Abstract
Normal plant development requires epigenetic regulation to enforce changes in developmental fate. Genomic imprinting is a type of epigenetic regulation in which identical alleles of genes are expressed in a parent-of-origin dependent manner. Deep sequencing of transcriptomes has identified hundreds of imprinted genes with scarce evidence for the developmental importance of individual imprinted loci. Imprinting is regulated through global DNA demethylation in the central cell prior to fertilization and directed repression of individual loci with the Polycomb Repressive Complex 2 (PRC2). There is significant evidence for transposable elements and repeat sequences near genes acting as cis-elements to determine imprinting status of a gene, implying that imprinted gene expression patterns may evolve randomly and at high frequency. Detailed genetic analysis of a few imprinted loci suggests an imprinted pattern of gene expression is often dispensable for seed development. Few genes show conserved imprinted expression within or between plant species. These data are not fully explained by current models for the evolution of imprinting in plant seeds. We suggest that imprinting may have evolved to provide a mechanism for rapid neofunctionalization of genes during seed development to increase phenotypic diversity of seeds.
Keywords: epigenetics, DNA methylation, histone modification, imprinting, genomics, seed development, maize endosperm, Arabidopsis endosperm
OVERVIEW OF ANGIOSPERM SEED DEVELOPMENT
In this review, we focus on the developmental role of epigenetic regulation, specifically genomic imprinting, in maize and Arabidopsis seeds. Imprinting, or parent-of-origin specific gene expression, has evolved convergently in mammals and angiosperms (Pires and Grossniklaus, 2014). Imprinted gene expression in angiosperms is found in developing seeds. Angiosperm seeds initiate with double fertilization of the megagametophyte (Peris et al., 2010). The pollen tube delivers two haploid sperm cells to the embryo sac. One sperm cell fuses with the haploid egg to generate a diploid embryo, and the other sperm cell fuses with the diploid central cell to form the triploid endosperm. The resulting embryo and endosperm are genetically identical except for their ploidy level with the endosperm having two maternal doses of the genome and one paternal dose. Although the endosperm and embryo have essentially the same genotype, they have markedly different developmental programs (Figure 1; Kiesselbach, 1949; Brown et al., 1999; Chandler et al., 2008; Peris et al., 2010).
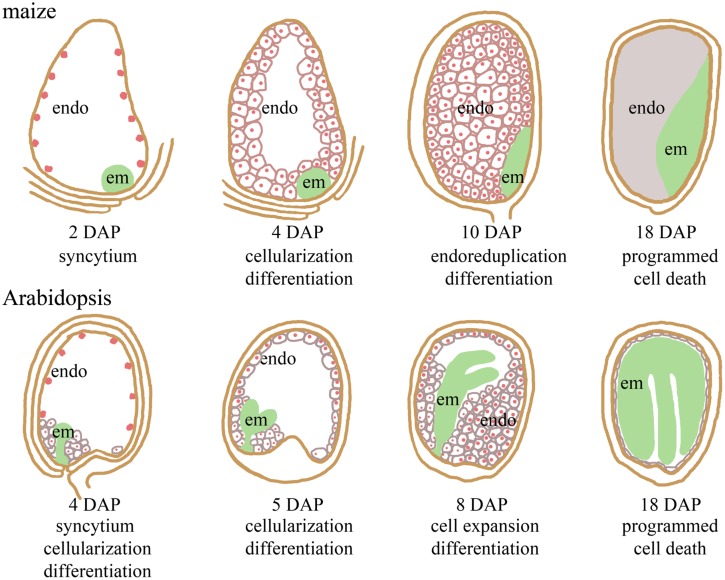
FIGURE 1.
Comparison of seed development in maize and Arabidopsis thaliana. The endosperm proliferates initially as a multi-nucleate syncytium, while the globular embryo develops. Endosperm cellularization precedes embryo transition. Endosperm cells expand and accumulate storage molecules once cellularization is complete. In maize, the endosperm is persistent and undergoes programmed cell death starting around 18 days after pollination (DAP). The Arabidopsis embryo consumes most of the endosperm prior to seed maturation. Endosperm (endo) nuclei are indicated in red. The embryo (em) is in green.
