Abstract
MicroRNAs (miRNAs) are a class of small RNAs, which typically function by guiding cleavage of target mRNAs. They are known to play roles in a variety of plant processes including development, responses to environmental stresses and senescence. To identify senescence regulation of miRNAs in Arabidopsis thaliana, eight small RNA libraries were constructed and sequenced at four different stages of development and senescence from both leaves and siliques, resulting in more than 200 million genome-matched sequences. Parallel analysis of RNA ends libraries, which enable the large-scale examination of miRNA-guided cleavage products, were constructed and sequenced, resulting in over 750 million genome-matched sequences. These large datasets led to the identification a new senescence-inducible small RNA locus, as well as new regulation of known miRNAs and their target genes during senescence, many of which have established roles in nutrient responsiveness and cell structural integrity. In keeping with remobilization of nutrients thought to occur during senescence, many miRNAs and targets had opposite expression pattern changes between leaf and silique tissues during the progression of senescence. Taken together, these findings highlight the integral role that miRNAs may play in the remobilization of resources and alteration of cellular structure that is known to occur in senescence.
Keywords: Arabidopsis thaliana, microRNA, nutrient remobilization, PARE, senescence, silique
Introduction
In plants, senescence is a broad term that describes whole plant as well as tissue-specific changes that take place over the course of aging. During senescence, the leaf's cellular structure, metabolic activities and physiological role are greatly altered. Chloroplasts degenerate and the photosynthetic apparatus disassembles (Hörtensteiner 2006; Thomas 2013). Senescence is also characterized by widespread and significant changes in the pattern of gene expression (Wagstaff et al. 2009; Breeze et al. 2011). The expression of many genes, such as those associated with photosynthesis, is repressed, while that of many other genes, termed senescence-associated genes (SAGs), is induced. The molecular mechanisms governing senescence regulation have been suggested to form a complex network responsible for activation of the different SAGs (Guo & Gan 2005). Various SAGs exhibit differential expression in different tissues and in response to different senescence-promoting stimuli, including hormones and abiotic stress (Park et al. 1998; Weaver et al. 1998; Lim et al. 2007; Fischer 2012). Senescence regulation involves the action of a large number of transcription factors and epigenetic programming via histone modifications (Gregersen & Holm 2007; Balazadeh et al. 2008; Ay et al. 2009; Breeze et al. 2011; Brusslan et al. 2012). Much less is known about post-transcriptional control during senescence.
Because of the tight regulatory control exerted during the progression of senescence, it can be seen as a final developmental stage (Gan & Amasino 1997). In annual plants, including Arabidopsis, leaf senescence is thought to be a response to dwindling nutrient availability as each growing season comes to a close. Faced with a limited supply of micro and macronutrients, plants direct senescence in leaves in order to mobilize resources, such as copper, phosphate and nitrate, into reproductive tissue to enhance seed yield (Himelblau & Amasino 2001; Hörtensteiner & Feller 2002; Diaz et al. 2008; Gregersen et al. 2008). This idea is known as the nutrient relocalization hypothesis (Gan & Amasino 1997; Guiboileau et al. 2010). Given their established role in responding to nutrient availability (Chiou et al. 2006; Abdel-Ghany & Pilon 2008; Kant et al. 2011), microRNAs (miRNAs) could easily play a crucial and currently underappreciated part in guiding this process.
miRNAs are a class of small RNAs (sRNAs) that typically function by guiding ARGONAUTE-mediated cleavage of target mRNAs. They have been shown to regulate a host of processes ranging from development to responses to nutrient levels, such as phosphate and nitrate (Chiou et al. 2006; Kant et al. 2011). miRNAs were directly implicated in regulating Arabidopsis senescence when it was shown that miR164 is a key player in the senescence regulatory pathway. Natural reduction of miR164 level during senescence results in increased cell death because of its control on its target, the senescence transcriptional activator gene ORE1 (Kim et al. 2009). However, no large-scale sequencing of sRNAs or analysis of their target genes over the course of senescence has been presented thus far.
In addition to changes in transcript abundance, senescence is also thought to result in broad changes in RNA decay through increased expression of ribonucleases (RNases; Taylor et al. 1993; Bariola et al. 1999). It has been proposed that this increase in RNA turnover may be an adaptive response to senescence-related changes in phosphate levels (Bariola et al. 1994; Green 1994). Construction and sequencing of parallel analysis of RNA ends (PARE) libraries enables the precise examination of decay intermediates generated by miRNA-guided cleavage events (German et al. 2008). When a miRNA guides an AGO protein to cleave its target mRNA, the downstream cleavage product has a monophosphorylated 5′ end which PARE is designed to capture. Thus, through the analysis of PARE data, a clearer picture of the RNA degradome during the course of senescence can be obtained.
In this study, we utilize high-throughput sequencing of sRNA libraries constructed from both senescing leaf and silique tissue to discover differential miRNA expression in Arabidopsis. Expression levels of target genes for many regulated miRNAs were also examined in detail, and often found to exhibit tissue-specific regulation as senescence progressed. This analysis was complemented by the construction and sequencing of PARE libraries, which enabled examination of miRNA-guided cleavage events during senescence. The combination of sRNA, target mRNA and PARE data led to a much more complete picture of the roles that miRNAs may play in Arabidopsis senescence.
Materials and Methods
Plant growth and stress treatments
Plant material was harvested from Arabidopsis Col-0 wild-type plants. Plants were grown in short days (8 h) for 2 weeks on 1/2 × MS 0.8% agar plates. Seedlings were transferred to artificial soil in 7 × 7 × 8 cm containers and grown with a light/dark cycle of 16/8 h (long days). For leaf tissue, leaves five and six were harvested from multiple plants and pooled at four different stages: young (20 d after transfer), mature (30 d after transfer), early senescence (35 d after transfer) and late senescence (40 d after transfer). Silique tissue was also harvested from multiple plants and pooled. It was staged based on visual morphology indicative of senescence and developmental stage, including increased diameter, progression of chlorotic tissue and splitting (Fig. 1a). All tissues were immediately frozen in liquid nitrogen and stored at −80 °C. RNA was extracted from leaves or siliques using Trizol reagent (siliques, Molecular Research Center, Cincinnati, Ohio, USA) or Plant RNA Isolation Reagent (leaves, Invitrogen, Carlsbad, CA, USA), according to the manufacturers' protocols.
Figure 1.

Leaves and siliques over the course of development and senescence. The course of development and senescence is represented by four stages. (a) Leaves at four stages: young (35 d), mature (45 d), early senescence (50 d), late senescence (55 d). (b) Siliques at four stages determined and selected based on visual inspection as described in methods.
sRNA library construction and sequencing
sRNA libraries were constructed as described previously with minor modifications (Lu et al. 2007). Briefly, sRNA was isolated from total RNAs using PEG8000/NaCl-mediated low molecular weight RNA precipitation and then run on a 15% denaturing PAGE gel to select and extract 20–30 nt RNAs. 5′ and 3′ adaptors were ligated, followed by additional PAGE purification after each reaction. RNA-containing adaptors were reverse transcribed and amplified via 18 cycles of PCR followed by PAGE purification to select the final product. RNA and DNA oligos used to prepare the libraries are listed in Supporting Information Table S1. Libraries were sequenced using an Illumina IIGX platform, Illumina, San Diego, CA, USA, with either one sample per lane (leaf sequencing data) or two barcoded samples per lane (silique sequencing data).
Analysis of sRNA sequencing data
Sequencing data from sRNA libraries were trimmed to remove adaptor sequences using a custom perl script, and then mapped to the Arabidopsis genome [The Arabidopsis Information Resource (TAIR) version 10; http://www.arabidopsis.org] with Bowtie, with no mismatches allowed (option -v 0). Reads matching perfectly to the genome and not matching tRNA, rRNA and snoRNA (as annotated in TAIR10) were retained for further analysis. These reads were normalized to transcripts per two million (TP2M) to account for varying library sequencing depth. Reads were then compared with known miRNAs from miRBase version 19 (http://www.mirbase.org) and only perfect matches to annotated miRNAs were used for quantification. Remaining sRNAs, which did not match to known miRNAs and were elevated threefold or more in early or late senescence leaf libraries compared with mature leaf libraries, were considered as possible new senescence-inducible sRNAs. These were further examined as described in the text and the RNA blot section below.
