Abstract
Efficient and effective HIV prevention measures for generalized epidemics in sub-Saharan Africa have not yet been validated at the population-level. Design and impact evaluation of such measures requires fine-scale understanding of local HIV transmission dynamics. The novel tools of HIV phylogenetics and molecular epidemiology may elucidate these transmission dynamics. Such methods have been incorporated into studies of concentrated HIV epidemics to identify proximate and determinant traits associated with ongoing transmission. However, applying similar phylogenetic analyses to generalized epidemics, including the design and evaluation of prevention trials, presents additional challenges. Here we review the scope of these methods and present examples of their use in concentrated epidemics in the context of prevention. Next, we describe the current uses for phylogenetics in generalized epidemics, and discuss their promise for elucidating transmission patterns and informing prevention trials. Finally, we review logistic and technical challenges inherent to large-scale molecular epidemiological studies of generalized epidemics, and suggest potential solutions.
Keywords: HIV-1, molecular epidemiology, phylogenetic, transmission networks, Sub-Saharan Africa
INTRODUCTION
Despite advances in HIV prevention over the past 30 years, an estimated 2.5 million persons were newly infected in 2011, bringing the number of people living with HIV worldwide to 34 million.1 The HIV burden continues to be greatest in sub-Saharan Africa; this region accounts for 75% of all HIV infections and the highest adult prevalence at nearly 5% overall,1 though this average mask extremes reported in subpopulations, some exceeding 50% infected.2 The use of antiretroviral treatment (ART) to reduce viral loads and associated transmission risk among sero-discordant couples (Treatment-as-Prevention, TasP) has garnered excitement as a means to curb the spread of the virus.3 A recent population-based cohort study conducted in a high prevalence region in KwaZulu-Natal found that the risk for HIV acquisition was lowest in areas with the highest ART coverage—providing ecological evidence for real-world effectiveness of TasP.4 As ART coverage has increased in Africa in the last decade, more HIV-infected individuals have been treated, and life-expectancy of infected individuals has increased.5 Meanwhile, new transmissions continue, and therefore overall prevalence of HIV can be expected to increase.6
To sustain an ongoing ART scale-up, and the potential widespread implementation of TasP in the future, expanded financial and public health resources will be required.7,8 However, the most biologically effective and financially efficient way to implement and evaluate prevention measures at the population level is unclear.9,10 Comprehensive knowledge about local epidemics will be required for successful prevention campaigns, including basic data about population demographics, transmission risk groups and viral subtypes, and complex estimates about transmission dynamics, social and sexual mixing networks, and patterns of geographic spread. Importantly, prospective information about success or failure of interventions is essential.
The tools of HIV phylogenetics and molecular epidemiology can be used to understand local transmission dynamics and assist in the design and evaluation of prevention trials. Since early in the epidemic these approaches have been employed to track HIV origin and geographic spread11–14 and in forensic studies evaluating small transmission chains.15–19 New developments, primarily led by the increased availability of viral sequences over the past 20 years, have allowed fine-scale transmission dynamics at the community, regional, and country level to be uncovered.20 The opportunity for such approaches has been facilitated by: 1) the increasing availability of large HIV sequence databases, driven by the routine provision of gene sequence based drug resistance testing, with sequences linked to epidemiological data (temporal, clinical, demographic, behavioral or geographic); 2) rapid advances in high-throughput sequencing technology and decreases in sequencing costs; and 3) theoretical and methodological advances in studies of viral transmission, using phylogenetics or genetic network analysis together with linking epidemiological and population genetic models. These advances provide a framework to identify individual traits or stages of infection that are associated with high relative infectiousness; the results of such studies can answer questions not easily resolved with standard epidemiological approaches. In effect, molecular epidemiology tools can help identify traits associated with ongoing HIV transmission (“who” is transmitting the virus?), rather than behaviors or demographic characteristics associated with high rates of infection (who is currently infected?). These analyses are increasing deployed for concentrated epidemics where HIV sequences are routinely available. However, the essential next step is to apply these tools to HIV prevention in generalized epidemics and resource-limited settings where the impact would be greatest. Further, the development of such approaches for HIV can provide a model for new approaches to the transmission dynamics of other pathogens.
SCOPE OF PHYLOGENETIC ANALYSIS TO INFORM MOLECULAR EPIDEMIOLOGY
Molecular epidemiology is the use of genetic data to inform disease etiology and distribution. In HIV, the term can encompass disparate approaches. For this review, we divide the use of genetic analysis to inform HIV epidemiology into three general categories: Molecular Epidemiology, Phylodynamics and Phylogeography (Table 1). While there is generally overlap across these categories within studies, the main questions associated with each category are distinct. Molecular Epidemiology allows understanding of the risk factors for HIV transmission and epidemic spread. Phylodynamics reconstructs epidemic history, and quantifies epidemic growth or decline, using viral genealogies and explicit population genetic models. Phylogeography describes the distribution of subtype diversity, estimates the impact of human migration on viral spread, and places historical and risk factor data into geographic context, in order to identify hubs of transmission.
Table 1.
Categories of genetic analysis related to HIV epidemiology, with examples provided from recent studies conducted in areas with generalized epidemics.
| Analysis | Description | Examples |
|---|---|---|
| Molecular Epidemiology | Quantification of risk factors for transmission and epidemic spread; Description of subtypes, recombinant forms and drug resistance mutations. | Population viral diversity in Mozambique,90 Togo,91 Gabon,92 Swaziland,96 Kenya,105 and South Africa;97 Identification of superinfection in Uganda138 and Zambia;139 Genetic linkage of seroconverters;107,110,111 Source of local infections;78,79 and Sexual network analysis106 |
| Phylodynamics | Quantification of epidemic growth or decline; and the cross-validation or enhancement of epidemiological model fitting | Epidemic growth in east Africa 67,68, Angola 69, and Cameroon72; Comparison of HIV-2 and HIV-1 population dynamics in west Africa73 |
| Phylogeography | Evaluation of the relationship between human migration (both temporary and permanent) and disease spread; identification of geographical hubs of transmission; and identification of local and imported cases | Subtype diversity and recombination;62–65,104 Spatial distribution in east Africa;67 Evolution of subtype C in Zimbabwe;66 Evolution of HIV-1 in Guinea-Bissau;71 Historical diversity in Kinshasa, DRC;14 Global spread13 |
As for any approach to elucidate HIV transmission patterns, gene sequences can only be generated for those already diagnosed and sampled, and therefore the contribution to transmission from those unsampled, undiagnosed individuals must be considered. Nevertheless, with a well-designed population sampling and sequencing strategy, with linkage to some key demographic, epidemiological and clinical data, questions from all three categories can be addressed for any given population (using the same sequence dataset).
PHYLOGENETIC STUDIES OF TRANSMISSION IN CONCENTRATED EPIDEMICS
Molecular epidemiological tools have been used to address a diverse array of research questions over the last two decades, predominantly in settings with concentrated epidemics. These approaches have advanced to describe transmission dynamics at the local, regional, and national level through the post-hoc utilization of large sequence datasets linked to epidemiological data. These sequence datasets are largely a consequence of routine antiretroviral drug resistance testing that accompanies initial HIV diagnosis, and contains partial HIV pol sequences (typically full protease and partial reverse transcriptase). The pol region has been shown to have sufficient variability to allow for phylogenetic reconstruction to the same extent as the more variable gag and env regions.21 Studies focused on transmission networks in concentrated epidemics have most commonly been conducted in Europe or North America, where large pol sequence datasets linked with epidemiological data exist (Table 2).
Table 2.
Studies in concentrated epidemics that used phylogenetics to assess HIV epidemic drivers or associations with transmission clusters
| Region | Study | Setting / Risk | Sequences / gene | Subtype B % | Transmission cluster definition | Inferred epidemic drivers or associations with cluster membership |
|---|---|---|---|---|---|---|
| Europe | ||||||
| Belgium | Chalmet et al. 201047 | Ghent / Mixed | 506 / pol | 60% | ≥3 sequences, bootstrap >90% | Younger age, MSM, Caucasian origin |
| Spain | Gonzalez-Alba et al. 2011140 | Madrid / Mixed | 2,792 + 1,822 reference / pol | 92% | bootstrap >70% | Non-B subtypes have outside sources, multiple introductions |
| Switzerland | Ambrosioni et al. 201255 | Geneva / Mixed | 1838 / pol | 66% | bootstrap >98% | MSM, recent infection, TDR |
| Kouyos et al. 201054 | Switzerland / Mixed | 5,700 + 5,700 reference / pol | 100% | Swiss clusters: ≥10 sequences with 80% of Swiss origin | IDU is driver of HET epidemic | |
| Yerly et al. 200950 | Geneva | 637 / pol | 49% | bootstrap >98%, at least two ancestors | Recent infection, TDR, high CD4, high HIV RNA | |
| UK | Fisher et al. 201024 | Brighton / MSM | 859 / pol | 100% | bootstrap >99%, genetic distance <1.5% | Younger age, recent infection, high VL, recent STI |
| Gifford et al. 2007141 | UK | 5,675 + 3,201 reference / pol | 74% | bootstrap >70% | non-B subtypes have sources outside the UK | |
| Hué et al. 200557 | UK / MSM | 1,645 + 1,784 reference / pol | 100% | UK cluster: ≥25 sequences with 90% of UK origin | Multiple separate MSM sub-epidemics / subtype-B introductions into the UK | |
| Hughes et al. 200958 | UK / HET | 11,071 / pol | 0% | <4.8% genetic distance, posterior probability of 1 | Smaller transmission clusters and slower dynamics than seen in MSM | |
| Lewis et al. 200856 | London / MSM | 2,126 / pol | 80% | <4.8% genetic distance, posterior probability of 1 | Episodic HIV transmission pattern of clustering over periods of 3–4 years | |
| Pao et al. 200528 | Brighton / MSM | 103 / pol | 100% | bootstrap >99%, genetic distance <1.5% | Younger age, high CD4, number of sexual contacts | |
| North America | ||||||
| Aldous et al. 201245 | 5 US CFAR sites/Mixed | 3,697 / pol | 98% | genetic distance ≤1.5% | Not on ART, high CD4, high HIV RNA | |
| Brenner et al. 200729 | Montreal/MSM | 717 / pol | 83% | bootstrap >98%, genetic distance <1.5% | Recent/early infection | |
| Dennis et al. 201244 | North Carolina/Mixed | 1,671 / pol | 100% | ≥3 sequences. posterior probability of 1 | Younger age, AHI, local residence, TDR | |
AHI, acute HIV infection; ART, antiretroviral therapy; CFAR, Center for AIDS Research; HET, heterosexual; IDU, injection drug use; MSM, men who have sex with men; TDR, Transmitted drug resistance
Basic methodological approach
The basic method to identify HIV transmission dynamics (traits associated with ongoing transmission) is straightforward (Figure 1): 1) HIV gene sequences are used to reconstruct phylogenetic trees or viral transmission networks; 2) transmission clusters are identified from the networks or phylogenies using ad-hoc thresholds for inclusion (e.g. minimum pairwise genetic distance between viral gene sequences, or by statistical robustness of the node that defines a phylogenetic clade); and 3) individual traits are evaluated for the strength of their association with cluster membership and used to infer underlying determinants of transmission.22,23 Cluster inclusion thresholds are ad-hoc in the sense that there is no widely accepted threshold definition or determining convention. Variation in threshold definitions and approaches can strongly impact the size and number of inferred clusters in a sequence data set, yet many studies do perform sensitivity analyses to quantitatively assess the impact of this variation on cluster identification in their respective data sets. A recent study of transmission dynamics in Brighton, UK, took this basic methodological approach one step further: clinical data were used to determine the most likely transmitter within transmission pairs or clusters, which allowed for likely onward transmission events (and not simply cluster membership based on pairwise genetic distance) to be evaluated for association with individual traits.24
Figure 1.
