Abstract
Sirtuin 1 (SIRT1), an NAD+-dependent protein deacetylase, regulates a host of target proteins, including peroxisome proliferator–activated receptor (PPAR)-γ coactivator-1α (PGC-1α), a transcriptional coregulator that binds to numerous transcription factors in response to deacetylation to promote mitochondrial biogenesis and oxidative metabolism. Our laboratory and others have shown that adipose triglyceride lipase (ATGL) increases the activity of the nuclear receptor PPAR-α, a PGC-1α binding partner, to promote fatty acid oxidation. Fatty acids bind and activate PPAR-α; therefore, it has been presumed that fatty acids derived from ATGL-catalyzed lipolysis act as PPAR-α ligands. We provide an alternate mechanism that links ATGL to PPAR-α signaling. We show that SIRT1 deacetylase activity is positively regulated by ATGL to promote PGC-1α signaling. In addition, ATGL mediates the effects of β-adrenergic signaling on SIRT1 activity, and PGC-1α and PPAR-α target gene expression independent of changes in NAD+. Moreover, SIRT1 is required for the induction of PGC-1α/PPAR-α target genes and oxidative metabolism in response to increased ATGL-mediated lipolysis. Taken together, this work identifies SIRT1 as a critical node that links β-adrenergic signaling and lipolysis to changes in the transcriptional regulation of oxidative metabolism.
Introduction
Sirtuin 1 (SIRT1), an NAD+-dependent protein deacetylase, has emerged as an important metabolic sensor that coordinates changes in energy metabolism. Upon activation, SIRT1 deacetylates target proteins to promote oxidative metabolism and stress resistance. SIRT1 ablation in specific tissues results in deranged oxidative metabolism and inflammation (1–3), whereas SIRT1 overexpression improves metabolic function and insulin sensitivity (4,5). In addition, SIRT1 is a key signaling node in life span determination (6) and is required for the effects of calorie restriction on life span extension as observed in numerous species (7–9).
SIRT1 is highly regulated through both transcriptional and posttranscriptional mechanisms. Regarding the latter, interaction of SIRT1 with active regulator of SIRT1 (AROS) promotes its activity (10), whereas interaction with deleted in breast cancer 1 (DBC1) is inhibitory (11,12). SIRT1 is regulated through numerous posttranscriptional covalent modifications, including phosphorylation. In response to β-adrenergic signaling, the cAMP-dependent protein kinase (PKA) pathway activates SIRT1 to promote downstream oxidative metabolism (13). SIRT1 is also highly regulated by the concentration of its substrate NAD+ (14), thereby coupling energy status to SIRT1 activity.
Peroxisome proliferator–activated receptor (PPAR)-γ coactivator-1α (PGC-1α) is a principal target of SIRT1 that coordinates transcriptional changes in response to SIRT1 activity (15). Upon deacetylation and activation, PGC-1α binds numerous transcription factors involved in regulating oxidative metabolism and mitochondrial biogenesis (16). One such transcription factor that partly mediates the downstream signaling effects of PGC-1α is PPAR-α, which is highly expressed in the liver and coordinates the induction of fatty acid oxidation in response to fasting (17). SIRT1 directly binds PPAR-α and facilitates PGC-1α/PPAR-α interactions (18). Consistent with these effects, SIRT1 is required for the induction of liver oxidative gene expression in response to fasting (18).
Our laboratory and others have shown that adipose triglyceride lipase (ATGL) increases the activity of PPAR-α to promote fatty acid oxidation (19–22). Fatty acids bind and activate PPAR-α (23,24); therefore, it has been presumed that fatty acids derived from ATGL-catalyzed lipolysis act as PPAR-α ligands. However, administration of a PPAR-α agonist to mice with ablated hepatic ATGL was unable to normalize oxidative gene expression (19), suggesting that ATGL regulates PPAR-α independent of ligand binding. In addition, ATGL does not influence hepatic free fatty acid levels and mediates PPAR-α signaling independent of liver fatty acid–binding protein, the major fatty acid carrier in the liver (25). Given the importance of SIRT1 in regulating PPAR-α, we tested whether ATGL manipulations alter SIRT1 activity as a mechanism to regulate the expression of oxidative genes. We characterize a novel axis involving β-adrenergic signaling, ATGL-catalyzed lipolysis, and SIRT1 activation that governs PGC-1α/PPAR-α signaling and oxidative metabolism.
Research Design and Methods
Mice and Adenovirus Administration
All animal protocols were approved by the University of Minnesota Institutional Animal Care and Use Committee. Male 6–8-week-old C57BL/6 mice were purchased from Harlan Laboratories and housed under controlled temperature and lighting (20–22°C; 12-h light-dark cycle). The mice were fed a purified control diet (TD 94045; Harlan Teklad Premier Laboratory) and acclimatized for 1 week before adenovirus injections. Adenoviruses to manipulate ATGL expression were provided by Andrew Greenberg (Tufts University) and generated as described previously (26,27), and adenovirus encoding mouse SIRT1 short hairpin RNA (shRNA) was provided by X. C. Dong (Indiana University School of Medicine). For single adenovirus treatments, mice were injected with 1 × 109 plaque-forming units adenovirus through the tail vein. Exactly 1 week after adenovirus injection, mice were sacrificed for tissue and serum collection after a 4-h fast (Ad-ATGL adenovirus treatments) or a 16-h fast (ATGL shRNA adenovirus treatments). For combination adenovirus treatments, four groups of mice were injected with either Ad-green fluorescent protein (GFP) and control shRNA, Ad-GFP and SIRT1 shRNA, Ad-ATGL and control shRNA, or Ad-ATGL and SIRT1 shRNA. Exactly 1 week after adenovirus injection, the mice were sacrificed for tissue and serum collection after a 4-h fast.