PARE RNA library construction, sequencing and processing
PARE libraries were constructed as previously described with minor modifications (German et al. 2008, 2009). Briefly, 300 μg of total RNA was poly-A selected using oligo d(T)25 magnetic beads (New England BioLabs, Inc., Beverly, MA, USA). A 5′ RNA adaptor (Supporting Information Table S1) was ligated onto the remaining RNA, which was subjected to a second poly-A selection. The ligated RNA was then reverse transcribed using an oligo d(T)18 primer (New England BioLabs, Inc.) and phenol-chloroform extracted to select for cDNA. The cDNA was subjected to a short (8-cycle) PCR amplification before being digested with MmeI (New England BioLabs, Inc.), resulting in a 42 nt fragment containing 5′ adaptor and 20 nt of the original poly-A selected mRNAs. An RNA duplex complimentary to the 5′ adaptor sequence (Supporting Information Table S1) was then ligated on and the resulting product was amplified by 20 cycles of PCR and size selected on a 6% PAGE gel. After purification from the gel, the final library product was sequenced using an Illumina IIGX platform. Sequencing data from PARE libraries were trimmed to remove adaptors using the same custom perl script that was used in sRNA trimming and then mapped to the Arabidopsis genome using Bowtie, with no mismatches allowed (option -v 0; TAIR version 10; http://www.arabidopsis.org). Reads matching perfectly to the genome were used for further analysis.
Prediction and validation of targets
Targets were predicted for the abundant sRNAs arising from the senescence-inducible sRNA (sen-sRNA) locus using psRNATarget (Dai & Zhao 2011). Targets with scores <4.0 were considered potential new targets. To validate predicted cleavage, senescence PARE sequencing data as well as PARE sequencing data from xrn4 flowers obtained from NCBI's Gene Expression Omnibus with accession number GSM280227 were utilized. Sequences matching to cleavage products starting between bases 10 and 11 from the 5′ end of the predicted sRNA pairings were considered as evidence for sRNA-guided cleavage and were obtained through use of a custom perl script.
RNA blot analysis
miRNAs and other sRNAs with potential expression changes in sRNA sequencing data were subjected to northern blotting in at least two biological replicates. One replicate corresponded to the same RNA used for sRNA sequencing in the majority of cases. sRNA analyses were carried out by running low molecular weight RNA on a 15% denaturing PAGE gel. RNA was transferred onto Hybond N + membrane (Amersham, Piscataway, NJ, USA) and fixed using ultraviolet (UV) cross linking. Membranes were then pre-hybridized for at least 2 h in Ultrahyb-Oligo hybridization buffer (Ambion, Austin, TX, USA). DNA oligos complementary to the miRNA of interest were labelled with [у32P]ATP using Optikinase (Amersham Biosciences), filtered using QIAquick nucleotide removal kit (Qiagen, Valencia, CA, USA). Labelled probes were added to the hybridization solution and allowed to hybridize for at least 12 h. mRNA blots were carried out by running 20 μg of total RNA onto 1.5% formaldehyde agarose gels, followed by transfer onto Hybond N+ membranes. Membranes were then pre-hybridized overnight using hybridization buffer (Church & Gilbert 1984) and probed with 32P labelled probes. The probes were generated by PCR using primers described in Supporting Information Table S1. mRNAs were visualized using a Typhoon phosphorimager (GE Healthcare Life Sciences, Piscataway, NJ, USA).
Splinted-ligation-mediated miRNA detection
miRNAs were examined by miRtect-IT miRNA labelling and detection kit (Affymetrix, Santa Clara, CA, USA) as described previously (Maroney et al. 2008; Jeong & Green 2012). In brief, miRNAs were captured from 2 μg of total RNA using a bridge oligonucleotide that is designed to specifically detect one specific miRNA and ligated to a 32P labelled detection oligo using T4 DNA ligase. The resulting products were separated via a 15% PAGE gel and visualized using a Typhoon phosphorimager (GE Healthcare Life Sciences).
GEO accession
sRNA and PARE data have been added to the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) (GSE55151).
Results
sRNA libraries from senescing Arabidopsis leaves and siliques
In order to examine the expression patterns of miRNAs during Arabidopsis senescence, sRNA libraries were constructed from leaves and siliques at four different developmental stages. Leaves five and six were selected based on age after germination (Fig. 1a): young leaves (35 d), mature leaves (45 d), early senescing leaves (50 d) and late senescing leaves (55 d). Siliques were selected based on visual morphology indicative of developmental and senescence stage, including diameter, chlorosis and splitting (Fig. 1b). After trimming of adaptors and removal of rRNA, tRNA and snoRNAs, these eight sRNA libraries yielded more than 200 million genome-matched sequences (Table 1), and represent the first massive sRNA sequencing from senescing plant tissues. After normalization to transcripts per two million (TP2M), the total abundance of sRNAs in each size class were plotted. In leaf, 21 nt sRNAs predominated, followed by 24 nt RNAs, while in silique, 24 nt sRNAs predominated, followed by 21 nt (Supporting Information Fig. S1). This distribution difference stems from leaf tissue possessing relatively more miRNAs, which are mostly 21 nt, compared with 24 nt siRNAs. The average total miRNA abundance for leaf libraries was over 1 000 000 TP2M, while in silique, it was only about 83 000 TP2M, indicating that miRNAs are the major type of sRNA in leaf but not silique tissues (Supporting Information Table S2).
Table 1.
Summary statistics of small RNA libraries
| Library | Tissue | Description | Raw trimmed | Genome matched | ||
|---|---|---|---|---|---|---|
| Reads | Reads | Distinct | t/r/sn/snoRNA | |||
| C0LY | Leaf | Young | 27 870 710 | 17 840 547 | 1 021 881 | 6 783 580 |
| C0LM | Leaf | Mature | 26 211 889 | 16 604 542 | 473 673 | 7 452 607 |
| C0LSeE | Leaf | Early senescence | 28 700 595 | 17 875 180 | 212 951 | 6 434 947 |
| C0LSeL | Leaf | Late senescence | 29 146 047 | 11 853 598 | 97 227 | 12 456 070 |
| C0SY | Silique | Young | 33 654 351 | 30 804 353 | 4 071 246 | 8 612 996 |
| C0SM | Silique | Mature | 43 310 733 | 38 683 184 | 6 700 592 | 7 112 309 |
| C0SSeE | Silique | Early senescence | 22 880 129 | 20 049 781 | 3 957 517 | 3 251 398 |
| C0SSeL | Silique | Late senescence | 74 829 453 | 64 627 925 | 8 320 789 | 13 003 515 |
Genome-matched abundance and distinct represent sequences, which remained after removal of t/r/sn/snoRNAs.
Differential expression of known miRNAs during senescence
Normalized miRNA expression levels from young, early senescence and late senescence leaf and silique libraries were then compared with expression in the respective mature tissue to determine if any miRNAs were differentially expressed (Supporting Information Table S2). As an initial screen for regulated candidates, the abundance of annotated miRNAs in mature leaf and silique libraries with abundance greater than 50 transcripts per two million (TP2M) in any library were plotted against their abundance in young, early senescence or late senescence libraries (Supporting Information Fig. S2). In general, the expression of miRNAs across libraries was highly similar, with mature versus young showing the strongest miRNA expression correlation in both leaf and silique libraries (R2 of 0.95 and 0.94, respectively) and in mature versus late senescence leaf and silique libraries showing the weakest correlation (R2 of 0.79 and 0.81, respectively). Despite mostly similar expression patterns, certain miRNAs did show marked increases or decreases in early and late senescing tissue (Supporting Information Table S2).
miRNAs showing expression changes greater than threefold from mature to early or late senescence were confirmed in two or more independent biological replicates via northern blotting or splinted ligation-mediated detection (Jeong & Green 2012). miR164, which targets the positive regulator of senescence-induced cell death ORE1 (AT5G39610), has been previously reported to decline in leaves as senescence progresses (Kim et al. 2009). Both sRNA sequencing data and sRNA northern blot validation reproduced this regulation, showing a steady decline from young to late senescence stages in leaves (Fig. 2). Interestingly, miR164 was reduced from young to mature siliques but did not continue to decline as senescence progressed in this tissue (Fig. 2) indicating that levels of senescence-responsive miRNAs may change in a tissue-dependent manner.
Figure 2.
