Abstract
Background
Tumor Necrosis Factor (TNF) signaling protects against ischemia-reperfusion-induced cardiomyocyte death, in-vitro, ex-vivo and in-vivo. TNF-Receptor Associated Factor-2 (TRAF2), an E3 ubiquitin ligase, coordinates cytoprotective signaling downstream of both TNF receptors, via unclear mechanisms. Noting that TRAF2 is recruited to mitochondria, and that autophagic removal of ubiquitin-tagged damaged mitochondria is cytoprotective, we tested the hypothesis that TRAF2 mediates mitochondrial autophagy.
Methods and Results
TRAF2 localizes to the mitochondria in neonatal rat cardiac myocytes (NRCMs) and TNF treatment transcriptionally upregulates TRAF2 abundance in the mitochondrial sub-fraction. TRAF2 co-localizes with ubiquitin, p62 adaptor protein, and mitochondria within LC3-bound autophagosomes; and exogenous TRAF2 enhances autophagic removal of mitochondria. TRAF2 knockdown with adenoviral shRNA transduction induces accumulation of depolarized mitochondria in resting NRCMs, as well as in those treated with TNF or uncoupling agent CCCP, suggesting an essential role for TRAF2 in homeostatic and stress-induced mitochondrial autophagy. TRAF2 also co-localizes with and interacts with PARKIN, a previously described E3 ubiquitin ligase and mitophagy effector, on depolarized mitochondria in NRCMs. Exogenous expression of TRAF2, but not its E3 ligase-deficient mutants, is sufficient to partially restore mitophagy in the setting of PARKIN knockdown, suggesting redundancy in their ubiquitin ligase roles. TRAF2 abundance increases in the mitochondrial sub-fraction of ischemia-reperfusion-modeled hearts; and exogenous TRAF2, but not its E3 ligase-deficient mutants, reduces depolarized mitochondria and rescues cell death in NRCMs subjected to hypoxia-reoxygenation.
Conclusions
Taken together, these data indicate an essential role for TRAF2 in concert with PARKIN as a mitophagy effector, which contributes to TRAF2-induced cytoprotective signaling.
Keywords: mitophagy, TNF signaling, TRAF2, hypoxia-reoxygenation
Emerging evidence indicates that activation of innate immunity signaling is critical for myocardial adaptation to stress (reviewed in 1). One such highly evolutionary conserved pathway is activated by Tumor Necrosis Factor (TNF), the prototypical member of the TNF superfamily of ligands.2 Indeed, TNF pretreatment, or activation of either TNFR1 or TNFR2 receptor, prevents hypoxia-reperfusion-induced cell death in mammalian cardiomyocytes, in-vitro;3 and transgenic expression or exogenous administration of low doses of TNF attenuates ex-vivo cardiac ischemia-reperfusion (IR) injury.4, 5 Also, absence of both TNFR1 and TNFR2 receptors increases IR-induced cardiomyocyte death, ex-vivo4 and results in marked increase in infarct size with in-vivo coronary ligation as compared with controls;6 pointing to a redundancy in cytoprotective signaling triggered by TNF via its cognate receptors. In this context, it is notable that TNF-receptor associated factor-2 (TRAF2), a scaffolding protein, is recruited to both TNF receptors upon their activation,7 and its transgenic expression (at low levels) attenuates cardiomyocyte death with experimental ex-vivo ischemia-reperfusion injury.4 While these data suggest that TRAF2 may facilitate cytoprotective signaling downstream of both TNF receptors, the underlying mechanisms remain largely unknown, despite extensive investigation.3-5
Ischemia-reperfusion injury results in generation of reactive oxygen species, which provoke mitochondrial permeabilization leading to programmed cardiomyocyte death.8 Autophagy is an evolutionarily conserved pro-survival pathway that sequesters damaged mitochondria within autophagosomes resulting in their intralysosomal degradation (by mitophagy), which is essential to protect against cardiomyocyte death in myocardial infarction.9, 10 Activation of mitophagy also plays a central role in ischemia preconditioning.11 TNF signaling is implicated in induction of cardiomyocyte autophagy, which is cytoprotective against LPS-induced cell death.12 Whether TNF induces mitophagy or signals via TRAF2 to promote mitochondrial autophagy, is not known.
Ubiquitination of mitochondrial proteins in response to mitochondrial damage is essential for their sequestration and degradation within the lysosomes.13 PARKIN, an E3 ubiquitin ligase is recruited to damaged mitochondria via activation by PINK1 (PTEN-induced putative kinase 1, a serine-heroine kinase), and ubiquitinates mitochondrial proteins.14, 15 However, while targeted ablation of PINK1 in cardiac myocytes results in mitochondrial abnormalities and cardiomyopathy,16 loss of PARKIN is well tolerated in the unstressed state,17 suggesting that other E3 ubiquitin ligases may be involved in removal of damaged mitochondria. Relevant to this discussion is the observation that TRAF2, an E3 ubiquitin ligase,18 is recruited to the mitochondria by MAVS (Mitochondrial Anti-Viral-Signalosome), a mitochondrially localized protein with multiple scaffolding domains,19-21 following activation of innate immune signaling pathways. Therefore, we tested the hypothesis that TRAF2 mediates mitochondrial autophagy. Here, we show for the first time that TRAF2 is present on the mitochondria in resting cardiac myocytes, and functions in concert with PARKIN as an E3 ubiquitin ligase to facilitate autophagic removal of damaged mitochondria, raising the intriguing possibility that TRAF2-mediated mitophagy may be an important determinant of cytoprotective TNF signaling in ischemia-reperfusion injury.
Methods
Neonatal rat cardiac myocyte (NRCM) cultures were prepared as described.22, 23 MHCsTNF mice with cardiomyocyte-specific overexpression of wild type (secretable) TNF were described previously.24 In-vivo ischemia-reperfusion injury was performed in adult male C57Bl6 mice, as described.22 All animal studies were approved by the Animal Studies Committee at Washington University School of Medicine; and by the Institutional Animal Care and Use Committee at the John Cochran VA Medical Center. Hypoxia-reoxygenation modeling, generation of adenoviral constructs, immunofluorescence and electron microscopy imaging, quantitative PCR analysis, flow cytometry, sub-cellular fractionation, assessment of mitochondrial DNA content, citrate synthase assays, immunoblotting and assessment of cell death were performed as previously described,22 with details described in supplementary methods.
Statistical analysis
post-hoc pairwise comparisonpost-hoc pairwise comparisonResults are expressed as mean ± SEM. Statistical differences were assessed with the unpaired 2-tailed Student’s t-test for two experimental groups, one-way ANOVA for multiple groups; and two-way ANOVA for testing two variables across multiple groups, with the SPSS software. Bonferroni’s post-hoc pairwise comparison adjustment was employed after ANOVA for testing for significant differences between groups. Assumptions of normality were verified with visual examination of the residuals via histograms and the Shapiro-Wilk test; and variance was assessed by employing the Levene’s test for equality of variances for t-test and the Levene’s homogeneity of variance test for ANOVA, using SPSS software. Non-parametric tests (Mann-Whitney and Kruskal-Wallis) were employed for data that was not normally distributed (in lieu of t-test and one-way ANOVA, respectively); and Dunnett’s T3 post-hoc analysis was applied after one-way ANOVA for data demonstrating unequal variance. A two-tailed P value of less than 0.05 was considered statistically significant.
Results
TNF induces mitochondrial autophagy with increased TRAF2 localization to mitochondria
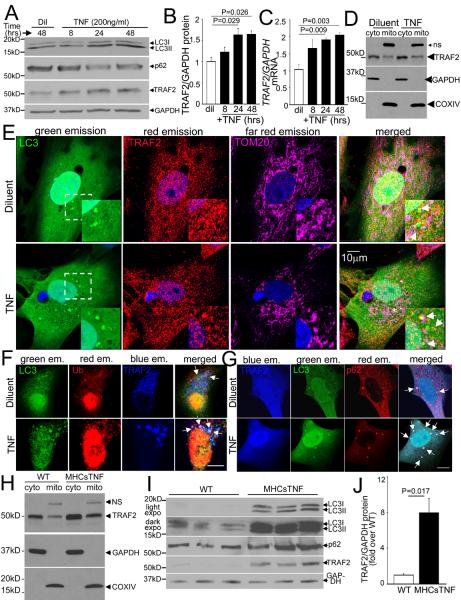
Previous studies indicate that TNF treatment is sufficient to induce autophagy in cardiomyocytes.12 We confirmed that TNF treatment induces autophagy in isolated neonatal rat cardiomyocytes (NRCMs) with progressive increase in levels of autophagosome-bound LC3-II (Fig 1A) and decline in p62 (an adaptor protein that gets consumed during autophagy, Fig. 1A; S1A); without alteration in their transcript levels (Fig. S1B, C). To evaluate the role of TRAF2 in TNF-induced autophagy, we examined TRAF2 expression and distribution in TNF-treated NRCMs. TNF-treatment induced a time-dependent upregulation of TRAF2 protein (Fig. 1A, B) and transcripts (Fig. 1C), paralleling the induction of autophagy. Guided by recent data demonstrating that TRAF2 is targeted to the mitochondria either as a part of the DISC (death-inducing signaling complex) upon TNF stimulation,25, 26 or via its interactions with a scaffolding protein MAVS (Mitochondria in Anti-Viral Signaling) after activation of innate immune responses,19-21 we examined whether TRAF2 localizes to mitochondria in cardiac myocytes. We found that endogenous TRAF2 co-segregates with cytochrome oxidase IV (COXIV), which is an inner mitochondrial membrane-localized respiratory chain protein (Fig. 1D) in unstimulated NRCMs; and co-localizes with TOM20 (Translocase of outer mitochondrial membrane 20 homologue; Fig. 1E, S2A), an outer mitochondrial membrane protein, as well as with COX-IV (Fig. S2B, C) on confocal microscopy, indicating that it is present on the mitochondria in the resting state. Notably, TNF treatment increased the relative abundance of TRAF2 in the mitochondrial sub-fraction (by 30% vs. diluent-treated group, n=3/group, P=0.026; Fig. 1D), with increased co-localization of endogenous TRAF2 and mitochondrial proteins in punctate GFP-LC3-labeled autophagosomes (Fig. 1E, S2A-C). TRAF2 also co-localizes with ubiquitin (Fig. 1F, S2D) and mitophagy adaptor protein, p62 (Fig. 1G, S2E),11 in GFP-LC3-labeled autophagosomes in unstimulated NRCMs; and this co-localization was further stimulated by TNF treatment suggesting that TRAF2 may participate in ubiquitination of mitochondrial proteins and autophagic sequestration of damaged mitochondria in both resting and TNF-treated cells.
