Abstract
Embryogenesis depends on a highly coordinated cascade of genetically encoded events. In animals, maternal factors contributed by the egg cytoplasm initially control development, while the zygotic nuclear genome is quiescent. Subsequently, the genome is activated, embryonic gene products are mobilized and maternal factors are cleared. This transfer of developmental control is called the maternal-to-zygotic transition (MZT). In this review, we discuss recent advances toward understanding the scope, timing and mechanisms that underlie zygotic genome activation at the MZT in animals. We describe high-throughput techniques to measure the embryonic transcriptome and explore how regulation of the cell cycle, chromatin and transcription factors together elicits specific patterns of embryonic gene expression. Finally, we discuss the interplay between zygotic transcription and maternal clearance, and show how these two activities combine to reprogram two terminally differentiated gametes into a totipotent embryo.
Keywords: Embryogenesis, Pluripotency, Cellular reprogramming, Pioneer factors, Maternal clearance
INTRODUCTION
Embryogenesis begins with a single cell composed of cytoplasm from the egg and DNA from both parents that fuses into a single zygotic nucleus. How a fully formed organism arises, far removed in appearance and function from the zygote, has long been the subject of scientific inquiry. Efforts to understand the principles underlying organismal development were closely tied to experiments in the nineteenth century by Theodor Boveri and others, using sea urchin embryos to investigate the relationship between cellular components and heredity (Laubichler and Davidson, 2008). Cross-fertilization between gametes of different species were found to yield larvae with intermediate characteristics of both parents, suggesting genetic determinants were encoded in the nuclear material contributed by the sperm. However, a range of hybrid characteristics was also observed in crosses using mechanically produced anucleate eggs, implying that some degree of genetic contribution was also conferred by the maternal cytoplasm (Laubichler and Davidson, 2008).
These observations have laid the groundwork for our current understanding of embryonic development, a process subject to both cytoplasmic and nuclear control. Initially, the embryo is transcriptionally quiescent, and development is directed exclusively by maternally provided proteins and RNAs from the egg cytoplasm. Subsequently, developmental control switches to the products of an activated nuclear genome, during a period called the maternal-to-zygotic transition (MZT).
The MZT encompasses two major molecular activities, which together "reprogram" the terminally differentiated oocyte and sperm into a totipotent embryo and beyond. One is maternal clearance, the deletion of maternal instructions -- mRNA and proteins -- that are necessary for oocyte maturation, homeostasis and the first stages of embryogenesis, but become unnecessary or deleterious as the embryo develops. The other is the installation of new zygotic instructions through gene expression, a process that is activated by the maternal program and is called zygotic genome activation (ZGA). Together these two activities dramatically remodel the embryonic gene expression landscape and cellular identities, a process that will be revisited throughout development and adulthood as cells differentiate and regenerate.
Recent reviews have provided extensive treatments of the mechanisms that regulate maternal clearance (Barckmann and Simonelig, 2013; Walser and Lipshitz, 2011). Here, we focus on the activation of zygotic gene expression. In the first section, we highlight recent advances in measuring the onset of zygotic transcription in animals, and present them in the context of seminal discoveries in the genetic control of embryogenesis. Next, we explore the mechanisms that drive ZGA and describe the interplay between the cell cycle, chromatin and transcription factors in regulating embryonic gene expression. Finally we discuss the functional consequences of ZGA, and show that maternal clearance and zygotic transcription are intimately linked and combine to give rise to a reprogrammed embryo.
MEASURING ZYGOTIC GENE EXPRESSION
The developmental context of ZGA
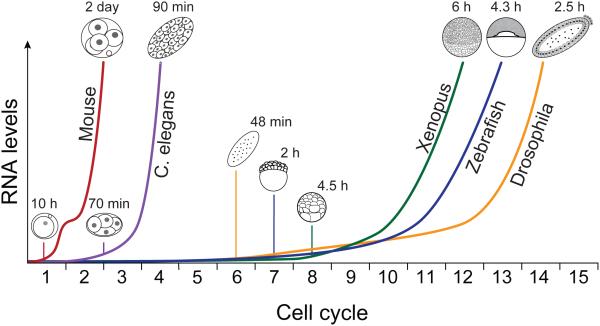
In most animals, the maternal contribution directs a series of synchronized mitoses while maintaining a relatively constant volume, as it forms a blastula. Subsequently, coordinated cell movements during gastrulation form distinct germ layers that specify the various tissues in the mature organism. Within this framework of embryogenesis, there is extensive variation in the timing and duration of these events among different species. The initial cell cycles of Xenopus (35 minutes), zebrafish (15 minutes), and Drosophila melanogaster (8 minutes) are synchronized and proceed in rapid succession, and gastrulation occurs within hours of fertilization (Foe and Alberts, 1983; Gerhart, 1980; Kane and Kimmel, 1993), while asymmetric divisions in Caenorhabditis elegans lead to a 28-cell gastrula after 100 minutes (Figure 1) (Sulston et al., 1983). In contrast, mouse and other mammals have relatively long cell cycles, with the first cleavage occurring about one day after fertilization (Figure 1) and gastrulation 5-6 days later (Oron and Ivanova, 2012).
Figure 1. Timing of zygotic genome activation across various model organisms.
Curves illustrate cumulative increases in zygotic gene expression as development progresses. In mouse, a minor wave of transcription during the first cell cycle is followed by a second wave during cycle 2. C. elegans divisions are asynchronous, with transcription detected by 4 cells. Zygotic transcription in Xenopus, zebrafish and D. melanogaster is detected several cell cycles later, and increases rapidly. Approximate times post fertilization are indicated.
The requirement for zygotic transcription for embryogenesis to proceed is universal across animals. Upon transcriptional inhibition, zebrafish and Xenopus embryos will continue to divide, but fail to undergo gastrulation (Kane et al., 1996; Newport and Kirschner, 1982a). Similarly, the C. elegans embryo experiences extreme morphological defects without zygotic transcription, despite reaching 100 cells before arresting (Edgar et al., 1994), and D. melanogaster, which does not complete cytokinesis for the first 13 cell cycles, requires zygotic transcripts for cellularization to occur (Edgar et al., 1986; Merrill et al., 1988). In mouse, development proceeds no further than the second mitosis, commonly referred to as the 2-cell block (Goddard and Pratt, 1983; Golbus et al., 1973; Warner et al., 1974). In each of these organisms, ZGA occurs well before these defects arise, suggesting that zygotic transcription does not merely coincide with a requirement to replenish RNAs for homeostasis, but is also essential to direct new developmental programs. Discovering the identity of these early zygotic RNAs is thus essential to understanding how development proceeds.
Distinguishing de novo zygotic transcription from the maternal contribution
From the earliest studies of embryonic RNA content, it was clear that maternally deposited RNA molecules greatly outnumber zygotic transcripts -- between 40% to 75% of all protein-coding genes across various species (Tadros and Lipshitz, 2009; Wang et al., 2004; Wei et al., 2006), and still amounting to ~60 to 70% of mRNA molecules at the peak of zygotic expression in zebrafish, for example (Lee et al., 2013) (Figure 2a). This large maternal contribution presents a challenge for detecting transcriptionally active genes, especially if maternal transcript copies greatly outnumber the zygotic contribution, or degradation of maternal copies occurs concurrently with de novo transcription, effectively canceling out the signal. This, coupled with dynamic regulation of maternal RNAs, including changes in poly(A) tail length, highlights the need to distinguish the maternal and zygotic contributions to the transcriptome pool.
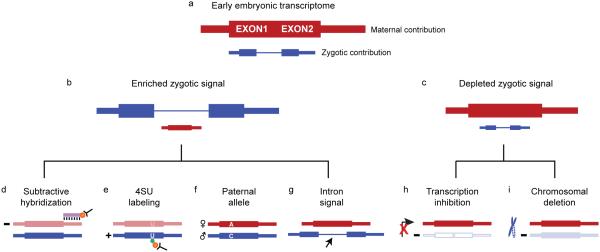
Figure 2. Identification of de novo zygotic transcription.
(a) In the early embryo, the maternal contribution (red) represents the majority of the transcripts present. This can mask the relatively small amount of de novo zygotic transcription (blue) and lead to detection difficulties. Experimental techniques that (b) enrich or (c) deplete the signal from zygotic RNAs relative to the maternal contribution can be used to identify the genes that are de novo activated. (d) Subtractive hybridization employs biotinylated (orange) antisense oligos constructed from oocyte cDNA libraries (purple) to selectively deplete complementary maternal transcripts. (e) De novo transcribed mRNAs can be labeled using the nucleoside analog 4-thiouridine (4SU) (green) and pulled down by biotinylation (orange). (f-g) Zygotic transcripts can be identified by unique properties present only in the zygotic form such as the presence of (f) the paternal genotype or (g) introns from pre-mRNAs, which should not be present in mature maternal mRNAs. (h) Transcription can be globally inhibited using chemicals such as a-amanitin. (i) Chromosomal deletion mutants remove the zygotic contribution for genes that fall within the deletion and can be used to identify genes that require both maternal and zygotic expression to reach wild type levels.
Microarrays and high-throughput sequencing (RNA-Seq) using Illumina and SOLiD technologies (Table 1) have greatly enhanced our ability to dissect the maternal and zygotic RNA populations. Time-course experiments have revealed transcriptome-wide changes in gene expression during the MZT in many different species (Aanes et al., 2011; Dobson et al., 2004; Hamatani et al., 2004; Harvey et al., 2013; Heyn et al., 2014; Lee et al., 2013; Paranjpe et al., 2013; Sirard et al., 2005; Tan et al., 2013; Vassena et al., 2011; Wang et al., 2004; Wei et al., 2006; Xie et al., 2010; Xue et al., 2013; Zhang et al., 2009); however, attributing these changes to active zygotic transcription can still be a challenge. Explicit techniques that distinguish bona fide zygotic gene expression from post-transcriptional regulation of the maternal contribution are thus invaluable for understanding the dynamics and extent of ZGA.
Table 1.
High-throughput technologies used to interrogate the embryonic transcriptome and genome
| Technique | Molecular target |
Measures | Examples |
|---|---|---|---|
| RNA-Seq, poly(A)+ | mRNA | Expression levels of polyadenylated (mature) mRNA. |
(Aanes et al., 2011) |
| RNA-Seq, total RNA | RNA | Expression levels of all RNAs (e.g., pre- mRNAs, introns). Typically performed in combination with ribosomal RNA depletion. |
(Lee et al., 2013; Paranjpe et al., 2013) |
| Ribosome profiling | coding mRNA |
Translation efficiency of mRNAs based on sequencing of protected footprints of active ribosomes. |
(Bazzini et al., 2012; Lee et al., 2013) |
| CAGE-Seq | mRNA | Gene promoter / TSS based on sequencing of mRNA fragments at the 5’ m7G cap. |
(Haberle et al., 2014) |
| ChIP-Seq | DNA | Chromatin binding sites for specific proteins of interest, e.g. modified histones or transcription factors. |
(Akkers et al., 2009; Harrison et al., 2011) |
| DNase-Seq | DNA | Chromatin binding sites for all proteins based on digestion of free DNA (DNase I hypersensitivity). |
(Harrison et al., 2011; Thomas et al., 2011) |
| MNase-Seq | DNA | Nucleosome positions on chromatin, based on digestion of surrounding DNA with MNase. |
(Zhang et al., 2014) |
| Bisulfite-Seq | DNA | Methylated and unmethylated cytosines in the genome, by assaying conversion to uracil by bisulfite treatment. |
(Jiang et al., 2013; Potok et al., 2013) |
Four general approaches have been used to more accurately assess the scope of ZGA, by emphasizing (Figure 2b) or removing (Figure 2c) the signal from zygotic transcripts. First, the maternal contribution can be depleted using subtractive hybridization techniques (Figure 2d). Zygotic samples hybridized to biotinylated oocyte cDNA can yield libraries where maternally contributed transcripts are underrepresented (Rothstein et al., 1992; Sive et al., 1988). This method has been effective for detecting rare zygotic transcripts from limited amounts of RNA (Zeng and Schultz, 2003), but less so for accurate measurement of genome-wide transcript levels.
Second, the zygotic contribution can be depleted (Figure 2c). Chemical treatments, such as a-amanitin and actinomycin D (Table 2), applied early during embryogenesis globally inhibit RNA polymerase activity (Figure 2h). Thus, only changes in RNA levels that are sensitive to these treatments can be considered products of zygotic transcription (Edgar and Schubiger, 1986; Edgar et al., 1994; Golbus et al., 1973; Hamatani et al., 2004; Kane et al., 1996; Lee et al., 2013; Newport and Kirschner, 1982a; Vassena et al., 2011). Using this approach in combination with microarrays, Hamatani, et al., were able to accurately measure the scope of distinct minor and major periods of zygotic transcription during mouse embryogenesis, and showed that apparent expression level increases in the early 1-cell stage were not due to transcription (Hamatani et al., 2004).
Table 2.
Chemical treatments used to study embryogenesis and ZGA
| Treatment | Target of inhibition | Examples |
|---|---|---|
| Actinomycin D | Transcription (Pol I, II, III) | (Golbus et al., 1973) |
| α-Amanitin | Transcription (Pol II, III) | (Golbus et al., 1973; Hamatani et al., 2004; Newport and Kirschner, 1982a) |
| Aphidicolin | DNA replication | (Aoki et al., 1997; Wiekowski et al., 1991) |
| 5-Azadeoxycytidine | DNA methylation | (Potok et al., 2013) |
| Butyrate | Histone deacetylases | (Adenot et al., 1997; Wiekowski et al., 1993) |
| Cordycepin (3′- deoxyadenosine) |
mRNA polyadenylation | (Aanes et al., 2011; Aoki et al., 2003) |
| Cycloheximide | Translation; cell cycle | (Edgar and Schubiger, 1986; Harvey et al., 2013; Kimelman et al., 1987; Lee et al., 2013) |
| Cytochalasins | Actin polymerization; cytokinesis | (Davis et al., 1996; Newport and Kirschner, 1982a) |
| Nocodazole | Microtubule polymerization; mitosis | (Kimelman et al., 1987) |
| Trichostatin A (TSA) | Histone deacetylases | (Adenot et al., 1997; Bultman et al., 2006), |
| U1 and U2 antisense morpholinos |
Spliceosome | (Lee et al., 2013) |
Selective loss of the zygotic contribution was achieved in D. melanogaster and C. elegans by measuring transcription in mutants with large chromosomal deletions (Figure 2i) (De Renzis et al., 2007; Merrill et al., 1988; Storfer-Glazer and Wood, 1994). Systematic removal of entire chromosomal arms revealed many maternally provided genes whose levels decreased when their genomic region was absent, thus demonstrating that their wild type levels are augmented by zygotically transcribed copies (De Renzis et al., 2007). In all, 20% of the D. melanogaster zygotic genome was found to contribute to embryonic development, two-thirds of which have a maternal contribution (De Renzis et al., 2007). These analyses helped to reveal shared mechanisms of activation, that involve concomitant maternal clearance activity as well as transcriptional regulation via shared cis regulatory elements, both of which are further discussed below (De Renzis et al., 2007).