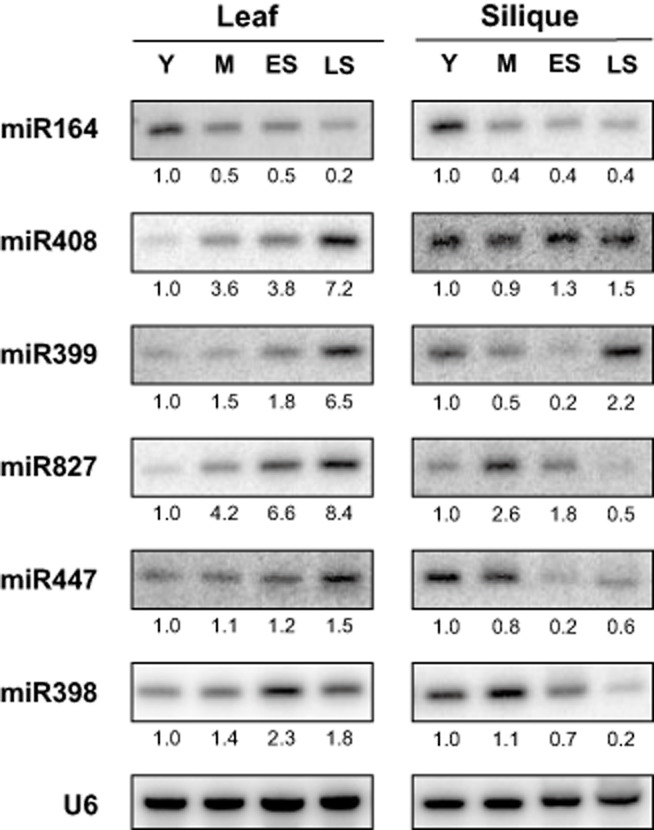
Expression of nutrient-responsive miRNAs in Arabidopsis leaf and silique over the course of senescence and development. miRNA expression level was examined via northern blotting. Stages Y (young), M (mature), ES (early senescence) and LS (late senescence) were selected as described in Fig. 1. U6-normalized quantification values are displayed below each stage. Data are representative of at least two biological replicates; one representative U6 is shown.
In addition to reproducing regulation of miR164, differential expression of seven other Arabidopsis miRNAs during the course of senescence was observed (Fig. 2). Interestingly, five out of these seven miRNAs have been reported to be nutrient responsive. miR408 targets a host of genes involved in pollen tube guidance, copper metabolism and cell wall integrity (Sunkar & Zhu 2004) and has been shown to be responsive to low copper levels (Abdel-Ghany & Pilon 2008). Both sRNA sequencing data and northern blot validation showed that this miRNA increased approximately sevenfold during senescence in leaf tissue (Fig. 2). However, the expression pattern of miR408 in siliques differed from leaves, only increasing very slightly (Fig. 2).
Several other nutrient-responsive miRNAs also showed different expression trends between leaf and silique during senescence. Both miR399, which targets the negative repressor of phosphate transport, PHOSPHATE2 (PHO2, AT2G33770) and miR827, which targets the nitrogen/phosphate balance regulator NITROGEN LIMITATION ADAPTATION (NLA, AT1G02860), are well known to be strongly induced by low phosphate conditions (Chiou et al. 2006; Kant et al. 2011). Both of these miRNAs increased as senescence progressed in leaves (Fig. 2) but had different expression patterns in siliques. In both cases, a reduction in expression occurred in early senescing siliques compared with mature. In late senescence, miR827 continued to decline, dropping to nearly undetectable levels. miR399, on the other hand, was strongly increased at the end of senescence.
The miR447 precursor has been demonstrated to increase under both phosphate and nitrate limiting conditions and the mature miRNA targets 2-PHOSPHOGLYCERATE KINASE (2PGK, AT5G60760), a gene involved in phytic acid metabolism (Allen et al. 2005; Pant et al. 2009; Kim & Tai 2011). Our results indicated that the mature miRNA increases slightly in leaves during senescence (Fig. 2). In siliques, however, miR447 is reduced at the outset of senescence and then increased in late senescence (Fig. 2). miR398 has been reported to respond to a host of nutrients including phosphate, nitrate and sucrose and targets genes involved in glucose metabolism and the response to oxidative stress (Sunkar et al. 2006; Dugas & Bartel 2008; Pant et al. 2009). miR398 showed a mild induction as senescence progressed in leaves and a much larger reduction in siliques (Fig. 2).
Two other regulated miRNA families without clear roles in nutrient responsiveness also showed senescence regulation (Fig. 3). The miR156/157 family is a highly conserved miRNA family, which targets SQUAMOSA PROMOTER LIKE BINDING (SPL) genes that are crucial for development (Gandikota et al. 2007). Intriguingly, miR157 showed a slight decrease during senescence in leaves while its close relative, miR156 was unchanged (Fig. 3a). Both miR156 and miR157 had different expression patterns in siliques compared with leaves, where they both increased strongly in late senescence. miR396a, which increased very slightly in both leaf and silique senescence (Fig. 3b), was recently found to produce an abundant miRNA*, miR396a-3p in certain tissues (Jeong et al. 2013). miR396a-3p had a larger increase in expression in leaves and a steep decline in siliques during late senescence (Fig. 3b), indicating that this newly annotated miRNA may play a previously unknown role during senescence.
Figure 3.
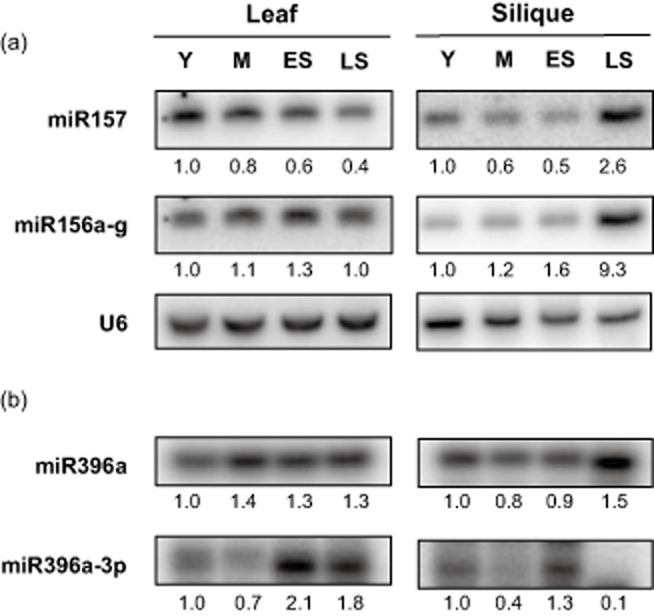
Expression of miRNAs in Arabidopsis leaf and silique. Stage selection and abbreviation as described in Fig. 1. (a) miR156/157 family members' expression level was examined via northern blotting. Data is representative of at least two biological replicates with U6-normalized quantification values are displayed below each stage. (b) miR396a and miR396a-3p expression was examined via splint-ligation-mediated miRNA detection. Quantification numbers are displayed below.
New senescence-inducible sRNA locus targets the alpha tubulin family
Given that no large-scale sequencing of sRNAs has been carried out in senescencing tissue, an analysis of senescence libraries could reveal sRNAs, which are only significantly expressed at this stage. To find new senescence-induced sRNAs, sRNA data were first filtered to remove tRNA, rRNA, snoRNA and previously annotated miRNAs. Regions surrounding the remaining sRNAs, which were elevated threefold or more in senescing leaf libraries compared with mature, were required to have a strand bias of greater than 90%. Remaining regions were then subjected to manual folding analysis using Centroidfold (Sato et al. 2009) to determine their ability to form stem-loop structures. After application of these filters, one new sRNA locus was discovered, and named sen-sRNA locus (Fig. 4a, Table 2). sen-sRNA precursor forms a nearly perfect inverted repeat and generates five abundant sRNAs, sen-sRNA1-5, which were confirmed to be senescence-regulated by northern blotting. All of these sRNAs from the sen-sRNA locus increased to their highest level in late senescing leaves (Fig. 4b). In siliques, these abundant sRNAs from the sen-sRNA precursor also showed their highest expression levels in late senescence (Fig. 4b).
Figure 4.
