Abstract
We present an innovative centrifugal microfluidic immunoassay platform (SpinDx) to address the urgent biodefense and public health need for ultrasensitive point-of-care/incident detection of botulinum toxin. The simple, sample-to-answer centrifugal microfluidic immunoassay approach is based on binding of toxins to antibody-laden capture particles followed by sedimentation of the particles through a density-media in a microfluidic disk and quantification by laser-induced fluorescence. A blind, head-to-head comparison study of SpinDx versus the gold-standard mouse bioassay demonstrates 100-fold improvement in sensitivity (limit of detection = 0.09 pg/mL), while achieving total sample-to-answer time of <30 min with 2-μL required volume of the unprocessed sample. We further demonstrate quantification of botulinum toxin in both exogeneous (human blood and serum spiked with toxins) and endogeneous (serum from mice intoxicated via oral, intranasal, and intravenous routes) samples. SpinDx can analyze, without any sample preparation, multiple sample types including whole blood, serum, and food. It is readily expandable to additional analytes as the assay reagents (i.e., the capture beads and detection antibodies) are disconnected from the disk architecture and the reader, facilitating rapid development of new assays. SpinDx can also serve as a general-purpose immunoassay platform applicable to diagnosis of other conditions and diseases.
Botulinum neurotoxin (BoNT), the most toxic substance known to man,1−3 remains one of the highest priority biological threat agents. It is easy to produce, and minute amounts are sufficient to kill a person. One gram of holotoxin, evenly dispersed, has the potential to incapacitate 100,000 adults.4 A 2001 Canadian report estimated that BoNT exposure to 100,000 people would result in 30,000 deaths and a total economic cost of $8.9 billion due to costly and long-term management of intoxication.5 It also remains a threat for public health in the form of foodborne, wound, and infant botulism. The extreme toxicity requires ultrasensitive diagnostic assays with detection limits well below the minimal lethal dose to allow timely administration of therapeutics.
Current diagnostic technology is limited to the live-mouse bioassay as the only FDA-approved metric for confirming the presence of active BoNT in a sample.4,6−8 The mouse bioassay, while sensitive, requires days for result confirmation thus rendering it ineffective for timely therapeutic mitigation. The mouse assay is also extremely costly, and public health laboratories spend millions of dollars every year to maintain and operate a mouse facility for screening. Moreover, only a few specialized laboratories in the country are capable of performing mouse bioassays. Therefore, in a suspected bioterrorism incident samples must be collected, preserved, and shipped to these laboratories for analysis, wasting precious time before a positive identification is made. Last, but not least, there are ethical concerns related to the use of mice for toxin testing.
There is an urgent need for rapid, sensitive, and accurate detection/diagnosis of BoNTs for swift and effective utilization of limited medical countermeasures to save lives and minimize socioeconomic impact. Passive immunization remains the only available therapeutic approach; timely administration of anti-BoNT antibodies to clear toxin from the bloodstream greatly improves the speed of patient recovery. To this end, several new in vitro assay platforms have been proposed in the literature9−19 as well as improvements to the existing in vivo assay.20,21 While many of these assays have the advantage of improved sensitivity, such as the ALISSA technique capable of detection of attomolar concentrations,15,22 the technology often relies on complex sample processing steps and long incubation times that do not meet usability requirements for point-of-care or point-of-incident diagnostic applications. A few field-deployable assays (strip or lateral-flow immunoassays) are available23−25 but suffer from major limitations including poor detection limit and inability to provide quantitative results.
We describe a centrifugal microfluidic platform (SpinDx) suitable for both laboratory-based as well as potential point-of-care/incident detection of BoNTs. Centrifugal microfluidics based biochemical analysis devices26−29 have attracted significant attention in the past few years as evidenced by numerous reports and commercial instruments. Madou’s group has implemented traditional ELISA chemistry in a homogeneous phase for detecting proteins including Dengue viral NS1 protein.30 The commercially available Gyrolab (Gyros, Inc.) utilizes a packed bed of streptavidin-coated particles with multiple reagent dispensing steps controlled by capillary and centrifugal forces to perform immunoassays and other methods.31 The reagents wash through the column through application of centrifugal force followed by interrogation by laser-based detection. The system developed by the Samsung Biomedical Research Institute utilizes a system of ferrowax valves individually actuated by heating with an IR laser to implement immunoassays using surface-immobilized affinity reagents.29 Zengerle et al. demonstrated picomolar detection of BoNT/A based on a luciferase reporter bound to microparticles in a centrifugal microfluidic system (LabDisk).32 An N-terminal HaloTag is used for bead attachment with two tandem repeats of SNAP-25 connecting the C-terminal luciferase. After cleavage of the SNAP-25 motif by the endopeptidase activity of the toxin, the bioluminescent signal can be detected in solution after removal of the solid support. We present a centrifugal microfluidic platform (Figure 1) for detection of botulinum toxin based on an innovative sedimentation-based immunoassay developed recently by our group.33 The sample is mixed with a detection cocktail consisting of a) capture beads coated with antibodies specific for the target(s) of interest and b) detection antibodies labeled with a fluorescent tag, which will be bound to the capture bead in the presence of the corresponding antigen (Figure 1A). Following incubation, the beads are separated from the sample via sedimentation through a density media that inherently washes the beads and removes any interfering agents as the beads stack at the end of the channel (periphery of the disk) (Figure 1B). The fluorescent signal of the resulting bead pellet is used to quantify the analyte present (Figure 1C). SpinDx was used for detection of botulinum toxin and outperformed the gold-standard mouse bioassay with respect to ease-of-use (completely automated requiring no manual sample preparation), sensitivity (100-fold limit of detection improvement), and time of assay (less than 30 min). The device is compatible with both clinical and nonclinical samples making it a universal platform for detection and quantification of BoNT and other protein analytes.
