Abstract
Ectopic overexpression of transcription factors has been used to reprogram cell fate. For example, virus-mediated overexpression of four transcription factors OCT4, SOX2, MYC, and KLF4, known as Yamanaka factors, can convert somatic cells to induced pluripotent stem (iPS) cells. However, gene-specific switch-on of endogenous gene production without the use of foreign DNA remains a challenge. The small RNA machinery that comprised small RNAs and Argonaute proteins is known to silence gene expression, but can be repurposed to activate gene expression when directed to gene promoters, a phenomenon known as RNA activation or RNAa. By screening of dsRNAs targeting OCT4 promoter, we identified a small activating RNA (saRNA) that activated OCT4 expression in several types of human mesenchymal stem cells (MSCs). We found that saRNA-induced OCT4 activation can be further enhanced by a histone deacetylase inhibitor, valproic acid. Furthermore, introducing OCT4 saRNA in combination with viruses encoding the remaining three Yamanaka factors (SOX2, MYC, and KLF4) into MSCs led to the derivation of partially reprogrammed iPS cells. Findings from this study suggest that, with further optimization, RNAa can be a powerful tool to reprogram cell fate by inducing the expression of endogenous genes.
Introduction
Small RNAs in complex with Argonaute proteins are powerful modulators of gene activity. Apart from silencing gene expression through the well-known RNAi and related mechanisms, the small RNA machinery has been found to positively affect gene expression at the epigenetic/transcriptional level [1,2]. This gene activation mechanism has been termed RNAa [3,4]. RNAa can be triggered by both artificially designed small activating RNAs (saRNAs) and endogenous small RNAs that target gene regulatory sequences or coding regions [2–8]. RNAa appears to be conserved in evolution, being present in animals ranging from Caenorhabditis elegans (C. elegans) to human [1,2,5]. Despite that the detailed mechanism for RNAa awaits to be elucidated, this unparalleled approach of sequence-specific activation of endogenous gene expression can obviously be harnessed for a number of purposes such as disease treatment [9,10] and cell fate reprogramming.
It has been widely demonstrated that overexpressing one or more factors using vector-based methods can change the identity of a committed cell type to another. Overexpression of OCT4, SOX2, MYC, and KLF4 (known as Yamanaka factors) can drive somatic cells back to a pluripotent state, known as induced pluripotent stem (iPS) cells [11,12]. Nonvector-based methods, including the use of small-molecule compounds [13], synthetic modified mRNA [14], miRNAs [15], and recombinant proteins [16], have also been employed either alone or in combination with vector-based overexpression to derive iPS cells with varying success. Safer and more efficient approaches for the generation of iPS cells are still being sought. We have previously shown that RNAa of NANOG, a pluripotency marker, in embryonic carcinoma cells is able to suppress retinoic acid-induced differentiation [17].
OCT4, also known as POU5F1 (POU domain, class 5, transcription factor 1), is a member of the POU transcription factor family. It is regarded as the most important and irreplaceable factor among the four Yamanaka factors. OCT4 alone has also been shown to be sufficient for directly reprogramming mouse neural stem cells and hair follicle dermal papilla cells (FDPCs) to pluripotent cells [18,19], highlighting its critical role in stem cell reprogramming. Transcription of OCT4 is controlled by a combination of transcription factors and epigenetic mechanisms [20,21]. Methods that either alter transcription factor binding or alleviate epigenetic repressors have been used to induce endogenous OCT4 expression [22,23].
In this study, we identified a saRNA, dsOCT4-622, which targets the promoter of OCT4 and activates OCT4 expression at both the mRNA and protein levels. OCT4 RNAa could be further enhanced by simultaneously treating cells with valproic acid (VPA), an epigenetic modifier. Replacing OCT4 virus in the Yamanaka iPS derivation protocol with OCT4 saRNA was able to convert human mesenchymal stem cells (MSCs) into a partially reprogrammed state.
Materials and Methods
Culture of human MSCs
Human adipose-derived stem cells (ADSCs), lot no. 38, were harvested and cultured, as previously described [24]. Human adipose tissues were obtained during abdominoplasty following informed patient consent and the guidelines set by the institutional committee on human research. Two additional human ADSC lines were purchased from Lonza (Cat. PT-5006, Lot. 7F4089) and ATCC (Cat. PCS-500-011, Lot. 59193163) and cultured in the MesenPRO RS Medium (Invitrogen). Human FDPCs established from two different donors (Cat. C-12071, Lot. 1011103.3 and Lot. 2030702.3) and MSCs from the umbilical cord matrix (UC-MSCs) (Cat. C-12971, Lot. 1112304.2) were purchased from PromoCell and cultured according to the manufacturer's instructions. All human MSC lines were expanded and maintained in a humidified atmosphere of 5% CO2 at 37°C and used at passage 5 throughout this study.
saRNA design and transfection
A 1-kb upstream region of the human OCT4A (POU5F1 transcript variant 1, NM_002701) promoter sequence was scanned for saRNA targets based on design rules, as previously described [5]. All of the dsRNAs screened are listed in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/scd). The day before transfection, cells were plated in a growth medium at a density of 60–70%. Transfection of dsRNA was carried out using Lipofectamine RNAiMax (Invitrogen) or HiPerFect (Qiagen) according to the reverse transfection protocol of the manufacturer's instructions. After a 5- to 6-h incubation, transfection complexes were removed and replaced with a fresh growth medium containing either chemical compounds or dimethyl sulfoxide (DMSO).