Figure 1. TNF upregulates endogenous TRAF2 expression and TRAF2 co-localizes with mitochondria in autophagosomes.
A, B) Representative immunoblots (A) depicting expression of LC3, p62, and TRAF2 (GAPDH as loading control); with quantitation of TRAF2 (B) in NRCMs treated with TNF (200ng/ml) or diluent for the indicated duration (n=3/time point). C) TRAF2 transcripts in NRCMs treated as in A (n=4/time point). P values are by post-hoc pairwise comparison vs. diluent after one-way ANOVA. D) Representative immunoblot demonstrating sub-cellular localization of endogenous TRAF2 in mitochondria-enriched (COXIV-expressing) and cytoplasmic fraction (GAPDH-expressing) in NRCMs treated with TNF (200ng/ml) or diluent for 24 hours. E-G) Representative confocal images demonstrating co-localization of endogenous TRAF2 with mitochondria (TOM20) and GFP-LC3 (arrows, magnified image of boxed area in inset in E); with GFP-LC3 and ubiquitin (arrows, in F); and with GFP-LC3 and p62 (arrows, in G) in NRCMs treated with TNF (200ng/ml) or diluent for 24 hours. Scale bar =10μm. H) Immunoblot demonstrating sub-cellular localization of endogenous TRAF2 in cardiac extracts from MHCsTNF and littermate mice fractionated into a mitochondria-enriched (COX IV expressing) and cytoplasmic fraction (GAPDH expressing). I, J) Representative immunoblot (I) of myocardial LC3-II, p62 and TRAF2 expression, with TRAF2 quantitation (J) in MHCsTNF and littermate controls (n=5-6/group). P value is by t-test.
In parallel studies, we also found that TRAF2 co-segregates with COXIV in mitochondria-enriched sub-fraction from wild-type mouse hearts (Fig. 1H); and its abundance is increased in the cardiac mitochondrial fraction of transgenic mice with cardiac restricted overexpression of TNF (fold change in TRAF2/COXIV over control: 3.2±0.4 fold in MHCsTNF mice vs. 1.0±0.1 in littermates, P=0.005, n=3/group; Fig 1H). Indeed, similar to the observations with TNF treatment of NRCMs, total TRAF2 abundance is transcriptionally induced in MHCsTNF transgenic hearts (Fig. 1I, J, S3A), along with increased autophagosome-bound LC3-II (Fig. 1I, S3B). Interestingly, the increase in total LC3 and p62 protein abundance (Fig. 1I, S3C, D) and their respective transcript levels (Fig. S3E, F), points to a transcriptional stimulation of autophagy with sustained TNF signaling in MHCsTNF hearts; whereas a transcriptional response is not observed with short-term TNF treatment, in-vitro (Fig. S1B, C). Importantly, ultrastructural examination of cardiomyocytes from MHCsTNF transgenic myocardium demonstrated evidence of mitophagy with abnormal mitochondria manifesting swelling and rarefaction of crista (Fig. S4A) enclosed within autophagic structures. This was accompanied by significantly reduced mitochondrial DNA content (Fig. S4B) and a trend towards reduced citrate synthase activity (Fig. S4C) in the MHCsTNF myocardium as compared with littermate controls, indicating reduced mitochondrial mass, likely as a result of upregulated mitochondrial autophagy. Taken together, these data confirm that TNF induces TRAF2 expression with evidence of autophagic sequestration of mitochondria, which raises the interesting possibility that TNF and/or TRAF2-induced mitophagy may contribute to the reduced mitochondrial mass observed in MHCsTNF mouse hearts.
Exogenous TRAF2 is sufficient to induce mitochondrial autophagy
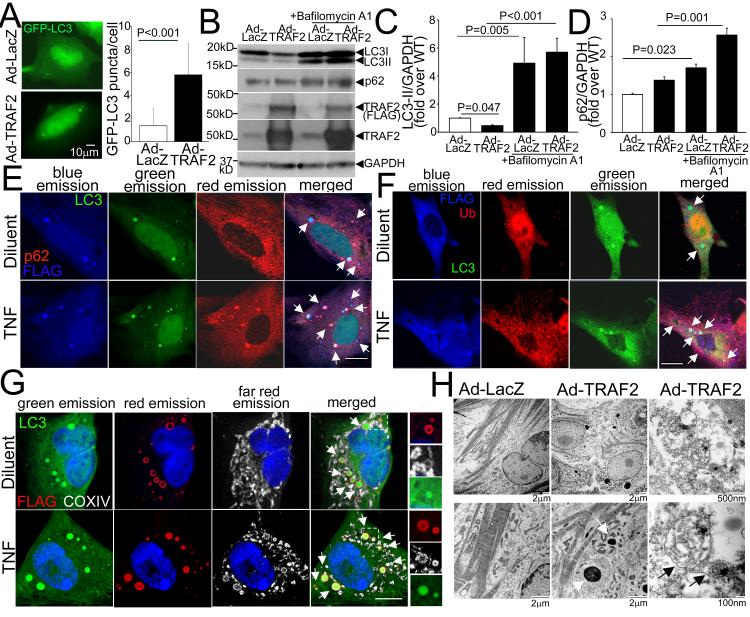
To determine if TRAF2 is sufficient to stimulate autophagy in cardiac myocytes, we adenovirally transduced NRCMs with TRAF2 and evaluated distribution of GFP-tagged LC3. As shown, exogenous TRAF2 expression induced autophagosome formation resulting in increased GFP-tagged LC3 puncta, as compared with control (Fig. 2A). TRAF2 overexpression (~10-fold over endogenous levels, Fig. 2B) induced a decline in endogenous LC3-II levels (Fig. 2B, C), with a decline in total LC3 abundance (fold change in total LC3/GAPDH over control: 0.51±0.09 in Ad-TRAF2 vs. 1.0±0.03 in Ad-LacZ-treated controls, P<0.001, N=7/group; see Figure 2B). Taken together with the accumulation of LC3-II and p62 (Fig. 2B-D) in presence of Bafilomycin A1 (to inhibit lysosomal acidification), these data indicate that TRAF2 stimulates autophagy with intact flux. Exogenous TRAF2 co-localized with GFP-tagged punctate LC3 (Fig. 2E-G, S2F), p62 (Fig. 2E) and ubiquitin (Fig. 2F) in ring-shaped structures, many of which enclose mitochondria (Fig. 2G and S2F (see magnified images)). Interestingly, this co-localization was observed with increased frequency in TNF-treated NRCMs (Fig. 2E-G, S2F) wherein the mitochondria appear increasingly fragmented (Fig. S2B, F) suggesting a role for exogenous TRAF2 in ubiquitination and autophagic sequestration of damaged mitochondria in TNF-treated cells. Ultrastructural analysis of TRAF2-treated NRCMs revealed multiple autophagic structures enclosing mitochondria (Fig. 2H). We also observed occasional amorphous dense deposits within TRAF2-transduced cardiac myocytes by EM analysis. Such deposits are observed in mice with transgenic expression of high levels of TRAF2 in the mouse myocardium,27 and may represent the aggregates of ubiquitinated cytoskeletal proteins observed in the MHCsTNF hearts.28 While this observation suggests a more generalized role for TRAF2 downstream of TNF signaling in protein ubiquitination; these data, taken together, indicate that TNF-induced upregulation of TRAF2 is also involved in ubiquitination of mitochondrial proteins targeted for autophagic sequestration.