Figure 1.
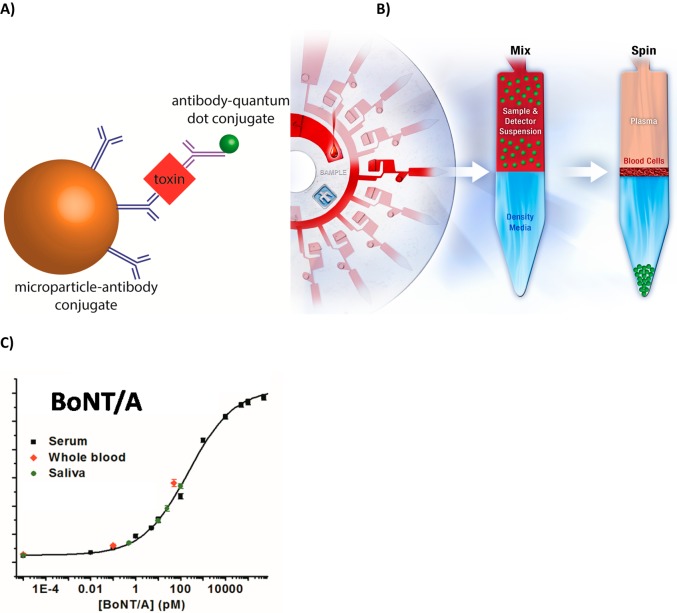
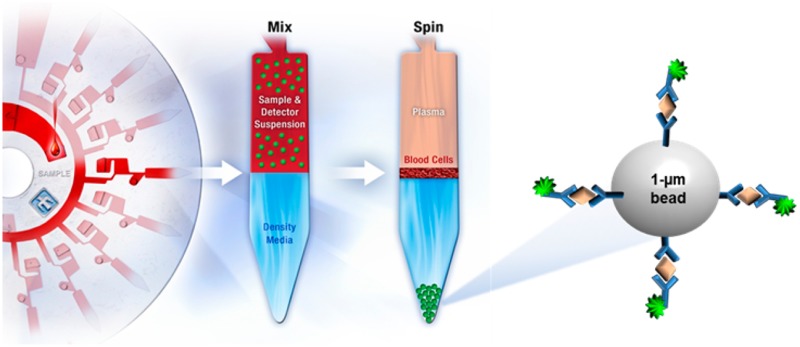
A) Schematic representation of the immunocomplex formed upon binding of the target analyte. B) SpinDx immunoassay schematic, depicted as multiplexed analysis of whole blood. Samples are mixed with antibody-conjugated capture beads and a fluorescent detection antibody in solution, loaded upon a preloaded density medium, and centrifuged such that beads sediment to the bottom of the channel where they form a rinsed pellet separated from background sample contaminants and unbound label. C) Dose–response quantification of purified BoNT/A in exogeneous clinical samples. For serum data: χ2 = 1.22, r2 = 0.999. LoD: 0.5 fM; LoQ: 20.5 fM.
Experimental Section
Materials and Methods
Detailed materials and methods can be found in the Supporting Information.
Biological Safety Considerations
Botulinum neurotoxins are the most deadly substance known. The amount necessary to cause harm (including paralysis and death) are minute (1 ng/kg). Extreme caution needs to be exercised while handling the toxin, particularly in powder form. Always use a Class 2 biosafety cabinet with appropriate personal protective equipment (safety glasses, gloves, lab coat) and avoid the use of sharps. Avoid generation of aerosols. Biological and sharps waste must be disposed according to federal, state, local, and institutional guidelines.
Human Samples
Whole human blood was purchased from Innovative Technologies (Novi, MI) and used without further treatment. Though the blood tested negative for a variety of blood-borne pathogens, it is important to observe universal precautions in the handling of potentially infectious materials. According to HHS regulations 45 CFR Part 46, publically available, commercially acquired, pooled, and deidentified human whole blood does not constitute human subjects research. As such, it is not subject to Institutional Review Board or Human Studies Board review. Protocols adhered to institutional guidelines approved by the Institutional Biosafety Committee.
Monoclonal Antibody Development (Western Regional Research Center, USDA)
Mouse anti-BoNT/A monoclonal antibodies (F1-2, F1-40, F1-51, and F2-43) used in this study were generated following immunization of Balb/c mice with BoNT toxoid.34 Animal protocols adhered to institutional guidelines approved by the Animal Care and Use Committee of the U.S. Department of Agriculture, Western Regional Research Center.
Antibody-Microparticle Conjugation
Conjugation of the capture antibody to the microparticle proceeded via standard carbodiimide chemistry. Silica microparticles prefunctionalized with carboxylic acid groups were activated equimolar amounts of N-ethyl-N′-(3-(dimethylamino)propyl)carbodiimide) (EDC) and of N-hydroxysuccinimide (NHS) at pH 6.4 in 100 mM 4-morpholinepropanesulfonic acid (MOPS) to form the succinimidyl ester. The capture antibody was added, and the solution was raised to pH 8.15 and reacted at 4 °C for 2 h. The particles were then twice blocked with 1% bovine serum albumin (BSA) for 30 min at 4 °C. The particles were then washed in wash buffer (138 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 0.05% (w/v) Tween-20, 0.05% (w/v) Pluronic F127, 0.05% (w/v) n-dodecyl β-d-maltoside, 7.6 mM NaN3, 0.1% (w/v) BSA) and resuspended in wash buffer to a concentration of 5% solids.