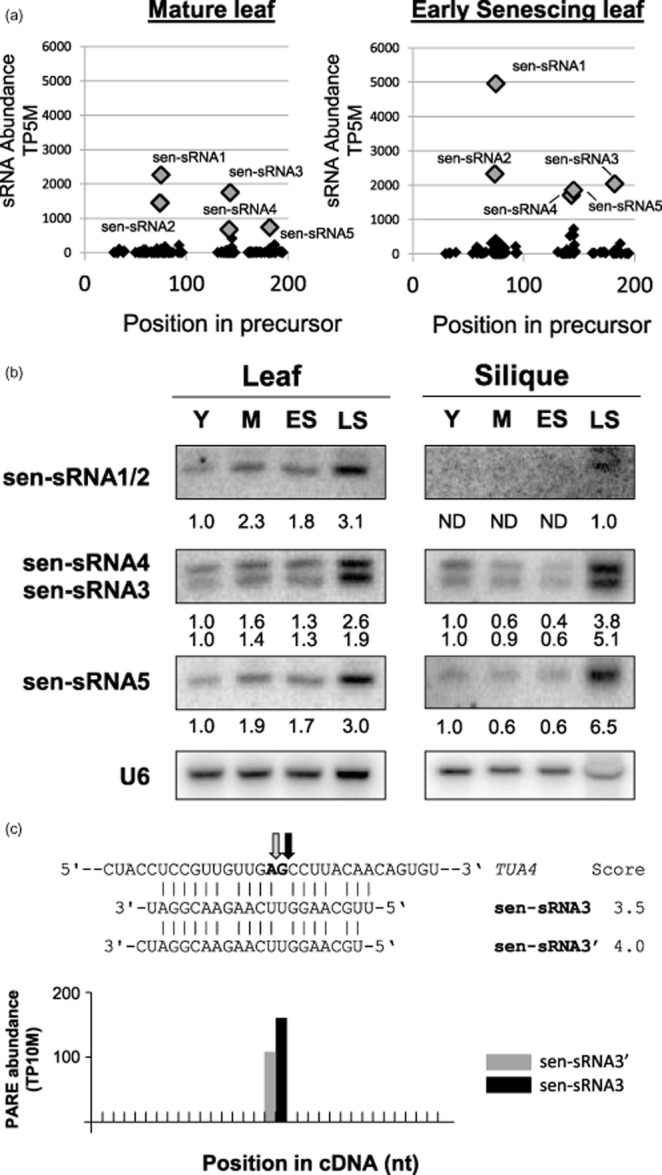
Expression and target cleavage activity of the sen-sRNA locus. (a) Small RNA plot of the sen-sRNA precursor. The abundance of small RNA sequences is plotted versus position within the predicted precursor of the sen-sRNA locus for both mature and early senescing leaf. (b) Expression of sRNAs from the sen-sRNA locus were examined via northern blotting. Data is representative of at least two biological replicates and U6-normalized quantification values are displayed below each stage. Stages were as described in Fig. 1. (c) Predicted cleavage sites of sen-sRNA3 and sen-sRNA3′ are indicated by arrows and bold 5′ nucleotides on target mRNAs. Target scores for each sRNA are also shown. Target cleavage data were obtained from PARE data of xrn4 flower (German et al. 2008). PARE abundance is shown in histograms. Grey bars indicates cleavage guided by sen-sRNA3′ while black bars indicate cleavage guided by sen-sRNA3.
Table 2.
Abundant small RNAs from the sen-sRNA precursor
| miRNA | Sequence | Size | Abundance (TP10M) |
|---|---|---|---|
| sen-sRNA1 | UUCUUGAACCUUGGAAGAAAA | 21 | 8361 |
| sen-sRNA2 | UCUUGAACCUUGGAAGAAAAC | 21 | 5840 |
| sen-sRNA3 | UGCAAGGUUCAAGAACGGAUC | 21 | 5435 |
| sen-sRNA4 | UCUUGCAAGGUUCAAGAACGGAUC | 24 | 2347 |
| sen-sRNA5 | AUUCGACAAAGUGAAGGGUUU | 21 | 2520 |
Sequence, size and sum of abundance across all eight small RNA libraries are shown.
Targets were predicted for all the abundant sRNAs arising from the sen-sRNA locus using psiRNA target prediction software (Dai & Zhao 2011). Published PARE data from xrn4 flowers were examined to search for evidence of cleavage for each potential target (German et al. 2008). The loss of the exoribonuclease XRN4 in this mutant background causes elevated levels of sRNA-guided cleavage intermediates to accumulate, enabling sRNA targets to be more easily identified (Souret et al., 2004; Rymarquis et al., 2011; Nagarajan et al., 2013). sen-sRNA3 was predicted to target a conserved region of the alpha tubulin family, pairing to TUA2, TUA4 and TUA6 with scores of 4.0, 3.5 and 4.0, respectively (Fig. 4c). Abundant cleavage products predicted for sen-sRNA3 and a variant that differs by one nucleotide were both found in this library. These decay intermediates could arise from cleavage of one, two or all three alpha tubulin family members, which cannot be distinguished because of the conserved nature of this region.
Analysis of PARE data from senescing leaves
In addition to utilizing sRNA and publicly available PARE libraries, six new PARE libraries were constructed from Arabidopsis leaves to examine targets of regulated miRNAs. After trimming of adaptors, these libraries yielded more than 750 million genome-matched sequences and represent the first large-scale analysis of the RNA degradome during Arabidopsis senescence (Table 3). Early and late senescing leaf libraries had significantly reduced genome-matched distinct sequences, despite a second round of library construction and sequencing from biological replicates (Table 3). This reproducible overall reduction in complexity is likely caused by the general increase in RNA decay associated with senescence progression (Taylor et al. 1993; Bariola et al. 1999). In keeping with this reduction in distinct sequences, the majority of miRNA target cleavage products were either missing or significantly reduced.
Table 3.
Summary statistics of PARE libraries
| Library | Tissue | Description | Raw trimmed | Genome matched | |
|---|---|---|---|---|---|
| Reads | Reads | Distinct | |||
| C0LY | Leaf | Young | 114 373 290 | 97 727 510 | 11 087 211 |
| C0LM | Leaf | Mature | 107 307 144 | 92 663 203 | 11 592 888 |
| C0LSeE1 | Leaf | Early senescence 1 | 380 719 524 | 335 224 824 | 1 689 330 |
| C0LSeL1 | Leaf | Late senescence 1 | 142 841 947 | 107 151 162 | 163 022 |
| C0LSeE2 | Leaf | Early senescence 2 | 127 367 565 | 114 143 220 | 1 192 825 |
| C0LSeL2 | Leaf | Late senescence 2 | 26 170 444 | 22 820 583 | 260 527 |
Initial analysis of these PARE libraries focused on examining targets of regulated miRNAs. Because of the reduction or absence of most miRNA target cleavage products in senescing leaves, it is not possible to draw conclusions about lower levels of cleavage products for targets whose miRNAs were shown to be reduced as senescence progresses. However, strong induction of miR408 in leaves was accompanied by an increase in the cleavage products generated for all four of its validated targets (Fig. 5). Similarly, NLA had a significant increase in the cleavage product generated by miR827-guided cleavage (data not shown).
Figure 5.
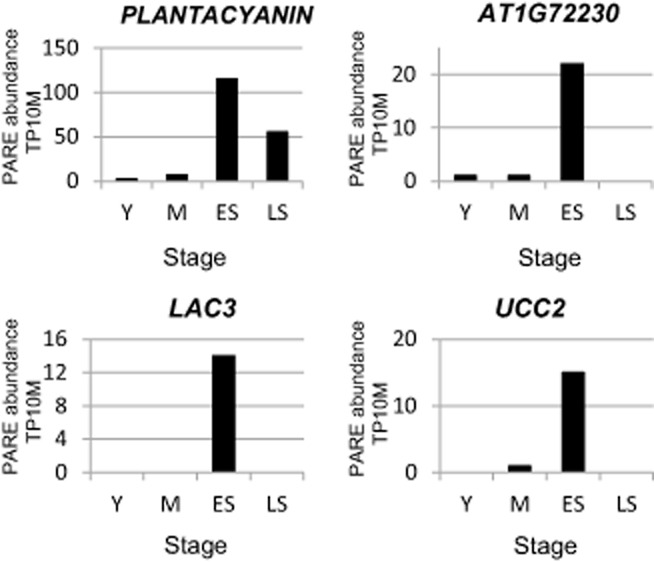
PARE sequences corresponding to precise miR408 target cleavage sites. PARE libraries data showing the level of decay intermediates generated by miR408-guided cleavage of PLATNACYANIN, AT1G72230, LAC3 and UCC2. Cleavage data were obtained from PARE libraries constructed at four stages during development and senescence. Bars represent the abundance of cleavage products guided by miR408.
Differential expression of senescence-regulated targets
Expression changes of the genes targeted by senescence-regulated miRNAs were also analysed. Although the majority of senescence-regulated miRNAs have previously been shown to be nutrient responsive, their targets possessed a wide variety of functions ranging from nutrient mobilization to cell wall integrity. Like many senescence-regulated miRNAs, these targets often showed different expression changes between leaf and silique tissue (Fig. 6). Some of these targets showed incoherent regulation (Jeong & Green 2013) relative to the miRNAs targeting them, that is, they increase despite a simultaneous increase in the miRNAs, which targets them (Figs. 2 & 6).
Figure 6.
