Abstract
Maturation of mRNA precursors often occurs simultaneously with their synthesis by RNA polymerase II (Pol II). The co-transcriptional nature of mRNA processing has permitted the evolution of coupling mechanisms that coordinate transcription with mRNA capping, splicing, editing and 3′ end formation. Recent experiments using sophisticated new methods for analysis of nascent RNA have provided important insights into the relative amount of co-transcriptional and post-transcriptional processing, the relationship between mRNA elongation and processing, and the role of the Pol II carboxy-terminal domain (CTD) in regulating these processes.
Messenger ribonucleoproteins (mRNPs) are made by a three-step process: first, the transcription of a DNA template by RNA polymerase II (Pol II) to make a pre-mRNA; second, the maturation of the pre-mRNA by processing factors; and third, packaging of the mature mRNA with proteins to make a particle that is competent for export to and translation in the cytoplasm. Maturation of most pre-mRNAs requires attachment of a 7-methylguanosine cap to the 5′ end, intron excision together with exon ligation, and formation of a 3′ end by cleavage and addition of a non-templated poly(A) tail (FIG. 1). Some mRNAs are also edited by selective deamination of adenosines and cytosines.
Figure 1. The major co-transcriptional mRNA processing steps.
Human protein names are given throughout. a | The RNA is shown in green; both GTP and the added guanosine cap (Gp) are shown in blue. The mRNA-capping enzyme in metazoans is bifunctional and has both triphosphatase and guanylyl-transferase activities that remove the γ-phosphate of the nascent transcript and transfer GMP from the GTP donor, respectively. The methyl donor S-adenosyl-l-methionine (SAM) is converted to S-adenosyl-l-homocysteine (SAH), which results in the 7-methylguanosine cap (shown in pink). b | Splicing clips out an intron or intervening sequences as a lariat and ligates the flanking exons together through two transesterification reactions. Conserved intronic splicing elements are indicated in red. Spliceosomal U1, U2, U4, U5 and U6 small nuclear ribonucleoprotein particles (snRNPs) and U2 auxiliary factor (U2AF) are shown, but numerous spliceosomal proteins are omitted79. c | 3′ ends of mRNAs are formed by coupled cleavage and polyadenylation. Cleavage of mammalian pre-mRNAs occurs ∼25 bases downstream of a consensus sequence (AAUAAA) and is carried out by the multisubunit complex (shown in purple), which comprises cleavage stimulation factor (CstF), cleavage and polyadenylation specificity factor (CPSF) that bears the endonuclease, and cleavage factors I and II (CFIm and CFIIm). Poly(A) polymerase (PAP) adds the poly(A) tail. 3′ ends of non-polyadenylated histone mRNAs (not shown) are also made co-transcriptionally by a cleavage complex that has many subunits in common with CstF and CPSF. The 5′-to-3′ RNA exonuclease 2 (XRN2) degrades RNA downstream of the cleavage site and facilitates transcription termination. d | Adenosine-to-inosine (A-to-I) editing is carried out by adenosine deaminases acting on RNA (ADAR), which deaminate adenosines into inosines. The folded GLUR-2 pre-mRNA substrate is shown with the exon in blue and intron in grey. Pi, inorganic phosphate. Part b is modified with permission from Chen & Cheng (2012) Biosci. Rep. 32, 345-359. © Biochemical Society.187
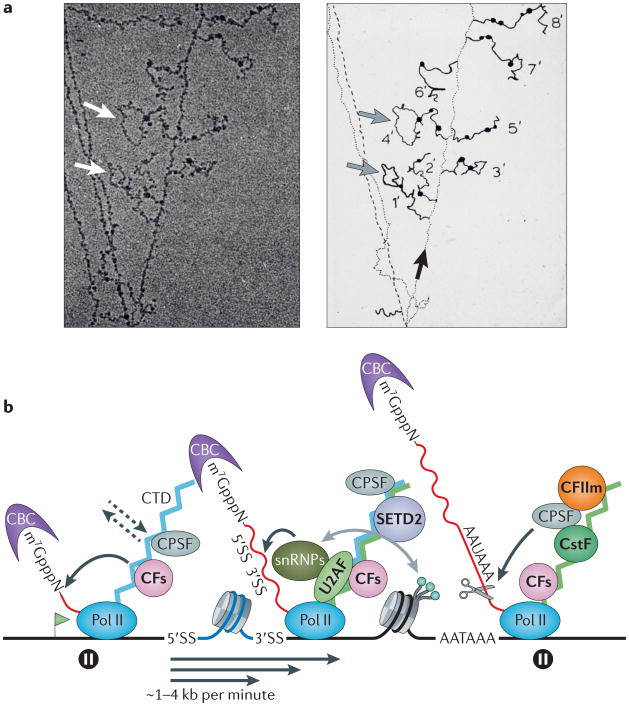
Textbooks often describe mRNA biogenesis as a pathway in which transcription is followed by capping, 3′ end formation and finally splicing. This scheme is consistent with the reconstitution of all of these reactions in vitro independently of one another. However, in living cells, transcription and processing are mostly not sequential but simultaneous; that is, processing is co-transcriptional rather than post-transcriptional. This is graphically shown by ‘Miller spread’ electron micrographs of introns being excised from nascent transcripts that are still attached to Pol II on the DNA template1 (FIG. 2a). Co-transcriptionality enhances the efficiency and the accuracy of pre-mRNA maturation2 and allows novel interactions with regulatory implications. These include communication between splicing and chromatin modifications3,4,5, as well as control of 3′ end processing by the spliceosomal U1 small nuclear ribonucleoprotein particle (snRNP)6. Powerful imaging and next-generation sequencing methods have recently yielded a wealth of new information about when and where transcripts are processed in the nucleus.
Figure 2. The co-transcriptional nature of pre-mRNA processing.
a | Transcription of a gene and co-transcriptional processing of nascent transcripts are shown.‘Miller spread’ electron micrograph (left) and its interpretation (right) are shown for a Drosophila melanogaster embryonic gene. The electron micrograph shows the DNA template with several engaged RNA polymerase II (Pol II) molecules and their associated nascent RNA transcripts with bound proteins (seen as dark blobs) that extend on either side of the DNA. Grey and white arrows mark introns that are spliced out co-transcriptionally. The black arrow indicates the direction of transcription along the DNA template. b | A co-transcriptional mRNA processing scheme is shown. A transcribed gene is shown with multiple polymerases, and the transcription start site is indicated with a green flag. Processing reactions (shown by curved black arrows) are carried out by Pol II-associated factors that act on the nascent transcript (shown in red) at 5′ and 3′ splice sites (5′SS and 3′SS) and the poly(A) site (AAUAAA). Capping factors (CFs, including the mRNA-capping enzyme and guanine-7-methyltransferase) and 3′ end processing/termination factors all bind directly to the carboxy-terminal domain (CTD) of Pol II (but not all factors that bind to the CTD are shown). Transcription elongation (shown by straight black arrows) occurs at variable rates with strong pauses near the transcription start site and downstream of the poly(A) site (shown by pause signs). Proteins that make direct contacts with the CTD are shown in bold, and the dynamic nature of binding is indicated by dashed arrows. CTD phosphorylation changes during the transcription cycle with high levels of phospho-Ser5 (represented by the CTD in blue) at the 5′end (which binds to CFs) and high levels of phospho-Ser2 (represented by the CTD in green) at the 3′end (which binds to the PCF11 subunit of cleavage factor II (CFIIm) (shown in orange)). Note that some 3′end processing factors — including cleavage and polyadenylation specificity factor (CPSF), CFs and the cap-binding complex (CBC) — are present on transcription elongation complexes throughout the length of genes. Nucleosomes that wrap the DNA template have different densities and methylation patterns in exons and introns3. Splicing and trimethylation of histone H3 lysine 36 (shown as green balls) by the methyltransferase SET domain-containing 2 (SETD2) may influence one another to establish and maintain splicing patterns (shown by the grey arrow). CstF, cleavage stimulation factor; snRNP, small nuclear ribonucleoparticle; U2AF, U2 auxiliary factor. Part a is reproduced, with permission, from REF. 1 © (1988) Cold Spring Harbor Laboratory Press.
Pre-mRNA processing is not only simultaneous with transcription but also mechanistically coupled to it, which means that synthesis and processing of the transcript are interdependent. Even post-transcriptional mRNA processing is not necessarily uncoupled from transcription, as commitment to a processing step such as splicing could occur co-transcriptionally even though actual intron excision is completed after release of pre-mRNA from a gene. Groundbreaking work showed that promoter elements can affect the decision to include an alternatively spliced exon7, and transcription initiation and elongation factors were subsequently found to influence capping8, splicing9–11 and 3′ end formation12,13. Conversely, processing factors have been implicated as effectors of transcription initiation or elongation14–18. In summary, the interdependence of transcription and processing has blurred the once clear distinction between transcription and processing factors.
The cellular transcription, processing and export machineries seem to have co-evolved to allow spatio-temporal coupling of the reactions that they carry out. Coupling in space is achieved by recruitment mechanisms that localize RNA packaging and processing factors to the right place to act on the nascent transcript. Coupling in time, or kinetic coupling, is achieved by coordinating the rates of elongation and processing of the transcript (BOX 1). The transcription elongation rate determines the delay between the appearances of upstream and downstream elements in the nascent pre-mRNA, which might compete with one another for RNA-binding and processing factors.
Box 1. Kinetic coupling and transcription elongation.
The notion that RNA polymerase II (Pol II) and the splicing apparatus have co-evolved to permit kinetic coupling is consistent with the fact that slow elongation seems to be a selected trait. Conserved amino acid residues with charged side chains that project into the side channel of Pol II are predicted to greatly impede diffusion of nucleoside triphosphates into the active site and thereby slow elongation163.
During the elongation phase of the transcription cycle (that is, initiation, elongation and termination), Pol II spends most of its time in a paused state. Pausing is detected experimentally in vivo as a local build-up of Pol II density by chromatin immunoprecipitation (ChIP) or by nuclear run-on assay. However, variations in apparent Pol II density should be interpreted cautiously, as its relationship to elongation rate in vivo is not fully understood164.