Cell Culture
Primary hepatocytes were isolated as described previously (28) and cultured in M199 media (Invitrogen) containing 23 mmol/L HEPES, 26 mmol/L sodium bicarbonate, 10% FBS, 50 IU/mL penicillin, 50 μg/mL streptomycin, 100 nmol/L dexamethasone, 100 nmol/L insulin, and 11 mmol/L glucose for 4 h followed by the same media with 10 nmol/L dexamethasone and no insulin or FBS. One hour before treatment with 1 mmol/L of the cAMP analog 8-bromoadenosine 3′,5′-cyclic monophosphate, the lipase inhibitors bromoenol lactone (BEL) (2 μmol/L) or Atglistatin (Astat) (30 μmol/L) were added. Hepatocytes with ATGL shRNA knockdown were isolated from mice exactly 3 days after adenovirus injection of ATGL shRNA through the tail vein. Hepatocytes isolated from mice treated with nontargeting shRNA were used as control. SIRT1 knockout mouse embryonic fibroblasts (MEFs) were provided by Michael McBurney (University of Ottawa).
SIRT1 Assays
SIRT1 activity was determined according to kit protocol (Enzo Life Sciences) with 0.5 mmol/L acetylated Fluor-de-Lys peptide and 10 mmol/L NAD+. Crude nuclear proteins were isolated by sucrose gradient centrifugation. The crude protein lysate and substrate were incubated for 15 min, and the reaction was quenched by the addition of a developer reagent. The developer was incubated for a further 45 min before reading with an excitation wavelength of 360 nm and emission at 460 nm. SIRT1 activity was normalized to protein content and expressed as percentage change over control.
PPAR-α and PGC-1α Reporter Assay
MEFs were transfected with indicated firefly luciferase reporter plasmids (TK-MH-UASluc), control Renilla luciferase (pRLSV40), and indicated GAL4-PPAR-α or PGC-1α constructs using Effectene Transfection Reagent (QIAGEN). For PPAR-α reporter activity, pSG5-GAL4-PPARα-LBD construct was transfected into cells. For PGC-1α reporter activity, pCMX-GAL4-PGC1α, provided by Brian Finck (Washington University in St. Louis), was transfected into cells. For ATGL overexpression studies, MEFs were transduced with 60 multiplicities of infection of Ad-ATGL or Ad-GFP for 18 h. To inhibit SIRT1 activity, MEFs were incubated for 18 h with 10 μmol/L EX-527. Cells were stimulated with 1 mmol/L cAMP analog for 4 h. Following treatments with indicated adenovirus or drugs, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase activity was normalized to the coexpressed Renilla luciferase activity.
Immunoprecipitation for Acetylated Proteins
PGC-1α acetylation was determined by immunoprecipitation of PGC-1α from mouse liver lysates using PGC-1α antibody (EMD Millipore) followed by immunobloting for acetylated lysine (Cell Signaling Technology, Danvers, MA). FOXO1 acetylation was determined by immunoprecipitation with acetylated lysine antibody followed by immunobloting for FOXO1 (Cell Signaling Technology).
Western Blotting
Protein isolation, electrophoresis, and immunoblotting were performed as described previously (19). ATGL, SIRT1, p38-mitogen-activated protein kinase (MAPK), and phospho-p38-MAPK antibodies were obtained from Cell Signaling Technology. β-Actin antibody was obtained from LI-COR Biotechnology (Lincoln, NE).
β-Hydroxybutyrate Assay
β-Hydroxybutyrate was determined from mouse serum samples using a β-hydroxybutyrate LiquiColor kit (Stanbio Laboratory, Boerne, TX) according to the manufacturer’s instructions.
NAD+ and NADH Measurements
NAD+ and NADH concentrations were determined using a colorimetric, enzyme-based NAD+/NADH quantification kit (Sigma-Aldrich) according to the manufacturer’s instructions.
RNA Isolation and RT-PCR Analysis
RNA was extracted with TRIzol from liver tissues followed by reverse transcription with SuperScript VILO cDNA Synthesis Kit (Invitrogen) to generate cDNA. Gene expression was quantified as described previously (19).
Mitochondrial DNA Copy Number Measurement
DNA was extracted from mouse liver tissues using the DNeasy Blood & Tissue Kit (QIAGEN). As described by Yatsuga and Suomalainen (29), 25 ng of total DNA was used as a template in RT-PCR, and the level of mitochondrial MTRNR1 (forward 5′-AGGAGCCTGTTCTATAATCGATAAA-3′ and reverse 5′-GATGGCGGTATATAGGCTGAA-3′) was normalized against the nuclear RBM15 (RNA-binding motif protein 15) gene (forward 5′-GGACACTTTTCTTGGGCAAC-3′ and reverse 5′-AGTTTGGCCCTGTGAGACAT-3′).
Statistical Analysis
Data are expressed as mean ± SEM. Statistical analyses were performed using Student t test or ANOVA where appropriate. Differences were considered significant at P < 0.05.