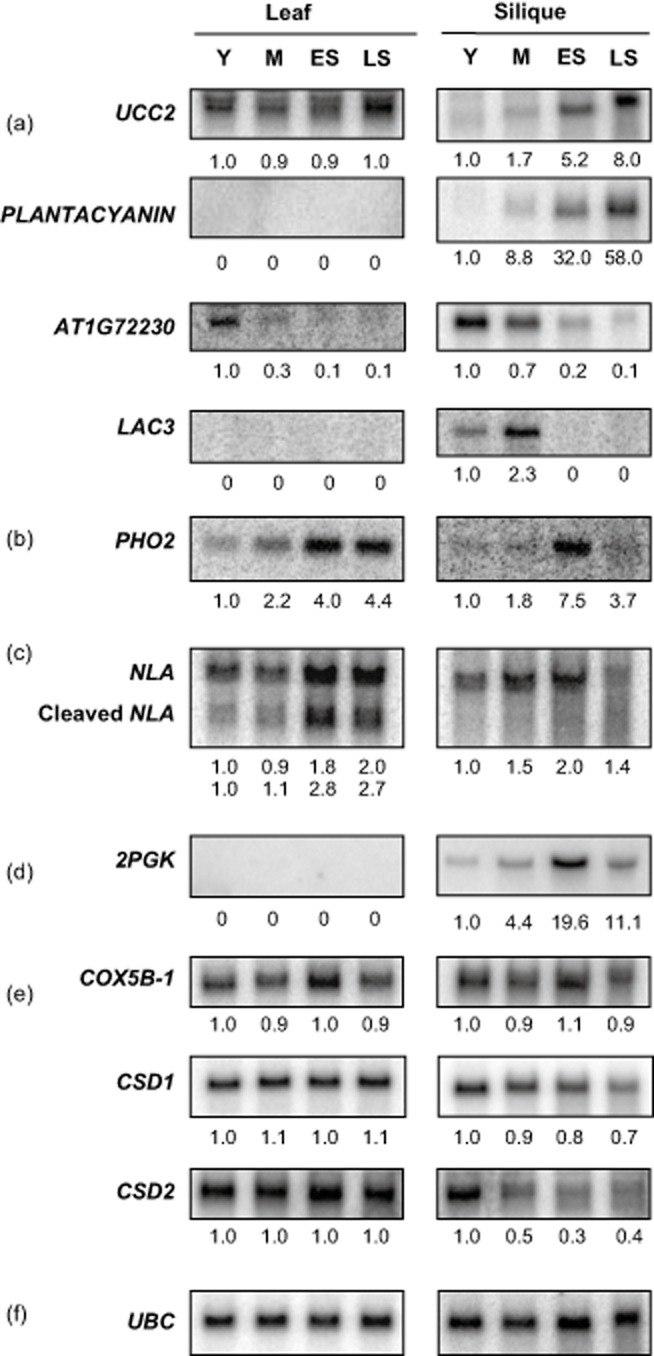
Senescence regulation of nutrient-responsive miRNA targets in leaf and silique. Northern blots showing differential expression of nutrient regulated miRNA targets in senescing leaf and silique. Data is representative of at least two biological replicates. Stages were selected as described in Fig. 1. UBC (At5g25760) normalized quantification values are displayed below each stage. (a) miR408 targets UCC2, PLANTACYANIN, AT1G72230 and LAC3; (b) miR399 target PHO2; (c) miR827 target NLA; (d) miR447 target 2PGK; (e) miR398 targets COX5B-1, CSD1; and CSD2 (f) Representative UBC.
Four of targets of miR408, which had increased cleavage products in senescence PARE data (Fig. 5), were analysed via northern blotting. AT1G72230, a member of the CUPREDOXIN superfamily, which binds copper and functions in the synthesis of xylan, a polysaccharide component of cell walls (Borner et al. 2002; Mutwil et al. 2009), decreased as aging progressed in both leaves and siliques (Fig. 6a). UCLACYANIN 2 (UCC2, AT2G44790) also contains a copper-binding domain but its function remains uncertain (Borner et al. 2002; Mutwil et al. 2009). Unlike AT1G72230, UCC2 was mostly unchanged during leaf senescence and actually increased in senescing siliques (Fig. 6a). LAC3 (AT2G30210) is a member of the laccase family that has been proposed to be involved in maintaining cell wall integrity (Turlapati et al. 2011). LAC3 expression was not detected via northern blotting in leaves, but it was highly expressed in young and mature siliques and decreased rapidly during senescence (Fig. 6a). PLANTACYANIN (AT2G02850), which is thought to function in pollen tube guiding, has only been reported to be expressed significantly in the pistil and root (Dong et al. 2005; Abdel-Ghany & Pilon 2008). Despite a highly abundant cleavage product present in leaf PARE libraries, no expression was detected in this tissue through northern blotting (Fig. 6a). However, PLANTACYANIN was detectable in early senescing siliques and increased further during late senescence, indicating incoherent regulation with miR408.
Both miR399, which targets PHO2, and miR827, which targets NLA, were strongly induced during senescence in leaves (Fig. 2). Interestingly, PHO2 and NLA were themselves increased in senescing leaves, showing incoherent regulation with the miRNAs which target them (Fig. 6b,c). While full-length NLA increased during leaf senescence, its putative miR827-guided cleavage product increased even more strongly (Fig. 6c). This result is consistent with an increase of miR827 and an accumulation of miR827 directed decay product in PARE results and implies that miR827 may be functioning to moderate NLA's expression increase during senescence instead of acting as an on/off switch. In siliques, PHO2 and NLA showed mostly coherent regulation, increasing to their highest levels in early senescence as miR399 and miR827 decreased.
2PGK, a target of miR447 has previously been reported to be flower and silique preferential (Kim & Tai 2011). In keeping with published work, 2PGK was undetectable in leaves at any point during senescence (Fig. 6d). In siliques, 2PGK increased from young to early senescence stages, reproducing earlier work demonstrating its role in increased phytic acid levels during silique maturation (Kim & Tai 2011). However, 2PGK decreased as siliques progressed to late senescence, again showing coherent regulation with miR447 at this stage.
miR398 targets a host of genes involved in a variety of different processes ranging from glucose metabolism to the oxidative stress response. COX5B-1 (AT3G15640), which is part of the glucose catabolic process (Comelli et al. 2009), was selected for northern blot analysis based on known macronutrient changes during senescence (Wingler & Roitsch 2008). Surprisingly, the expression of COX5B-1 was relatively unchanged in leaves or siliques during senescence despite regulation of miR398 itself (Fig. 6e). Other targets of miR398, SUPEROXIDE DISMUTASE 1(CSD1) and SUPEROXIDE DISMUTASE 2 (CSD2), which are responsible for reducing reactive oxygen species (ROS) during oxidative stress, were unchanged in leaves but declined in senescing siliques (Fig. 6e).
Targets of sen-sRNA3 include three members of the alpha tubulin family, TUA2, TUA4 and TUA6 (Fig. 4c), which were examined via northern blotting. All three of these targets declined to approximately half of their normal expression level in late senescing leaves (Fig. 7). In siliques, this decline was substantially stronger, with all targets of sen-sRNA3 declining to nearly undetectable levels by late senescence (Fig. 7).
Figure 7.
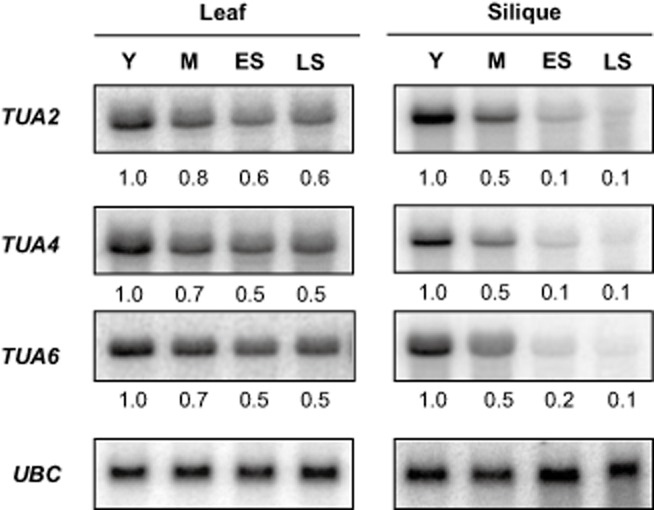
Senescence regulation of sen-sRNA3 targets. Northern blots showing differential expression of sen-sRNA3 targets in senescing leaf and silique. Data is representative of at least two biological replicates. Stages were selected as described in Fig. 1. UBC (At5g25760) normalized quantification values. One representative UBC is shown.
Discussion
In this study, we report the first sRNA and PARE libraries from senescing leaves and siliques, which enabled large-scale examination of miRNA expression and miRNA target cleavage in these tissues. Seven previously annotated miRNAs were demonstrated to change as senescence progressed, in a tissue-specific manner. Deep sequencing data also enabled identification of a new regulated sRNA locus, the sen-sRNA locus, which is most strongly expressed in senescing leaves and siliques (Fig. 4). Examination of the expression levels and decay intermediates of the targets of these known miRNAs and the new sen-sRNAs revealed that they also undergo tissue-specific changes during the course of senescence. Instances of coherent and incoherent regulation were observed between the miRNAs and their targets, a number of which are associated with nutrient limitation.