Zygotic genes can also be distinguished according to how they are regulated. Genes that are directly activated by the maternal contribution can be seen as a “first wave” of transcription, compared to “subsequent wave” genes that depend on factors encoded by the zygotic genome for their expression. In zebrafish, inhibitors of zygotic protein activity were used to isolate the first-wave genes. Cycloheximide applied prior to ZGA to inhibit translation of zygotic mRNAs (Harvey et al., 2013; Lee et al., 2013), as well as antisense inactivating morpholinos directed against spliceosomal RNAs to inhibit splicing and maturation of zygotic transcripts (Table 2) (Lee et al., 2013), led to the identification of 269 genes directly activated by maternal factors, constituting the first layer of zygotic gene transcription.
Third, the zygotic contribution can be labeled using modified ribonucleotides. Early on, radioactive nucleotide incorporation using [3]H or [32]P was widely used to measure zygotic genome activity. Newly produced radiolabeled RNA could be detected as early as the first two divisions in the sea urchin embryo (Nemer, 1963; Poccia et al., 1985), during late cleavage stages in Xenopus (Brown and Littna, 1964; Newport and Kirschner, 1982a), blastoderm stage (2hpf) in D. melanogaster (Edgar and Schubiger, 1986; Zalokar, 1976) and 2-cell stage in mouse (Knowland and Graham, 1972). Using BrUTP, transcription was detected even earlier, mainly deriving from the male pronucleus in the 1-cell mouse embryo (Aoki et al., 1997; Bouniol et al., 1995). These analyses revealed general characteristics of the first zygotic genes based on molecular weight, which include heterogenous mRNAs, as well as ribosomal RNA (Edgar and Schubiger, 1986) and small RNAs transcribed by RNA Polymerase III (Knowland and Graham, 1972; Newport and Kirschner, 1982a). However, determining the individual identity of transcribed genes using these techniques was a challenge.
Nucleoside analogs such as 4-thiouridine (4SU) are more amenable to recovering gene identity. Heyn, et al., (Heyn et al., 2014) injected 4SU into 1-cell zebrafish embryos, which was incorporated into new transcripts and selectively biotinylated at the thiol moieties (Figure 2e). Pull down with Illumina sequencing revealed enrichment of 592 genes transcribed in the early blastula, when cell cycle progression is still rapid. Accordingly, these genes are short, presumably in order for transcription to transpire in the limited period between rapid cell divisions (Heyn et al., 2014), a property that was also found in early D. melanogaster genes (De Renzis et al., 2007; Rothe et al., 1992). In addition, transcription by mitochondrial RNA polymerase was detected immediately after fertilization, indicating that the mitochondrial genome does not have a quiescent period, a result that echoes observations of cytoplasmic RNA synthesis in early sea urchin embryos (Craig, 1970; Selvig et al., 1970).
Finally, sensitive RNA-Seq techniques can detect features of zygotic transcripts that distinguish them from the maternal contribution. In most cases, de novo transcription is expected to arise from both the maternal and paternal alleles, while the maternal contribution should consist of only maternal alleles. Assuming minimal or no RNA contribution from sperm, the appearance of single-nucleotide polymorphisms (SNPs) specific to the paternal genome can be used to gauge the activity of the zygotic genome (Figure 2f) (Sawicki et al., 1981). Using this approach, Harvey et al. (Harvey et al., 2013) performed RNA-seq on embryos generated from two different zebrafish strains in parallel with whole-exome sequencing on the parental genomes. They found that 61% of expressed genes with informative SNPs had transcripts bearing both maternal and paternal genotypes, showing that a large proportion of the embryonic transcriptome has both a maternal and zygotic contribution. Similar approaches were used to identify genes transcribed in early D. melanogaster embryos (Ali-Murthy et al., 2013), and to measure parent-of-origin gene expression by following the maternal and paternal genotypes in single-cell embryonic transcriptomes (Xue et al., 2013) -- 39% of polymorphic genes in the human 8-cell embryo displayed biallelic expression, which increased to 70% by morula stage (Xue et al., 2013).
Because many genes do not contain informative polymorphisms, Lee and Bonneau, et al. (Lee et al., 2013), leveraged the capacity to sequence unspliced pre-mRNA molecules in ribosomal RNA-depleted total RNA libraries, as an alternative to traditional poly(A)+ selected libraries (Table 1), which enrich for mature mRNAs (Figure 2g). Using intron sequence coverage as an unambiguous signal to distinguish de novo zygotic transcription, they were able to identify >7000 transcribed genes from the active zygotic genome in the late blastula, 74% of which had a maternal contribution that had previously obscured detection using exonic signal alone, due to low levels of expression (Lee et al., 2013). Thus, the scale of ZGA in zebrafish now appears to be at least an order of magnitude larger than has previously been reported, involving not only embryo-specific genes, but also a large fraction of genes that were already represented in the maternal contribution. (Harvey et al., 2013; Lee et al., 2013).
Comparisons between total and poly(A)+ sequencing libraries can also help distinguish de novo transcribed and polyadenylated RNAs from maternal RNAs that have been subject to post-transcriptional regulation of the poly(A) tail. New zygotic expression is well correlated between the two sequencing strategies (Lee et al., 2013; Paranjpe et al., 2013). In contrast, maternal mRNAs with short poly(A) tails are less efficiently sequenced using poly(A)+ selection, resulting in apparent elevated expression levels when compared with total RNA preparations (Paranjpe et al., 2013). In this way, Paranjpe, et al., were able to detect cytoplasmic polyadenylation of maternal mRNAs in Xenopus cycle 6 embryos and distinguish it from de novo zygotic transcription at a later stage, based on the asymmetry of total and poly(A)+ sequencing signals (Paranjpe et al., 2013).
Together, these approaches have progressively uncovered earlier and more extensive activation of the zygotic genome, and have identified the first genes that are transcribed during the MZT, providing insight into their mechanisms of co-activation.
Dynamics of activation
The analyses described above have revealed extensive variability in the timing and dynamics of zygotic gene expression. Among vertebrates, mouse embryos experience the earliest ZGA, with respect to cell cycle count (Aoki et al., 1997; Bouniol et al., 1995; Hamatani et al., 2004; Park et al., 2013; Xue et al., 2013). The punctuated bursts of zygotic transcription at 1-cell and 2-cell stages constitute a minor and major wave of ZGA (Figure 1) (Hamatani et al., 2004), now thought to involve as many as ~800 and ~3500 genes, respectively (Park et al., 2013; Xue et al., 2013). These two waves of ZGA are in line with a global shift in chromatin organization spanning the first cleavage (see below). Other mammals also seem to experience minor and major waves, with a trend toward slightly delayed expression patterns when compared to mouse (Dobson et al., 2004; Vassena et al., 2011; Xue et al., 2013; Zhang et al., 2009), though there is now evidence that human embryos are also transcriptionally active by 1-cell stage (Xue et al., 2013), prior to the major wave at 4-8 cell stages (Braude et al., 1988; Dobson et al., 2004; Vassena et al., 2011; Zhang et al., 2009).
In other vertebrates, peak zygotic genome activity occurs after several cell cycles have elapsed, but from an absolute time perspective this often occurs much sooner than in mammals. Lower levels of de novo transcribed genes appear earlier, though whether this constitutes a distinct early phase of activation, rather than a gradual ramping up of transcription, is unclear. In Xenopus embryos, high levels of transcription are observed at the mid-blastula transition (MBT) (Box 1), approximately 6-7 hours and 12 cell cycles into development (Newport and Kirschner, 1982a; Paranjpe et al., 2013; Tan et al., 2013), with early transcripts appearing between cycles 6 and 9 (Figure 1) (Blythe et al., 2010; Paranjpe et al., 2013; Yang et al., 2002). In zebrafish, multiple recent analyses place the onset of transcription after the 64-cell stage (cycle 7) (Aanes et al., 2011; Harvey et al., 2013; Heyn et al., 2014; Lee et al., 2013), starting with several hundred transcripts (Heyn et al., 2014) and increasing to several thousand prior to gastrulation (Figure 1) (Harvey et al., 2013; Lee et al., 2013). Previous observations of an earlier wave of activation at 4-cell stage (Mathavan et al., 2005) were likely due to cytoplasmic polyadenylation and not zygotic transcription (Harvey et al., 2013).
BOX 1.
The mid-blastula transition (MBT) was originally defined with respect to amphibian development, and refers to the approximate midpoint of the blastula stage, after 12 cleavages in Xenopus (Gerhart, 1980; Newport and Kirschner, 1982a). At this stage, the cell cycle lengthens and becomes asynchronous, cells gain motility, and zygotic transcription is active and required, thus marking the time when nuclear control of the embryo begins. In this way, the MBT was the morphological embodiment of the MZT in Xenopus, as well as in other species such as zebrafish and D. melanogaster (Blankenship et al., 2001; Kane et al., 1993). However, given that zygotic genome activity precedes the MBT in many organisms -- including Xenopus (Skirkanich et al., 2011; Yang et al., 2002), but most notably mouse, which is already transcriptionally active at the 1-cell stage (Aoki et al., 1997; Bouniol et al., 1995; Hamatani et al., 2004b; Park et al., 2013; Xue et al., 2013) -- the MBT may not be such a widely applicable concept with respect to ZGA (Yasuda and Schubiger, 1992).
The patterns in invertebrates are even more variable. In D. melanogaster, the bulk of zygotic transcription does not occur until the long cell cycle pause that accompanies cellularization (Benoit et al., 2009; De Renzis et al., 2007; Foe and Alberts, 1983; Lécuyer et al., 2007), though transcription begins as early as cycle 6 (Figure 1) (Ali-Murthy et al., 2013; Karr et al., 1985). The expression dynamics leading up to cellularization remain to be resolved in a high-throughput manner. Among nematodes, C. elegans transcription levels increase steadily from 4-cell stage until gastrulation, all within ~1.5 hours (Figure 1) (Baugh et al., 2003; Edgar et al., 1994). This is in stark contrast to recent findings in the parasitic worm Ascaris suum, which remains at 1-cell stage for 36 hours and already has an active zygotic genome from fertilization, transcribing ~2500 genes deriving from both pronuclei prior to fusion (Wang et al., 2014). Finally, sea urchin embryos also seem to be transcriptionally active at 1-cell (Poccia et al., 1985), reaching a peak in the early blastula after 15 hours (Wei et al., 2006).
These variable patterns reveal that multiple, diverse mechanisms are in place to regulate ZGA. Activation is widespread, though not ubiquitous across the genome, suggesting that one component of ZGA may be a global attainment of genome competency, but with additional layers of regulation to account for gene-specific expression timing and magnitude. We explore many of these potential mechanisms in the following section.
MECHANISMS OF GENOME ACTIVATION
General models of activation
Traditionally, two contrasting models of activation have been the focus of research on the ZGA. On the one hand, the "nucleocytoplasmic (N/C) ratio" model posits that the increasing quantity of nuclear material relative to a constant cytoplasm volume, through progressive cell divisions, alleviates transcriptional repression (Newport and Kirschner, 1982a, b). Under this formulation, the barrier to ZGA is maternally provided factors, whose relative levels must be diminished before transcription can occur (Figure 3a).
Figure 3. General models for ZGA.
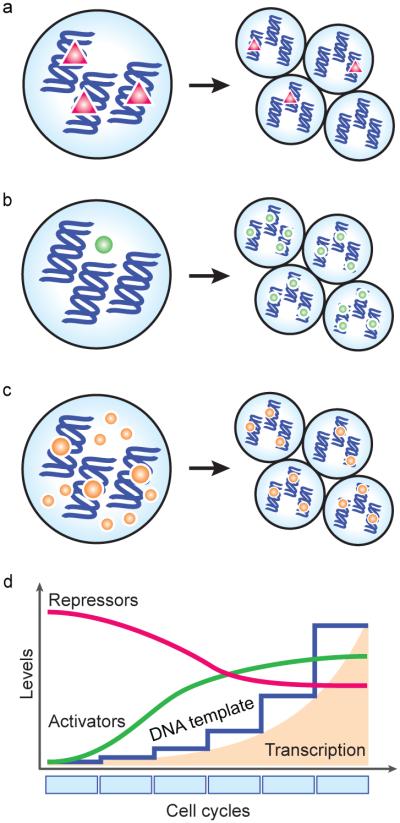
(a-c) Schematics illustrating the relationship between maternal factors and the zygotic genome as cells divide. (a) Early during embryogenesis, a maternal repressor (red triangles) prevents zygotic transcription. After several divisions, the repressor is titrated away as the ratio of nuclear material to cytoplasm increases (the nucleocytoplasmic ratio), thus allowing transcription to initiate. (b) Developmental time may be necessary for sufficient levels of an activator to accumulate, e.g., resulting from activated translation of a maternal mRNA. (c) Alternatively, the embryo may already possess transcriptional activators (orange circles) and be transcriptionally competent early, but a threshold amount of DNA template is required for transcription to be detected. (d) All of these general mechanisms could combine to elicit increasing transcription levels over developmental time.