On many metazoan genes, transcription is punctuated by 5′ and 3′ pauses, which causes Pol II to pile up at the 5′ end near the transcription start site and at the 3′ end 1–2 kb downstream of the poly(A) site. The 5′ pause is imposed by negative elongation factor (NELF) and DRB sensitivity-inducing factor (DSIF), and it is relieved by positive transcription elongation factor b (P-TEFb), which phosphorylates NELF and the carboxy-terminal domain of Pol II165. In budding yeast, which lacks NELF, there is a much less prominent 5′ pause72. In addition to two major pauses, Pol II pauses for shorter periods at numerous sites within genes.
The average net rate of transcription (∼2 kb per minute on human genes) is therefore a function of the maximum elongation rate between pauses (which is estimated at 4.3 kb per minute59), and the frequency and duration of pauses. By using global run-on sequencing (Gro-seq)166 to study the progress of Pol II along genes following gene activation, one study uncovered a remarkable variation in elongation rate by more than fourfold between genes and cell types. Elongation rate even accelerated markedly within a gene58. Similarly, a threefold change in elongation rate was observed at a yeast gene in different phases of the cell cycle167. Therefore, kinetic coupling mechanisms probably need to be able to adjust to considerable variation in elongation rate.
In this Review, I discuss mRNP biogenesis from the perspective of the recruitment and kinetic competition models for coupling transcription with mRNA capping, splicing, editing and 3′ end formation. I highlight recent experiments that illuminate the following questions: how much mRNA processing is co-transcriptional and how much of it is post-transcriptional? What is the relationship between elongation of the nascent RNA transcript and mRNA processing? And what is the role of the Pol II carboxy-terminal domain (CTD) and its phosphorylation ‘code’ in controlling recruitment of processing factors to the transcription elongation complex (TEC)?
Mechanism of co-transcriptional coupling
The recruitment model
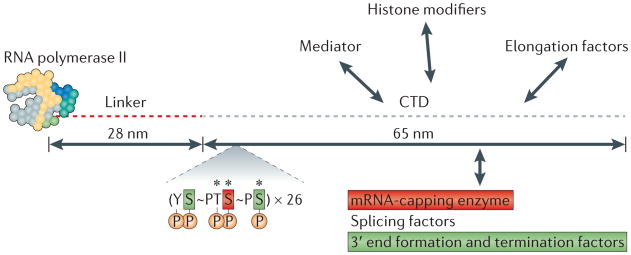
Transcription by an ‘in appropriate’ RNA polymerase, such as Pol I or Pol III, impairs pre-mRNA maturation19,20. The ‘fate’ of a transcript is therefore determined by the polymerase that made it, and RNA Pol II is unique in that it supports coupling and hence efficient pre-mRNA processing. For the purpose of spatial coupling of transcription and pre-mRNA processing, Pol II is uniquely equipped with an appendage — the conserved CTD of the large subunit, which comprises heptad repeats with the consensus sequence YSPTSPS21 (BOX 2). The CTD acts as a ‘landing pad’ (REF. 22) that recruits processing factors to the TEC and binds directly to capping factors, splicing factors and 3′ end processing factors (FIGS 2b, 3; reviewed in REFS 23–25). Recruitment of these factors to the CTD enhances mRNA maturation by the mass action effect of concentrating factors to their site of action, by allosteric activation26 and possibly also by facilitating co-transcriptional assembly of multisubunit processing factors27,28. In vitro, this domain stimulates all three major mRNA processing reactions26,29,30. The advantage of processing in close proximity to the CTD is shown by the fact that processing is inhibited if the CTD is deleted31 or if the transcript is cut loose from the polymerase prematurely by ribozyme cleavage32,33. The CTD interaction surface is modified in a programmed way during the cycle of transcription initiation, elongation and termination by reversible phosphorylation at multiple positions24,34 (BOX 2; FIG. 3). It has been suggested that these changes constitute a CTD code that directs the traffic of factors on and off the landing pad.
Box 2. The carboxy-terminal domain ‘landing pad’ and its code.
The carboxy-terminal domain (CTD) of the largest RNA polymerase II (Pol II) subunit comprises a series of heptad repeats with the consensus sequence YSPTSPS that is conserved from fungi to humans but is absent from Pol I and Pol III. The CTD is the target of numerous modifications that are remodelled in a synchronized manner with the transcription cycle24,168. The human CTD has 52 repeats, and each heptad can be modified by phosphorylation at five positions (Tyr1, Ser2, Thr4, Ser5 and Ser7)24,169, by peptidyl-prolyl bond isomerization at two positions and by O-GlcNacylation at three positions (Thr4, Ser5 and Ser7)170 (FIG. 2b). As a result, the potential number of CTD isoforms is far greater than the number of atoms in the universe. Each CTD phospho-isoform has a profile along the length of the average gene that reflects its particular dynamics. On a typical gene, the levels of phospho-Ser5 and phospho-Ser7 decrease in a 5′→3′ direction; conversely, the levels of phospho-Ser2, phospho-Thr4 and phospho-Tyr1 increase in a 5′→3′ direction110,111,143,169,171 (FIG. 2b). It should be borne in mind that studies that map phosphorylated CTDs rely on reactivity with antibodies that are specific to each phospho-isoform, which do not necessarily give a linear readout. This is because phospho-epitopes could be masked by interacting proteins, and when they are accessible, they may not be recognized equally well in all contexts within the tandem heptad array169.
Whereas 5′→3′ profiles of Ser2 and Ser5 phosphorylation in the CTD are fairly uniform in yeast, there is more variation among metazoan genes. For example, at the 5′ ends of some human genes there are non-canonical peaks of phospho-Ser2, the functional importance of which is unknown107 but could be related to recruitment of cleavage and polyadenylation factors that can occur at 5′ ends50. On polycistronic genes in Caenorhabditis elegans, peaks of phospho-Ser2 in the CTD are reiterated at each poly(A) site172. Whether poly(A) sites dictate where hyperphosphorylation of Ser2 occurs or whether phospho-Ser2 dictates processing at a poly(A) site remains to be resolved.
Phosphates in the CTD are attached and detached during transcription by multiple kinases and phosphatases. In metazoans, phospho-Ser2 is added by cyclin-dependent kinase 11 (CDK11) and CDK12 (REF. 173), in addition to CDK9, positive transcription elongation factor b (P-TEFb) and several super-elongator complexes with distinct activities174,175. Additions of phosphate groups to Ser5 and Ser7 are mediated by the CDK7 subunit of transcription factor IIH (TFIIH), but CDK9 can also modify these residues, at least in vitro176,177,178. Several CTD phosphatases probably work both co-transcriptionally and post-transcriptionally to remodel the ‘landing pad’; an example is SSU72, which is coupled to mRNA 3′ end formation179 through interaction with the cleavage and polyadenylation factor symplekin180. In summary, cycles of CTD phosphorylation and dephosphorylation integrate with the cycle of transcription initiation, elongation and termination, and prepare the landing pad to receive and release proteins.
In yeast, almost 100 proteins bind to the phosphorylated CTD either directly or indirectly181. The CTD interaction domain (CID) is conserved among several binding proteins from yeast to mammals, but many other protein domains are able to contact the CTD138,182. The CTD is remarkably flexible, and CIDs have adapted to bind to it in multiple conformations28,183,184. CTD phosphorylation can either enhance or inhibit binding interactions. Therefore, phospho-Ser2 enhances binding of the yeast 3′ end processing/termination factors Pcf11 and Rtt103 (REFS 145,185), and probably also the mammalian splicing factor U2 auxiliary factor 65 kDa subunit (U2AF65)121,147. Phospho-Ser5 promotes binding of the mRNA-capping enzyme26 and the yeast termination factor Nrd1 (REFS 144,184), whereas phospho-Ser7 binds to the integrator complex that processes the 3′ ends of mammalian small nuclear RNAs186. Contacts with the aromatic side chain of Tyr1 stabilize most complexes with the CTD, and phosphorylation of this residue probably triggers release of CTD-bound factors171,182. The tandem repeat of heptads in the CTD provides an excess of binding sites over what is absolutely required149 and also permits cooperative binding to adjacent sites, which may promote co-transcriptional assembly of the 3′ end processing machinery28.
Many proteins that land on the CTD do not remain there but ‘hop off’ onto other proteins or onto the transcript. As reactions that exchange these proteins are important for coordinated biogenesis of messenger ribonucleoproteins, interactions with the CTD have probably evolved to be easily reversible23, which is consistent with the observation that mutation of a conserved residue in the CID of Pcf11 actually increases its affinity for the CTD28.
Figure 3. The carboxy-terminal domain ‘landing pad’ of RNA polymerase II.
The core yeast RNA polymerase II structure (which consists of ten subunits and has a mass of ∼500 kDa) and its extended carboxy-terminal domain (CTD, which contains 26 heptads with the consensus sequence YSPTSPS) are shown to scale. The human CTD has 52 heptads with the consensus sequence YSPTSPS. The five phosphorylation (P) sites, two peptidyl-prolyl bonds (∼) that are converted between cis and trans conformations and three sites of O-GlcNAcylation (*) are indicated. Heptads with phospho-Ser5 make conserved direct interactions with the mRNA-capping enzyme, whereas heptads with phospho-Ser2 and phospho-Ser7 directly interact with the 3′ end processing/termination factor Pcf11, and the integrator complex, respectively. The figure is modified, with permission, from REF. 188 © (2001) American Association for the Advancement of Science.