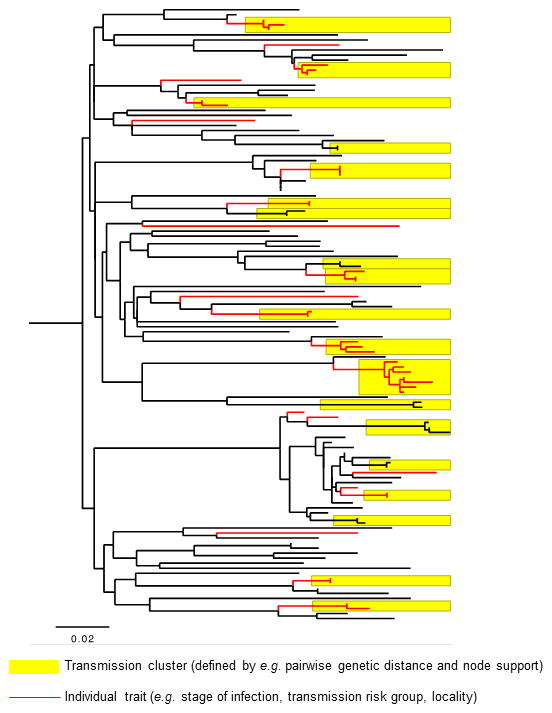
Example population-level HIV phylogeny reconstructed from HIV pol sequences, to illustrate the basic approach to identifying traits associated with transmission using a phylogeny. Putative clusters of linked transmissions are identified using (ad-hoc) criteria such as pairwise genetic distance and/or nodal support (yellow boxes).137 Individual clinical or demographic traits are then examined for significant association with linked or unlinked individuals in the phylogeny (red lineages designate individuals with a certain trait, e.g. a particular transmission risk group). Note that not all individuals are included in transmission clusters, and that transmission clusters do not include all individuals with the “red” trait.
What is the contribution of acute and early infection to ongoing transmissions?
Acute HIV infection is generally defined as the time after HIV acquisition but before seroconversion.25 Early HIV infection represents the first few months of infection, sometimes using a staging system reported by Feibig et al.26 Most recent results suggest HIV transmission is increased during acute and early HIV infection, for a period of uncertain duration that may range from weeks to months.27 Multiple phylogenetic studies have focused on acute and early (primary) HIV infection to assess the contribution of acute infection transmissions to overall HIV incidence in a population.24,28–34 The majority of these studies analyzed only sequences from patients diagnosed during acute or early HIV infection, and found variable rates of cluster formation, ranging from 13%33 to greater than 50%29,30 of the study population. These results suggest that acute and early infections are responsible for a disproportionate number of onward transmissions, relative to chronic infections. This might be expected, as: 1) individuals with newly acquired infections remain sexually active;35,36 2) such individuals are unlikely to know their HIV status;37 3) HIV viral load is exceptionally high for weeks to months after infection;38 and 4) the transmitted/founder viruses that establish successful infections may have transmission advantages.39–41 However, the proportion of transmission events attributed to acute or primary infection is dependent on definitions used for these stages of infection; more rigorous definitions of early-stage infection, and focusing on individuals with estimated infection dates31 or known dates of seroconversion,32 can help to resolve this issue. It is essential to understand the contribution of early-stage infection to onward transmission, as a high frequency may compromise TasP strategies.42
Other factors linked to transmissions
Trends or patterns associated with onward transmission have been described in greater detail by incorporating sequences from chronically infected patients thereby increasing the study population sampling density. Using molecular epidemiology tools, transmission network characteristics have been described by various factors such as race/ancestry and ethnicity,43–46 transmission risk group,44,45 HIV subtype,47–49 transmitted drug resistance mutations,50–52 and transmission cluster growth.53–55 A detailed longitudinal study of MSM sought to find the most likely transmitter for new infections and assess risk factors at the time of transmission.24 Even with 75% of all diagnosed HIV-infected individuals from the local clinic providing pol sequences, the most likely transmitter could be identified in only 25% of those recently infected. This implies that even high sampling fractions of local epidemics may not completely reveal underlying transmission linkage due to, for instance, extra-community or undiagnosed infections as major sources of new transmissions.
Large scale analyses can delineate sub-epidemics
Large scale analyses of pol sequences have been used to assess the potential influence of viral subtype and regional scale on HIV transmission dynamics. The United Kingdom HIV Drug Resistance Database (UKRDB) (containing >85,000 pol sequences, http://www.hivrdb.org.uk/) has provided data for several transmission studies. Using phylodynamics and a relaxed molecular clock approach, transmission dynamics were reconstructed among MSM56,57 and heterosexuals.58 Among MSM, six large transmission clusters were identified, representing separate introductions of subtype B into the U.K. in the 1980s.57 Additional analysis within the large MSM clusters (reconstructed with ~2,000 individual sequences) indicated that 25% of transmissions likely occurred within six months of infection, with most clusters arising over periods of 3–4 years—an episodic epidemic with multiple clusters of transmission.56 However, in heterosexuals, where the epidemic is dominated by non-B subtypes, much slower transmission dynamics were found.58 This demonstrates that phylogenetic approaches actually reveal the differing dynamics between different risk groups.
The Swiss HIV Cohort includes large repositories of pol sequences that have been used to assess changes in subtype B and non-B transmission clusters over time. Largely independent epidemics of MSM and heterosexuals/injection drug users were noted in the subtype B clusters, but heterosexuals alone did not dominate any of the clusters.54 Over time, the contribution of injection drug use to the heterosexual epidemic notably decreased. The effect of migration was investigated by analyzing non-B subtypes sampled both in and outside Switzerland. Less than 25% of the non-B subtypes sampled in Switzerland were found in clusters with other Swiss sequences, suggesting that most non-B infections in the country could not be prevented through national prevention measures targeting individuals of only Swiss origin.49
Transmission network analysis
The framework of social network analysis provides an alternative method to understand HIV transmission dynamics, which can be supplemental to the explicitly evolutionary approach of phylogenetic analysis. Transmission clusters are reconstructed similar to contact tracing, but based on genetic distance metrics. Transmission network parameters among MSM were estimated using phylodynamics of over 14,000 UKRDB sequences (representing ~60% of UK MSM).59 Using an inferred network distribution and associated parameter values, the HIV epidemic was characterized by preferential association, which predicted that the epidemic would persist even under conditions of poor overall transmission following a randomly distributed intervention. Cluster growth also differed across the MSM and heterosexual transmission risk groups. However, the relationship between phylogenetic clusters and sexual networks is not direct (they are not one and the same), and the use of phylogenies to understand the underlying transmission network is complex and requires further elucidation.60
PHYLOGENETICS AND TRANSMISSION DYNAMICS IN GENERALIZED EPIDEMICS OF SUB-SAHARAN AFRICA: WHAT QUESTIONS CAN BE ADDRESSED?
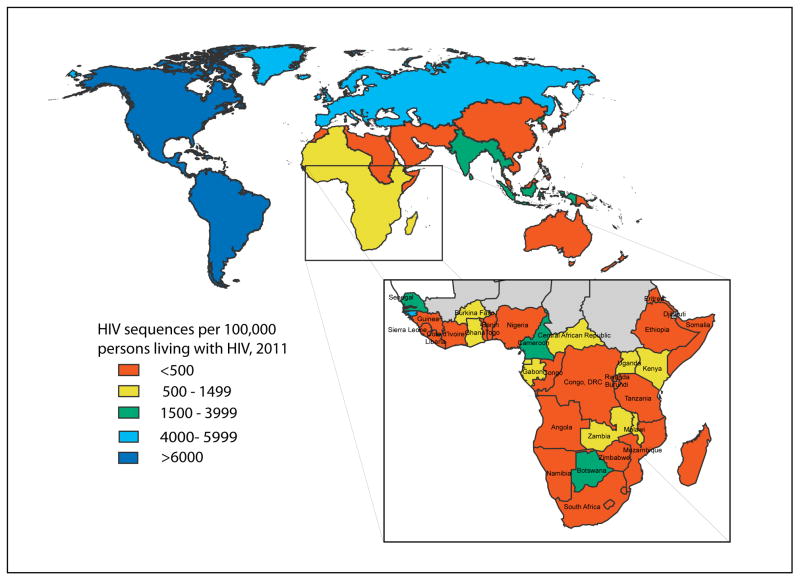
HIV transmission dynamic studies in concentrated epidemics largely involve the post-hoc use of HIV drug resistance screening datasets whose coverage can reach a substantial proportion of the HIV-infected population. Large datasets of this type are rarely found in Africa—despite sub-Saharan Africa accounting for two-thirds of the global HIV infections, only 24% of sequences deposited in the LANL HIV Sequence Database (http://www.hiv.lanl.gov, which receives all HIV sequences deposited in GenBank) are derived from the region (Figure 2). Notably, the Southern African Treatment and Resistance Network (SATuRN) has a growing database currently with >7,000 HIV sequences, albeit sampled from a very large HIV-infected population.61 Nonetheless, informative phylogenetic analyses of generalized epidemics will still require significant de novo sequencing effort. To enable detailed molecular epidemiological studies to inform prevention, clinical, demographic and behavioral data must be linked to each sequence. This represents a major logistical challenge, given that current sequence databases are generally based on opportunistic approaches to sequence acquisition. Several important questions surrounding HIV epidemiology and prevention can be addressed in the region with available molecular epidemiology tools, particularly when linked to traditional epidemiologic data.
