Abstract
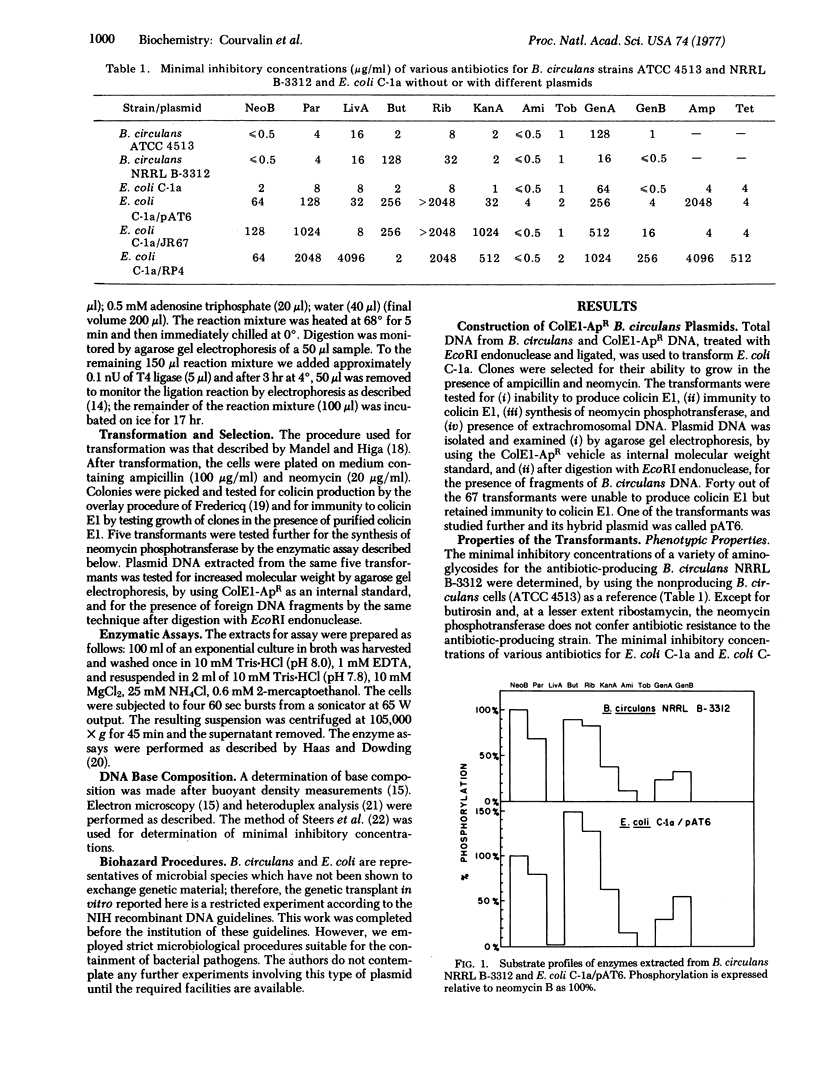
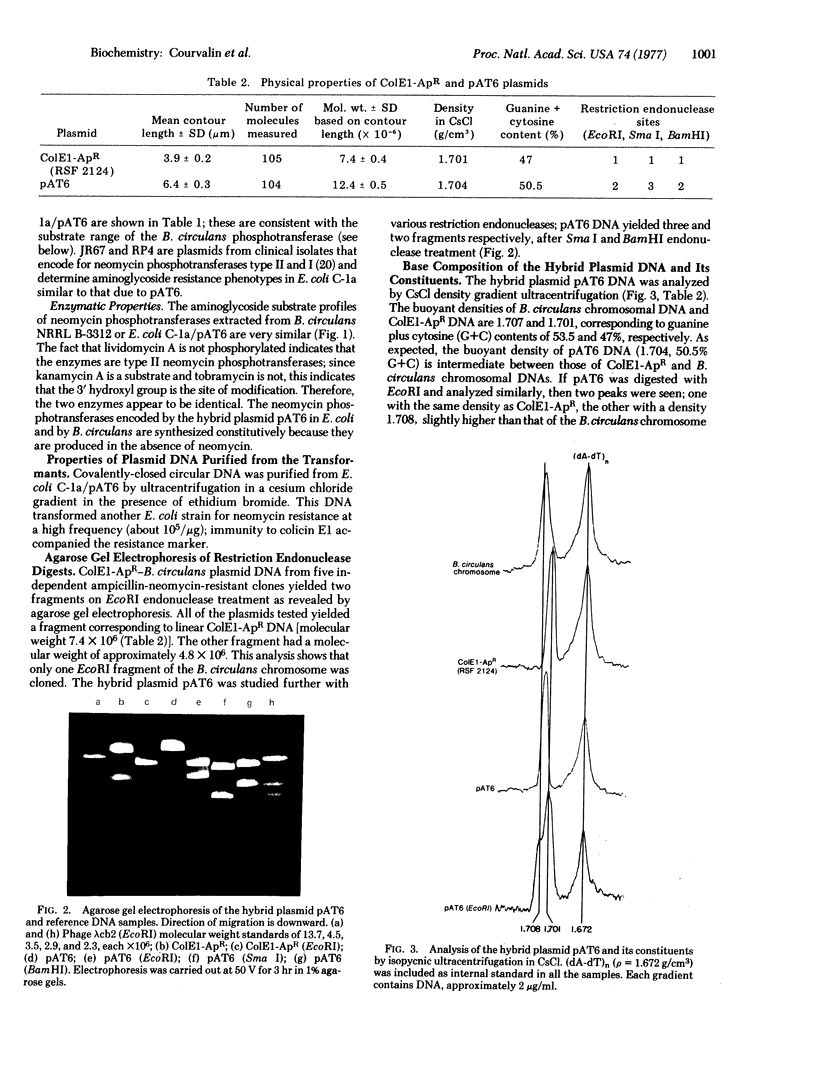
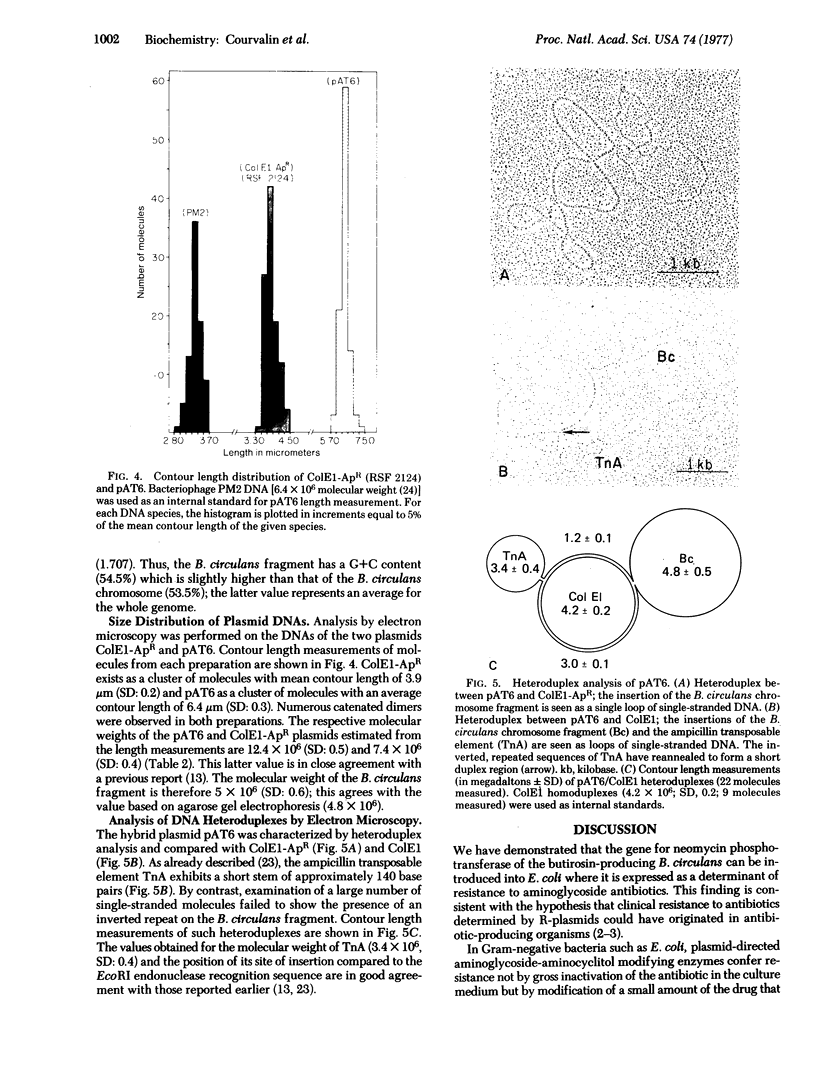
Bacillus circulans NRRL B-3312, a nonpathogenic bacterium that produces the aminoglycoside antibiotic butirosin, is known to contain an aminoglycoside phosphotransferase that is similar to the neomycin phosphotransferases of clinically isolated antibiotic-resistant bacteria. Purified DNAs from B. circulans and the plasmid ColE1-ApR were digested with EcoRI endonuclease and the resulting fragments covalently joined with polynucleotide ligase. The recombined DNA was used to transform E. coli and ampicillin-neomycin resistant colonies were selected. Analysis of several clones indicated that neomycin resistance in the E. coli transformants was due to the presence of the B. circulans phosphotransferase gene. This observation is consistent with the notion that anitbiotic-modifying enzymes from antibiotic-producing organisms may be the sources of antibiotic resistance in plasmid-containing bacteria.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Argoudelis A. D., Coats J. H. Microbial transformation of antibiotics. VI. Acylation of chloramphenicol by Streptomyces coelicolor. J Antibiot (Tokyo) 1971 Mar;24(3):206–208. doi: 10.7164/antibiotics.24.206. [DOI] [PubMed] [Google Scholar]
- Benveniste R., Davies J. Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2276–2280. doi: 10.1073/pnas.70.8.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D. E., Davies J., Allet B., Rochaix J. D. Transposition of R factor genes to bacteriophage lambda. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3628–3632. doi: 10.1073/pnas.72.9.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courvalin P. M., Carlier C., Croissant O., Blangy D. Identification of two plasmids determining resistance to tetracycline and to erythromycin in group D streptococcus. Mol Gen Genet. 1974;132(3):181–192. doi: 10.1007/BF00269391. [DOI] [PubMed] [Google Scholar]
