Abstract
Methamphetamine use disorder is characterized by recurrent binge episodes. Humans addicted to methamphetamine experience various degrees of cognitive deficits and show evidence of neurodegenerative processes in the brain. Binge injections of METH to rodents also cause significant toxic changes in the brain. In addition, this pattern of METH injections can alter gene expression in the dorsal striatum. Gene expression is regulated, in part, by histone deacetylation. We thus tested the possibility that METH toxic doses might cause changes in the mRNA levels of histone deacetylases (HDACs). We found that METH did produce significant decreases in the mRNA expression of HDAC8, which is a class I HDAC. METH also decreased expression of HDAC6, HDAC9, and HDAC10 that are class II HDACs. The expression of the class IV HDAC, HDAC11, was also suppressed by METH. The expression of Sirt2, Sirt5, and Sirt6 that are members of class III HDACs was also downregulated by METH injections. Our findings implicate changes in HDAC expression may be an early indicator of impending METH-induced neurotoxicity in the striatum. This idea is consistent with the accumulated evidence that some HDACs are involved in neurodegenerative processes in the brain.
Keywords: Epigenetics, sirtuins, neurotoxicity, gene expression
1. Introduction
Methamphetamine (METH) is a widely abused psychostimulant because it is cheap, readily accessible, and addictive (Cadet et al, 2003). In humans, large doses of METH induce cardiac arrhythmia, strokes, seizures, and hyperthermia (Krasnova and Cadet, 2009). Acute effects of METH in the brain are mediated by release of dopamine (DA) from DA terminals (O’Dell et al, 1993) and stimulation of DA receptors in dopaminergic projection areas (Jayanthi et al, 2005; Xu et al, 2005). These large increases in DA levels are associated with substantial depletion of dopaminergic markers in the dorsal striatum (Kuhn et al, 2008; Sonsalla et al, 1986). METH can also cause death of neurons located post-synaptic to monoaminergic terminals (Deng et al, 1999; 2001; Jayanthi et al, 2001; 2005; Thiriet et al, 2005). Injections of multiple doses of METH are also accompanied by significant changes in gene expression in the dorsal striatum (Beauvais et al, 2011; 2010)). Multiple high doses of the drug also affect the expression of immediate early genes (IEGs) in the brain (Beauvais et al, 2011). Specifically, this pattern of drug injections induces significant time-dependent increases in IEG mRNA and protein expression (Beauvais et al, 2011). Binge METH injections also caused time-dependent increases in the mRNA expression of heat shock proteins, Gadd34, CHOP/Gadd153, and Bad but decreases in Bcl2 expression (Beauvais et al, 2011). These transcriptional changes appear to occur via stimulation of the DA D1-like receptor subtypes that are found in high concentration in the dorsal striatum (Cadet et al, 2010).
Gene transcription is regulated by complex epigenetic changes that include post-translational histone modifications and DNA methylation (Murr, 2010). The N-tails of histones possess lysine residues that can be reversibly acetylated by histone acetyltransferases (HATs) or deacetylated by several histone deacetylases (HDACs). HDACs remove acetyl groups from lysine residues on histones and stimulate the recruitment of several repressor complexes that mediate transcriptional changes (Thiagalingam et al, 2003). HDACs are divided into four classes based on sequence similarities (Mottet and Castronovo, 2008). These include Class I (HDAC1, HDAC2, HDAC3, and HDAC8), Class II (HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, HDAC10), Class III (Sirtuins1-7) and Class IV (HDAC11) HDACs (Mottet and Castronovo, 2008). Class I, II and IV HDACs are referred to as “classical” HDACs and are Zn2+-dependent enzymes (Codd et al, 2009) whereas the sirtuins require NAD+ as a cofactor (Neugebauer et al, 2008). Because HDACs participate in gene regulation, we investigated whether binge METH injections might influence the expression of these HDACs in the brain.
2. Materials and methods
2.1. Animals
Male Sprague–Dawley rats (Charles River Labs, Raleigh, NC, USA), weighing 250–300 g were used in the present study. The animals were maintained in a room at temperature of 22 °C and had free access to food and water. They were divided into two groups of animals: 1) Saline and 2) METH (10 mg/kg × 4 intraperitoneal injections every 2 h). The animals were euthanized at 16 hours after the last injection. All animal use procedures were according to the NIH Guide for the Care and Use of Laboratory Animals and were approved by Animal Care Committee of NIDA, NIH.
2.2. RNA Isolation
Dorsal striatal tissues were rapidly dissected, put on dry ice, and kept frozen at −80 C. Total RNA was extracted using Qiagen RNeasy Midi kit (Qiagen, Valencia, CA, USA) according to the company’s protocol. The RNA quality and quantity were assessed using an Agilent 2100 Bioanalyzer 2 (Agilent, Palo Alto, CA, USA) and showed no degradation.
2.3. Quantitative RT-PCR
Individual total RNA obtained from 6–8 rats per group was reverse-transcribed with oligo dT primers and RT for PCR kit (Clontech, Palo Alto, CA). PCR experiments were performed on Lightcycler 480 II (Roche, Indianapolis, IN), using and iQ SYBR Green Supermix (BioRad, Hercules, CA) according to the manufacturer’s protocol. Sequences for rat gene-specific primers corresponding to PCR targets were obtained using LightCycler Probe Design software (Roche). The primers were synthesized and HPLC-purified at the Synthesis and Sequencing Facility of Johns Hopkins University (Baltimore, MD). The sequences for these primers are shown in Table 1. Quantitative PCR values were normalized using OAZ1 (ornithine decarboxylase antizyme 1) based on the paper by de Jonge et al. (2007) who had reported that OAZ1 showed very stable expression in the mouse based on their analyses of 2,543 tissue samples hybridized to Affymetrix Mouse GeneChips after exposure to various experimental manipulations. The results are reported as relative changes calculated as the ratios of normalized gene expression data of METH-treated group compared to the control group.
Table 1.