RNA isolation and mRNA expression analysis
Total RNA was isolated by using the RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. RNA (1 μg) was reverse transcribed with oligo(dT) primers. The resultant cDNA was amplified by polymerase chain reaction (PCR) using gene-specific primers (Supplementary Table S2) in conjunction with Fast SYBR Green or TaqMan PCR Master Mix (Applied Biosystems). GAPDH was amplified as an internal control.
Chemical compounds
Trichostatin A (TSA) and 5-Azacytidine (5-Aza) were obtained from Sigma. BIX01294, VPA, sodium butyrate (NaB), and Tranylcypromine hydrochloride (Parnate) were purchased from Stemgent. All chemical compounds were dissolved in DMSO.
Immunoblot analysis
Cell lysates were prepared from ADSCs using a standard protocol. Equivalent amounts of proteins were resolved on sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels and transferred to nitrocellulose membranes. The resulting blots were blocked with nonfat milk and incubated with anti-OCT4A (C-10) (1:1,000 dilution, sc-5279; Santa Cruz Biotechnology) and anti-GAPDH (1:5,000 dilution, #2118; Cell Signaling Technology). Blots were washed and subsequently incubated with horseradish peroxidase-conjugated secondary antibodies and visualized by chemiluminescence (Thermo Scientific).
Immunocytochemical analysis
For immunocytochemical analysis of OCT4A on ADSCs, cells were seeded on coverslips in six-well plates with the growth medium. Four days after transfection, the cells were fixed, permeabilized, blocked with normal horse serum, and then incubated with anti-OCT4A (C-10) (1:100 dilution, sc-5279; Santa Cruz Biotechnology) at 4°C overnight. After overnight incubation, cells were washed and incubated with the Alexa 488-conjugated secondary antibody (Invitrogen) for visualization. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Immunocytochemical analysis of Tra-1-60 was performed by using the StainAlive Tra-1-60 antibody (DyLight 488) (Stemgent) and following the manufacturer's instructions. Fluorescent images were obtained using a Nikon E600 microscope equipped with a digital camera.
Cell proliferation assay
Cell proliferation of ADSCs 7F4089 and FDPCs 1011103.3 was determined by using the CellTiter96 Aqueous one solution cell proliferation assay kit (Promega) containing the MTS [3-(4, 5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] reagent, as previously described [17].
iPS reprogramming
Human ADSCs (8×104 cells per well of a six-well plate) were transfected with Mock, dsControl, or dsOCT4-622 on day 1, following the reverse transfection protocol of RNAiMax (Invitrogen). On day 0 and 1, cells were transduced twice with OSKM (OCT4, SOX2, KLF4, and MYC) or SKM (SOX2, KLF4, and MYC) retroviral particles generated, as previously described [25]. From day 2 to 14, the cells were sequentially transfected with Mock, dsControl, or dsOCT4-622 every 3 days for five more times according to the forward transfection protocol of RNAiMax (Invitrogen). Four days after initial infection (day 4), the cells were replated onto irradiated mouse embryonic fibroblast (MEF) feeder layer in the ADSC growth medium. The medium was replaced the next day (day 5) with the human embryonic stem (ES) cell medium (KnockOut Dulbecco's modified Eagle's medium: Nutrient Mixture F-12 (DMEM/F-12) medium supplemented with 20% knockout serum replacement serum, 2 mM GlutaMAX, 100 μM nonessential amino acids, 100 μM β-mercaptoethanol, and 10 ng/mL basic fibroblast growth factor; Invitrogen). iPS-like colonies were picked and expanded onto MEF feeders in the human ES medium. VPA (1 mM) was added in the ADSC growth medium and human ES medium from day 1 to 16 and both media were changed daily.