The kinetic coupling model
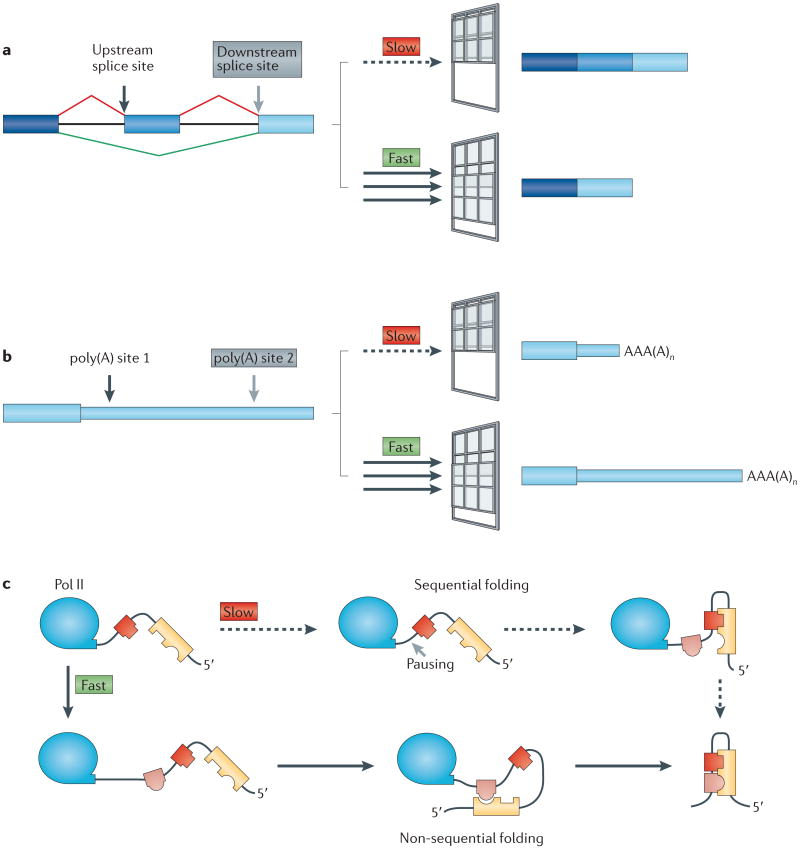
Kinetic coupling (also known as kinetic competition)35,36 is a generalization of the ‘first come, first served model’ of splicing, which proposed that 5′ introns will be removed before 3′ introns because they get a ‘head start’ on spliceosome assembly37. As transcription proceeds, RNA sequence targets on the nascent transcript become available to bind to proteins and complementary RNA elements in a progression, the timing of which is set by the rate of elongation. If these RNA targets compete with one another for binding partners, then the elongation rate could affect the outcomes of co-transcriptional RNA processing events, because slow elongation lengthens the ‘window of opportunity’ for an upstream event to occur on the nascent transcript before facing competition from a downstream RNA sequence element (FIG. 4). Kinetic competition could have a major influence on alternative splicing, which occurs at ∼100,000 sites on >95% of human pre-mRNAs and is a major source of proteome diversity36. If a weak upstream 3′ splice site that borders an alternative exon competes with a strong downstream 3′ splice site, then slow elongation and a long window of opportunity for recognition of the upstream site should favour inclusion of the alternative exon (FIG. 4a). Consistent with this prediction, slow Pol II elongation enhanced inclusion of some, but not all, alternative exons, including exon 33 of the human fibronectin 1 gene (FN1)38.
Figure 4. Kinetic coupling of transcription with folding and processing of the pre-mRNA.
Transcription elongation rate determines the length of the ‘window of opportunity’ for an upstream event to occur on the nascent RNA before it must compete with a downstream event. Slow elongation widens the window of opportunity for commitment to processing at upstream splice sites and poly(A) sites. This could lead to inclusion of alternative exons (part a) and 3′end formation at upstream poly(A) sites (part b), which results in mRNAs with shorter 3′untranslated regions. Slow elongation also favours RNA folding by base-pairing of proximal complementary sequences (represented by interlocking shapes) (part c), which results in sequential rather than non-sequential folding72,126. Part c is modified, with permission, from REF. 134 © (2006) Annual Reviews.
Co-transcriptional capping
The first step in the maturation of a pre-mRNA is attachment of a 7-methylguanosine cap by a 5′–5′ linkage (FIG. 1a). Nascent transcripts as short as 20–30 nucleotides become capped39 shortly after their 5′ ends emerge from the RNA exit channel, which is adjacent to the CTD attachment point on Pol II. This reaction was thought to proceed to completion on all Pol II transcripts, but there is now reason to doubt the infallibility of capping. Newly discovered nuclear and cytoplasmic quality control mechanisms in yeast and human cells specifically remove unmethylated caps and then degrade the transcripts in a 5′→3′ direction40–42. Moreover, a cytoplasmic capping activity has been reported, which is consistent with the possibility of post-transcriptional capping43,44. The extent of co-transcriptional capping has yet to be determined by a genome-wide analysis of nascent transcripts, but these recent findings suggest that it is not as universal as previously suspected and may even be regulated45. Capping is the only processing step that can be reversed in vivo, and recent evidence suggests that decapping occurs not only in the cytoplasm but also co-transcriptionally in the nucleus, where it provokes premature termination of transcription46,47. The CTD with phospho-Ser5 residues in the heptad repeats has a crucial role in co-transcriptional capping by directly interacting with both guanine-7-methyltransferase and the mRNA-capping enzyme, which it then activates26,48,49 (FIG. 3).
Capping and transcriptional pausing
Capping of short nascent transcripts coincides approximately with the promoter-proximal pause in transcription elongation at many metazoan genes39 (BOX 1). The human mRNA-capping enzyme, guanine-7-methyltransferase and the nuclear cap-binding complex (CBC) are all recruited to genes at this pause and can remain associated with Pol II all the way to the 3′ ends of genes50,51 (FIG. 2b). The mRNA-capping enzyme interacts with the elongation factor DSIF (also known as SPT4/5) (BOX 1), and DSIF-dependent pausing may be a checkpoint to ensure proper capping before transcription resumes in mammals52–54. Interestingly, the HIV-1 Tat protein, which activates transcription elongation by binding to positive transcription elongation factor b (P-TEFb), is one of the few transcription factors that have been reported to stimulate co-transcriptional capping directly8. The capped nascent transcript can also enhance elongation indirectly through interaction of CBC with P-TEFb15. An important future challenge is to clarify the cause-and-effect relationships that link capping with transcriptional pausing and elongation.
Co-transcriptional splicing
Assessing the extent of co-transcriptional splicing
Splicing can happen co-transcriptionally because it occurs over a period of several minutes, which coincides with the time needed to transcribe a gene of average length. On the basis of intron half-lives (0.4–7 minutes55–57) and transcription rates (1.8–4.0 kb per minute57–60) in vivo, it is predicted that splicing will occur by the time Pol II has elongated ∼5 kb past a 3′ splice site, and this is exactly what was seen in seminal electron microscopy studies in Drosophila melanogaster and Chironomus tentans1,61. Processing of non-coding microRNAs (miRNAs) from within introns in mammalian cells can also occur co-transcriptionally62. Moreover, classic pulse-labelling studies detected a proportion of large heterogeneous nuclear RNA (hnRNA) molecules, which were presumably unspliced, with 3′ ends that had already been polyadenylated63. Assuming that hnRNAs are precursors of mRNAs, splicing of these transcripts must occur post-transcriptionally. Only in the past two years has the extent of co-transcriptional splicing been compared with that of post-transcriptional splicing in comprehensive genome-wide studies.
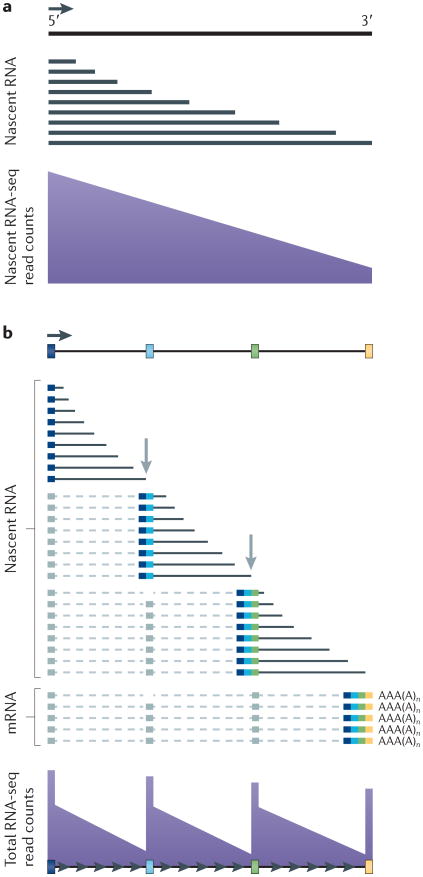
This breakthrough was achieved by deep sequencing of RNA populations (RNA-seq) of human, mouse and fly samples that were enriched for nascent transcripts (reviewed in REF. 64). When interpreting these experiments, it is important to recall that, unlike mature mRNA, a clean nascent RNA preparation is enriched in sequences from 5′ ends of genes because most transcripts are incomplete (FIG. 5a). Several studies sequenced RNA from urea-washed chromatin fractions65. However, chromatin-associated RNA is not precisely equivalent to nascent RNA, as these preparations can contain transcripts with mature polyadenylated 3′ ends66,67, and it is also difficult to eliminate the possibility of artefactual RNA interactions that are formed during cell extraction68.
Figure 5. Global co-transcriptional splicing detected by deep sequencing of nascent RNA.
a | Nascent transcripts are mostly incomplete, and 5′ sequences are therefore over-represented relative to 3′ sequences across the gene in nascent RNA sequencing (RNA-seq) profiles. b | Co-transcriptional splicing of nascent transcripts causes a saw-tooth profile in read counts from sequencing of total RNA owing to rapid removal and degradation of sequences from the 3′ ends of introns shortly after the 3′ splice site has been transcribed. Figure is modified, with permission, from REF. 74 © (2011) Macmillian Publishers Ltd. All rights reserved.
Notwithstanding this caveat, sequencing of chromatin-associated RNA from human, mouse and fly samples66,69–71 detected an enrichment of exon over neighbouring intron sequences as expected if splicing is substantially co-transcriptional. In budding yeast, extensive co-transcriptional splicing was also detected by native elongating transcript sequencing (NET-seq; that is, sequencing of the 3′ ends of RNAs that co-immunoprecipitated with Pol II)72 and by micro-array analysis of chromatin-associated RNA73. In D. melanogaster S2 cells and fly heads, introns were more frequently spliced co-transcriptionally than post-transcriptionally in >75% of cases70. Similarly, at a subset of constitutively expressed genes in mouse macrophages, most introns were spliced co-transcriptionally most of the time66; in a human erythropoietic cell line, the median level of co-transcriptional splicing was 75%71. However, the frequency of co-transcriptional splicing in mouse liver cells was reported to be only about half of that69.
