Abstract
DNA glycosylases perform a genome-wide search to locate damaged nucleotides among a great excess of undamaged nucleotides. Many glycosylases are capable of facilitated diffusion, whereby multiple sites along the DNA are sampled during a single binding encounter. Electrostatic interactions between positively charged amino acids and the negatively charged phosphate backbone are crucial for facilitated diffusion, but the extent to which diffusing proteins rely on the double-helical structure DNA is not known. Kinetic assays were used to probe the DNA searching mechanism of human alkyladenine DNA glycosylase (AAG) and to test the extent to which diffusion requires B-form duplex DNA. Although AAG excises εA lesions from single-stranded DNA, it is not processive on single-stranded DNA because dissociation is faster than N-glycosidic bond cleavage. However, the AAG complex with single-stranded DNA is sufficiently stable to allow for DNA annealing when a complementary strand is added. This observation provides evidence of nonspecific association of AAG with single-stranded DNA. Single-strand gaps, bubbles, and bent structures do not impede the search by AAG. Instead, these flexible or bent structures lead to the capture of a nearby site of damage that is more efficient than that of a continuous B-form duplex. The ability of AAG to negotiate these helix discontinuities is inconsistent with a sliding mode of diffusion but can be readily explained by a hopping mode that involves microscopic dissociation and reassociation. These experiments provide evidence of relatively long-range hops that allow a searching protein to navigate around DNA binding proteins that would serve as obstacles to a sliding protein.
Human alkyladenine DNA glycosylase (AAG) is a 36 kDa monomeric enzyme that recognizes a wide variety of alkylated and deaminated purines.1−5 One of the most efficiently recognized lesions is 1,N6-ethenoadenine (εA), which is formed under conditions of lipid peroxidation or upon exposure to vinyl chlorides.6 Crystal structures of AAG bound to εA-DNA reveal that the specific recognition of εA requires nucleotide flipping.7 AAG binds and flips out εA very rapidly, and the specific recognition complex is extremely stable.8 Although the rate of N-glycosidic bond hydrolysis is relatively slow, almost every specific binding event results in excision.8,9 These properties make εA an excellent substrate for characterizing the searching mechanism of AAG.
Previous work showed that AAG uses electrostatic interactions to diffuse along DNA and search for sites of damage.9,10 The searching of many different sites in a single binding encounter is commonly termed facilitated diffusion and can involve sliding, whereby the protein maintains continuous contact with the DNA backbone, and hopping, which involves microscopic dissociation and reassociation.11,12 Many different DNA binding proteins, including several DNA glycosylases, have been found to use combinations of these modes. Although sliding could be more efficient for a local search of DNA, because every site is sampled, this mode of diffusion has two major limitations. First, a sliding monomeric protein could sample only one member of a base pair, and damaged nucleotides on the opposing strand would not be sampled. Second, tightly bound proteins would block sliding and effectively trap a searching protein, slowing the search for damaged sites. Nevertheless, sliding has been invoked for several different proteins on the basis of a variety of different observations.13−20 The previous work with AAG has shown that it searches both strands of the duplex and is able to bypass a bound protein, providing strong evidence of a hopping mode.9 However, these observations do not address whether AAG is capable of short-range sliding. Extensive studies of uracil DNA glycosylase have favored a model of rapid short-range sliding, accompanied by more infrequent hops.13,14,21,22
We probed the DNA structural requirements for DNA searching by AAG, testing whether AAG is processive on single-stranded DNA (ssDNA) and whether helix discontinuities act as a barrier to diffusion. The results from multiple-turnover processivity assays establish that facilitated diffusion is not hindered by encounter with single-stranded gaps, bubbles, or bulges (kinks) in the DNA. AAG associates with ssDNA, and single-stranded segments appear to decrease the rate of macroscopic dissociation. However, AAG is not processive on ssDNA because of the low efficiency of excision. These observations suggest that AAG does not maintain continuous contact with the DNA while diffusing along it but rather makes frequent microscopic hops.
Materials and Methods
Proteins and Oligonucleotides
Full-length and truncated (Δ80) human AAG were purified, and the concentration of active glycosylase was determined by burst analysis as previously described.10 Oligonucleotides were synthesized by Integrated DNA Technologies or the Keck Center at Yale University and purified by denaturing PAGE as previously described.10 The concentration of ssDNA was determined from the absorbance at 260 nm using the calculated extinction coefficients. For the 5′ or dual fluorescein-labeled DNA, we determined the labeling efficiency by comparing the absorbance at 260 nm with that at 495 nm, and it was greater than 90% in all cases. Unless otherwise stated, the duplex substrates were obtained by annealing a labeled εA-containing strand to a 2-fold excess of the unlabeled complement strand in annealing buffer [10 mM NaMES (pH 6.5) and 50 mM NaCl] by heating the DNA to 95 °C for 5 min and then slowly cooling it to room temperature.
Glycosylase Activity Assays
Reactions were conducted at 37 °C in 50 mM NaMES (pH 6.1), 1 mM EDTA, 1 mM DTT, 10% (v/v) glycerol, and 0.1 mg/mL BSA, and the ionic strength was adjusted with NaCl to obtain the desired concentration of sodium ions. Reactions were initiated by the addition of enzyme to obtain a reaction volume of 50–100 μL that typically contained 200 nM fluorescein-labeled DNA. Aliquots were withdrawn at various times and reactions quenched with NaOH (final concentration of 0.2 M). Samples were heated at 70 °C for 15 min; formamide was added to a final concentration of 65%, and the DNA fragments were resolved on 14% (w/v) polyacrylamide gels containing 8 M urea. Gels were scanned with a Typhoon Trio+ fluorescence imager (GE Healthcare) to detect fluorescein (excitation at 532 nm and emission with a 526SP filter). The resulting fluorescence signal was quantified with ImageQuant TL and corrected for background signal. The intensity of each DNA band was converted into a fraction of the total DNA by dividing its intensity by the sum of the intensities for all of the DNA species present.