On the other hand, a "maternal clock" independent of the number of cell divisions may determine the timing of gene expression (Howe et al., 1995). The molecular instantiation of this model can be seen as an increase in quantity or activity of a maternal factor, which must reach a critical level in order to trigger transcription (Figure 3b). This model is particularly appealing, given the prevalence of cytoplasmic polyadenylation of maternally provided mRNAs (Aanes et al., 2011; Harvey et al., 2013; Richter and Lasko, 2011) and correlated increases in translation efficiency, which can be seen through polysome profiling (Qin et al., 2007) and high-throughput ribosome footprinting (Table 1) (Lee et al., 2013). Thus, maternally provided mRNAs encoding critical components such as transcription factors and chromatin modifiers may be gradually mobilized over time.
These models are not mutually incompatible, or necessarily independent, and there is evidence in D. melanogaster that both modes of regulation exist simultaneously for the activation of roughly equal numbers of genes (Lu et al., 2009). However, it is also important to consider that all of these mechanisms also depend on the relative amount of DNA template available, which increases exponentially after each cell cycle. Reaching a threshold quantity of DNA, as well as allowing sufficient time for transcript numbers to accumulate, is a factor in achieving detectable levels of transcriptional output (Figure 3c,d). The mechanisms that regulate ZGA may in fact act at an earlier time than when the effects can be measured using current techniques.
In the following subsections, rather than focusing on these models, we explore control of ZGA from the perspective of three aspects of the developing embryo: the cell cycle; changes in chromatin structure, histone marks and epigenetic prepatterning; and the activity of transcription factors.
Cell cycle regulation
In Xenopus, zygotic transcriptional activation accompanies the loss of cell cycle synchrony at the MBT (Gerhart, 1980). In investigating the mechanisms that influence the timing of these events, Newport and Kirschner discovered that an increasing N/C ratio controls the MBT and zygotic transcription, rather than the number of cleavages or rounds of DNA synthesis (Newport and Kirschner, 1982a, b).
In a series of experiments that altered the N/C ratio using cleavage inhibitors (Table 2), mechanical constriction of the cytoplasm, induced polyspermy and injections of exogenous nonspecific DNA, they found that transcription could be prematurely activated when the DNA content equaled that found in wild type cycle 13 embryos, independent of cell cycle count (Newport and Kirschner, 1982a, b). From these observations, Newport and Kirschner proposed that titration of some maternally provided repressive factor, relative to an exponentially increasing amount of DNA, ultimately determined the timing of ZGA (Newport and Kirschner, 1982a, b). Such factors could include heterochromatin-promoting histones or transcription inhibitors, both of which are discussed in following subsections.
This role of the N/C ratio was subsequently observed in the zebrafish mutant futile cycle (fue), in which failure of chromosomal segregation leads to polyploid cells with locally higher N/C ratios and premature RNA polymerase II (Pol II) initiation (Dekens et al., 2003). Zygotic transcription in D. melanogaster embryos also depends on an increasing N/C-ratio; however it seemed to strongly affect only a subset of genes, suggesting that other mechanisms are in place to regulate ZGA (Edgar et al., 1986; Lu et al., 2009; Yasuda et al., 1991).
Other evidence now suggests that the N/C ratio affects transcription activation indirectly, through regulation of cell cycle rate. Edgar, et al, found that D. melanogaster mitotic cycle length prior to cellularization slows according to the N/C ratio (Edgar et al., 1986). Recently, the Cdc25 homolog Twine has been implicated in effecting this cell cycle pause: degradation of Twine, possibly in an N/C ratio-dependent manner, stabilizes Cdk1 phosphorylation, thus preventing entry into mitosis and allowing zygotic transcription to occur (Figure 4a) (Di Talia et al., 2013; Farrell and O’Farrell, 2013). However, early zygotic transcription prior to cellularization is in turn required for Twine degradation (Farrell and O’Farrell, 2013; Sung et al., 2013), indicating that this mechanism cannot account for all of ZGA.
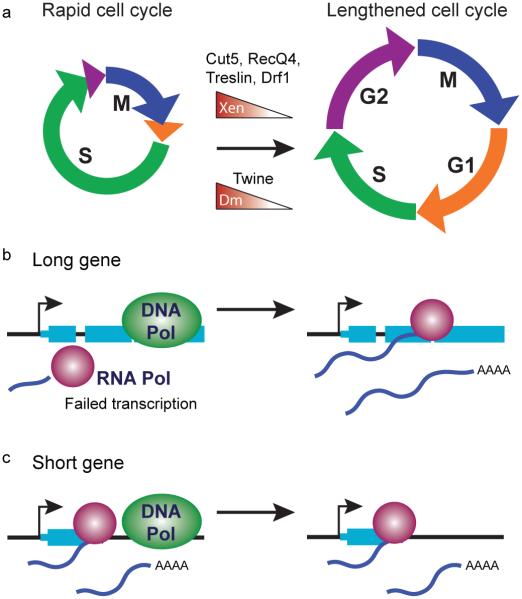
Figure 4. Influence of cell cycle on zygotic gene transcription.
(a) In some species, early cleavage stages progress through mitosis (M) and DNA synthesis (S) phases with little to no gap phases (G1 and G2). As the embryo divides, maternal DNA replication factors -- Cut5, RecQ4, Treslin and Drf1 in Xenopus (Xen), and Twine in D. melanogaster (Dm) -- are titrated away as the nucleocytoplasmic ratio increases, resulting in cell cycle lengthening. (b) Rapid cell cycles may be incompatible with transcription of longer genes, resulting in failed or abortive RNA polymerase engagement. Longer cell cycles would allow more time for elongation and the accumulation of zygotic transcripts. (c) However, rapid cell cycles do not appear to be prohibitive for transcription of short genes.
In Xenopus, the four replication factors Cut5, RecQ4, Treslin and Drf1 have been found to control the cell cycle, and thus guide the onset of zygotic transcription (Collart et al., 2013). Titration of these factors causes DNA replication to slow, leading to the onset of asymmetric cell divisions at the MBT (Figure 4a). Overexpression resulted in an increase in replication initiation and cell cycle count, and as a result delayed the transcription of a large number of genes. Additionally, early activation of the replication checkpoint kinase Chk1 was observed (Collart et al., 2013). Chk1 regulates entry into S and G2 phases via repression of cyclin-dependent kinase activity (reviewed in (Sorensen and Syljuasen, 2012)), thus together these mechanisms may contribute to the lengthening of the cell cycle at the Xenopus MBT and the associated increase in zygotic transcription.
These results indicate that cell cycle length plays a role in at least the later stages of zygotic gene activation. Xenopus, zebrafish and D. melanogaster all experience rapid, synchronous cell cycles during the first cleavages, in contrast to the slow pace of cell cycles in mouse. Congruently, the bulk of zygotic gene activation occurs in the former species only after the cell cycle begins to slow, whereas it has already occurred in the 2-cell mouse embryo. Consistent with this observation, chemical inhibition of either DNA replication or the cell cycle results in precocious expression of zygotic transcripts in Xenopus (Table 2) (Kimelman et al., 1987). Similarly, in D. melanogaster, interphase arrest induces premature transcription at cycle 10; however, earlier treatment inhibits transcription altogether, suggesting that a critical supply of protein activators is also necessary, and that cell cycle length is only permissive for transcription (Edgar and Schubiger, 1986).
Thus, short cell cycles appear to be incompatible with the time it takes to transcribe and process longer genes, which would be prematurely disrupted by the DNA replication machinery (Figure 4b,c). Three lines of evidence support this observation. First, in D. melanogaster, failure to complete transcription during early mitoses has been detected at individual gene loci using in situ nuclear labeling of mRNA 5’ and 3’ ends. Abortive transcription followed by degradation is detected for ultrabithorax, until the G2 pause of cycle 14 (Shermoen et al., 1991). Second, key components of RNA biogenesis are inhibited during mitosis, including polyadenylation and splicing (Colgan et al., 1996; Shin and Manley, 2002). Third, early-transcribed genes tend to be short. Rothe et al. showed that early expression of the gap gene knirps (kni) was possible due to its short 3 kilobase (kb) length (Rothe et al., 1992). In contrast, homologous knirps-related (knrl) is 20kb longer, and was subject to abortive transcription. In an elegant demonstration of the significance of gene length, they were able to rescue the segmentation defects in kni mutants using krnl, but only when expressed as an intronless minigene and not when the 19kb intron was included (Rothe et al., 1992). The trend toward shorter genes with fewer introns appears to be a general feature of the earliest transcripts across different organisms (De Renzis et al., 2007; Heyn et al., 2014; Swinburne and Silver, 2008).
Taken together, cell cycle dynamics appear to define a hierarchy of zygotic gene activation, where transcription of shorter genes is compatible with rapid cell cycles, while expression of longer genes is delayed until the cell cycle lengthens. In this way, the cell cycle length is permissive for ZGA, which implies that additional, instructive mechanisms are required for proper activation to occur.
Chromatin competency
When Newport and Kirschner microinjected a plasmid containing a yeast tRNA gene into fertilized Xenopus eggs, they found that the RNA was immediately synthesized, as soon as 10 minutes after injection, well before endogenous zygotic transcription takes place (Newport and Kirschner, 1982b). Similar results were obtained in mouse (Wiekowski et al., 1993) and zebrafish (Harvey et al., 2013), and together indicate that many components of the transcription complex are already competent in the early vertebrate embryo. However, Newport and Kirschner went on to show that transcription of the plasmid was subsequently repressed, only to resume again in the late blastula (Newport and Kirschner, 1982b). Since co-injecting non-specific DNA was able to rescue expression, presumably by competing away the repression, they hypothesized that the effect was due to assembly of the plasmid DNA into closed chromatin.
Interplay between transcriptional machinery and chromatin in the early embryo likely regulates the timing of ZGA (Prioleau et al., 1994). Chromatin is composed of DNA wound around nucleosomes, octamers of the core histone proteins H2A, H2B, H3 and H4, which are in turn joined together by histone linker proteins, e.g. H1, to form compacted heterochromatin. As transcription factors bind to regulatory regions in the DNA sequence, access to a particular gene locus can be occluded by this closed structure.
Chromatin accessibility is regulated through nucleosome position and configuration, which is influenced by histone variants as well as post-transcriptional modifications of histone N-terminal tails (“marks”), such as methylation and acetylation. In mouse, hyper-accessible chromatin seems to underlie ZGA (CHO et al., 2002). Prior to fusion into the zygotic nucleus, the uncondensed male pronucleus is transcriptionally competent, and low levels of endogenous transcription during mid to late S phase contribute to the minor wave of ZGA (Aoki et al., 1997; Bouniol et al., 1995; Park et al., 2013; Ram and Schultz, 1993; Wiekowski et al., 1993; Xue et al., 2013). In contrast, the female pronucleus remains transcriptionally silent (Wiekowski et al., 1993). This asymmetry is likely due to a transient open chromatin state induced by the repackaging of the paternal genome. Sperm DNA is bound by arginine-rich protamines, which are exchanged for maternal histones prior to S phase (Nonchev and Tsanev, 1990). These new histones are subject to a transcriptionally permissive pattern of modifications, including H4 hyperacetylation (Adenot et al., 1997; van der Heijden et al., 2006) and H3K9 and H3K27 monomethylation (Santos et al., 2005). Protamines, interspersed with paternal histones, are also found in human, Xenopus, and D. melanogaster sperm DNA (Hammoud et al., 2009; Jayaramaiah Raja and Renkawitz-Pohl, 2005; Shechter et al., 2009), but surprisingly not in zebrafish (Wu et al., 2011).
Histone exchange is a general mechanism during embryogenesis, as gamete-specific variants are replaced by somatic versions. This process could mediate gradual nucleosome unpacking prior to ZGA, as maternal histones are diluted in favor of permissive zygotic versions (Figure 5a). The repressive H2A variant macroH2A is found preferentially in the mouse female pronucleus, and appears to contribute to its transcriptional silence; macroH2A is progressively lost as the embryo becomes transcriptionally active (Chang et al., 2005). In contrast, embryonic H2A.Z is required for development (Faast et al., 2001; Whittle et al., 2008). In C. elegans, H2A.Z (HTZ-1) was found to be translated from maternal mRNAs and incorporated proximal to developmentally critical genes, suggesting that it influences expression specificity (Whittle et al., 2008). H3.3 incorporation is required for male pronuclear formation in D. melanogaster (Torres-Padilla et al., 2006), while H3.3 knockdown in mouse induces chromosomal over condensation by 2-cell stage and impaired zygotic transcription in the morula (Lin et al., 2013). Finally, repressive oocyte-specific H1 linker histone variants are replaced by somatic versions coincidentally with the alleviation of transcriptional quiescence. (Fu et al., 2003; Perez-Montero et al., 2013; Smith et al., 1988; Tanaka et al., 2001). In D. melanogaster, loss of embryonic H1 variant dBigH1 leads to premature Pol II activity and gene expression (Perez-Montero et al., 2013), showing that one barrier to ZGA is the early prevalence of higher-order chromatin.
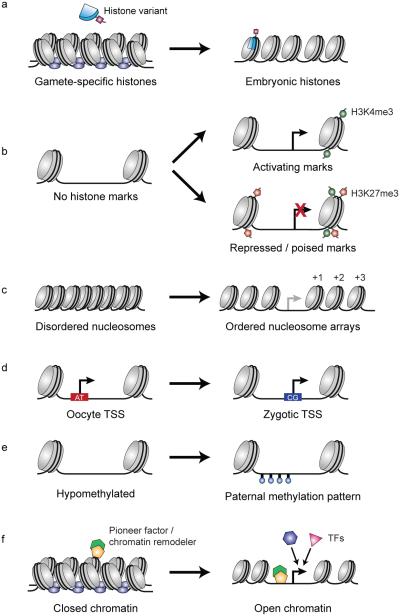
Figure 5. Modes of chromatin regulation and establishing competency for zygotic gene activation.
(a) Gamete-specific repressive histone variants are replaced with zygotic variants after fertilization, which often bear permissive modifications. (b) Selective post-transcriptional modifications of histone tails are acquired during the MZT, including activating H3K4me3 and repressing H3K27me3 over gene promoters. Some promoters are marked bivalently (both H3K4me3 and H3K27me3), indicating a poised transcriptional state. (c) The organization of nucleosomes around gene promoters is associated with transcriptional competence. Ordered arrays of nucleosomes (labeled +1, +2, +3) form during ZGA around both active promoters and promoters driving later developmental expression. (d) Transcription start site (TSS) utilization on embryonic genes shifts from oocyte-specific A/T-rich regions to G/C-rich sequences. (e) DNA methylation, which is associated with repression, is selectively applied during the MZT in a manner that matches the sperm methylation pattern. (f) Widespread compacted chromatin in the early embryo is thought to be prohibitive for transcription. Maternal pioneer transcription factors may bind closed chromatin in a sequence-specific manner and induce open chromatin through recruitment of chromatin remodelers, allowing access to gene promoters.