Antibody-Quantum Dot Conjugation
Detection antibodies were labeled with quantum dots using standard EDC/NHS carbodiimide chemistry. Carboxylic acid-terminated quantum dots were conjugated to the detection antibody with equimolar amounts of EDC/NHS at room temperature for 30 min with stirring. The reaction was spun through a desalting column made of Sephacryl S400HR, and the first fraction was taken as the quantum dot-antibody conjugate. Degree of labeling was determined using the published value for UV absorption of the quantum dot, and the protein concentration was determined using a bicinchoninic acid protein assay (BCA).
SpinDx Immuonassay Protocol
Immunoassays were performed in technical triplicate. Standard curves were collected by diluting BoNT/A in fetal bovine serum (FBS) as the sample matrix. To 7 μL of a 5% solids suspension of capture particles was added 1 μL of a 300 nM solution of quantum dot-labeled detection antibody. To this suspension was added 7 μL of the BoNT/A-spiked FBS to yield 20 nM final concentration of detection antibody. The suspension was incubated with mixing for 20 min at room temperature. Each channel of the disk was preloaded with 3 μL of a density medium consisting of 90% Percoll in phosphate buffered saline (PBS [138 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, pH 7.4]) with 0.05% (w/v) Tween 20, 0.1% (w/v) BSA, and 0.1% (w/v) F127. After incubation, 4 μL of the suspension was added to the channel, and the disk was spun at 8000 rpm for 45 s. The bead pellet was analyzed on an Olympus IX-70 fluorescence microscope with 405 nm excitation and 705 nm emission, a CoolSnap HQ interline CCD camera (Roper Scientific, Trenton, NJ), and Image-Pro Plus imaging software (MediaCybernetics, Bethesda, MD). The average fluorescence of each bead pellet was measured and compared with calibration curves generated in parallel with standard dilutions to quantify the target analyte.
Mouse Intranasal Intoxication and Sample Collection (UMass Dartmouth)
Swiss-Webster female mice (22 to 25- g) were purchased from Charles River Laboratories, Inc. (Wilmington, MA). Intranasal toxin application was carried out by first lightly anesthetizing mice with isoflurane (Isothesia, Abbott Laboratories North, Chicago, IL). Toxin was administered by single application of 20 μL solution to the nares at doses of 50-ng, 500-ng, and 5-μg. Care was taken to avoid generation of aerosol during the administration of the toxin. The heads of animals were maintained in an upright position to minimize drainage into the posterior pharynx. Blood samples were collected at 30 min and 60 min postintoxication via retro-orbital bleeding. Serum was separated by centrifugation at 1665xg for 16 min, stored at −20 °C, and shipped to Sandia National Laboratories for analysis. All procedures involving animals were reviewed and approved by the Institutional Animal Care and Use Committee, University of Massachusetts Dartmouth (2003).
Mouse Intravenous and Oral Intoxication and Sample Collection (Western Regional Research Center, USDA)
For oral dosing, 4–5 week old female Swiss-Webster mice were treated with 100 μL of BoNT/A complex (dose levels 500, 5000 or 50,000 ng/mL) or with the same volume of control phosphate gelatin buffer via gavage using Popper feeding needles. Surviving mice were monitored for at least 7 days following experiments for signs of intoxication. For intravenous mouse treatments, mice were injected with 100 μL of BoNT/A holotoxin (500, 5000, or 50,000 pg/mL) or with the same volume of phosphate gelatin buffer control via the lateral tail vein. Blood and samples were incubated on ice for at least 30 min before centrifugation at 3000 g for 10 min to separate sera from the cellular fraction. Samples were then aliquoted and stored at −80 °C before analysis. Animal protocols adhere to institutional guidelines approved by the Animal Care and Use Committee of the U.S. Department of Agriculture, Western Regional Research Center.
Mouse Bioassay (Western Regional Research Center, USDA)
Samples consisting of serial dilutions of BoNT/A holotoxin in phosphate gelatin buffer were prepared, blinded, and shared with Sandia National Laboratories for parallel analysis via SpinDx, ELISA, and the live mouse bioassay on the same day. Random groups of 10 Swiss Webster mice (females 4–5 weeks old) were injected with 500 μL of each dosage level intraperitoneally. Animals were monitored for 7 days for signs of intoxication (wasp-waist phenotype, labored breathing, and paralysis) or death. Moribund animals were humanely euthanized and counted as dead.
ELISA Protocol
The capture ELISA used here was previously described.34 Results from a typical analysis of standards are shown in Figure S1.
Statistical Analysis
All data were subjected to statistical analysis with OriginPro 9.1 (OriginLabs, Cambridge, MA). Goodness of fit is reported as χ-squared and r-squared values, and limits of detection and quantification are formally defined through IUPAC convention: 3-times the standard deviation of the blank and 10-times the standard deviation of the blank, respectively.
Results and Discussion
SpinDx Immunoassay
An overview of the centrifugal sedimentation assay protocol is shown in Figure 1A and B, depicted for multiplexed analysis of a drop of whole blood. An equal volume of the sample is mixed and incubated at room temperature with a suspension comprising capture antibody-functionalized beads (1-μm silica microparticles) and an unbound fluorescently labeled detection antibody. The sample/detection suspension is loaded on top of a preloaded density medium in a channel embedded on the disk. During centrifugation, the microparticles (with density greater than that of the density medium) sediment through the density medium and pellet at the bottom of the channel (periphery of the disk). The fluorescence of the microparticle pellet is measured to quantify concentration of the target analyte in the sample. The entire assay requires less than 30 min (compared to several hours for other in vitro assay approaches or days for the live-mouse bioassay). Furthermore, the scale of the device allows for small samples sizes (2-μL per sample), whereas other assays typically use much larger volumes (100-μL for ELISA or 500-μL for the mouse bioassay). Figures 1C and 4B show calibration curves for BoNT/A spiked in serum and other matrices. The SpinDx assay is both sensitive (limit of detection ∼0.09 pg/mL, defined as 3 standard deviation above background signal) and has a wide dynamic range (8 orders of magnitude) for BoNT/A spiked in fetal bovine serum.
