SpaD, a backbone-pilin fragment from L. rhamnosus GG, was crystallized and X-ray diffraction data were collected to a resolution of 2.0 Å. In-house iodide-derivative data were collected to 2.2 Å resolution for phase determination by the single-wavelength anomalous dispersion method.
Keywords: Lactobacillus rhamnosus GG, SpaFED pili, fimbria, adhesion, in-house iodide SAD phasing, limited proteolysis
Abstract
SpaD is the predicted backbone-pilin subunit of the SpaFED pilus, whose loci are encoded by the fimbrial spaFED operon in Lactobacillus rhamnosus GG, a Gram-positive gut-adapted commensal strain with perceived probiotic benefits. In this study, soluble recombinant SpaD protein was overproduced in Escherichia coli and then purified by Ni2+-chelating affinity and gel-filtration chromatography. After limited proteolysis with α-chymotrypsin, good-quality crystals of SpaD were obtained which diffracted beyond 2.0 Å resolution. These crystals belonged to the orthorhombic space group P212121, with unit-cell parameters a = 50.11, b = 83.27, c = 149.65 Å. For phasing, sodium iodide-derivatized crystals were prepared using the halide quick-soaking method and diffraction data were collected in-house to a resolution of 2.2 Å. An interpretable electron-density map was successfully obtained using single-wavelength anomalous diffraction (SAD).
1. Introduction
Until very recently, studies of sortase-assembled pili in Gram-positive bacteria have mainly dealt with a variety of different pathogenic strains. These pili are protein appendages that contain two or three subunits and emanate from the cell surface. They take part in cellular adhesion and colonization processes, and as such are considered to be key virulence factors during host pathogenesis (for reviews, see Kang & Baker, 2012 ▶; Mandlik et al., 2008 ▶; Vengadesan & Narayana, 2011 ▶). With the discovery of pili in nonpathogenic lactic acid bacteria (LAB; Kankainen et al., 2009 ▶; Gorbach, 2000 ▶), this particular view of surface piliation has been expanded to also include a role as a niche-adaptation factor.
Pilus-like structures have been confirmed as sortase-catalyzed assemblies in the gut-adapted probiotic Lactobacillus rhamnosus strain GG (Kankainen et al., 2009 ▶). These pili (called SpaCBA) are encoded by the fimbrial spaCBA operon and have a backbone structure consisting of SpaA pilins. Additionally, two ancillary pilins with differing locations and functions are incorporated into this backbone structure, with SpaC at the pilus tip for adhesion and SpaB at the pilus base for anchoring (Reunanen et al., 2012 ▶). The SpaCBA pilus has been shown to have substrate-binding specificities for mucus (Kankainen et al., 2009 ▶; von Ossowski et al., 2010 ▶) and collagen (Tripathi et al., 2013 ▶). These properties apparently help to extend the transient colonization of the gut by L. rhamnosus GG cells. Presumably, this is an advantage over other nonpiliated probiotic bacteria.
Aside from the spaCBA operon, the L. rhamnosus GG genome also contains loci for another type of pilus structure (Kankainen et al., 2009 ▶; Kant et al., 2014 ▶). The corresponding fimbrial operon (called spaFED) encodes pilin proteins that are presumed to form the pilus backbone (SpaD), the pilus tip (SpaF) and the base (SpaE), as well as a putative sortase enzyme (SrtC2) that is required for pilus assembly. Recombinant SpaF has been shown to bind intestinal mucus (von Ossowski et al., 2010 ▶). However, the genes associated with the fimbrial spaFED operon are not constitutively expressed but rather are seemingly silent (Reunanen et al., 2012 ▶). Thus, the natural role of the SpaFED pilus remains hypothetical, not only in L. rhamnosus GG but also in other strains carrying the spaFED operon and in two taxonomically related Lactobacillus species (Kant et al., 2014 ▶; Reunanen et al., 2012 ▶). However, L. rhamnosus GG SpaFED pili can readily be produced as an assembled structure in recombinant L. lactis (Rintahaka et al., 2014 ▶).
As part of our ongoing structural investigations of piliation in beneficial gut commensal bacteria (Singh et al., 2013 ▶), here we report the purification, crystallization and preliminary X-ray diffraction analysis of SpaD, the backbone-pilin subunit of the pilus encoded by the L. rhamnosus GG spaFED operon. The full-length protein yielded only poor-quality crystals. Diffraction-quality crystals were successfully grown only after proteolytic cleavage of a 16 kDa fragment at the N-terminus.