Results and Discussion
OCT4 saRNA design and screening
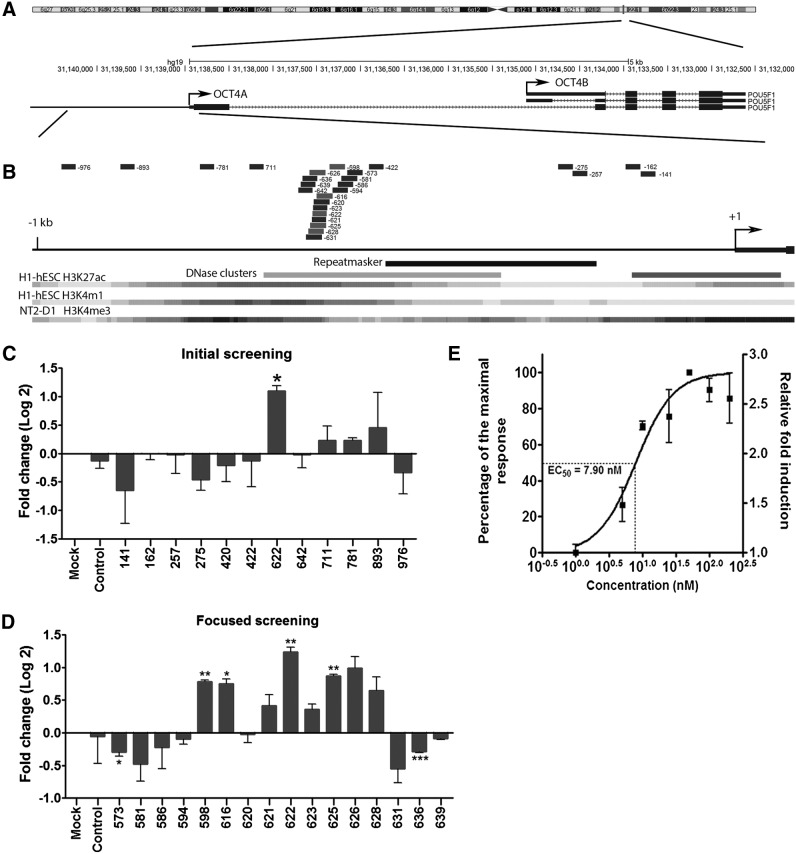
To investigate the feasibility of RNAa-mediated OCT4 activation in primary cells, we designed a series of dsRNAs to target the 1 kb upstream region of the human OCT4A gene, the only OCT4 variant with stemness properties [21,26] (Fig. 1A, B). For the initial screening, 12 candidate dsRNAs targeting the OCT4A promoter at sites ranging from −976 to −141 relative to the transcription start site (TSS) (Fig. 1B and Supplementary Table S1) were designed according to rules previously described [5]. A repeat region in the proximal promoter was avoided (Fig. 1A, B). These saRNA candidates were chemically synthesized and individually transfected into human ADSCs, which express a low level of endogenous OCT4 compared to human embryonic stem cells ([27] and Supplementary Fig. S1). Compared to mock treatment, one of the candidates, dsOCT4-622 which targeted the −622 location, significantly induced OCT4 mRNA expression (2.1-fold, P<0.05) as assessed at 72 h following transfection. A control dsRNA (dsControl) or other candidates had negligible effects on OCT4 levels (Fig. 1C). We then performed a second round of focused screening by manually designing 15 additional dsRNAs, which covered a 66-bp region surrounding the dsOCT4-622 target (−639 to −573) (Supplementary Table S1). Of the 15 candidates, four dsRNAs (dsOCT4-598, dsOCT4-616, dsOCT4-625, and dsOCT4-626) induced OCT4 mRNA expression by at least 1.8-fold, whereas two dsRNAs (dsOCT4-573 and dsOCT4-636) reduced OCT4 mRNA level by 13–18% relative to mock (Fig. 1B, D). Among all the candidates tested, dsOCT4-622 is the most potent activator of OCT4 in human ADSCs and was chosen to be used in all subsequent experiments.
FIG. 1.
Design and screening of OCT4 small activating RNAs (saRNAs) in human adipose-derived stem cells (ADSCs). (A) The genomic locus of human OCT4 and OCT4 alternatively spliced variants are shown. Two major OCT4 variants OCT4A and OCT4B as annotated by the UCSC Genome Browser are indicated. (B) The location of dsRNAs targeting OCT4A promoter. The long solid black line in the middle denotes a 1-kb OCT4A promoter sequence. Short bars (black and gray) above are the saRNA candidates with their target location indicated by the number to their right. Gray bars are candidates with RNAa activity. Shown below are selected genomic annotation tracks, including Repeatmasker, DNase clusters, and three types of histone marks. (C, D) Quantitative reverse transcription–polymerase chain reaction (qRT-PCR) results of the initial (C) and focused (D) dsRNA screening. ADSCs lot no. 38 were transfected with 50 nM of the indicated OCT4 dsRNAs or dsControl (scrambled dsRNA control) for 72 h. Mock samples were transfected in the absence of dsRNA. Results are shown as log twofold change relative to mock. (E) ADSCs (lot no. 38) were transfected with increasing concentrations of dsOCT4-622 for 72 h. Maximum and minimum levels of induction corresponded to 100% and 0% response, respectively. Nonlinear regression analysis was utilized to generate best fit curves. The concentrations of dsOCT4-622 (nM) are shown in log scale. Dotted lines illustrate approximate half maximal effective concentration value. Total OCT4 mRNA expression was analyzed by qRT-PCR. Expression values were normalized to GAPDH levels [mean±standard deviation (SD) of two independent experiments]. *P<0.05; **P<0.01; ***P<0.001.
RNAa is known to be sensitive to target location at promoters [3,4,9,17]. After two rounds of screening 27 saRNA candidates, we were able to identify a hot spot on the OCT4 promoter (−628 to −598 relative to OCT4A TSS), where multiple saRNAs can be identified. Such positional effects could be attributed to target accessibility determined by the local chromatin structure and the presence or absence of nucleosomes [28]. Interestingly, this hot spot happens to overlap a DNase hypersensitive region, which is annotated based on the ENCODE DNase I hypersensitivity data from 125 cell types (Fig. 1B). Perhaps, this region represents a nucleosome-free sequence with its DNA readily accessible for targeting. Nevertheless, target accessibility is not the only determinant for the RNAa activity. Other factors such as sequence context and thermodynamic properties of corresponding dsRNAs could also play important roles since shifting even one base pair from a saRNA site could lead to an unfunctional saRNA or even result in gene silencing instead. Further research of high-resolution screening may help unveil additional rules for optimal saRNA design.