Figure 4.
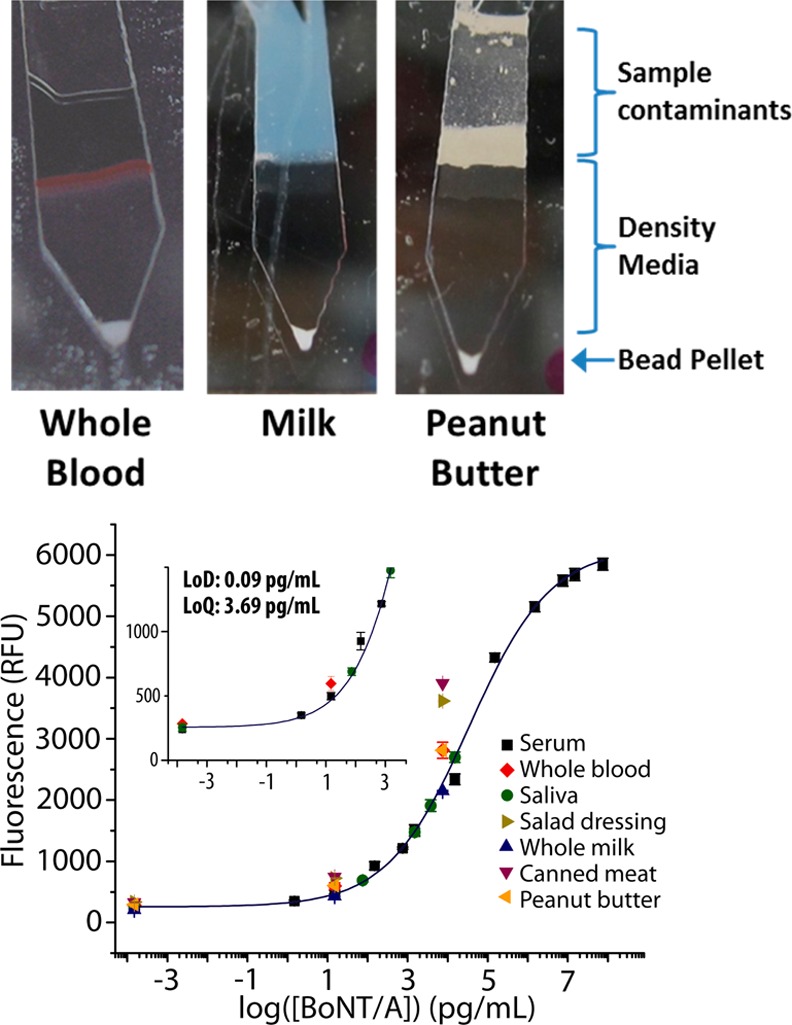
Matrix independence of SpinDx measurements. A) Photographs of assay channels after centrifugation on the SpinDx platform. Background interfering agents from the sample are clearly separated from the tip of the channel where the assay is read. (B) Botulinum neurotoxin A immunoassays results from a wide variety of sample matrices. Good linearity is observed from all matrices shown. χ2 = 1.48, r2 = 0.997. LoD: 0.09 pg/mL; LoQ: 3.69 pg/mL.
SpinDx Assays Are Ultrasensitive
Due to their extreme toxicities, high sensitivity is an essential requirement for detection of botulinum toxin in both clinical and food samples. We achieved subfemtomolar sensitivity that greatly surpassed conventional diagnostic methods (100-fold more sensitive than the ELISAs developed by the USDA researchers34,35 and National Biodefense Analysis and Countermeasures Center (NBACC)). The enhanced sensitivity of the SpinDx assay is attributed to several unique features of the sedimentation approach, including the following: a) 1-μm beads provide a capture surface ∼320× larger than a standard 96-well microtiter plate; b) isolating the capture beads from the sample and excess label during the sedimentation step inherently washes the beads with several hundred times the particle volume significantly reducing the background signal without requiring separate wash steps; c) pelleting the beads at the end of the channel permits averaging of signal over thousands of beads; and d) the use of quantum dots as the detection label provides a large (300 nm) Stokes shift thereby further reducing autofluorescence and background noise. Quantum dots are also resistant to photobleaching allowing for longer signal acquisition times to improve signal. Quantum dot-antibody conjugates are known to enhance fluorescent assay sensitivities while maintaining antibody reactivity.36−38 Intra- and interassay coefficients of variation are low (intra: 4.3%, inter: 7.2%) indicating that the antibodies exhibit good consistency.