List of RT-PCR Primers.
| Gene Name | Forward | Reverse |
|---|---|---|
| HDAC1 | GCC CTT CCA ATA TGA CTA AC | GAG CAG ATG GAA ATT CGT |
| HDAC2 | TGT TAA GGA AGA AGA CAA ATC CA | ACA GCG AAG GTT TCT TAT C |
| HDAC3 | ATG AAA CAT CTC TGC TGG TA | GGC GGA TCT GGT CTA GAT A |
| HDAC4 | GAA CAA GGA GAA GGG CA | TGT CTT CCC ATA CCA GTA G |
| HDAC5 | TGG ACT GGG ACA TTC AC | CAC GCC ACA TTT ACG TT |
| HDAC6 | GTC TCA TCC TAC CTG CTC | GGC AGA TGT AGA TGG ACT |
| HDAC7 | CTG CTT TCA GGA TAG TGG | CAG CTG CTG TGT CAT GTA |
| HDAC8 | CTC AGG CTG AGT CTG AAA | CTT CAC AAG GGA ATC GCA |
| HDAC9 | TCT GAA CAT CAC TCA CTA CT | GTG CAG CTC ATT CCA AA |
| HDAC10 | GTG CCC TGG AGT CTA TC | CCA AGG CAA CAG CTA TG |
| HDAC11 | TCA CAC TGG CTA TCA AGT T | GTA GAT GTG GCG GTT GTA AA |
| SIRT1 | CCA GAT CCT CAA GCC ATG TT | CCA AAA TTG CTT TCC TTC CA |
| SIRT2 | TTG AAG GAG TGA CAC GCT A | GTA TGG AAG GTG GTA TTT CT |
| SIRT3 | CAA TGT CGC TCA CTA CT | GCA CGT AGC TGA TAC AAA |
| SIRT4 | TCC GGT TAC AGG TTC AT | CTG TCA CTG TGG GTC TA |
| SIRT5 | TGT ACC TCG TGT GGC AAT GT | CAG GAT CCA GGT TTT CTC CA |
| SIRT6 | GCT GAG AGA CAC CAT TC | GTT GAC AAT GAC CAG ACG |
| SIRT7 | CAG CCT CTA TCC CAG ATT | TGT TGC ACC AGC TTA TG |
2.4. Statistical analysis
Statistical analysis for the qRT-PCR data was performed by StatView (SAS Institute, Cary, NC, USA) using unpaired student’s T-test. The null hypothesis was rejected at p < 0.05.
3. Results
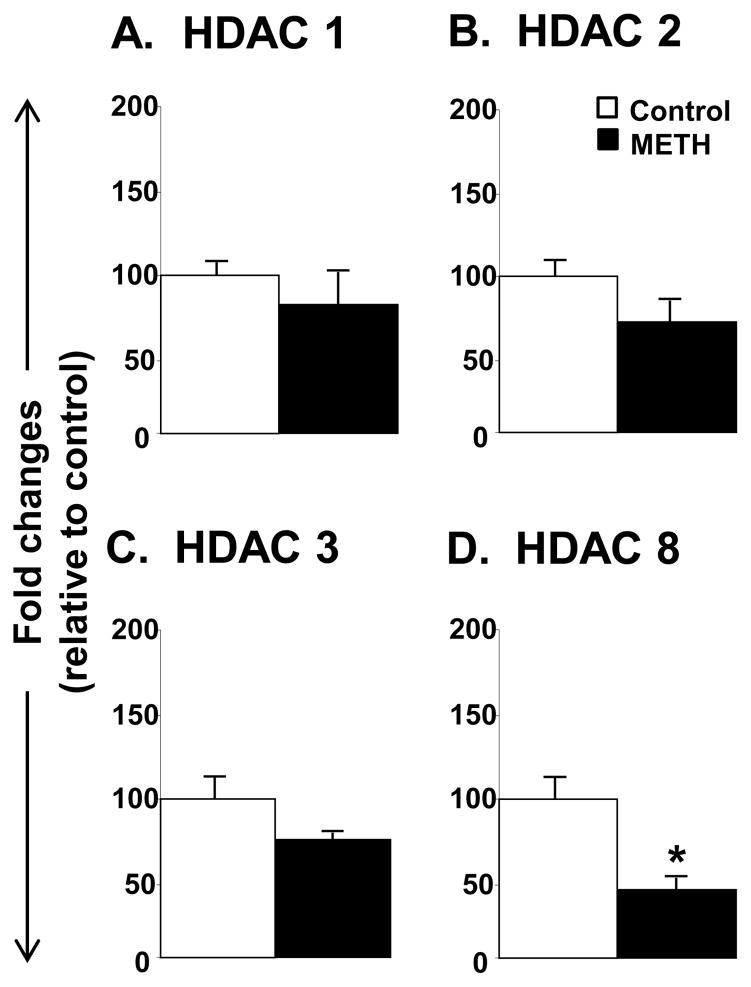
Figure 1 shows the effects of METH on the expression of class I HDACs. METH did not cause any changes in the expression of HDAC1, HDAC2 or HDAC3 (Fig. 1A–1C). However, there were significant changes in the expression of HDAC8 (−51%, p = 0.018).
Figure 1.
Toxic METH binges caused decreased HDAC8 mRNA levels in the rat striatum. Rats were injected with METH (10 mg/kg) given 4 times at 2-hr interval within one day. The animals were euthanized 16 hours after the last injection. RNA extraction and quantitative PCR were conducted as described in the text. Statistical analysis was done by Student’s T test. Key to statistics: * < 0.05, in comparison to control animals.
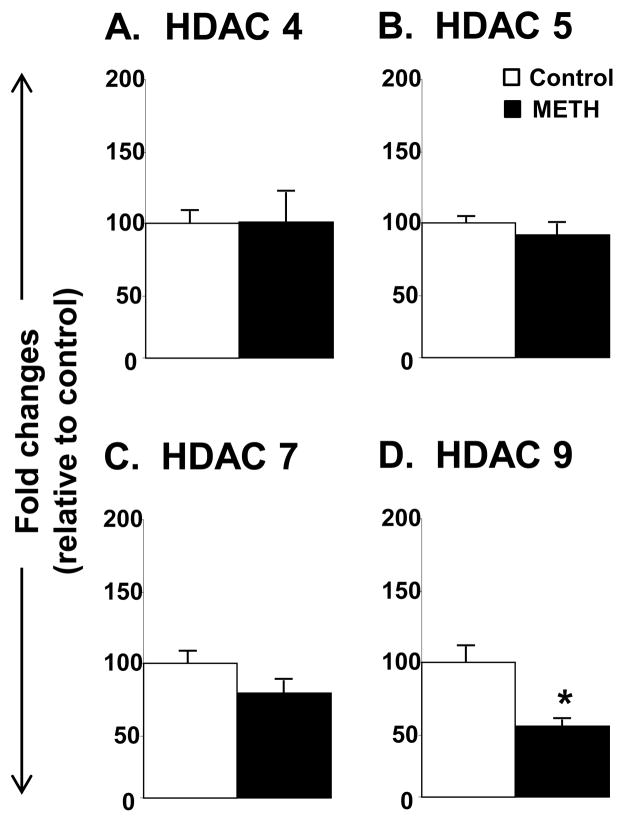
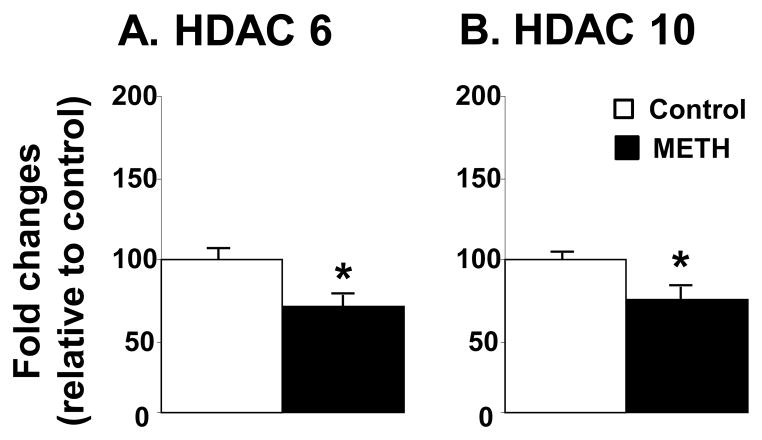
Figure 2 shows the effects of METH on the mRNA levels for Class IIA HDACs. There were no significant METH effects on HDAC4, 5, or 7 (Fig. 2A–2C). In contrast, METH caused significant decreases in HDAC9 mRNA levels (−42%, p= 0.031). The results of class IIB HDACs are reported in Fig. 3. Both HDAC6 (−31%, p = 0.022) and HDAC10 (−25%, p = 0.042) were significantly impacted by the toxic dose of METH (Fig. 3).