To characterize dsOCT4-622 potency with respect to gene activation, we determined its half maximal effective concentration (EC50) value by measuring OCT4 expression levels in ADSCs treated with 1–200 nM dsOCT4-622 for 72 h. As shown in Fig. 1E, OCT4 expression was induced in a dose-dependent manner by dsOCT4-622 at concentrations ranging from 1 to 50 nM, while concentrations higher than 50 nM did not further enhance OCT4 expression. The estimated EC50 of dsOCT4-622 was ∼7.9 nM in ADSCs (Fig. 1E).
Epigenetic modifiers enhance OCT4 RNAa
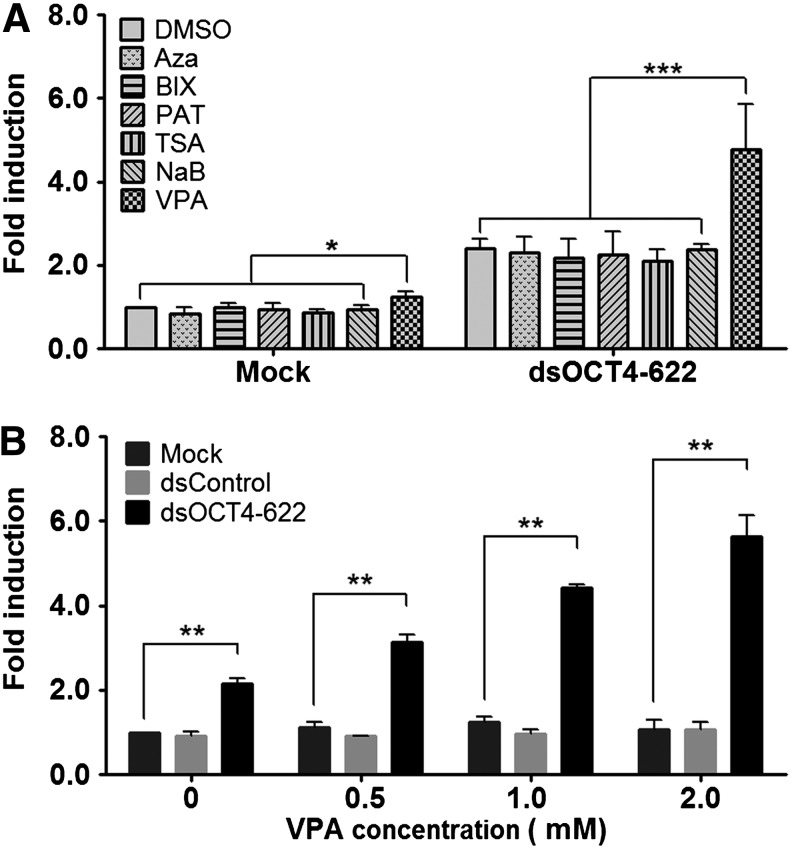
It is known that epigenetic mechanisms play critical roles in cell fate decision and epigenetic modifiers have been employed to enhance reprogramming efficiency or increase lineage plasticity of somatic cells and adult stem cells by reducing epigenetic barriers [29,30]. Epigenetic mechanisms also participate in gene activation induced by saRNA during RNAa [31]. We have previously shown that 5-aza-2′-deoxycytidine, a DNA methyltransferase (DNMT) inhibitor, can sensitize methylation-silenced promoters to RNAa [3]. To determine if epigenetic modifiers could facilitate OCT4 activation, human ADSCs (lot no. 38) were transfected with dsOCT4-622 in the presence of one of the following small molecules known to enhance iPS cell induction: DNMT inhibitor (5-Aza), histone methyltransferase inhibitor (BIX), histone demethylase inhibitor (PAT), and histone deacetylase inhibitors (TSA, VPA, and NaB) (Fig. 2A). Of these compounds, only VPA caused a modest but statistically significant OCT4 induction when used alone. In combination with 5-Aza, BIX, PAT, TSA, or NaB, the induction level of OCT4 mRNA by dsOCT4-622 was comparable to that of combined with vehicle (Fig. 2A), suggesting that these compounds have no apparent effect on OCT4 expression nor on dsOCT4-622 activity. However, in the presence of 1 mM VPA, dsOCT4-622 caused a 4.8-fold induction of OCT4 (Fig. 2A) (P<0.001), a twofold increase in activity compared to dsOCT4-622 alone (Fig. 1C), suggesting that VPA can sensitize the OCT4 gene to dsOCT4-622-mediated activation. We also determined the dose-dependent effect of VPA on the activity of dsOCT4-622 and found that in the presence of increasing VPA concentrations, dsOCT4-622 could further increase OCT4 expression (Fig. 2B). However, a higher concentration (2 mM) of VPA caused considerable cytotoxicity, thus, 1 mM VPA was selected as the appropriate concentration for subsequent experiments. This concentration of VPA possessed negligible cytotoxicity for ADSCs (data not shown) and has been used as a supplement in iPS cell derivation culture medium [14,29].
FIG. 2.