Detection of Botulinum Toxin in Intoxicated Animal Models
Rapid and sensitive detection of BoNT/A from intoxicated patients is extremely important for accurate diagnosis, effective therapeutic intervention, and public health awareness.39,40 However, it is impractical and unethical to perform human exposure studies with biotoxins. Hence, we explored BoNT/A quantification in the peripheral serum collected from intoxicated mice. Swiss-Webster mice (4–5 week old female) were intoxicated with BoNT/A by three routes of exposure: intravenous and intranasal intoxication (simulating aerosolized exposure) with BoNT/A holotoxin and oral intoxication with BoNT/A complex (simulating food-borne contamination). Exposure doses were chosen to bracket the LD50 for each intoxication route based on previous studies.6,41,42 Blood was drawn at 30 min and 60 min postintoxication for the intravenous and intranasal exposure groups and 1-h and 7-h postintoxication for the oral exposure groups. As shown in Figures 2A–C, SpinDx was able to detect the presence of BoNT/A in the serum from all three groups. Furthermore, the assay’s low limit of detection detected trace concentrations of BoNT/A not previously measured in mouse serum, such as in mice orally exposed to 500-ng BoNT/A complex (a nonlethal dose). As expected, the relative amount of BoNT/A in the serum of orally exposed mice was lower than that from the intravenous and intranasal routes due to destruction of the toxin by the digestive system; previously, it has been shown that the LD50 from oral exposure is several orders of magnitude higher than from other routes of intoxication.6,43,44 Also note the rapid decrease in BoNT/A concentration in the peripheral serum of the intravenous and intranasal exposure groups due to clearance and translocation to internal organs and tissues. This result highlights the importance of rapid diagnostic analysis following exposure to the toxin.
Figure 2.
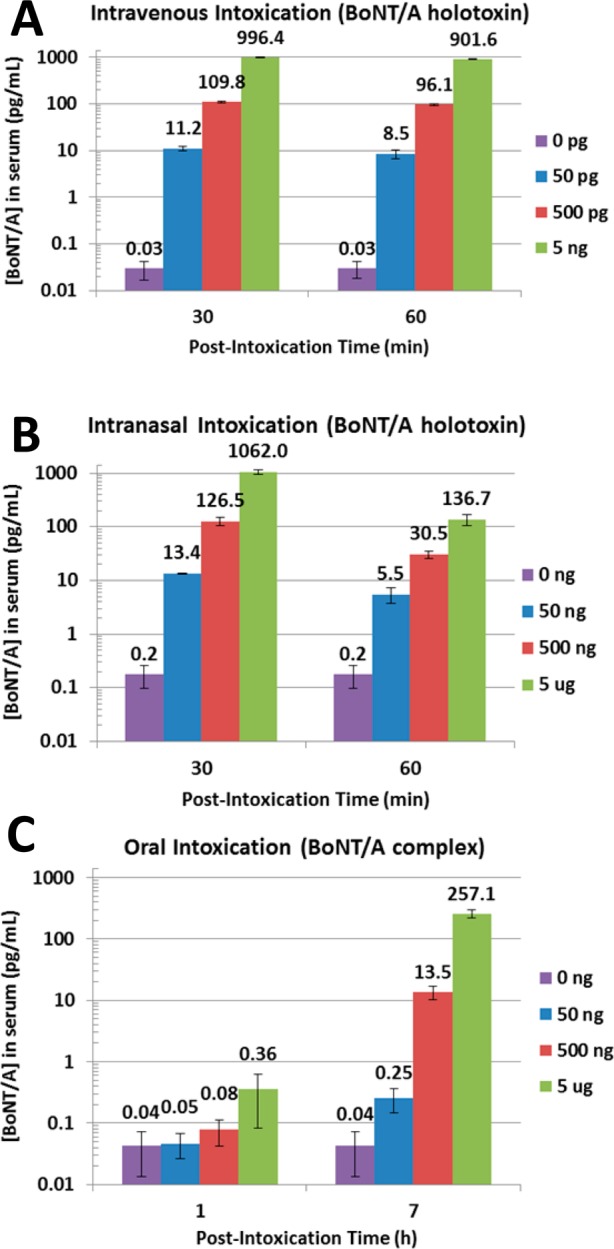
SpinDx quantification of BoNT/A holotoxin in peripheral serum of intoxicated mice due to (A) intravenous intoxication with BoNT/A holotoxin, (B) intranasal intoxication with BoNT/A holotoxin, and (C) oral intoxication with BoNT/A complex. Samples were each run in triplicate; error bars represent standard error of the mean (SEM).
Immunoassays Results Correlate Well with Gold Standard Mouse Bioassays and ELISAs
Botulinum Immunoassays Exceed Sensitivity of Gold Standard Mouse Bioassays
We also conducted a direct, head-to-head comparison of the SpinDx botulinum assay with the mouse bioassay (n = 10 animals per dilution). Freshly prepared serial dilutions of purified BoNT/A toxin in gelatin-phosphate diluent were blinded, shared, and analyzed on the same day by each assay. 4–5 week old Swiss Webster mice were observed for 4 days postexposure (following the standard mouse bioassay protocol,35 whereas results from the SpinDx platform were available after 30 min). Only the highest 4 concentrations (2, 10, 50, and 100 pg/mL) were tested by the mouse bioassay to reduce animal usage. Results from the study are shown in Figure 3A. The mouse bioassay limit of detection was found to be 50 pg/mL, with visual detection at 10 pg/mL based on the appearance of a distinctive wasp waist phenotype in mice as a subjective metric to judge intoxication (Figure 3B). Lot-to-lot variability of the toxin as well as individual variation between the mice is known to confound the mouse bioassay; previous work has shown the mouse bioassay to be sensitive to approximately 20 pg/mL.8 In contrast, the SpinDx platform in this assay was sensitive to 0.09 pg/mL, more than 100-fold more sensitive than the best-reported mouse assay while using ∼250-fold less sample volume.
Figure 3.