Results
ATGL Regulates Hepatic PGC-1α and Mitochondrial Biogenesis
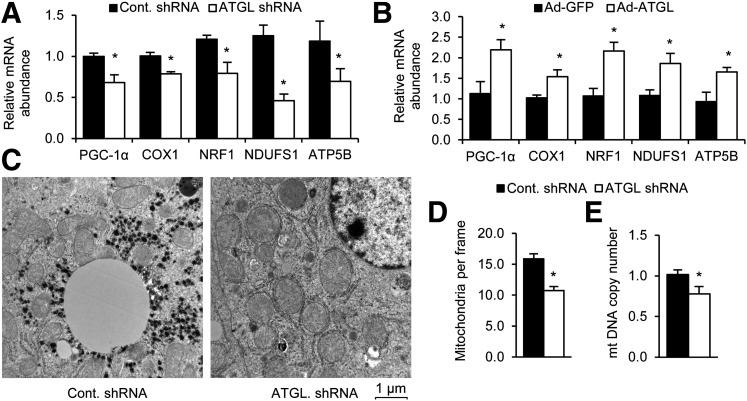
Given the previous links between lipolysis and mitochondrial biogenesis, we first tested whether hepatic ATGL could influence PGC-1α, which is well documented to govern mitochondrial biogenesis. Indeed, previous studies have shown that ATGL influences PGC-1α in the heart (21). Knocking down ATGL in the liver resulted in a significant reduction in the expression of PGC-1α and its target genes COX1, NRF1, NDUFS1, and ATP5B (Fig. 1A). Consistent with these data, ATGL overexpression increased PGC-1α and its target genes (Fig. 1B). The ATGL shRNA used here has been shown to successfully reduce ATGL mRNA in the liver by ∼70% and ATGL protein by ∼80% (19). Furthermore, administration of adenoviral vectors to overexpress ATGL successfully increases protein expression of hepatic ATGL in mice approximately threefold (25). We further tested the effects of ATGL on tissue mitochondria content. Quantification of mitochondria from electron micrographs revealed that livers treated with the ATGL knockdown adenovirus had fewer mitochondria (Fig. 1C and D). Hepatic mitochondrial DNA copy number was also lower following ATGL knockdown (Fig. 1E). Taken together, these results demonstrate that ATGL regulates PGC-1α and its downstream effects on mitochondrial biogenesis.
Figure 1.
ATGL affects PGC-1α target genes and mitochondrial biogenesis. A: Hepatic ATGL knockdown in vivo leads to changes in hepatic PGC-1α target gene expression (n = 6). B: Hepatic ATGL overexpression in vivo increases PGC-1α target gene expression (n = 6). C and D: ATGL knockdown leads to a decrease in mitochondrial biogenesis. Livers from mice treated with the indicated adenovirus were analyzed by electron microscopy, and the number of mitochondria per frame were quantitated (n = 2). E: Mitochondial DNA (mtDNA) copy number are decreased in ATGL knockdown livers (n = 6). *P < 0.05.
ATGL Activates SIRT1
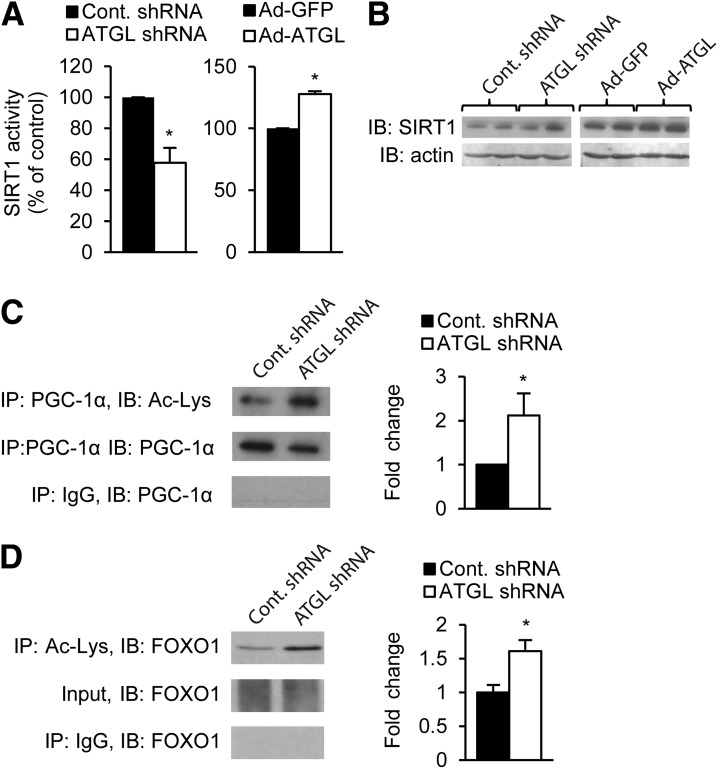
We next pursued mechanisms through which ATGL could influence PGC-1α. SIRT1 is known to positively regulate both PGC-1α and PPAR-α signaling; thus, we tested whether ATGL affected SIRT1 deacetylase activity. Hepatic SIRT1 activity was suppressed by ∼50% in mice treated with adenovirus harboring shRNA targeted to ATGL, whereas adenoviral-mediated overexpression of ATGL increased hepatic SIRT1 activity (Fig. 2A). ATGL overexpression or knockdown did not influence SIRT1 protein abundance, suggesting that the changes in SIRT1 activity were not due to altered SIRT1 expression (Fig. 2B). We next explored whether the changes in SIRT1 activity coincided with altered acetylation of the SIRT1 substrates PGC-1α and FOXO1. Consistent with the decrease in SIRT1 deacetylase activity, PGC-1α and FOXO1 were hyperacetylated in response to ATGL knockdown (Fig. 2C and D). Thus, these data identify for the first time to our knowledge that ATGL influences SIRT1 activity and target protein acetylation.
Figure 2.
ATGL activates SIRT1. A: ATGL alters SIRT1 deacetylase activity. SIRT1 activity was determined by fluorometric kinetic assay from nuclear lysates from liver samples of mice treated with knockdown or overexpression adenoviruses (n = 6). B: ATGL manipulation does not affect total SIRT1 protein levels. Liver samples were analyzed by Western blot. C and D: PGC-1α and FOXO1 are hyperacetylated in response to ATGL knockdown. The left panel shows representative Western blots of whole-liver tissue homogenate (input) or protein precipitates. The right panel shows fold change over control in densitometry of acetylated proteins, normalized to total protein (n = 3). *P < 0.05. IB, immunoblot; IP, immunoprecipitation.