Epigenetic modifiers enhance OCT4 activation by saRNAs. (A) Chemical compound screening. ADSCs were mock transfected or transfected with 50 nM dsOCT4-622 for 96 h in the presence of the indicated compounds 1 μM Aza, 1 μM BIX, 1 μM PAT, 20 nM trichostatin A (TSA), 0.25 mM sodium butyrate (NaB) and 1 mM valproic acid (VPA). Dimethyl sulfoxide (DMSO) served as a vehicle control. Total OCT4 mRNA expression was analyzed by qRT-PCR. (B) VPA facilitates dsOCT4-622-mediated OCT4 activation. Human ADSCs were transfected with the indicated saRNAs as in (A) in the presence of VPA at the indicated concentrations. Total OCT4 mRNA expression was analyzed by qRT-PCR. Expression values were normalized to GAPDH levels (mean±SD of two independent experiments). *P<0.05; **P<0.01; ***P<0.001.
Upregulation of OCT4 expression in human mesenchymal stem by RNAa
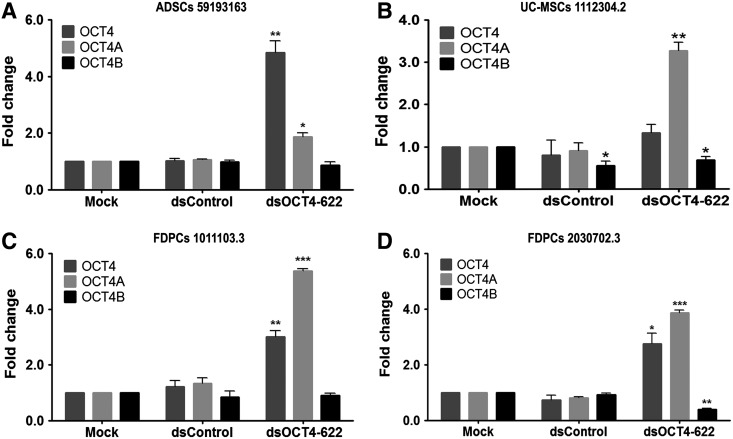
It has been previously reported that RNAa of certain genes could be cell-type specific, depending on the basal level of target gene expression and genomic context of the target promoter [3,32]. To determine if OCT4 expression was susceptible to dsOCT4-622 in ADSCs from different individuals, we treated ADSCs from two additional donors (7F4089 and 59193163) with dsOCT4-622+VPA and assessed the total OCT4 mRNA levels. Consistent with the screening results from ADSCs (lot no. 38), dsOCT4-622 was also able to upregulate OCT4 mRNA expression in these donor ADSCs, with an induction level of ∼4.5-fold (P<0.001) and ∼4.8-fold (P<0.01) in ADSCs 7F4089 and ADSCs 59193163, respectively (Figs. 3A and 4A). OCT4 transcript variants can be generated through alternative promoter usage. We further evaluated the two major transcript variants of OCT4 (ie, OCT4A and OCT4B) by quantitative reverse transcription–polymerase chain reaction (qRT-PCR) using variant-specific TaqMan primers. As shown in Fig. 3A and 4A, dsOCT4-622 activated OCT4A by 2- to 2.9-fold (P<0.01) in ADSCs, while OCT4B was only slightly increased by ∼15% compared to mock. In addition to ADSCs, we also tested dsOCT4-622 in other human MSCs derived from FDPCs or UC-MSCs, which express low levels of endogenous OCT4 (Supplementary Fig. S1). Similar with ADSCs, FDPCs and UC-MSCs were also responsive to RNAa-mediated gene activation of OCT4 (Fig. 3), with moderately inhibited cell proliferation (Supplementary Fig. S2). Interestingly, OCT4A was activated by dsOCT4-622 to a greater extent in FDPCs of different donors (3.9- to 5.4-fold) and UC-MSCs (3.3-fold) than in ADSCs (1.9- to ∼2.9-fold) (Fig. 3). Our results indicate that RNAa-mediated OCT4 activation works consistently in a panel of human MSCs that we tested. Other groups have also shown that promoter-targeted saRNAs could consistently induce endogenous KLF4 and c-MYC expressions in human MSCs derived from bone marrow [33]. Taken together, these observations suggest that saRNA-mediated gene activation could effectively upregulate endogenous pluripotent factors in human MSCs.
FIG. 3.
Induction of OCT4 expression by dsOCT4-622 in additional human mesenchymal stem cells (MSCs). (A–D) Cells were mock transfected or transfected with 50 nM of the indicated saRNAs in the presence of 1 mM VPA for 96 h. Total OCT4, OCT4A, and OCT4B mRNA expression was assessed by qRT-PCR using TaqMan primers. Expression values were normalized to GAPDH levels. *P<0.05; **P<0.01; ***P<0.001. ADSCs 59193163 (A), umbilical cord (UC)-MSCs 1112304.2 (B), follicle dermal papilla cells (FDPCs) 1011103.3 (C), FDPCs 2030702.3 (D). n=2.