Differential expression of miRNAs and their target mRNAs in leaves and siliques
It has been reported that a significant change in the levels of both micro- and macronutrients occurs in senescing leaves (Bleecker & Patterson 1997; Gan & Amasino 1997; Himelblau & Amasino 2001). These changes are thought to be the result of a mobilization of resources away from vegetative tissue and towards developing reproductive tissue and lead to a host of expression changes in both tissues (Leopold 1961; Guiboileau et al. 2010). Our analysis of miRNA expression during the course of senescence revealed different, and often completely opposite, expression patterns between leaves and siliques (Figs. 2 & 3). In keeping with the nutrient remobilization hypothesis, five out of the seven miRNAs (miR408, miR399, miR827, miR447 and miR398), which were senescence regulated, have been previously reported to be nutrient responsive (Fujii et al. 2005; Abdel-Ghany & Pilon 2008; Dugas & Bartel 2008; Pant et al. 2009; Kant et al. 2011), indicating that they may play a currently underappreciated role in controlling the flow of nutrients from vegetative to reproductive tissue.
miR399 and miR827 were both strongly induced during senescence in leaves but showed more complex pattern of regulation in siliques, where their expression was initially lower in early senescence (Fig. 2). The correlation between these two miRNAs in both leaves and siliques is consistent with their reported response to phosphate levels (Kant et al. 2011). However, their target genes PHO2 and NLA, which are both negative repressors of phosphate transport, responded differently to senescence than they do to phosphate deficiency (Fig. 6b,c; Fujii et al. 2005). In leaves, PHO2 gradually increased during senescence, showing incoherent regulation with miR399 and an opposite expression change than has been reported in non-senescence-induced phosphate deficiency. However, it has been shown that while miR399 and PHO2 are co-expressed in vascular tissue, miR399 alone is expressed in mesophyll cells, where it may have a currently unidentified target (Aung et al. 2006), which could be repressed during senescence. Additionally, Arabidopsis expresses target mimics that sequester miR399, which could explain how both miR399 and PHO2 levels could increase under the same conditions (Franco-Zorrilla et al. 2007). The miR827 target NLA also increased in senescing leaves, showing incoherent regulation and an opposite pattern than has been reported in phosphate deficient Arabidopsis. However, both PARE (data not shown) and northern blot (Fig. 6c) showed an even stronger accumulation of the miR827 directed cleavage product, which is consistent with elevated target and miRNA levels. Given NLA's role in regulating phosphate transport in a nitrate-dependent manner, it is likely that miR827 may be functioning to fine tune this relationship during senescence instead of acting as an on/off switch. Additionally, NLA has been shown to positively affect anthocyanin production, which is a well-known senescence phenotype (Peng et al. 2007). The incoherent regulation of miR399 and miR827's target genes, which does not occur under whole-plant nutrient starvation, implies that nutrient-responsive miRNAs and their target genes may play additional roles in adaptation to shifting nutrient balance during natural senescence.
miR447, which has been shown to be induced under nitrate-deficient conditions (Pant et al. 2009), was mildly increased during senescence in leaves (Fig. 2), where it is known that nitrate levels decrease with age (Diaz et al. 2008). Its target 2PGK, however, has been reported to be strongly silique and flower preferential and was undetectable in leaves at any stage. In siliques, miR447 had the lowest level of expression in early senescence where its target 2PGK showed its highest expression (Fig. 6). The increase of 2PGK as siliques transitioned from young to early senescence is consistent with earlier findings, which implicate increases of this gene in the silique maturation-induced accumulation of phytic acid, a phosphate storage compound found in seeds, which is inaccessible to human absorption and considered an anti-nutrient (Stevenson-Paulik et al. 2005). Knockouts of 2PGK have been shown to have reduced phytic acid, a desirable agricultural trait, but also have reduced seed germination under stress (Kim & Tai 2011). The natural repression of 2PGK by miR447, which is shown to be reduced in late senescence, may be more amenable to manipulation in future work.
Of the five nutrient-responsive miRNAs, which changed during the course of aging, only miR398 responded in an opposite manner in senescence compared with its demonstrated response to nutrient deficiency. While sucrose, phosphate and nitrate deficiencies have been reported to repress miR398 (Dugas & Bartel 2008; Pant et al. 2009), its levels were actually increased during aging in leaves. Surprisingly COX5b-1, a target of miR398, was unchanged during senescence (Fig. 6e) despite its reported responses to sucrose levels (Comelli et al. 2009). Other targets of miR398, CSD1 and CSD2, however, were down-regulated during senescence in siliques. This is surprising, given the reduction of miR398 during senescence in siliques (Fig. 2) and the known increase in ROS during senescence (Woo et al. 2004) and indicates that the role of miR398 in senescence warrants further investigation. Like miR399 and miR827, this miRNA may also play a different role in senescence than it does in whole-plant nutrient deficiency and oxidative stress.
The majority of senescence-regulated miRNAs have previously been reported to respond to changes in nutrient levels. However, members of the miR156/157 family and miR396a-3p have not been shown to change significantly under nutrient starvation. miR396a itself has been reported to target the GROWTH REGULATING TRANSCRIPTION FACTOR (GRF) family (Jones-Rhoades & Bartel 2004), but increased very slightly in senescing leaves and siliques. Interestingly, the miR396a precursor was recently reported to produce an abundant miR star, miR396a-5p (Jeong et al. 2013), which was induced during senescence in leaves and repressed in siliques (Fig. 3b). This miRNA is currently without any validated targets, making it difficult to speculate on its potential role in senescence. All members of the miR156/157 family are predicted to target members of the SPL transcription factor family, which control development. Despite overlapping predicted targets, it has recently been reported that miR156 and miR157 family members do mediate differential cleavage of specific SPL transcripts (Meng et al. 2012). The senescence repression of miR157 in leaves without any change in miR156 therefore may be a fine-tuning mechanism to control expression of specific SPLs as senescence progresses in leaves. In siliques, both miRNAs were strongly induced in late senescence, which could lead to repression of multiple SPL family members in this tissue.
miR408 targets genes involved in many different biological processes and its senescence regulation could potentially have a variety of effects. Although UCC2 and PLANTACYANIN are both validated targets of miR408, they showed incoherent regulation by increasing as senescence progressed (Fig. 6), and are involved in processes, which have no known involvement in the senescence process (Dong et al. 2005). In contrast, two other targets, CUPREDOXIN and LAC3, both showed the expected reduction, which correlated with an increase of miR408 during senescence (Fig. 6). CUPREDOXIN has been proposed to be involved in xylan biosynthesis (Mutwil et al. 2009) and its down-regulation by miR408 could therefore affect cell wall integrity during senescence. Similarly LAC3, which was also reduced during senescence, is thought to act on the cell wall. Transgenic knockdowns of LAC3 have been reported to have altered cell wall integrity and more readily detachable xylem fibre cell walls in poplar (Ranocha et al. 1999). Senescence is known to bring about a host of cell wall changes (Bleecker & Patterson 1997) and the regulation of CUPREDOXIN and LAC3 by miR408 imply that this miRNA may be playing an important role in the process.
Sen-sRNAs are induced by senescence and target the alpha tubulin gene family
The discovery of the senescence-inducible sen-sRNA locus through deep sequencing of sRNAs in senescing tissues highlights the importance of continued investigation of the sRNA transcriptome under diverse sets of conditions. It is interesting that the sen-sRNA precursor can form a stem-loop structure, passes strand bias criteria for miRNAs and sRNAs from it guide cleavage of target mRNAs similar to miRNAs. Thus, although sen-sRNAs have miRNA-like function, the sen-sRNA precursor fails the abundance cut-offs for precise excision required for formal annotation as a miRNA locus (Meyers et al. 2008).
The expression of nearly all members of the alpha tubulin gene family has been reported to decline during senescence (Wagstaff et al. 2009; Keech et al. 2010; Breeze et al. 2011). Intriguingly, we show that all alpha tubulin mRNAs that are sen-sRNA3 targets decline slightly in leaves and drastically in siliques (Fig. 7), which has the potential to lead to massive changes in cellular integrity. sen-sRNA3 may therefore be working in concert with miR408 to alter cellular structure by targeting alpha tubulins and represents an interesting candidate for future studies investigating senescence-associated changes in cell structure. Both miR408 and sen-sRNA3 are the first indications that sRNAs, which have demonstrated roles in many developmental processes (Gandikota et al. 2007), are also involved in what is often termed the final developmental stage, senescence.