Figure 2. Exogenous TRAF2 localizes to the mitochondria in autophagosomes.
A) Representative immunofluorescence images demonstrating GFP-LC3 expression (left) with quantitation (right) of punctate GFP-LC3 in NRCMs adenovirally transduced with FLAG-TRAF2 or LacZ as control (multiplicity of infection, MOI=100, 48hrs). N=45-52 nuclei/group. P value is by Mann-Whitney test. B-D) Representative immunoblot (B) and quantitation of LC3-II (C) and p62 (D) expression (with GAPDH as loading control) in NRCMs adenovirally transduced with TRAF2 or LacZ (MOI=100, 48hrs) and treated with Bafilomycin A1 (100nmol/l, 24hrs) to assess autophagic flux. N=7/group. P values are by post-hoc pairwise comparisons after Kruskal-Wallis test in C and D. E-G) Representative confocal images demonstrating co-localization of adenovirally-transduced FLAG-tagged TRAF2 with GFP-LC3 and p62 (arrows, in E); with GFP-LC3 and ubiquitin (arrows, in F); and with GFP-LC3 and mitochondria (identified by COX IV immunostaining; arrows, in G) in NRCMs treated with TNF (200ng/ml) or diluent. Boxed area is shown with magnification of individual channels to demonstrate co-localization. Scale bar =10μm. H) Representative transmission electron micrographs of NRCMs adenovirally transduced with TRAF2 or LacZ as control (MOI=100) demonstrating increased autophagic structures enclosing mitochondrial remnants, with TRAF2 overexpression. White arrows point to amorphous proteinaceous deposits; and black arrows point to autophagic structures.
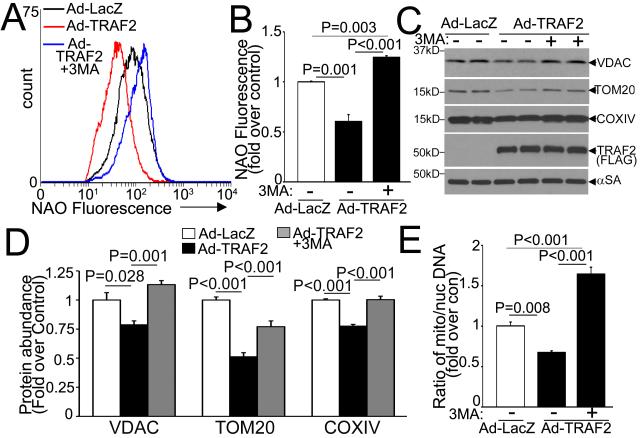
Exogenous TRAF2 provoked a decline in mitochondrial mass assessed by cardiolipin content (with expression of nonyl-acridine orange, NAO; Fig. 3A, B), mitochondrial protein abundance (Fig. 3C, D) and mitochondrial DNA content (Fig. 3E); which was prevented by treatment with 3-methyl adenine (3MA; a PI3Kinase III inhibitor,29 which inhibits autophagosome formation). Also, TRAF2 overexpression led to reduced citrate synthase activity (100.7±7.8 vs. 147.8±4.1 nmol/mg/min in LacZ-treated controls, P=0.006, n=3/group) indicating that exogenous TRAF2 is sufficient to provoke autophagic removal of mitochondria.
Figure 3. Exogenous TRAF2 facilitates autophagic removal of mitochondria.
A, B) Representative flow cytometric tracings (A) with quantitation (B) of NAO expression in NRCMs adenovirally transduced with TRAF2 or LacZ (MOI=100, 48hrs) and treated with 3MA (7mmol/l, 24hrs) or diluent (n=3-9/group). C, D) Representative immunoblot (C) with quantitation (D) of selected mitochondrial proteins in NRCMs treated as in A (n=4/group). α-Sarcomeric actin (αSA) is shown as loading control. E) Mitochondrial DNA content in NRCMs treated as in A (n=4/group). All P values are by post-hoc pairwise comparison after one-way ANOVA.
Endogenous TRAF2 is essential for removal of mitochondria with TNF treatment
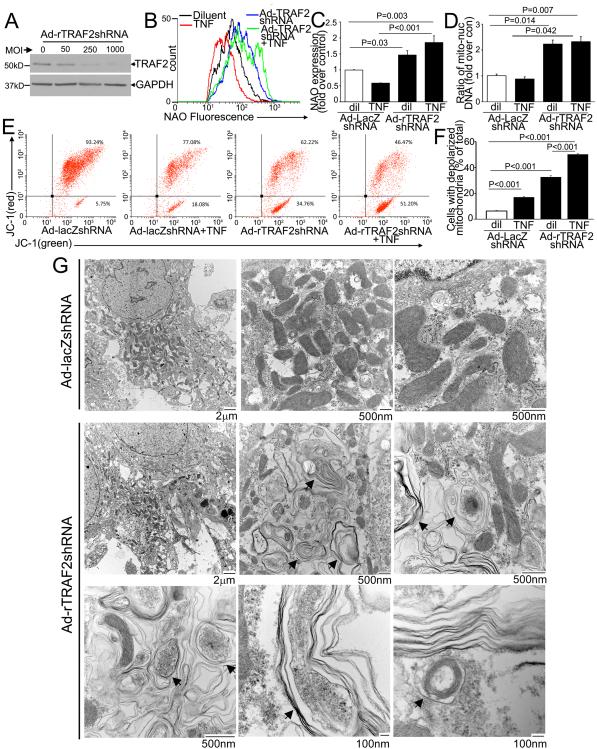
TNF-treatment in NRCMs induced a decline in mitochondrial mass assessed with NAO expression (Fig. S5A, B) and mitochondrial DNA content (Fig. S5C) as compared with control; and concomitant inhibition of autophagy with 3MA prevented this decline (Fig. S5A-C) indicating that TNF induces mitophagy, in-vitro, mirroring the observations in MHCsTNF transgenic mice (Fig. S4). To examine the role of endogenous TRAF2 in TNF-induced mitophagy, we adenovirally transduced NRCMs with shRNA targeting rat TRAF2 (or LacZ as non-targeting control, Fig. 4A) and assessed total mitochondrial mass, and relative content of depolarized mitochondria. Interestingly, knockdown of endogenous TRAF2 provoked a significant increase in NAO expression and mitochondrial DNA content not only in TNF-treated, but also in diluent-treated NRCMs (Fig. 4B-D); with increased citrate synthase activity (310.6±20.1 with TRAF2shRNA vs. 171.7±12.8 nmol/mg/min in LacZshRNA control, P=0.005, n=3/group) in diluent-treated NRCMs, indicating that TRAF2 knockdown results in increased mitochondrial mass. To assess if increased mitochondrial biogenesis was contributing to the observed increase in mitochondrial mass, we examined the transcript levels for PGC1α and PGC1β, two transcriptional co-activators essential for mitochondrial biogenesis in cardiac myocytes.30 TRAF2 knockdown was associated with a significant decline in the expression of PGC1α and PGC1β (Fig. S6) as compared with controls, suggesting suppression of the mitochondrial biogenesis program, likely in response to accumulation of damaged mitochondria with TRAF2 knockdown. Indeed, TRAF2 knockdown resulted in a relative increase in depolarized mitochondria in diluent-treated NRCMs, evidenced by a significant increase in expression of JC-1 monomers (that fluoresce green, right lower quadrant in Fig. 4E, F) with concomitant loss of JC-1 aggregates (cells that fluoresce both red and green, right upper quadrant) indicating loss of mitochondrial inner membrane potential. These observations with TRAF2 knockdown mirror the effect of 3MA treatment to inhibit basal autophagy in resting cells, which also results in accumulation of depolarized mitochondria (Fig. S7) and increased mitochondrial mass (Fig. S5A-C). 31
Figure 4. Endogenous TRAF2 is essential for removal of damaged mitochondria.
A) Immunoblot depicting knockdown of endogenous TRAF2 protein with increasing dose (MOI) of viral transduction after 72 hours. Adenoviral particles coding for shRNA targeting LacZ (as control) were added to equalize viral dose. B, C) NAO expression with representative flow cytometric tracings (B) and quantitation of mean fluorescence (C) in NRCMs adenovirally transduced with shRNA targeting rat TRAF2 or LacZ as non-targeting control (MOI=200; 72 hrs) and treated with TNF (200ng/ml; 24 hrs). N=7/group. P values are by pair-wise comparison after Kruskal-Wallis test. D) Mitochondrial DNA (n=5-6/group); and E, F) JC-1 expression (n=3/group) in NRCMs treated as in B. P values are by post-hoc pairwise comparison after one-way ANOVA. G) Representative TEM images of NRCMs transduced with shRNA targeting rat TRAF2 or LacZ for 72 hours. Black arrows point to membrane bound structures, often with multiple layers of membranes akin to onion skin appearance, enclosing remnants of mitochondrial cristae.