An unambiguous indicator of co-transcriptional splicing is the specific under-representation of sequences at the 3′ ends of introns due to the excision and degradation of these sequences shortly after transcription of the 3′ splice site74. As a result, the total RNA-seq read counts have saw-tooth profiles (FIG. 5b). Using this criterion, the frequency of co-transcriptional splicing was shown to differ between tissues; in humans, it is higher in the liver than in the brain and higher in adult brain than in fetal brain74.
Independent support for widespread co-transcriptional splicing was provided by sequencing of pulse-labelled RNA from human B lymphocytes, which showed that 65% of intron sequences had already been spliced out and degraded after only five minutes75. Furthermore, by using an antibody against a specific phosphorylated isoform of the splicing protein SF3B to detect active spliceosomes, researchers found that >80% of active spliceosomes were bound to chromatin in HeLa cells27, which suggests that they are working co-transcriptionally. The 10–20% of active spliceosomes that act post-transcriptionally localize at nuclear speckles76. In summary, several independent approaches show that, in multiple species and cell types, co-transcriptional splicing operates on a large proportion of introns most of the time. However, a substantial amount of splicing is also completed post-transcriptionally, and such delayed splicing can control the timing of gene activation77,78.
Kinetic regulation of splicing
By its nature, the degree of co-transcriptional splicing for each intron depends on how quickly it is spliced relative to the time for Pol II to travel from the 3′ splice site to the end of the gene and for the transcript to be cut loose at the poly(A) site. Hence, relative rates of splicing, transcription elongation and poly(A) site cleavage can all affect the extent of co-transcriptional splicing. Consistent with this model, upstream introns that have longer windows of opportunity are, with some exceptions70, spliced more co-transcriptionally than downstream introns69–71 as predicted by the first come, first served model37.
One general conclusion from nascent RNA-seq is that alternatively spliced introns are removed more slowly than constitutive introns70,71,79,80. In two well-studied cases — human FN1 exon 33 and SRC exon N1 — splicing that skips the alternative exon is slower than splicing that includes the alternative exon, but all of these events are completed co-transcriptionally most of the time79. The reasons that alternative splicing tends to be slower are unclear, but the relative weakness of their splice sites is probably a contributing factor81.
Elongation rate can affect alternative splicing decisions38 by determining the duration of windows of opportunity for competing upstream and downstream events on the nascent transcript36 (FIG. 4). Elongation rate could influence where spliceosome components assemble and where sequence-specific splicing regulators bind, which may in turn determine whether they enhance or inhibit exon inclusion (reviewed in REF. 82).
The first come, first served model predicts that when an alternatively spliced exon is included, the 5′ flanking intron should be removed before the 3′ flanking intron (FIG. 4a) but, unexpectedly, examination of nascent RNA intermediates showed that when human FN1 exon 25 or exon 33 is included, it is actually the 3′ flanking intron that is removed first79,83. One can rationalize these findings if a stable commitment complex84 forms on the 5′ intron before the 3′ intron is recognized, even though excision of the 5′ intron is delayed83. Formation of a commitment complex on the 5′ intron would be favoured by slow transcription elongation that affords a longer window of opportunity for upstream events (FIG. 4a). Currently, little is known about the stability of committed splicing complexes in vivo, but U2 and U5 snRNPs dwell at the site of transcription for 15–30 seconds85 and intermediates can persist in vitro for >10 minutes86,87. An important future challenge is to gain a genome-wide perspective on how elongation rate affects constitutive and alternative splicing. In this regard, it is interesting to note that, in fly and yeast cells, slow elongation is associated with enhanced intron removal70,88.
Kinetic coupling implies that factors that coordinate elongation with mRNA processing exist. Such coupling factors are predicted to directly modulate both splicing and transcription elongation. Of note, nascent RNA-binding factors can also indirectly affect transcription elongation by preventing formation of R-loops (that is, extended RNA–DNA hybrids) that slow down Pol II89. One candidate kinetic coupling factor is the abundant serine/arginine-rich splicing factor 2 (SRSF2; also known as SC35) that acts as a positive effector of elongation by interacting with P-TEFb18,90. Another candidate coupling factor is the DBC1–ZIRD (DBIRD) complex, which associates with hnRNPs, enhances elongation and promotes selective exon skipping91. The RNA-binding protein KHDRBS1 (KH domain-containing, RNA-binding, signal transduction-associated protein 1; also known as SAM68) is another possible coupling regulator that influences alternative splicing and impairs transcription elongation in collaboration with BRM (also known as SMARCA2), which is the ATPase subunit of the SWI/SNF chromatin remodeller. However, this effect of BRM is independent of chromatin remodelling92. An intriguing question is how kinetic coupling factors could modulate elongation rate to affect splicing of specific exons. One possibility is that it is achieved by local re-decoration of phosphate groups in the CTD of Pol II92,93, and another possibility is by local ‘opening’ of chromatin by hyperacetylation94 to accelerate elongation or ‘closing’ of chromatin by heterochromatinization95,96 to decelerate elongation.
Interdependent pausing and splicing
The kinetic coupling model has provoked great interest in the relationship between splicing and transcriptional pausing. In yeast, in which most genes are short, Pol II pausing at 3′ ends can provide time for co-transcriptional splicing73. Alternatively, there is evidence in yeast that the splicing machinery can manipulate transcription to induce pausing downstream of introns and to allow time for splicing to occur co-transcriptionally93. Whether splicing can induce pausing in metazoan cells has yet to be demonstrated97. Pausing imposed by a protein ‘speed bump’ that binds to a specific DNA sequence can also influence alternative splicing. Thus, binding of CCCTC-binding factor CTCF within genes correlates with the inclusion of alternatively spliced exons98. CTCF binding is prevented by CpG methylation, which indicates a potential mechanism for regulation of alternative splicing by DNA methylation.
Widespread pausing at the front edge of positioned nucleosomes was revealed by NET-seq in yeast72. Pausing also often occurs at the beginning of mammalian exons99 either because of a higher density of nucleosomal speed bumps100 or because splicing induces pausing93. A strongly positioned nucleosome can slow down an elongating Pol II molecule by approximately six seconds relative to naked DNA; this delay is slightly reduced by histone acetylation101, which is implicated in splicing regulation94,102. It remains to be established whether such pauses, which are usually measured in seconds, have a meaningful effect on splicing that usually occurs on a timescale of minutes.
Co-transcriptional versus post-transcriptional splicing: does it matter?
Are there functional consequences associated with whether introns are spliced co-transcriptionally or post-transcriptionally? Whereas co-transcriptional splicing takes place at the gene, post-transcriptional splicing can occur either close to the gene in a chromatin environment or at distant sites throughout the nucleoplasm80, including at the periphery of nuclear speckles76. The location of splicing in the nucleus could be important if different RNA-binding proteins, which can determine RNA stability and cellular localization, were associated with transcripts that are spliced at different locations. Whether post-transcriptional splicing occurs close to the gene or elsewhere depends on how rapidly the RNA is released from the chromatin after cleavage at the poly(A) site. Estimated release times range from a few seconds to many minutes. Retention at the gene is characteristic of transcripts with processing defects that are destined for degradation by the exosome103–105, but such retention might also facilitate completion of post-transcriptional processing66,67. It remains to be clarified whether retention at the gene following cleavage at the poly(A) site is common for mRNAs that are processed in a normal way. It will also be important to establish whether the local environment of splicing — be it at the gene, a speckle or elsewhere in the nucleus — can influence the ultimate fate of the transcript.
Some introns have evolved to be spliced slowly, such as many alternatively spliced introns70,71, introns that harbour small nucleolar RNAs (snoRNAs)75, and those that are substrates for RNA editing106. The hairy and enhancer of split 7 gene (Hes7), which controls somite segmentation in the mouse embryo, has a 1.8-kb intron that causes a 19-minute delay in expression of the protein when it is spliced. Hes7 regulates itself by a negative-feedback loop, and the splicing-dependent delay causes its expression to oscillate in a way that is abolished when the intron is deleted78.
Most evidence suggests that slow splicing, which tends to be post-transcriptional, is specific to individual introns, but an apparent exception occurs when FUS expression is disrupted. FUS is a protein that binds to both RNA and the Pol II CTD107 and that is mutated in some patients with familial amyotrophic lateral sclerosis. The knockdown of fus in Xenopus laevis embryos specifically inhibited splicing of 3–5% of mRNAs, including those encoding regulators of gastrulation. Remarkably, all introns in the affected transcripts were retained to the same extent when expression of the fus protein was compromised, which suggests that, in these cases, the entire gene functions as an integrated unit with respect to splicing activity108. How FUS stimulates splicing throughout an entire pre-mRNA and whether it works co-transcriptionally or post-transcriptionally is still a mystery, but it could be related to its influence on phosphorylation of the Pol II CTD107.
Adenosine-to-inosine editing
RNA editing by deamination of adenosine residues to inosine residues is widespread in multicellular organisms and is necessary for maturation of a few mRNAs (including those encoding ion channels) and many non-coding RNAs. Adenosine-to-inosine (A-to-I) editing is carried out by adenosine deaminases acting on RNA (ADAR), the substrates of which are double-stranded guide RNA structures that are often formed by introns folding back on themselves and adjacent exon sequences (FIG. 1d). Such editing must therefore precede splicing, and introns that participate in A-to-I editing are characterized by slow splicing106. Nascent RNA-seq in D. melanogaster confirmed that editing is mainly co-transcriptional and that slow splicing of edited introns is an intrinsic property that does not require ADAR106. The Pol II CTD is implicated in delaying splicing of some edited introns by an unknown mechanism that ensures that editing can precede splicing109. In summary, slow splicing can enhance proper gene expression in several ways and seems to be a selected characteristic of specific introns.
Co-transcriptional 3′ end formation
Indirect evidence suggests that co-transcriptional poly(A) site cleavage is the major pathway of mRNA 3′ end maturation. Hence, yeast cleavage and polyadenylation factors localize at the 3′ ends of transcribed genes throughout the genome110,111. Moreover, termination of transcription is a poly(A) site-dependent event112–114, which means that poly(A) site recognition precedes or accompanies termination and must therefore be co-transcriptional. At most (but not all) genes115, cleavage at the poly(A) site probably provides the entry point for the RNA 5′-to-3′ exonuclease XRN2 (or its yeast homologue Rat1) (FIG. 1c), which acts as a ‘torpedo’ to promote polymerase eviction from the DNA template116. In this scenario, both recognition and RNA cleavage at the poly(A) site must precede termination and therefore occur co-transcriptionally.