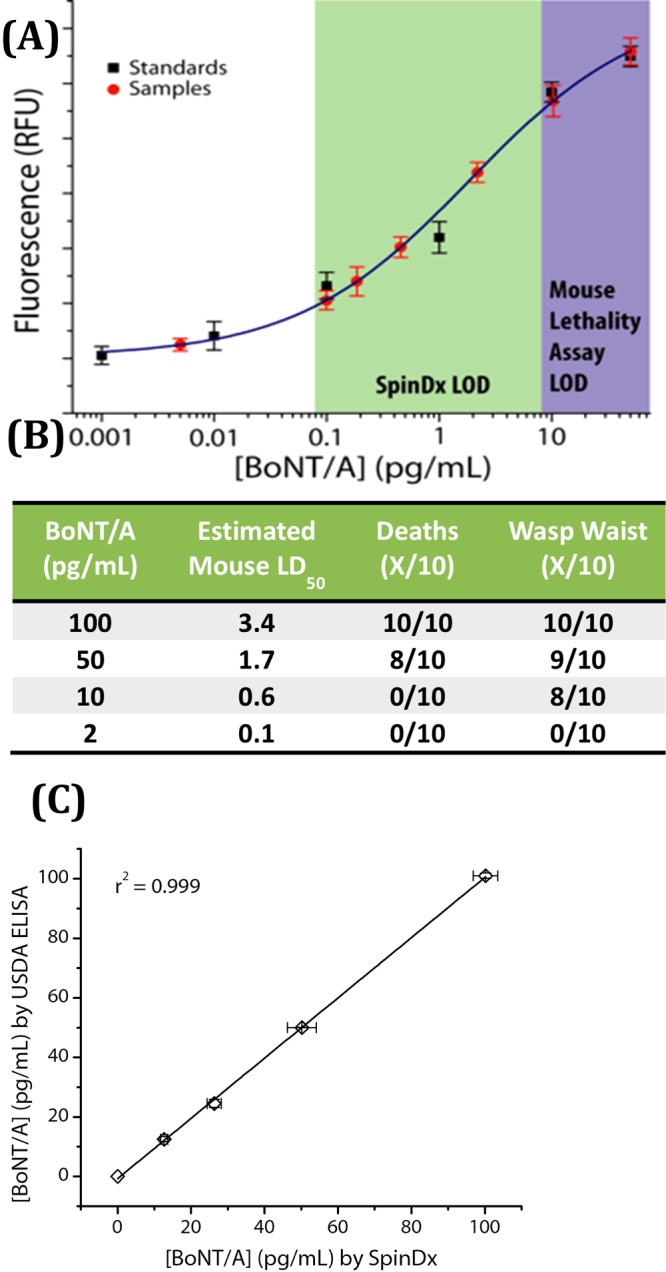
Head-to-head comparison of SpinDx with mouse bioassay and ELISA. Spiked, blinded samples were analyzed on the same day by each method. (A) SpinDx analysis of blinded samples plotted against standard concentrations. Shaded areas highlight the LODs of each assay. (B) Results from mouse bioassay with 4 blinded concentrations (2, 10, 50, and 100 pg/mL). Calculated LD50 = 0.68 ng/kg. (C) Comparison of SpinDx assays with a published protocol by USDA researchers.34 Spiked, blinded samples were analyzed on the same day by each method. Phosphate buffered gelatin solution was used as the sample matrix.
Comparison with Conventional ELISAs
We compared results of our SpinDx immunoassay for BoNT/A with ELISA protocols established and used by the National Biodefense Analysis and Countermeasures Center (NBACC) and a published protocol by USDA researchers.34 The analysis shows exceptional agreement for BoNT/A detection, y = 1.02x + 5.3886, χ2 = 0.986 (Figure 3C).
No off-Device Sample Preparation Is Required for Environmental, Food or Clinical Samples
A key advantage of this approach is its compatibility with complex sample matrices without requiring additional sample preparation. We demonstrate detection in a wide variety of sample matrices in this study, ranging from solids to liquids to colloids. Figure 4A shows photographs of the on-disk detection channels following assay completion with contaminants such as plasma and cells from blood, caseins from milk, and lipids from peanut butter isolated from the bead pellet by the density medium. Figure 4B shows results from direct analysis of several important clinical and food sample matrices. BoNT/A was detected in clinical matrices such as whole blood, serum, and saliva with minimal matrix interference compared to the response from buffered systems alone. Food samples such as milk, canned meat, canned vegetables, juice, and salad dressing were also compatible with minimal deviation from a linear relationship. Foods which have low pH and are minimally cooked are most vulnerable to BoNT/A contamination.45−47 The detection suspension is buffered by PBS, minimizing the effect of acidic sample matrices such as fruit and vegetable juices. Note that the two major outbreaks of BoNT/A intoxication in the United States were traced to canned green beans and carrot juice.23,48,49 However, while the food-based detection results in Figure 4A were obtained using spiked BoNT/A holotoxin, the toxin is often found in its natural complex form in environmental and food samples (and subsequently dissociated following intestinal absorption). Therefore, we also include dose–response quantification of hemagglutinin 70 (HA70) – a protein found in the BoNT/A complex – using HA70-specific monoclonal antibodies (see Figure S2 in the Supporting Information). New assays are easily developed upon the SpinDx platform by simple substitution of sandwich immunoassay affinity reagents.
Finally, our diagnostic approach is highly adaptable as the assay reagents (i.e., the capture beads and detection antibodies) are disconnected from the disk architecture, facilitating rapid development of new assays. Multiplexed assays measuring a number of targets (up to 64 parallel assays using the current disk architecture) may be readily developed and demonstrated within hours given availability of sandwich immunoassay affinity reagents.