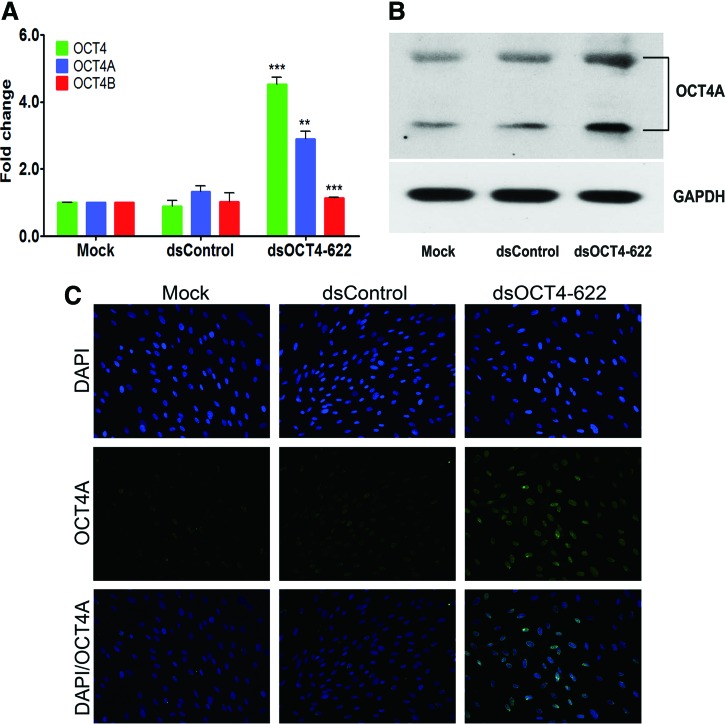
FIG. 4.
RNAa-mediated OCT4 activation in ADSCs. ADSCs 7F4089 were transfected with 50 nM dsOCT4-622 in the presence of 1 mM VPA for 96 h. (A) Total OCT4, OCT4A, and OCT4B mRNA expression was assessed by qRT-PCR using variant-specific TaqMan primers. Expression values were normalized to GAPDH levels. **P<0.01; ***P<0.001. n=3. (B) OCT4 protein levels in the transfected cells were determined by immunoblot analysis with anti-OCT4 (C-10), a monoclonal antibody specific for OCT4A isoform. GAPDH served as a loading control. (C) Immunofluorescence analysis of OCT4 expression with the anti-OCT4A antibody (C-10). Top panel: 4′,6-diamidino-2-phenylindole (DAPI) nuclear staining (blue); middle panel: OCT4A staining (green); bottom panel: merged images of DAPI and OCT4A staining. Fluorescent images were obtained using a Nikon E600 microscope equipped with a digital camera (200×). Color images available online at www.liebertpub.com/scd
Recent reports have demonstrated that the human OCT4 gene can generate multiple transcripts and pseudogenes [21,22,26], causing potential artifacts in RT-PCR. Immunoblot and immunocytochemical analyses were performed to confirm RNAa-mediated OCT4 activation by using OCT4A variant-specific antibodies [34]. As shown in Fig. 4B, immunoblot analysis revealed that the OCT4A protein level was significantly induced by dsOCT4-622 in ADSCs, which is consistent with the qRT-PCR results. Immunocytochemical analysis showed a modest staining of OCT4A in a subpopulation of ADSCs transfected with either mock or dsControl (Fig. 4C). In contrast, a remarkable increase in the fluorescence signal of OCT4A protein was detected in nearly all ADSCs transfected with dsOCT4-622 (Fig. 4C). It is worth noting that the fluorescence signal of OCT4A in ADSCs exhibited a nuclear pattern as evident from its colocalization with DAPI (Fig. 4C), which is consistent with the functional role of OCT4A as a transcription factor. These results suggest that RNAa was able to induce endogenous OCT4A protein overexpression while retaining a proper subcellular localization.
Replacing OCT4 virus by RNAa in cell reprogramming
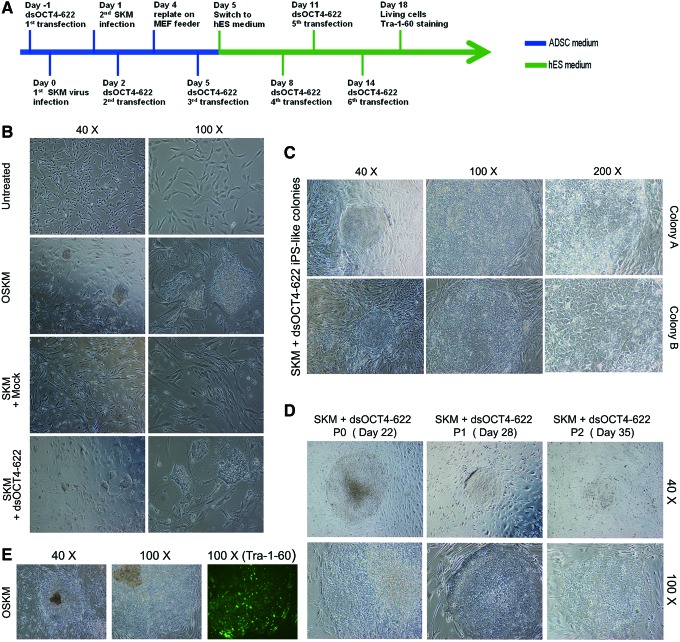
To assess whether the identified OCT4 saRNA could be used to manipulate cell fate, we replaced the OCT4 virus in OSKM (ie, OCT4, SOX2, KLF4, and MYC)-mediated iPS derivation with dsOCT4-622 (SKM+dsOCT4-622) and examined whether SKM+dsOCT4-622 could still induce iPS cells. We transfected ADSCs with dsOCT4-622 before the transduction of SKM virus and repeated the transfection of dsOCT4-622 for five more times (Fig. 5A). As expected, iPS-like colonies started to appear in standard four-factor (OSKM) transduced cells from day 9 to 10. Around the same time, similar iPS-like colonies also appeared in cells treated with SKM+dsOCT4-622, although fewer in numbers compared to OSKM treatment. However, SKM in combination with control treatments (mock or dsControl) did not give rise to iPS-like colonies (Fig. 5B). The resulted iPS colonies (OSKM or SKM+dsOCT4-622) exhibited typical iPS colony morphology characteristic of tightly packed cells with clearly defined borders. Cells in the colonies had a high nuclear-to-cytoplasm ratio with enlarged and prominent nucleoli (Fig. 5C). Since ADSCs express high levels of the endogenous alkaline phosphatase, Tra-1-60 has been used to isolate ADSC-derived iPS-like colonies [27]. Immunocytochemical analysis conducted at day 16 indicated that the iPS-like colonies generated by OSKM transduction expressed stem cell surface marker Tra-1-60, whereas SKM+dsOCT4-622 colonies did not (Fig. 5E). Although iPS-like colonies induced by dsOCT4-622 (Tra-1-60 negative) were able to expand on MEF feeders with iPS-like morphology for 2–3 passages (6 days per passage), they eventually lost iPS-like morphology and became differentiated after 3 weeks of subculture (Fig. 5D). Our observation was consistent with previous reports that some partially reprogrammed human iPS-like colonies could even be expanded/maintained with the flattened compact morphology typical of human ESC-like iPSC colonies for more than 10 passages, without expressing human ESC-specific cell surface antigens, such as Tra-1-60 [35]. Our results suggest that replacing OCT4 virus in the OSKM recipe could partially dedifferentiate somatic cells to a partially reprogrammed stage.
FIG. 5.