ATGL Is Required for the β-Adrenergic Induction of SIRT1 and PGC1-α/PPAR-α Target Gene Expression
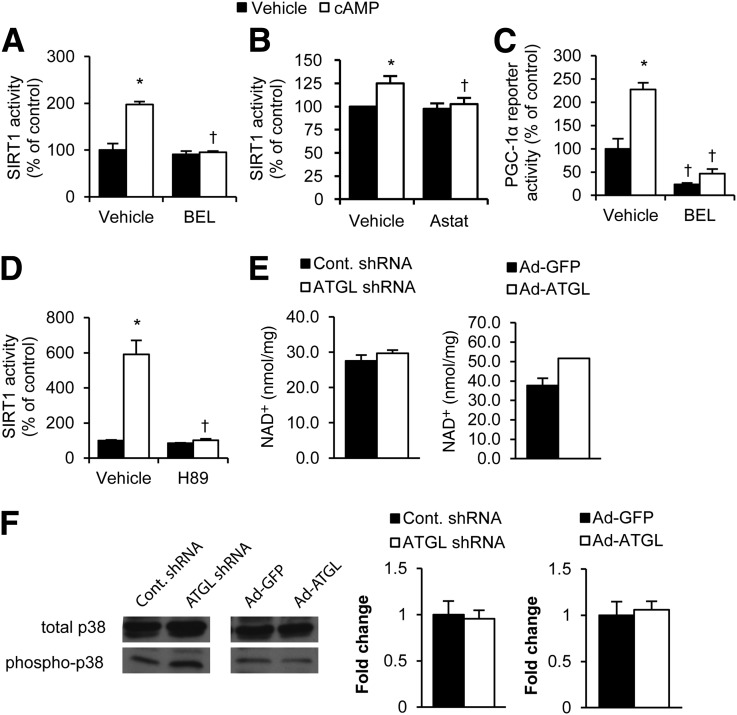
β-Adrenergic signaling and the downstream activation of PKA are robust activators of lipolysis (30). In addition, the PKA pathway activates SIRT1, leading to the induction of oxidative gene expression (13,31). Given that both pathways are induced by PKA and the aforementioned data showing that ATGL activates SIRT1, we tested whether lipolysis could influence the effects of cAMP/PKA on SIRT1 activation. For these studies, we examined primary mouse hepatocytes in the presence of the cell-permeable cAMP analog. We found that cAMP stimulation of SIRT1 activity was attenuated by pharmacological inhibition of ATGL. Pretreatment of primary mouse hepatocytes with ATGL inhibitors, either BEL or Astat, blocked the ability of cAMP to stimulate SIRT1 activity (Fig. 3A and B). Consistent with its inhibition of SIRT1, BEL also blocked the induction PGC-1α activity in response to cAMP in MEFs (Fig. 3C). Attenuating cAMP/PKA signaling with a PKA inhibitor (H89) blocked SIRT1 activation by cAMP in primary hepatocytes (Fig. 3D), suggesting that the PKA arm of the β-adrenergic signaling cascade is responsible for the observed effects. We found no changes in NAD+ levels when ATGL was knocked down or overexpressed (Fig. 3E), supporting previous studies showing that SIRT1 can be activated independent of NAD+ concentrations (13,31). Phosphorylation of p38-MAPK, a robust activator of PGC-1α (32), was unaltered in response to ATGL knockdown or overexpression (Fig. 3F), suggesting that it is not involved in the ATGL-mediated regulation of SIRT1 and PGC-1α signaling.
Figure 3.
ATGL is required for the β-adrenergic induction of SIRT1 and PGC1-α activity. A and B: Inhibition of ATGL blocks cAMP induction of SIRT1 activity. Primary hepatocytes were treated with or without 1 mmol/L cAMP analog (8-bromoadenosine 3′,5′-cyclic monophosphate) for 10 min after a 1-h pretreatment with 2 μmol/L ATGL inhibitor BEL (A) or 30 μmol/L ATGL inhibitor Astat (B). SIRT1 activity was determined by fluorometric kinetic assay from hepatocyte nuclear lysates (n = 3). C: SIRT1 activity is inhibited by a PKA inhibitor. Primary hepatocytes were treated with cAMP analog following a 1-h preincubation with 30 μmol/L H89 (n = 3). D: Inhibition of ATGL blocks cAMP induction of PGC-1α reporter expression. PGC-1α reporter was transfected into primary hepatocytes, and reporter activity was determined by dual-luciferase reporter assay (n = 3). E: NAD+ levels in mouse liver tissue treated with indicated adenoviruses for 7 days as determined by enzyme-based NAD/NADH quantification kit (n = 4). F: p38-MAPK phosphorylation in livers of mice with overexpressed or knocked down ATGL. Graphs reflect fold change over control in densitometry of acetylated proteins, normalized to total protein (n = 3). *P < 0.05, vehicle vs. cAMP; †P < 0.05, vehicle vs. BEL, Astat, or H89.
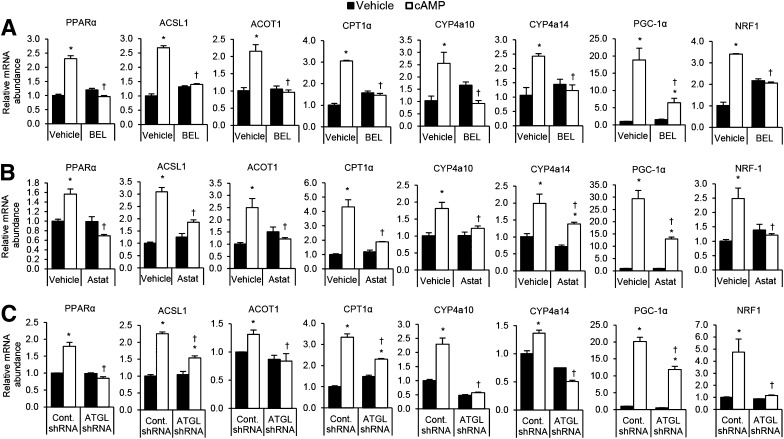
We next evaluated whether lipolysis mediated the effects of cAMP on oxidative gene expression. Similarly to the effects on SIRT1 and PGC-1α activity, BEL blocked the induction of PPAR-α and PGC-1α and their target genes in cultured primary hepatocytes in response to cAMP (Fig. 4A). These effects appear to be specifically due to ATGL inhibition because an ATGL-specific inhibitor, Astat, and targeted shRNA knockdown of ATGL also attenuated cAMP-induced expression of PPAR-α and PGC-1α target genes (Fig. 4B and C). These findings demonstrate that ATGL-catalyzed lipolysis is required for the cAMP/PKA-mediated activation of SIRT1 and induction of PGC-1α/PPAR-α target genes.