- Datta N., Hedges R. W., Shaw E. J., Sykes R. B., Richmond M. H. Properties of an R factor from Pseudomonas aeruginosa. J Bacteriol. 1971 Dec;108(3):1244–1249. doi: 10.1128/jb.108.3.1244-1249.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. E., Benveniste R. E. Enzymes that inactivate antibiotics in transit to their targets. Ann N Y Acad Sci. 1974 May 10;235(0):130–136. doi: 10.1111/j.1749-6632.1974.tb43262.x. [DOI] [PubMed] [Google Scholar]
- Franklin R. M. Structure and synthesis of bacteriophage PM2, with particular emphasis on the viral lipid bilayer. Curr Top Microbiol Immunol. 1974;(68):107–159. doi: 10.1007/978-3-642-66044-3_5. [DOI] [PubMed] [Google Scholar]
- Haas M. J., Dowding J. E. Aminoglycoside-modifying enzymes. Methods Enzymol. 1975;43:611–628. doi: 10.1016/0076-6879(75)43124-x. [DOI] [PubMed] [Google Scholar]
- Heffron F., Rubens C., Falkow S. Translocation of a plasmid DNA sequence which mediates ampicillin resistance: molecular nature and specificity of insertion. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3623–3627. doi: 10.1073/pnas.72.9.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Sano H., Feix G. Ribonucleic acid ligase activity of deoxyribonucleic acid ligase from phage T4 infected Escherichia coli. Biochemistry. 1974 Dec 3;13(25):5110–5115. doi: 10.1021/bi00722a009. [DOI] [PubMed] [Google Scholar]
- Sasaki I., Bertani G. Growth abnormalities in Hfr derivatives of Escherichia coli strain C. J Gen Microbiol. 1965 Sep;40(3):365–376. doi: 10.1099/00221287-40-3-365. [DOI] [PubMed] [Google Scholar]
