmasker.org) and then used to design primers to confirm the overlaps of BAC clones and to obtain the extended segments in the next round of clone screening. The target BACs were subjected to shotgun sequencing and assembled by Majorbio (Shanghai, China). The MHC genomic sequences were deposited in GenBank (accession number: KP182407–KP182409).
To avoid mistakes arising from commercial assembly, we performed manual assembly of sequencing reads by using Lasergene software (DNASTAR Inc.; Infogen Bioinformatics, Broxburn, UK). Furthermore, we performed a series of La-PCRs using BAC-positive clones as template to further verify specific regions with complex structures. We adopted a two-step method using LA Taq (Takara, Dalian, China) or KOD DNA polymerase (Toyobo) with annealing at 68°C for product length-dependent extension times (Table S3). The PCR products were further cloned and sequenced (for both clones and PCR products). Each La fragment was confirmed by at least three additional independent rounds of La-PCRs.
Two-color FISH
We prepared cell metaphases from the peripheral blood of the crested ibis as described by Moorhead et al.51 and performed two-color FISH according to our previous protocol27. We isolated plasmid DNA from BAC clones by using the Qiagen Large-Construct Kit (Qiagen, Hilden, Germany) and labeled the plasmids by using red fluorescence (ChromaTide Alexa Fluor 488-5-dUTP; Molecular Probes, Eugene, Oregon) and green fluorescence (Alexa Fluor 594-5-dUTP, Molecular Probes). The three BAC plasmids involved were from the routine (110G1 and 1189C6) and reverse-4D (M23-1B5D8) libraries, respectively.
Gene prediction and annotation
We predicted the genes by using GENSCAN (http://genes.mit.edu/GENSCAN.html) and Softberry FGENESH (http://linux1.softberry.com/berry.phtml), and confirmed the reliability of coding sequences by BLAST-based identification of homologous domain to known genes. For class I and II genes, the annotation follows the nomenclature suggested by Klein et al.52 and meanwhile incorporates the information of gene order, phylogenetic relationship, and sequence similarity. We employed RepeatMasker and tRNAScan (http://lowelab.ucsc.edu/tRNAscan-SE/) to find repetitive and tRNA elements, respectively. CpG islands were marked by Softberry CpGFinder (http://linux1.softberry.com/berry.phtml?topic=cpgfinder&group=programs), and GC content was analyzed using Isochore (http://www.ebi.ac.uk/Tools/seqstats/emboss_isochore/). Identity dots of Nini-MHC were generated in PipMaker53 against the chicken (AB268588) and quail (AB078884) sequences.
qRT-PCR expression detection and full-length cDNA amplification
Total RNA from the nine different tissues was separately extracted using TRIzol reagent (Invitrogen, Carlsbad, California) and reverse-transcribed to cDNA with the PrimeScript™ First Strand cDNA Synthesis Kit (Takara). Then, we detected the expression pattern of class I genes in the nine different tissues using qRT-PCR. The housekeeping gene GAPDH (F: 5′-AAGGCTGAGAATGGGAAAC-3′, R: 5′-TTCAGGGACTTGTCATACTTC-3′) was used to control for variations in the amounts of cDNA template. All the qRT-PCR assays were performed on a fluorescence thermal cycler (7500 Fast Real-Time PCR System, Applied Biosystems, USA) using SYBR® Premix Ex Taq Premix (Takara) and locus-specific primers (Table S4). Since the full-length sequences of UCA1 and UCA2 are highly similar with only a few SNPs, they were simultaneously amplified by a common pair of primers, and we used the average values as the potential expression levels of each gene. Each sample was run in triplicate for two independent reactions to obtain the sample mean value. The relative quantification 2−ΔΔCT method54 was adopted to analyze the relative expression level of the five class I genes.
The liver total RNA was also used to acquire the full-length cDNA with the GeneRacer™ Kit (Invitrogen). 5′ rapid amplification of cDNA ends (RACE) (5′UTR–exon 3) and 3′ RACE (exon 2–3′UTR) for the MHC class I and II genes were conducted with the Nini-MHC gene-specific primers (Table S4) and universal 5′/3′-primers provided in the kit. PCR products were cloned, and ten positive clones per primer set were sequenced. Each fragment was verified by at least three independent rounds of PCRs. Full-length cDNA sequences were obtained by assembling the two overlapping fragments in Lasergene and deposited in deposited in GenBank (accession number: KP182410–KP182423).