Figure 2.
The regional distribution of HIV sequences deposited in the LANL HIV Sequence Database scaled to the estimated number of persons living with HIV, by WHO region and sub-Saharan Africa (inset). Map generated through query of the LANL database for number of sequences sampled by geographic region and country (http://www.hiv.lanl.gov; queried on June 26, 2013). Numbers were scaled to the 2011 WHO estimates on numbers of persons living with HIV by region and country (http://apps.who.int/gho/data/).
Historical pattern of growth or decline for a given HIV epidemic
Molecular epidemiology and phylogeography can be used to understand historical patterns in epidemic origin, spread, and growth over time and space. The high genetic diversity of HIV-1 Group M has given rise to 9 genetically divergent subtypes (A–D, F–H and J–K), intersubtype recombinants and circulating recombinant forms (CRFs).62 Sub-Saharan Africa has the highest HIV genetic diversity, and is also characterized by distinct geographical subtype distributions that have remained relatively stable over the past decade.63 Surveillance of genetic diversity has historically supported tracking global epidemiology64 and public health strategies to slow further viral spread.65
The integration of time-stamped sequence (those with known dates of sampling) data with phylogenetics, coalescent models and molecular clock models allows inferences to be made on the timing of epidemic origin and spread in Africa.13,66–74 These analyses can contribute to the design of intervention strategies through better understanding of epidemic growth potential, or factors contributing to historical geographic spread of subtypes or recombinants. Phylogeographic approaches showed that HIV-1C in Zimbabwe expanded through multiple introductions originating in southern Africa and localized exponential growth in the 1980s corresponding to demographic and political change.66 Travel accessibility and infrastructure is felt to be critical in epidemic spread, particularly the rapid growth of HIV-A and D into east Africa,67 and the expansion of HIV-1C into east Africa68 and Angola.69
Geographic source of local epidemics or outbreaks
HIV prevalence in Africa is heterogeneously distributed within countries and often communities; epidemics can be overlapping sub-epidemics defined by geography, time, and a complex interplay of local epidemic drivers. This variation in HIV spread through populations requires that prevention efforts be tailored to characteristics of local epidemics.1,75 For example, the geographic clustering of HIV along roadways in KwaZulu-Natal indicated the need for more intensified interventions within these communities; this study also indicated that many infections were imported from outside local communities.76 High transport connectivity and mobile populations may explain the hyper-endemic outbreaks experienced in eastern and southern Africa.77 Understanding the degree of transmission that occurs from outside communities may have a substantial impact on the design and success of targeted prevention efforts such as TasP.
Communities with high transport connectivity may have local HIV epidemics supported by a significant proportion of transmissions from outside communities. Among 153 HIV pol sequences from patients in one community in rural coastal Kenya, multiple subtypes and significant recombination were documented.78 In a phylogeographic analysis, many of these sequences were related to different regions in Africa, suggesting multiple introductions into this community, likely reflecting its extensive transport links. In the Rakai district in Uganda, 14,595 individuals in 46 communities underwent extensive HIV surveillance including spatial and phylogenetic transmission linkage analysis.79 Of 189 HIV-incident cases, an estimated 39% of new cases were infected by household partners, and many new cases that were infected by an extra-household partner were from outside the community. The high degree of external HIV introductions into these communities suggests that the ability of test-and-treat strategies to reduce HIV transmissions may be difficult to measure in small populations unless external introductions are reduced, or unless geographic TasP coverage is sufficiently widespread that migration no longer becomes a relevant problem.
Results from phylogeographic studies must be interpreted in the context of the viral sequences analyzed. Studies based on convenience samples with limited temporal or spatial scales could lead to erroneous inferences about transmission rates between populations (e.g. between the studied community and the extra-community). While this potential limitation can be addressed to some extent with extensive sampling of the community of interest, the use of simulation to validate inferences can be useful.80
Tracking the transmission and evolution of drug resistance mutations
The decreases in morbidity and mortality following ART provision81,82 have led to widespread ART rollout over the past decade.83 With increased ART access in Africa, the prevalence of transmitted drug resistance (TDR) has duly increased.84,85 High levels of acquired drug resistance, often unrecognized when laboratory monitoring for virologic failure is unavailable, and the failure of ART to prevent secondary transmission, contribute to increasing TDR in the population.86 Simulations suggest that TDR could have a significant impact on mortality,87 thus highlighting the need for ongoing attention for tracking and preventing TDR. Further, with TasP programmes expanding, and WHO guidelines88 recommending treatment initiation at CD4 lymphocyte counts<500 cells/mL, then it is likely that an increasing proportion of new infections will be with resistant viruses, despite an overall drop in incidence.89
Although TDR prevalence in sub-Saharan Africa is moderate at an estimated 5.7%,86 it is projected to have increased by 14–29% per year in southern and east Africa following ART rollout.84 Phylogenetic analyses have been employed in TDR prevalence studies primarily to characterize the extensive genetic diversity in specific regions90–94 and the dominance of HIV-1C in southern Africa.95–99 Few studies have evaluated transmission lineages of drug resistant strains through transmission cluster analysis to support or refute epidemiological linkages.90,96,100 Transmission clusters combined with participant life histories revealed a high degree of sexual partner mixing in Ugandan fishing communities and uncovered clusters sharing similar TDR mutations.94 Only one study incorporated antiretroviral history in the probable transmitting partners to further characterize linkages among individuals with TDR.100
In contrast to resource-rich settings, drug resistance testing is rarely performed at entry to clinical care, thereby limiting the number of sequences available for phylogenetics. However, sequences will become increasingly available as drug resistance monitoring strategies continue to expand and through ongoing or pending TasP protocols. Phylogenetics could be used to evaluate trends in genetic diversity, further the understanding of sexual networks or transmission clusters harboring resistant strains, track the evolution of drug resistance on a population-level, and to help assess the effect of TDR on transmissibility by subtype.
Understanding transmission patterns to help design targeted prevention measures
Targeting epidemic drivers, or core groups, for enhanced prevention may increase the effectiveness of an intervention, but requires detailed epidemiological understanding of viral transmission in the community. This is challenging because HIV transmission dynamics of generalized African epidemics are largely unknown.101–103 Molecular epidemiological approaches can help uncover local HIV epidemic drivers by contributing the links between overlapping sub-epidemics that are characterized by geography, time and social/sexual interaction.
Characterizing HIV subtypes and including linkage analysis within epidemiological studies can shed light on transmission patterns between high risk groups and the general population.104–107 Among MSM in Senegal, HIV phylogenetics revealed different subtype distributions compared to the general population.104 Most of these men reported sex with women, thus likely contributing to bridging between these groups that may modify the subtype distribution. In coastal Kenya, local subtypes among MSM predominated, including those frequent among female sex workers, indicating an epidemic of local origin and confirmation of observed behavioral links between MSM and the general population.105 By combining partnership histories and phylogenetic analysis among Ugandan female sex workers, partial sexual networks and multiple infections were observed, confirming high-risk networks.106 Among HIV concordant heterosexual couples in Senegal, most couples had phylogenetically linked sequences. When combined with interview data, the male partner was often the most likely index, implying concurrency associated transmission among stable partners.107
In the design of prevention measures, these phylogenetic tools could be expanded to other questions; currently unknown is whether epidemic growth in local epidemics of sub-Saharan Africa is driven primarily by those with high viremia or consistent low-level transmission from chronically infected individuals in concurrent partnerships. Determining epidemic drivers allows for targeted prevention in a more cost effective manner. For example, Avahan, the India AIDS Initiative, focused prevention on groups at high risk for transmission and acquisition in India (sex workers, their clients, and injection drug users; identified without phylogenetic analyses) leading to over 100,000 estimated HIV infections prevented in the general population between 2003 to 2008.108,109 The success of these types of approaches, however, depends on having an in-depth understanding of local epidemic drivers in the HIV epidemics of sub-Saharan Africa.
Assessing the impact of an intervention
Molecular epidemiology approaches also hold promise in evaluating HIV prevention interventions. At the individual level phylogenetics has been used to confirm or refute transmission linkages among seroconverters and their partners in heterosexual serodiscordant partnership trials.110,111 By determining genetic linkages between enrolled partners, the primary efficacy of the intervention can be better assessed (if transmission occurred between partners despite the intervention, or did the transmission arise from an outside partnership). The Partners in Prevention HSV/HIV Transmission study (PiP) assessed the efficacy of genital herpes suppression in reducing HIV transmission among serodiscordant couples in east and southern Africa.110 Nearly 27% of couples were found to be unlinked through phylogenetic analyses, showing that a substantial number of transmissions occurred through outside partnerships. In HPTN 052, pol and env sequences among index-partner pairs and controls were also evaluated with phylogenetic methods.111 Similar to PiP, 24% of the index cases were not linked to their partner. There was a strong association between linked transmission and the delayed ART initiation study arm: 28 of 29 linked transmission events were in the delayed arm. The association between early ART initiation and transmission reduction became stronger when only the linked events are included in the analysis; this emphasizes the importance of genetic linkage analysis to assess seroconversion events in prevention studies.
Several combination prevention trials that will assess the impact of TasP on a population level are either planned or are currently in progress in sub-Saharan Africa. Most of these trials have planned or are considering integration of molecular epidemiology analyses (Table 3).
Table 3.