Taken together, these findings highlight the integral, and currently underappreciated, role that miRNAs play during senescence. The high-percentage senescence-regulated miRNAs, which have previously been noted as nutrient responsive, and their predominantly antagonistic regulation between leaves and siliques, implies they may function in the regulatory pathways, which govern mobilization of nutrients away from vegetative tissue and towards reproductive tissue during the course of senescence. Beyond the mobilization of nutrients, several miRNAs have target genes that could be of strong interest agriculturally, such as miR447, which controls phytic acid accumulation, or miR408 and sen-sRNA3, which have the potential to alter cell structure and integrity. This study greatly expands upon the existing knowledge of the roles of miRNAs in senescence and provides a number of new avenues to explore.
Acknowledgments
We are very grateful to Blake Meyers for providing databases and database support for this work. The assistance of Monica Accerbi and Sunhee Park is appreciated. We also thank Dong-Hoon Jeong for very helpful discussions. This research was supported by grant no. IS-4180-08 from BARD, the United States–Israel Binational Agricultural Research and Development Fund to A.L. and P.J.G. S.R.T. was supported, in part, by NIH/NIGMS CBI Training Grant 5T32 GM 08550-15. C.W. was supported, in part, by an undergraduate fellowship from the Howard Hughes Medical Institute and funds from the DuPont Distinguished Scholars Program from the University of Delaware.
Supporting Information
Figure S1. Small RNA size distribution plots. Genome-matched small RNA data were normalized to transcripts per two million (TP2M) and the total abundance of each size class of small RNA was plotted. (A) Size distributions in young, mature, early senescing and late senescing leaf. (B) Size distributions in young, mature, early senescing and late senescing silique.
Figure S2. Comparison of miRNA abundance between mature and other stages.
miRNA abundance comparison between mature and other stages. Correlation coefficients for each comparison are also shown. (A) Mature leaves compared with young leaves, early senescing leaves and late senescing leaves. (B) Mature siliques compared with young siliques, early senescing siliques and late senescing siliques.
Table S1. Oligomers used in this study.
Table S2. miRNA quantification from leaf and silique small RNA libraries. Values represent the abundance of the miRNA sequence in the indicated library (abbreviations as in Table 1) normalized to TP2M (transcripts per two million) trimmed genome-matched reads.
References
- Abdel-Ghany SE. Pilon M. MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. The Journal of Biological Chemistry. 2008;283:15932–15945. doi: 10.1074/jbc.M801406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM. Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Aung K, Lin SI, Wu CC, Huang YT, Su CL. Chiou TJ. pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiology. 2006;141:1000–1011. doi: 10.1104/pp.106.078063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ay N, Irmler K, Fischer A, Uhlemann R, Reuter G. Humbeck K. Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. The Plant Journal: For Cell and Molecular Biology. 2009;58:333–346. doi: 10.1111/j.1365-313X.2008.03782.x. [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Riano-Pachon DM. Mueller-Roeber B. Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biology. 2008;10:63–75. doi: 10.1111/j.1438-8677.2008.00088.x. [DOI] [PubMed] [Google Scholar]
- Bariola PA, Howard CJ, Taylor CB, Verburg MT, Jaglan VD. Green PJ. The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. The Plant Journal: For Cell and Molecular Biology. 1994;6:673–685. doi: 10.1046/j.1365-313x.1994.6050673.x. [DOI] [PubMed] [Google Scholar]
- Bariola PA, MacIntosh GC. Green PJ. Regulation of S-like ribonuclease levels in Arabidopsis. Antisense inhibition of RNS1 or RNS2 elevates anthocyanin accumulation. Plant Physiology. 1999;119:331–342. doi: 10.1104/pp.119.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker AB. Patterson SE. Last exit: senescence, abscission, and meristem arrest in Arabidopsis. The Plant Cell. 1997;9:1169–1179. doi: 10.1105/tpc.9.7.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GH, Sherrier DJ, Stevens TJ, Arkin IT. Dupree P. Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A genomic analysis. Plant Physiology. 2002;129:486–499. doi: 10.1104/pp.010884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C. Buchanan-Wollaston V. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. The Plant Cell. 2011;23:873–894. doi: 10.1105/tpc.111.083345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusslan JA, Alvarez-Canterbury AMR, Nair NU, Rice JC, Hitchler MJ. Pellegrini M. Genome-wide evaluation of histone methylation changes associated with leaf senescence in Arabidopsis. PLoS ONE. 2012;7:e33151. doi: 10.1371/journal.pone.0033151. doi:10.1371/journal.pone.033151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF. Su CL. Regulation of phosphate homeostasis by microRNA in Arabidopsis. The Plant Cell. 2006;18:412–421. doi: 10.1105/tpc.105.038943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. Gilbert W. Genomic sequencing. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comelli RN, Viola IL. Gonzalez DH. Characterization of promoter elements required for expression and induction by sucrose of the Arabidopsis COX5b-1 nuclear gene, encoding the zinc-binding subunit of cytochrome c oxidase. Plant Molecular Biology. 2009;69:729–743. doi: 10.1007/s11103-008-9451-0. [DOI] [PubMed] [Google Scholar]
- Dai X. Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Research. 2011;39(Suppl. 2):W155–W159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz C, Lemaître T, Christ A, Azzopardi M, Kato Y, Sato F. Masclaux-Daubresse C. Nitrogen recycling and remobilization are differentially controlled by leaf senescence and development stage in Arabidopsis under low nitrogen nutrition. Plant Physiology. 2008;147:1437–1449. doi: 10.1104/pp.108.119040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Kim ST. Lord EM. Plantacyanin plays a role in reproduction in Arabidopsis. Plant Physiology. 2005;138:778–789. doi: 10.1104/pp.105.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas DV. Bartel B. Sucrose induction of Arabidopsis miR398 represses two Cu/Zn superoxide dismutases. Plant Molecular Biology. 2008;67:403–417. doi: 10.1007/s11103-008-9329-1. [DOI] [PubMed] [Google Scholar]
- Fischer AM. The complex regulation of senescence. Critical Reviews in Plant Sciences. 2012;31:124–147. [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I. Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nature Genetics. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- Fujii H, Chiou T-J, Lin S-I, Aung K. Zhu J-K. A miRNA involved in phosphate-starvation response in Arabidopsis. Current Biology. 2005;15:2038–2043. doi: 10.1016/j.cub.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Gan S. Amasino RM. Making sense of senescence (molecular genetic regulation and manipulation of leaf senescence) Plant Physiology. 1997;113:313–319. doi: 10.1104/pp.113.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandikota M, Birkenbihl RP, Hohmann S, Cardon GH, Saedler H. Huijser P. The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. The Plant Journal: For Cell and Molecular Biology. 2007;49:683–693. doi: 10.1111/j.1365-313X.2006.02983.x. [DOI] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P. Green PJ. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nature Biotechnology. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- German MA, Luo S, Schroth G, Meyers BC. Green PJ. Construction of parallel analysis of RNA ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nature Protocols. 2009;4:356–362. doi: 10.1038/nprot.2009.8. [DOI] [PubMed] [Google Scholar]
- Green PJ. The ribonucleases of higher plants. Annual Review of Plant Physiology and Plant Molecular Biology. 1994;45:421–445. [Google Scholar]
- Gregersen PL. Holm PB. Transcriptome analysis of senescence in the flag leaf of wheat (Triticum aestivum L.) Plant Biotechnology Journal. 2007;5:192–206. doi: 10.1111/j.1467-7652.2006.00232.x. [DOI] [PubMed] [Google Scholar]
- Gregersen PL, Holm PB. Krupinska K. Leaf senescence and nutrient remobilisation in barley and wheat. Plant Biology. 2008;10:37–49. doi: 10.1111/j.1438-8677.2008.00114.x. [DOI] [PubMed] [Google Scholar]
- Guiboileau A, Sormani R, Meyer C. Masclaux-Daubresse C. Senescence and death of plant organs: nutrient recycling and developmental regulation. Comptes Rendus Biologies. 2010;333:382–391. doi: 10.1016/j.crvi.2010.01.016. [DOI] [PubMed] [Google Scholar]
- Guo YF. Gan SS. Leaf senescence: signals, execution, and regulation. In: Schatten GP, editor; Current Topics in Developmental Biology. Vol. 71. San Diego: Elsevier Academic Press Inc; 2005. pp. 83–112. [DOI] [PubMed] [Google Scholar]