In mammalian cells at least, mRNA 3′ end processing can be fairly slow, with reaction times of 1–5 minutes60. Consequently, the rate of poly(A) site processing could be an important determinant of whether there is time for splicing to be completed co-transcriptionally. Splicing of the last intron is facilitated by recognition of the poly(A) site117; conversely, recognition of the last intron facilitates cleavage and polyadenylation118. This is one reason that introns stimulate gene expression119. Coupling of splicing with 3′ end processing occurs through direct interaction of the U1 and U2 snRNPs and U2 auxiliary factor 65 kDa subunit (U2AF65) with cleavage factor I (CFIm) and with cleavage and polyadenylation specificity factor (CPSF)120, all of which localize to transcribed genes50,121,122.
Co-transcriptional coupling of terminal intron splicing and 3′ end processing means that these reactions can occur in quick succession, but the order of events can differ between genes. At the C. tentans BR1 gene, most chromatin-associated polyadenylated transcripts are already spliced123. Similarly, splicing precedes 3′ end processing at various human cellular genes55. By contrast, adenovirus transcripts are processed in the reverse order119. In summary, although 3′ end processing and terminal intron splicing are coupled reactions, they do not occur in an obligatory order but in a gene-specific way that is probably determined by the strengths of the processing sites.
Kinetic competition and alternative polyadenylation
The decision between processing alternative poly(A) sites controls the sequences that are present in the 3′ untranslated regions (3′UTRs) of >60% of human mRNAs. As 3′UTR sequences are recognized by miRNAs and regulatory RNA-binding proteins, alternative polyadenylation can modulate both mRNA abundance and translational efficiency124. In some cancer cells, the balance between alternative poly(A) sites becomes biased in favour of upstream or proximal sites, which results in production of mRNAs with truncated 3′UTRs that can evade normal regulatory mechanisms125,126. As most mRNA 3′ end processing occurs co-transcriptionally, it is likely that the same is true for alternative polyadenylation decisions. Indeed, an important negative regulator of 3′ end processing is the U1 snRNP6, which can access poly(A) sites in the nascent transcript through its association with Pol II127.
The kinetic competition model predicts that if alternative poly(A) sites in the nascent transcript compete for processing factors, then slow elongation will lengthen the window of opportunity for processing at proximal sites128,129 (FIG. 4b). At several genes in mutant D. melanogaster with slow Pol II, proximal poly(A) sites were indeed favoured relative to wild type130. Similarly, a slow Pol II mutant in yeast favoured proximal termination sites for non-coding RNAs131. Kinetic competition is therefore likely to modulate the choices of co-transcriptional poly(A) sites and termination sites. It will be interesting to determine whether slow elongation contributes to the shift to the proximal poly(A) site in cancer cells.
RNA folding and processing
The extent of co-transcriptional RNA processing depends on the position of the processing site relative to the end of the gene, the elongation rate and the sequence determinants that control reaction rate, including accessibility to processing factors. The way the nascent RNA folds can affect how it is processed, as such folding determines which elements are sequestered within secondary structures and which are available for interaction with proteins and other RNA sequences80,132. Sequestration of the intronic polypyrimidine tract (FIG. 1b) by folding into an RNA secondary structure impedes splicing and makes it post-transcriptional80. Folding of a nascent RNA depends on its rate of transcription (FIG. 4c). Slower transcription favours base-pairing between elements in the order in which they are transcribed (that is, sequential folding). Conversely, faster transcription permits more base-pairing between distant complementary sequences (that is, non-sequential folding), which favours the formation of branched structures over rodlike structures133,134. RNA folding at the 3′ ends of yeast introns affects 3′ splice site selection by sequestering cryptic 3′ splice sites135. The way the RNA folds can also feedback on the rate of elongation, as regions of high secondary structure can favour faster transcription in vitro by impeding backtracking136. The physiological importance of the interplay between elongation rate, nascent RNA folding and mRNA processing remains an intriguing topic for future investigations.
Recruiting RNA processing factors
Recruitment by the Pol II CTD
The CTD facilitates the two processing steps that are common to all Pol II transcripts — capping and 3′ end formation — by making protein–protein contacts that recruit, and in some cases allosterically activate, processing factors that work co-transcriptionally (BOX 2; FIG. 2b; reviewed in REFS 137,138). By contrast, splicing is not a universal processing step, and splicing factors could, in principle, be recruited either constitutively or on an ‘as needed’ basis to intron-containing genes.
In budding yeast and mammalian cells, the U2, U5 and U6 snRNPs in the activated spliceosome are specifically located at intron-containing genes, which is consistent with recruitment by recognition of splicing signals in the nascent transcript51,97,139,140. However, the U1 snRNP is recruited to some mammalian genes regardless of intron status97,140, which is consistent with the fact that it co-purifies with Pol II124,140. The core splicing factor with the best characterized direct CTD interaction is U2AF65 (REF. 121) (FIG. 2b), which is not found at intron-less genes97,140. This indicates that it may be recruited through the CTD in an intron-specific way. In vitro, however, U2AF contacts short nascent transcripts that lack splice sites soon after they emerge from the Pol II exit channel141. In summary, there is rather heavy traffic of processing factors going to and from the TEC. A remaining problem is how this traffic is controlled to ensure that the right factors will be present when they are required.
Satisfyingly, in some cases a CTD-binding factor colocalizes on genes in vivo with the CTD phosphoisoform that it recognizes in vitro (FIG. 2b). For example, the mRNA-capping enzyme and Nrd1 colocalize with peaks of CTD phosphorylated at Ser5 at 5′ ends of yeast genes142–144, and Pcf11 colocalizes with CTD phosphorylated at Ser2 at 3′ ends145. These correlations are consistent with a code in which modifications of the CTD direct the binding and release of protein passengers on the TEC146 but, of course, correlation does not necessarily imply causation. It is also possible that, conversely, CTD-binding proteins modulate phosphorylation of the CTD as suggested for FUS, which regulates Ser2 phosphorylation107. Phosphorylation of Ser2 and Ser5 has also been implicated in the control of transcription elongation, which suggests that the recruitment and kinetic coupling mechanisms do not operate entirely independently147,148.
The CTD code has been elegantly tested in Schizosaccharomyces pombe using mutants that substitute each of the phosphorylation sites (Tyr1, Ser2, Thr4, Ser5 and Ser7) in every heptad repeat149,150. Remarkably, the only phosphorylated residue required for viability is Ser5, which recruits the mRNA-capping enzyme. As few as two repeats that comprise ten residues with the sequence YSPT[pS]PSYSP (where pS denotes phosphoserine) are sufficient for viability. In a compelling experiment, the lethal Ser5Ala mutation was rescued by fusing the mRNA-capping enzyme to the carboxyl terminus of the Pol II large subunit, proving that the essential function of Ser5 phosphorylation in S. pombe is to recruit the mRNA-capping enzyme149.
Although the phospho-Ser5 CTD code word for the mRNA-capping enzyme seems to be universal, other coded signals affect mRNA processing in gene-specific ways. The major Ser2 kinase in budding yeast Ctk1 is dispensable for viability151, but its inactivation causes gene-specific defects in transcription elongation and/or termination and in 3′ end processing110,152. Thr4 and Ser7 phosphorylation of the CTD also have gene-specific effects on maturation of the non-polyadenylated 3′ ends of histone mRNAs and small nuclear RNAs, respectively24.
CTD-independent recruitment
In addition to binding specified by the CTD code, proteins are also recruited to the TEC by other means. The yeast 3′ end formation and termination factor Nrd1 binds to phospho-Ser5 heptads through the CTD interaction domain (CID) that is located in its amino terminus. However, this domain is not required for viability144, presumably because the protein can also bind to the nascent transcript through its RNA-binding domain. The CTD is not the only protein surface on the TEC that can recruit factors. The yeast mRNA localization factor She2 is passed to the nascent transcript after initial recruitment to the elongation factor Spt4/5, which travels with Pol II153. The mRNA-capping enzyme can also interact with a surface on Pol II that is distinct from the CTD154.
Another potential platform for recruitment of processing factors to the site of transcription is the chromatin itself. Histone H3 trimethylated at lysine 36 (H3K36me3) can recruit RNA-binding proteins that are presumed to act on the nascent transcript when a polymerase passes by155,156. In this way, histone modifications could modulate alternative splicing (FIG. 2b). Similarly, H3K4me3 may indirectly recruit the U2 snRNP and thereby promote splicing157. How histone modifications are established at appropriate places to influence splicing is poorly understood, but this might involve binding of histone modifiers, such as deacetylases158 and the H3K36 methyltransferase SET domain-containing 2 (SETD2)159, to the phosphorylated CTD. Conversely, splicing promotes local H3K36 methylation, and histone modifications could therefore help to both establish and maintain stable patterns of alternative splicing160,161.
Conclusions
Recent work has established a framework for understanding the spatiotemporal integration of mRNA processing with transcription by mechanisms of recruitment and kinetic competition. However, many questions about how these coupling mechanisms work to achieve accurate and regulated mRNP biogenesis remain to be resolved.
We know little about how these coupling mechanisms intersect through factors (such as CTD phosphorylation) that affect both recruitment of processing factors and transcription elongation. The important CTD phosphorylation code words that regulate processing factor recruitment in metazoans have yet to be defined precisely, and how they are interpreted in gene-specific ways is still quite mysterious. Kinetic competition suggests ways in which elongation rate could affect alternative processing outcomes, such as whether to add a cap or not, to include or exclude an exon or a retained intron, to splice co-transcriptionally or post-transcriptionally, and to use an upstream or downstream poly(A) site. The model remains an intriguing but incompletely tested one. We do not yet know how widespread the effect of transcription elongation rate is on alternative mRNA processing decisions nor do we have a grasp on whether elongation rates within a gene are modulated to affect alternative processing. In vivo elongation rates seem to be far more variable than previously suspected, which raises the question of whether kinetic coupling can adjust rates of processing to compensate for changes in elongation rate. It remains possible that aspects of transcription other than elongation rate influence co-transcriptional mRNA processing, such as whether initiation of RNA synthesis occurs randomly or in bursts162. We also know little about whether cellular signalling pathways target coupling mechanisms to regulate whether splicing occurs co-transcriptionally or post-transcriptionally. Furthermore, the functional consequences of whether a transcript is processed co-transcriptionally or post-transcriptionally remain unexplored.