Sequence and phylogenetic analysis
We performed sequence alignments using the ClustalW algorithm with manual modifications in MEGA 5.155. Nucleotide diversity values (π) along pairwise full-length genomic sequences of class II genes were calculated in DnaSP 5.056 with a 5-bp sliding window (a 2-bp overlap), based on which the line charts of nucleotide variation distributions (considering gap sites in the alignments) were created using MATLAB 7.0. Besides, we employed MegAlign in Lasergene to calculate pairwise sequence identities and divergences among full-length genomic sequences and intron 1 sequences of IIβ genes. Pairwise identity is computed by comparing sequences directly with no consideration of phylogenetic relationships, while pairwise divergence is estimated by dividing the sum of two branch lengths of a sequence pair by the sum of all branch lengths in the constructed phylogeny.
Phylogenetic trees were constructed by the Bayesian inference method in MrBayes 3.257 and the maximum likelihood (ML) method in PhyML58. The most appropriate evolutionary models of nucleotide substitution were inferred from jModelTest 2.1.159 by following the Akaike information criterion60. For Bayesian tree construction, we performed two independent runs of four Markov chain Monte Carlo chains, with 100,0000 generations sampled every 100 generations, and we abandoned the first 25% as “burn-in”. For the ML tree, we employed the subtree pruning and regrafting approach with five random starts to estimate tree topologies and performed 1,000 bootstrap replicates. We built phylogenetic trees of exons 2 and 3 for Nini-IIβ genes and exons 2–3 for Nini-I genes. Since most avian studies failed to assign MHC-I sequences to individual loci, we only used one representative sequence from those species. We first constructed an intra-species Bayesian tree using a lizard class I gene (DQ145788) as the outgroup and then selected the most primitive branch as the representative sequence. For Nini-I genes, we did not construct phylogenetic trees of non-PBR exon 4 for comparison because very few exon 4 sequences are available.
Author Contributions
Conceived and designed the research: Q.H.W. Performed the research: L.C.C., H.L., L.S., Y.L.D., K.Y.T. and Q.H.W. Wrote the manuscript: L.C.C., H.L. and Q.H.W. All authors read and approved the final manuscript.
Supplementary Material
Supplementary Tables S1-S4 and Figures S1-S2
Acknowledgments
This work was supported by a grant from the National Natural Science Foundation of China (No. 31270423), a special grant from the State Forestry Administration, and the Fundamental Research Funds for the Central Universities of the P. R. China.
References
- Klein J. Natural history of the major histocompatibility complex. 1–775 (John Wiley and Sons, New York, 1986). [Google Scholar]
- Geraghty D. E., Daza R., Williams L. M., Vu Q. & Ishitani A. Genetics of the immune response: identifying immune variation within the MHC and throughout the genome. Immunol. Rev. 190, 69–85 (2002). [DOI] [PubMed] [Google Scholar]
- Spurgin L. G. & Richardson D. S. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc. R. Soc. Lond., Ser. B: Biol. Sci. 277, 979–988 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkman P. J. & Parham P. Structure, function, and diversity of class I major histocompatibility complex molecules. Annu. Rev. Biochem. 59, 253–288 (1990). [DOI] [PubMed] [Google Scholar]
- Villadangos J. A. Presentation of antigens by MHC class II molecules: getting the most out of them. Mol. Immunol. 38, 329–346 (2001). [DOI] [PubMed] [Google Scholar]
- Hughes A. L. & Nei M. Evolution of the major histocompatibility complex: independent origin of nonclassical class I genes in different groups of mammals. Mol. Biol. Evol. 6, 559–579 (1989). [DOI] [PubMed] [Google Scholar]
- Moon D. A., Veniamin S. M., Parks-Dely J. A. & Magor K. E. The MHC of the duck (Anas platyrhynchos) contains five differentially expressed class I genes. J. Immunol. 175, 6702–6712 (2005). [DOI] [PubMed] [Google Scholar]
- Kaufman J. Co-evolving genes in MHC haplotypes: the “rule” for nonmammalian vertebrates? Immunogenetics 50, 228–236 (1999). [DOI] [PubMed] [Google Scholar]
- Kaufman J. The avian MHC. Avian Immunology, 159–181; 10.1016/B978-012370634-8.50011-1 (2008). [DOI] [Google Scholar]
- Gorer P. A., Lyman S. & Snell G. D. Studies on the genetic and antigenic basis of tumour transplantation. Linkage between a histocompatibility gene and ‘fused’ in mice. Proc. R. Soc. Lond., Ser. B: Biol. Sci. 135, 499–505 (1948). [Google Scholar]