Multiple-Turnover Kinetics
The multiple-turnover kinetics with the processivity substrates were measured with a 100-fold excess of substrate (200 nM) over enzyme (2 nM). The initial rates were measured from the first 10% of the reaction and were linear in all cases. The velocity was determined from the depletion of substrate, and the fraction processive (Fp) was determined from the formation of products (Vp) and intermediates (Vi) that still contained an εA lesion according to eq 1.10,23 Approximately 2% of εA nucleotides are in a ring-opened form that is not a substrate for AAG, and therefore, the two-lesion substrate has ∼4% of the molecules with one ring-opened site. This leads to a maximal observed processivity value of 0.92.10 With a 100-fold excess of DNA over enzyme, the minimal processivity value that can be observed is 0.05 for a completely distributive mechanism due to stochastic rebinding of AAG. This value of 0.05 was estimated from the effect of having 5% intermediate formed via a purely distributive mechanism (the midpoint of the initial 10% reaction that was used for measurement of initial rates), and the non-negligible probability that AAG might encounter a previously released intermediate. The presence of the intermediate does not change the velocity for product formation, because product is formed from either substrate or intermediate, but the velocity for intermediate formation must be adjusted to reflect the fraction of binding events in which the enzyme rebinds to an intermediate and converts it into a product (Vi,corrected = 0.95Vp – 0.05Vp = 0.9Vp). Putting this corrected velocity for intermediate formation into eq 1 gives a value of 0.05 for a purely distributive mechanism [Fp = (Vp – 0.9Vp)/(Vp + 0.9Vp) = 0.052]. Therefore, the effective range of experimental Fp values is 0.87 (0.92–0.05). To facilitate comparison with previously published results, we have corrected all of the observed processivity values according to eq 2.9
| 1 |
| 2 |
Single-Turnover Kinetics
Single-turnover kinetic analysis for excision of εA from either single- or double-stranded oligonucleotides containing a single lesion was measured as previously described.10 The concentration of AAG was varied in excess over the oligonucleotide substrates (TEC) (Figure S1 of the Supporting Information) to ensure single-turnover conditions. In all cases, the reactions followed single-exponential kinetics according to eq 3, in which F is the fraction product, A is the amplitude, and k is the observed single-turnover rate constant. At a saturating enzyme concentration, this observed rate constant reaches a maximal rate constant (kmax). We varied the concentration of enzyme by at least 3-fold and found no difference in the observed rate constant, confirming that kmax was measured.
| 3 |
Pulse–Chase Assays To Measure the Efficiency of Excision
The efficiency of AAG-catalyzed excision of εA from ssDNA was determined by using the pulse–chase assays under the standard glycosylase reaction condition at 100 mM NaCl. Typically, 100 nM fluorescein-labeled ssDNA substrate (25-mer TEC) was mixed with 400 nM AAG for 20 s before the addition of chase DNA [1 or 2 μM unlabeled TEC duplex DNA or 5 μM unlabeled TEG duplex DNA (see Figure S1 of the Supporting Information for DNA sequences)]. Pulse–chase assays on dsDNA were performed using slightly decreased substrate and enzyme concentrations, as AAG binds more tightly to dsDNA. Briefly, 50 nM labeled TEC duplex substrate was incubated with 100 nM AAG for 20 s, and then a chase of 5 μM unlabeled TEC DNA was added. Pulse–chase control assays were performed by preincubating substrate and chase DNA before adding enzyme to initiate the reaction. At various time points, a sample was removed from the reaction mixture and analyzed as described above (Glycosylase Activity Assays).
AAG-catalyzed base excision yields labeled product, whereas dissociation of AAG releases unreacted labeled substrate. The data were converted to fraction product [fraction product = product/(product + substrate)] and fit by a single exponential followed by a steady-state phase. After the exponential rate constant (kobs) and burst amplitude (Aobs) values had been obtained from the data fitting, the efficiency of excision was determined as compared to the same experiment without chase (kmax and Amax) by eq 4. The pulse–chase control data were fit by linear regression.
| 4 |
Native Gel Electrophoresis
Annealed oligonucleotides were prepared in standard pH 6.1 glycosylase reaction buffer with an ionic strength of 200 mM adjusted with NaCl. Samples were loaded onto a 10% native polyacrylamide gel (19:1 acrylamide:bisacrylamide with 0.5× TBE buffer) and run at a constant voltage of 10 V/cm. The fluorescein-labeled DNA was visualized as described for denaturing PAGE analysis.
Results
Experimental Approach and Design of Oligonucleotides with Helix Discontinuities
Multiple-turnover processivity assays provide a quantitative assay for the ability of a DNA glycosylase to locate sites of damage on an oligonucleotide substrate.10,14,24 This is a kinetic partitioning assay that operates on the principle that an enzyme bound to its abasic product can either dissociate to leave an oligonucleotide containing one abasic and one lesion site or successfully diffuse to locate the second lesion site on the oligonucleotide and excise it, to create an oligonucleotide containing two abasic sites (Figure 1). Alkaline cleavage of abasic sites and denaturing polyacrylamide gel analysis allow the different products to be quantified to determine the fraction of processive events [Fp = kcapture/(koff + kcapture)]. A highly efficient (processive) search will yield a Fp value approaching 1, whereas a distributive search corresponds to an Fp value of 0 (koff ≫ kcapture). In the work presented here, we have tested helix discontinuities to evaluate whether they serve as barriers to the diffusion of AAG.
Figure 1.
Multiple-turnover processivity assay to quantify facilitated diffusion by AAG. After excising one of the two εA (E) lesions, AAG will either dissociate (koff) or diffuse to find the other lesion site (kcapture). The shaded box denotes the position of alternative DNA structures that were tested as potential blocks of a sliding enzyme.
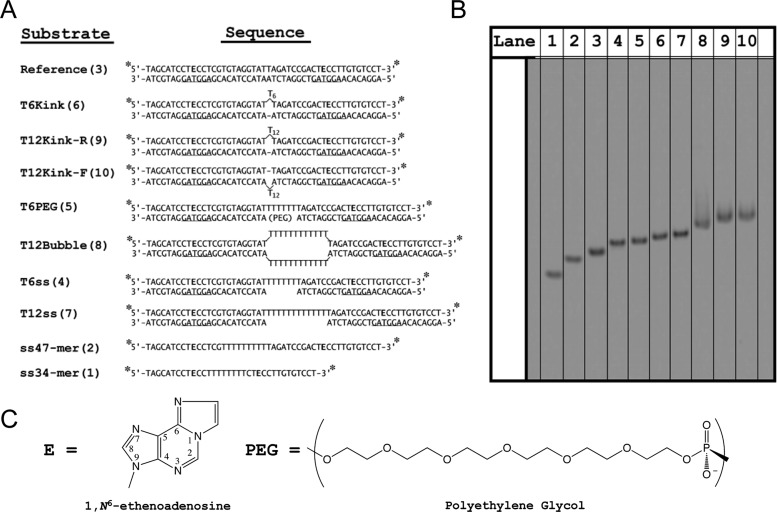
To generate different types of DNA structures, we prepared a series of oligonucleotides that systematically vary from the previously characterized duplex oligonucleotide containing two εA lesions separated by 25 bp, which serves as a reference substrate (Figure 1).10 Helix discontinuities were introduced midway between the two sites, because structural and functional studies suggest that this location is well outside of the DNA binding footprint of AAG when it is bound to either of the lesion sites.7,25 This ensures that the efficiency of excision is the same for all substrates under a given set of conditions, and any changes in processivity can be attributed to the probability that the enzyme locates the second site of damage. To simplify the secondary structure of single-stranded portions, we used runs of either 6 or 12 thymidine nucleotides. Duplexes with kinks in them were generated by inserting the polyT segments onto either the top or bottom strand. Previous characterization of a six-nucleotide bulge showed an ∼90° bend angle (Figure S2 of the Supporting Information).26 The 12-nucleotide bulge is expected to adopt a similar sharply bent structure. Native gel analysis confirms that the T12 bulge migrates more slowly than a T12 gap DNA, consistent with the T12 bulge forming a kinked structure (Figure 2B, lane 7 vs lanes 9 and 10). Bubbles were generated by placing two T12 strands opposite each other or by placing a T6 strand opposite a polyethylene glycol spacer containing an 18-atom linker (Figure 2). Finally, gap molecules were generated in which the lesion-containing strand is continuous but the opposite strand leaves a central single-strand gap of T6 or T12. If sliding contributes to the AAG search, then these discontinuities are expected to cause decreased processivity.