Importantly, the relative increase in NAO expression with TRAF2 knockdown (as compared with LacZshRNA-treated control) was significantly more in TNF-treated as compared with diluent-treated controls (3.4±0.4 vs. 1.5±0.1 fold over respective LacZshRNA group; P<0.001, N=7/group; Fig. 4B, C) with a trend towards increase in mitochondrial DNA content (2.7±0.2 vs. 2.2±0.1 fold over respective LacZshRNA-treated group, P=0.07, N=6/group; Fig. 4D). Also, TRAF2 knockdown resulted in significantly increased content of depolarized mitochondria in TNF-treated as compared with diluent-treated NRCMs (Fig. 4E, F), indicating further accumulation of depolarized mitochondria with TRAF2 deficiency in TNF-treated cells. Remarkably, a gain of function strategy with exogenous TRAF2 expression had the opposite effect. Exogenous TRAF2 significantly reduced the relative proportion of depolarized mitochondria in TNF-treated cells (Fig. S5D, E) with a trend towards a further reduction in NAO expression (as compared with Ad-LacZ control, Fig. S5A left, B). And, while exogenous TRAF2 did not further reduce mitochondrial DNA in TNF-treated cells (Fig. S5C), examination of flux with simultaneous 3MA treatment to prevent mitochondrial autophagy32, 33 revealed that TNF treatment increased flux through mitochondrial autophagy pathway which was further upregulated by concomitant exogenous TRAF2 expression, as evidenced by fold increase in NAO expression with 3MA treatment (1.2±0.1, 2.5±0.3, and 4.7±0.1 fold increase over the respective diluent-treated group in 3MA only, TNF+3MA and TNF+TRAF2+3MA, respectively; P<0.05 by post-hoc pairwise comparison for all comparisons; N=5/group; Fig. S5B) and fold increase in mitochondrial DNA content with 3MA treatment (1.2±0.1, 1.4±0.1, and 1.6±0.1 fold increase over respective diluent-treated group in 3MA only, TNF+3MA and TNF+TRAF2+3MA, respectively; P<0.05 by post-hoc pairwise comparison for all comparisons; N=5-11/group; Fig. S5C). Taken together, these data point to an essential role for TRAF2 in facilitating TNF-induced mitophagy.
Endogenous TRAF2 is essential for homeostatic clearance of depolarized mitochondria
The observation that TRAF2 knockdown increases mitochondrial mass and depolarization without inducing the mitochondrial biogenic program in resting cells (vide supra) indicates accumulation of damaged mitochondria with TRAF2 deficiency. To confirm this finding, we examined the mitochondrial ultrastructure in NRCMs transduced with TRAF2shRNA, and found a striking accumulation of degenerating structures containing mitochondria-like crista within multiple membranes, often in an onion skin-like whorled appearance (Fig. 4G). Conceivably, these structures represent damaged mitochondria in various stages of degeneration, when their well-orchestrated autophagic removal is prevented in the absence of TRAF2.
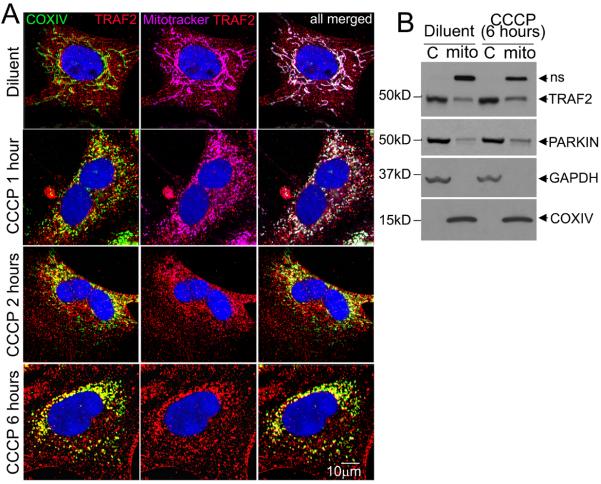
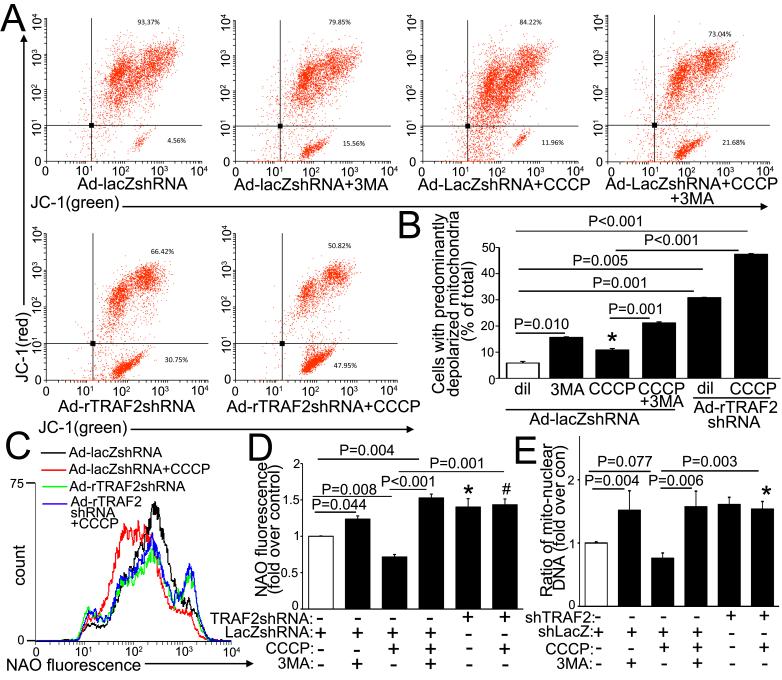
Previous studies have demonstrated a central role for PINK1 in sensing mitochondrial damage to facilitate recruitment of an E3 ubiquitin ligase, PARKIN, to ubiquitinate mitochondrial proteins and target damaged mitochondria for autophagic degradation.15 However, the lack of alteration in cardiac structure and function with detection of normally functioning cardiomyocyte mitochondria in unstressed PARK2 null young adult mice (with loss of PARKIN)17, suggests redundancy with other ubiquitin ligases in their role in homeostatic mitochondrial autophagy in cardiac myocytes. Given our observations with accumulation of depolarized mitochondria with TRAF2 knockdown (Fig. 4), we tested the hypothesis that TRAF2, which is also an E3 ubiquitin ligase, plays an important role in autophagic removal of depolarized mitochondria in cardiac myocytes. We treated NRCMs with carbonyl cyanide m-chlorophenyl hydrazone (CCCP), an ionophore that provokes loss of mitochondrial membrane potential and targets mitochondria for autophagic degradation.15 Treatment with CCCP provoked progressive mitochondrial fragmentation with loss of mitochondrial potential-dependent mitotracker staining (pink, Fig. 5A) as described previously;14, 15 and increased co-localization of endogenous TRAF2 (red) with depolarized mitochondria (identified by COXIV, green; with co-localization in yellow, Fig. 5A). Also, CCCP treatment provoked an increase in TRAF2 abundance in the mitochondria-enriched sub-cellular fraction paralleling a relative increase in mitochondrial PARKIN abundance (Fig. 5B). CCCP treatment provoked mitochondrial depolarization (Fig. 6A, B), and a decline in mitochondrial mass, as evidenced by a reduction in NAO fluorescence (Fig. 6C, D) with a trend towards reduced mitochondrial DNA content (Fig. 6E). Notably, all parameters were markedly increased with concomitant 3MA treatment indicating accumulation of CCCP-damaged mitochondria, when their autophagic removal is impaired. Importantly, TRAF2 knockdown also provoked an increase in depolarized mitochondria (Fig. 6A, B) with increased mitochondrial mass (assessed by NAO and mitochondrial DNA content; Fig. 6D-E) in CCCP-treated NRCMs, mimicking the effects of 3MA treatment. Taken together, these data indicate that TRAF2 plays an essential role in homeostatic removal of depolarized mitochondria in cardiomyocytes.
Figure 5. Endogenous TRAF2 localizes to CCCP-depolarized mitochondria.
A) Representative confocal images demonstrating increased co-localization of endogenous TRAF2 with COX IV on mitochondria, with loss of mitotracker (Deep Red) staining in NRCMs treated with CCCP (50μmol/l) or diluent for indicated duration. B) Immunoblot demonstrating sub-cellular localization of endogenous TRAF2 in NRCMs treated with CCCP or diluent and separated into a mitochondria-enriched (COX IV expressing) and cytoplasmic (GAPDH expressing, ‘C’) fraction. Representative of n=2 experiments for A, B.
Figure 6. Endogenous TRAF2 is essential for removal of CCCP-depolarized mitochondria.
A, B) Representative flow cytometric analysis of JC-1 expression (A) with quantitation (B) of cells with depolarized mitochondria (n=3/group) in NRCMs adenovirally transduced with shRNA targeting TRAF2 or LacZ (MOI=200, 72hrs) and treated with CCCP (50μmol/l, 24hrs) +/− 3MA (7mmol/l, 24hrs) or diluent (n=5-6/group). P values are by post-hoc pairwise comparison after one-way ANOVA. ‘*’ indicates P =0.033 vs. control (white bar). C, D) Representative flow cytometric tracings (C) with quantitation (D) of NAO expression (n=3-6/group) in NRCMs treated as in A. P values are by post-hoc pairwise comparison after one-way ANOVA. ‘*’ and ‘#’ indicate P=0.048 and 0.025 vs. control (open bars), respectively. E) Mitochondrial DNA content in NRCMs treated as in A (n=3-6/group). P values reported are by pair-wise comparisons after Kruskal-Wallis test. * indicates P=0.030 vs. control (open bars).