The endosperm starts development by dividing nuclei without completing cytokinesis (reviewed in Olsen, 2004; Sabelli and Larkins, 2009b). This syncytial development transitions to cellularization in which the nuclei become enclosed in cell walls (Figure 1). As the endosperm cellularizes, the cells begin to take on differentiated fates with internal endosperm cells accumulating nutrient storage reserves (Kiesselbach, 1949; Brown et al., 1994, 1996, 1999; Stangeland et al., 2003). In many eudicots, like Arabidopsis, the embryo consumes the endosperm reserves as it develops resulting in most of the endosperm degenerating by seed maturity. By contrast, the internal storage cells in the maize endosperm persist through seed development and the storage reserves are used during seedling growth (Kiesselbach, 1949). Epidermal endosperm cells take on different fates from the internal storage cell types. In Arabidopsis, there are distinct endosperm cell morphologies at the micropyllar and chalazal ends of the embryo sac (Brown et al., 1999). In maize, epidermal endosperm cells differentiate into basal transfer cells, embryo surrounding region, and aleurone (Kiesselbach, 1949). All maize endosperm cells, except the aleurone, undergo programmed cell death prior to seed maturation (Figure 1; Young et al., 1997; Young and Gallie, 2000).
Embryo development starts with asymmetric cell division of the zygote to form an apical-basal axis (Chandler et al., 2008; Peris et al., 2010). Basal cells divide to develop the suspensor and contribute to the root meristem. Apical cells initially develop a globular embryo, which transitions to form the shoot and root apical meristems along with cotyledons in Arabidopsis or a scutellum, coleoptile, and embryonic leaves in maize (i.e., transition stage). The genetic programs controlling meristem specification and lateral organ initiation have been extensively reviewed (Chandler et al., 2008; De Smet et al., 2010; Wendrich and Weijers, 2013).
Imprinted genes primarily show parent-of-origin expression patterns in the endosperm although there are imprinted genes also in the developing embryo (Jahnke and Scholten, 2009; Raissig et al., 2013). Endosperm growth has a significant impact on final seed size, and imprinting has been hypothesized to regulate seed size (Arnaud and Feil, 2006; Xiao et al., 2006; Li and Berger, 2012; Fatihi et al., 2013). However, there is significant data arguing that the endosperm has developmental functions beyond providing nutrition for the developing embryo. Embryo transition occurs soon after endosperm cell differentiation, and recent evidence indicates differentiated endosperm is important for embryo developmental programs. For example, the embryo surrounding endosperm in Arabidopsis secretes the ESF1 signaling peptide to promote normal basal embryo development (Costa et al., 2014). Failure to differentiate the embryo surrounding endosperm in maize causes an embryo developmental block at the transition stage suggesting a similar function for this cell type in maize (Fouquet et al., 2011). Later in Arabidopsis seed development, the ZHOUPI basic-helix-loop-helix transcription factor is expressed exclusively in the embryo surrounding region and activates a signaling pathway required for normal epidermal differentiation in the embryo (Yang et al., 2008; Xing et al., 2013). These data show that the endosperm plays an active role in promoting embryo development and argue that epigenetic regulation of endosperm gene expression could have consequences for seed size as well as embryo developmental programs.
WHAT IS IMPRINTING?
Genomic imprinting in plants is an epigenetic phenomenon by which genetically identical alleles are differentially expressed in a parent-of-origin dependent manner. Imprinted gene expression primarily occurs in the endosperm and there is strong data for imprinted genes controlling early endosperm cell divisions as well as regulating the transfer of nutrients to the seed (Gutierrez-Marcos et al., 2004; Day et al., 2008; Sabelli and Larkins, 2009a; Tiwari et al., 2010; Shirzadi et al., 2011; Costa et al., 2012). Imprinting is an exception from Mendel’s Laws on the expression and inheritance of the two parental alleles in which dominant alleles express phenotypes over recessive alleles irrespective of the parental source of the allele. Instead, imprinted genes will express either the maternal or paternal allele even though the primary sequences of these alleles may be identical.