Figure 2.
Oligonucleotide substrates for studying facilitated diffusion. (A) Each substrate contains two εA (E) lesions and is labeled at the 5′ and 3′ ends with fluorescein (asterisk). The number in parentheses after the substrate name refers to the lane number in panel B, native gel electrophoresis of the DNA substrates. (C) Structures of εA and polyethylene glycol (PEG).
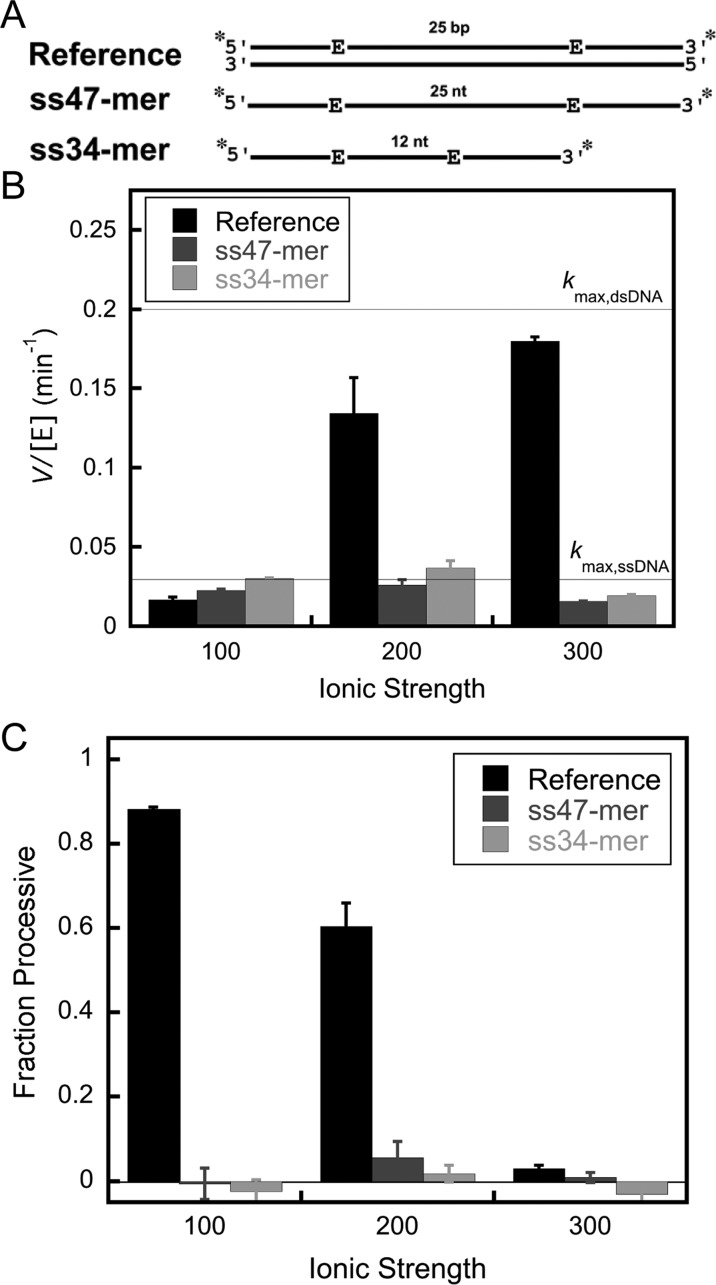
AAG is able to excise εA from ssDNA,4,12 and therefore, it may be able to slide or otherwise diffuse along ssDNA. Our initial experiments with the previously characterized processivity substrate (Figure 2A, reference substrate)10 revealed that the lesion-containing strand forms alternative base-pairing structure when the complement is omitted, and therefore, it is not a suitable substrate for testing the ability of AAG to search ssDNA (data not shown). Therefore, we designed two DNA sequences that were predicted to be strongly single-stranded, and in which two εA lesions are separated by either 25 or 12 nucleotides (Figure 2A, ss47-mer and ss34-mer). Native gel electrophoresis was used to confirm that these oligonucleotides do not form any detectable secondary structure (Figure 2B, lanes 1 and 2).
Bypass of Kinks, Bubbles, and Gaps
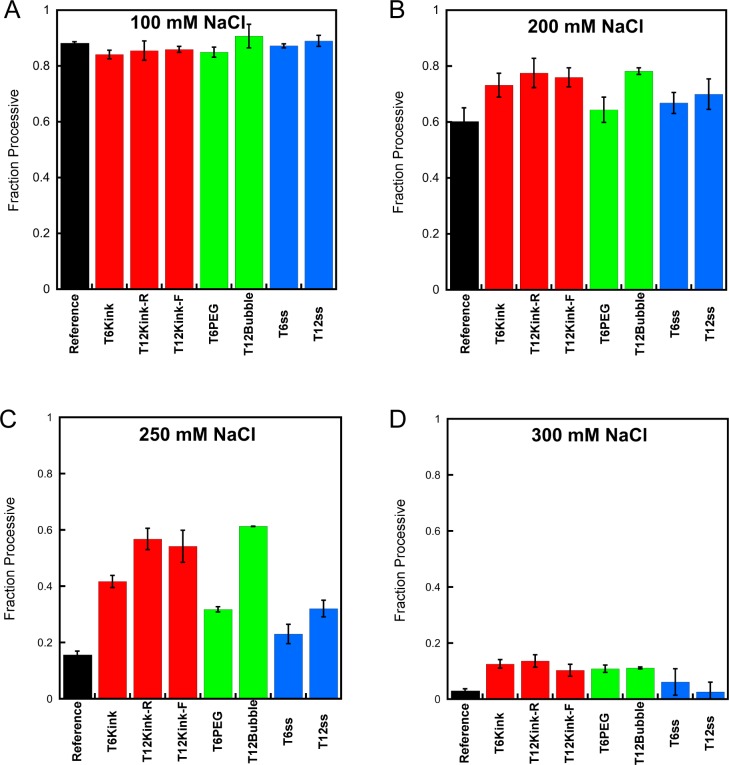
The ability of AAG to perform facilitated diffusion is strongly dependent upon electrostatic interactions, and therefore, we measured the processivity of AAG at several different concentrations of sodium ions. When AAG searches dsDNA, it is highly processive at 100 mM NaCl but becomes fully distributive at 300 mM NaCl.10 Under the conditions of 100 mM NaCl, the kinks, bubbles, and single-strand gaps exhibited processivity almost identical to that of the reference substrate that contained a continuous DNA duplex (Figure 3A; see Table S1 of the Supporting Information for values). This observation suggests that these disruptions in the DNA duplex do not dramatically impede the searching ability of AAG. When the concentration of NaCl was increased to 200 mM, a small increase in the processivity of AAG with the kinked DNA molecules and the longer gap and bubble structures was observed (Figure 3B). This trend was more pronounced with 250 mM NaCl, in which case there is a significant increase in the processivity of AAG with the kinked DNA molecules and the longer gap and bubble substrates, relative to the processivity observed under the same condition for the substrate with a continuous duplex (Figure 3C). At a slightly higher NaCl concentration (300 mM), AAG becomes mostly distributive for all of the substrates that were tested, but there is a small but detectable increase for the kink and bubble substrates even at this high salt concentration (Figure 3D). The helix discontinuities were not found to cause a decrease in the searching efficiency of AAG under any conditions that were tested.