Figure 2.
Differential effects of toxic doses of METH on Class IIA HDAC mRNA levels. Rats were treated as described in the text and in Figure 1. Key to statistics: * < 0.05 in comparison to control rats.
Figure 3.
METH caused decreased expression of HDAC6 and HDAC10 mRNA expression. Rats were treated as described in the text and in Figure 1. Key to statistics: * < 0.05 in comparison to control rats.
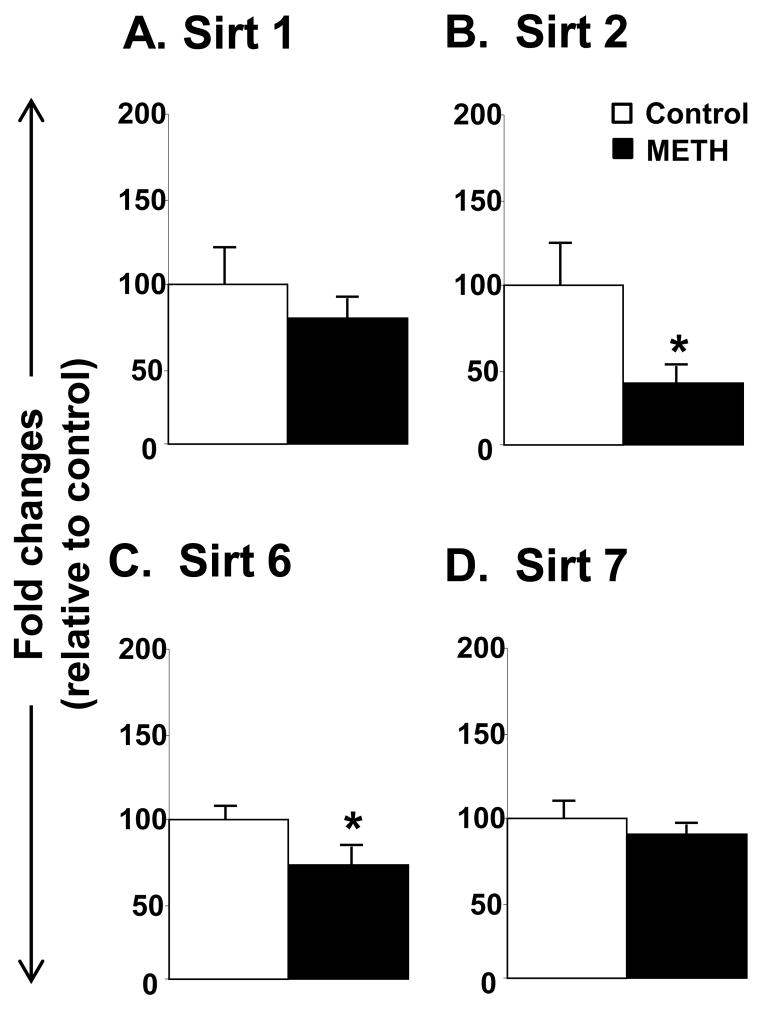
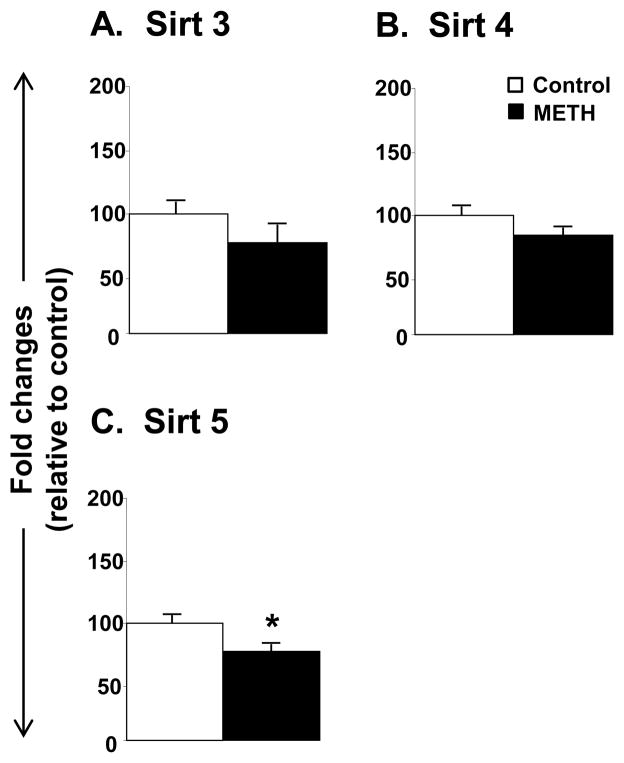
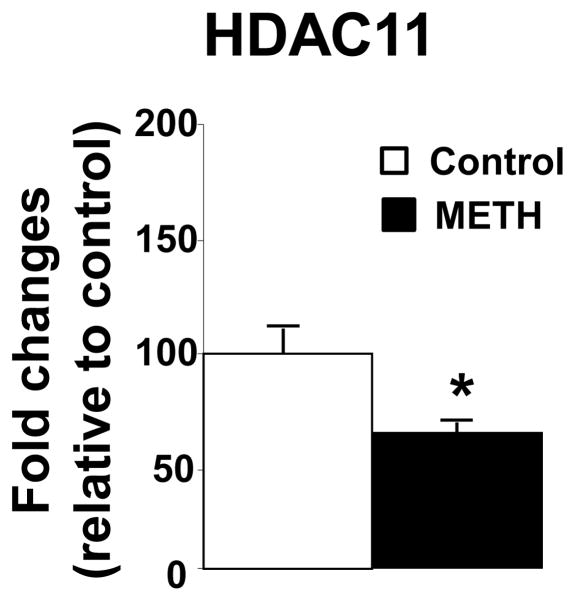
The METH injections also influenced the expression of class III HDACs called sirtuins (Figs. 4 and 5). The data on the sirtuins are presented in terms of their subcellular localization. The effects of METH on cytoplasmic and nuclear sirtuins are shown in figure 4. METH did not cause any changes in the expression of Sirt1 (Fig. 4A) or Sirt7 (Fig. 4D). However, METH caused significant decreases in Sirt2 (−75%, p =0.041, Fig. 4B) and Sirt6 (−27%, p = 0.029, Fig. 4C) mRNA levels. The effects of METH on the mitochondrial sirtuins are shown in Figure 5. There were no significant changes in Sirt3 and Sirt4 expression (Figs 5A and 5B). However, METH caused significant decreases in Sirt5 mRNA levels (−24%, p = 0.026, Fig. 5C). Finally, figure 6 shows that the METH injections caused significant decreases (−33%, p = 0.024) in the mRNA expression of the class IV HDAC, HDAC11.