TRAF2 functions in concert with PARKIN to remove damaged mitochondria
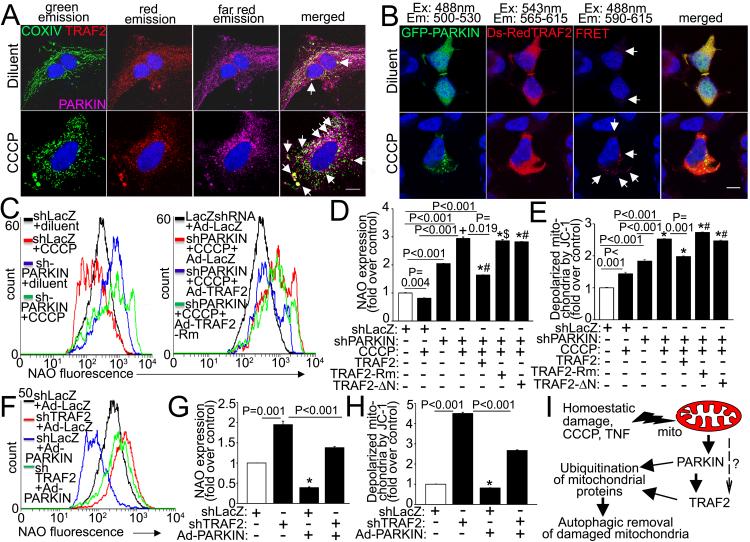
Analogous to the observation that TRAF2 abundance is increased in the mitochondrial compartment with TNF stimulation (Fig. 1D, E, S2B) and in MHCsTNF hearts (Fig. 1H), TNF treatment also stimulates increased PARKIN abundance (Fig. S8A) and mitochondrial localization (Fig. S8B) in NRCMs, and MHCsTNF hearts have increased PARKIN levels (Fig. S8C) in the mitochondrial sub-fraction (Fig. S8D). Viewed together with the observation that both TRAF2 and PARKIN localize to the mitochondrial fraction with CCCP treatment (Fig. 5B), these data indicate that as observed with PARKIN,14 TRAF2 is also recruited to damaged mitochondria. Indeed, TRAF2 co-localizes with PARKIN on mitochondria in resting cardiomyocytes, presumably on damaged mitochondria (Fig. 7A, arrows; S9A); and this co-localization is markedly enhanced with CCCP treatment suggesting that TRAF2 and PARKIN are recruited together to depolarized mitochondria (Fig. 7A, S9A). Interestingly, previous studies suggest that PARKIN physically interacts with TRAF2 and ubiquitinates it independent of TRAF2’s endogenous E3 ubiquitin ligase activity, to activate TRAF2-mediated signaling.34 We have confirmed this physical interaction with a FRET assay employing a DsRed and AcGFP FRET pair-tagged TRAF2 and PARKIN, respectively (with controls in Fig. S9B). The interaction between TRAF and PARKIN is minimal in the resting state (arrows, Fig. 7B) and is markedly enhanced in presence of CCCP-induced mitochondrial depolarization (arrows, Fig. 7B), suggesting increased interaction between these proteins upon mitochondrial damage; which was confirmed via co-immunoprecipitation studies in the mitochondrial sub-fraction of CCCP-treated cells (Fig. S9C, D).
Figure 7. TRAF2 interacts with PARKIN and coordinately facilitates removal of depolarized mitochondria.
A) Representative confocal images demonstrating co-localization of TRAF2 with PARKIN on mitochondria in NRCMs transfected with DsRed-TRAF2 for 24 hours and then treated with CCCP (50μmol/l) or diluent for 8 hrs (arrows). Scale bar = 10μm. B) Representative confocal images demonstrating FRET interaction between AcGFP-tagged PARKIN and DsRed-tagged TRAF2 in HEK293 cells co-transfected with both for 24hrs and then treated with CCCP (10μmol/l) or diluent for 1 hr (arrows). Scale bar = 10μm. C, D) Flow cytometric assessment of NAO expression with representative tracings (E) and quantification (F) in NRCMs adenovirally transduced with shRNA targeting rat PARKIN or LacZ (MOI=10, 72hrs), and CCCP (50μmol/l, 24hrs) in the presence of adenoviruses expressing TRAF2, or its mutants, namely TRAF2-Rm and TRAF2-ΔN (tracing not shown); and LacZ (each at MOI=10, 24hrs). N=4-6/group. ‘*’ indicates P<0.001 vs. control (white bar); and ‘+’, ‘#’, and ‘$’ indicate P=0.002, P<0.001 and P=0.001, respectively vs. PARKINshRNA group (3rd bar from the left). E) Relative abundance of cells expressing predominantly depolarized mitochondria assessed by flow cytometry for JC-1, in cells treated as in E (n=3/group). ‘*’ indicates P<0.001 vs. control (white bar) and ‘#’ indicates P<0.001 vs. PARKINshRNA group (3rd bar from the left). F, G) Flow cytometric assessment of NAO expression with representative tracings (G) and quantitation (H) in NRCMs transduced with shRNA targeting rat TRAF2 or LacZ (MOI=200, 72hrs); and adenoviruses coding for HA-tagged rat PARKIN or LacZ as control (MOI=100, 48hrs). N=6/group. * indicates P<0.001 vs. control (open bar). H) Relative abundance of cells expressing depolarized mitochondria in cells treated as in G (n=3/group). * indicates P<0.05 vs. control (open bar). * indicates P=0.035 vs. control (open bar). All P values are by post-hoc pairwise comparison after one-way ANOVA (for D, E, G and H). I) Schematic depicting proposed role of TRAF2 in mitochondrial autophagy. Mitochondria are damaged in resting cells under homeostatic conditions, and depolarization is also triggered by CCCP (an uncoupler) and with TNF signaling. With mitochondrial damage, PARKIN is recruited and interacts with TRAF2 on the damaged mitochondria, with both acting in concert as E3 ubiquitin ligases to ubiquitinate mitochondrial proteins and target the damaged mitochondria for autophagic removal. “?” indicates a putative PARKIN-independent pathway for recruitment of TRAF2 to damaged mitochondria, experimental evidence for which is lacking at this time.
To examine the role of TRAF2 vis-à-vis PARKIN in mitochondrial autophagy, we transduced NRCMs with adenoviruses coding for shRNA targeting PARKIN (Fig. S10A) in concert with exogenous TRAF2; and examined mitochondrial mass (by NAO expression) and prevalence of depolarized mitochondria (by JC-1). PARKIN knockdown provoked accumulation of depolarized mitochondria with increased NAO expression (Fig. 7C, D) and of JC-1 monomers (Fig. 7E) in the resting state; with further increase observed with CCCP- induced depolarization. Exogenous expression of full length TRAF2, but not TRAF2 mutants deficient in E3 ubiquitin ligase activity (TRAF2-Rm and ΔN-TRAF2, Fig. S11),7, 35, 36 partially attenuated the increase in NAO expression (Fig. 7C, D) and abundance of depolarized mitochondria (Fig. 7E) in CCCP-treated NRCMs transduced with PARKINshRNA (by ~44% and 22%, respectively). Similarly, expression of exogenous TRAF2, but not the E3 ubiquitin ligase deficient mutants, was sufficient to partially attenuate the accumulation of mitochondria in TNF-treated NRCMs transduced with PARKINshRNA (Fig. S12A, B). Therefore, TRAF2 can partially complement PARKIN deficiency as an E3 ubiquitin ligase to promote mitochondrial autophagy.
We next evaluated whether PARKIN can substitute for TRAF2 in promoting mitochondrial autophagy. Exogenous PARKIN (Fig. S10B) is sufficient to provoke a decline in NAO expression (Fig. 7F, G) with a reduction in depolarized mitochondria in resting NRCMs (Fig. 7H) indicating that PARKIN overexpression is sufficient to facilitate autophagic removal of damaged mitochondria. Interestingly, exogenous PARKIN is also sufficient to partially restore mitochondrial autophagy in the setting of TRAF2 knockdown, as evidenced by a reduction in NAO expression (by ~29%; Fig. 7F, G) and depolarized mitochondria (by ~40%; Fig. 7H) when compared with TRAF2-deficient cells treated with Ad-LacZ as control. Notably, endogenous levels of TRAF2 are only modestly increased in the setting of PARKIN knockdown; whereas PARKIN levels are not significantly altered when TRAF2 is knocked down, in-vitro in NRCMs (Fig. S13A-C). This observation suggests that at least in-vitro, regulation of endogenous TRAF2 and PARKIN may be unable to compensate for each other’s deficiency, resulting in a defect in mitophagy when either is knocked down. Interestingly, mitochondrial proteins accumulate differentially with deficiency of TRAF2 or PARKIN suggesting independent roles in ubiquitination and degradation of specific mitochondrial proteins for these E3 ubiquitin ligases (Fig. S13A, D-F). Taken together, these findings suggest that TRAF2 functions as an E3 ubiquitin ligase in concert with PARKIN, to facilitate ubiquitination of proteins on damaged mitochondria and assist in their removal through the process of autophagy (Fig. 7I).