Figure 3.
Effect of helix discontinuities on the searching efficiency of AAG. The processivity of AAG was determined for oligonucleotides containing kinks, bubbles, and single-strand gaps using the multiple-turnover processivity assay with 2 nM AAG and the indicated substrate (200 nM) at 100 (A), 200 (B), 250 (C), and 300 mM NaCl (D). Each value is the mean ± the standard deviation (n ≥ 4).
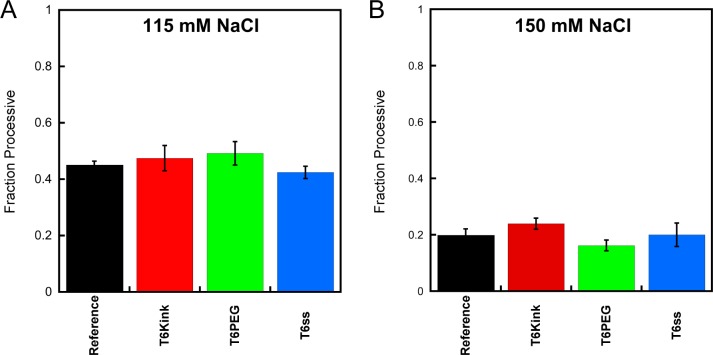
The amino terminus of AAG is poorly conserved, and the human protein has a high density of basic amino acids in this region. The amino terminus is dispensable for catalytic activity, but the truncated protein (Δ80 AAG) has been found to have reduced affinity for DNA and exhibits less processive behavior than the full-length enzyme at a physiological salt concentration.9,10 The full-length AAG has not been crystallized, but it has the same DNA binding footprint as the truncated protein.25 To gain further insight into the contribution from the amino terminus, we measured the fraction processive for Δ80 AAG with a subset of the kink, bubble, and gap substrates. Under the conditions tested, these helix discontinuities had no significant effect on the searching ability of Δ80 AAG (Figure 4; see Table S2 of the Supporting Information for values). These observations confirm that the ability of AAG to diffuse on DNA does not require the amino terminus and that diffusion of the truncated protein is not blocked by these DNA structures. From these data, it is not clear if the amino terminus is responsible for the more efficient searching of duplexes containing discontinuities, because the truncated protein is not processive at the higher salt concentration where this phenomenon is observed.
Figure 4.
Effect of helix discontinuities on the searching efficiency of truncated AAG. The processivity of Δ80 AAG was determined for a subset of oligonucleotide substrates containing a kink, bubble, and single-strand gap using a multiple-turnover processivity assay and compared to the results with a continuous duplex (reference). Data were collected at (A) 115 and (B) 150 mM NaCl with 2 nM Δ80 AAG and the indicated dsDNA substrate (200 nM). Values reflect the mean ± the standard deviation (n ≥ 4).
Glycosylase Activity of AAG on ssDNA
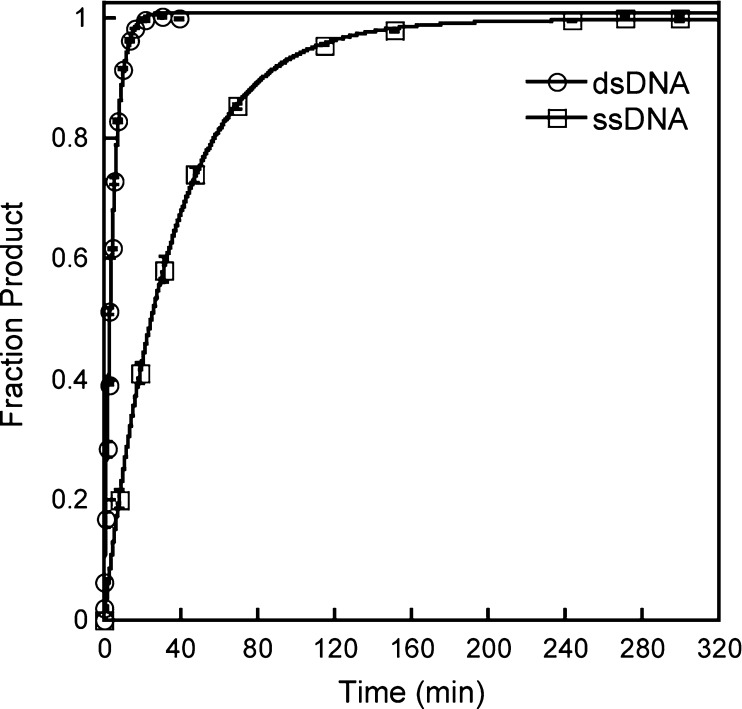
To evaluate if AAG is capable of diffusing on ssDNA, we performed processivity experiments with oligonucleotides that were designed to be single-stranded. As previous experiments with ssDNA have not used the full-length AAG protein, we first compared single-turnover excision with a ssDNA substrate (25-mer) that contained a single εA lesion with that of the dsDNA (εA·T). In all cases, the reactions with ssDNA substrates proceeded to completion and were well fit by a single exponential (Figure 5). The rate of the single-turnover reaction is independent of the concentration of AAG (data not shown). It is almost certain that the measured value of kmax reflects the rate constant for N-glycosidic bond cleavage on ssDNA, and it is approximately 8-fold lower than the rate constant for the comparable reaction on dsDNA. This observation confirms that dsDNA is the preferred substrate for AAG.
Figure 5.
Comparison of single-turnover glycosylase activity of AAG on single- and double-stranded DNA. Single-turnover glycosylase activity of AAG was measured for a 25-mer substrate with 100 nM single-stranded (□) or double-stranded (○) 25-mer oligonucleotide containing a single εA lesion site (εA·T base pair for the duplex). The concentration of AAG was 200 nM for dsDNA and 400 nM for ssDNA. Each data point corresponds to the mean ± the standard deviation of two to four independent reactions. Reaction progress curves were fit to a single exponential to give the observed rate constant. The single-turnover rate constant is independent of the concentration of AAG under these conditions, giving a maximal single-turnover rate constant (kmax) of 0.23 ± 0.01 min–1 for dsDNA and 0.029 ± 0.001 min–1 for ssDNA.