- Shaw W. V., Hopwood D. A. Chloramphenicol acetylation in Streptomyces. J Gen Microbiol. 1976 May;94(1):159–166. doi: 10.1099/00221287-94-1-159. [DOI] [PubMed] [Google Scholar]
- So M., Gill R., Falkow S. The generation of a ColE1-Apr cloning vehicle which allows detection of inserted DNA. Mol Gen Genet. 1975 Dec 30;142(3):239–249. doi: 10.1007/BF00425649. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Weisblum B. Construction of a colicin E1-R factor composite plasmid in vitro: means for amplification of deoxyribonucleic acid. J Bacteriol. 1975 Jan;121(1):354–362. doi: 10.1128/jb.121.1.354-362.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. B., Skorvaga M. Phosphorylation of streptomycin and dihydrostreptomycin by Streptomyces. Enzymatic synthesis of different diphosphorylated derivatives. J Biol Chem. 1973 Apr 10;248(7):2435–2440. [PubMed] [Google Scholar]
- Walker M. S., Walker J. B. Streptomycin biosynthesis and metabolism. Enzymatic phosphorylation of dihydrostreptobiosamine moieties of dihydro-streptomycin-(streptidino) phosphate and dihydrostreptomycin by Streptomyces extracts. J Biol Chem. 1970 Dec 25;245(24):6683–6689. [PubMed] [Google Scholar]
- Watanabe T. The problems of drug-resistant pathogenic bacteria. The origin of R factors. Ann N Y Acad Sci. 1971 Jun 11;182:126–140. doi: 10.1111/j.1749-6632.1971.tb30652.x. [DOI] [PubMed] [Google Scholar]
- Westmoreland B. C., Szybalski W., Ris H. Mapping of deletions and substitutions in heteroduplex DNA molecules of bacteriophage lambda by electron microscopy. Science. 1969 Mar 21;163(3873):1343–1348. doi: 10.1126/science.163.3873.1343. [DOI] [PubMed] [Google Scholar]
- Wilson G. A., Young F. E. Isolation of a sequence-specific endonuclease (BamI) from Bacillus amyloliquefaciens H. J Mol Biol. 1975 Sep 5;97(1):123–125. doi: 10.1016/s0022-2836(75)80028-3. [DOI] [PubMed] [Google Scholar]