Figure 4.
Differential effects of METH on cytoplasmic and nuclear Class III HDAC mRNA levels. Rats were treated as described in the text and in Figure 1. Key to statistics: * < 0.05 in comparison to control rats.
Figure 5.
METH caused decreased mRNA expression of the mitochondrial Class III HDAC, Sirt5. Rats were treated as described in the text and in Figure 1. Key to statistics: * < 0.05 in comparison to control rats.
Figure 6.
METH decreases HDAC11 mRNA expression. Rats were treated as described in the text and in Figure 1. Key to statistics: * < 0.05 in comparison to control rats.
4. Discussion
The main findings are that injections of toxic METH doses can cause decreased mRNA levels of several HDACs that include HDAC6, HDAC8, HDAC9, and HDAC10. In addition, the METH injections caused substantial changes in Sirt2, Sirt5, and Sirt6. These observations suggest that toxic doses of METH that are known to cause DA depletion in the striatum can also impact genes that are involved in epigenetic regulation of gene expression and in the maintenance of metabolic homeostasis.
Class I HDACs including HDAC8 are known to be involved in the regulation of gene expression (Hayakawa and Nakayama, 2011). Previous studies from this laboratory had reported that chronic administration of non-toxic METH doses for two weeks increased the mRNA expression of HDAC1 and HDAC2 in the striatum (Jayanthi et al, 2014). In the present study, we did not detect any significant changes in HDAC1, HDAC2, or HDAC3 in animals euthanized 16 hours after toxic METH injections. Together, these observations suggest that the patterns of METH administration can significantly impact the presence and direction of METH-induced alterations in epigenetic markers. In contrast to the lack of changes in these three genes, we found marked decreases in HDAC8 expression. The cloning and characterization of HDAC8 was first published by several investigators in 2000 and was shown to share homologies with other known Class I HDACs (Buggy et al, 2000; Hu et al, 2000). HDAC8 is located in the nucleus and has histone deacetylase activity that can be blocked by β-hydroxybutyrate and trichostatin A (Buggy et al, 2000; Hu et al, 2000). HDAC8 preferentially deacetylates histones H3 and H4 to regulate gene expression. HDAC8 can be phosphorylated by protein kinase A, resulting in a significant inhibition of its activity (Lee et al, 2004). In the central nervous system (CNS), HDAC8-like immunoreactivity was reported to show a nucleo-cytoplasmic staining pattern in neurons located in various brain regions (Takase et al, 2013). HDAC8 is also found in the cytoplasm of smooth muscle cells (de Leval et al, 2006).
Phosphorylated HDAC8 recruits the chaperone proteins, HSP70/HSP90, to a complex that can block an E3 ligase-mediated degradative pathway (Lee et al, 2006). Interestingly, overexpression of HDAC8 was shown to cause cell proliferation and inhibition of liver cancer cells (Wu et al, 2013). In contrast, HDAC8 inhibition causes apoptosis of T-cell lymphomas (Balasubramanian et al, 2008) and of hepatocellular carcinoma (Wu et al, 2013). These observations suggest that the METH-induced decreased HDAC8 expression might play a role in promoting cell death in this model of METH toxicity (Deng et al, 1999; Krasnova and Cadet, 2009).
Another gene that showed METH-induced decreased expression was HDAC9. HDAC9 was cloned in 2001 and reported to show similarities of class IIA HDACs (Zhou et al, 2001). Class IIA HDACs shuttle between the cytosol and the nucleus (Lahm et al, 2007; Martin et al, 2007). HDAC9 is highly expressed in the brain and skeletal muscle where it serves to repress myocyte enhancer factor 2 (MEF2)-mediated gene transcription (Zhou et al, 2001). HDAC9 co-localizes and binds to several transcription repressors, co-repressors, and transcription factors including mSin3A, mSin3B, and N-CoR (Petrie et al, 2003). HDAC9 also interacts with HDAC3 and HDAC4 (Petrie et al, 2003). HDAC9 is also a signal-responsive repressor that is downregulated in the denervated muscle, leading to increased MEF2-dependent gene expression (Mejat et al, 2005). Thus, the METH-induced downregulation suggests that there might be an increase in the expression of specific MEF2-regulated genes in intrinsic striatal cells since members of the MEF2 family of transcription factors are also highly expressed in the brain and participate in the regulation of genes involved in neuronal survival (Akhtar et al, 2012; Dietrich, 2013), a process that is regulated, in part, by the cAMP-PKA signaling pathway (Wang et al, 2005). Interestingly, MEF2 family members have been reported to regulate depolarization-induced BDNF expression (Lyons et al, 2012). MEF2 also regulates the expression of activity-dependent genes including Arc (Flavell et al, 2006). Other genes of interest include Homer1, JNK and adenyl cyclase 8 (Flavell et al, 2008), some of which have been shown to be regulated by METH in various contexts (Cadet et al, 2014; Martin et al, 2012). Taken together, these observations implicate HDAC9 and MEF2 in the regulation of the survival of specific classes of striatal neurons after toxic doses of METH since enkephalinergic neurons seem to be more susceptible than other striatal cells to the toxic effects of the drug (Jayanthi et al, 2005; Thiriet et al, 2005).