Acknowledgments
The author apologizes to colleagues whose work was not referenced because of space limitations. Work in D.L.B's laboratory is supported by the US National Institutes of Health grants GM58163 and GM063873. The author thanks Y. Shav-Tal, D. Licatalosi, T. Blumenthal, R. Davis, D. Ish-Horowitz and members of his laboratory for discussions, and K. A. Nasmyth, J. Cooper and Cancer Research UK for their hospitality during preparation of the manuscript.
Glossary
- Carboxy-terminal domain (CTD)
Located in the large subunit of RNA polymerase II, the CTD is a signature feature of this polymerase that contains conserved heptad repeats (52 YSPTSPS repeats in humans) and is not found in RNA polymerases I and III
- Transcription elongation complex (TEC)
A complex of RNA polymerase that is stably bound to the DNA template, polymerase-bound proteins and the nascent RNA chain
- Alternative splicing
The most important mechanism by which the transcriptome is diversified through production of multiple mRNAs from a single gene. Alternatively spliced mRNAs differ in their coding and non-coding sequences as a result of selective inclusion and exclusion of exon and intron sequences
- Transcription start site
The site on the DNA where the first phosphodiester bond in a RNA transcript is formed
- Commitment complex
A stable complex formed between an intron-containing pre-mRNA, the U1 small nuclear ribonucleoprotein particle bound to the 5′ splice site, branch point-binding protein bound to the branch point and U2 auxiliary factor bound to the 3′ splice site. Once formed, it remains committed to the completion of splicing even when challenged with an excess of pre-mRNA substrate
- R-loops
Structures that are formed by the hybridization of RNA transcripts to double-stranded DNA in which the displaced non-template DNA strands are looped out. Their recombinogenic nature causes genomic instability
- Alternative polyadenylation
The decision to process at one of multiple poly(A) sites that are present at the 3′ ends of most human genes. This decision can have important functional consequences because it determines the sequence content of the 3′ untranslated region, which controls mRNA stability and translational efficiency
Footnotes
Competing interests statement: The author declares no competing interests.
References
- 1.Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. This is an early graphic demonstration of co-transcriptional splicing. [DOI] [PubMed] [Google Scholar]
- 2.Bird G, Zorio DA, Bentley DL. RNA polymerase II carboxy-terminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3′-end formation. Mol Cell Biol. 2004;24:8963–8969. doi: 10.1128/MCB.24.20.8963-8969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolasinska-Zwierz P, et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nature Genet. 2009;41:376–381. doi: 10.1038/ng.322. This paper is the first to show that histone modifications correlate with splicing activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieberstein NI, Carrillo Oesterreich F, Straube K, Neugebauer KM. First exon length controls active chromatin signatures and transcription. Cell Rep. 2012;2:62–68. doi: 10.1016/j.celrep.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 5.Gomez Acuna LI, Fiszbein A, Allo M, Schor IE, Kornblihtt AR. Connections between chromatin signatures and splicing. Wiley Interdiscip Rev RNA. 2013;4:77–91. doi: 10.1002/wrna.1142. [DOI] [PubMed] [Google Scholar]
- 6.Berg MG, et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150:53–64. doi: 10.1016/j.cell.2012.05.029. This study shows a powerful effect of the U1 snRNA, which is a subunit of the spliceosome, in repressing premature polyadenylation and potentially regulating alternative poly(A) site choice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cramer P, Pesce C, Baralle F, Kornblihtt A. Functional association between promoter structure and transcripts alternative splicing. Proc Natl Acad Sci USA. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. This paper provides early key evidence that transcription and splicing are mechanistically coupled. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu YL, et al. Tat stimulates cotranscriptional capping of HIV mRNA. Mol Cell. 2002;10:585–597. doi: 10.1016/s1097-2765(02)00630-5. [DOI] [PubMed] [Google Scholar]
- 9.Monsalve M, et al. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol Cell. 2000;6:307–316. doi: 10.1016/s1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 10.Huang Y, et al. Mediator complex regulates alternative mRNA processing via the MED23 subunit. Mol Cell. 2012;45:459–469. doi: 10.1016/j.molcel.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benson MJ, et al. Heterogeneous nuclear ribonucleoprotein L-like (hnRNPLL) and elongation factor, RNA polymerase II, 2 (ELL2) are regulators of mRNA processing in plasma cells. Proc Natl Acad Sci USA. 2012;109:16252–16257. doi: 10.1073/pnas.1214414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosonina E, Bakowski MA, McCracken S, Blencowe BJ. Transcriptional activators control splicing and 3′-end cleavage levels. J Biol Chem. 2003;278:43034–43040. doi: 10.1074/jbc.M307289200. [DOI] [PubMed] [Google Scholar]
- 13.Nagaike T, et al. Transcriptional activators enhance polyadenylation of mRNA precursors. Mol Cell. 2011;41:409–418. doi: 10.1016/j.molcel.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schroeder SC, Zorio DA, Schwer B, Shuman S, Bentley D. A function of yeast mRNA cap methyltransferase, Abd1, in transcription by RNA polymerase II. Mol Cell. 2004;13:377–387. doi: 10.1016/s1097-2765(04)00007-3. [DOI] [PubMed] [Google Scholar]
- 15.Lenasi T, Peterlin BM, Barboric M. Cap-binding protein complex links pre-mRNA capping to transcription elongation and alternative splicing through positive transcription elongation factor b (P-TEFb) J Biol Chem. 2011;286:22758–22768. doi: 10.1074/jbc.M111.235077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martins SB, et al. Spliceosome assembly is coupled to RNA polymerase II dynamics at the 3′ end of human genes. Nature Struct Mol Biol. 2011;18:1115–1123. doi: 10.1038/nsmb.2124. [DOI] [PubMed] [Google Scholar]
- 17.Andersen PK, Lykke-Andersen S, Jensen TH. Promoter-proximal polyadenylation sites reduce transcription activity. Genes Dev. 2012;26:2169–2179. doi: 10.1101/gad.189126.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji X, et al. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013;153:855–868. doi: 10.1016/j.cell.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sisodia SS, Sollner WB, Cleveland DW. Specificity of RNA maturation pathways: RNAs transcribed by RNA polymerase III are not substrates for splicing or polyadenylation. Mol Cell Biol. 1987;7:3602–3612. doi: 10.1128/mcb.7.10.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smale ST, Tjian R. Transcription of herpes simplex virus tk sequences under the control of wild-type and mutant human RNA polymerase I promoters. Mol Cell Biol. 1985;5:352–362. doi: 10.1128/mcb.5.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corden JL, Cadena DL, Ahearn JM, Jr, Dahmus ME. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc Natl Acad Sci USA. 1985;82:7934–7938. doi: 10.1073/pnas.82.23.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenleaf AL. Positive patches and negative noodles: linking RNA processing to transcription? Trends Biochem Sci. 1993;18:117–119. doi: 10.1016/0968-0004(93)90016-g. This paper presents a prescient speculation that the CTD functions in pre-mRNA processing. [DOI] [PubMed] [Google Scholar]
- 23.Perales R, Bentley D. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol Cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Egloff S, Dienstbier M, Murphy S. Updating the RNA polymerase CTD code: adding gene-specific layers. Trends Genet. 2012;28:333–341. doi: 10.1016/j.tig.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho C, Shuman S. Distinct effector roles for Ser2 and Ser5 phosphorylation of the RNA polymerase II CTD in the recruitment and allosteric activation of mammalian capping enzyme. Mol Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- 27.Johnson SA, Kim H, Erickson B, Bentley DL. The export factor Yra1 modulates mRNA 3′ end processing. Nature Struct Mol Biol. 2011;18:1164–1171. doi: 10.1038/nsmb.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lunde BM, et al. Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain. Nature Struct Mol Biol. 2010;17:1195–1201. doi: 10.1038/nsmb.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirose Y, Manley JL. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 30.Hirose Y, Tacke R, Manley JL. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 1999;13:1234–1239. doi: 10.1101/gad.13.10.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCracken S, et al. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. References 29–31 show the importance of the Pol II CTD for mRNA processing both in vitro and in vivo. [DOI] [PubMed] [Google Scholar]
- 32.Rigo F, Kazerouninia A, Nag A, Martinson HG. The RNA tether from the poly(A) signal to the polymerase mediates coupling of transcription to cleavage and polyadenylation. Mol Cell. 2005;20:733–745. doi: 10.1016/j.molcel.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 33.Bird G, Fong N, Gatlin JC, Farabaugh S, Bentley DL. Ribozyme cleavage reveals connections between mRNA release from the site of transcription and pre-mRNA processing. Mol Cell. 2005;20:747–758. doi: 10.1016/j.molcel.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 34.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dujardin G, et al. Transcriptional elongation and alternative splicing. Biochim Biophys Acta. 2013;1829:134–140. doi: 10.1016/j.bbagrm.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aebi M, Hornig H, Padgett RA, Reiser J, Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986;47:555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- 38.de la Mata M, et al. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. This study proposes the influential window of opportunity model to explain how transcription elongation rate could affect the outcome of alternative splicing decisions. [DOI] [PubMed] [Google Scholar]
- 39.Rasmussen EB, Lis JT. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci USA. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiao X, et al. Identification of a quality-control mechanism for mRNA 5′-end capping. Nature. 2010;467:608–611. doi: 10.1038/nature09338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang JH, et al. Dxo1 is a new type of eukaryotic enzyme with both decapping and 5′-3′ exoribonuclease activity. Nature Struct Mol Biol. 2012;19:1011–1017. doi: 10.1038/nsmb.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiao X, Chang JH, Kilic T, Tong L, Kiledjian M. A mammalian pre-mRNA 5′ end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing. Mol Cell. 2013;50:104–115. doi: 10.1016/j.molcel.2013.02.017. References 40–42 show a conserved function that monitors the quality of mRNA 5′ cap structures and that destroys transcripts that do not meet quality control standards. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otsuka Y, Kedersha NL, Schoenberg DR. Identification of a cytoplasmic complex that adds a cap onto 5′-monophosphate RNA. Mol Cell Biol. 2009;29:2155–2167. doi: 10.1128/MCB.01325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fejes-Toth K, et al. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernandez-Sanchez ME, Gonatopoulos-Pournatzis T, Preston G, Lawlor MA, Cowling VH. S-adenosyl homocysteine hydrolase is required for Myc-induced mRNA cap methylation, protein synthesis, and cell proliferation. Mol Cell Biol. 2009;29:6182–6191. doi: 10.1128/MCB.00973-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brannan K, et al. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol Cell. 2012;46:311–324. doi: 10.1016/j.molcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davidson L, Kerr A, West S. Co-transcriptional degradation of aberrant pre-mRNA by Xrn2. EMBO J. 2012;31:2566–2578. doi: 10.1038/emboj.2012.101. References 46 and 47 suggest that capping, which is a co-transcriptional mRNA processing step, can be reversed by decapping, which leads to premature transcriptional termination. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCracken S, et al. 5′-capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho EJ, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. References 26, 48 and 49 show recruitment of mRNA-capping enzyme to the Pol II CTD and consequent allosteric activation of the guanylyltransferase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nature Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nature Struct Mol Biol. 2006;13:815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- 52.Wen Y, Shatkin AJ. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev. 1999;13:1774–1779. doi: 10.1101/gad.13.14.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mandal SS, et al. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc Natl Acad Sci USA. 2004;101:7572–7577. doi: 10.1073/pnas.0401493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pei Y, Shuman S. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J Biol Chem. 2002;277:19639–19648. doi: 10.1074/jbc.M200015200. [DOI] [PubMed] [Google Scholar]