We next measured multiple-turnover glycosylase activity with ssDNA substrates containing two εA lesions (Figure 6A). The initial rate for the disappearance of the substrate is plotted for three different salt concentrations for ssDNA and dsDNA (reference) substrates (Figure 6B). Multiple-turnover glycosylase activity on dsDNA substrates increases with an increasing concentration of NaCl as the rate-limiting step changes from product release at low salt concentrations to the rate of base excision at higher salt concentrations.10 The multiple-turnover rate constant in the presence of 300 mM NaCl is approximately equal to the single-turnover rate constant, indicating that product release is much faster than base excision (Figure 6B). For the ssDNA, the initial rates are insensitive to the concentration of NaCl, suggesting that the base excision step is rate-limiting across the range of conditions that were tested (Figure 6B). Indeed, the multiple-turnover rate constant is almost identical to the maximal single-turnover rate constant that was independently measured for a substrate with a single εA site (Figure 5). AAG shows faster multiple-turnover reactions on single-stranded than on duplex DNA when the salt concentration is low [100 mM NaCl (Figure 6B)]. As discussed above, this reflects a change in the rate-limiting step, with product release limiting the reaction of dsDNA and N-glycosidic bond cleavage limiting the reaction of ssDNA. At higher salt concentrations, the N-glycosidic bond cleavage step is rate-limiting for both contexts and the preference for dsDNA is apparent.
Figure 6.
Multiple-turnover processivity assays to investigate the ability of AAG to search single-stranded DNA. (A) Schematic for the processivity substrates containing terminal fluorescein labels (asterisk) and two εA lesions (see Figure 2 for sequences). (B) Initial rates of substrate disappearance were measured for ssDNA and dsDNA processivity substrates with 2 nM full-length AAG and the indicated DNA (200 nM). (C) Processivity values for ssDNA and dsDNA (reference) were calculated from the initial rates of formation of the products and intermediates as described in Materials and Methods. Values are means ± the standard deviation (n ≥ 4). The action of AAG is distributive on ssDNA under every condition tested.
Although the εA-containing ssDNA is a reasonable substrate for AAG, the Fp values indicate that the enzyme is distributive under all of the conditions tested (Figure 6C). This is true whether the sites are 12 or 25 nucleotides apart. This difference between ssDNA and dsDNA contexts could be explained by differences in searching ability or by the decreased efficiency of excision in the single-strand context. Therefore, it is important to independently test the extent to which AAG is committed to catalysis once it has bound to the εA site.
Pulse–chase assays were used to measure the excision efficiency for εA-DNA in the context of both ssDNA and dsDNA. The experimental design is outlined in Figure 7A. In the pulse–chase experiment, AAG is mixed with labeled substrate to form the specific complex and then is chased with unlabeled dsDNA(εA·T) competitor. Control experiments, in which AAG is added to a mixture of labeled substrate and unlabeled chase, confirmed that the concentration of chase DNA was sufficient to capture all AAG that dissociated. An experiment with dsDNA is shown in Figure 7B. In the presence of the chase, there is a slight decrease in the amplitude and a slight increase in the rate constant. This corresponds to an efficiency of excision of 0.95 under these conditions. Importantly, the control reaction shows no burst, confirming that sufficient chase DNA was used (Figure 7B, blue line). As the chase contains unlabeled substrate that is otherwise identical to the labeled DNA, a slow steady-state rate is observed. When the pulse–chase experiment was repeated with εA-containing ssDNA, a very different result was obtained (Figure 7C). In this case, all of the AAG dissociates prior to excision of εA (red line). We estimate an efficiency of <0.01 for excision of εA from ssDNA based upon these data.
Figure 7.
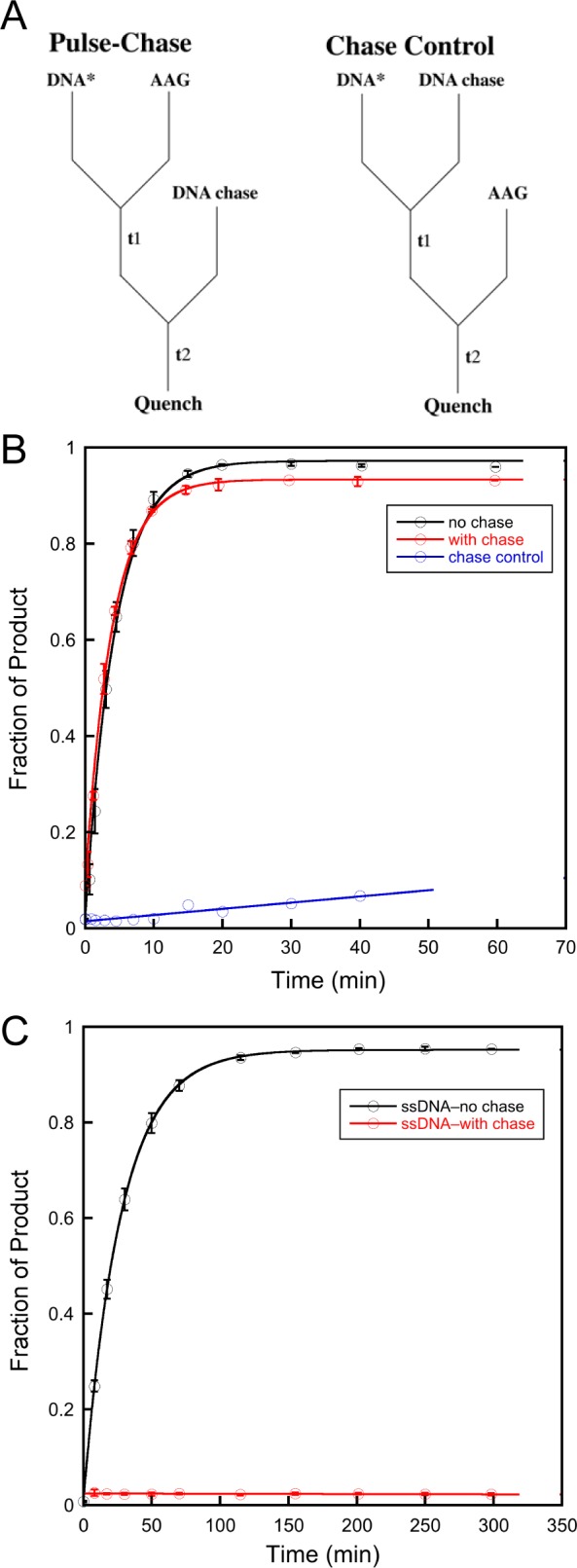
Pulse–chase experiments to measure the efficiency of excision. (A) Experimental design. (B) Pulse–chase to measure the efficiency of excision with dsDNA(εA·T). (C) Pulse–chase to measure efficiency of excision with ssDNA. The standard conditions with 100 mM NaCl were used (see Materials and Methods for details).