The two class IIB HDACs, HDAC6 and HDAC10, were also impacted by the binge METH injections. HDAC6 is a microtubule-associated cytoplasmic deacetylase (Grozinger and Schreiber, 2000; Hubbert et al, 2002). Interestingly, tubulin deacetylase have been reported to modulate the formation of aggresomes and the accumulation of misfolded proteins, processes that might serve to maintain cellular viability (Boyault et al, 2007; Kawaguchi et al, 2003). Indeed, HDAC6 provides an important link between autophagy and the unfolded protein response (UPR) (Pandey et al, 2007). These observations are of interest because toxic doses of METH cause significant changes in the expression of proteins involved in the UPR (Beauvais et al, 2011; Jayanthi et al, 2004; Jayanthi et al, 2009) and suggest that METH-induced decreased HDAC6 expression might serve to increase acetylation of some chaperone proteins (Bali et al, 2005). The other class IIB member, HDAC10, was identified in 2002 (Guardiola and Yao, 2002; Kao et al, 2002; Tong et al, 2002). HDAC10 is located in the cytoplasm and nucleus, possesses TCA-sensitive deacetylase activity, and is involved in transcription regulation (Guardiola and Yao, 2002; Kao et al, 2002; Tong et al, 2002). Interestingly, HDAC10 inhibition causes accumulation of reactive oxygen species in gastric cancer cells (Lee et al, 2010), findings that are consistent with the demonstration that HDAC10 can promote autophagy-induced cell survival (Oehme et al, 2013). Thus, METH-induced decreases in HDAC10 mRNA expression might also serve to promote toxicity in the striatum.
The METH injections also caused decreased HDAC11 mRNA expression. The class IV HDAC, HDAC11, was cloned and characterized by Gao et al.(Gao et al, 2002) and is the only member of that class at present. HDAC11 contains residues within its catalytic domain that are shared by classes I and II HDACs (Gao et al, 2002). HDAC11 is highly expressed in the brain (Takase et al, 2013) and is differentially regulated during development (Liu et al, 2008). HDAC11 is also involved in the regulation of gene expression in maturing oligodendrocyte cultures that showed increased expression of the enzyme (Liu et al, 2009). Increased HDAC11 expression was also associated with decreased abundance of histone H3 acetylated at lysine 9 (H3K9) and at lysine 14 (H3K14). Suppressing HDAC11 expression produced increased H3K9Ac and H3K14Ac (Liu et al, 2009). These results suggest that toxic doses of METH might cause similar increases in histone H3 acetylation by suppressing HDAC11 expression, as shown in our results. Interestingly, a recent study has reported that acute injection of ethanol (3 g/kg), a known neurotoxin (Jacobus and Tapert, 2013), also caused significant decreases in HDAC11 expression (Finegersh and Homanics, 2014). Taken together, these results suggest that other neurotoxic compounds might impact HDAC11 expression in the brain.
The injections of METH also caused significant decreases in the expression of 3 class III HDACs, Sirt2, Sirt5, and Sirt6. Sirt2 is localized predominantly in the cytoplasm (Afshar and Murnane, 1999) where it deacetylates FOXO3 in response to oxidative stress (Wang et al, 2007). Sirt2 is also an alpha-tubulin deacetylase (North et al, 2003), a function that appears to be dependent, in part, on its interaction with the class IIB deacetylase, HDAC6 (Nahhas et al, 2007). Thus, the downregulation of both of these HDACs by METH suggest the possibility that these doses of the drug might increase tubulin acetylation, thereby influencing microtubule dynamics in the rat striatum. Also of interest is the fact that Sirt2 also shuttles from the cytoplasm to the nucleus where it is involved in deacetylation of histone H4 at lysine 16 (H4K16ac) (Vaquero et al, 2006). METH-induced decreased Sirt2 expression might thus lead to increased H4K16 acetylation, with substantial impact on chromatin structure and function (Dion et al, 2005).
Sirt5 is a mitochondrial protein that is involved in regulating metabolic adaptations (Gertz and Steegborn, 2010). Mitochondrial proteins that are involved in oxidative phosphorylation, nucleotide metabolism, and in the urea cycle are subject to substantial lysine modifications (Kim et al, 2006; Newman et al, 2012). Sirt5 also deacetylates and activates carbamoyl phosphate synthase (CPS1) to modulate the function of the urea cycle (Nakagawa et al, 2009; Ogura et al, 2010). Sirt5 is also known to be a NAD-dependent remover of malonyl and succinyl groups from specific lysines (Du et al, 2011). These actions of Sirt5 repress cellular respiration through its effects of the pyruvate dehydrogenase protein complex (Park et al, 2013). Together, these observations suggest that METH injections can significantly impact mitochondrial functions, an idea supported by the observed effects of the drug on mitochondrial proteins and enzymatic activities (Beauvais et al, 2011; Brown et al, 2005; Jayanthi et al, 2001; Jayanthi et al, 2004).
Sirt6 is a class III HDAC that is located mainly in the nucleus (Michishita et al, 2005). It is a NAD+-dependent deacetylase that associates with heterochromatin (Michishita et al, 2005). The enzyme is involved in DNA repair and its absence in mice causes premature aging (Mostoslavsky et al, 2006). Sirt6 exerts this function by forming a macromolecular complex with the DNA double-strand break (DSB) repair factor DNA-dependent protein kinase (PK), thereby promoting DNA DSB repair (McCord et al, 2009). Aging-induced decline in homologous recombination (HR) is accompanied by decreased Sirt6 expression, with this decline in HR being rescuable by overexpression of Sirt6 in pre-senescent cells (Mao et al, 2012). These findings suggest that binge METH injections can promote aging-dependent processes in the rat striatum by interfering with the normal Sirt6 functions. This suggestion is supported by a recent report that showed that METH exposure led to impaired recognition memory and abnormalities in monoaminergic systems that were similar to those observed in aged animals (Melo et al, 2012).