Figure 4.
ATGL is required for the β-adrenergic induction PGC1-α/PPAR-α target genes. A and B: Chemical inhibition of ATGL blocks cAMP induction of PPAR-α, PGC-1α, and their target genes. Primary hepatocytes were treated with either 2 μmol/L ATGL inhibitor BEL (A) or 30 μmol/L ATGL inhibitor Astat (B) followed by 1 mmol/L cAMP analog for 4 h (n = 3). C: ATGL knockdown blocks cAMP induction of PPAR-α, PGC-1α, and their target genes. Primary hepatocytes from ATGL knockdown or control-treated mice were treated with 1 mmol/L cAMP analog for 4 h (n = 3). *P < 0.05 vs. vehicle; †P < 0.05 vs. vehicle or control shRNA.
SIRT1 Mediates the Effects of ATGL on PGC-1α/PPAR-α Signaling and Oxidative Metabolism
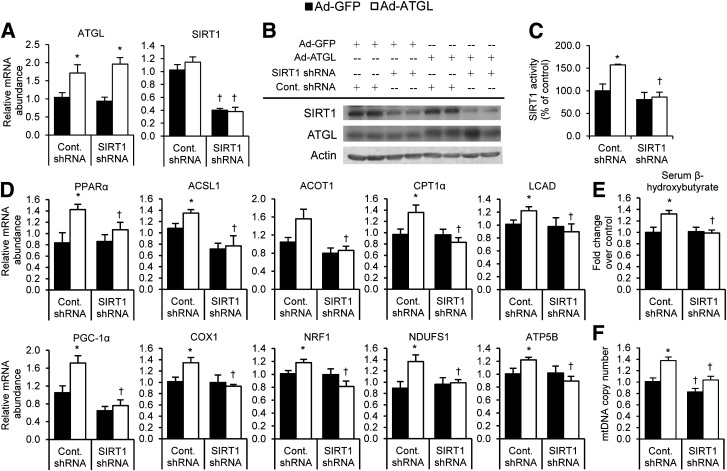
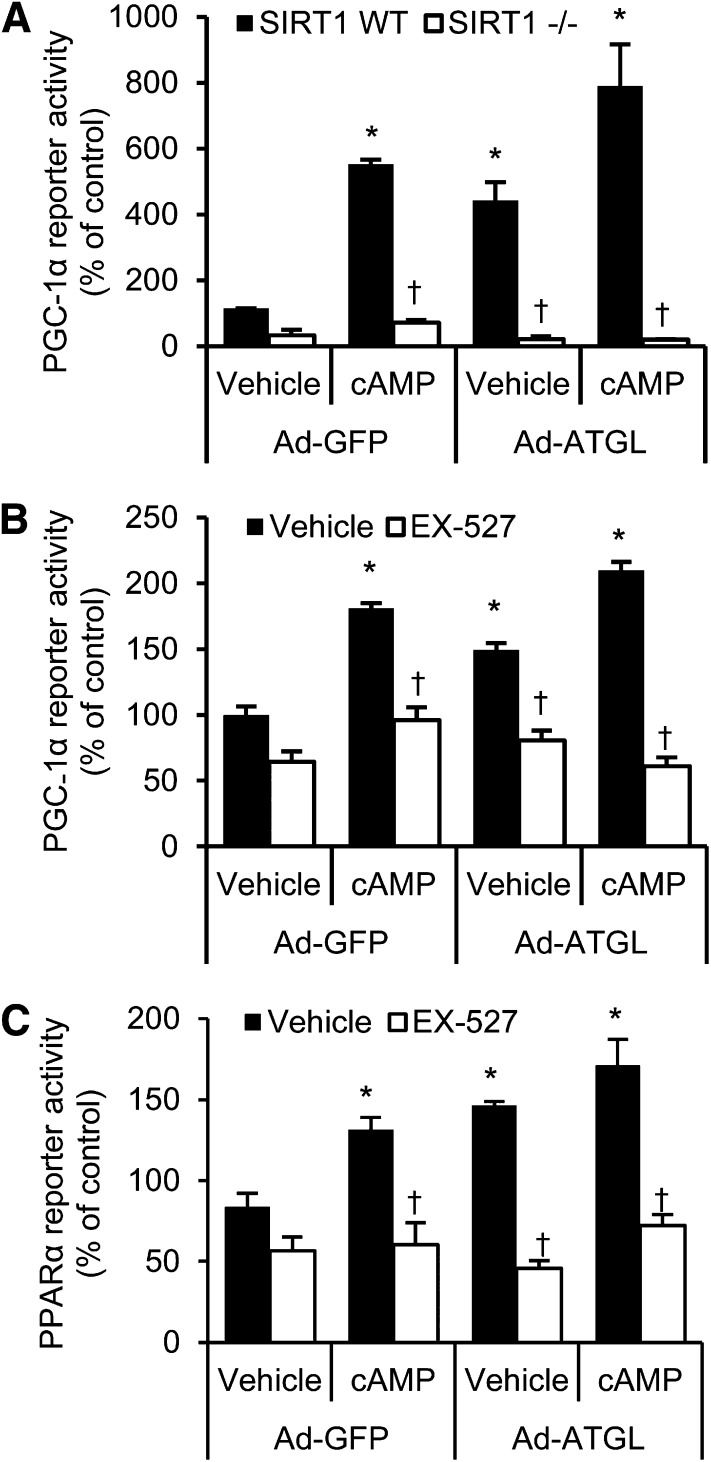
The aforementioned studies show that ATGL-catalyzed lipolysis is an important regulator of SIRT1 activity, but they do not show the importance of SIRT1 in mediating the effects of ATGL on PGC-1α/PPAR-α signaling and oxidative gene expression. To further test the importance of SIRT1 in this signaling axis, we used a combination of adenoviruses to simultaneously overexpress ATGL and knockdown SIRT1 in vivo (Fig. 5A and B). We sacrificed mice after a short-term 4-h fast for these studies because this is a time when ATGL is not yet overexpressed (33) and when NAD/NADH levels, which profoundly influence SIRT1 activity, are unaltered (34). The latter likely explains why SIRT1 activity was not altered in response to SIRT1 knockdown despite reduced protein abundance. As expected, overexpression of hepatic ATGL increased SIRT1 activity (Fig. 5C) and PGC-1α/PPAR-α target gene expression (Fig. 5D), but this effect was abolished when SIRT1 was knocked down. Indicative of liver fatty acid oxidation, serum β-hydroxybutyrate levels were increased with ATGL overexpression, and this increase was blocked with SIRT1 knockdown (Fig. 5E). Consistent with changes on PGC-1α target genes, ATGL overexpression increased mitochondrial DNA copy number, but this induction was attenuated in mice treated with SIRT1 shRNA (Fig. 5F). We further tested whether SIRT1 was required to mediate the effects of cAMP and/or ATGL on PGC-1α and PPAR-α activity in MEFs. As expected, PGC-1α activity was stimulated by both cAMP and ATGL overexpression and even more so by a combination of the two (Fig. 6A). However, this induction in response to cAMP or ATGL overexpression was abolished in SIRT1 knockout MEFs. Similarly, administration of the SIRT1 inhibitor EX-527 completely blocked the effects of cAMP and/or ATGL overexpression on PGC-1α (Fig. 6B) or PPAR-α (Fig. 6C) activity. Taken together, these data demonstrate that SIRT1 is required for ATGL-mediated induction of PGC-1α/PPAR-α signaling to control oxidative metabolism.
Figure 5.