Although AAG is not committed to catalysis when it is bound to ssDNA, it clearly can bind to ssDNA and exhibits glycosylase activity. To further explore the lifetime of the AAG·ssDNA complex, we repeated the pulse–chase experiment and added complementary ssDNA along with a duplex chase oligonucleotide (Figure 8A). If the complement strand were able to anneal in the AAG·ssDNA complex to form an AAG·dsDNA complex, then it should be committed to the excision of εA because AAG has a high efficiency of excision of εA from dsDNA. Consistent with this model, we observed a burst of product formation that was approximately 90% of the value observed for AAG bound to dsDNA (Figure 8B, red line). If the order of addition is reversed (chase control), then no burst of product formation is observed, confirming that sufficient chase DNA was used to sequester all of the AAG in the experiment (Figure 8B, blue line). This control shows the expected velocity for multiple-turnover excision of εA from dsDNA, indicating that the labeled DNA annealed during the first mixing step and AAG acts on both labeled and unlabeled dsDNA. Figure 8C illustrates the two pathways, with 10% of the enzyme molecules dissociating from ssDNA and 90% remaining bound during the rapid DNA annealing step.
Figure 8.
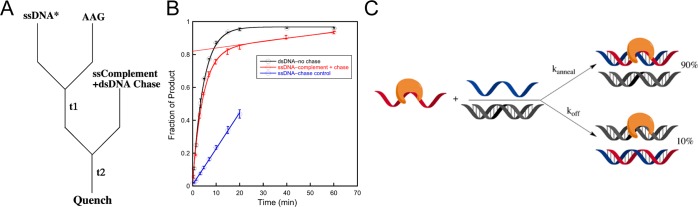
Kinetic experiment to interrogate the lifetime of the complex between AAG and ssDNA. (A) Experimental design. (B) The pulse–chase experiment was performed as depicted in panel A (red), and the line indicates the extrapolated burst amplitude. A control in which the order of addition of the enzyme and chase was switched (blue line) gives the expected steady-state turnover, with no burst. (C) Cartoon illustrating that annealing occurs within the AAG·ssDNA complex and allows for efficient excision of εA.
Discussion
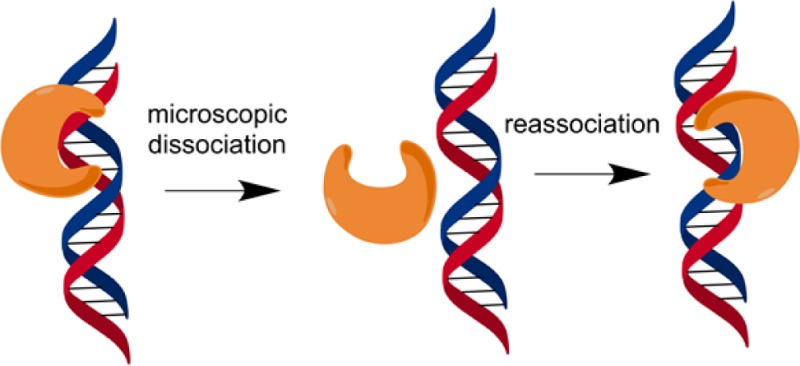
We investigated the extent to which helix discontinuities affect the searching process of AAG. In considering these experiments, we thought it was important that we revisit the activity and searching ability of AAG on ssDNA. Our initial premise was that sliding, which has been invoked for most previously studied DNA binding proteins, would be sensitive to single-stranded or kinked structures. By inserting these structures between two lesion sites, we were able to use multiple-turnover processivity assays to quantify the effect of these structures on DNA searching. In no case do these perturbations in the B-form helix cause a decrease in the searching efficiency of AAG. Unexpectedly, we find a wide range of conditions under which these structures increase the probability that AAG can locate a site of damage that is located nearby on the same DNA segment. These results cannot easily be interpreted in the context of a sliding model but are consistent with a mode of diffusion that is dominated by hopping (Figure 9).
Figure 9.
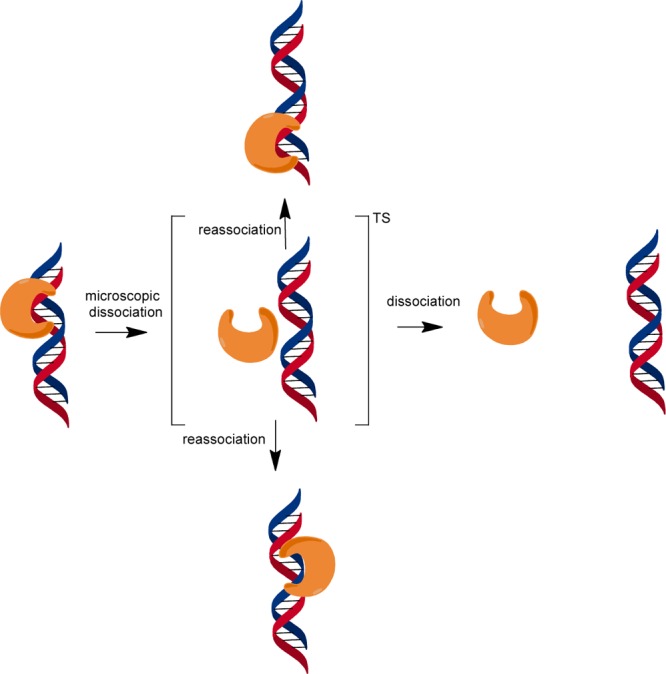
Model for facilitated diffusion by AAG. Diffusion appears to be dominated by microscopic dissociation events in which the protein becomes at least partially solvated but is in the proximity of the DNA and is likely to reassociate. This state would resemble a transition state for dissociation.13,22 At a high salt concentration, this species has a high probability of macroscopic dissociation that would be followed by three-dimensional diffusion. At lower salt concentrations, the vertical pathways dominate with hopping to either the same strand (top) or the opposing strand (bottom). Diffusion on duplex DNA is shown, but AAG can also diffuse along ssDNA.31
Characterizing the Activity and Processivity of AAG on ssDNA
DNA glycosylases vary in their preference for dsDNA versus ssDNA. Whereas many have a strong preference for dsDNA, uracil DNA glycosylase shows similar catalytic efficiencies on ssDNA and dsDNA substrates.13,21 Enzymes of the endoVIII superfamily, exemplified by mammalian NEIL1–3, also show comparable activity on ssDNA and dsDNA.27−29 Previous experiments show that AAG generally prefers dsDNA over ssDNA.3,30 For example, single-turnover glycosylase assays with deoxyinosine as a substrate show ∼6000-fold lower catalytic efficiency toward ssDNA than toward dsDNA.30 In contrast, it has been reported that AAG will excise both oxanine and εA with reasonable efficiency in either ssDNA or dsDNA context.3 The results presented in this work demonstrate that the single-turnover rate constant for base excision is only 8-fold lower in a ssDNA context than in an εA·T dsDNA context (Figure 5). The fact that AAG has a relatively high catalytic activity toward εA-containing ssDNA is consistent with much tighter binding of εA than of deoxyinosine. Nevertheless, the catalytic activity toward ssDNA is significantly worse than toward dsDNA even with the εA lesion.