Conclusion
We have developed a platform that can serve as a rapid, reliable detection device for use in public health laboratories and field-laboratories designated for testing of environmental and clinical samples for botulinum toxin. We envision SpinDx to be a simple-to-use device in which manual intervention is limited to introducing the sample into a disk, loading the disk into a reader, and hitting the start button. It meets the stringent sensitivity, ease of use, and short assay time requirements for point-of-care and point-of-incidence applications. Furthermore, it can perform direct analysis of samples (blood, food, etc.) with no additional sample prep required. Unique to the platform are signal enrichment and background suppression elements inherent to the assay approach enable sensitivities unmatched by conventional approaches. The proposed device not only meets an urgent unmet need for biodefense but also provides revolutionary instrumentation and capabilities for the public health community.
Acknowledgments
This work was funded by the National Institute of Allergies and Infectious Disease (NIAID) Grants U01A1075441 and R01AI098853. L.W.C. and L.H.S. were funded by the United States Department of Agriculture, Agricultural Research Service, National Program project NP108, CRIS 5325-42000-043-00D and the National Institute of Allergy and Infectious Diseases Service Grant U54 AI065359. Sandia National Laboratories is a multiprogram laboratory managed and operated by Sandia Corporation, a wholly owned subsidiary of Lockheed Martin Corporation, for the U.S. Department of Energy’s National Nuclear Security Administration under contract DE-AC04-94AL85000 - SAND2014-20313J.
Supporting Information Available
Detailed experimental methods and supplemental figures. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
∥ Sandstone Diagnostics, Livermore, CA.
Author Present Address
⊥ Cepheid, Sunnyvale, CA.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Arnon S. S.; Schechter R.; Maslanka S. E.; Jewell N. P.; Hatheway C. L. N. Engl. J. Med. 2006, 354, 462–471. [DOI] [PubMed] [Google Scholar]
- Balali-Mood M.; Moshiri M.; Etemad L. Toxicon 2013, 69, 131–142. [DOI] [PubMed] [Google Scholar]
- Gill D. M. Microbiol Rev. 1982, 46, 86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon S. S.; Schechter R.; Inglesby T. V.; Henderson D. A.; Bartlett J. G.; Ascher M. S.; Eitzen E.; Fine A. D.; Hauer J.; Layton M.; Lillibridge S.; Osterholm M. T.; O’Toole T.; Parker G.; Perl T. M.; Russell P. K.; Swerdlow D. L.; Tonat K.; Biodefense W. G. C. JAMA, J. Am. Med. Assoc. 2001, 285, 1059–1070. [DOI] [PubMed] [Google Scholar]
- St. John R.; Finlay B.; Blair C. Can. J. Infect. Dis. 2001, 12, 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L. W.; Henderson T. D. 2nd Toxicon 2011, 58, 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K.; Cai S.; Singh B. R. Expert Opin. Drug Discovery 2014, 9, 319–333. [DOI] [PubMed] [Google Scholar]
- Lindstrom M.; Korkeala H. Clin. Microbiol. Rev. 2006, 19, 298–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.; Baudys J.; Kalb S. R.; Barr J. R. Anal. Biochem. 2011, 412, 67–73. [DOI] [PubMed] [Google Scholar]
- Vidal D.; Taggart M. A.; Badiola I.; Mateo R. J. Vet. Diagn. Invest. 2011, 23, 942–946. [DOI] [PubMed] [Google Scholar]
- Ruge D. R.; Dunning F. M.; Piazza T. M.; Molles B. E.; Adler M.; Zeytin F. N.; Tucker W. C. Anal. Biochem. 2011, 411, 200–209. [DOI] [PubMed] [Google Scholar]
- Swain M. D.; Anderson G. P.; Zabetakis D.; Bernstein R. D.; Liu J. L.; Sherwood L. J.; Hayhurst A.; Goldman E. R. Anal. Bioanal. Chem. 2010, 398, 339–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillehoj P. B.; Wei F.; Ho C. M. Lab Chip 2010, 10, 2265–2270. [DOI] [PubMed] [Google Scholar]
- Goldman E. R.; Anderson G. P.; Conway J.; Sherwood L. J.; Fech M.; Vo B.; Liu J. L.; Hayhurst A. Anal. Chem. 2008, 80, 8583–8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagramyan K.; Barash J. R.; Arnon S. S.; Kalkum M. PLoS One 2008, 3, e2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. K.; Harrison S. H.; Schoeniger J. S. Anal. Chem. 2000, 72, 6019–6024. [DOI] [PubMed] [Google Scholar]
- Mason J. T.; Xu L.; Sheng Z.-m.; O’Leary T. J. Nat. Biotechnol. 2006, 24, 555–557. [DOI] [PubMed] [Google Scholar]
- Garber E. A. E.; Venkateswaran K. V.; O’Brien T. W. J. Agric. Food Chem. 2010, 58, 6600–6607. [DOI] [PubMed] [Google Scholar]