TRAF2 facilitates mitophagy to confer cytoprotection in hypoxia-reoxygenation injury
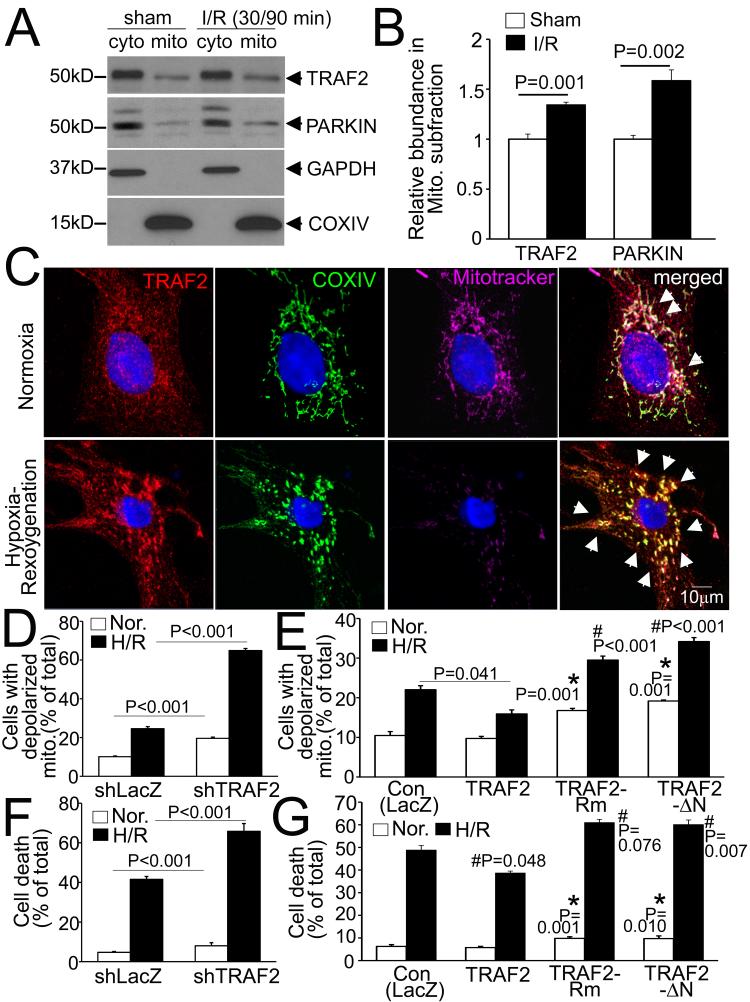
ROS-induced mitochondrial permeabilization is implicated in provoking cardiomyocyte death in IR injury.8 To examine if TRAF2 is recruited to the mitochondria with IR-induced mitochondrial damage, we subjected mice to in-vivo IR modeling and performed sub-cellular fractionation on injured myocardial tissue. Interestingly, we observed a relative increase in TRAF2 abundance in the mitochondrial sub-fraction from the injured myocardium, early after IR injury, paralleling that of PARKIN (Fig. 8A, B). To determine the role of TRAF2 in mitochondrial autophagy in this setting, we examined sub-cellular location of TRAF2 and performed loss-of-function and gain-of-function modeling for TRAF2 in NRCMs subjected to hypoxia-reoxygenation injury, in-vitro. Endogenous TRAF2 increasingly co-localized with depolarized mitochondria (with loss of Mitotracker signal) in hypoxia-reoxygenation-injured NRCMs (Fig. 8C). Knockdown of endogenous TRAF2 resulted in accumulation of depolarized mitochondria (Fig. 8D) and increased cell death (Fig. 8F) under normoxic conditions, which was markedly exacerbated with hypoxia-reoxygenation injury (Fig. 8D, F). Conversely, exogenous TRAF2, but not its E3-ligase deficient mutants (which are also mitophagy-deficient, vide supra) was sufficient to attenuate hypoxia-reoxygenation-induced cell death (Fig. 8G) with a significant reduction in relative content of depolarized mitochondria (Fig. 8E). Interestingly, exogenous expression of the E3-ligase deficient mutants enhanced hypoxia-reoxygenation induced cell death with increased content of depolarized mitochondria, paralleling our recent observations in ex-vivo IR injury,37 indicating a dominant negative effect. Taken together, these data suggest a central role for TRAF2 as an E3 ubiquitin ligase in facilitating mitophagy as a mechanism for its observed cytoprotection with ischemia-reperfusion injury.
Figure 8. TRAF2-induced mitophagy confers cytoprotection to hypoxia-reoxygenation injury.
A) Representative immunoblot demonstrating sub-cellular localization of endogenous TRAF2 and PARKIN in cardiac extracts from hearts subjected to in-vivo reversible left anterior descending (LAD) coronary artery ligation, to induce ischemia (30 minutes) followed by reperfusion (90 minutes) or a sham procedure; and subjected to sub-cellular fractionation into a mitochondria-enriched (COX IV expressing) and cytoplasmic fraction (GAPDH expressing). B) Quantitation of TRAF2 and PARKIN (normalized to COXIV) in the mitochondrial sub-fraction from hearts treated as in A, expressed as fold over sham-treated control. N=4/group. P is by t-test. C) Representative confocal images demonstrating increased co-localization of endogenous TRAF2 with COX IV on mitochondria (arrows) with loss of mitotracker (Deep Red) staining in NRCMs subjected to hypoxia (6 hours) followed by reoxygenation (18 hours) or cultured under normoxic conditions, as control. D, E) Relative abundance of cells expressing predominantly depolarized mitochondria assessed by flow cytometry for JC-1, in NRCMs transduced with shTRAF2 (or shLacZ, MOI=200, 72 hrs; D) and Ad-TRAF2, its E3 ligase-deficient mutants or Ad-LacZ (MOI=10, 48 hours; E) and subjected to hypoxia-reoxygenation or normoxic conditions, as in C. F, G) Cell death in NRCMs treated as in D, E. For D-G: P values shown are by post-hoc pairwise comparison after two-way ANOVA. All parameters for hypoxia-reoxygenation were significantly differently vs. normoxia within the same group. * indicates P vs. control for Normoxia; and # indicates P vs. control for hypoxia-reoxygenation. N=3-4/group for JC-1 and N=8-16/group for cell death assays.
Discussion
The results of the present study suggest that TRAF2 mediates mitochondrial autophagy through an E3 ligase dependent mechanism in both unstressed and stressed mammalian cardiomyocytes. The following lines of evidence support this conclusion. First, TRAF2 localizes to the mitochondria in quiescent cardiac myocytes and in native wild-type hearts. In the setting of TNF-induced mitochondrial damage, TRAF2 expression is transcriptionally upregulated with increased mitochondrial localization, both in-vitro and in-vivo (Fig. 1). Second, knockdown of endogenous TRAF2 results in the accumulation of depolarized mitochondria in resting cells, as well as in cells treated with TNF (Fig. 4), the uncoupling agent CCCP (Fig. 6) and in cells subjected to hypoxia-reoxygenation injury (Fig. 8); whereas exogenous TRAF2 is sufficient to facilitate autophagic removal of mitochondria in these settings (Fig. 3, 4 and 8). Third, TRAF2 localizes to CCCP-depolarized mitochondria, where it interacts with PARKIN, a previously described mitophagy effector (Fig. 5, 7). Fourth, exogenous TRAF2 can partially restore mitophagy in the setting of PARKIN deficiency (Fig. 7), while over-expression of PARKIN can partially overcome the effects of TRAF2 deficiency with respect to mitophagy; suggesting a partial redundancy in the E3 ligase activity of TRAF2 and PARKIN to facilitate ubiquitination and degradation of mitochondrial proteins. Fifth, endogenous TRAF2 increasingly localizes to the mitochondria in hearts subjected to in-vivo cardiac ischemia-reperfusion injury and in cardiac myocytes subjected to hypoxia-reoxygenation, in-vitro (Fig. 8). Lastly, endogenous TRAF2 signaling is essential to prevent cardiomyocyte death with hypoxia-reoxygenation injury; and expression of exogenous TRAF2, but not its E3 ligase mutants, results in decreased abundance of depolarized mitochondria and enhanced cytoprotection with hypoxia-reoxygenation injury (Fig. 8).