Randomized controlled trials for combination HIV prevention and HIV cohort studies with ongoing or planned integration of molecular epidemiology in Africa.
| Study | Location | Population | Est. HIV prevalence | Study arms/Description | Status | Phylogenetics component |
|---|---|---|---|---|---|---|
| BCPP: Botswana Combination Prevention Project1* | Botswana | 30 rural communities in 3 geographic areas in Botswana (~6,027 individuals per community). The estimated number of targeted adults aged 16–64 years is ~105,000 | 25% |
|
Starting | Env gp120 V1C5 sequencing to address: (1) the proportion of new infections in 20% of households that cannot be phylogenetically linked to HIV-infected persons in the same study arm; (2) an adjusted analysis of Combination Prevention package efficacy that includes only linked incident HIV infections per study arm; and (3) the proportion of recent HIV-1 infections that can be linked to HIV-infected adults in intervention communities. |
| PopART: Population Effects of Antiretroviral Therapy to Reduce HIV Transmission (HPTN 071)3* | Zambia & South Africa | 21 clusters (60,000 individuals per cluster, with 2,500 enrolled in observation cohort) | 15% |
|
Starting | Planned in 12/21 clusters (4 triplets). Samples to be collected from ~30,000 untreated individuals. Sequencing method to be determined. |
| TasP study (ANRS 1249)4 | KwaZulu Natal, South Africa | 34 clusters (2,000 adults/cluster) | 20% |
|
Ongoing | Pol sequences for all individuals with virologic failure or seroconversion within the trial; phylodynamic analysis |
| Africa Centre Demographic Surveillance Site5 | KwaZulu Natal, South Africa | 90,000 people from 11,000 households | 24% | Observational study; longitudinal prospective cohort. Annual HIV-testing of all resident adults and a sample of non-resident adults | Ongoing | Pol sequences for all seroconverters in the surveillance area (N>700) between 2004 and 2009 |
| MP3: An HIV Prevention Program for Mochudi, Botswana6 | Botswana | Northeastern sector of Mochudi, total population ~15,000 | 21% | ART at CD4 ≥250/350 cells/mL and HIV-1 RNA ≥50,000 copies/mL | Completed | Env gp120 V1C5 sequences from 785 individuals to assess general clustering patterns and proportions of clustered vs. non- clustered sequences among prevalent and incident HIV infections. |
| Rakai Community Cohort Study7 | Uganda | 12–15,000 total individuals from 46 different communities (general Rakai cohort) | 12% | Observational study; longitudinal prospective cohort. 14 near-annual surveys, since 1994. Recent expansion into 4 hotspot communities along Lake Victoria | Ongoing | Gag and env sequenced for >1,000 ART-naïve individuals from 3 survey rounds (1994, 2002, 2009) in general cohort communities |
ART, Antiretroviral therapy; HCT, HIV testing and counseling; PMTCT, Prevention of mother to child transmission; MC, medical circumcision; IEC, Information Education Communication campaign ; STI, sexually transmitted infection
Partially adapted from Boily (2012)142
Harvard/CDC/Botswana Harvard AIDS Institute Partnership/Botswana Ministry of Health, U01 GH000447; PI’s: M. Essex & V. DeGruttola
Johns Hopkins/USAID
NIH/Harvard/NIH/LSHTM/Imperial College London, ZAMBART, DTTC of Public Health
ANRS
Wellcome Trust
NIH/Harvard, R01 AI083036; PI: M. Essex;
NIH
At the population level, analysis of genetic data can potentially supplement standard approaches to evaluate the impact of an intervention. Comparative phylogenetic analyses of a baseline trial population and the population over the course of a trial can reveal the emergence or disappearance of clusters associated with particular traits. This approach was used to assess the impact of targeted hepatitis B vaccination in The Netherlands, and showed that resulting decreases in HBV incidence were due largely to declines in intravenous drug or heterosexual (but not MSM) risk groups.112 However, a follow-up study with increased sample size (n=894, versus n=85) suggested that reduced HBV transmission was in fact due to reduced incidence in MSM, highlighting the importance of sample size and extended sampling periods for studies of this type.113 Alternatively, gene sequence data can be used to reconstruct HIV transmission networks rather than phylogenies, and cluster size distributions (CSD) can be compared over the course of a prevention trial or intervention.59,114,115 In theory, CSDs will be dominated by larger clusters in populations where epidemic drivers (individuals with relatively high infectiousness) persist. Changes in CSD can reflect an intervention’s impact on particular subgroups or the overall transmission patterns. These population level approaches may be most suitable for trials in which clear incidence outcomes are equivocal, or in which there are clear decreases in incidence but the underlying cause is unknown. While the methods can be applied to a trial with any targeting strategy, clinical and demographic data from sequenced individuals are required.
Phylodynamics is the use of pathogen sequences to reconstruct epidemic history using viral genealogies and explicit population genetic models.114,116,117 Despite great methodological potential and scientific interest, phylodynamics has to date been rarely used for impact evaluation. This is likely related to a lack of consensus about the interpretation of the estimated parameter Ne, (nominally the effective population size, a quantity proportional to the number of infected individuals, and estimated by coalescent approaches within the product Ne * tau, where tau is the mean (viral) generation time). Estimates of both Ne and tau can be strongly affected by epidemic stage, transmission dynamics, or population sampling,11,118,119 and come with large variances. There is poor resolution of population size changes in the recent past (~5 years).119 Additionally, simulation studies have shown that it may be difficult to disentangle the effects of changing incidence and changing transmission networks on phylodynamic parameters;119,120 information on transmission network structure might be required for accurate parameter estimation. On a positive note, there is a growing base of modeling approaches to understand the relationships between epidemic models, phylogenetics, and transmission networks, which could be utilized to better understand how transmission network structure affects phylogenetic trees, and to model outcomes of specific prevention trial designs.60,114,121–125 Of particular interest is the incorporation of stochastic birth-death processes into phylodynamic estimation of epidemic parameters in lieu of standard coalescent models, allowing for more realistic assumptions about changes in epidemic size through time.126–130
CHALLENGES AND SOLUTIONS FOR IMPLEMENTING PHYLOGENETIC STUDIES IN GENERALIZED EPIDEMICS
Phylogenetic analyses of HIV transmission and other epidemiological questions hold great promise to further our understanding of generalized epidemics and inform prevention efforts. However, we must consider how differences between concentrated and generalized epidemics will affect the design and implementation of such studies. Below we note several key challenges that must be met, followed by potential solutions.
HIV transmission networks will be difficult to detect with limited population sampling
As seen in phylogenetic studies of concentrated HIV epidemics, a large sampling fraction (e.g. >25% of infected individuals in a community) is needed to identify transmission pairs or clusters. This result is seen empirically24 and in simulations.121,122 For phylogenetic studies in generalized HIV epidemics, especially in regions where prevalence can exceed 10%, a substantial number of individuals will need to be sampled and sequenced.
Potential solution 1
Population sampling that is biased toward sequencing of incident cases can both decrease the required sample fraction and increase the probability of identifying transmission linkages. This “targeted” sampling strategy contrasts with the opportunistic sampling strategy generally found in standard phylogenetic studies. The approach is suited for studies that seek to identify HIV transmissions, rather than reconstruct viral evolutionary history. As such, phylogenetic studies of transmission will be most informative and efficient when epidemiological questions are not simply overlaid onto ad-hoc phylogenies reconstructed from randomly sampled individuals in a population (the standard approach for phylogenetic studies in e.g. systematics, biogeography and phylodynamics).
Potential solution 2
Population sampling conducted over multiple time periods, with the initial sample completed prior to the intervention, increases the probability of identifying transmission pairs or clusters with phylogenetic analysis. Transmission studies in concentrated epidemics generally involve post-hoc use of sequence datasets which allows for retrospective analyses of epidemic history or transmission dynamics. The utility of HIV phylogenetics to inform prevention trials cannot be fully realized, however, based solely on retrospective analyses. The sampling frequency required to improve transmission cluster detection is unclear, and will likely vary according to local incidence rates.
HIV sequencing requirements may be extensive
As large sample fractions will be required for informative phylogenetic analyses of generalized epidemics, significant de novo sequencing effort will be necessary, even with “targeted” sampling of incident infections. This will require extensive technical capability and ability for large volumes of sequences to be generated relatively rapidly. Except for the SATuRN database,61 there are no standing HIV sequence databases in the region that can be readily used for molecular epidemiological studies, in contrast to resource-rich settings that have a larger proportion of sequences deposited compared to their epidemic size (Figure 2).
Potential solution
Developing a sequencing pipeline is necessary for large scale HIV phylogenetic and molecular epidemiology projects. Five features of this pipeline are required: 1) the pipeline must work on clinical samples; 2) it must scale across multiple diverse HIV genomes (different subtypes), based on a universal primer set, or alternative methods of genome enrichment that produce equivalent amplification rates across diverse samples; 3) it must scale from 100s to 1000s of samples with ~75% sequencing success rate across a wide range of genome copy number (viral loads of the individual samples); 4) it must produce accurate consensus sequences with no manual editing; 5) it must detect and accurately quantify minority variants; and 6) it must maximize the informative phylogenetic signal, by extending sequencing from one gene (typically the partial pol gene sequenced to around 1kbase length), to the whole viral genome (9.8kbase in length).131 A component of this solution may be the development in Africa of the capacity for high-throughput full genome HIV sequencing.
The development of a pan-African sequence database (analogous to the LANL HIV Sequence Database), will provide an important resource for future studies. The few phylogenetic studies of HIV transmission in Africa to date79,132 suggest that extra-community HIV transmission sources are common. The potential role of a large pan-African database would be to provide sets of African outgroup sequences, in order to identify extra-community sources and to clarify their impact vs. undiagnosed local sources. The database would also be useful for studying the spread of inter-subtype recombinants and for characterizing the diversity of regional epidemics in Africa, i.e. for phylodynamics and phylogeography of the African HIV epidemic.
Methodological challenges: Integrating molecular epidemiology into phylodynamics
Molecular epidemiology and phylodynamics have both been areas of active methodological development, as evidenced by the articles referenced above. Nonetheless, current studies in molecular epidemiology that define risk factors for transmission do not make full use of the data available, and do not adequately account for the uncertainty and arbitrariness inherent in clustering.
Potential solution
A preferable approach would be to integrate the estimation of transmission risk factors (and other statistics of molecular epidemiology) directly into a phylodynamic inference framework, so that all of the data available could be used and arbitrary clustering would not be a prerequisite step. Some authors have begun this process,133 but further methodological development and validation is needed. Additionally, the development of consistent quantitative definitions of transmission clusters, e.g. based on tree shape characteristics such as average branch lengths or nodal support, can make the identification of clusters more rigorous;134 this includes assessing the statistical significance of trait clusters via simulation procedures similar to those used to examine gene flow among populations.135
Ethical challenges: risks to individual privacy and stigmatization
Ethical challenges in studies involving transmission dynamics in HIV epidemics extend beyond those faced by randomized HIV control trials136 and apply to both concentrated and generalized epidemics. Phylogenetic studies conducted at the village or small community-level may involve collection of HIV sequences and individual clinical and demographic data, and in some studies may include geographic location data. The risk of individual identification could result in the loss of privacy and, in locations where HIV transmission or reckless exposure is a criminal offence, prosecution. Additionally, an important goal in phylogenetic studies of transmission is to identify traits associated with individuals and groups responsible for onward HIV transmission. Thus there is the potential for stigmatization of individuals linked with or have common features of transmission network members, either underlying (e.g. socioeconomic or demographic group) or proximate (e.g. injection-drug use or sexual practice).22,23 In contrast to the stigma related to HIV-infection, this challenge will include HIV-negative individuals as well.