- 55.Schmidt U, et al. Real-time imaging of cotranscriptional splicing reveals a kinetic model that reduces noise: implications for alternative splicing regulation. J Cell Biol. 2011;193:819–829. doi: 10.1083/jcb.201009012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Audibert A, Weil D, Dautry F. In vivo kinetics of mRNA splicing and transport in mammalian cells. Mol Cell Biol. 2002;22:6706–6718. doi: 10.1128/MCB.22.19.6706-6718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nature Struct Mol Biol. 2009;16:1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Danko CG, et al. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol Cell. 2013;50:212–222. doi: 10.1016/j.molcel.2013.02.015. This study uses powerful global run-on sequencing analysis to measure transcription elongation rates on many gene in vivo and shows remarkable variation in Pol II speed between genes and between 5′ and 3′ regions within genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Darzacq X, et al. In vivo dynamics of RNA polymerase II transcription. Nature Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boireau S, et al. The transcriptional cycle of HIV-1 in real-time and live cells. J Cell Biol. 2007;179:291–304. doi: 10.1083/jcb.200706018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bauren G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. doi: 10.1016/0092-8674(94)90182-1. [DOI] [PubMed] [Google Scholar]
- 62.Morlando M, et al. Primary microRNA transcripts are processed co-transcriptionally. Nature Struct Mol Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Darnell JE., Jr Reflections on the history of pre-mRNA processing and highlights of current knowledge: a unified picture. RNA. 2013;19:443–460. doi: 10.1261/rna.038596.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brugiolo M, Herzel L, Neugebauer KM. Counting on co-transcriptional splicing. F1000Prime Rep. 2013;5:9. doi: 10.12703/P5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol Cell Biol. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bhatt DM, et al. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pandya-Jones A, et al. Splicing kinetics and transcript release from the chromatin compartment limit the rate of Lipid A-induced gene expression. RNA. 2013;19:811–827. doi: 10.1261/rna.039081.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khodor YL, Menet JS, Tolan M, Rosbash M. Cotranscriptional splicing efficiency differs dramatically between Drosophila and mouse. RNA. 2012;18:2174–2186. doi: 10.1261/rna.034090.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khodor YL, et al. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev. 2011;25:2502–2512. doi: 10.1101/gad.178962.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tilgner H, et al. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 2012;22:1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell. 2010;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 74.Ameur A, et al. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nature Struct Mol Biol. 2011;18:1435–1440. doi: 10.1038/nsmb.2143. [DOI] [PubMed] [Google Scholar]
- 75.Windhager L, et al. Ultrashort and progressive 4sU-tagging reveals key characteristics of RNA processing at nucleotide resolution. Genome Res. 2012;22:2031–2042. doi: 10.1101/gr.131847.111. References 69–75 show that co-transcriptional splicing is widespread in vivo using genome-wide sequencing of nascent RNA populations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Girard C, et al. Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nature Commun. 2012;3:994. doi: 10.1038/ncomms1998. [DOI] [PubMed] [Google Scholar]
- 77.Hao S, Baltimore D. RNA splicing regulates the temporal order of TNF-induced gene expression. Proc Natl Acad Sci USA. 2013;110:11934–11939. doi: 10.1073/pnas.1309990110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takashima Y, Ohtsuka T, Gonzalez A, Miyachi H, Kageyama R. Intronic delay is essential for oscillatory expression in the segmentation clock. Proc Natl Acad Sci USA. 2011;108:3300–3305. doi: 10.1073/pnas.1014418108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. RNA. 2009;15:1896–1908. doi: 10.1261/rna.1714509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vargas DY, et al. Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell. 2011;147:1054–1065. doi: 10.1016/j.cell.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yeo GW, Van Nostrand E, Holste D, Poggio T, Burge CB. Identification and analysis of alternative splicing events conserved in human and mouse. Proc Natl Acad Sci USA. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nature Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de la Mata M, Lafaille C, Kornblihtt AR. First come, first served revisited: factors affecting the same alternative splicing event have different effects on the relative rates of intron removal. RNA. 2010;16:904–912. doi: 10.1261/rna.1993510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 85.Huranova M, et al. The differential interaction of snRNPs with pre-mRNA reveals splicing kinetics in living cells. J Cell Biol. 2010;191:75–86. doi: 10.1083/jcb.201004030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hoskins AA, et al. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331:1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abelson J, et al. Conformational dynamics of single pre-mRNA molecules during in vitro splicing. Nature Struct Mol Biol. 2010;17:504–512. doi: 10.1038/nsmb.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Braberg H, et al. From structure to systems: high-resolution, quantitative genetic analysis of RNA polymerase II. Cell. 2013;154:775–788. doi: 10.1016/j.cell.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 90.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nature Struct Mol Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Close P, et al. DBIRD complex integrates alternative mRNA splicing with RNA polymerase II transcript elongation. Nature. 2012;484:386–389. doi: 10.1038/nature10925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nature Struct Mol Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 93.Alexander RD, Innocente SA, Barrass JD, Beggs JD. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell. 2010;40:582–593. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou HL, et al. Hu proteins regulate alternative splicing by inducing localized histone hyperacetylation in an RNA-dependent manner. Proc Natl Acad Sci USA. 2011;108:E627–E635. doi: 10.1073/pnas.1103344108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schor IE, Rascovan N, Pelisch F, Allo M, Kornblihtt AR. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc Natl Acad Sci USA. 2009;106:4325–4330. doi: 10.1073/pnas.0810666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Saint-Andre V, Batsche E, Rachez C, Muchardt C. Histone H3 lysine 9 trimethylation and HP1γ favor inclusion of alternative exons. Nature Struct Mol Biol. 2011;18:337–344. doi: 10.1038/nsmb.1995. [DOI] [PubMed] [Google Scholar]
- 97.Brody Y, et al. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol. 2011;9:e1000573. doi: 10.1371/journal.pbio.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shukla S, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kwak H, Fuda NJ, Core LJ, Lis JT. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nature Struct Mol Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 101.Bintu L, et al. Nucleosomal elements that control the topography of the barrier to transcription. Cell. 2012;151:738–749. doi: 10.1016/j.cell.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gunderson FQ, Merkhofer EC, Johnson TL. Dynamic histone acetylation is critical for cotranscriptional spliceosome assembly and spliceosomal rearrangements. Proc Natl Acad Sci USA. 2011;108:2004–2009. doi: 10.1073/pnas.1011982108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- 104.de Almeida SF, Garcia-Sacristan A, Custodio N, Carmo-Fonseca M. A link between nuclear RNA surveillance, the human exosome and RNA polymerase II transcriptional termination. Nucleic Acids Res. 2010;38:8015–8026. doi: 10.1093/nar/gkq703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Eberle AB, et al. Splice-site mutations cause Rrp6-mediated nuclear retention of the unspliced RNAs and transcriptional down-regulation of the splicing-defective genes. PLoS ONE. 2010;5:e11540. doi: 10.1371/journal.pone.0011540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rodriguez J, Menet JS, Rosbash M. Nascent-seq indicates widespread cotranscriptional RNA editing in Drosophila. Mol Cell. 2012;47:27–37. doi: 10.1016/j.molcel.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schwartz JC, et al. FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev. 2012;26:2690–2695. doi: 10.1101/gad.204602.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dichmann DS, Harland RM. fus/TLS orchestrates splicing of developmental regulators during gastrulation. Genes Dev. 2012;26:1351–1363. doi: 10.1101/gad.187278.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ryman K, Fong N, Bratt E, Bentley DL, Ohman M. The C-terminal domain of RNA Pol II helps ensure that editing precedes splicing of the GluR-B transcript. RNA. 2007;13:1071–1078. doi: 10.1261/rna.404407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim H, et al. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nature Struct Mol Biol. 2010;17:1279–1286. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mayer A, et al. Uniform transitions of the general RNA polymerase II transcription complex. Nature Struct Mol Biol. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 112.Zaret KS, Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982;28:563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]
- 113.Logan J, Falck PE, Darnell JEJ, Shenk T. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse β maj-globin gene. Proc Natl Acad Sci USA. 1987;84:8306–8310. doi: 10.1073/pnas.84.23.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Whitelaw E, Proudfoot N. α-thalassaemia caused by a poly(A) site mutation reveals that transcriptional termination is linked to 3′ end processing in the human α2 globin gene. EMBO J. 1986;5:2915–2922. doi: 10.1002/j.1460-2075.1986.tb04587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nojima T, Dienstbier M, Murphy S, Proudfoot NJ, Dye MJ. Definition of RNA polymerase II CoTC terminator elements in the human genome. Cell Rep. 2013;3:1080–1092. doi: 10.1016/j.celrep.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Connelly S, Manley JL. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 1988;2:440–452. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- 117.Niwa M, Berget SM. Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes Dev. 1991;5:2086–2095. doi: 10.1101/gad.5.11.2086. [DOI] [PubMed] [Google Scholar]