Mitophagy removes damaged mitochondria to protect against cell death in myocardial infarction10 and neurodegeneration;38 and is required for ischemic preconditioning in the myocardium.11 Our findings indicate that TRAF2-mediated mitophagy may be a mechanism whereby endogenous TRAF2 signals downstream of either TNF receptor activation7 to prevent cardiomyocyte death with hypoxia-reoxygenation injury, in-vitro3 and with myocardial infarction, in-vivo.6 Also, TRAF2-mediated upregulation of mitophagy may further attenuate IR-induced cardiomyocyte death, as observed in transgenic mouse hearts with cardiomyocyte-specific expression of TRAF2 and TNF (at low levels) subjected to ex-vivo ischemia-reperfusion injury;4 and facilitate the preconditioning effects of TNF administration on the myocardium, in-vivo.5 Conversely, the observed critical role for TRAF2 in mediating homeostatic mitophagy may explain the early lethality in TRAF2 null mice in the neonatal period, which is preceded by marked runting and elevated circulating TNF levels, with markedly increased cell death in various cell types. 39, 40
Our data point to an essential role for TRAF2 in parallel with PARKIN in facilitating mitophagy in cardiac myocytes. Activation of PARKIN, an E3 ubiquitin ligase, by PINK1 (a serine-threoine kinase) on damaged mitochondria has been ascribed an essential role in orchestrating their autophagic removal; and impaired homeostatic mitophagy has been implicated as the underlying mechanism for development of Parkinson’s disease in individuals harboring mutations in their respective genes.13-15 However, studies in the heart reveal that while mice with cardiomyocyte-specific ablation of PINK1 demonstrate cardiomyopathy with markedly abnormal mitochondria,16 loss of PARKIN (with PARK2 ablation) does not alter cardiac structure or function in young adult mice,10 or increase susceptibility to in-vivo ischemia-reperfusion injury.11 These observations suggest that PARKIN deficiency may be complemented by other proteins in the heart; and its deficiency becomes apparent only with a surge in mitochondrial damage as occurs with prolonged ischemia;10 or cumulatively with aging.17 Our data demonstrate that TRAF2 is present on the mitochondria in resting cardiomyocytes. Additionally, TRAF2 has also been demonstrated to localize to the mitochondria upon TNF receptor signaling, associated with other proteins namely TRADD, FADD, RIP1 and procaspase-8 in the DISC26 or ‘complex II’;41 and recent studies indicate that TRAF2 interacts with mitochondrial scaffolding proteins such as MAVS,19-21 whereby it would be positioned to participate in mitochondrial autophagy. Also, PARKIN interacts with TRAF2 (Fig. 7B) and PARKIN-mediated ubiquitination has been demonstrated to activate TRAF2 in fibroblasts.34 Given our observation that damaged mitochondria accumulate with TRAF2 deficiency, mimicking the effects of PARKIN knockdown, we posit that TRAF2 functions in concert with PARKIN in ubiquitinating mitochondrial proteins upon mitochondrial damage (as proposed in Fig. 7I). Notably, while both exogenous PARKIN and TRAF2 are sufficient to partially restore mitophagy with each other’s deficiency, in vitro (Figure 7C-H); mitochondrial proteins accumulate differentially with individual deficiencies of TRAF2 and PARKIN (Fig. S13), suggesting both unique and redundant roles for TRAF2 vis-a-vis PARKIN. Further studies are required to clarify the molecular mechanism for recruitment of TRAF2 to damaged mitochondria, and determine whether PARKIN-mediated ubiquitination of TRAF2 is necessary to activate TRAF2 signaling; or TRAF2 functions independently of and in parallel to PARKIN in cardiomyocyte mitophagy. Additionally, whether TRAF2-mediated ubiquitination is also required for removal of non-mitochondrial proteins via autophagy or ubiquitin-proteasome pathway, needs to be explored.
Recent studies indicate that preventing removal of damaged mitochondria and mitochondrial DNA induces pro-inflammatory signaling.42, 43 In this context, it is notable that TRAF2 null mice die of colitis with widespread bowel wall inflammation within the first three weeks of life;39 and treatment with antibiotics or concomitant TNF receptor 1 ablation only partially rescues the inflammatory colitis39 with a modest prolongation of lifespan.44 Additionally, concomitant ablation of TNF does not attenuate the chronic inflammatory state observed in various organ systems of the TRAF2 null mice45 indicating that the primary driver of the pathogenesis is downstream of the inflammatory mediators activated with loss of TRAF2. Intriguingly, mutations in genes that play critical roles in autophagic signaling have been implicated in loss of autophagy in enterocytes, leading to development of colitis and Crohn’s disease.46-48 Thereby, impairment in homeostatic removal of damaged mitochondria may be the underlying mechanism for development of colitis and multi-organ inflammation in TRAF2 deficient mice, a premise which requires further experimental validation.
TRAF2 plays a key pro-survival role as an adaptor protein to transduce activation of kinases and transcription factors downstream of multiple innate immunity receptors.1, 7, 41 The results of the present study extend these prior observations4 and suggest a novel cytoprotective role for TRAF2 in mediating mitochondrial autophagy through an E3 ligase-dependent mechanism. Unraveling the role for innate immunity pathways with respect to regulation of mitochondrial mass and mitochondrial quality control may provide important insights in understanding the molecular mechanisms of human disease, as well as developing targeted therapies to counteract untoward effects of sustained inflammatory signaling.
Acknowledgments
We wish to thank Jeffrey E. Saffitz, M.D., Ph.D., Chairman, Department of Pathology at Beth Israel Deaconess Medical Center for assistance with interpretation of electron micrographs. We thank Eric Novak, M.S., Cardiovascular Division at Washington University, for expert input on statistical analyses methodology. We also acknowledge Joan Avery and Lora Staloch from Center of Cardiovascular Research for technical assistance.
Sources of Funding
This study was supported by grants from Department of Veterans Affairs (I01BX000448, I01BX001969) and National Institutes of Health (HL107594) to A.D; NIH (HL89543, HL58081, and 111094) to D.L.M.; and Mentors in Medicine award from Department of Internal Medicine at Washington University to K-C. Y.
Footnotes
Disclosures
None.
References
- 1.Mann DL. The emerging role of innate immunity in the heart and vascular system: for whom the cell tolls. CircRes. 2011;108:1133–1145. doi: 10.1161/CIRCRESAHA.110.226936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ueland T, Aukrust P, Damas JK, Gullestad L, Yndestad A. The tumor necrosis factor superfamily in heart failure. FutureCardiol. 2006;2:101–111. doi: 10.2217/14796678.2.1.101. [DOI] [PubMed] [Google Scholar]
- 3.Nakano M, Knowlton AA, Dibbs Z, Mann DL. Tumor necrosis factor-alpha confers resistance to hypoxic injury in the adult mammalian cardiac myocyte. Circulation. 1998;97:1392–1400. doi: 10.1161/01.cir.97.14.1392. [DOI] [PubMed] [Google Scholar]
- 4.Burchfield JS, Dong JW, Sakata Y, Gao F, Tzeng HP, Topkara VK, Entman ML, Sivasubramanian N, Mann DL. The cytoprotective effects of tumor necrosis factor are conveyed through tumor necrosis factor receptor-associated factor 2 in the heart. CircHeart Fail. 2010;3:157–164. doi: 10.1161/CIRCHEARTFAILURE.109.899732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lecour S, Suleman N, Deuchar GA, Somers S, Lacerda L, Huisamen B, Opie LH. Pharmacological preconditioning with tumor necrosis factor-alpha activates signal transducer and activator of transcription-3 at reperfusion without involving classic prosurvival kinases (Akt and extracellular signal-regulated kinase) Circulation. 2005;112:3911–3918. doi: 10.1161/CIRCULATIONAHA.105.581058. [DOI] [PubMed] [Google Scholar]
- 6.Kurrelmeyer KM, Michael LH, Baumgarten G, Taffet GE, Peschon JJ, Sivasubramanian N, Entman ML, Mann DL. Endogenous tumor necrosis factor protects the adult cardiac myocyte against ischemic-induced apoptosis in a murine model of acute myocardial infarction. ProcNatlAcadSciUSA. 2000;97:5456–5461. doi: 10.1073/pnas.070036297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rothe M, Sarma V, Dixit VM, Goeddel DV. TRAF2-mediated activation of NF-kappa B by TNF receptor 2 and CD40. Science. 1995;269:1424–1427. doi: 10.1126/science.7544915. [DOI] [PubMed] [Google Scholar]
- 8.Hausenloy DJ, Duchen MR, Yellon DM. Inhibiting mitochondrial permeability transition pore opening at reperfusion protects against ischaemia-reperfusion injury. CardiovascRes. 2003;60:617–625. doi: 10.1016/j.cardiores.2003.09.025. [DOI] [PubMed] [Google Scholar]