2. Materials and methods
2.1. Macromolecule production
Details of the cloning and expression of the L. rhamnosus GG spaD locus as a recombinant protein in Escherichia coli have been reported elsewhere (von Ossowski et al., 2010 ▶). A protein construct consisting of residues 36–495 of SpaD (UniProtKB code C7T6S8; 517 residues) lacking the N-terminal signal peptide (residues 1–35) and C-terminal LPXTG sorting signal (residues 496–517) was generated in the E. coli BL21 (DE3) pLysS strain (Table 1 ▶). Purification of recombinant SpaD by Ni2+-chelating affinity chromatography and gel-filtration chromatography was performed at 277 K using an ÄKTA FPLC system (GE Healthcare). The cell-free lysate was applied onto a 5 ml NiCl2-charged HiTrap Chelating HP column (GE Healthcare) which had previously been equilibrated with buffer A (50 mM NaH2PO4 pH 7.4, 150 mM NaCl). Any loosely bound proteins were rinsed away with buffer A containing 10 mM imidazole. SpaD protein was eluted from the column resin using a linear gradient of buffer B (50 mM NaH2PO4 pH 7.4, 150 mM NaCl, 400 mM imidazole). Eluted fractions were monitored by SDS–PAGE for SpaD proteins. SpaD-containing fractions were pooled and dialyzed overnight at 277 K against 20 mM HEPES pH 7.5 buffer containing 150 mM NaCl and 1 mM EDTA. Dialyzed SpaD protein was concentrated using an Amicon ultrafiltration device fitted with a 10 kDa molecular-weight cutoff and then purified further on a Sephacryl 200 26/60 gel-filtration column (GE Healthcare) using the dialysis buffer mentioned above.
Table 1. Macromolecule-production information.
| Source organism | L. rhamnosus GG (ATCC 53103) |
| DNA source | L. rhamnosus GG (ATCC 53103) |
| Forward primer† | 5-ACCCGTACAGAATTCGACAACGACTGTG |
| Reverse primer‡ | 5-GTCCGATTCCGCCCTCGAGCGGCAATAATTG |
| Cloning vector | pET-28b+ (Novagen) |
| Expression vector | pET-28b+ (Novagen) |
| Expression host | E. coli BL21(DE3)/pLysS |
| Complete amino-acid sequence of the construct produced§ | MGRDPNSTTTVDFTLHKIEQTSDEQIQNTGHDLGLTGRKPVQGAQFKIFNVTDAFYQLLENHDKTTAASMISQNLGQYVNLQDPNAATVTTDADGLAAFKGLAAKTNGRHSVYAFHEAVTPQPYQKAADMIVSLPVRQDDGSDLTNIHLYPKDSLVTKNLTEINEQAVATKDLHDVAVGDVLTYQVQFQIPHDIGALADHSQDTFKYNQFKVLDYMTKEGLTFKALTAITVDGQDILKALTGKMAFMSSNDAAWQQTHNYPFGFELDFLGGTDPDAVRNLLTQYAGKRVTVAYTGIVNEKMIPDQKVGNTAEVSFDPDSKITVNGPEIQTGGIRFFKHEAGSSKSLANATFILQRMNGNVREYAVLEGVNGMAGTYQPTKITWTTNQDAATRLKTSGAETANLTIQGLLPGRYTLVETAAPEGYEILDPTTDFEVIAGTWGTKTIRIANTPVNQLLPLEHHHHHH |
The EcoRI site is underlined.
The XhoI site is underlined.
The cloning artifact is underlined. The N-terminal residues of the 34kDa fragment are shown in italics.
For limited proteolysis, the purified full-length protein in 20 mM HEPES pH 7.5 buffer containing 150 mM NaCl was incubated with α-chymotrypsin at a ratio of 1:100 (α-chymotrypsin:SpaD) at 310 K for 20 h. The protein was subsequently purified by gel filtration after adding 1 mM phenylmethylsulfonyl fluoride (PMSF). N-terminal sequencing of the digested product was performed using an in-house protein-sequencer facility (PPSQ-33A, Shimadzu Corporation). The intact mass of purified proteins were determined by LC-ESI-MS on Shimadzu Ultra HPLC coupled to an AB SCIEX TripleTOF 5600 system equipped with a dual electrospray ion source.