In summary, we are reporting, for the first time, that toxic METH doses can have substantial but differential effects on the mRNA expression of several proteins that are members of several HDAC classes. Specifically, while we found significant changes in HDAC6, HDAC8, HDAC9, HDAC10 and HDAC11, the other class I, II, and IV HDACs were not significantly affected, with HDAC4 not even showing any trend for being affected by the toxic METH dose. We also showed significant decreases in Sirt2, Sirt5, and Sirt6 expression. Interestingly, some of these HDACs are involved in the regulation of gene expression whereas others are involved in the regulation of microtubule dynamics. Importantly, several of these HDACs have been shown to participate in degenerative processes in the brain (Simoes-Pires et al, 2013). Of significant interest to the field of METH toxicity, our observations of METH-induced decreased Sirt5 expression are consistent with the accumulated evidence that the drug can negatively impact mitochondrial functions in the brain (Jayanthi et al, 2001). The effects on Sirt6 expression also support the notion that METH can affect DNA repair functions (Cadet et al, 2003). Finally, these results support, in part, the idea that METH may cause substantial transcriptional changes in the brain (Cadet et al, 2014). Future studies will investigate which specific roles that these METH-induced alterations in HDAC expression might play in drug-induced neurotoxicity.
Highlights.
Methamphetamine (METH) is an addictive and toxic agent.
Toxic doses of METH cause significant decreased HDAC6, 8, 9, 10 and 11 mRNA levels in the rat striatum.
METH also decreases the expression of the mitochondrial HDAC, SIRT5.
In addition, the expression of the nuclear HDACs, SIRT2 and SIRT6, is also decreased by the METH injections.
The present observations add to the accumulating evidence that METH can substantially influence epigenetic landscapes in the brain.
Acknowledgments
This research was supported by funds of the Intramural Research Program of the DHHS/NIH/NIDA. Pawaris Wongprayoon and Dr. Piyarat Govitrapong were supported by funds from the Thailand Research Fund (TRF) - Royal Golden Jubilee Ph.D. Program and Mahidol University. The authors also thank the comments of two reviewers who help make this a better paper.
Footnotes
Conflict of interest
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afshar G, Murnane JP. Characterization of a human gene with sequence homology to Saccharomyces cerevisiae SIR2. Gene. 1999;234:161–168. doi: 10.1016/s0378-1119(99)00162-6. [DOI] [PubMed] [Google Scholar]
- Akhtar MW, Kim MS, Adachi M, Morris MJ, Qi X, Richardson JA, et al. In vivo analysis of MEF2 transcription factors in synapse regulation and neuronal survival. PLoS One. 2012;7:e34863. doi: 10.1371/journal.pone.0034863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Ramos J, Luo W, Sirisawad M, Verner E, Buggy JJ. A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas. Leukemia. 2008;22:1026–1034. doi: 10.1038/leu.2008.9. [DOI] [PubMed] [Google Scholar]
- Bali P, Pranpat M, Bradner J, Balasis M, Fiskus W, Guo F, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280:26729–26734. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- Beauvais G, Atwell K, Jayanthi S, Ladenheim B, Cadet JL. Involvement of dopamine receptors in binge methamphetamine-induced activation of endoplasmic reticulum and mitochondrial stress pathways. PLoS One. 2011;6:e28946. doi: 10.1371/journal.pone.0028946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauvais G, Jayanthi S, McCoy MT, Ladenheim B, Cadet JL. Differential effects of methamphetamine and SCH23390 on the expression of members of IEG families of transcription factors in the rat striatum. Brain Res. 2010;1318:1–10. doi: 10.1016/j.brainres.2009.12.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyault C, Zhang Y, Fritah S, Caron C, Gilquin B, Kwon SH, et al. HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes Dev. 2007;21:2172–2181. doi: 10.1101/gad.436407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Quinton MS, Yamamoto BK. Methamphetamine-induced inhibition of mitochondrial complex II: roles of glutamate and peroxynitrite. J Neurochem. 2005;95:429–436. doi: 10.1111/j.1471-4159.2005.03379.x. [DOI] [PubMed] [Google Scholar]
- Buggy JJ, Sideris ML, Mak P, Lorimer DD, McIntosh B, Clark JM. Cloning and characterization of a novel human histone deacetylase, HDAC8. Biochem J. 2000;350(Pt 1):199–205. [PMC free article] [PubMed] [Google Scholar]
- Cadet JL, Brannock C, Jayanthi S, Krasnova IN. Transcriptional and Epigenetic Substrates of Methamphetamine Addiction and Withdrawal: Evidence from a Long-Access Self-Administration Model in the Rat. Mol Neurobiol. 2014 doi: 10.1007/s12035-014-8776-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet JL, Jayanthi S, Deng X. Speed kills: cellular and molecular bases of methamphetamine-induced nerve terminal degeneration and neuronal apoptosis. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2003;17:1775–1788. doi: 10.1096/fj.03-0073rev. [DOI] [PubMed] [Google Scholar]
- Cadet JL, Jayanthi S, McCoy MT, Beauvais G, Cai NS. Dopamine D1 receptors, regulation of gene expression in the brain, and neurodegeneration. CNS Neurol Disord Drug Targets. 2010;9:526–538. doi: 10.2174/187152710793361496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codd R, Braich N, Liu J, Soe CZ, Pakchung AA. Zn(II)-dependent histone deacetylase inhibitors: suberoylanilide hydroxamic acid and trichostatin A. Int J Biochem Cell Biol. 2009;41:736–739. doi: 10.1016/j.biocel.2008.05.026. [DOI] [PubMed] [Google Scholar]
- de Jonge HJ, Fehrmann RS, de Bont ES, Hofstra RM, Gerbens F, Kamps WA, et al. Evidence based selection of housekeeping genes. PLoS One. 2007;2:e898. doi: 10.1371/journal.pone.0000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leval L, Waltregny D, Boniver J, Young RH, Castronovo V, Oliva E. Use of histone deacetylase 8 (HDAC8), a new marker of smooth muscle differentiation, in the classification of mesenchymal tumors of the uterus. Am J Surg Pathol. 2006;30:319–327. doi: 10.1097/01.pas.0000188029.63706.31. [DOI] [PubMed] [Google Scholar]