- Hurt P. et al. The genomic sequence and comparative analysis of the rat major histocompatibility complex. Genome Res. 14, 631–639 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuhki N. et al. Comparative genome organization of human, murine, and feline MHC class II region. Genome Res. 13, 1169–1179 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Q. H., Zeng C. J., Ni X. W., Pan H. J. & Fang S. G. Giant panda genomic data provide insight into the birth-and-death process of mammalian major histocompatibility complex class II genes. PLos One 4, e4147; 10.1371/journal.pone.0004147 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman J. et al. The chicken B locus is a minimal essential major histocompatibility complex. Nature 401, 923–925 (1999). [DOI] [PubMed] [Google Scholar]
- Salomonsen J. et al. The properties of the single chicken MHC classical class II α chain (B-LA) gene indicate an ancient origin for the DR/E-like isotype of class II molecules. Immunogenetics 55, 605–614 (2003). [DOI] [PubMed] [Google Scholar]
- Chaves L. D., Krueth S. B. & Reed K. M. Defining the turkey MHC: sequence and genes of the B locus. J. Immunol. 183, 6530–6537 (2009). [DOI] [PubMed] [Google Scholar]
- Ye Q., He K., Wu S. Y. & Wan Q. H. Isolation of a 97-kb minimal essential MHC B locus from a new reverse-4D BAC library of the golden pheasant. PLoS One 7, e32154; 10.1371/journal.pone.0032154 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina T. et al. Comparative genomic analysis of two avian (quail and chicken) MHC regions. J. Immunol. 172, 6751–6763 (2004). [DOI] [PubMed] [Google Scholar]
- Balakrishnan C. N. et al. Gene duplication and fragmentation in the zebra finch major histocompatibility complex. BMC Biol. 8, 29; 10.1186/1741-7007-8-29 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcaide M., Edwards S. V., Cadahía L. & Negro J. J. MHC class I genes of birds of prey: isolation, polymorphism and diversifying selection. Conserv. Genet. 10, 1349–1355 (2009). [Google Scholar]
- Alcaide M., Edwards S. V. & Negro J. J. Characterization, polymorphism, and evolution of MHC class II B genes in birds of prey. J. Mol. Evol. 65, 541–554 (2007). [DOI] [PubMed] [Google Scholar]
- Ekblom R., Grahn M. & Höglund J. Patterns of polymorphism in the MHC class II of a non-passerine bird, the great snipe (Gallinago media). Immunogenetics 54, 734–741 (2003). [DOI] [PubMed] [Google Scholar]
- Burri R., Hirzel H. N., Salamin N., Roulin A. & Fumagalli L. Evolutionary patterns of MHC class II B in owls and their implications for the understanding of avian MHC evolution. Mol. Biol. Evol. 25, 1180–1191 (2008). [DOI] [PubMed] [Google Scholar]
- Burri R., Salamin N., Studer R. A., Roulin A. & Fumagalli L. Adaptive divergence of ancient gene duplicates in the avian MHC class II beta. Mol. Biol. Evol. 27, 2360–2374 (2010). [DOI] [PubMed] [Google Scholar]
- Hughes C. R., Miles S. & Walbroehl J. M. Support for the minimal essential MHC hypothesis: a parrot with a single, highly polymorphic MHC class II B gene. Immunogenetics 60, 219–231 (2008). [DOI] [PubMed] [Google Scholar]
- Ren L. M. et al. Characterization of the MHC class II α-chain gene in ducks. Immunogenetics 63, 667–678 (2011). [DOI] [PubMed] [Google Scholar]
- Wan Q. H. et al. Genome analysis and signature discovery for diving and sensory properties of the endangered Chinese alligator. Cell Res. 23, 1091–1105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- BirdLife International. Nipponia nippon. The IUCN Red List of Threatened Species (2014) Avaiable at: www.iucnredlist.org (Accessed: 28/01/2014). [Google Scholar]
- Li X. H. & Li D. M. Current state and the future of the crested ibis (Nipponia nippon): a case study by population viability analysis. Ecol. Res. 13, 323–333 (1998). [Google Scholar]
- Ding C. Q. Research on the Crested Ibis. 1–388 (Shanghai Scientific and Technological Publishing House, 2004) (in Chinese). [Google Scholar]
- He L. P., Wan Q. H., Fang S. G. & Xi Y. M. Development of novel microsatellite loci and assessment of genetic diversity in the endangered Crested Ibis, Nipponia Nippon. Conserv. Genet. 7, 157–160 (2006). [Google Scholar]
- Zhang B., Fang S. G. & Xi Y. M. Low genetic diversity in the endangered crested ibis Nipponia nippon and implications for conservation. Bird Conserv. Int. 14, 183–190 (2004). [Google Scholar]