SIRT1 is required for the effects of ATGL on oxidative gene expression and metabolism in vivo. A: Adenovirus treatments successfully raise ATGL and lower SIRT1 expression. Livers from mice treated with the indicated adenoviruses were homogenized, and mRNA abundance levels were analyzed by RT-PCR (n = 6–8). B: Protein expression levels were analyzed by SDS-PAGE and immunoblotting using the indicated antibodies. C: Raising ATGL levels increases SIRT1 activity, and this increase is abolished when SIRT1 expression is suppressed. Data are representative of three independent blots. D: SIRT1 ablation blocks the effects of ATGL overexpression on PGC-1α/PPAR-α target genes. mRNA abundance of PPAR-α and its targets (top row) and PGC-1α and its targets (bottom row) are shown (n = 6–8). E: ATGL-induced serum β-hydroxybutyrate requires SIRT1 (n = 6). F: SIRT1 knockdown attenuates the ATGL-mediated induction of mitochondrial DNA (mtDNA) copy number (n = 6). *P < 0.05 vs. Ad-GFP; †P < 0.05 vs. control shRNA.
Figure 6.
SIRT1 mediates the effects of ATGL on PGC-1α and PPAR-α reporter activity. A: Wild-type (WT) or SIRT1 knockout MEFs were transfected with PGC-1α reporter constructs, transduced with indicated adenoviruses, and stimulated with cAMP, and reporter activity was determined (n = 3). B and C: Chemical inhibition of SIRT1 blocks the effect of cAMP and ATGL on PGC-1α activity. WT MEFs were transfected with PGC-1α (B) or PPAR-α (C) reporter constructs and transduced with indicated adenoviruses followed by incubation with 10 μmol/L EX-527 for 18 h. Cells were then treated with or without 1 mmol/L cAMP analog for 4 h before cell lysis and analysis for reporter activity (n = 3). *P < 0.05 vs. Ad-GFP vehicle; †P < 0.05 vs. vehicle or WT MEFs.
Discussion
Numerous studies have linked lipolysis, mediated through manipulations of ATGL or other lipid droplet proteins, to changes in PPAR-α and oxidative gene expression in multiple tissues, including heart, liver, adipose, and intestinal (20–22,26,29,35,36). Because fatty acids are ligands for PPAR-α, it has been presumed that fatty acids generated from lipolysis act as agonists for PPAR-α (36). However, we have shown that administration of fenofibrate, a PPAR-α agonist, results in a similar fold induction of PPAR-α target genes in the livers of mice treated with control or ATGL shRNA adenoviruses but was unable to normalize gene expression between the two groups (19). These data suggest that a mechanism other than PPAR-α agonism accounts for the effects of ATGL and lipolysis on oxidative gene expression. The present data showing that ATGL activates SIRT1 provide strong evidence of an alternate signaling mechanism linking lipolysis to PPAR-α and PGC-1α activity. The present data also show that SIRT1 is required for ATGL-mediated induction of PGC-1α/PPAR-α signaling and downstream changes in oxidative metabolism. Consistent with these data, administration of a PPAR-α agonist could not normalize PPAR-α target gene expression in fasted mice with ablated hepatic SIRT1 (18). In contrast to the present studies, administration of the PPAR-α agonist WY-14643 normalizes cardiac mitochondrial function and prevents the cardiomyopathy that occurs in ATGL knockout mice (21). It is unclear whether tissue-specific differences in ATGL/SIRT1/PGC-1α signaling or whether different PPAR-α ligands and dosages account for these discrepancies.