Thus, a globally permissive chromatin conformation is a prerequisite for ZGA, which is shaped in part by dynamic incorporation of embryonic histone variants. However, the specificity of activation -- i.e., the genes that are eventually transcribed from the competent genome -- likely requires local changes to chromatin accessibility. In the following two subsections, we explore the roles that histone marks and epigenetic prepatterning may play in guiding these changes.
Histone modifications
Two types of histone modifications have been implicated in shaping gene expression during the MZT: lysine acetylation and lysine (tri)methylation. In mouse, permissive H4 acetylation distinguishes the transcriptionally active male pronucleus from the silent female pronucleus (Adenot et al., 1997; van der Heijden et al., 2006). Following the minor wave of ZGA, histone deacetylase (HDAC) activity contributes to a transient period of transcriptional repression, such that injected plasmids as well as endogenous genes that are normally expressed at 1-cell stage are not readily transcribed in the 2-cell embryo (Martinez-Salas et al., 1989; Wiekowski et al., 1991). Inhibition of HDACs (Davis et al., 1996) or DNA synthesis (Table 2) (Christians et al., 1995) relieves this repression, suggesting that replication-dependent deacetylation provides transcriptional specificity for the major wave of ZGA. Widespread H4 deacetylation leading up to the MBT has also been observed in Xenopus (Dimitrov et al., 1993), consistent with the creation of a “default off” transcriptional state coupled with gene-specific regulation of chromatin accessibility. Chromatin remodelers that induce or respond to acetylation are likely involved in this process. Two maternally provided components of the ATP-dependent chromatin remodeling SWI/SNF complex, Brg1 and SRG3, are required for mouse embryogenesis (Bultman et al., 2006; Sun et al., 2007), with loss of Brg1 resulting in reduced levels of 30% of zygotic genes and arrest at 2-cell (Bultman et al., 2006).
H3 lysine methylation also influences the timing of gene activation during the MZT (Akkers et al., 2009; Chen et al., 2013; Lindeman et al., 2011; Schuettengruber et al., 2009; Vastenhouw et al., 2010), though their effects vary widely across different species and contexts. The opposing marks H3K4 and H3K27 trimethylation (H3K4me3 and H3K27me3) at gene promoter regions have received considerable attention, for their association with activate and repressed gene expression, respectively (Figure 5b). In mouse, the transcriptionally inactive female pronucleus is associated with H3K27me3, with the transcriptionally competent male pronucleus only acquiring H3K27me3 toward the end of the minor ZGA (Santos et al., 2005). However, activating H3K4me3 is also found preferentially in the female pronucleus despite its transcriptional quiescence, and this asymmetry rapidly diminishes as the male pronucleus incorporates maternal H3 histones (Lepikhov and Walter, 2004). Thus, H3K27me3, but not H3K4me3, seems to affect early mouse gene activity.
In other organisms where large numbers of embryos are more readily available, high-throughput chromatin immunoprecipitation (ChIP) assays (Table 1) show greater association of H3K4me3 with transcription, but its role in guiding ZGA is not straightforward. In D. melanogaster, H3K4me3 becomes strongly enriched in zygotic-transcribed genes that have a maternal contribution (Chen et al., 2013; Schuettengruber et al., 2009), but only later in development. Neither H3K4me3 nor H3K27me3 is present prior to cellularization, even among early expressed genes (Chen et al., 2013).
For both zebrafish and Xenopus, H3K4me3 is found across embryonic gene promoters, with a preference for those with housekeeping roles expressed soon after ZGA (Akkers et al., 2009; Lindeman et al., 2011; Vastenhouw et al., 2010). In contrast, H3K27me3 preferentially associates with genes encoding specific developmental functions (Lindeman et al., 2011; Vastenhouw et al., 2010) and subject to differential spatial regulation (Akkers et al., 2009). Thus, in these species, H3K4me3 and H3K27me3 appear to distinguish earlier versus later zygotic transcription.
However, H3K4me3 is also found on inactive genes in zebrafish embryos, based on lack of H3K36me3 signal (which marks Pol II elongation) or evidence of transcription in RNA-Seq experiments (Lindeman et al., 2011; Vastenhouw et al., 2010). These genes do tend to be expressed at later stages, suggesting that H3K4 trimethylation also marks promoters that are poised for rapid mobilization in the appropriate developmental context (Lindeman et al., 2011; Vastenhouw et al., 2010). In addition, many of these promoters are also simultaneously occupied by inactivating H3K27me3 (Figure 5b) (Vastenhouw et al., 2010), reminiscent of so-called "bivalent" domains that have been described in embryonic stem (ES) cells. In ES cells, bivalent marks are associated with genes with imminent roles in differentiated lineages, but whose expression is repressed during pluripotency (Bernstein et al., 2006). Analogously, during zebrafish embryogenesis, bivalent marks are found in the promoters of lineage-specific genes that are not expressed immediately at ZGA (Lindeman et al., 2011; Vastenhouw et al., 2010) and could allow for rapid gene activation at the appropriate time. Xenopus embryos also have genes that are marked by both H3K4me3 and H3K27me3, but in contrast to zebrafish, the marks do not co-exist in the same cell, and appear to be providing temporal-spatial specificity of expression rather than bivalency (Akkers et al., 2009).
But still other genes marked by H3K4me3 are not transcribed at all (at detectable levels), and eventually lose the mark during gastrulation (Lindeman et al., 2011). So perhaps H3K4me3 has a broader role that extends beyond the immediate transcriptional needs at ZGA, and is instead involved in a larger-scale remodeling of embryonic chromatin to a non-oocyte state. In support of this model, a recent study measuring genome-wide nucleosome positioning, using micrococcal nuclease (MNase) digestion (Table 1), found distinct patterns of occupancy on gene promoters marked by H3K4me3 (Zhang et al., 2014). At ZGA in zebrafish, well-ordered arrays of nucleosomes form, precisely positioned at the transcription start site (TSS) of genes, and independently of active transcription or Pol II binding (Figure 5c) (Zhang et al., 2014).
These results together show that histone modifications inform the timing and spatial specificity of gene expression during the MZT, which is correlated with localized changes to chromatin conformation at zygotic genes. How this specificity is established remains largely unknown, though evidence suggests that epigenetic inheritance from the gametes plays a role.
Epigenetic prepatterning
Epigenetic features from both egg and the sperm chromatin appear to strongly influence the patterns of chromatin modifications acquired in the embryo. In mouse oocytes, Polycomb activity was shown to be required both for correct specification of histone marks in the embryo, as well as shaping the maternal contribution (Posfai et al., 2012; Puschendorf et al., 2008). The Polycomb Repressive Complexes 1 and 2 (PRC1 and PRC2) together apply and maintain H3K27 trimethylation and repress transcription (Simon and Kingston, 2009). Mouse embryos lacking the maternal contribution of Ring1 and Rnf2, two core components of PRC1, arrest at 2-cell stage and experience aberrant gene expression at ZGA (Posfai et al., 2012). Similarly, Mll2 activity in the oocyte is required to establish H3K4me3 patterns and normal ZGA expression, though other histone methyltransferases are likely involved (Andreu-Vieyra et al., 2010).
In both mouse and human sperm DNA, a small number of modified histones are present among the protamines, which are transmitted to the male pronucleus (Brykczynska et al., 2010; Hammoud et al., 2009). H3K4me2, H3K4me3 and H3K27me3 are found at developmental promoters in sperm, with the latter occurring preferentially at genes that are initially repressed in the embryo (Brykczynska et al., 2010; Hammoud et al., 2009). The chromatin in zebrafish sperm has also been reported to contain both H3K4me3 and H3K27me3, along with several other activating and repressing marks organized in large multivalent chromosomal regions that contain genes active late in embryogenesis (Wu et al., 2011).
However, early embryos appear to be completely devoid of distinguishing histone marks (Vastenhouw et al., 2010; Zhang et al., 2014), suggesting that these sperm modifications are initially lost, but then reapplied by a mechanism that retains epigenetic memory. De novo nucleosome repositioning and histone marks may be guided by intrinsic signals encoded by the DNA sequence. In zebrafish, a global switch in promoter usage occurs at ZGA, revealed by high-throughput cap analysis of gene expression (CAGE) (Table 1) (Haberle et al., 2014). TSS positions shift from oocyte-specific A/T-rich sequence to nearby G/C-rich regions, which are the sites of H3K4me3 marks and nucleosome repositioning prior to gene activation (Figure 5d) (Haberle et al., 2014). Thus, specific DNA motifs recognized by chromatin remodelers or pioneer transcription factors (see below) (Figure 5f) could delineate the embryonic-specific program of gene expression, in a manner that aligns with sperm patterns.
G/C-rich regions are also significant in that they are sites for 5-methylcytosine modifications, which are associated with regions of transcriptional quiescence. Cytosine methylation at CpG dinucleotides is catalyzed by DNA methyltransferases such as Dnmt1, and inhibition of methyltransferase activity causes premature expression of many zygotic genes during the MZT (Table 2) (Potok et al., 2013; Stancheva and Meehan, 2000), though for Xenopus Dnmt1 only the DNA binding function independent of the actual methylation appears to be necessary to repress transcription (Dunican et al., 2008). Repressive interactions with methyl-CpG-binding domain proteins such as Xenopus Kaiso are necessary to regulate transcriptional timing at ZGA (Ruzov et al., 2004). Conversely, DNA hypomethylation is generally predictive of genes expressed at ZGA (Potok et al., 2013; Stancheva et al., 2002) and the deposition of H3K4me3 marks (Andersen et al., 2012), though again in Xenopus the relationship between methylation and gene expression may be more complex (Bogdanovic et al., 2011).
As with histone marks, specific methylation patterns are present in sperm DNA (Hammoud et al., 2009; Wu et al., 2011), but these are largely lost upon fertilization, only to reappear in the blastula (Mhanni and McGowan, 2004; Oswald et al., 2000; Santos et al., 2002; Smith et al., 2012; Stancheva et al., 2002). Two high-throughput assays of DNA methylation during zebrafish embryogenesis using bisulfite sequencing (Table 1) have revealed that nearly the exact paternal pattern of differential methylation reemerges in the blastula, despite having adopted a more oocyte-specific pattern during cleavage stages (Figure 5e) (Jiang et al., 2013; Potok et al., 2013). This is achieved independent of a sperm methylation template, as parthenogenic embryos produced by fertilization with UV-irradiated sperm, which lack functional chromatin, still adopt paternal methylation patterns on chromatin derived solely from the oocyte (Potok et al., 2013), implicating intrinsic signals in the genome that guide these modifications.
In summary, transcriptional competence as well as the specificity of gene expression are achieved through regulation of chromatin accessibility, in a manner informed by both parental epigenomes. The acquisition of an open nucleosome arrangement, histone modifications and DNA methylation combine to license transcription at the MZT (Figure 5). Engagement of this permissive chromatin to elicit gene expression is the role of transcriptional machinery and accessory factors, but as we describe in the final subsection, some of these factors may in fact establish accessible chromatin for themselves.
Transcriptional repressors and activators
During early characterizations of transcriptional competency in mouse embryos, a change in the cis regulatory requirements was observed from 1 to 2-cell stage, such that enhancer sequences became necessary for efficient transcription (Martinez-Salas et al., 1989). Sequence-specific transcription factors that bind gene loci to recruit the transcriptional machinery were thus hypothesized to help navigate a repressive chromatin landscape during the major ZGA (Majumder and DePamphilis, 1995).
Across all species, the availability of both general and specific transcription factors are expected to help regulate gene expression at ZGA. Components of the TFIID complex, which binds the core promoter to activate basal transcription, are one point of regulation. In C. elegans, TAF-4 is sequestered in the cytoplasm by phosphorylated OMA-1 and OMA-2, thus preventing TFIID from assembling onto nuclear DNA during the first cleavages (Guven-Ozkan et al., 2008). Phosphorylation is mediated by fertilization through activation of the kinase MBK-2, which interestingly is also responsible for phosphorylating other maternal proteins to mark them for degradation (Stitzel et al., 2006). Pol II inactivity is maintained until the 4-cell embryo, when OMA-1/2 themselves are degraded and ZGA begins (Guven-Ozkan et al., 2008).
Another TFIID subunit, TATA-binding protein (TBP), along with its paralogs TBP2 and TBP-like (TLF), was found to be essential for embryonic development in Xenopus, C. elegans and zebrafish (Bártfai et al., 2004; Dantonel et al., 2000; Ferg et al., 2007; Jallow et al., 2004; Kaltenbach et al., 2000; Martianov et al., 2002; Müller et al., 2001; Prioleau et al., 1994; Veenstra et al., 2000; Veenstra et al., 1999), with semi-redundant roles in transcription activation. In Xenopus, TBP is rate-limiting for embryonic transcription (Prioleau et al., 1994; Veenstra et al., 1999). Although TBP mRNA is maternally provided, protein levels are undetectable until the early blastula (Veenstra et al., 1999). This is in contrast to other basal transcription factors such as TGIIB and TFIIF RAP74 (Veenstra et al., 1999), suggesting that specific translational repression of TBP, likely mediated by delayed cytoplasmic polyadenylation, contributes to early transcriptional quiescence. In mouse, TBP loss results in failed transcriptional activation in the blastocyst, though the effect was only observed for Pol I and III transcription (Martianov et al., 2002). Additionally, in D. melanogaster, TBP is strongly associated with the earliest transcribed genes (Chen et al., 2013). Overall, differential TATA box usage in gene promoters coupled with post-transcriptional regulation of maternal TATA-binding factors may be a mechanism to define temporally specific patterns of activation for different sets of genes during the MZT.