- 118.Niwa M, Rose SD, Berget SM. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 1990;4:1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- 119.Lu S, Cullen BR. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA. 2003;9:618–630. doi: 10.1261/rna.5260303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Martinson HG. An active role for splicing in 3′-end formation. Wiley Interdiscip Rev RNA. 2011;2:459–470. doi: 10.1002/wrna.68. [DOI] [PubMed] [Google Scholar]
- 121.David CJ, Boyne AR, Millhouse SR, Manley JL. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65–Prp 19 complex. Genes Dev. 2011;25:972–983. doi: 10.1101/gad.2038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Venkataraman K, Brown KM, Gilmartin GM. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19:1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bauren G, Belikov S, Wieslander L. Transcriptional termination in the Balbiani ring 1 gene is closely coupled to 3′-end formation and excision of the 3′-terminal intron. Genes Dev. 1998;12:2759–2769. doi: 10.1101/gad.12.17.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 2013;27:2380–2396. doi: 10.1101/gad.229328.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mayr C, Bartel DP. Widespread shortening of 3′ UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Das R, et al. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 128.Enriquez HP, Levitt N, Briggs D, Proudfoot NJ. A pause site for RNA polymerase II is associated with termination of transcription. EMBO J. 1991;10:1833–1842. doi: 10.1002/j.1460-2075.1991.tb07709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Peterson ML, Bertolino S, Davis F. An RNA polymerase pause site is associated with the immunoglobulin mus poly(A) site. Mol Cell Biol. 2002;22:5606–5615. doi: 10.1128/MCB.22.15.5606-5615.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pinto PA, et al. RNA polymerase II kinetics in polo polyadenylation signal selection. EMBO J. 2011;30:2431–2444. doi: 10.1038/emboj.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hazelbaker DZ, Marquardt S, Wlotzka W, Buratowski S. Kinetic competition between RNA polymerase II and Sen1-dependent transcription termination. Mol Cell. 2013;49:55–66. doi: 10.1016/j.molcel.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Eperon LP, Graham IR, Griffiths AD, Eperon IC. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988;54:393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- 133.Zhao P, Zhang W, Chen SJ. Cotranscriptional folding kinetics of ribonucleic acid secondary structures. J Chem Phys. 2011;135:245101. doi: 10.1063/1.3671644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pan T, Sosnick T. RNA folding during transcription. Annu Rev Biophys Biomol Struct. 2006;35:161–175. doi: 10.1146/annurev.biophys.35.040405.102053. [DOI] [PubMed] [Google Scholar]
- 135.Meyer M, Plass M, Perez-Valle J, Eyras E, Vilardell J. Deciphering 3′ss selection in the yeast genome reveals an RNA thermosensor that mediates alternative splicing. Mol Cell. 2011;43:1033–1039. doi: 10.1016/j.molcel.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 136.Zamft B, Bintu L, Ishibashi T, Bustamante C. Nascent RNA structure modulates the transcriptional dynamics of RNA polymerases. Proc Natl Acad Sci USA. 2012;109:8948–8953. doi: 10.1073/pnas.1205063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 138.Jasnovidova O, Stefl R. The CTD code of RNA polymerase II: a structural view. Wiley Interdiscip Rev RNA. 2013;4:1–16. doi: 10.1002/wrna.1138. [DOI] [PubMed] [Google Scholar]
- 139.Tardiff DF, Lacadie SA, Rosbash M. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly posttranscriptional. Mol Cell. 2006;24:917–929. doi: 10.1016/j.molcel.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Spiluttini B, et al. Splicing-independent recruitment of U1 snRNP to a transcription unit in living cells. J Cell Sci. 2010;123:2085–2093. doi: 10.1242/jcs.061358. [DOI] [PubMed] [Google Scholar]
- 141.Ujvari A, Luse DS. Newly initiated RNA encounters a factor involved in splicing immediately upon emerging from within RNA polymerase II. J Biol Chem. 2004;279:49773–49779. doi: 10.1074/jbc.M409087200. [DOI] [PubMed] [Google Scholar]
- 142.Schroeder SC, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. This study shows that dynamic changes in Pol II CTD phosphorylation are coordinated with transcription initiation, elongation and termination. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1–Nab3–Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nature Struct Mol Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Licatalosi DD, et al. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- 146.Buratowski S. The CTD code. Nature Struct Biol. 2003;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- 147.Gu B, Eick D, Bensaude O. CTD serine-2 plays a critical role in splicing and termination factor recruitment to RNA polymerase II in vivo. Nucleic Acids Res. 2013;41:1591–1603. doi: 10.1093/nar/gks1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Munoz MJ, et al. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell. 2009;137:708–720. doi: 10.1016/j.cell.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 149.Schwer B, Shuman S. Deciphering the RNA polymerase II CTD code in fission yeast. Mol Cell. 2011;43:311–318. doi: 10.1016/j.molcel.2011.05.024. This paper uses an elegant genetic test of the CTD code hypothesis to show that the essential function of Ser5 phosphorylation is to recruit the mRNA-capping enzyme. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Schwer B, Sanchez AM, Shuman S. Punctuation and syntax of the RNA polymerase II CTD code in fission yeast. Proc Natl Acad Sci USA. 2012;109:18024–18029. doi: 10.1073/pnas.1208995109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lee JM, Greenleaf AL. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr. 1991;1:149–167. [PMC free article] [PubMed] [Google Scholar]
- 152.Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 153.Shen Z, St-Denis A, Chartrand P. Cotranscriptional recruitment of She2p by RNA Pol II elongation factor Spt4–Spt5/DSIF promotes mRNA localization to the yeast bud. Genes Dev. 2010;24:1914–1926. doi: 10.1101/gad.1937510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Suh MH, et al. A dual interface determines the recognition of RNA polymerase II by RNA capping enzyme. J Biol Chem. 2010;285:34027–34038. doi: 10.1074/jbc.M110.145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Luco RF, et al. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet. 2012;8:e1002717. doi: 10.1371/journal.pgen.1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sims RJ, 3rd, et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Govind CK, et al. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell. 2010;39:234–246. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kizer KO, et al. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.de Almeida SF, et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nature Struct Mol Biol. 2011;18:977–983. doi: 10.1038/nsmb.2123. [DOI] [PubMed] [Google Scholar]
- 161.Kim S, Kim H, Fong N, Erickson B, Bentley DL. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci USA. 2011;108:13564–13569. doi: 10.1073/pnas.1109475108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lionnet T, Singer RH. Transcription goes digital. EMBO Rep. 2012;13:313–321. doi: 10.1038/embor.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Batada NN, Westover KD, Bushnell DA, Levitt M, Kornberg RD. Diffusion of nucleoside triphosphates and role of the entry site to the RNA polymerase II active center. Proc Natl Acad Sci USA. 2004;101:17361–17364. doi: 10.1073/pnas.0408168101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ehrensberger AH, Kelly GP, Svejstrup JQ. Mechanistic interpretation of promoter-proximal peaks and RNAPII density maps. Cell. 2013;154:713–715. doi: 10.1016/j.cell.2013.07.032. [DOI] [PubMed] [Google Scholar]
- 165.Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science. 2011;332:475–478. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Payne JM, Laybourn PJ, Dahmus ME. The transition of RNA polymerase II from initiation to elongation is associated with phosphorylation of the carboxyl-terminal domain of subunit IIA. J Biol Chem. 1989;264:19621–19629. [PubMed] [Google Scholar]
- 169.Heidemann M, Hintermair C, Voss K, Eick D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim Biophys Acta. 2013;1829:55–62. doi: 10.1016/j.bbagrm.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 170.Ranuncolo SM, Ghosh S, Hanover JA, Hart GW, Lewis BA. Evidence of the involvement of O-GlcNAc-modified human RNA polymerase II CTD in transcription in vitro and in vivo. J Biol Chem. 2012;287:23549–23561. doi: 10.1074/jbc.M111.330910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mayer A, et al. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science. 2012;336:1723–1725. doi: 10.1126/science.1219651. [DOI] [PubMed] [Google Scholar]
- 172.Garrido-Lecca A, Blumenthal T. RNA polymerase II C-terminal domain phosphorylation patterns in Caenorhabditis elegans operons, polycistronic gene clusters with only one promoter. Mol Cell Biol. 2010;30:3887–3893. doi: 10.1128/MCB.00325-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Bartkowiak B, et al. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010;24:2303–2316. doi: 10.1101/gad.1968210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- 175.Lin C, et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Roy R, et al. The MO15 cell-cycle kinase is associated with the TFIIH transcription DNA-repair factor. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 177.Akhtar MS, et al. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell. 2009;34:387–393. doi: 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Czudnochowski N, Bosken CA, Geyer M. Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nature Commun. 2012;3:842. doi: 10.1038/ncomms1846. [DOI] [PubMed] [Google Scholar]
- 179.He X, et al. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev. 2003;17:1030–1042. doi: 10.1101/gad.1075203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Xiang K, et al. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature. 2010;467:729–733. doi: 10.1038/nature09391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Phatnani HP, Jones JC, Greenleaf AL. Expanding the functional repertoire of CTD kinase I and RNA polymerase II: novel phosphoCTD-associating proteins in the yeast proteome. Biochemistry. 2004;43:15702–15719. doi: 10.1021/bi048364h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ghosh A, Shuman S, Lima CD. Structural insights to how mammalian capping enzyme reads the CTD code. Mol Cell. 2011;43:299–310. doi: 10.1016/j.molcel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Meinhart A, Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature. 2004;430:223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- 184.Kubicek K, et al. Serine phosphorylation and proline isomerization in RNAP II CTD control recruitment of Nrd1. Genes Dev. 2012;26:1891–1896. doi: 10.1101/gad.192781.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kim M, et al. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 186.Egloff S, et al. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chen HC, Cheng SC. Functional roles of protein splicing factors. Biosci Rep. 2012;32:345–359. doi: 10.1042/BSR20120007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]