Replacing OCT4 virus with OCT4 saRNA leads to the derivation of induced pluripotent stem (iPS)-like cells. (A) Schematic representation of iPS cell induction protocol in ADSCs. Detailed protocol is described in section “Materials and Methods”. (B) Representative phase-contrast images taken on day 12 show typical iPS-like morphology of colonies in OSKM treatment and SKM+dsOCT4-622 treatment (40× and 100×). (C) Representative phase-contrast images of two colonies (SKM+dsOCT4-622) at three different magnifications (40×, 100×, and 200×) were taken on day 16. (D) iPS-like colonies (P0) were mechanically picked up from the original wells and expanded on fresh mouse embryonic fibroblast feeders. Phase-contrast images of a representative iPS-like colony taken on day 22, 28, and 35 (40×and 100×). (E) Representative phase-contrast images (40×and 100×, left and middle panels) and Tra-1-60 immunofluorescence image (100×, right panel) of OSKM colonies were taken on day 16. Color images available online at www.liebertpub.com/scd
Taken together, our study identified a potent OCT4 saRNA, dsOCT4-622, which can induce endogenous OCT4 expression in different types of MSCs. We showed that the use of the OCT4 saRNA in combination with the remaining Yamanaka factors through virus-mediated overexpression led to the derivation of iPS-like cells with partial pluripotency. Of interest, recent studies have shown that ectopic overexpression of OCT4 alone with the use of lineage-specific culture conditions could directly convert fibroblasts to cardiomyocytes [36], blood progenitors [37], and neural stem cells [38]. With improvement in target selection and saRNA design, RNAa-mediated endogenous gene activation may offer an alternative approach to converting cell fate.
Supplementary Material
Acknowledgments
This work was supported by the California Institute for Regenerative Medicine (RL1-00660-1 to L.-C.L.) and the National Institutes of Health (1R01GM090293-0109 to L.-C.L).
Author Disclosure Statement
The authors have no competing financial interests to declare.
References
- 1.Turner MJ, Jiao AL. and Slack FJ. (2014). Autoregulation of lin-4 microRNA transcription by RNA activation (RNAa) in C. elegans. Cell Cycle 13:772–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seth M, Shirayama M, Gu W, Ishidate T, Conte D, Jr., and Mello CC. (2013). The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev Cell 27:656–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H. and Dahiya R. (2006). Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci U S A 103:17337–17342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE. and Corey DR. (2007). Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol 3:166–173 [DOI] [PubMed] [Google Scholar]
- 5.Huang V, Qin Y, Wang J, Wang X, Place RF, Lin G, Lue TF. and Li LC. (2010). RNAa is conserved in mammalian cells. PLoS One 5:e8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang V, Place RF, Portnoy V, Wang J, Qi Z, Jia Z, Yu A, Shuman M, Yu J. and Li LC. (2012). Upregulation of cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res 40:1695–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Place RF, Li LC, Pookot D, Noonan EJ. and Dahiya R. (2008). MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A 105:1608–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dharap A, Pokrzywa C, Murali S, Pandi G. and Vemuganti R. (2013). MicroRNA miR-324-3p induces promoter-mediated expression of RelA gene. PLoS One 8:e79467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Place RF, Wang J, Noonan EJ, Meyers R, Manoharan M, Charisse K, Duncan R, Huang V, Wang X. and Li LC. (2012). Formulation of small activating RNA into lipidoid nanoparticles inhibits xenograft prostate tumor growth by inducing p21 expression. Mol Ther Nucleic Acids 1:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren S, Kang MR, Wang J, Huang V, Place RF, Sun Y. and Li LC. (2013). Targeted induction of endogenous NKX3-1 by small activating RNA inhibits prostate tumor growth. Prostate 73:1591–1601 [DOI] [PubMed] [Google Scholar]
- 11.Takahashi K. and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676 [DOI] [PubMed] [Google Scholar]
- 12.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K. and Yamanaka S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872 [DOI] [PubMed] [Google Scholar]
- 13.Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, et al. (2013). Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science 341:651–654 [DOI] [PubMed] [Google Scholar]
- 14.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, et al. (2010). Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7:618–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, et al. (2011). Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 8:633–638 [DOI] [PubMed] [Google Scholar]