Establishing critical levels of other activating and repressive transcription factors likely plays a significant role during ZGA; however, to date few such factors have been identified. In D. melanogaster, Tramtrack (TTK) is a maternal inhibitor of fushi tarazu (ftz) expression (Brown et al., 1991; Pritchard et al., 1996). Nuclear concentration of TTK is simultaneously reduced by an increasing N/C ratio and regulated degradation of ttk mRNA by the maternal clearance factor Smaug (Benoit et al., 2009; Tadros et al., 2007). Loss of Smaug has a widespread effect on zygotic gene expression, likely beyond the effect of sustained TTK activity, suggesting that Smaug is responsible for inactivating a number of other unknown repressive factors that are involved in ZGA (Benoit et al., 2009; Chen et al., 2014).
Once repression is overcome, early gene activation in D. melanogaster is driven by Zelda (also known as Vielfältig) (Liang et al., 2008), a maternally deposited transcription factor with ubiquitous expression in the pre-blastoderm embryo and specific patterns of subcellular localization over the course of the cell cycle (Staudt et al., 2006). Zelda binds promoter regions of early zygotic genes via a heptamer DNA motif called the TAGteam (De Renzis et al., 2007; ten Bosch et al., 2006) that is conserved in other insects (Biedler et al., 2012). Loss of Zelda causes mitotic defects (Staudt et al., 2006) and a failure to activate 120 early zygotic genes during cycles 8-13 (Liang et al., 2008). However, Zelda has also been shown to bind hundreds of additional gene promoters by cycle 8, prior to their eventual activation (Harrison et al., 2011), suggesting a broader role in licensing zygotic transcription.
In zebrafish, early zygotic genes are enriched in binding sites for the pluripotency-inducing factors Nanog, Pou5f1 (also known as Oct4) and Sox19b (an ortholog of Sox2 in the SoxB1 family) (Lee et al., 2013; Leichsenring et al., 2013). These three factors are maternally provided (Burgess et al., 2002; Okuda et al., 2010; Onichtchouk et al., 2010; Xu et al., 2012) and are the most highly translated sequence-specific transcription factors prior to ZGA, as determined by ribosome profiling (Lee et al., 2013). Combined loss of these factors has a profound effect on transcriptional output and development, with 82% of genes deficient in the late blastula and a corresponding failure to initiate gastrulation, similar to an a-amanitin phenotype (Lee et al., 2013). Binding of the factors appears to be coordinated and widespread (Leichsenring et al., 2013; Xu et al., 2012), reminiscent of their behaviors in ES cells and induced pluripotent stem cells (iPS cells), where they guide specific gene expression patterns to induce or maintain pluripotency (reviewed in (Young, 2011)).
Nanog, Sox2 and Pou5f1 are associated with chromatin remodeling activities in ES and iPS cells (Orkin et al., 2011), and have been proposed to have "pioneering" activity, by binding to regions of repressed chromatin to induce nucleosome repositioning and allow other factors to bind in a cooperative manner (Figure 5f) (Zaret et al., 2011). In fact, they may play similar roles during ZGA (Lee et al., 2013; Leichsenring et al., 2013), and would thus provide a mechanism by which the silent embryonic genome is initially engaged. In Xenopus, beta-Catenin has been shown to induce epigenetic modifications at genes prior to their eventual expression during early embryogenesis, implicating it as another maternal pioneer factor (Blythe et al., 2010).
The widespread binding pattern of Zelda on gene promoters suggests that it too may have pioneering activity. Zelda binding is correlated with accessible chromatin as measured by DNaseI hypersensitivity (Table 1) (Harrison et al., 2011; Thomas et al., 2011), and TAGteam sequences are additionally bound by other factors (Satija et al., 2012) that include Bicoid Stability Factor (BSF) (De Renzis et al., 2007), STAT92E (Tsurumi et al., 2011) and Grainyhead (Harrison et al., 2010). In this way, gene activation may be the product of combinatorial binding of Zelda, which primes the chromatin, and other transcription factors, which would provide the temporal-spatial specificity and magnitude of the transcriptional output (Figure 5f).
In sum, maternal transcription factors have widespread roles in guiding ZGA at specific gene loci. However, specific factors with pioneering activity may ultimately be the determinants of ZGA, by binding repressed chromatin and inducing remodeling, thus allowing the transcriptional machinery to access gene promoters and drive zygotic gene expression.
DEVELOPMENTAL SIGNIFICANCE OF THE FIRST ZYGOTIC GENES
Functions of the first zygotic genes
Zygotic gene expression during the MZT provides the embryo with what the maternal contribution lacks. To some extent, these genes will encode transcription factors and signaling pathway components that prepare the embryo for gastrulation and cell specification and differentiation, but there will also be genes that carry out basic functions, common to all cell types and tissues, so-called "housekeeping" genes. Increased cell number, protein turnover rates or the need to establish specific spatial expression patterns may explain why it is necessary to supplement or replenish large parts of the maternal contribution.
The relative proportion of these two classes of genes varies widely among different organisms, likely correlated with the different times that ZGA occurs in relation to developmental milestones. In mouse, where both the minor and major waves of ZGA occur prior to two cleavages, spanning two days of development, activated genes predominantly encode basic cellular mechanisms including protein and RNA metabolism; transcription factors and patterning genes seem to activate slightly later (Hamatani et al., 2004; Park et al., 2013; Xue et al., 2013). In contrast, zebrafish zygotic transcripts appear after 2-2.5 hours of development supported by the maternal contribution, and ~2 hours prior to the onset of gastrulation; accordingly, ZGA involves roughly equal numbers of housekeeping genes and genes encoding transcription factors and chromatin modifiers (Aanes et al., 2011; Lee et al., 2013). Signaling and patterning genes appear early in Xenopus (Paranjpe et al., 2013; Yang et al., 2002), as well as in D. melanogaster (Ali-Murthy et al., 2013; Karr et al., 1985), where early genes are specifically required for proper cell division synchrony (Karr et al., 1985), establishing the anterior-posterior axis (Blankenship et al., 2001) and cellularization (Lecuit et al., 2002).
Beyond these broad themes of gene function, there is not a strong coherence in the patterns of zygotic expression for individual genes. Among more closely related species, orthologous zygotic genes vary greatly in expression timing (Xue et al., 2013) and levels (Yanai et al., 2011), which suggests a rapidly evolving regulatory landscape (Xie et al., 2010). Across longer evolutionary distances, zygotic genes appear to be almost completely divergent (Heyn et al., 2014). As an extreme example, much of the maternal contribution in C. elegans is in fact transcribed de novo in fellow nematode A. suum, including critical regulators of the MZT such as oma (Wang et al., 2014).
The intimate relationship between ZGA and maternal clearance
One notable function that does seem to be more conserved, in both theme and mechanism, is maternal clearance. Two forms of maternal mRNA decay are found across animals: the “maternal mode,” which relies on maternally provided factors, and the “zygotic mode,” which depends on de novo zygotic transcription (Bashirullah et al., 1999; Walser and Lipshitz, 2011). In many species, failure to properly activate zygotic gene expression leads to loss of the zygotic mode of maternal clearance and stabilization of maternal messages (Figure 6a,b) (Bashirullah et al., 1999; De Renzis et al., 2007; Edgar et al., 1996; Hamatani et al., 2004; Lee et al., 2013; Surdej and Jacobs-Lorena, 1998; Tadros et al., 2007).
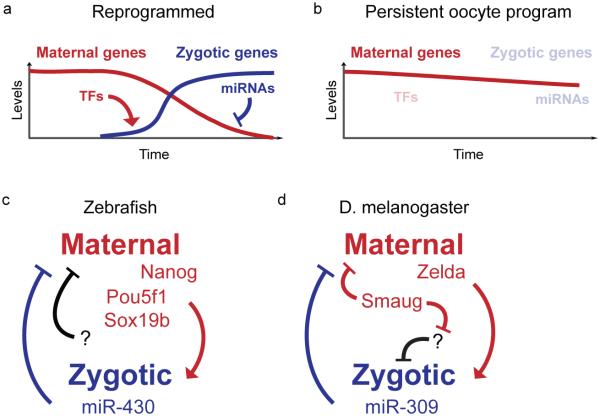
Figure 6. Activation of the zygotic genome and maternal clearance together reprogram the embryo during the MZT.
(a) Maternally provided factors (red) activate transcription of zygotic genes (blue) through the action of transcription factors (TFs) that engage the early embryonic genome. In turn, zygotic genes, including microRNAs, direct clearance of maternal RNAs. (b) Failure to activate the zygotic genome by inhibiting maternal TF activity results in stabilization of the maternal program and arrested development. (c) In zebrafish, the TFs Nanog, Pou5f1 and Sox19b are required for widespread zygotic gene expression, which includes miR-430. miR-430, along with other unknown factors, induces clearance of a large subset of maternal RNAs. (d) In D. melanogaster, Zelda plays a similar role, and is responsible for activating miR-309, which in turn mediates maternal clearance. Maternal Smaug activity is also required for maternal clearance, as well as for miR-309 expression through an unknown intermediate.
Generally, both modes can be found in a given species. In mouse, there is a strong maternal mode that is active shortly after fertilization (Hamatani et al., 2004; Pikó and Clegg, 1982), but a second round of clearance overlaps with the major ZGA and is abrogated by transcription inhibition (Hamatani et al., 2004). A prominent maternal mode is also found in Xenopus embryos, partially mediated by AU-rich elements (ARE) (Voeltz et al., 1998) and embryonic deadenylation elements (EDEN) (Bouvet et al., 1994; Paillard et al., 1998) in the 3' untranslated regions (UTR) of maternal mRNAs. Fertilization-induced activation of maternal factors including EDEN-Binding Protein (Paillard et al., 1998) causes deadenylation of target mRNAs (Duval et al., 1990). However, zygotic transcription is required for these deadenylated transcripts to eventually be degraded (Audic et al., 1997; Duval et al., 1990). D. melanogaster Smaug also induces maternal mRNA deadenylation, by recruiting the CCR4/POP2/NOT-deadenylase complex (Semotok et al., 2005), but this activity likely does not depend on zygotic transcription. However, other zygotic mechanisms activate later in embryogenesis that accelerate decay for targets of the maternal mode (Bashirullah et al., 1999), in addition to destabilizing transcripts that are only subject only to the zygotic mode (Surdej and Jacobs-Lorena, 1998).
In the zygotic mode of clearance, microRNAs (miRNAs) appear to be major players. miRNAs are ~22nt small RNAs that are incorporated into silencing complexes (miRISC) that target mRNAs to induce translation repression, deadenylation and decay (Huntzinger and Izaurralde, 2011). The role of miRNAs in maternal clearance was first identified in zebrafish with the aid of a maternal-zygotic mutant for dicer (MZdicer) (Giraldez et al., 2006), which encodes an RNaseIII enzyme required for canonical miRNA biogenesis. MZdicer embryos are deficient in mature miR-430, which results in the stabilization of hundreds of maternal mRNAs that are normally translationally repressed and degraded in the wild type embryo (Bazzini et al., 2012; Giraldez et al., 2006). The miR-430 gene cluster is one of the earliest and most highly transcribed genes from the zygotic genome (Chen et al., 2005; Heyn et al., 2014; Lee et al., 2013), and is directly activated by ZGA transcription factors Nanog, Pou5f1 and SoxB1 (Figure 6c) (Lee et al., 2013). Loss of Nanog in particular results in severely reduced miR-430 levels and maternal clearance activity (Lee et al., 2013), thus illustrating the tight coordination of ZGA and maternal clearance (Figure 6)
miR-430 has conserved zygotic expression in another teleost fish, medaka (Tani et al., 2010), and has sequence similarity to several other embryonic miRNAs, including miR-295 in mouse; miR-302, miR-327 and miR-516-520 in human (Chen et al., 2005; Giraldez, 2010); and miR-427 in Xenopus (Lund et al., 2009). miR-427 has similar expression dynamics to miR-430 in the early Xenopus embryo, and was likewise shown to mediate maternal clearance activity (Lund et al., 2009), while miR-302 was found to be activated by Pou5f1 and Sox2 in ES cells (Card et al., 2008).
Beyond vertebrates, the miR-309 cluster plays an analogous role in D. melanogaster embryogenesis, suggesting that flies have independently evolved a similar strategy to clear maternal mRNAs (Bushati et al., 2008). miR-309 is activated by Zelda along with other early zygotic genes (Liang et al., 2008), and loss of miR-309 function leads to maternal transcript stabilization (Bushati et al., 2008). Interestingly, mRNA clearance activity mediated by maternally provided Smaug is also required for normal miR-309 expression (Benoit et al., 2009) (Figure 6d). Thus, distinct pathways of maternal clearance may be functionally linked through the regulation of zygotic transcription (Figure 6).
CONCLUDING REMARKS
Zygotic gene expression at the MZT is a multifaceted process
In conclusion, we have described the context, mechanisms and function of zygotic genome activation and gene expression during the MZT, a process that is fundamentally conserved across animals, and yet in many ways is surprisingly variable among different species. High-throughput transcriptome analyses show widespread, but specific, patterns of zygotic gene transcription, much of which overlaps with the maternal contribution of RNAs. Regulation of the cell cycle, chromatin and transcriptional machinery by maternal factors together elicits transcriptional output with individual-gene specificity. Zygotic genes span the functional requirements of the developing embryo, including the clearance of maternal instructions as the embryonic genome assumes developmental control. Despite the diversity of mechanisms found across different species to regulate gene expression, unifying themes are emerging, including the roles of key transcription factors and miRNAs in directing the MZT (Figure 6). We anticipate that other common regulatory paradigms will soon emerge.
Although great advances have been made in understanding the function of each of these aspects of the MZT and ZGA, we still lack a unified picture of how all of these components combine to precisely activate transcription. Does ZGA arise from the cumulative effects of several independent mechanisms, or is there a cascade of molecular events that has a single point of control? Specifically, understanding how maternal transcription factors influence the embryonic chromatin will be key to distinguishing these two scenarios, and it may be that the activity of pioneer factors is ultimately what induces genome competency and initiates the zygotic program.
We also lack a complete understanding of how zygotic gene activation and maternal clearance complement each other and feed back on themselves. Zygotic transcription is already known to be required for some maternal clearance, and vice versa, but how this is accomplished and what factors are involved are still largely unknown. It is tempting to speculate that there are mechanisms that directly link these two RNA metabolic activities.