2.2. Crystallization
For crystallization screening trials, purified recombinant SpaD protein was concentrated to ∼50 mg ml−1 in 20 mM HEPES pH 7.5 buffer containing 150 mM NaCl and 1 mM EDTA. Roughly 600 different crystallization screening conditions were initially tested with the assistance of a Mosquito Crystal automated liquid-dispensing system (TTP Labtech). After about one week of incubation, SpaD protein crystals were observed under several different conditions, and some of the conditions were further optimized manually to obtain large crystals. One of the conditions is listed in Table 2 ▶. Single crystals of the full-length protein of diffractable size were tested on the in-house X-ray source. They yielded anisotropic and streaky diffraction patterns, which precluded data processing (data not shown). Therefore, limited proteolysis was attempted; this has become a standard strategy for modifying crystallization behaviour and increasing the likelihood of crystallization by removing flexible regions (Vengadesan et al., 2010 ▶). After limited proteolysis with α-chymotrypsin, improved diffraction-quality crystals were grown in two conditions by the hanging-drop vapour-diffusion method at 295 K (Table 2 ▶). Here, equal volumes (1 µl) of protein solution and reservoir solution [either 0.25 M ammonium chloride, 22%(w/v) PEG 3350 or 0.17 M lithium sulfate, 25%(w/v) PEG 3350] were mixed and equilibrated against 1 ml reservoir solution for 2 d. Crystals from the condition described in Table 2 ▶ were used in the diffraction experiments.
Table 2. Crystallization.
| Full length | Chymotrypsin-treated | |
|---|---|---|
| Method | Hanging-drop vapour diffusion | Hanging-drop vapour diffusion |
| Plate type | VDX plate (Hampton Research) | VDX plate (Hampton Research) |
| Temperature (K) | 295 | 295 |
| Protein concentration (mgml1) | 50 | 35 |
| Buffer composition of protein solution | 20mM HEPES pH 7.5, 150mM NaCl, 1mM EDTA | 20mM HEPES pH 7.5, 150mM NaCl |
| Composition of reservoir solution | 100mM HEPES pH 7.5, 16% PEG 4000, 250mM ammonium sulfate, 12% 2-propanol | 170mM lithium sulfate, 25% PEG 3350 |
| Volume and ratio of drop | 2l, 1:1 | 2l, 1:1 |
| Volume of reservoir (l) | 1000 | 1000 |
2.3. Data collection and processing
Native X-ray diffraction data sets were collected from crystals of α-chymotrypsin-digested SpaD protein at the in-house facility using a Xenocs GeniX3D Cu HF (high-flux) microbeam X-ray generator operating at 0.6 mA and 50 kV and a MAR 345 image-plate detector (MAR Research). Native data were collected from a single crystal over a range of 180° with 0.5° oscillation steps, a 160 mm crystal-to-detector distance and 10 min exposure time (Table 3 ▶). Crystals in mother liquor supplemented with 30%(v/v) ethylene glycol as a cryoprotectant were flash-cooled in a nitrogen-gas stream at 100 K. Diffraction data were indexed and integrated using XDS (Kabsch, 2010 ▶) and scaled with AIMLESS (Evans & Murshudov, 2013 ▶) from the CCP4 suite (Winn et al., 2011 ▶).
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Native | NaI derivative | |
|---|---|---|
| Diffraction source | In-house | In-house |
| Wavelength () | 1.5418 | 1.5418 |
| Temperature (K) | 100 | 100 |
| Detector | MAR 345 image plate | MAR 345 image plate |
| Crystal-to-detector distance (mm) | 160 | 180 |
| Rotation range per image () | 0.5 | 0.5 |
| Total rotation range () | 180 | 186 |
| Exposure time per image (s) | 600 | 600 |
| Space group | P212121 | P212121 |
| a, b, c () | 50.1, 83.3, 149.6 | 50.2, 82.9, 149.2 |
| , , () | 90, 90, 90 | 90, 90, 90 |
| Mosaicity () | 0.16 | 0.18 |
| Resolution range () | 47.52.0 (2.112.00) | 47.62.2 (2.322.20) |
| Total No. of reflections | 304709 | 232743 |
| No. of unique reflections | 42983 | 31851 |
| Completeness (%) | 99.9 (99.5) | 98.8 (96.4) |
| Multiplicity | 7.1 (7.0) | 7.3 (6.9) |
| I/(I)† | 21.5 (8.8) | 25.4 (11.1) |
| R meas | 0.07 (0.25) | 0.07 (0.20) |
| Overall B factor from Wilson plot (2) | 18.9 | 28.2 |
| DelAnom correlation between half sets | 0.59 (0.26) |
The crystal diffracted beyond 2.0 resolution, but the data-set resolution is limited to 2.0 by the detector edge.