Potential solution
Although sampling from generalized epidemics in regions of high prevalence might make such identifications unlikely, principles and governance on patient identifiable data will be necessary. Data from phylogenetic studies of transmission should be reported in ways where individuals cannot be identified. For example, the UKRDB and the Swiss HIV Cohort Study have adopted the strategy where only a minority (e.g. 10%) of the sequences collected will be released, with these sequences chosen at random. This includes location data; one approach is to reduce the resolution of location data by including a set number of individuals (e.g. 200) in the location set, such that individual identification by location is not possible. These data security strategies will also be addressed in the patient consent process and the tight restriction needed on data release must be recognized by funding agencies and scientific journals.
CONCLUSIONS
Opportunities to implement phylogenetic methods at the inception of HIV prevention studies should not be lost. Progress in computational and analytic techniques for reconstructing HIV phylogenies is ongoing. The costs associated with HIV gene or genome sequencing will continue to decrease; rapid, high-throughput sequencing will produce more sequences and larger databases. These advances make it all but certain that sequences or specimens collected in broad reaching studies will eventually be sequenced and used for phylogenetic analyses. However, planning for such analyses from the beginning will maximize their usefulness and the likelihood that phylogenetic analyses can be used in impact evaluations. Additionally, implementing these analyses prospectively will help in identifying hidden sub-populations or core-transmitter groups, as well as monitoring the spread of transmitted drug resistance, especially among rapidly transmitting networks or clusters.
Acknowledgments
Source of Funding: AD reports receiving support from the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant KL2TR000084. JH reports receiving support from the University of Washington Center for AIDS Research, an NIH funded program under award number P30AI027757 which is supported by the following NIH Institutes and Centers (NIAID, NCI, NIMH, NIDA, NICHD, NHLBI, NIA, NIGMS, NIDDK).
We thank Ward Cates and Nancy Padian for their careful review of earlier versions of the manuscript and Vladimir Novitsky and Mary Kate Grabowski for their helpful discussions. This work stemmed from discussions held during meetings sponsored by the NIH HIV Prevention Trials Network (Feb 21-22, 2012) and the Bill and Melinda Gates Foundation (BMGF) [Nov 1-2, 2012]. We thank participants of these meetings and specifically David Burns at the NIH and Gina Dallabeta at the BMGF. We would also like to acknowledge members of the Phylogenetic and Networks for Generalised HIV Epidemics in Africa (PANGEA), a consortium sponsored by the BMGF.
Footnotes
Conflicts of Interest: We declare that we have no conflicts of interest.
References
- 1.UNAIDS. UNAIDS World AIDS Day report, 2012. Geneva, Switzerland: 2012. [Accessed July 2, 2013]. Joint United Nations Programme on HIV/AIDS. Available at http://www.unaids.org/en/media/unaids/contentassets/documents/epidemiology/2012/gr2012/20121120_UNAIDS_Global_Report_2012_en.pdf. [Google Scholar]
- 2.Karim QA, Kharsany AB, Frohlich JA, et al. Stabilizing HIV prevalence masks high HIV incidence rates amongst rural and urban women in KwaZulu-Natal, South Africa. International Journal of Epidemiology. 2011 Aug 1;40(4):922–930. doi: 10.1093/ije/dyq176. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen MS, Chen YQ, McCauley M, et al. Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med. 2011 Aug 11;365(6):493–505. doi: 10.1056/NEJMoa1105243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanser F. High coverage of ART associated with decline in risk of HIV acquisition in rural KwaZulu-Natal, South Africa. Science (New York, NY) 2013;339(6122):966–971. doi: 10.1126/science.1228160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bor J, Herbst AJ, Newell ML, Barnighausen T. Increases in adult life expectancy in rural South Africa: valuing the scale-up of HIV treatment. Science. 2013 Feb 22;339(6122):961–965. doi: 10.1126/science.1230413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaidi J, Grapsa E, Tanser F, Newell ML, Barnighausen T. Dramatic increases in HIV prevalence after scale-up of antiretroviral treatment: a longitudinal population-based HIV surveillance study in rural kwazulu-natal. AIDS. 2013 May 10; doi: 10.1097/QAD.0b013e328362e832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer-Rath G, Over M. HIV treatment as prevention: modelling the cost of antiretroviral treatment--state of the art and future directions. PLoS Med. 2012;9(7):e1001247. doi: 10.1371/journal.pmed.1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnighausen T, Salomon JA, Sangrujee N. HIV treatment as prevention: issues in economic evaluation. PLoS Med. 2012;9(7):e1001263. doi: 10.1371/journal.pmed.1001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garnett GP, Becker S, Bertozzi S. Treatment as prevention: translating efficacy trial results to population effectiveness. Curr Opin HIV AIDS. 2012;7(2):157–163. doi: 10.1097/COH.0b013e3283504ab7. [DOI] [PubMed] [Google Scholar]
- 10.Eaton JW, Johnson LF, Salomon JA, et al. HIV Treatment as Prevention: Systematic Comparison of Mathematical Models of the Potential Impact of Antiretroviral Therapy on HIV Incidence in South Africa. PLoS Med. 2012;9(7):e1001245. doi: 10.1371/journal.pmed.1001245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmes EC, Nee S, Rambaut A, Garnett GP, Harvey PH. Revealing the history of infectious disease epidemics through phylogenetic trees. Philos Trans R Soc Lond B Biol Sci. 1995 Jul 29;349(1327):33–40. doi: 10.1098/rstb.1995.0088. [DOI] [PubMed] [Google Scholar]
- 12.Rambaut A, Robertson DL, Pybus OG, Peeters M, Holmes EC. Human immunodeficiency virus. Phylogeny and the origin of HIV-1. Nature. 2001 Apr 26;410(6832):1047–1048. doi: 10.1038/35074179. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert MT, Rambaut A, Wlasiuk G, Spira TJ, Pitchenik AE, Worobey M. The emergence of HIV/AIDS in the Americas and beyond. Proceedings of the National Academy of Sciences of the United States of America. 2007 Nov 20;104(47):18566–18570. doi: 10.1073/pnas.0705329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Worobey M, Gemmel M, Teuwen DE, et al. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature. 2008 Oct 2;455(7213):661–664. doi: 10.1038/nature07390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ou CY, Ciesielski CA, Myers G, et al. Molecular epidemiology of HIV transmission in a dental practice. Science. 1992 May 22;256(5060):1165–1171. doi: 10.1126/science.256.5060.1165. [DOI] [PubMed] [Google Scholar]
- 16.Scaduto DI, Brown JM, Haaland WC, Zwickl DJ, Hillis DM, Metzker ML. Source identification in two criminal cases using phylogenetic analysis of HIV-1 DNA sequences. Proc Natl Acad Sci U S A. 2010 Dec 14;107(50):21242–21247. doi: 10.1073/pnas.1015673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leitner T, Escanilla D, Franzen C, Uhlen M, Albert J. Accurate reconstruction of a known HIV-1 transmission history by phylogenetic tree analysis. Proceedings of the National Academy of Sciences of the United States of America. 1996 Oct 1;93(20):10864–10869. doi: 10.1073/pnas.93.20.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemey P, Van Dooren S, Van Laethem K, et al. Molecular testing of multiple HIV-1 transmissions in a criminal case. AIDS (London, England) 2005 Oct 14;19(15):1649–1658. doi: 10.1097/01.aids.0000187904.02261.1a. [DOI] [PubMed] [Google Scholar]
- 19.Pistello M, Del Santo B, Buttò S, Bargagna M, Domenici R, Bendinelli M. Genetic and phylogenetic analyses of HIV-1 corroborate the transmission link hypothesis. Journal of Clinical Virology. 2004;30(1):11–18. doi: 10.1016/j.jcv.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 20.Brenner BG, Wainberg MA. Future of Phylogeny in HIV Prevention. J Acquir Immune Defic Syndr. 2013 Jul;63 (Suppl 2):S248–254. doi: 10.1097/QAI.0b013e3182986f96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hue S, Clewley JP, Cane PA, Pillay D. HIV-1 pol gene variation is sufficient for reconstruction of transmissions in the era of antiretroviral therapy. AIDS. 2004 Mar 26;18(5):719–728. doi: 10.1097/00002030-200403260-00002. [DOI] [PubMed] [Google Scholar]
- 22.Boerma JT, Weir SS. Integrating demographic and epidemiological approaches to research on HIV/AIDS: the proximate-determinants framework. J Infect Dis. 2005 Feb 1;191 (Suppl 1):S61–67. doi: 10.1086/425282. [DOI] [PubMed] [Google Scholar]
- 23.Lewis JJ, Donnelly CA, Mare P, Mupambireyi Z, Garnett GP, Gregson S. Evaluating the proximate determinants framework for HIV infection in rural Zimbabwe. Sex Transm Infect. 2007 Aug;83 (Suppl 1):i61–69. doi: 10.1136/sti.2006.023671. [DOI] [PubMed] [Google Scholar]
- 24.Fisher M, Pao D, Brown AE, et al. Determinants of HIV-1 transmission in men who have sex with men: a combined clinical, epidemiological and phylogenetic approach. AIDS. 2010 Jul 17;24(11):1739–1747. doi: 10.1097/QAD.0b013e32833ac9e6. [DOI] [PubMed] [Google Scholar]
- 25.Pilcher CD, Eron JJ, Jr, Galvin S, Gay C, Cohen MS. Acute HIV revisited: new opportunities for treatment and prevention. The Journal of Clinical Investigation. 2004;113(7):937–945. doi: 10.1172/JCI21540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiebig EW, Wright DJ, Rawal BD, et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS. 2003 Sep 5;17(13):1871–1879. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 27.Powers KA, Ghani AC, Miller WC, et al. The role of acute and early HIV infection in the spread of HIV and implications for transmission prevention strategies in Lilongwe, Malawi: a modelling study. Lancet. 2011 Jul 16;378(9787):256–268. doi: 10.1016/S0140-6736(11)60842-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pao Da, Fisher Ma, Hue Sbe, et al. Transmission of HIV-1 during primary infection: relationship to sexual risk and sexually transmitted infections. AIDS. 2005;19(1):85–90. doi: 10.1097/00002030-200501030-00010. [DOI] [PubMed] [Google Scholar]