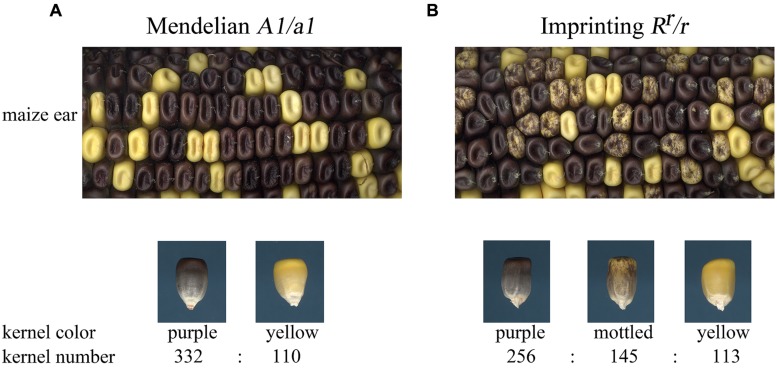
It is easiest to understand imprinted inheritance through an example. The A1 locus of maize encodes a structural gene for anthocyanin biosynthesis (O’Reilly et al., 1985), while the R locus encodes a transcription factor that induces anthocyanin biosynthesis (Ludwig et al., 1989; Perrot and Cone, 1989). The A1 locus shows Mendelian inheritance, while certain haplotypes of the R locus are imprinted (Kermicle, 1969). Indeed, R was the first imprinted locus described in plants. The Rr allele shows altered expression when Rr is inherited from pollen. The expression pattern of paternally inherited Rr can be seen by contrasting self-pollinations of heterozygous individuals for A1/a1 or Rr/r (Figure 2). When a plant heterozygous for A1/a1 is self-pollinated, the seeds segregate in a 3:1 ratio for full color to yellow kernels (Figure 2). Self-pollination of Rr/r yields three kernel color types in a 2:1:1 ratio of purple to yellow to mottled purple kernels. These mottled purple kernels have an endosperm genotype of r r/Rr where the dominant allele inherited from the male is repressed in a stochastic pattern in the seed. The same paternal Rr allele is not affected in the embryo and plants from mottled kernels will yield full color kernels if crossed as female to an r/r plant and mottled kernels if crossed as a male to the r/r genotype.
FIGURE 2.
Comparison of Mendelian genetic inheritance and imprinted inheritance. (A) The a1 locus shows Mendelian inheritance in self-pollinations of A1/a1 individuals. Full purple color kernels are dominant over yellow kernels and the progeny segregate in a 3:1 ratio. (B) The Rr allele shows imprinted inheritance. Self-pollination of Rr/r yields three kernel color types in a 2:1:1 ratio of purple to mottled to yellow kernels. Purple kernels inherited the Rr allele from the megagametophyte. Mottled kernels are heterozygous individuals that inherited the Rr allele from the pollen. Kernel counts are given for the ears shown in the upper panels.
One interpretation of the mottled r r/Rr kernel phenotype is that it is due to an insufficient dosage as a consequence of the endosperm fusing a diploid maternal central cell with a haploid paternal sperm cell. However, introducing multiple copies of Rr with translocation and trisomic stocks does not alter the anthocyanin phenotypes. When more than two doses are inherited maternally, the kernel is always full color, and when multiple Rr alleles are inherited paternally, the mottled phenotype always results (Kermicle, 1970). These and more recent data indicate that imprinting is an independent phenomenon from dosage effects.
The r locus is an example of maternal bias of gene expression. Imprinted genes can show bias for either the maternal or paternal allele and consequently are classified into maternally expressed genes (MEGs) and paternally expressed genes (PEGs). MEGs and PEGs can be identified molecularly by examining allele-specific expression in reciprocal crosses. Single nucleotide polymorphisms and small insertion-deletions within transcripts from diverse parents are used to identify the expression level of both the maternal and paternal allele. By carrying out the same gene expression analysis on reciprocal crosses, it is possible to identify genes that express only the maternal or only the paternal allele irrespective of the polymorphisms found within the alleles of the genes. A variety of molecular strategies have been employed to identify individual imprinted genes such as Maternally expressed gene1 (Meg1) and Maternally expressed in embryo1 (Mee1) in maize (Gutierrez-Marcos et al., 2004; Jahnke and Scholten, 2009).