The halide quick-soaking method (Dauter et al., 2000 ▶) was used to prepare sodium iodide-derivative crystals for SAD phasing. Crystals of α-chymotrypsin-digested SpaD protein were soaked in the cryo-solution supplemented with 400 mM NaI for about 5 min just prior to data collection. The data were processed as described above for the native data, and SAD phase calculations were performed using SHARP/autoSHARP (Vonrhein et al., 2007 ▶). An initial search identified 19 iodide sites with occupancies of greater than 0.2 [correlation coefficient CC(all/weak) = 38.2/20.4; Supplementary Table S1].
3. Results and discussion
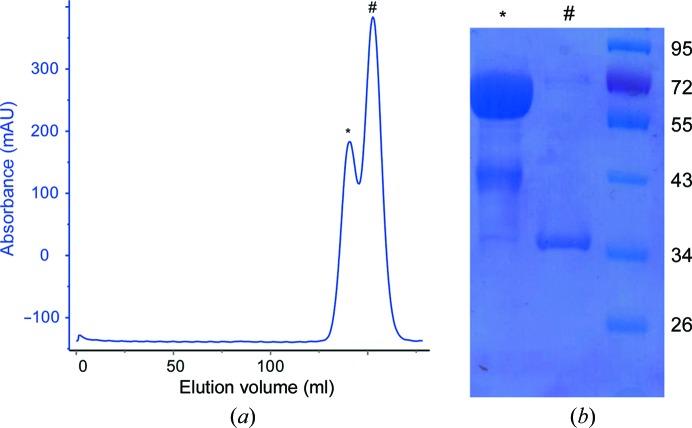
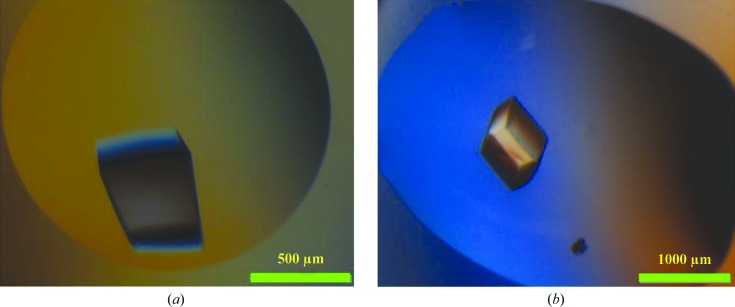
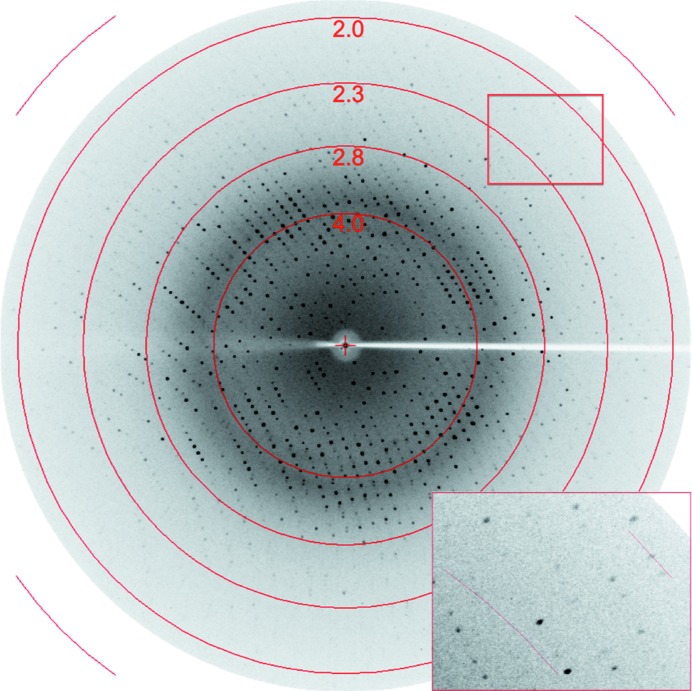
The recombinant full-length L. rhamnosus GG SpaD36–495 pilin and its stable fragment which was derived from limited proteolysis with α-chymotrypsin were both purified (Fig. 1 ▶). As determined by mass spectroscopy, the intact molecular mass of the full-length SpaD protein is 50 120 Da, with that of its chymotrypsin-digested product being 33 894 Da. The first residue of the digested product, as determined from N-terminal sequencing, is the tyrosine at position 178 (178YPKDS182). The theoretical molecular weight of this fragment including the C-terminal His tag is 34 873 Da, which suggests that proteolysis could have removed an approximately 16 kDa fragment at the N-terminus and the His tag at the C-terminus. As mentioned earlier, the recombinant full-length SpaD protein did not yield good diffraction-quality crystals (Fig. 2 ▶ a), whereas its 34 kDa fragment yielded much better crystals (Fig. 2 ▶ b) that diffracted to beyond 2 Å resolution at the home source (Fig. 3 ▶).