- 29.Brenner BG, Roger M, Routy JP, et al. High rates of forward transmission events after acute/early HIV-1 infection. The Journal of Infectious Diseases. 2007 Apr 1;195(7):951–959. doi: 10.1086/512088. [DOI] [PubMed] [Google Scholar]
- 30.Brenner BG, Roger M, Stephens D, et al. Transmission clustering drives the onward spread of the HIV epidemic among men who have sex with men in Quebec. J Infect Dis. 2011 Oct 1;204(7):1115–1119. doi: 10.1093/infdis/jir468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown AE, Gifford RJ, Clewley JP, et al. Phylogenetic reconstruction of transmission events from individuals with acute HIV infection: toward more-rigorous epidemiological definitions. The Journal of infectious diseases. 2009 Feb 1;199(3):427–431. doi: 10.1086/596049. [DOI] [PubMed] [Google Scholar]
- 32.Chibo D, Kaye M, Birch C. HIV transmissions during seroconversion contribute significantly to new infections in men who have sex with men in Australia. AIDS Res Hum Retroviruses. 2012 May;28(5):460–464. doi: 10.1089/AID.2011.0137. [DOI] [PubMed] [Google Scholar]
- 33.Frange P, Meyer L, Deveau C, et al. Recent HIV-1 infection contributes to the viral diffusion over the French territory with a recent increasing frequency. PLoS One. 2012;7(2):e31695. doi: 10.1371/journal.pone.0031695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bezemer D, van Sighem A, Lukashov VV, et al. Transmission networks of HIV-1 among men having sex with men in the Netherlands. AIDS. 2010 Jan 16;24(2):271–282. doi: 10.1097/QAD.0b013e328333ddee. [DOI] [PubMed] [Google Scholar]
- 35.Weinhardt LS, Kelly JA, Brondino MJ, et al. HIV transmission risk behavior among men and women living with HIV in 4 cities in the United States. Journal of Acquired Immune Deficiency Syndromes. 2004 Aug 15;36(5):1057–1066. doi: 10.1097/00126334-200408150-00009. [DOI] [PubMed] [Google Scholar]
- 36.Avants SK, Warburton LA, Hawkins KA, Margolin A. Continuation of high-risk behavior by HIV-positive drug users. Treatment implications. Journal of substance abuse treatment. 2000 Jul;19(1):15–22. doi: 10.1016/s0740-5472(99)00092-6. [DOI] [PubMed] [Google Scholar]
- 37.Daar ES. Clinical presentation and diagnosis of primary HIV-1 infection. Current opinion in HIV & AIDS. 2008;3(1):10–15. doi: 10.1097/COH.0b013e3282f2e295. [DOI] [PubMed] [Google Scholar]
- 38.Vergis EN, Mellors JW. Natural history of HIV-1 infection. Infect Dis Clin North Am. 2000 Dec;14(4):809–825. v–vi. doi: 10.1016/s0891-5520(05)70135-5. [DOI] [PubMed] [Google Scholar]
- 39.Lythgoe KA, Fraser C. New insights into the evolutionary rate of HIV-1 at the within-host and epidemiological levels. Proceedings Biological sciences / The Royal Society. 2012 Aug 22;279(1741):3367–3375. doi: 10.1098/rspb.2012.0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alizon S, Fraser C. Within-host and between-host evolutionary rates across the HIV-1 genome. Retrovirology. 2013;10:49. doi: 10.1186/1742-4690-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parrish NF, Gao F, Li H, et al. Phenotypic properties of transmitted founder HIV-1. Proceedings of the National Academy of Sciences of the United States of America. 2013 Apr 23;110(17):6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen MS, Dye C, Fraser C, Miller WC, Powers KA, Williams BG. HIV Treatment as Prevention: Debate and Commentary—Will Early Infection Compromise Treatment-as-Prevention Strategies? PLoS Med. 2012;9(7):e1001232. doi: 10.1371/journal.pmed.1001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oster AM, Pieniazek D, Zhang X, et al. Demographic but not geographic insularity in HIV transmission among young black MSM. AIDS. 2011 Nov 13;25(17):2157–2165. doi: 10.1097/QAD.0b013e32834bfde9. [DOI] [PubMed] [Google Scholar]
- 44.Dennis AM, Hue S, Hurt CB, et al. Phylogenetic insights into regional HIV transmission. AIDS. 2012 Sep 10;26(14):1813–1822. doi: 10.1097/QAD.0b013e3283573244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aldous JL, Pond SK, Poon A, et al. Characterizing HIV Transmission Networks Across the United States. Clin Infect Dis. 2012 Oct;55(8):1135–1143. doi: 10.1093/cid/cis612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kramer MA, Cornelissen M, Paraskevis D, et al. HIV transmission patterns among The Netherlands, Suriname, and The Netherlands Antilles: a molecular epidemiological study. AIDS Res Hum Retroviruses. 2010 Feb;27(2):123–130. doi: 10.1089/aid.2010.0115. [DOI] [PubMed] [Google Scholar]
- 47.Chalmet K, Staelens D, Blot S, et al. Epidemiological study of phylogenetic transmission clusters in a local HIV-1 epidemic reveals distinct differences between subtype B and non-B infections. BMC Infectious Diseases. 2010;10(1):262. doi: 10.1186/1471-2334-10-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Callegaro A, Svicher V, Alteri C, et al. Epidemiological network analysis in HIV-1 B infected patients diagnosed in Italy between 2000 and 2008. Infection, Genetics, and Evolution. 2011 Apr;11(3):624–632. doi: 10.1016/j.meegid.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 49.von Wyl V, Kouyos RD, Yerly S, et al. The role of migration and domestic transmission in the spread of HIV-1 non-B subtypes in Switzerland. Journal of Infectious Diseases. 2011;204(7):1095–1103. doi: 10.1093/infdis/jir491. [DOI] [PubMed] [Google Scholar]
- 50.Yerly S, Junier T, Gayet-Ageron A, et al. The impact of transmission clusters on primary drug resistance in newly diagnosed HIV-1 infection. AIDS. 2009;23(11):1415–1423. doi: 10.1097/QAD.0b013e32832d40ad. [DOI] [PubMed] [Google Scholar]
- 51.Kaye M, Chibo D, Birch C. Phylogenetic investigation of transmission pathways of drug-resistant HIV-1 utilizing pol sequences derived from resistance genotyping. Journal of acquired immune deficiency syndromes (1999) 2008 Sep 1;49(1):9–16. doi: 10.1097/QAI.0b013e318180c8af. [DOI] [PubMed] [Google Scholar]
- 52.Hue S, Gifford RJ, Dunn D, Fernhill E, Pillay D on Behalf of the UK Collaborative Group on HIV Drug Resistance. Demonstration of Sustained Drug-Resistant Human Immunodeficiency Virus Type 1 Lineages Circulating among Treatment-Naive Individuals. Journal of Virology. 2009;83(6):2645–2654. doi: 10.1128/JVI.01556-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ragonnet-Cronin M, Ofner-Agostini M, Merks H, et al. Longitudinal Phylogenetic Surveillance Identifies Distinct Patterns of Cluster Dynamics. Journal of Acquired Immune Deficiency Syndromes. 2010;55(1):102–108. doi: 10.1097/QAI.0b013e3181e8c7b0. [DOI] [PubMed] [Google Scholar]
- 54.Kouyos RD, von Wyl V, Yerly S, et al. Molecular epidemiology reveals long-term changes in HIV type 1 subtype B transmission in Switzerland. J Infect Dis. 2010 May 15;201(10):1488–1497. doi: 10.1086/651951. [DOI] [PubMed] [Google Scholar]
- 55.Ambrosioni J, Junier T, Delhumeau C, et al. Impact of highly active antiretroviral therapy on the molecular epidemiology of newly diagnosed HIV infections. AIDS. 2012 Oct 23;26(16):2079–2086. doi: 10.1097/QAD.0b013e32835805b6. [DOI] [PubMed] [Google Scholar]
- 56.Lewis F, Hughes GJ, Rambaut A, Pozniak A, Leigh Brown AJ. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS medicine. 2008 Mar 18;5(3):e50. doi: 10.1371/journal.pmed.0050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hue S, Pillay D, Clewley JP, Pybus OG. Genetic analysis reveals the complex structure of HIV-1 transmission within defined risk groups. Proceedings of the National Academy of Sciences of the United States of America. 2005 Mar 22;102(12):4425–4429. doi: 10.1073/pnas.0407534102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hughes GJ, Fearnhill E, Dunn D, et al. Molecular phylodynamics of the heterosexual HIV epidemic in the United Kingdom. PLoS pathogens. 2009 Sep;5(9):e1000590. doi: 10.1371/journal.ppat.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leigh Brown AJ, Lycett SJ, Weinert L, Hughes GJ, Fearnhill E, Dunn DT. Transmission network parameters estimated from HIV sequences for a nationwide epidemic. J Infect Dis. 2011 Nov;204(9):1463–1469. doi: 10.1093/infdis/jir550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robinson K, Fyson N, Cohen T, Fraser C, Colijn C. How the dynamics and structure of sexual contact networks shape pathogen phylogenies. PLoS computational biology. 2013 Jun;9(6):e1003105. doi: 10.1371/journal.pcbi.1003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Oliveira T, Shafer RW, Seebregts C. Public database for HIV drug resistance in southern Africa. Nature. 2010;464(7289):673. doi: 10.1038/464673c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robertson DL, Anderson JP, Bradac JA, et al. HIV-1 nomenclature proposal. Science. 2000 Apr 7;288(5463):55–56. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- 63.Lihana RW, Ssemwanga D, Abimiku A, Ndembi N. Update on HIV-1 diversity in Africa: a decade in review. AIDS Rev. 2012 Apr-Jun;14(2):83–100. [PubMed] [Google Scholar]
- 64.Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global trends in molecular epidemiology of HIV-1 during 2000-2007. AIDS. 2011 Mar 13;25(5):679–689. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salemi M. Toward a robust monitoring of HIV subtypes distribution worldwide. AIDS. 2011 Mar 13;25(5):713–714. doi: 10.1097/QAD.0b013e32834543e7. [DOI] [PubMed] [Google Scholar]