- 16.Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, Bien G, Yao S, et al. (2009). Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell 4:381–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Wang J, Huang V, Place RF. and Li LC. (2012). Induction of NANOG expression by targeting promoter sequence with small activating RNA antagonizes retinoic acid-induced differentiation. Biochem J 443:821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim JB, Sebastiano V, Wu G, Arauzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, et al. (2009). Oct4-induced pluripotency in adult neural stem cells. Cell 136:411–419 [DOI] [PubMed] [Google Scholar]
- 19.Tsai SY, Bouwman BA, Ang YS, Kim SJ, Lee DF, Lemischka IR. and Rendl M. (2011). Single transcription factor reprogramming of hair follicle dermal papilla cells to induced pluripotent stem cells. Stem Cells 29:964–971 [DOI] [PubMed] [Google Scholar]
- 20.Pardo M, Lang B, Yu L, Prosser H, Bradley A, Babu MM. and Choudhary J. (2010). An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell 6:382–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X. and Dai J. (2010). Concise review: isoforms of OCT4 contribute to the confusing diversity in stem cell biology. Stem Cells 28:885–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hawkins PG. and Morris KV. (2010). Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcription 1:165–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li W, Tian E, Chen ZX, Sun G, Ye P, Yang S, Lu D, Xie J, Ho TV, et al. (2012). Identification of Oct4-activating compounds that enhance reprogramming efficiency. Proc Natl Acad Sci U S A 109:20853–20858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ning H, Liu G, Lin G, Garcia M, Li LC, Lue TF. and Lin CS. (2009). Identification of an aberrant cell line among human adipose tissue-derived stem cell isolates. Differentiation 77:172–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye L, Chang JC, Lin C, Sun X, Yu J. and Kan YW. (2009). Induced pluripotent stem cells offer new approach to therapy in thalassemia and sickle cell anemia and option in prenatal diagnosis in genetic diseases. Proc Natl Acad Sci U S A 106:9826–9830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liedtke S, Enczmann J, Waclawczyk S, Wernet P. and Kogler G. (2007). Oct4 and its pseudogenes confuse stem cell research. Cell Stem Cell 1:364–366 [DOI] [PubMed] [Google Scholar]
- 27.Sun N, Panetta NJ, Gupta DM, Wilson KD, Lee A, Jia F, Hu S, Cherry AM, Robbins RC, Longaker MT. and Wu JC. (2009). Feeder-free derivation of induced pluripotent stem cells from adult human adipose stem cells. Proc Natl Acad Sci U S A 106:15720–15725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Portnoy V, Huang V, Place RF. and Li LC. (2011). Small RNA and transcriptional upregulation. Wiley Interdiscip Rev RNA 2:748–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W. and Melton DA. (2008). Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol 26:1269–1275 [DOI] [PubMed] [Google Scholar]
- 30.Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K. and Ding S. (2010). Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell 7:651–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li LC. (2014). Chromatin remodeling by the small RNA machinery in mammalian cells. Epigenetics 9:45–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR. and Janowski BA. (2008). Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol 15:842–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voutila J, Saetrom P, Mintz P, Sun G, Alluin J, Rossi JJ, Habib NA. and Kasahara N. (2012). Gene expression profile changes after short-activating RNA-mediated induction of endogenous pluripotency factors in human mesenchymal stem cells. Mol Ther Nucleic Acids 1:e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marti M, Mulero L, Pardo C, Morera C, Carrio M, Laricchia-Robbio L, Esteban CR. and Izpisua Belmonte JC. (2013). Characterization of pluripotent stem cells. Nat Protoc 8:223–253 [DOI] [PubMed] [Google Scholar]
- 35.Yu J, Chau KF, Vodyanik MA, Jiang J. and Jiang Y. (2011). Efficient feeder-free episomal reprogramming with small molecules. PLoS One 6:e17557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Cao N, Spencer CI, Nie B, Ma T, Xu T, Zhang Y, Wang X, Srivastava D. and Ding S. (2014). Small molecules enable cardiac reprogramming of mouse fibroblasts with a single factor, Oct4. Cell Rep 6:951–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szabo E, Rampalli S, Risueno RM, Schnerch A, Mitchell R, Fiebig-Comyn A, Levadoux-Martin M. and Bhatia M. (2010). Direct conversion of human fibroblasts to multilineage blood progenitors. Nature 468:521–526 [DOI] [PubMed] [Google Scholar]
- 38.Zhu S, Ambasudhan R, Sun W, Kim HJ, Talantova M, Wang X, Zhang M, Zhang Y, Laurent T, et al. (2014). Small molecules enable OCT4-mediated direct reprogramming into expandable human neural stem cells. Cell Res 24:126–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.