The ability to measure AAG-catalyzed excision from a ssDNA context suggested the possibility of monitoring the diffusion of AAG on ssDNA using processivity assays. Under all conditions that were tested, AAG showed a distributive pattern of cleavage (Figure 6). This is markedly different from the highly processive pattern of cleavage for dsDNA substrates. Pulse–chase experiments revealed that this difference is explained by the absence of a commitment to excision on ssDNA [efficiency < 0.01 (Figure 7C)]. This is in sharp contrast to the strong commitment to excision on dsDNA [efficiency = 0.95 (Figure 7B)]. This indicates that binding of AAG to ssDNA is in rapid equilibrium relative to the slow N-glycosidic bond hydrolysis step. Nevertheless, the lifetime of AAG bound to DNA is significant. The AAG·ssDNA complex efficiently anneals upon being exposed to a complementary oligonucleotide even in the presence of a dsDNA trap (Figure 8). Such a long-lived complex with ssDNA is consistent with the observation that a ssDNA segment can serve as a conduit to increase the number of productive binding encounters with AAG.31 We conclude that AAG is able to diffuse along both dsDNA and ssDNA but shows distributive repair activity on ssDNA caused by rapidly reversible binding of the lesion site in this context.
In contrast to the inefficient search of ssDNA by AAG, other enzymes have been reported to efficiently search ssDNA. For example, uracil DNA glycosylase exhibits better searching ability on ssDNA than on dsDNA.13,21 This could be important for activity on viral DNA and on transcription bubbles in the context of somatic hypermutation. Similarly, the ssDNA binding proteins replication protein A and RecA also have been shown to diffuse along ssDNA.32,33 In each case, the physiological role of the protein involves binding to ssDNA. It appears that the normal physiological role of AAG is restricted to dsDNA, given the vast excess of dsDNA in the genome.
Effect of Helix Discontinuities on DNA Searching by AAG
We investigated the effect of kinks, bubbles, and gaps on the searching ability of AAG. As each substrate contained two εA lesions in identical duplex contexts, differences in the fraction processive (Fp) directly report on the probability that AAG can diffuse past the region in question to locate a site of damage (Figure 1). Under all conditions tested, these introduced discontinuities failed to block the searching performed by AAG (Figure 3). We were intrigued to observe that AAG showed an increased probability of finding the second site of damage when kinks or bubbles were introduced under high-salt conditions (200–300 mM NaCl). This behavior is inconsistent with a sliding model but can be explained by a hopping model in which AAG diffuses via rapid dissociation and reassociation events (Figure 9).
It is tempting to speculate that the flexibility of bubble or kinked DNA decreases the distance between the lesion sites and thereby increases the probability of an intersegmental transfer event between the two duplex arms of the DNA. However, this model is unlikely because previous experiments in which flexible PEG linkers, which cannot be bound by AAG, were used to bring two duplex regions into the proximity of each other showed no increase in Fp values relative to that of the continuous duplex under the same conditions (250 mM NaCl).31 Furthermore, searching by AAG is not dependent on the distance between two sites separated by either 25 or 50 bp of rigid duplex (an increase of ∼85 Å).9 The observation of identical searching parameters on different sized DNA segments also rules out a model in which the kink or bubble makes the search more efficient merely by restricting the number of sites that need to be searched. We are left with the idea that the ssDNA segment is itself responsible for the increased processivity. One model that would take advantage of the ability of AAG to bind to ssDNA is that it provides a larger three-dimensional volume of DNA sites such that a greater fraction of microscopic dissociation events results in reassocation. A similar model can be invoked to explain why uracil DNA glycosylase is more efficient at searching ssDNA than dsDNA.13,21
Implications for Searching by AAG
All of the available information regarding the searching mechanism of AAG is consistent with a hopping model in which rapid dissociation and/or reassociation allows the protein to randomly sample sites in the search for sites of damage (Figure 9). We envision a hopping conformation that resembles a transition state for dissociation in which the interactions with the DNA have been fully broken and there may be partial hydration of the protein and DNA surfaces and rebinding of cations.22 Most of the time these microscopic dissociation events do not proceed to full macroscopic dissocation and instead culminate in rebinding to a nearby site on either strand of the duplex. In studies of other proteins, it is common to invoke DNA sliding, whereby the protein maintains continuous contact with the DNA. For example, there is evidence that uracil DNA glycosylase exists in a closed conformation that cannot bind to the uracil product from solution but is capable of finding and excising a uracil from a nearby site.13 It was suggested that OGG1 prefers a sliding mode that would involve rotation-coupled diffusion,15,16 but more recent work favors a hopping mechanism.34 These studies suggest that single-molecule imaging studies that measure diffusion constants lack the resolution to distinguish between hopping and sliding and that assays employing roadblocks offer more direct mechanistic tests that could support the sliding mechanism.9,18,20,35,36 Although we cannot rule out the possibility that AAG might utilize both sliding and hopping modes, we note that the sliding mode presents significant challenges that would have to be overcome in the physiological context. This is because a sliding protein monomer can detect damage in only one of the two strands and would be restricted in its search by tightly bound proteins, such as nucleosomes. In contrast, a hopping mode easily overcomes these challenges, because a protein can rapidly switch strands and has a significant probability of bypassing bound proteins.
Acknowledgments
We thank members of the O’Brien laboratory for helpful discussions and comments on the manuscript.
Glossary
Abbreviations
- AAG
alkyladenine DNA glycosylase, also known as methylpurine DNA glycosylase (MPG)
- BER
base excision repair
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- εA
1,N6-ethenoadenine
- NaHEPES
sodium N-(2-hydroxyethyl)piperazine-N′-(2-ethanesulfonate)
- NaMES
sodium 2-(N-morpholino)ethanesulfonate
- PAGE
polyacrylamide gel electrophoresis.
Supporting Information Available
Two supporting figures showing additional oligonucleotide sequences and dimensions of kink oligonucleotides and two tables of processivity values measured for full-length and Δ80 AAG. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
§ M.H.: Department of Chemistry, The Pennsylvania State University, University Park, PA 16802.
Author Contributions
Y.Z. and M.H. contributed equally and are considered cofirst authors.
This work was supported by a Rackham International Fellowship and a Barbour Fellowship from the University of Michigan to Y.Z. and by grants from the National Institutes of Health to P.J.O. (CA122254 and GM108022). P.J.O. received additional financial support from the American Cancer Society (RSG-11-148-01-DMC).