- Deng X, Ladenheim B, Tsao LI, Cadet JL. Null mutation of c-fos causes exacerbation of methamphetamine-induced neurotoxicity. J Neurosci. 1999;19:10107–10115. doi: 10.1523/JNEUROSCI.19-22-10107.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Wang Y, Chou J, Cadet JL. Methamphetamine causes widespread apoptosis in the mouse brain: evidence from using an improved TUNEL histochemical method. Brain Res Mol Brain Res. 2001;93:64–69. doi: 10.1016/s0169-328x(01)00184-x. [DOI] [PubMed] [Google Scholar]
- Dietrich JB. The MEF2 family and the brain: from molecules to memory. Cell Tissue Res. 2013;352:179–190. doi: 10.1007/s00441-013-1565-2. [DOI] [PubMed] [Google Scholar]
- Dion MF, Altschuler SJ, Wu LF, Rando OJ. Genomic characterization reveals a simple histone H4 acetylation code. Proc Natl Acad Sci U S A. 2005;102:5501–5506. doi: 10.1073/pnas.0500136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegersh A, Homanics GE. Acute ethanol alters multiple histone modifications at model gene promoters in the cerebral cortex. Alcohol Clin Exp Res. 2014;38:1865–1873. doi: 10.1111/acer.12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, et al. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Cueto MA, Asselbergs F, Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Biol Chem. 2002;277:25748–25755. doi: 10.1074/jbc.M111871200. [DOI] [PubMed] [Google Scholar]
- Gertz M, Steegborn C. Function and regulation of the mitochondrial sirtuin isoform Sirt5 in Mammalia. Biochim Biophys Acta. 2010;1804:1658–1665. doi: 10.1016/j.bbapap.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Grozinger CM, Schreiber SL. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc Natl Acad Sci U S A. 2000;97:7835–7840. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardiola AR, Yao TP. Molecular cloning and characterization of a novel histone deacetylase HDAC10. J Biol Chem. 2002;277:3350–3356. doi: 10.1074/jbc.M109861200. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Nakayama J. Physiological roles of class I HDAC complex and histone demethylase. J Biomed Biotechnol. 2011;2011:129383. doi: 10.1155/2011/129383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu E, Chen Z, Fredrickson T, Zhu Y, Kirkpatrick R, Zhang GF, et al. Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor. J Biol Chem. 2000;275:15254–15264. doi: 10.1074/jbc.M908988199. [DOI] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- Jacobus J, Tapert SF. Neurotoxic effects of alcohol in adolescence. Annu Rev Clin Psychol. 2013;9:703–721. doi: 10.1146/annurev-clinpsy-050212-185610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayanthi S, Deng X, Bordelon M, McCoy MT, Cadet JL. Methamphetamine causes differential regulation of pro-death and anti-death Bcl-2 genes in the mouse neocortex. FASEB J. 2001;15:1745–1752. doi: 10.1096/fj.01-0025com. [DOI] [PubMed] [Google Scholar]
- Jayanthi S, Deng X, Ladenheim B, McCoy MT, Cluster A, Cai NS, et al. Calcineurin/NFAT-induced up-regulation of the Fas ligand/Fas death pathway is involved in methamphetamine-induced neuronal apoptosis. Proc Natl Acad Sci U S A. 2005;102:868–873. doi: 10.1073/pnas.0404990102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayanthi S, Deng X, Noailles PA, Ladenheim B, Cadet JL. Methamphetamine induces neuronal apoptosis via cross-talks between endoplasmic reticulum and mitochondria-dependent death cascades. FASEB J. 2004;18:238–251. doi: 10.1096/fj.03-0295com. [DOI] [PubMed] [Google Scholar]
- Jayanthi S, McCoy MT, Beauvais G, Ladenheim B, Gilmore K, Wood W, et al. Methamphetamine induces dopamine D1 receptor-dependent endoplasmic reticulum stress-related molecular events in the rat striatum. PLoS One. 2009;4:e6092. doi: 10.1371/journal.pone.0006092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayanthi S, McCoy MT, Chen B, Britt JP, Kourrich S, Yau HJ, et al. Methamphetamine downregulates striatal glutamate receptors via diverse epigenetic mechanisms. Biol Psychiatry. 2014;76:47–56. doi: 10.1016/j.biopsych.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HY, Lee CH, Komarov A, Han CC, Evans RM. Isolation and characterization of mammalian HDAC10, a novel histone deacetylase. J Biol Chem. 2002;277:187–193. doi: 10.1074/jbc.M108931200. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Krasnova IN, Cadet JL. Methamphetamine toxicity and messengers of death. Brain Res Rev. 2009;60:379–407. doi: 10.1016/j.brainresrev.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn DM, Francescutti-Verbeem DM, Thomas DM. Dopamine disposition in the presynaptic process regulates the severity of methamphetamine-induced neurotoxicity. Ann N Y Acad Sci. 2008;1139:118–126. doi: 10.1196/annals.1432.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahm A, Paolini C, Pallaoro M, Nardi MC, Jones P, Neddermann P, et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci U S A. 2007;104:17335–17340. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Rezai-Zadeh N, Seto E. Negative regulation of histone deacetylase 8 activity by cyclic AMP-dependent protein kinase A. Mol Cell Biol. 2004;24:765–773. doi: 10.1128/MCB.24.2.765-773.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Sengupta N, Villagra A, Rezai-Zadeh N, Seto E. Histone deacetylase 8 safeguards the human ever-shorter telomeres 1B (hEST1B) protein from ubiquitin-mediated degradation. Mol Cell Biol. 2006;26:5259–5269. doi: 10.1128/MCB.01971-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Jeong EG, Choi MC, Kim SH, Park JH, Song SH, et al. Inhibition of histone deacetylase 10 induces thioredoxin-interacting protein and causes accumulation of reactive oxygen species in SNU-620 human gastric cancer cells. Mol Cells. 2010;30:107–112. doi: 10.1007/s10059-010-0094-z. [DOI] [PubMed] [Google Scholar]
- Liu H, Hu Q, D’Ercole AJ, Ye P. Histone deacetylase 11 regulates oligodendrocyte-specific gene expression and cell development in OL-1 oligodendroglia cells. Glia. 2009;57:1–12. doi: 10.1002/glia.20729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Hu Q, Kaufman A, D’Ercole AJ, Ye P. Developmental expression of histone deacetylase 11 in the murine brain. J Neurosci Res. 2008;86:537–543. doi: 10.1002/jnr.21521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons MR, Schwarz CM, West AE. Members of the myocyte enhancer factor 2 transcription factor family differentially regulate Bdnf transcription in response to neuronal depolarization. J Neurosci. 2012;32:12780–12785. doi: 10.1523/JNEUROSCI.0534-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Tian X, Van Meter M, Ke Z, Gorbunova V, Seluanov A. Sirtuin 6 (SIRT6) rescues the decline of homologous recombination repair during replicative senescence. Proc Natl Acad Sci U S A. 2012;109:11800–11805. doi: 10.1073/pnas.1200583109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Kettmann R, Dequiedt F. Class IIa histone deacetylases: regulating the regulators. Oncogene. 2007;26:5450–5467. doi: 10.1038/sj.onc.1210613. [DOI] [PubMed] [Google Scholar]