β-Agonism is a major signal responsible for inducing lipolysis (37). Although downstream PKA activation is not believed to regulate ATGL directly, phosphorylation of the ATGL coactivator CGI-58 and perilipin proteins facilitates access of ATGL to lipid droplets to promote triglyceride hydrolysis (38). Of note, we found that blocking ATGL through shRNA or chemical inhibition prevented the induction of PGC-1α/PPAR-α/ target gene expression in response to cAMP. Knocking down ATGL in brown adipocytes also reduced β-adrenergic induction of PGC-1α/PPAR-α target genes (36), whereas ATGL had an opposite effect in white adipocytes (39). Previous work has shown that β-adrenergic signaling activates SIRT1 independent of changes in NAD+ (13,31). Consistent with these findings, we also did not observe changes in NAD+ in response to ATGL manipulations. Although these data suggest that ATGL regulates SIRT1 through an NAD+-independent mechanism, we cannot exclude a role for NAD+. Ongoing studies are attempting to elucidate the biological mechanism through which ATGL-catalyzed lipolysis regulates SIRT1 deacetylase activity.
When ATGL was discovered, it was generally assumed that inhibitors of ATGL would be viable targets to reduce adipose tissue lipolysis, the subsequent increase in serum free fatty acids, and their ectopic deposition in nonadipose tissue (40). However, adipose-specific overexpression of ATGL does not alter serum free fatty acid levels; instead, it reduces body fat, promotes resistance to diet-induced obesity and insulin resistance, and increases adipose mitochondrial biogenesis and whole-body O2 consumption (41). Consistent with these findings, adipose-specific ATGL ablation reduces brown fat UCP1 expression, O2 consumption, and mitochondrial biogenesis (22). Work from other groups also points toward a beneficial role of ATGL in insulin sensitivity. Overexpression of hepatic ATGL improves insulin signaling in the liver (42), and cardiac ATGL overexpression alleviates diabetes-induced cardiomyopathy (43). The lone study to show no benefit of ATGL overexpression is the recently described muscle-specific ATGL transgenic mouse (44). However, this study did not test mice under exercised conditions when β-adrenergic signaling would be activated. Thus, in general, studies that evaluated tissue-specific ATGL overexpression highlighted very positive outcomes on insulin sensitivity. These data are also consistent with the therapeutic benefits of SIRT1 activation, although the importance of SIRT1 in mediating the effects of ATGL on metabolic disease risk has yet to be determined.
One of the most well-studied roles of SIRT1 is life span extension. Overexpression or activation of SIRT1, or its homologs, increases life span, and SIRT1 ablation results in reduced life span (45). Consistent with a link between ATGL and SIRT1, studies in fly and worm models have implicated lipolysis as a pathway that regulates life span. Specifically, inhibiting lipolysis, such as through the ablation of the Drosophila ATGL homolog brummer, reduces life span, whereas activating lipolysis extends life span (46–48). Although the mechanism through which altering lipolysis affects life span has not been tested, the aforementioned data strongly suggest that lipolysis-mediated activation of SIRT1 may be a primary signaling node underlying these effects.
In summary, these data identify a novel link between ATGL-catalyzed lipolysis and SIRT1 activation that explains the growing body of literature linking lipolysis to oxidative metabolism. Given the broad benefits of SIRT1 activation, these results highlight ATGL and lipolysis as an important signaling node that influences cell function well beyond simply altering lipid catabolism.
Article Information
Acknowledgments. The authors thank Ellen Fischer and Joe O’Brien (University of Minnesota) for technical support and Brian Finck (Washington University in St. Louis) and David Bernlohr and Howard Towle (University of Minnesota) for helpful discussions.
Funding. The authors acknowledge financial support for these studies from the National Institutes of Health to D.G.M. (DK-090364) and the Minnesota Obesity Center (DK-050456).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. S.A.K. contributed to the experiments and data analysis and writing and editing of the manuscript. A.S., M.T.M., K.T.O., and E.E.W.-H. contributed to the experiments and data analysis. D.G.M. contributed to the writing and editing of the manuscript. D.G.M. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
References
- 1.Yoshizaki T, Schenk S, Imamura T, et al. SIRT1 inhibits inflammatory pathways in macrophages and modulates insulin sensitivity. Am J Physiol Endocrinol Metab 2010;298:E419–E428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang R-H, Kim H-S, Xiao C, Xu X, Gavrilova O, Deng C-X. Hepatic Sirt1 deficiency in mice impairs mTorc2/Akt signaling and results in hyperglycemia, oxidative damage, and insulin resistance. J Clin Invest 2011;121:4477–4490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schug TT, Xu Q, Gao H, et al. Myeloid deletion of SIRT1 induces inflammatory signaling in response to environmental stress. Mol Cell Biol 2010;30:4712–4721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banks AS, Kon N, Knight C, et al. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab 2008;8:333–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Xu S, Giles A, et al. Hepatic overexpression of SIRT1 in mice attenuates endoplasmic reticulum stress and insulin resistance in the liver. FASEB J 2011;25:1664–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 1999;13:2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen HY, Miller C, Bitterman KJ, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 2004;305:390–392 [DOI] [PubMed] [Google Scholar]
- 8.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A 2004;101:15998–16003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boily G, Seifert EL, Bevilacqua L, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS One 2008;3:e1759. [DOI] [PMC free article] [PubMed]
- 10.Kim E-J, Kho J-H, Kang M-R, Um S-J. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell 2007;28:277–290 [DOI] [PubMed] [Google Scholar]
- 11.Kim J-E, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature 2008;451:583–586 [DOI] [PubMed] [Google Scholar]
- 12.Zhao W, Kruse J-P, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature 2008;451:587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerhart-Hines Z, Dominy JE Jr Blättler SM, et al. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+). Mol Cell 2011;44:851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Revollo JR, Li X. The ways and means that fine tune Sirt1 activity. Trends Biochem Sci 2013;38:160–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 2005;434:113–118 [DOI] [PubMed] [Google Scholar]