- Himelblau E. Amasino R. Nutrients mobilized from leaves of Arabidopsis thaliana during leaf senescence. Journal of Plant Physiology. 2001;158:1317–1323. [Google Scholar]
- Hörtensteiner S. Chlorophyll degradation during senescence. Annual Review of Plant Biology. 2006;57:55–77. doi: 10.1146/annurev.arplant.57.032905.105212. [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S. Feller U. Nitrogen metabolism and remobilization during senescence. Journal of Experimental Botany. 2002;53:927–937. doi: 10.1093/jexbot/53.370.927. [DOI] [PubMed] [Google Scholar]
- Jeong DH. Green PJ. Methods for validation of miRNA sequence variants and the cleavage of their targets. Methods (San Diego, Calif.) 2012;58:135–143. doi: 10.1016/j.ymeth.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Jeong DH. Green PJ. The role of rice microRNAs in Abiotic stress responses. Journal of Plant Biology. 2013;56:187–197. [Google Scholar]
- Jeong DH, Thatcher SR, Brown RS, Zhai J, Park S, Rymarquis LA. Green PJ. Comprehensive investigation of microRNAs enhanced by analysis of sequence variants, expression patterns, ARGONAUTE loading, and target cleavage. Plant Physiology. 2013;162:1225–1245. doi: 10.1104/pp.113.219873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW. Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Molecular Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Kant S, Peng M. Rothstein SJ. Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in arabidopsis. PLoS Genetics. 2011;7:e1002021. doi: 10.1371/journal.pgen.1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech O, Pesquet E, Gutierrez L, Ahad A, Bellini C, Smith SM. Gardeström P. Leaf senescence is accompanied by an early disruption of the microtubule network in Arabidopsis. Plant Physiology. 2010;154:1710–1720. doi: 10.1104/pp.110.163402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH. Nam HG. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science. 2009;323:1053–1057. doi: 10.1126/science.1166386. [DOI] [PubMed] [Google Scholar]
- Kim SI. Tai TH. Identification of genes necessary for wild-type levels of seed phytic acid in Arabidopsis thaliana using a reverse genetics approach. Molecular Genetics and Genomics. 2011;286:119–133. doi: 10.1007/s00438-011-0631-2. [DOI] [PubMed] [Google Scholar]
- Leopold AC. Senescence in plant development: the death of plants or plant parts may be of positive ecological or physiological value. Science. 1961;134:1727–1732. doi: 10.1126/science.134.3492.1727. [DOI] [PubMed] [Google Scholar]
- Lim PO, Kim HJ. Nam HG. Leaf senescence. Annual Review of Plant Biology. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- Lu C, Meyers BC. Green PJ. Construction of small RNA cDNA libraries for deep sequencing. Methods (San Diego, Calif.) 2007;43:110–117. doi: 10.1016/j.ymeth.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Maroney PA, Chamnongpol S, Souret F. Nilsen TW. Direct detection of small RNAs using splinted ligation. Nature Protocols. 2008;3:279–287. doi: 10.1038/nprot.2007.530. [DOI] [PubMed] [Google Scholar]
- Meng Y, Shao C, Ma X, Wang H. Chen M. Expression-based functional investigation of the organ-specific MicroRNAs in Arabidopsis. PLoS ONE. 2012;7:e50870. doi: 10.1371/journal.pone.0050870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL. Zhu JK. Criteria for annotation of plant MicroRNAs. The Plant Cell. 2008;20:3186–3190. doi: 10.1105/tpc.108.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M, Ruprecht C, Giorgi FM, Bringmann M, Usadel B. Persson S. Transcriptional wiring of cell wall-related genes in Arabidopsis. Molecular Plant. 2009;2:1015–1024. doi: 10.1093/mp/ssp055. [DOI] [PubMed] [Google Scholar]
- Nagarajan VK, Jones CI, Newbury SF, Green PJ. XRN 5′ -> 3′ exoribonucleases: structure, mechanisms and functions. Biochim and Biophys Acta. 2013;1829:590–603. doi: 10.1016/j.bbagrm.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant BD, Musialak-Lange M, Nuc P, May P, Buhtz A, Kehr J. Scheible WR. Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiology. 2009;150:1541–1555. doi: 10.1104/pp.109.139139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Oh SA, Kim YH, Woo HR. Nam HG. Differential expression of senescence-associated mRNAs during leaf senescence induced by different senescence-inducing factors in Arabidopsis. Plant Molecular Biology. 1998;37:445–454. doi: 10.1023/a:1005958300951. [DOI] [PubMed] [Google Scholar]
- Peng M, Hannam C, Gu H, Bi YM. Rothstein SJ. A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. The Plant Journal: For Cell and Molecular Biology. 2007;50:320–337. doi: 10.1111/j.1365-313X.2007.03050.x. [DOI] [PubMed] [Google Scholar]
- Ranocha P, McDougall G, Hawkins S, Sterjiades R, Borderies G, Stewart D. Goffner D. Biochemical characterization, molecular cloning and expression of laccases – a divergent gene family – in poplar. European Journal of Biochemistry. 1999;259:485–495. doi: 10.1046/j.1432-1327.1999.00061.x. [DOI] [PubMed] [Google Scholar]
- Rymarquis LA, Souret FF, Green PJ. Evidence that XRN4, an Arabidopsis homolog of exoribonuclease XRN1, preferentially impacts transcripts with certain sequences or in particular functional categories. RNA. 2011;17:501–511. doi: 10.1261/rna.2467911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Hamada M, Asai K. Mituyama T. CENTROIDFOLD: a web server for RNA secondary structure prediction. Nucleic Acids Research. 2009;37:W277–W280. doi: 10.1093/nar/gkp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souret FF, Kastenmayer JP, Green PJ. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Molecular Cell. 2004;15:173–183. doi: 10.1016/j.molcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Stevenson-Paulik J, Bastidas RJ, Chiou ST, Frye RA. York JD. Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12612–12617. doi: 10.1073/pnas.0504172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R. Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. The Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A. Zhu JK. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. The Plant Cell. 2006;18:2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CB, Bariola PA, delCardayre SB, Raines RT. Green PJ. RNS2: a senescence-associated RNase of Arabidopsis that diverged from the S-RNases before speciation. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:5118–5122. doi: 10.1073/pnas.90.11.5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H. Senescence, ageing and death of the whole plant. The New Phytologist. 2013;197:696–711. doi: 10.1111/nph.12047. [DOI] [PubMed] [Google Scholar]
- Turlapati PV, Kim KW, Davin LB. Lewis NG. The laccase multigene family in Arabidopsis thaliana: towards addressing the mystery of their gene function(s) Planta. 2011;233:439–470. doi: 10.1007/s00425-010-1298-3. [DOI] [PubMed] [Google Scholar]
- Wagstaff C, Yang TJ, Stead AD, Buchanan-Wollaston V. Roberts JA. A molecular and structural characterization of senescing Arabidopsis siliques and comparison of transcriptional profiles with senescing petals and leaves. The Plant Journal: For Cell and Molecular Biology. 2009;57:690–705. doi: 10.1111/j.1365-313X.2008.03722.x. [DOI] [PubMed] [Google Scholar]
- Weaver LM, Gan SS, Quirino B. Amasino RM. A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Molecular Biology. 1998;37:455–469. doi: 10.1023/a:1005934428906. [DOI] [PubMed] [Google Scholar]
- Wingler A. Roitsch T. Metabolic regulation of leaf senescence: interactions of sugar signalling with biotic and abiotic stress responses. Plant Biology. 2008;10(Suppl. 1):50–62. doi: 10.1111/j.1438-8677.2008.00086.x. [DOI] [PubMed] [Google Scholar]
- Woo HR, Kim JH, Nam HG. Lim PO. The delayed leaf senescence mutants of Arabidopsis, ore1, ore3, and ore9 are tolerant to oxidative stress. Plant and Cell Physiology. 2004;45:923–932. doi: 10.1093/pcp/pch110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Small RNA size distribution plots. Genome-matched small RNA data were normalized to transcripts per two million (TP2M) and the total abundance of each size class of small RNA was plotted. (A) Size distributions in young, mature, early senescing and late senescing leaf. (B) Size distributions in young, mature, early senescing and late senescing silique.
Figure S2. Comparison of miRNA abundance between mature and other stages.
miRNA abundance comparison between mature and other stages. Correlation coefficients for each comparison are also shown. (A) Mature leaves compared with young leaves, early senescing leaves and late senescing leaves. (B) Mature siliques compared with young siliques, early senescing siliques and late senescing siliques.
Table S1. Oligomers used in this study.
Table S2. miRNA quantification from leaf and silique small RNA libraries. Values represent the abundance of the miRNA sequence in the indicated library (abbreviations as in Table 1) normalized to TP2M (transcripts per two million) trimmed genome-matched reads.