- Martin TA, Jayanthi S, McCoy MT, Brannock C, Ladenheim B, Garrett T, et al. Methamphetamine causes differential alterations in gene expression and patterns of histone acetylation/hypoacetylation in the rat nucleus accumbens. PLoS One. 2012;7:e34236. doi: 10.1371/journal.pone.0034236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord RA, Michishita E, Hong T, Berber E, Boxer LD, Kusumoto R, et al. SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging (Albany NY) 2009;1:109–121. doi: 10.18632/aging.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejat A, Ramond F, Bassel-Duby R, Khochbin S, Olson EN, Schaeffer L. Histone deacetylase 9 couples neuronal activity to muscle chromatin acetylation and gene expression. Nat Neurosci. 2005;8:313–321. doi: 10.1038/nn1408. [DOI] [PubMed] [Google Scholar]
- Melo P, Magalhaes A, Alves CJ, Tavares MA, de Sousa L, Summavielle T, et al. Methamphetamine mimics the neurochemical profile of aging in rats and impairs recognition memory. Neurotoxicology. 2012;33:491–499. doi: 10.1016/j.neuro.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Mottet D, Castronovo V. Histone deacetylases: target enzymes for cancer therapy. Clin Exp Metastasis. 2008;25:183–189. doi: 10.1007/s10585-007-9131-5. [DOI] [PubMed] [Google Scholar]
- Murr R. Interplay between different epigenetic modifications and mechanisms. Adv Genet. 2010;70:101–141. doi: 10.1016/B978-0-12-380866-0.60005-8. [DOI] [PubMed] [Google Scholar]
- Nahhas F, Dryden SC, Abrams J, Tainsky MA. Mutations in SIRT2 deacetylase which regulate enzymatic activity but not its interaction with HDAC6 and tubulin. Mol Cell Biochem. 2007;303:221–230. doi: 10.1007/s11010-007-9478-6. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugebauer RC, Sippl W, Jung M. Inhibitors of NAD+ dependent histone deacetylases (sirtuins) Curr Pharm Des. 2008;14:562–573. doi: 10.2174/138161208783885380. [DOI] [PubMed] [Google Scholar]
- Newman JC, He W, Verdin E. Mitochondrial protein acylation and intermediary metabolism: regulation by sirtuins and implications for metabolic disease. J Biol Chem. 2012;287:42436–42443. doi: 10.1074/jbc.R112.404863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- O’Dell SJ, Weihmuller FB, Marshall JF. Methamphetamine-induced dopamine overflow and injury to striatal dopamine terminals: attenuation by dopamine D1 or D2 antagonists. J Neurochem. 1993;60:1792–1799. doi: 10.1111/j.1471-4159.1993.tb13405.x. [DOI] [PubMed] [Google Scholar]
- Oehme I, Linke JP, Bock BC, Milde T, Lodrini M, Hartenstein B, et al. Histone deacetylase 10 promotes autophagy-mediated cell survival. Proc Natl Acad Sci U S A. 2013;110:E2592–2601. doi: 10.1073/pnas.1300113110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura M, Nakamura Y, Tanaka D, Zhuang X, Fujita Y, Obara A, et al. Overexpression of SIRT5 confirms its involvement in deacetylation and activation of carbamoyl phosphate synthetase 1. Biochem Biophys Res Commun. 2010;393:73–78. doi: 10.1016/j.bbrc.2010.01.081. [DOI] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell. 2013;50:919–930. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie K, Guidez F, Howell L, Healy L, Waxman S, Greaves M, et al. The histone deacetylase 9 gene encodes multiple protein isoforms. J Biol Chem. 2003;278:16059–16072. doi: 10.1074/jbc.M212935200. [DOI] [PubMed] [Google Scholar]
- Simoes-Pires C, Zwick V, Nurisso A, Schenker E, Carrupt PA, Cuendet M. HDAC6 as a target for neurodegenerative diseases: what makes it different from the other HDACs? Mol Neurodegener. 2013;8:7. doi: 10.1186/1750-1326-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonsalla PK, Gibb JW, Hanson GR. Roles of D1 and D2 dopamine receptor subtypes in mediating the methamphetamine-induced changes in monoamine systems. J Pharmacol Exp Ther. 1986;238:932–937. [PubMed] [Google Scholar]
- Takase K, Oda S, Kuroda M, Funato H. Monoaminergic and neuropeptidergic neurons have distinct expression profiles of histone deacetylases. PLoS One. 2013;8:e58473. doi: 10.1371/journal.pone.0058473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiagalingam S, Cheng KH, Lee HJ, Mineva N, Thiagalingam A, Ponte JF. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann N Y Acad Sci. 2003;983:84–100. doi: 10.1111/j.1749-6632.2003.tb05964.x. [DOI] [PubMed] [Google Scholar]
- Thiriet N, Deng X, Solinas M, Ladenheim B, Curtis W, Goldberg SR, et al. Neuropeptide Y protects against methamphetamine-induced neuronal apoptosis in the mouse striatum. J Neurosci. 2005;25:5273–5279. doi: 10.1523/JNEUROSCI.4893-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong JJ, Liu J, Bertos NR, Yang XJ. Identification of HDAC10, a novel class II human histone deacetylase containing a leucine-rich domain. Nucleic Acids Res. 2002;30:1114–1123. doi: 10.1093/nar/30.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Nguyen M, Qin FX, Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell. 2007;6:505–514. doi: 10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Tang X, Li M, Marshall J, Mao Z. Regulation of neuroprotective activity of myocyte-enhancer factor 2 by cAMP-protein kinase A signaling pathway in neuronal survival. J Biol Chem. 2005;280:16705–16713. doi: 10.1074/jbc.M501819200. [DOI] [PubMed] [Google Scholar]
- Wu J, Du C, Lv Z, Ding C, Cheng J, Xie H, et al. The up-regulation of histone deacetylase 8 promotes proliferation and inhibits apoptosis in hepatocellular carcinoma. Dig Dis Sci. 2013;58:3545–3553. doi: 10.1007/s10620-013-2867-7. [DOI] [PubMed] [Google Scholar]
- Xu W, Zhu JP, Angulo JA. Induction of striatal pre- and postsynaptic damage by methamphetamine requires the dopamine receptors. Synapse. 2005;58:110–121. doi: 10.1002/syn.20185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Marks PA, Rifkind RA, Richon VM. Cloning and characterization of a histone deacetylase, HDAC9. Proc Natl Acad Sci U S A. 2001;98:10572–10577. doi: 10.1073/pnas.191375098. [DOI] [PMC free article] [PubMed] [Google Scholar]