Cellular reprogramming during embryogenesis and beyond
We close by revisiting the idea that the MZT is a cellular reprogramming event in vivo, by deletion of the old, maternal program and installation and maintenance of the new zygotic program. Maternal clearance, zygotic gene expression and the crosstalk between them combine to induce a transformation in cellular identity, away from the terminally differentiated gametes toward transient stages of totipotency, pluripotency and eventually re-differentiation (Giraldez, 2010). Aspects of this process and the mechanisms regulating it are found beyond embryogenesis, most notably in stem cells, which have simultaneous requirements to give rise to differentiated cell types, and to self renew in order to maintain an undifferentiated state.
iPS cells are the in vitro instantiation of this process. Reprogramming of mouse somatic cells to a pluripotent state was achieved through heterologous expression of the four so-called Yamanaka factors Oct4 (Pou5f1), Sox2, Klf4 and c-Myc (Takahashi et al., 2006). Although the mechanisms leading to induced pluripotency are poorly understood, numerous other transcription factors, chromatin modifiers and miRNAs are known to be involved, and manipulation of many of these can enhance reprogramming, or even substitute for some of the four factors (Young, 2011). However, these various players all seem to lead back to a regulatory circuit consisting of Oct4, Sox2 and Nanog, which are endogenously activated during reprogramming and are at the core of the pluripotency gene network (reviewed in (Oron and Ivanova, 2012; Young, 2011)).
It is likely that core transcription factors are central to embryogenesis as well, which is already apparent in two organisms -- Zelda in D. melanogaster, and orthologs of Nanog, Oct4 and Sox2 themselves in zebrafish. In this way, the post-fertilization mobilization of maternal factors is analogous to the introduction of Yamanaka factors into somatic cells, with a similar end result: activation of a pluripotency network through expression of the core regulatory circuit. If iPS cells are in fact a good model for the MZT, then there potentially exist many possible paths to pluripotency that will lead to the same core regulatory circuit, which may account for some of the species-specific differences that have been observed in the maternal contribution and targets of ZGA. As we continue to elucidate the mechanisms that feed into this genetic network, we will develop a greater understanding of the principles that guide cellular reprogramming both in vivo and in vitro, and more generally how gene regulation can induce large-scale changes in cellular identity.
SUMMARY POINTS.
* Zygotic gene activation and maternal clearance are conserved activities during the maternal-to-zygotic transition that combine to reprogram terminally differentiated gametes to embryonic totipotency / pluripotency.
* Recent high-throughput interrogation of the embryonic transcriptome, coupled with dissection of the maternal and zygotic contributions, has revealed earlier and broader patterns of zygotic transcription than have been previously observed.
* Cell cycle dynamics and chromatin structure combine to license gene expression during the MZT.
* Maternally provided pioneer transcription factors may initiate ZGA, by inducing open chromatin and recruiting other transcriptional machinery.
* Zygotic miRNAs transcribed soon after ZGA are key players in the zygotic mode of maternal clearance across different species.
FUTURE ISSUES.
* How do broadly conserved mechanisms that regulate the MZT integrate with species-specific gene expression to direct developmental programs across different organisms?
* What is the role of genome quiescence, and does a truly transcriptionally quiescent period actually exist in different species, or are we limited by current detection technologies?
* How are maternal factors post-transcriptionally regulated during oogenesis versus during embryogenesis, and does differential regulation control the timing of ZGA?
* What are the biochemical determinants of maternal mRNA regulation, e.g. structural elements, ribonucleotide modifications and RNA binding proteins?
* What changes to embryonic chromatin are directly required for ZGA, versus instructive for gene expression beyond the MZT?
* What is the role of pioneer transcription factors in inducing open chromatin prior to ZGA?
* How is maternal clearance informative for ZGA and what other mechanisms regulate maternal mRNA stability?
ACRONYMS AND DEFINITIONS
- Maternal-to-zygotic transition (MZT)
The period during embryogenesis when developmental control transitions from maternally provided factors to ones produced by zygotic (embryonic) transcription
- Zygotic genome activation (ZGA)
The period during the MZT when the embryonic genome first begins to transcribe RNA
- Totipotency, pluripotency
The capacity of a cell to give rise to all (totipotent) or most (pluripotent) differentiated cells in an organism
- Maternal clearance
The process of regulated degradation of maternally provided RNAs and proteins during the MZT
- Cellularization
The partitioning of a multi-nucleated cytoplasm (syncytium) into individual cells, as observed during D. melanogaster embryogenesis
- RNA-Seq
Massively parallel RNA sequencing to measure gene expression levels in a high-throughput manner
- Transcriptome
The set of all RNA transcripts in a cell, tissue or organism
- RNA polymerase II (Pol II)
The enzyme responsible for transcribing messenger RNAs in eukaryotes
- Pronucleus
The haploid nucleus of the sperm or egg following fertilization, prior to fusion into a single zygotic nucleus
- Single-nucleotide polymorphism (SNP)
Variation of a single base position in the genome that differs between individuals in a population
- Nucleocytoplasmic (N/C) ratio
The ratio of nuclear to cytoplasmic volume in a cell. Gap phase: The period between DNA replication (S) and mitosis (M) during the cell cycle. G1 follows M phase, G2 follows S phase
- Chromatin
The physical structure of a chromosome, consisting of DNA bound by nucleosomes and other proteins
- Epigenetic
Refers to heritable changes affecting gene expression that are not encoded in the DNA sequence
- H3K4me3 & H3K27me3
Post-transcriptional modifications of histone H3 at lysine 4 and 27, consisting of trimethylation of the epsilon amino group
- Embryonic stem cells (ESC)
Pluripotent cells derived from the blastocyst, capable of differentiating into any of the three germ layers
- Transcription start site (TSS)
The site of transcription initiation proximal to the promoter that defines the 5’ end of a gene transcript
- Transcription factor
A protein that binds to DNA sequences to regulate gene expression
- Induced pluripotent (iPS) cell
A pluripotent cell that is derived from a differentiated cell by reprogramming its gene expression
- Pioneer factor
A transcription factor that can bind closed chromatin to induce chromatin remodeling and accessibility for other factors
REFERENCES
- Aanes H, et al. Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome research. 2011;21(8):1328–1338. doi: 10.1101/gr.116012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adenot PG, et al. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development (Cambridge, England) 1997;124(22):4615–4625. doi: 10.1242/dev.124.22.4615. [DOI] [PubMed] [Google Scholar]
- Akkers RC, et al. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Developmental cell. 2009;17(3):425–434. doi: 10.1016/j.devcel.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali-Murthy Z, et al. An essential role for zygotic expression in the pre-cellular Drosophila embryo. PLoS genetics. 2013;9(4):e1003428. doi: 10.1371/journal.pgen.1003428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen IS, et al. Developmental features of DNA methylation during activation of the embryonic zebrafish genome. Genome biology. 2012;13(7):R65. doi: 10.1186/gb-2012-13-7-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu-Vieyra CV, et al. MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing. PLoS biology. 2010;8(8) doi: 10.1371/journal.pbio.1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki F, Hara KT, Schultz RM. Acquisition of transcriptional competence in the 1-cell mouse embryo: requirement for recruitment of maternal mRNAs. Molecular reproduction and development. 2003;64(3):270–274. doi: 10.1002/mrd.10227. [DOI] [PubMed] [Google Scholar]
- Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Developmental biology. 1997;181(2):296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- Audic Y, Omilli F, Osborne HB. Postfertilization deadenylation of mRNAs in Xenopus laevis embryos is sufficient to cause their degradation at the blastula stage. Molecular and cellular biology. 1997;17(1):209–218. doi: 10.1128/mcb.17.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barckmann B, Simonelig M. Control of maternal mRNA stability in germ cells and early embryos. RNA Decay Mechanisms. 2013;1829(6–7):714–724. doi: 10.1016/j.bbagrm.2012.12.011. [DOI] [PubMed] [Google Scholar]
- Bashirullah A, et al. Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster. The EMBO journal. 1999;18(9):2610–2620. doi: 10.1093/emboj/18.9.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, et al. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development (Cambridge, England) 2003;130(5):889–900. doi: 10.1242/dev.00302. [DOI] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science (New York, N.Y.) 2012;336(6078):233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bártfai R, et al. TBP2, a Vertebrate-Specific Member of the TBP Family, Is Required in Embryonic Development of Zebrafish. Current Biology. 2004;14(7):593–598. doi: 10.1016/j.cub.2004.03.034. [DOI] [PubMed] [Google Scholar]
- Benoit B, et al. An essential role for the RNA-binding protein Smaug during the Drosophila maternal-to-zygotic transition. Development (Cambridge, England) 2009;136(6):923–932. doi: 10.1242/dev.031815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Biedler JK, et al. Identification of early zygotic genes in the yellow fever mosquito Aedes aegypti and discovery of a motif involved in early zygotic genome activation. PloS one. 2012;7(3):e33933. doi: 10.1371/journal.pone.0033933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship JT, Wieschaus E. Two new roles for the Drosophila AP patterning system in early morphogenesis. Development (Cambridge, England) 2001;128(24):5129–5138. doi: 10.1242/dev.128.24.5129. [DOI] [PubMed] [Google Scholar]
- Blythe SA, et al. beta-Catenin primes organizer gene expression by recruiting a histone H3 arginine 8 methyltransferase, Prmt2. Developmental cell. 2010;19(2):220–231. doi: 10.1016/j.devcel.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanovic O, et al. Temporal uncoupling of the DNA methylome and transcriptional repression during embryogenesis. Genome research. 2011;21(8):1313–1327. doi: 10.1101/gr.114843.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Bosch JR, Benavides JA, Cline TW. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development (Cambridge, England) 2006;133(10):1967–1977. doi: 10.1242/dev.02373. [DOI] [PubMed] [Google Scholar]
- Bouniol C, Nguyen E, Debey P. Endogenous transcription occurs at the 1-cell stage in the mouse embryo. Experimental cell research. 1995;218(1):57–62. doi: 10.1006/excr.1995.1130. [DOI] [PubMed] [Google Scholar]
- Bouvet P, et al. The deadenylation conferred by the 3' untranslated region of a developmentally controlled mRNA in Xenopus embryos is switched to polyadenylation by deletion of a short sequence element. Molecular and cellular biology. 1994;14(3):1893–1900. doi: 10.1128/mcb.14.3.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braude P, Bolton V, Moore S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature. 1988;332(6163):459–461. doi: 10.1038/332459a0. [DOI] [PubMed] [Google Scholar]
- Brown DD, Littna E. RNA synthesis during the development of Xenopus laevis, the South African clawed toad. Journal of molecular biology. 1964;8(5):669–687. doi: 10.1016/s0022-2836(64)80116-9. [DOI] [PubMed] [Google Scholar]
- Brown JL, et al. Repression of the Drosophila fushi tarazu (ftz) segmentation gene. The EMBO journal. 1991;10(3):665–674. doi: 10.1002/j.1460-2075.1991.tb07995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska U, et al. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nature structural & molecular biology. 2010;17(6):679–687. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- Bultman SJ, et al. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes & development. 2006;20(13):1744–1754. doi: 10.1101/gad.1435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, et al. The zebrafish spiel-ohne-grenzen (spg) gene encodes the POU domain protein Pou2 related to mammalian Oct4 and is essential for formation of the midbrain and hindbrain, and for pre-gastrula morphogenesis. Development (Cambridge, England) 2002;129(4):905–916. doi: 10.1242/dev.129.4.905. [DOI] [PubMed] [Google Scholar]
- Bushati N, et al. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Current biology : CB. 2008;18(7):501–506. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- Card DAG, et al. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Molecular and cellular biology. 2008;28(20):6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-C, et al. A maternal store of macroH2A is removed from pronuclei prior to onset of somatic macroH2A expression in preimplantation embryos. Developmental biology. 2005;278(2):367–380. doi: 10.1016/j.ydbio.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Chen K, et al. A global change in RNA polymerase II pausing during the Drosophila midblastula transition. eLife. 2013;2:e00861. doi: 10.7554/eLife.00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, et al. Global regulation of mRNA translation and stability in the early Drosophila embryo by the Smaug RNA-binding protein. Genome biology. 2014;15(1):R4. doi: 10.1186/gb-2014-15-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, et al. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes & development. 2005;19(11):1288–1293. doi: 10.1101/gad.1310605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho T, et al. Involvement of chromatin structure in the regulation of mouse zygotic gene activation. Animal Science Journal. 2002;73(2):113–122. [Google Scholar]
- Christians E, et al. Expression of the HSP 70.1 gene, a landmark of early zygotic activity in the mouse embryo, is restricted to the first burst of transcription. Development (Cambridge, England) 1995;121(1):113–122. doi: 10.1242/dev.121.1.113. [DOI] [PubMed] [Google Scholar]
- Colgan DF, et al. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature. 1996;384(6606):282–285. doi: 10.1038/384282a0. [DOI] [PubMed] [Google Scholar]
- Collart C, et al. Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science (New York, N.Y.) 2013;341(6148):893–896. doi: 10.1126/science.1241530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig SP. Synthesis of RNA in non-nucleate fragments of sea urchin eggs. Journal of molecular biology. 1970;47(3):615–618. doi: 10.1016/0022-2836(70)90331-1. [DOI] [PubMed] [Google Scholar]
- Dantonel J-C, et al. TBP-like Factor Is Required for Embryonic RNA Polymerase II Transcription in C. elegans. Molecular cell. 2000;6(3):715–722. doi: 10.1016/s1097-2765(00)00069-1. [DOI] [PubMed] [Google Scholar]