- 66.Dalai SC, de Oliveira T, Harkins GW, et al. Evolution and molecular epidemiology of subtype C HIV-1 in Zimbabwe. AIDS. 2009 Sep 16; doi: 10.1097/QAD.0b013e3283320ef3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gray RR, Tatem AJ, Lamers S, et al. Spatial phylodynamics of HIV-1 epidemic emergence in east Africa. AIDS. 2009 Sep 10;23(14):F9–F17. doi: 10.1097/QAD.0b013e32832faf61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Delatorre EO, Bello G. Phylodynamics of HIV-1 subtype C epidemic in east Africa. PLoS One. 2012;7(7):e41904. doi: 10.1371/journal.pone.0041904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Afonso JM, Morgado MG, Bello G. Evidence of multiple introductions of HIV-1 subtype C in Angola. Infect Genet Evol. 2012 Oct;12(7):1458–1465. doi: 10.1016/j.meegid.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 70.Bello G, Afonso JM, Morgado MG. Phylodynamics of HIV-1 subtype F1 in Angola, Brazil and Romania. Infect Genet Evol. 2012 Jul;12(5):1079–1086. doi: 10.1016/j.meegid.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 71.Esbjornsson J, Mild M, Mansson F, Norrgren H, Medstrand P. HIV-1 molecular epidemiology in Guinea-Bissau, West Africa: origin, demography and migrations. PLoS One. 2011;6(2):e17025. doi: 10.1371/journal.pone.0017025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Faria NR, Suchard MA, Abecasis A, et al. Phylodynamics of the HIV-1 CRF02_AG clade in Cameroon. Infect Genet Evol. 2012 Mar;12(2):453–460. doi: 10.1016/j.meegid.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Silva TI, van Tienen C, Onyango C, et al. Population dynamics of HIV-2 in rural West Africa: comparison with HIV-1 and ongoing transmission at the heart of the epidemic. AIDS. 2013 Jan 2;27(1):125–134. doi: 10.1097/QAD.0b013e32835ab12c. [DOI] [PubMed] [Google Scholar]
- 74.Lemey P, Pybus OG, Rambaut A, et al. The molecular population genetics of HIV-1 group O. Genetics. 2004 Jul;167(3):1059–1068. doi: 10.1534/genetics.104.026666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hankins CA, de Zalduondo BO. Combination prevention: a deeper understanding of effective HIV prevention. AIDS. 2010;24:S70–S80. doi: 10.1097/1001.aids.0000390709.0000304255.fd. [DOI] [PubMed] [Google Scholar]
- 76.Tanser F, Barnighausen T, Cooke GS, Newell ML. Localized spatial clustering of HIV infections in a widely disseminated rural South African epidemic. Int J Epidemiol. 2009 Aug;38(4):1008–1016. doi: 10.1093/ije/dyp148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tatem AJ, Hemelaar J, Gray RR, Salemi M. Spatial accessibility and the spread of HIV-1 subtypes and recombinants. AIDS. 2012 Nov 28;26(18):2351–2360. doi: 10.1097/QAD.0b013e328359a904. [DOI] [PubMed] [Google Scholar]
- 78.Hue S, Hassan AS, Nabwera H, et al. HIV type 1 in a rural coastal town in Kenya shows multiple introductions with many subtypes and much recombination. AIDS Res Hum Retroviruses. 2012 Feb;28(2):220–224. doi: 10.1089/aid.2011.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Grabowski MK, Lessler J, Redd AD, et al. The Role of Viral Introductions in Sustaining Community-Based HIV Epidemics in Rural Uganda: Evidence from Spatial Clustering, Phylogenetics, and Egocentric Transmission Models. PLoS Med. 2014 Mar;11(3):e1001610. doi: 10.1371/journal.pmed.1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carnegie NB, Wang R, Novitsky V, De Gruttola V. Linkage of viral sequences among HIV-infected village residents in Botswana: estimation of linkage rates in the presence of missing data. PLoS computational biology. 2014 Jan;10(1):e1003430. doi: 10.1371/journal.pcbi.1003430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hogg RS, Yip B, Chan KJ, et al. Rates of Disease Progression by Baseline CD4 Cell Count and Viral Load After Initiating Triple-Drug Therapy. JAMA. 2001;286(20):2568–2577. doi: 10.1001/jama.286.20.2568. [DOI] [PubMed] [Google Scholar]
- 82.Egger M, May M, Chene G, et al. Prognosis of HIV-1-infected patients starting highly active antiretroviral therapy: a collaborative analysis of prospective studies. Lancet. 2002;360(9327):119. doi: 10.1016/s0140-6736(02)09411-4. [DOI] [PubMed] [Google Scholar]
- 83.Gilks CF, Crowley S, Ekpini R, et al. The WHO public-health approach to antiretroviral treatment against HIV in resource-limited settings. Lancet. 2006 Aug 5;368(9534):505–510. doi: 10.1016/S0140-6736(06)69158-7. [DOI] [PubMed] [Google Scholar]
- 84.Gupta RK, Jordan MR, Sultan BJ, et al. Global trends in antiretroviral resistance in treatment-naive individuals with HIV after rollout of antiretroviral treatment in resource-limited settings: a global collaborative study and meta-regression analysis. Lancet. 2012 Oct 6;380(9849):1250–1258. doi: 10.1016/S0140-6736(12)61038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hamers RL, Sigaloff KC, Kityo C, Mugyenyi P, de Wit TF. HIV-1 drug resistance in antiretroviral-naive patients in sub-Saharan Africa. Lancet Infect Dis. 2013 Mar;13(3):196–197. doi: 10.1016/S1473-3099(13)70012-4. [DOI] [PubMed] [Google Scholar]
- 86.Stadeli KM, Richman DD. Rates of emergence of HIV drug resistance in resource-limited settings: a systematic review. Antivir Ther. 2013;18(1):115–123. doi: 10.3851/IMP2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cambiano V, Bertagnolio S, Jordan MR, Lundgren JD, Phillips A. Transmission of Drug Resistant HIV and Its Potential Impact on Mortality and Treatment Outcomes in Resource-Limited Settings. J Infect Dis. 2013 Jun;207 (Suppl 2):S57–62. doi: 10.1093/infdis/jit111. [DOI] [PubMed] [Google Scholar]
- 88.World Health Organization. Consolidated guidelines on general HIV care and the use of antiretroviral drugs for treating and preventing HIV infection: recommendations for a public health approach. 2013. [PubMed] [Google Scholar]
- 89.Cambiano V, Bertagnolio S, Jordan MR, et al. Predicted levels of HIV drug resistance in South Africa: potential impact of expanding diagnosis, retention and eligibility criteria for antiretroviral therapy initiation. Aids. doi: 10.1097/QAD.0000000000000082. In Press. [DOI] [PubMed] [Google Scholar]
- 90.Bartolo I, Casanovas J, Bastos R, et al. HIV-1 genetic diversity and transmitted drug resistance in health care settings in Maputo, Mozambique. J Acquir Immune Defic Syndr. 2009 Jul 1;51(3):323–331. doi: 10.1097/qai.0b013e3181a24906. [DOI] [PubMed] [Google Scholar]
- 91.Yaotse DA, Nicole V, Roch NF, Mireille PD, Eric D, Martine P. Genetic characterization of HIV-1 strains in Togo reveals a high genetic complexity and genotypic drug-resistance mutations in ARV naive patients. Infect Genet Evol. 2009 Jul;9(4):646–652. doi: 10.1016/j.meegid.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 92.Caron M, Lekana-Douki SE, Makuwa M, et al. Prevalence, genetic diversity and antiretroviral drugs resistance-associated mutations among untreated HIV-1-infected pregnant women in Gabon, central Africa. BMC Infect Dis. 2012;12:64. doi: 10.1186/1471-2334-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sigaloff KC, Mandaliya K, Hamers RL, et al. Short communication: High prevalence of transmitted antiretroviral drug resistance among newly HIV type 1 diagnosed adults in Mombasa, Kenya. AIDS Res Hum Retroviruses. 2012 Sep;28(9):1033–1037. doi: 10.1089/AID.2011.0348. [DOI] [PubMed] [Google Scholar]
- 94.Nazziwa J, Njai HF, Ndembi N, et al. Short Communication: HIV Type 1 Transmitted Drug Resistance and Evidence of Transmission Clusters Among Recently Infected Antiretroviral-Naive Individuals from Ugandan Fishing Communities of Lake Victoria. AIDS Res Hum Retroviruses. 2013 May;29(5):788–795. doi: 10.1089/aid.2012.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bessong PO, Mphahlele J, Choge IA, et al. Resistance mutational analysis of HIV type 1 subtype C among rural South African drug-naive patients prior to large-scale availability of antiretrovirals. AIDS Res Hum Retroviruses. 2006 Dec;22(12):1306–1312. doi: 10.1089/aid.2006.22.1306. [DOI] [PubMed] [Google Scholar]
- 96.Deho L, Walwema R, Cappelletti A, et al. Subtype assignment and phylogenetic analysis of HIV type 1 strains in patients from Swaziland. AIDS Res Hum Retroviruses. 2008 Feb;24(2):323–325. doi: 10.1089/aid.2007.0233. [DOI] [PubMed] [Google Scholar]
- 97.Jacobs GB, Laten A, van Rensburg EJ, et al. Phylogenetic diversity and low level antiretroviral resistance mutations in HIV type 1 treatment-naive patients from Cape Town, South Africa. AIDS Res Hum Retroviruses. 2008 Jul;24(7):1009–1012. doi: 10.1089/aid.2008.0028. [DOI] [PubMed] [Google Scholar]
- 98.Bussmann H, de la Hoz Gomez F, Roels TH, et al. Prevalence of transmitted HIV drug resistance in Botswana: lessons learned from the HIVDR-Threshold Survey conducted among women presenting for routine antenatal care as part of the 2007 national sentinel survey. AIDS Res Hum Retroviruses. 2011 Apr;27(4):365–372. doi: 10.1089/aid.2009.0299. [DOI] [PubMed] [Google Scholar]
- 99.Nwobegahay J, Bessong P, Masebe T, et al. Prevalence of drug-resistant mutations in newly diagnosed drug-naive HIV-1-infected individuals in a treatment site in the waterberg district, limpopo province. S Afr Med J. 2011 May;101(5):335–337. doi: 10.7196/samj.4391. [DOI] [PubMed] [Google Scholar]
- 100.Price MA, Wallis CL, Lakhi S, et al. Transmitted HIV type 1 drug resistance among individuals with recent HIV infection in East and Southern Africa. AIDS Res Hum Retroviruses. 2011 Jan;27(1):5–12. doi: 10.1089/aid.2010.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chemaitelly H, Shelton JD, Hallett TB, Abu-Raddad LJ. Only a fraction of new HIV infections occur within identifiable stable discordant couples in sub-Saharan Africa. AIDS. 2013 Jan 14;27(2):251–260. doi: 10.1097/QAD.0b013e32835ad459. [DOI] [PubMed] [Google Scholar]