- Zhang B., Fang S. G. & Xi Y. M. Major histocompatibility complex variation in the endangered crested ibis Nipponia nippon and implications for reintroduction. Biochem. Genet. 44, 110–120 (2006). [DOI] [PubMed] [Google Scholar]
- Lan H. et al. The first report of a Pelecaniformes defensin cluster: Characterization of β-defensin genes in the crested ibis based on BAC libraries. Sci. Rep. 4, 6923; 10.1038/srep06923 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. W., Payne R. B. & Mindell D. P. Nuclear DNA does not reconcile ‘rocks’ and ‘clocks’ in Neoaves: a comment on Ericson et al. Biol. Lett. 3, 257–260 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkman P. et al. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature 329, 512–518 (1987). [DOI] [PubMed] [Google Scholar]
- Saper M., Bjorkman P. & Wiley D. Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 Å resolution. J. Mol. Biol. 219, 277–319 (1991). [DOI] [PubMed] [Google Scholar]
- Wallny H. J. et al. Peptide motifs of the single dominantly expressed class I molecule explain the striking MHC-determined response to Rous sarcoma virus in chickens. Proc. Natl. Acad. Sci. USA 103, 1434–1439 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman J., Salomonsen J. & Flajnik M. Evolutionary conservation of MHC class I and class II molecules—different yet the same. Semin. Immunol. 6, 411–424 (1994). [DOI] [PubMed] [Google Scholar]
- Betts M. J. & Russell R. B. in Bioinformatics for geneticists (eds Barnes Michael R. & Gray Ian C.) 289–316 (John Wiley and Sons, Chichester, 2003). [Google Scholar]
- Grossberger D. & Parham P. Reptilian class I major histocompatibility complex genes reveal conserved elements in class I structure. Immunogenetics 36, 166–174 (1992). [DOI] [PubMed] [Google Scholar]
- Shiina T., Hosomichi K. & Hanzawa K. Comparative genomics of the poultry major histocompatibility complex. Anim. Sci. J. 77, 151–162 (2006). [Google Scholar]
- Ohta Y., Goetz W., Hossain M. Z., Nonaka M. & Flajnik M. F. Ancestral organization of the MHC revealed in the amphibian Xenopus. J. Immunol. 176, 3674–3685 (2006). [DOI] [PubMed] [Google Scholar]
- Kumánovics A., Takada T. & Lindahl K. F. Genomic organization of the mammalian MHC. Annu. Rev. Immunol. 21, 629–657 (2003). [DOI] [PubMed] [Google Scholar]
- Germain R. N., Bentley D. M. & Quill H. Influence of allelic polymorphism on the assembly and surface expression of class II MHC (Ia) molecules. Cell 43, 233–242 (1985). [DOI] [PubMed] [Google Scholar]
- Nonaka M. et al. Major histocompatibility complex gene mapping in the amphibian Xenopus implies a primordial organization. Proc. Natl. Acad. Sci. USA 94, 5789–5791 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takami K., Zaleska-Rutczynska Z., Figueroa F. & Klein J. Linkage of LMP, TAP, and RING3 with Mhc class I rather than class II genes in the zebrafish. J. Immunol. 159, 6052–6060 (1997). [PubMed] [Google Scholar]
- Antoniou A. N., Powis S. J. & Elliott T. Assembly and export of MHC class I peptide ligands. Curr. Opin. Immunol. 15, 75–81 (2003). [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- Zeng C. J. et al. Giant panda BAC library construction and assembly of a 650-kb contig spanning major histocompatibility complex class II region. BMC Genomics 8, 315; 10.1186/1471-2164-8-315 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead P. S., Nowell P. C., Mellman W. J., Battips D. M. & Hungerford D. A. Chromosome preparations of leukocytes cultured from human peripheral blood. Exp. Cell Res. 20, 613–616 (1960). [DOI] [PubMed] [Google Scholar]
- Klein J. et al. Nomenclature for the major histocompatibility complexes of different species: a proposal. Immunogenetics 31, 217–219 (1990). [DOI] [PubMed] [Google Scholar]
- Schwartz S. et al. PipMaker—a web server for aligning two genomic DNA sequences. Genome Res. 10, 577–586 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P. & Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009). [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J. P. & Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 (2001). [DOI] [PubMed] [Google Scholar]
- Guindon S. & Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 (2003). [DOI] [PubMed] [Google Scholar]
- Darriba D., Taboada G. L., Doallo R. & Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772–772 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. & Buckley T. R. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 53, 793–808 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables S1-S4 and Figures S1-S2