- Davis WJ, De Sousa PA, Schultz RM. Transient expression of translation initiation factor eIF-4C during the 2-cell stage of the preimplantation mouse embryo: identification by mRNA differential display and the role of DNA replication in zygotic gene activation. Developmental biology. 1996;174(2):190–201. doi: 10.1006/dbio.1996.0065. [DOI] [PubMed] [Google Scholar]
- De Renzis S, et al. Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS biology. 2007;5(5):e117. doi: 10.1371/journal.pbio.0050117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekens MPS, et al. The maternal-effect gene futile cycle is essential for pronuclear congression and mitotic spindle assembly in the zebrafish zygote. Development (Cambridge, England) 2003;130(17):3907–3916. doi: 10.1242/dev.00606. [DOI] [PubMed] [Google Scholar]
- Di Talia S, et al. Posttranslational Control of Cdc25 Degradation Terminates Drosophila's Early Cell-Cycle Program. Current Biology. 2013;23(2):127–132. doi: 10.1016/j.cub.2012.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov S, et al. Chromatin transitions during early Xenopus embryogenesis: changes in histone H4 acetylation and in linker histone type. Developmental biology. 1993;160(1):214–227. doi: 10.1006/dbio.1993.1299. [DOI] [PubMed] [Google Scholar]
- Dobson AT, et al. The unique transcriptome through day 3 of human preimplantation development. Human molecular genetics. 2004;13(14):1461–1470. doi: 10.1093/hmg/ddh157. [DOI] [PubMed] [Google Scholar]
- Dunican DS, et al. xDnmt1 regulates transcriptional silencing in pre-MBT Xenopus embryos independently of its catalytic function. Development (Cambridge, England) 2008;135(7):1295–1302. doi: 10.1242/dev.016402. [DOI] [PubMed] [Google Scholar]
- Duval C, et al. Stability of maternal mRNA in Xenopus embryos: role of transcription and translation. Molecular and cellular biology. 1990;10(8):4123–4129. doi: 10.1128/mcb.10.8.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Schubiger G. Parameters controlling transcriptional activation during early drosophila development. Cell. 1986;44(6):871–877. doi: 10.1016/0092-8674(86)90009-7. [DOI] [PubMed] [Google Scholar]
- Edgar BA, et al. Zygotic degradation of two maternal Cdc25 mRNAs terminates Drosophila's early cell cycle program. Genes & development. 1996;10(15):1966–1977. doi: 10.1101/gad.10.15.1966. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Kiehle CP, Schubiger G. Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development. Cell. 1986;44(2):365–372. doi: 10.1016/0092-8674(86)90771-3. [DOI] [PubMed] [Google Scholar]
- Edgar LG, Wolf N, Wood WB. Early transcription in Caenorhabditis elegans embryos. Development (Cambridge, England) 1994;120(2):443–451. doi: 10.1242/dev.120.2.443. [DOI] [PubMed] [Google Scholar]
- Faast R, et al. Histone variant H2A.Z is required for early mammalian development. Current Biology. 2001;11(15):1183–1187. doi: 10.1016/s0960-9822(01)00329-3. [DOI] [PubMed] [Google Scholar]
- Farrell JA, O'Farrell PH. Mechanism and Regulation of Cdc25/Twine Protein Destruction in Embryonic Cell-Cycle Remodeling. Current Biology. 2013;23(2):118–126. doi: 10.1016/j.cub.2012.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferg M, et al. The TATA-binding protein regulates maternal mRNA degradation and differential zygotic transcription in zebrafish. The EMBO journal. 2007;26(17):3945–3956. doi: 10.1038/sj.emboj.7601821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe VE, Alberts BM. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. Journal of cell science. 1983;61(1):31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- Fu G, et al. Mouse oocytes and early embryos express multiple histone H1 subtypes. Biology of reproduction. 2003;68(5):1569–1576. doi: 10.1095/biolreprod.102.012336. [DOI] [PubMed] [Google Scholar]
- Gerhart JC. Mechanisms Regulating Pattern Formation in the Amphibian Egg and Early Embryo. In: Goldberger R, editor. Biological Regulation and Development. Springer US; Boston, MA: 1980. pp. 133–316. 316. [Google Scholar]
- Giraldez AJ. microRNAs, the cell's Nepenthe: clearing the past during the maternal-to-zygotic transition and cellular reprogramming. Current opinion in genetics & development. 2010;20(4):369–375. doi: 10.1016/j.gde.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science (New York, N.Y.) 2006;312(5770):75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Goddard MJ, Pratt HPM. Control of events during early cleavage of the mouse embryo: an analysis of the “2-cell block. Journal of embryology and experimental morphology. 1983;73(1):111–133. [PubMed] [Google Scholar]
- Golbus MS, Calarco PG, Epstein CJ. The effects of inhibitors of RNA synthesis (alpha-amanitin and actinomycin D) on preimplantation mouse embryogenesis. The Journal of experimental zoology. 1973;186(2):207–216. doi: 10.1002/jez.1401860211. [DOI] [PubMed] [Google Scholar]
- Guven-Ozkan T, et al. Global Transcriptional Repression in C. elegans Germline Precursors by Regulated Sequestration of TAF-4. Cell. 2008;135(1):149–160. doi: 10.1016/j.cell.2008.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V, et al. Two independent transcription initiation codes overlap on vertebrate core promoters. Nature. 2014 doi: 10.1038/nature12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamatani T, et al. Dynamics of Global Gene Expression Changes during Mouse Preimplantation Development. Developmental cell. 2004;6(1):117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460(7254):473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, et al. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS genetics. 2011;7(10):e1002266. doi: 10.1371/journal.pgen.1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Botchan MR, Cline TW. Grainyhead and Zelda compete for binding to the promoters of the earliest-expressed Drosophila genes. Developmental biology. 2010;345(2):248–255. doi: 10.1016/j.ydbio.2010.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey SA, et al. Identification of the zebrafish maternal and paternal transcriptomes. Development (Cambridge, England) 2013;140(13):2703–2710. doi: 10.1242/dev.095091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn P, et al. The Earliest Transcribed Zygotic Genes Are Short, Newly Evolved, and Different across Species. Cell reports. 2014;6(2):285–292. doi: 10.1016/j.celrep.2013.12.030. [DOI] [PubMed] [Google Scholar]
- Howe JA, et al. Identification of a developmental timer regulating the stability of embryonic cyclin A and a new somatic A-type cyclin at gastrulation. Genes & development. 1995;9(10):1164–1176. doi: 10.1101/gad.9.10.1164. [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nature reviews. Genetics. 2011;12(2):99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- Jallow Z, et al. Specialized and redundant roles of TBP and a vertebrate-specific TBP paralog in embryonic gene regulation in Xenopus. Proceedings of the National Academy of Sciences. 2004;101(37):13525–13530. doi: 10.1073/pnas.0405536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaramaiah Raja S, Renkawitz-Pohl R. Replacement by Drosophila melanogaster Protamines and Mst77F of Histones during Chromatin Condensation in Late Spermatids and Role of Sesame in the Removal of These Proteins from the Male Pronucleus. Molecular and cellular biology. 2005;25(14):6165–6177. doi: 10.1128/MCB.25.14.6165-6177.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, et al. Sperm, but Not Oocyte, DNA Methylome Is Inherited by Zebrafish Early Embryos. Cell. 2013;153(4):773–784. doi: 10.1016/j.cell.2013.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbach L, et al. The TBP-like Factor CeTLF Is Required to Activate RNA Polymerase II Transcription during C. elegans Embryogenesis. Molecular cell. 2000;6(3):705–713. doi: 10.1016/s1097-2765(00)00068-x. [DOI] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB. The zebrafish midblastula transition. Development (Cambridge, England) 1993;119(2):447–456. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- Kane DA, et al. The zebrafish epiboly mutants. Development (Cambridge, England) 1996:47–55. doi: 10.1242/dev.123.1.47. [DOI] [PubMed] [Google Scholar]
- Karr TL, et al. The engrailed locus of D. melanogaster provides an essential zygotic function in precellular embryos. Cell. 1985;43(3):591–601. doi: 10.1016/0092-8674(85)90231-4. Pt 2. [DOI] [PubMed] [Google Scholar]
- Kimelman D, Kirschner M, Scherson T. The events of the midblastula transition in Xenopus are regulated by changes in the cell cycle. Cell. 1987;48(3):399–407. doi: 10.1016/0092-8674(87)90191-7. [DOI] [PubMed] [Google Scholar]
- Knowland J, Graham C. RNA synthesis at the two-cell stage of mouse development. Journal of embryology and experimental morphology. 1972;27(1):167–176. [PubMed] [Google Scholar]
- Laubichler MD, Davidson EH. Boveri's long experiment: Sea urchin merogones and the establishment of the role of nuclear chromosomes in development. Developmental biology. 2008;314(1):1–11. doi: 10.1016/j.ydbio.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit T, Samanta R, Wieschaus E. slam encodes a developmental regulator of polarized membrane growth during cleavage of the Drosophila embryo. Developmental cell. 2002;2(4):425–436. doi: 10.1016/s1534-5807(02)00141-7. [DOI] [PubMed] [Google Scholar]
- Lee MT, et al. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature. 2013;503(7476):360–364. doi: 10.1038/nature12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichsenring M, et al. Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science (New York, N.Y.) 2013;341(6149):1005–1009. doi: 10.1126/science.1242527. [DOI] [PubMed] [Google Scholar]
- Lepikhov K, Walter J. Differential dynamics of histone H3 methylation at positions K4 and K9 in the mouse zygote. BMC developmental biology. 2004;4:12. doi: 10.1186/1471-213X-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lécuyer E, et al. Global Analysis of mRNA Localization Reveals a Prominent Role in Organizing Cellular Architecture and Function. Cell. 2007;131(1):174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Liang H-L, et al. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456(7220):400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C-J, Conti M, Ramalho-Santos M. Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development (Cambridge, England) 2013;140(17):3624–3634. doi: 10.1242/dev.095513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman LC, et al. Prepatterning of Developmental Gene Expression by Modified Histones before Zygotic Genome Activation. Developmental cell. 2011;21(6):993–1004. doi: 10.1016/j.devcel.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Lu X, et al. Coupling of zygotic transcription to mitotic control at the Drosophila mid-blastula transition. Development (Cambridge, England) 2009;136(12):2101–2110. doi: 10.1242/dev.034421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, et al. Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. RNA (New York, N.Y.) 2009;15(12):2351–2363. doi: 10.1261/rna.1882009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder S, DePamphilis ML. A unique role for enhancers is revealed during early mouse development. BioEssays : news and reviews in molecular, cellular and developmental biology. 1995;17(10):879–889. doi: 10.1002/bies.950171010. [DOI] [PubMed] [Google Scholar]
- Martianov I, Viville S, Davidson I. RNA Polymerase II Transcription in Murine Cells Lacking the TATA Binding Protein. Science (New York, N.Y.) 2002;298(5595):1036–1039. doi: 10.1126/science.1076327. [DOI] [PubMed] [Google Scholar]
- Martinez-Salas E, et al. The need for enhancers in gene expression first appears during mouse development with formation of the zygotic nucleus. Genes & development. 1989;3(10):1493–1506. doi: 10.1101/gad.3.10.1493. [DOI] [PubMed] [Google Scholar]
- Mathavan S, et al. Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS genetics. 2005;1(2):260–276. doi: 10.1371/journal.pgen.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill PT, Sweeton D, Wieschaus E. Requirements for autosomal gene activity during precellular stages of Drosophila melanogaster. Development (Cambridge, England) 1988;104(3):495–509. doi: 10.1242/dev.104.3.495. [DOI] [PubMed] [Google Scholar]
- Mhanni AA, McGowan RA. Global changes in genomic methylation levels during early development of the zebrafish embryo. Development genes and evolution. 2004;214(8):412–417. doi: 10.1007/s00427-004-0418-0. [DOI] [PubMed] [Google Scholar]
- Muller F, et al. TBP is not universally required for zygotic RNA polymerase II transcription in zebrafish. Current Biology. 2001;11(4):282–287. doi: 10.1016/s0960-9822(01)00076-8. [DOI] [PubMed] [Google Scholar]
- Nemer M. Old and new rna in the embryogenesis of the purple sea urchin. Proceedings of the National Academy of Sciences of the United States of America. 1963;50(2):230. doi: 10.1073/pnas.50.2.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982a;30(3):675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982b;30(3):687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- Nonchev S, Tsanev R. Protamine-histone replacement and DNA replication in the male mouse pronucleus. Molecular reproduction and development. 1990;25(1):72–76. doi: 10.1002/mrd.1080250113. [DOI] [PubMed] [Google Scholar]
- Okuda Y, et al. B1 SOX coordinate cell specification with patterning and morphogenesis in the early zebrafish embryo. PLoS genetics. 2010;6(5):e1000936. doi: 10.1371/journal.pgen.1000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onichtchouk D, et al. Zebrafish Pou5f1-dependent transcriptional networks in temporal control of early development. Molecular systems biology. 2010;6:354. doi: 10.1038/msb.2010.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Hochedlinger K. Chromatin Connections to Pluripotency and Cellular Reprogramming. Cell. 2011;145(6):835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oron E, Ivanova N. Cell fate regulation in early mammalian development. Physical biology. 2012;9(4):045002. doi: 10.1088/1478-3975/9/4/045002. [DOI] [PubMed] [Google Scholar]
- Oswald J, et al. Active demethylation of the paternal genome in the mouse zygote. Current Biology. 2000;10(8):475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- Paillard L, et al. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. The EMBO journal. 1998;17(1):278–287. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjpe SS, et al. A genome-wide survey of maternal and embryonic transcripts during Xenopus tropicalis development. BMC genomics. 2013;14:762. doi: 10.1186/1471-2164-14-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-J, et al. Inferring the choreography of parental genomes during fertilization from ultralarge-scale whole-transcriptome analysis. Genes & development. 2013;27(24):2736–2748. doi: 10.1101/gad.227926.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Montero S, et al. The embryonic linker histone H1 variant of Drosophila, dBigH1, regulates zygotic genome activation. Developmental cell. 2013;26(6):578–590. doi: 10.1016/j.devcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- Pikó L, Clegg KB. Quantitative changes in total RNA, total poly(A), and ribosomes in early mouse embryos. Developmental biology. 1982;89(2):362–378. doi: 10.1016/0012-1606(82)90325-6. [DOI] [PubMed] [Google Scholar]
- Poccia D, et al. RNA synthesis in male pronuclei of the sea urchin. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1985;824(4):349–356. doi: 10.1016/0167-4781(85)90042-9. [DOI] [PubMed] [Google Scholar]