Figure 1.
(a) A representative gel-filtration chromatogram of the α-chymotrypsin-digested SpaD product. A Sephacryl 200 26/60 gel-filtration column (GE Healthcare) was used. The two peaks at volumes of 139 ml (*) and 152 ml (#) correspond to SpaD36–495 (∼50 kDa) and its α-chymotrypsin-digested product (∼34 kDa), respectively, as monitored by SDS–PAGE and analyzed by mass-spectrometric analysis. (b) SDS–PAGE of purified full-length SpaD36–495 (*) and its α-chymotrypsin-digested product (#) used for crystallization. The lane on the right contains molecular-weight marker (labelled in kDa).
Figure 2.
(a) Crystal of full-length SpaD. (b) Crystal of chymotrypsin-digested SpaD.
Figure 3.
X-ray diffraction pattern of a single crystal of chymotrypsin-digested SpaD. The inset is a magnification showing diffraction spots at 2.0 Å resolution.
The related backbone pilin structures from Gram-positive pathogenic bacteria (Supplementary Table S1) often consist of tandem immunoglobulin (Ig)-like domains with an autocatalytically generated intradomain isopeptide bond between Lys and Asn/Asp residues (Kang & Baker, 2012 ▶). The backbone pilins form the pilus shaft with the help of the pilin-specific sortase, which covalently links the C-terminus of each pilin to a Lys residue on the next pilin (Mandlik et al., 2008 ▶). These backbone pilins generally have a conserved Lys residue-containing pilin motif (YPKN) in a flexible linker that connects the first and second domains at the N-terminus and a sortase-recognition (LPXTG) motif at the C-terminus for facilitating pilus polymerization (Vengadesan & Narayana, 2011 ▶). The pilin motif-containing linker appears to be susceptible to proteolysis and labile by having a Lys residue that is involved in the intermolecular isopeptide bond during pilus polymerization and a neighbouring Asn/Asp residue that participates in an intramolecular isopeptide bond in the N-terminal (∼16 kDa) domain (Vengadesan & Narayana, 2011 ▶). Hence, the removal of the N-terminal (∼16 kDa) domain from SpaD is likely to have improved the crystal diffraction quality, much as has been observed with some backbone-pilin structures from pathogenic bacteria (GBS80 from Streptococcus agalactiae, BcpA from Bacillus cereus, RrgB from S. pneumoniae and FimA from Actinomyces oris), where the N-terminal domain was either lost during crystallization or removed by limited proteolysis to obtain diffraction-quality crystals (Vengadesan & Narayana, 2011 ▶). This was also evident in the recent structural study of SpaD from Corynebacterium diphtheriae (Kang et al., 2014 ▶).
The α-chymotrypsin-treated SpaD crystals belonged to the orthorhombic space group P212121, with unit-cell parameters a = 50.11, b = 82.27, c = 149.65 Å (Table 3 ▶). The calculated Matthews coefficient V M of 2.3 Å3 Da−1 indicated the presence of two molecules in the asymmetric unit, equivalent to a solvent content of 46% and a molecular mass of 34 kDa. Attempts to obtain the correct phases by molecular replacement were initially unsuccessful, as SpaD has only limited sequence identity (<25%) to any of the related structures (Supplementary Table S1) deposited in the Protein Data Bank (PDB). The best homologous model, with 24% sequence identity, is GBS80, a backbone pilin from S. agalactiae (Vengadesan et al., 2011 ▶). We used the halide quick-soaking method (Dauter et al., 2000 ▶) to prepare sodium iodide-derivatized crystals (Table 3 ▶). NaI-derivative data collected to 2.2 Å resolution at the home source were used for SAD phasing (Supplementary Table S2). An interpretable electron-density map (Supplementary Fig. S1) was obtained from the identified iodide sites with occupancies of greater than 0.2. Model building and refinement are currently under way.