- 102.Morris M. Barking up the wrong evidence tree. Comment on Lurie & Rosenthal, “Concurrent partnerships as a driver of the HIV epidemic in sub-Saharan Africa? The evidence is limited”. AIDS Behav. 2010 Feb;14(1):31–33. doi: 10.1007/s10461-009-9639-6. discussion 34-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lurie M, Rosenthal S, Williams B. Concurrency driving the African HIV epidemics: where is the evidence? Lancet. 2009 Oct 24;374(9699):1420. doi: 10.1016/S0140-6736(09)61860-2. author reply 1420-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ndiaye HD, Toure-Kane C, Vidal N, et al. Surprisingly High Prevalence of Subtype C and Specific HIV-1 Subtype/CRF Distribution in Men Having Sex With Men in Senegal. JAIDS Journal of Acquired Immune Deficiency Syndromes. 2009;52(2):249–252. doi: 10.1097/QAI.0b013e3181af70a4. 210.1097/QAI.1090b1013e3181af1070a1094. [DOI] [PubMed] [Google Scholar]
- 105.Tovanabutra S, Sanders EJ, Graham SM, et al. Evaluation of HIV type 1 strains in men having sex with men and in female sex workers in Mombasa, Kenya. AIDS Res Hum Retroviruses. 2010 Feb;26(2):123–131. doi: 10.1089/aid.2009.0115. [DOI] [PubMed] [Google Scholar]
- 106.Ssemwanga D, Ndembi N, Lyagoba F, et al. HIV type 1 subtype distribution, multiple infections, sexual networks, and partnership histories in female sex workers in Kampala, Uganda. AIDS Res Hum Retroviruses. 2012 Apr;28(4):357–365. doi: 10.1089/aid.2011.0024. [DOI] [PubMed] [Google Scholar]
- 107.Jennes W, Kyongo JK, Vanhommerig E, et al. Molecular epidemiology of HIV-1 transmission in a cohort of HIV-1 concordant heterosexual couples from Dakar, Senegal. PLoS One. 2012;7(5):e37402. doi: 10.1371/journal.pone.0037402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ng M, Gakidou E, Levin-Rector A, Khera A, Murray CJ, Dandona L. Assessment of population-level effect of Avahan, an HIV-prevention initiative in India. Lancet. 2011 Nov 5;378(9803):1643–1652. doi: 10.1016/S0140-6736(11)61390-1. [DOI] [PubMed] [Google Scholar]
- 109.Boily MC, Pickles M, Lowndes CM, et al. Positive impact of a large-scale HIV prevention program among female sex workers and clients in Karnataka state, India. AIDS. 2013 Mar 3; doi: 10.1097/QAD.0b013e32835fba81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Campbell MS, Mullins JI, Hughes JP, et al. Viral linkage in HIV-1 seroconverters and their partners in an HIV-1 prevention clinical trial. PLoS One. 2011;6(3):e16986. doi: 10.1371/journal.pone.0016986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Eshleman SH, Hudelson SE, Redd AD, et al. Analysis of genetic linkage of HIV from couples enrolled in the HIV Prevention Trials Network 052 trial. J Infect Dis. 2011 Dec 15;204(12):1918–1926. doi: 10.1093/infdis/jir651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.van Houdt R, Bruisten SM, Koedijk FD, et al. Molecular epidemiology of acute hepatitis B in the Netherlands in 2004: nationwide survey. J Med Virol. 2007 Jul;79(7):895–901. doi: 10.1002/jmv.20820. [DOI] [PubMed] [Google Scholar]
- 113.Hahne S, van Houdt R, Koedijk F, et al. Selective hepatitis B virus vaccination has reduced hepatitis B virus transmission in the Netherlands. PLoS One. 2013;8(7):e67866. doi: 10.1371/journal.pone.0067866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Volz EM, Kosakovsky Pond SL, Ward MJ, Leigh Brown AJ, Frost SD. Phylodynamics of infectious disease epidemics. Genetics. 2009 Dec;183(4):1421–1430. doi: 10.1534/genetics.109.106021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wertheim JO, Kosakovsky Pond SL, Little SJ, De Gruttola V. Using HIV transmission networks to investigate community effects in HIV prevention trials. PLoS One. 2011;6(11):e27775. doi: 10.1371/journal.pone.0027775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Grenfell BT, Pybus OG, Gog JR, et al. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004 Jan 16;303(5656):327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- 117.Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005 May;22(5):1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- 118.Frost SD, Volz EM. Viral phylodynamics and the search for an ‘effective number of infections’. Philos Trans R Soc Lond B Biol Sci. 2010 Jun 27;365(1548):1879–1890. doi: 10.1098/rstb.2010.0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.de Silva E, Ferguson NM, Fraser C. Inferring pandemic growth rates from sequence data. JR Soc Interface. 2012;9(73):1797–1808. doi: 10.1098/rsif.2011.0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.O’Dea EB, Wilke CO. Contact heterogeneity and phylodynamics: how contact networks shape parasite evolutionary trees. Interdiscip Perspect Infect Dis. 2011;2011:238743. doi: 10.1155/2011/238743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Volz EM, Koopman JS, Ward MJ, Brown AL, Frost SD. Simple epidemiological dynamics explain phylogenetic clustering of HIV from patients with recent infection. PLoS Comput Biol. 2012;8(6):e1002552. doi: 10.1371/journal.pcbi.1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Frost SD, Volz EM. Modelling tree shape and structure in viral phylodynamics. Philos Trans R Soc Lond B Biol Sci. 2013 Mar 19;368(1614):20120208. doi: 10.1098/rstb.2012.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Leventhal GE, Kouyos R, Stadler T, et al. Inferring epidemic contact structure from phylogenetic trees. PLoS Comput Biol. 2012;8(3):e1002413. doi: 10.1371/journal.pcbi.1002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Magiorkinis G, Sypsa V, Magiorkinis E, et al. Integrating phylodynamics and epidemiology to estimate transmission diversity in viral epidemics. PLoS Comput Biol. 2013;9(1):e1002876. doi: 10.1371/journal.pcbi.1002876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ratmann O, Donker G, Meijer A, Fraser C, Koelle K. Phylodynamic inference and model assessment with approximate bayesian computation: influenza as a case study. PLoS Comput Biol. 2012;8(12):e1002835. doi: 10.1371/journal.pcbi.1002835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Volz EM. Complex population dynamics and the coalescent under neutrality. Genetics. 2012 Jan;190(1):187–201. doi: 10.1534/genetics.111.134627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rasmussen DA, Ratmann O, Koelle K. Inference for nonlinear epidemiological models using genealogies and time series. PLoS computational biology. 2011 Aug;7(8):e1002136. doi: 10.1371/journal.pcbi.1002136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Leventhal GE, Gunthard HF, Bonhoeffer S, Stadler T. Using an epidemiological model for phylogenetic inference reveals density dependence in HIV transmission. Molecular biology and evolution. 2014 Jan;31(1):6–17. doi: 10.1093/molbev/mst172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kuhnert D, Stadler T, Vaughan TG, Drummond AJ. Simultaneous reconstruction of evolutionary history and epidemiological dynamics from viral sequences with the birth-death SIR model. Journal of the Royal Society, Interface / the Royal Society. 2014 May 6;11(94):20131106. doi: 10.1098/rsif.2013.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Stadler T. On incomplete sampling under birth-death models and connections to the sampling-based coalescent. Journal of theoretical biology. 2009 Nov 7;261(1):58–66. doi: 10.1016/j.jtbi.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 131.Gall A, Ferns B, Morris C, et al. Universal amplification, next-generation sequencing, and assembly of HIV-1 genomes. J Clin Microbiol. 2012 Dec;50(12):3838–3844. doi: 10.1128/JCM.01516-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Novitsky V, Wang R, Logan A, Moyo S. The Role of HIV-1 Viral Linkage and Viral Load in Treatment-as-Prevention Studies. Paper presented at: HIV Dynamics and Evolution; 2012; Asheville, NC, USA. [Google Scholar]
- 133.Stadler T, Kuhnert D, Bonhoeffer S, Drummond AJ. Birth-death skyline plot reveals temporal changes of epidemic spread in HIV and hepatitis C virus (HCV) Proceedings of the National Academy of Sciences of the United States of America. 2013 Jan 2;110(1):228–233. doi: 10.1073/pnas.1207965110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chevenet F, Jung M, Peeters M, de Oliveira T, Gascuel O. Searching for virus phylotypes. Bioinformatics. 2013 Mar 1;29(5):561–570. doi: 10.1093/bioinformatics/btt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Slatkin M, Maddison WP. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics. 1989 Nov;123(3):603–613. doi: 10.1093/genetics/123.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Osrin D, Azad K, Fernandez A, et al. Ethical challenges in cluster randomized controlled trials: experiences from public health interventions in Africa and Asia. Bull World Health Organ. 2009 Oct;87(10):772–779. doi: 10.2471/BLT.08.051060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.FigTree v 1.4 [computer program] 2009. [Google Scholar]
- 138.Redd AD, Collinson-Streng A, Martens C, et al. Identification of HIV superinfection in seroconcordant couples in Rakai, Uganda, by use of next-generation deep sequencing. J Clin Microbiol. 2011 Aug;49(8):2859–2867. doi: 10.1128/JCM.00804-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kraft CS, Basu D, Hawkins PA, et al. Timing and source of subtype-C HIV-1 superinfection in the newly infected partner of Zambian couples with disparate viruses. Retrovirology. 2012;9:22. doi: 10.1186/1742-4690-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.González-Alba JM, Holguín Á, Garcia R, et al. Molecular Surveillance of HIV-1 in Madrid, Spain: a Phylogeographic Analysis. Journal of Virology. 2011 Oct 15;85(20):10755–10763. doi: 10.1128/JVI.00454-11. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gifford RJ, de Oliveira T, Rambaut A, et al. Phylogenetic surveillance of viral genetic diversity and the evolving molecular epidemiology of human immunodeficiency virus type 1. Journal of Virology. 2007 Dec;81(23):13050–13056. doi: 10.1128/JVI.00889-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Boily MC, Masse B, Alsallaq R, et al. HIV treatment as prevention: considerations in the design, conduct, and analysis of cluster randomized controlled trials of combination HIV prevention. PLoS Med. 2012;9(7):e1001250. doi: 10.1371/journal.pmed.1001250. [DOI] [PMC free article] [PubMed] [Google Scholar]