- Hill B. J.; Skerry J. C.; Smith T. J.; Arnon S. S.; Douek D. C. BMC Microbiol. 2010, 10, 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder-Kofie T. D.; Luquez C.; Adler M.; Dykes J. K.; Coleman J. D.; Maslanka S. E. Comp. Med. 2011, 61, 235–242. [PMC free article] [PubMed] [Google Scholar]
- Dressler D.; Lange M.; Bigalke H. Mov. Disord. 2005, 20, 1617–1619. [DOI] [PubMed] [Google Scholar]
- Bagramyan K.; Kalkum M. Methods Mol. Biol. (Clifton, N.J.) 2011, 739, 23–36. [DOI] [PubMed] [Google Scholar]
- Sharma S. K.; Whiting R. C. J. Food Prot. 2005, 68, 1256–1263. [DOI] [PubMed] [Google Scholar]
- Hodge D. R.; Prentice K. W.; Ramage J. G.; Prezioso S.; Gauthier C.; Swanson T.; Hastings R.; Basavanna U.; Datta S.; Sharma S. K.; Garber E. A.; Staab A.; Pettit D.; Drumgoole R.; Swaney E.; Estacio P. L.; Elder I. A.; Kovacs G.; Morse B. S.; Kellogg R. B.; Stanker L.; Morse S. A.; Pillai S. P. Biosecur. Bioterrorism 2013, 11, 237–250. [DOI] [PubMed] [Google Scholar]
- Ching K. H.; Lin A.; McGarvey J. A.; Stanker L. H.; Hnasko R. J. Immunol. Methods 2012, 380, 23–29. [DOI] [PubMed] [Google Scholar]
- Gorkin R.; Park J.; Siegrist J.; Amasia M.; Lee B. S.; Park J. M.; Kim J.; Kim H.; Madou M.; Cho Y. K. Lab Chip 2010, 10, 1758–1773. [DOI] [PubMed] [Google Scholar]
- Lai S.; Wang S.; Luo J.; Lee L. J.; Yang S. T.; Madou M. J. Anal. Chem. 2004, 76, 1832–1837. [DOI] [PubMed] [Google Scholar]
- Madou M.; Zoval J.; Jia G.; Kido H.; Kim J.; Kim N. Annu. Rev. Biomed. Eng. 2006, 8, 601–628. [DOI] [PubMed] [Google Scholar]
- Lee B. S.; Lee J. N.; Park J. M.; Lee J. G.; Kim S.; Cho Y. K.; Ko C. Lab Chip 2009, 9, 1548–1555. [DOI] [PubMed] [Google Scholar]
- Aeinehvand M. M.; Ibrahim F.; Harun S. W.; Djordjevic I.; Hosseini S.; Rothan H. A.; Yusof R.; Madou M. J. Biosens. Bioelectron. 2014, 10.1016/j.bios.2014.08.076. [DOI] [PubMed] [Google Scholar]
- Fraley K. J.; Abberley L.; Hottenstein C. S.; Ulicne J. J.; Citerone D. R.; Szapacs M. E. Bioanalysis 2013, 5, 1765–1774. [DOI] [PubMed] [Google Scholar]
- van Oordt T.; Stevens G. B.; Vashist S. K.; Zengerle R.; von Stetten F. RSC Adv. 2013, 3, 22046–22052. [Google Scholar]
- Schaff U. Y.; Sommer G. J. Clin Chem. 2011, 57, 753–761. [DOI] [PubMed] [Google Scholar]
- Stanker L. H.; Merrill P.; Scotcher M. C.; Cheng L. W. J. Immunol. Methods 2008, 336, 1–8. [DOI] [PubMed] [Google Scholar]
- Scotcher M. C.; Cheng L. W.; Stanker L. H. PLoS One 2010, 5, e11047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakovich T. Y.; Mahfoud O. K.; Mohamed B. M.; Prina-Mello A.; Crosbie-Staunton K.; Van Den Broeck T.; De Kimpe L.; Sukhanova A.; Baty D.; Rakovich A.; Maier S. A.; Alves F.; Nauwelaers F.; Nabiev I.; Chames P.; Volkov Y. ACS Nano 2014, 8, 5682–5695. [DOI] [PubMed] [Google Scholar]
- Kotagiri N.; Li Z.; Xu X.; Mondal S.; Nehorai A.; Achilefu S. Bioconjugate Chem. 2014, 25, 1272–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao U. L.; Mulchandani A.; Chen W. J. Am. Chem. Soc. 2006, 128, 14756–14757. [DOI] [PubMed] [Google Scholar]
- Rivera V. R.; Gamez F. J.; Keener W. K.; White J. A.; Poli M. A. Anal. Biochem. 2006, 353, 248–256. [DOI] [PubMed] [Google Scholar]
- Cai S.; Singh B. R.; Sharma S. Crit. Rev. Microbiol. 2007, 33, 109–125. [DOI] [PubMed] [Google Scholar]
- Schantz E. J.; Johnson E. A. Microbiol. Rev. 1992, 56, 80–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Saleem F. H.; Ancharski D. M.; Joshi S. G.; Elias M.; Singh A.; Nasser Z.; Simpson L. L. Infect. Immun. 2012, 80, 4133–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi I. Infect. Immun. 1984, 43, 487–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugii S.; Ohishi I.; Sakaguchi G. Jpn. J. Med. Sci. Biol. 1977, 30, 70–73. [PubMed] [Google Scholar]
- Rasooly R.; Stanker L. H.; Carter J. M.; Do P. M.; Cheng L. W.; He X.; Brandon D. L. Int. J. Food Microbiol. 2008, 126, 135–139. [DOI] [PubMed] [Google Scholar]
- Sachdeva A.; Defibaugh-Chavez S. L.; Day J. B.; Zink D.; Sharma S. K. Appl. Environ. Microbiol. 2010, 76, 7653–7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenicia L.; Fach P.; van Rotterdam B. J.; Anniballi F.; Segerman B.; Auricchio B.; Delibato E.; Hamidjaja R. A.; Wielinga P. R.; Woudstra C.; Agren J.; De Medici D.; Knutsson R. Int. J. Food Microbiol. 2011, 145Suppl 1S152–157. [DOI] [PubMed] [Google Scholar]
- Wictome M.; Newton K. A.; Jameson K.; Dunnigan P.; Clarke S.; Gaze J.; Tauk A.; Foster K. A.; Shone C. C. Dev. Biol. Stand. 1999, 101, 141–145. [PubMed] [Google Scholar]
- Yoon S. Y.; Chung G. T.; Kang D. H.; Ryu C.; Yoo C. K.; Seong W. K. Microbiol. Immunol. 2005, 49, 505–511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.