- 9.Carreira RS, Lee Y, Ghochani M, Gustafsson AB, Gottlieb RA. Cyclophilin D is required for mitochondrial removal by autophagy in cardiac cells. Autophagy. 2010;6:462–472. doi: 10.4161/auto.6.4.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kubli DA, Zhang X, Lee Y, Hanna RA, Quinsay MN, Nguyen CK, Jimenez R, Petrosyan S, Murphy AN, Gustafsson AB. Parkin protein deficiency exacerbates cardiac injury and reduces survival following myocardial infarction. JBiolChem. 2013;288:915–926. doi: 10.1074/jbc.M112.411363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang C, Andres AM, Ratliff EP, Hernandez G, Lee P, Gottlieb RA. Preconditioning involves selective mitophagy mediated by Parkin and p62/SQSTM1. PLoSOne. 2011;6:e20975. doi: 10.1371/journal.pone.0020975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan H, Perry CN, Huang C, Iwai-Kanai E, Carreira RS, Glembotski CC, Gottlieb RA. LPS-induced autophagy is mediated by oxidative signaling in cardiomyocytes and is associated with cytoprotection. AmJPhysiol Heart CircPhysiol. 2009;296:H470–H479. doi: 10.1152/ajpheart.01051.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. JCell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. JCell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoSBiol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Billia F, Hauck L, Konecny F, Rao V, Shen J, Mak TW. PTEN-inducible kinase 1 (PINK1)/Park6 is indispensable for normal heart function. ProcNatlAcadSciUSA. 2011;108:9572–9577. doi: 10.1073/pnas.1106291108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubli DA, Quinsay MN, Gustafsson AB. Parkin deficiency results in accumulation of abnormal mitochondria in aging myocytes. CommunIntegrBiol. 2013;6:e24511. doi: 10.4161/cib.24511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alvarez SE, Harikumar KB, Hait NC, Allegood J, Strub GM, Kim EY, Maceyka M, Jiang H, Luo C, Kordula T, Milstien S, Spiegel S. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 2010;465:1084–1088. doi: 10.1038/nature09128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lad SP, Yang G, Scott DA, Chao TH, Correia JS, de la Torre JC, Li E. Identification of MAVS splicing variants that interfere with RIGI/MAVS pathway signaling. MolImmunol. 2008;45:2277–2287. doi: 10.1016/j.molimm.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 21.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 22.Ma X, Liu H, Foyil SR, Godar RJ, Weinheimer CJ, Hill JA, Diwan A. Impaired autophagosome clearance contributes to cardiomyocyte death in ischemia/reperfusion injury. Circulation. 2012;125:3170–3181. doi: 10.1161/CIRCULATIONAHA.111.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma X, Godar RJ, Liu H, Diwan A. Enhancing lysosome biogenesis attenuates BNIP3-induced cardiomyocyte death. Autophagy. 2012;8:297–309. doi: 10.4161/auto.18658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sivasubramanian N, Coker ML, Kurrelmeyer KM, MacLellan WR, DeMayo FJ, Spinale FG, Mann DL. Left ventricular remodeling in transgenic mice with cardiac restricted overexpression of tumor necrosis factor. Circulation. 2001;104:826–831. doi: 10.1161/hc3401.093154. [DOI] [PubMed] [Google Scholar]
- 25.Kim JJ, Lee SB, Park JK, Yoo YD. TNF-alpha-induced ROS production triggering apoptosis is directly linked to Romo1 and Bcl-X(L) Cell DeathDiffer. 2010;17:1420–1434. doi: 10.1038/cdd.2010.19. [DOI] [PubMed] [Google Scholar]
- 26.Schneider-Brachert W, Tchikov V, Neumeyer J, Jakob M, Winoto-Morbach S, Held-Feindt J, Heinrich M, Merkel O, Ehrenschwender M, Adam D, Mentlein R, Kabelitz D, Schutze S. Compartmentalization of TNF receptor 1 signaling: internalized TNF receptosomes as death signaling vesicles. Immunity. 2004;21:415–428. doi: 10.1016/j.immuni.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 27.Divakaran VG, Evans S, Topkara VK, Diwan A, Burchfield J, Gao F, Dong J, Tzeng HP, Sivasubramanian N, Barger PM, Mann DL. Tumor necrosis factor receptor-associated factor 2 signaling provokes adverse cardiac remodeling in the adult mammalian heart. CircHeart Fail. 2013;6:535–543. doi: 10.1161/CIRCHEARTFAILURE.112.000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panagopoulou P, Davos CH, Milner DJ, Varela E, Cameron J, Mann DL, Capetanaki Y. Desmin mediates TNF-alpha-induced aggregate formation and intercalated disk reorganization in heart failure. JCell Biol. 2008;181:761–775. doi: 10.1083/jcb.200710049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. CurrOpinCell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai L, Leone TC, Zechner C, Schaeffer PJ, Kelly SM, Flanagan DP, Medeiros DM, Kovacs A, Kelly DP. Transcriptional coactivators PGC-1alpha and PGC-lbeta control overlapping programs required for perinatal maturation of the heart. Genes Dev. 2008;22:1948–1961. doi: 10.1101/gad.1661708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rouschop KM, Ramaekers CH, Schaaf MB, Keulers TG, Savelkouls KG, Lambin P, Koritzinsky M, Wouters BG. Autophagy is required during cycling hypoxia to lower production of reactive oxygen species. RadiotherOncol. 2009;92:411–416. doi: 10.1016/j.radonc.2009.06.029. [DOI] [PubMed] [Google Scholar]
- 32.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. JBiolChem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Henn IH, Bouman L, Schlehe JS, Schlierf A, Schramm JE, Wegener E, Nakaso K, Culmsee C, Berninger B, Krappmann D, Tatzelt J, Winklhofer KF. Parkin mediates neuroprotection through activation of IkappaB kinase/nuclear factor-kappaB signaling. JNeurosci. 2007;27:1868–1878. doi: 10.1523/JNEUROSCI.5537-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Habelhah H, Takahashi S, Cho SG, Kadoya T, Watanabe T, Ronai Z. Ubiquitination and translocation of TRAF2 is required for activation of JNK but not of p38 or NF-kappaB. EMBO J. 2004;23:322–332. doi: 10.1038/sj.emboj.7600044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Yan J, Jiang S, Wen J, Chen L, Zhao Y, Lin A. Site-specific ubiquitination is required for relieving the transcription factor Miz1-mediated suppression on TNF-alpha-induced JNK activation and inflammation. ProcNatlAcadSciUSA. 2012;109:191–196. doi: 10.1073/pnas.1105176108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tzeng HP, Evans S, Gao F, Chambers K, Topkara VK, Sivasubramanian N, Barger PM, Mann DL. Dysferlin mediates the cytoprotective effects of TRAF2 following myocardial ischemia reperfusion injury. J Am Heart Assoc. 2014;3:e000662. doi: 10.1161/JAHA.113.000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsui H, Gavinio R, Asano T, Uemura N, Ito H, Taniguchi Y, Kobayashi Y, Maki T, Shen J, Takeda S, Uemura K, Yamakado H, Takahashi R. PINK1 and Parkin complementarily protect dopaminergic neurons in vertebrates. HumMolGenet. 2013;22:2423–2434. doi: 10.1093/hmg/ddt095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piao JH, Hasegawa M, Heissig B, Hattori K, Takeda K, Iwakura Y, Okumura K, Inohara N, Nakano H. Tumor necrosis factor receptor-associated factor (TRAF) 2 controls homeostasis of the colon to prevent spontaneous development of murine inflammatory bowel disease. JBiolChem. 2011;286:17879–17888. doi: 10.1074/jbc.M111.221853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeh WC, Shahinian A, Speiser D, Kraunus J, Billia F, Wakeham A, de la Pompa JL, Ferrick D, Hum B, Iscove N, Ohashi P, Rothe M, Goeddel DV, Mak TW. Early lethality, functional NF-kappaB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity. 1997;7:715–725. doi: 10.1016/s1074-7613(00)80391-x. [DOI] [PubMed] [Google Scholar]
- 41.Hsu H, Shu HB, Pan MG, Goeddel DV. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 42.Oka T, Hikoso S, Yamaguchi O, Taneike M, Takeda T, Tamai T, Oyabu J, Murakawa T, Nakayama H, Nishida K, Akira S, Yamamoto A, Komuro I, Otsu K. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485:251–255. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen LT, Duncan GS, Mirtsos C, Ng M, Speiser DE, Shahinian A, Marino MW, Mak TW, Ohashi PS, Yeh WC. TRAF2 deficiency results in hyperactivity of certain TNFR1 signals and impairment of CD40-mediated responses. Immunity. 1999;11:379–389. doi: 10.1016/s1074-7613(00)80113-2. [DOI] [PubMed] [Google Scholar]
- 45.Lin WJ, Su YW, Lu YC, Hao Z, Chio II, Chen NJ, Brustle A, Li WY, Mak TW. Crucial role for TNF receptor-associated factor 2 (TRAF2) in regulating NFkappaB2 signaling that contributes to autoimmunity. ProcNatlAcadSciUSA. 2011;108:18354–18359. doi: 10.1073/pnas.1109427108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, Stone CD, Brunt EM, Xavier RJ, Sleckman BP, Li E, Mizushima N, Stappenbeck TS, Virgin HW. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJ, Campbell BJ, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. NatMed. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 48.McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH, Duerr RH, Silverberg MS, Taylor KD, Rioux JD, Altshuler D, Daly MJ, Xavier RJ. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. NatGenet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]