- 16.Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol 2000;20:1868–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest 1999;103:1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab 2009;9:327–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ong KT, Mashek MT, Bu SY, Greenberg AS, Mashek DG. Adipose triglyceride lipase is a major hepatic lipase that regulates triacylglycerol turnover and fatty acid signaling and partitioning. Hepatology 2011;53:116–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obrowsky S, Chandak PG, Patankar JV, et al. Adipose triglyceride lipase is a TG hydrolase of the small intestine and regulates intestinal PPARα signaling. J Lipid Res 2013;54:425–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haemmerle G, Moustafa T, Woelkart G, et al. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-α and PGC-1. Nat Med 2011;17:1076–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmadian M, Abbott MJ, Tang T, et al. Desnutrin/ATGL is regulated by AMPK and is required for a brown adipose phenotype. Cell Metab 2011;13:739–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ. Proc Natl Acad Sci U S A 1997;94:4312–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kliewer SA, Forman BM, Blumberg B, et al. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc Natl Acad Sci U S A 1994;91:7355–7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ong KT, Mashek MT, Davidson NO, Mashek DG. Hepatic ATGL mediates PPAR-α signaling and fatty acid channeling through an L-FABP independent mechanism. J Lipid Res 2014;55:808–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyoshi H, Perfield JW 2nd Souza SC, et al. Control of adipose triglyceride lipase action by serine 517 of perilipin A globally regulates protein kinase A-stimulated lipolysis in adipocytes. J Biol Chem 2007;282:996–1002 [DOI] [PubMed] [Google Scholar]
- 27.Miyoshi H, Perfield JW 2nd Obin MS, Greenberg AS. Adipose triglyceride lipase regulates basal lipolysis and lipid droplet size in adipocytes. J Cell Biochem 2008;105:1430–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bu SY, Mashek MT, Mashek DG. Suppression of long chain acyl-CoA synthetase 3 decreases hepatic de novo fatty acid synthesis through decreased transcriptional activity. J Biol Chem 2009;284:30474–30483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yatsuga S, Suomalainen A. Effect of bezafibrate treatment on late-onset mitochondrial myopathy in mice. Hum Mol Genet 2012;21:526–535 [DOI] [PubMed] [Google Scholar]
- 30.Jaworski K, Sarkadi-Nagy E, Duncan RE, Ahmadian M, Sul HS. Regulation of triglyceride metabolism. IV. Hormonal regulation of lipolysis in adipose tissue. Am J Physiol Gastrointest Liver Physiol 2007;293:G1–G4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nin V, Escande C, Chini CC, et al. Role of deleted in breast cancer 1 (DBC1) protein in SIRT1 deacetylase activation induced by protein kinase A and AMP-activated protein kinase. J Biol Chem 2012;287:23489–23501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knutti D, Kressler D, Kralli A. Regulation of the transcriptional coactivator PGC-1 via MAPK-sensitive interaction with a repressor. Proc Natl Acad Sci U S A 2001;98:9713–9718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palou M, Priego T, Sánchez J, et al. Sequential changes in the expression of genes involved in lipid metabolism in adipose tissue and liver in response to fasting. Pflugers Arch 2008;456:825–836 [DOI] [PubMed] [Google Scholar]
- 34.Adamovich Y, Shlomai A, Tsvetkov P, et al. The protein level of PGC-1α, a key metabolic regulator, is controlled by NADH-NQO1. Mol Cell Biol 2013;33:2603–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sapiro JM, Mashek MT, Greenberg AS, Mashek DG. Hepatic triacylglycerol hydrolysis regulates peroxisome proliferator-activated receptor alpha activity. J Lipid Res 2009;50:1621–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mottillo EP, Bloch AE, Leff T, Granneman JG. Lipolytic products activate peroxisome proliferator-activated receptor (PPAR) α and δ in brown adipocytes to match fatty acid oxidation with supply. J Biol Chem 2012;287:25038–25048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coleman RA, Mashek DG. Mammalian triacylglycerol metabolism: synthesis, lipolysis, and signaling. Chem Rev 2011;111:6359–6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greenberg AS, Coleman RA, Kraemer FB, et al. The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest 2011;121:2102–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mottillo EP, Granneman JG. Intracellular fatty acids suppress β-adrenergic induction of PKA-targeted gene expression in white adipocytes. Am J Physiol Endocrinol Metab 2011;301:E122–E131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langin D. Adipose tissue lipolysis as a metabolic pathway to define pharmacological strategies against obesity and the metabolic syndrome. Pharmacol Res 2006;53:482–491 [DOI] [PubMed] [Google Scholar]
- 41.Ahmadian M, Duncan RE, Varady KA, et al. Adipose overexpression of desnutrin promotes fatty acid use and attenuates diet-induced obesity. Diabetes 2009;58:855–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turpin SM, Hoy AJ, Brown RD, et al. Adipose triacylglycerol lipase is a major regulator of hepatic lipid metabolism but not insulin sensitivity in mice. Diabetologia 2011;54:146–156 [DOI] [PubMed] [Google Scholar]
- 43.Pulinilkunnil T, Kienesberger PC, Nagendran J, et al. Myocardial adipose triglyceride lipase overexpression protects diabetic mice from the development of lipotoxic cardiomyopathy. Diabetes 2013;62:1464–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sitnick MT, Basantani MK, Cai L, et al. Skeletal muscle triacylglycerol hydrolysis does not influence metabolic complications of obesity. Diabetes 2013;62:3350–3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baur JA, Ungvari Z, Minor RK, Le Couteur DG, de Cabo R. Are sirtuins viable targets for improving healthspan and lifespan? Nat Rev Drug Discov 2012;11:443–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grönke S, Mildner A, Fellert S, et al. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab 2005;1:323–330 [DOI] [PubMed] [Google Scholar]
- 47.Wang MC, O’Rourke EJ, Ruvkun G. Fat metabolism links germline stem cells and longevity in C. elegans. Science 2008;322:957–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lapierre LR, Gelino S, Meléndez A, Hansen M. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr Biol 2011;21:1507–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]