- Posfai E, et al. Polycomb function during oogenesis is required for mouse embryonic development. Genes & development. 2012;26(9):920–932. doi: 10.1101/gad.188094.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potok ME, et al. Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell. 2013;153(4):759–772. doi: 10.1016/j.cell.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioleau MN, et al. Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell. 1994;77(3):439–449. doi: 10.1016/0092-8674(94)90158-9. [DOI] [PubMed] [Google Scholar]
- Pritchard DK, Schubiger G. Activation of transcription in Drosophila embryos is a gradual process mediated by the nucleocytoplasmic ratio. Genes & development. 1996;10(9):1131–1142. doi: 10.1101/gad.10.9.1131. [DOI] [PubMed] [Google Scholar]
- Puschendorf M, et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nature Genetics. 2008;40(4):411–420. doi: 10.1038/ng.99. [DOI] [PubMed] [Google Scholar]
- Qin X, et al. Global analyses of mRNA translational control during early Drosophila embryogenesis. Genome biology. 2007;8(4):R63. doi: 10.1186/gb-2007-8-4-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram PT, Schultz RM. Reporter gene expression in G2 of the 1-cell mouse embryo. Developmental biology. 1993;156(2):552–556. doi: 10.1006/dbio.1993.1101. [DOI] [PubMed] [Google Scholar]
- Richter JD, Lasko P. Translational control in oocyte development. Cold Spring Harbor perspectives in biology. 2011;3(9):a002758. doi: 10.1101/cshperspect.a002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe M, et al. Loss of gene function through rapid mitotic cycles in the Drosophila embryo. Nature. 1992;359(6391):156–159. doi: 10.1038/359156a0. [DOI] [PubMed] [Google Scholar]
- Rothstein JL, et al. Gene expression during preimplantation mouse development. Genes & development. 1992;6(7):1190–1201. doi: 10.1101/gad.6.7.1190. [DOI] [PubMed] [Google Scholar]
- Ruzov A, et al. Kaiso is a genome-wide repressor of transcription that is essential for amphibian development. Development (Cambridge, England) 2004;131(24):6185–6194. doi: 10.1242/dev.01549. [DOI] [PubMed] [Google Scholar]
- Santos F, et al. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Developmental biology. 2005;280(1):225–236. doi: 10.1016/j.ydbio.2005.01.025. [DOI] [PubMed] [Google Scholar]
- Santos F, et al. Dynamic reprogramming of DNA methylation in the early mouse embryo. Developmental biology. 2002;241(1):172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- Satija R, et al. The TAGteam motif facilitates binding of 21 sequence-specific transcription factors in the Drosophila embryo. Genome research. 2012;22(4):656–665. doi: 10.1101/gr.130682.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki JA, Magnuson T, Epstein CJ. Evidence for expression of the paternal genome in the two-cell mouse embryo. Nature. 1981;294(5840):450–451. doi: 10.1038/294450a0. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, et al. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS biology. 2009;7(1):e13. doi: 10.1371/journal.pbio.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvig SE, Gross PR, Hunter AL. Cytoplasmic synthesis of RNA in the sea urchin embryo. Developmental biology. 1970;22(2):343–365. doi: 10.1016/0012-1606(70)90158-2. [DOI] [PubMed] [Google Scholar]
- Semotok JL, et al. Smaug Recruits the CCR4/POP2/NOT Deadenylase Complex to Trigger Maternal Transcript Localization in the Early Drosophila Embryo. Current Biology. 2005;15(4):284–294. doi: 10.1016/j.cub.2005.01.048. [DOI] [PubMed] [Google Scholar]
- Shechter D, et al. Analysis of histones in Xenopus laevis. I. A distinct index of enriched variants and modifications exists in each cell type and is remodeled during developmental transitions. The Journal of biological chemistry. 2009;284(2):1064–1074. doi: 10.1074/jbc.M807273200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shermoen AW, O'Farrell PH. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell. 1991;67(2):303–310. doi: 10.1016/0092-8674(91)90182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C, Manley JL. The SR protein SRp38 represses splicing in M phase cells. Cell. 2002;111(3):407–417. doi: 10.1016/s0092-8674(02)01038-3. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nature reviews. Molecular cell biology. 2009;10(10):697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Sirard M-A, et al. Potential and limitations of bovine-specific arrays for the analysis of mRNA levels in early development: preliminary analysis using a bovine embryonic array. Reproduction, fertility, and development. 2005;17(1-2):47–57. doi: 10.1071/rd04113. [DOI] [PubMed] [Google Scholar]
- Sive HL, St John T. A simple subtractive hybridization technique employing photoactivatable biotin and phenol extraction. Nucleic acids research. 1988;16(22):10937. doi: 10.1093/nar/16.22.10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirkanich J, et al. An essential role for transcription before the MBT in Xenopus laevis. Developmental biology. 2011;357(2):478–491. doi: 10.1016/j.ydbio.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RC, Dworkin-Rastl E, Dworkin MB. Expression of a histone H1-like protein is restricted to early Xenopus development. Genes & development. 1988;2(10):1284–1295. doi: 10.1101/gad.2.10.1284. [DOI] [PubMed] [Google Scholar]
- Smith ZD, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484(7394):339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancheva I, Meehan RR. Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes & development. 2000;14(3):313–327. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Stancheva I, et al. DNA methylation at promoter regions regulates the timing of gene activation in Xenopus laevis embryos. Developmental biology. 2002;243(1):155–165. doi: 10.1006/dbio.2001.0560. [DOI] [PubMed] [Google Scholar]
- Staudt N, et al. Mutations of the Drosophila zinc finger-encoding gene vielfältig impair mitotic cell divisions and cause improper chromosome segregation. Molecular biology of the cell. 2006;17(5):2356–2365. doi: 10.1091/mbc.E05-11-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitzel ML, Pellettieri J, Seydoux G. The C. elegans DYRK Kinase MBK-2 Marks Oocyte Proteins for Degradation in Response to Meiotic Maturation. Current biology : CB. 2006;16(1):56–62. doi: 10.1016/j.cub.2005.11.063. [DOI] [PubMed] [Google Scholar]
- Storfer-Glazer FA, Wood WB. Effects of chromosomal deficiencies on early cleavage patterning and terminal phenotype in Caenorhabditis elegans embryos. Genetics. 1994;137(2):499–508. doi: 10.1093/genetics/137.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, et al. The embryonic cell lineage of the nematode Caenorhabditis elegans. Developmental biology. 1983;100(1):64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Sun F, et al. Expression of SRG3, a chromatin-remodelling factor, in the mouse oocyte and early preimplantation embryos. Zygote (Cambridge, England) 2007;15(2):129–138. doi: 10.1017/S096719940600400X. [DOI] [PubMed] [Google Scholar]
- Sung H-W, et al. Number of nuclear divisions in the Drosophila blastoderm controlled by onset of zygotic transcription. Current biology : CB. 2013;23(2):133–138. doi: 10.1016/j.cub.2012.12.013. [DOI] [PubMed] [Google Scholar]
- Surdej P, Jacobs-Lorena M. Developmental regulation of bicoid mRNA stability is mediated by the first 43 nucleotides of the 3' untranslated region. Molecular and cellular biology. 1998;18(5):2892–2900. doi: 10.1128/mcb.18.5.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinburne IA, Silver PA. Intron delays and transcriptional timing during development. Developmental cell. 2008;14(3):324–330. doi: 10.1016/j.devcel.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen CS, Syljuåsen RG. Safeguarding genome integrity: the checkpoint kinases ATR, CHK1 and WEE1 restrain CDK activity during normal DNA replication. Nucleic acids research. 2012;40(2):477–486. doi: 10.1093/nar/gkr697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development (Cambridge, England) 2009;136(18):3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Tadros W, et al. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Developmental cell. 2007;12(1):143–155. doi: 10.1016/j.devcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tan MH, et al. RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. Genome research. 2013;23(1):201–216. doi: 10.1101/gr.141424.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, et al. A mammalian oocyte-specific linker histone gene H1oo: homology with the genes for the oocyte-specific cleavage stage histone (cs-H1) of sea urchin and the B4/H1M histone of the frog. Development (Cambridge, England) 2001;128(5):655–664. doi: 10.1242/dev.128.5.655. [DOI] [PubMed] [Google Scholar]
- Tani S, et al. Genomic organization and embryonic expression of miR- 430 in medaka (Oryzias latipes): insights into the post-transcriptional gene regulation in early development. Gene. 2010;449(1-2):41–49. doi: 10.1016/j.gene.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Thomas S, et al. Dynamic reprogramming of chromatin accessibility during Drosophila embryo development. Genome biology. 2011;12(5):R43. doi: 10.1186/gb-2011-12-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Padilla M-E, et al. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. The International journal of developmental biology. 2006;50(5):455–461. doi: 10.1387/ijdb.052073mt. [DOI] [PubMed] [Google Scholar]
- Tsurumi A, et al. STAT is an essential activator of the zygotic genome in the early Drosophila embryo. PLoS genetics. 2011;7(5):e1002086. doi: 10.1371/journal.pgen.1002086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden GW, et al. Transmission of modified nucleosomes from the mouse male germline to the zygote and subsequent remodeling of paternal chromatin. Developmental biology. 2006;298(2):458–469. doi: 10.1016/j.ydbio.2006.06.051. [DOI] [PubMed] [Google Scholar]
- Vassena R, et al. Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development (Cambridge, England) 2011;138(17):3699–3709. doi: 10.1242/dev.064741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, et al. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464(7290):922–926. doi: 10.1038/nature08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra GJ, Destrée OH, Wolffe AP. Translation of maternal TATA-binding protein mRNA potentiates basal but not activated transcription in Xenopus embryos at the midblastula transition. Molecular and cellular biology. 1999;19(12):7972–7982. doi: 10.1128/mcb.19.12.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra GJC, Weeks DL, Wolffe AP. Distinct Roles for TBP and TBP-Like Factor in Early Embryonic Gene Transcription in Xenopus. Science (New York, N.Y.) 2000;290(5500):2312–2315. doi: 10.1126/science.290.5500.2312. [DOI] [PubMed] [Google Scholar]
- Voeltz GK, Steitz JA. AUUUA Sequences Direct mRNA Deadenylation Uncoupled from Decay during Xenopus Early Development. Molecular and cellular biology. 1998;18(12):7537–7545. doi: 10.1128/mcb.18.12.7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser CB, Lipshitz HD. Transcript clearance during the maternal-to-zygotic transition. Current opinion in genetics & development. 2011;21(4):431–443. doi: 10.1016/j.gde.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Wang J, Garrey J, Davis RE. Transcription in pronuclei and one- to four-cell embryos drives early development in a nematode. Current biology : CB. 2014;24(2):124–133. doi: 10.1016/j.cub.2013.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QT, et al. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Developmental cell. 2004;6(1):133–144. doi: 10.1016/s1534-5807(03)00404-0. [DOI] [PubMed] [Google Scholar]
- Warner CM, et al. In vivo and in vitro effect of alpha-amanitin on preimplantation mouse embryo RNA polymerase. Nature. 1974;248(450):678–680. doi: 10.1038/248678a0. [DOI] [PubMed] [Google Scholar]
- Wei Z, Angerer RC, Angerer LM. A database of mRNA expression patterns for the sea urchin embryo. Developmental biology. 2006;300(1):476–484. doi: 10.1016/j.ydbio.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle CM, et al. The genomic distribution and function of histone variant HTZ-1 during C. elegans embryogenesis. PLoS genetics. 2008;4(9):e1000187. doi: 10.1371/journal.pgen.1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiekowski M, Miranda M, DePamphilis ML. Regulation of gene expression in preimplantation mouse embryos: Effects of the zygotic clock and the first mitosis on promoter and enhancer activities. Developmental biology. 1991;147(2):403–414. doi: 10.1016/0012-1606(91)90298-h. [DOI] [PubMed] [Google Scholar]
- Wiekowski M, Miranda M, DePamphilis ML. Requirements for promoter activity in mouse oocytes and embryos distinguish paternal pronuclei from maternal and zygotic nuclei. Developmental biology. 1993;159(1):366–378. doi: 10.1006/dbio.1993.1248. [DOI] [PubMed] [Google Scholar]
- Wu S-F, et al. Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm. Genome research. 2011;21(4):578–589. doi: 10.1101/gr.113167.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D, et al. Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome research. 2010;20(6):804–815. doi: 10.1101/gr.100594.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, et al. Nanog-like regulates endoderm formation through the Mxtx2-Nodal pathway. Developmental cell. 2012;22(3):625–638. doi: 10.1016/j.devcel.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500(7464):593–597. doi: 10.1038/nature12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, et al. Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Developmental cell. 2011;20(4):483–496. doi: 10.1016/j.devcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, et al. Beta-catenin/Tcf-regulated transcription prior to the midblastula transition. Development (Cambridge, England) 2002;129(24):5743–5752. doi: 10.1242/dev.00150. [DOI] [PubMed] [Google Scholar]
- Yasuda GK, Schubiger G. Temporal regulation in the early embryo: is MBT too good to be true? Trends in genetics : TIG. 1992;8(4):124–127. doi: 10.1016/0168-9525(92)90369-F. [DOI] [PubMed] [Google Scholar]
- Yasuda GK, Baker J, Schubiger G. Temporal regulation of gene expression in the blastoderm Drosophila embryo. Genes & development. 1991;5(10):1800–1812. doi: 10.1101/gad.5.10.1800. [DOI] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144(6):940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalokar M. Autoradiographic study of protein and RNA formation during early development of Drosophila eggs. Developmental biology. 1976;49(2):425–437. doi: 10.1016/0012-1606(76)90185-8. [DOI] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes & development. 2011;25(21):2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F, Schultz RM. Gene expression in mouse oocytes and preimplantation embryos: use of suppression subtractive hybridization to identify oocyte- and embryo-specific genes. Biology of reproduction. 2003;68(1):31–39. doi: 10.1095/biolreprod.102.007674. [DOI] [PubMed] [Google Scholar]
- Zhang P, et al. Transcriptome profiling of human pre-implantation development. PloS one. 2009;4(11):e7844. doi: 10.1371/journal.pone.0007844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. Canonical nucleosome organization at promoters forms during genome activation. Genome research. 2014;24(2):260–266. doi: 10.1101/gr.157750.113. [DOI] [PMC free article] [PubMed] [Google Scholar]