Supplementary Material
Supporting Information.. DOI: 10.1107/S2053230X14027216/hc5185sup1.pdf
Acknowledgments
We thank Dr Dinakar Salunke for his support and critical reviews. Financial support from the Regional Centre for Biotechnology (RCB) and the Department of Biotechnology (DBT) is gratefully acknowledged. We acknowledge the in-house X-ray facility for X-ray diffraction data collection and the central instrumentation facility at RCB for the N-terminal sequencing mass-spectrometric analysis. Funding for the cloning of the recombinant SpaD construct at the University of Helsinki was through an Academy of Finland general research grant (No. 118165) and the Center of Excellence in Microbial Food Safety (CoE-MiFoSa) research program (Grant Nos. 118602 and 141140).
References
- Dauter, Z., Dauter, M. & Rajashankar, K. R. (2000). Acta Cryst. D56, 232–237. [DOI] [PubMed]
- Evans, P. R. & Murshudov, G. N. (2013). Acta Cryst. D69, 1204–1214. [DOI] [PMC free article] [PubMed]
- Gorbach, S. L. (2000). Am. J. Gastroenterol. 95, S2–S4. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kang, H. J. & Baker, E. N. (2012). Curr. Opin. Struct. Biol. 22, 200–207. [DOI] [PubMed]
- Kang, H. J., Paterson, N. G., Kim, C. U., Middleditch, M., Chang, C., Ton-That, H. & Baker, E. N. (2014). Acta Cryst. D70, 1190–1201. [DOI] [PMC free article] [PubMed]
- Kankainen, M. et al. (2009). Proc. Natl Acad. Sci. USA, 106, 17193–17198.
- Kant, R., Rintahaka, J., Yu, X., Sigvart-Mattila, P., Paulin, L., Mecklin, J. P., Saarela, M., Palva, A. & von Ossowski, I. (2014). PLoS One, 9, e102762. [DOI] [PMC free article] [PubMed]
- Mandlik, A., Swierczynski, A., Das, A. & Ton-That, H. (2008). Trends Microbiol. 16, 33–40. [DOI] [PMC free article] [PubMed]
- Ossowski, I. von, Reunanen, J., Satokari, R., Vesterlund, S., Kankainen, M., Huhtinen, H., Tynkkynen, S., Salminen, S., de Vos, W. M. & Palva, A. (2010). Appl. Environ. Microbiol. 76, 2049–2057. [DOI] [PMC free article] [PubMed]
- Reunanen, J., von Ossowski, I., Hendrickx, A. P., Palva, A. & de Vos, W. M. (2012). Appl. Environ. Microbiol. 78, 2337–2344. [DOI] [PMC free article] [PubMed]
- Rintahaka, J., Yu, X., Kant, R., Palva, A. & von Ossowski, I. (2014). PLoS One, 9, e113922. [DOI] [PMC free article] [PubMed]
- Singh, D., von Ossowski, I., Palva, A. & Krishnan, V. (2013). Acta Cryst. F69, 1182–1185. [DOI] [PMC free article] [PubMed]
- Tripathi, P., Beaussart, A., Alsteens, D., Dupres, V., Claes, I., von Ossowski, I., de Vos, W. M., Palva, A., Lebeer, S., Vanderleyden, J. & Dufrêne, Y. F. (2013). ACS Nano, 7, 3685–3697. [DOI] [PubMed]
- Vengadesan, K., Ma, X., Dwivedi, P., Ton-That, H. & Narayana, S. V. L. (2010). Acta Cryst. F66, 1666–1669. [DOI] [PMC free article] [PubMed]
- Vengadesan, K., Ma, X., Dwivedi, P., Ton-That, H. & Narayana, S. V. L. (2011). J. Mol. Biol. 407, 731–743. [DOI] [PMC free article] [PubMed]
- Vengadesan, K. & Narayana, S. V. L. (2011). Protein Sci. 20, 759–772. [DOI] [PMC free article] [PubMed]
- Vonrhein, C., Blanc, E., Roversi, P. & Bricogne, G. (2007). Methods Mol. Biol. 364, 215–230. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information.. DOI: 10.1107/S2053230X14027216/hc5185sup1.pdf