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Engelward B. P.; Weeda G.; Wyatt M. D.; Broekhof J. L.; de Wit J.; Donker I.; Allan J. M.; Gold B.; Hoeijmakers J. H.; Samson L. D. (1997) Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase. Proc. Natl. Acad. Sci. U.S.A. 94, 13087–13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien P. J.; Ellenberger T. (2004) Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase. J. Biol. Chem. 279, 9750–9757. [DOI] [PubMed] [Google Scholar]
- Hitchcock T. M.; Dong L.; Connor E. E.; Meira L. B.; Samson L. D.; Wyatt M. D.; Cao W. (2004) Oxanine DNA glycosylase activity from mammalian alkyladenine glycosylase. J. Biol. Chem. 279, 38177–38183. [DOI] [PubMed] [Google Scholar]
- Miao F.; Bouziane M.; O’Connor T. R. (1998) Interaction of the recombinant human methylpurine-DNA glycosylase (MPG protein) with oligodeoxyribonucleotides containing either hypoxanthine or abasic sites. Nucleic Acids Res. 26, 4034–4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor T. R. (1993) Purification and characterization of human 3-methyladenine-DNA glycosylase. Nucleic Acids Res. 21, 5561–5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenberg J. A.; Fedtke N.; Ciroussel F.; Barbin A.; Bartsch H. (1992) Etheno adducts formed in DNA of vinyl chloride-exposed rats are highly persistent in liver. Carcinogenesis 13, 727–729. [DOI] [PubMed] [Google Scholar]
- Lau A. Y.; Wyatt M. D.; Glassner B. J.; Samson L. D.; Ellenberger T. (2000) Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG. Proc. Natl. Acad. Sci. U.S.A. 97, 13573–13578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe A. E.; O’Brien P. J. (2009) Kinetic mechanism for the flipping and excision of 1,N(6)-ethenoadenine by human alkyladenine DNA glycosylase. Biochemistry 48, 11357–11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedglin M.; O’Brien P. J. (2010) Hopping enables a DNA repair glycosylase to search both strands and bypass a bound protein. ACS Chem. Biol. 5, 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedglin M.; O’Brien P. J. (2008) Human alkyladenine DNA glycosylase employs a processive search for DNA damage. Biochemistry 47, 11434–11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg O. G.; Winter R. B.; von Hippel P. H. (1981) Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry 20, 6929–6948. [DOI] [PubMed] [Google Scholar]
- Halford S. E.; Marko J. F. (2004) How do site-specific DNA-binding proteins find their targets?. Nucleic Acids Res. 32, 3040–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonhoft J. D.; Stivers J. T. (2012) Timing facilitated site transfer of an enzyme on DNA. Nat. Chem. Biol. 8, 205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porecha R. H.; Stivers J. T. (2008) Uracil DNA glycosylase uses DNA hopping and short-range sliding to trap extrahelical uracils. Proc. Natl. Acad. Sci. U.S.A. 105, 10791–10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blainey P. C.; Luo G.; Kou S. C.; Mangel W. F.; Verdine G. L.; Bagchi B.; Xie X. S. (2009) Nonspecifically bound proteins spin while diffusing along DNA. Nat. Struct. Mol. Biol. 16, 1224–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blainey P. C.; van Oijen A. M.; Banerjee A.; Verdine G. L.; Xie X. S. (2006) A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl. Acad. Sci. U.S.A. 103, 5752–5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givaty O.; Levy Y. (2009) Protein sliding along DNA: Dynamics and structural characterization. J. Mol. Biol. 385, 1087–1097. [DOI] [PubMed] [Google Scholar]
- Pluciennik A.; Modrich P. (2007) Protein roadblocks and helix discontinuities are barriers to the initiation of mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 104, 12709–12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata-Sogawa K.; Shimamoto N. (2004) RNA polymerase can track a DNA groove during promoter search. Proc. Natl. Acad. Sci. U.S.A. 101, 14731–14735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J.; Chowdhury A.; Surtees J. A.; Shimada J.; Reichman D. R.; Alani E.; Greene E. C. (2007) Dynamic basis for one-dimensional DNA scanning by the mismatch repair complex Msh2-Msh6. Mol. Cell 28, 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonhoft J. D.; Stivers J. T. (2013) DNA translocation by human uracil DNA glycosylase: The case of single-stranded DNA and clustered uracils. Biochemistry 52, 2536–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonhoft J. D.; Kosowicz J. G.; Stivers J. T. (2013) DNA translocation by human uracil DNA glycosylase: Role of DNA phosphate charge. Biochemistry 52, 2526–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry B. J.; Jack W. E.; Modrich P. (1985) Facilitated diffusion during catalysis by EcoRI endonuclease. Nonspecific interactions in EcoRI catalysis. J. Biol. Chem. 260, 13130–13137. [PubMed] [Google Scholar]
- Francis A. W.; David S. S. (2003) Escherichia coli MutY and Fpg utilize a processive mechanism for target location. Biochemistry 42, 801–810. [DOI] [PubMed] [Google Scholar]
- Baldwin M. R.; O’Brien P. J. (2012) Defining the functional footprint for recognition and repair of deaminated DNA. Nucleic Acids Res. 40, 11638–11647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohlke C.; Murchie A. I.; Lilley D. M.; Clegg R. M. (1994) Kinking of DNA and RNA helices by bulged nucleotides observed by fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U.S.A. 91, 11660–11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H.; Mitra S.; Hazra T. K. (2003) Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J. Biol. Chem. 278, 49679–49684. [DOI] [PubMed] [Google Scholar]
- Zhao X.; Krishnamurthy N.; Burrows C. J.; David S. S. (2010) Mutation versus repair: NEIL1 removal of hydantoin lesions in single-stranded, bulge, bubble, and duplex DNA contexts. Biochemistry 49, 1658–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.; Imamura K.; Averill A. M.; Wallace S. S.; Doublie S. (2013) Structural characterization of a mouse ortholog of human NEIL3 with a marked preference for single-stranded DNA. Structure 21, 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons D. M.; O’Brien P. J. (2009) Efficient recognition of an unpaired lesion by a DNA repair glycosylase. J. Am. Chem. Soc. 131, 17742–17743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedglin M.; Zhang Y.; O’Brien P. J. (2013) Isolating contributions from intersegmental transfer to DNA searching by alkyladenine DNA glycosylase. J. Biol. Chem. 288, 24550–24559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell J. C.; Plank J. L.; Dombrowski C. C.; Kowalczykowski S. C. (2012) Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA. Nature 491, 274–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen B.; Sokoloski J.; Galletto R.; Elson E. L.; Wold M. S.; Lohman T. M. (2014) Diffusion of human replication protein A along single-stranded DNA. J. Mol. Biol. 426, 3246–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland M. M.; Schonhoft J. D.; McKibbin P. L.; David S. S.; Stivers J. T. (2014) Microscopic mechanism of DNA damage searching by hOGG1. Nucleic Acids Res. 42, 9295–9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeltsch A.; Alves J.; Wolfes H.; Maass G.; Pingoud A. (1994) Pausing of the restriction endonuclease EcoRI during linear diffusion on DNA. Biochemistry 33, 10215–10219. [DOI] [PubMed] [Google Scholar]
- Gorman J.; Plys A. J.; Visnapuu M. L.; Alani E.; Greene E. C. (2010) Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat. Struct. Mol. Biol. 17, 932–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.