RNA-seq transcriptomics allows global analysis of imprinted gene expression at a much larger scale. Maize is particularly well suited for allele identification in RNA-seq experiments, because maize inbred lines show high levels of polymorphism allowing for a large number of genes to be assayed for imprinting in a single experiment (Chia et al., 2012; Jiao et al., 2012). Initial experiments in maize examined reciprocal crosses between the reference genome inbred line, B73, and Mo17 (Waters et al., 2011; Zhang et al., 2011). These studies identified hundreds of MEGs and PEGs with relatively little overlap between them. By examining additional time-points during seed development and additional inbred crosses more than 500 genes show statistically significant bias for gene expression based on the parent of origin (Waters et al., 2013; Zhang et al., 2014). Many imprinted genes only show parent-of-origin bias transiently. For example, Meg1 is maternally expressed early during seed development and is biallelic, expressed from both maternal and paternal alleles, by mid-seed development (Gutierrez-Marcos et al., 2004). Attempts to apply transcriptomics to early stages of developing maize seeds did not effectively isolate endosperm or embryo tissue from maternal tissue, so most identified maize MEGs and PEGs show imprinted expression patterns after embryo transition (Xin et al., 2013). The overlap of imprinted genes between all data sets is low. For example, Waters et al. (2013) found that only 5–10% of the imprinted genes in a survey of four inbred lines showed imprinting in all genotype combinations. These results have raised questions about whether sequencing depth, statistical approaches, allele-specific effects, or environmental factors have significant impact on the genes detected as imprinted.
Similar transcriptomic approaches have been applied to identify imprinted genes in Arabidopsis. Endosperm and embryo transcripts from reciprocal crosses between Col-0 and Ler accessions identified over 200 imprinted genes (Gehring et al., 2011; Hsieh et al., 2011). Expanding these studies to additional accessions found that, like in maize, only a small number of genes consistently show imprinting in all accessions (Wolff et al., 2011; Pignatta et al., 2014). For example, Pignatta et al. (2014) found about 10% of MEGs and 5% of PEGs are shared between the three accessions they surveyed. It seems surprising that both maize and Arabidopsis transcriptome surveys have found only a few conserved imprinted genes within each species. Considering both the maize and Arabidopsis observations, the data suggest that relatively few loci are stably selected for imprinted gene expression.
Transcriptomic studies have also identified allele-specific imprinting in both maize and Arabidopsis (Waters et al., 2013; Pignatta et al., 2014). Allele-specific imprinted genes have MEG or PEG expression for a single allele from a single accession or inbred line, much like the Rr allele of maize. Using kernel phenotypes, allele-specific imprinting has been observed in other maize loci including the dzr1Mo17 and B-Boliva alleles that control zein and anthocyanin accumulation, respectively (Chaudhuri and Messing, 1994; Selinger and Chandler, 2001). These older examples indicate that allele-specific imprinting can have significant effects on kernel phenotypes. However, phenotypes have not been associated with the more recently discovered allele-specific imprints.
With hundreds of imprinted genes identified, annotation of these genes suggests that imprinted loci do function in processes proximate to developmental programs. Imprinted genes include proteins involved in chromatin modification, transcription factors, hormone signaling, ubiquitin-targeted protein degradation, and RNA processing (Gehring et al., 2011; Hsieh et al., 2011; Wolff et al., 2011; Zhang et al., 2011; Xin et al., 2013; Pignatta et al., 2014). MEGs show some enrichment for transcription factors, such as MYB family genes (Hsieh et al., 2011; Pignatta et al., 2014), while PEGs show enrichment for chromatin and transcriptional modifiers (Waters et al., 2013; Pignatta et al., 2014). However, only two genes show conserved imprinted expression between Arabidopsis, rice, and maize (Waters et al., 2013). A loss of function allele of one of these PEGs, ZmYuc1, is tightly linked to the recessive defective endosperm18 locus of maize, suggesting that residual maternal expression is sufficient to confer normal seed development (Bernardi et al., 2012). Both allele-specific imprinting and the low conservation of imprinted gene expression across angiosperms suggest that deeply conserved developmental circuits have not been selected for this type of epigenetic regulation in angiosperms. Based on these and additional arguments below, we suggest that imprinting is primarily a form of regulation to enable rapid diversifying selection of seed phenotypes.
MOLECULAR MECHANISMS OF IMPRINTING
Altering the expression state of an allele depending upon the parent-of-origin requires epigenetic modification of the alleles inherited by the male and female gametes. The mechanisms by which MEGs and PEGs are identified and programmed have been extensively reviewed (Köhler et al., 2012; Gehring, 2013; Zhang et al., 2013). As a brief overview, both histone modification and DNA methylation have essential roles in setting imprinted patterns of gene expression. The Arabidopsis model for establishing contrasting epigenetic states in the male and female gametes starts with differential demethylation of the genome. The DNA glycosylase gene, DEMETER (DME), is expressed in the central cell of the megagametophyte but not the sperm cells of the pollen (Choi et al., 2002; Schoft et al., 2011). DME activity removes 5-methylcytosine predominantly from transposable element and repeat sequences leading to most repetitive sequences having reduced methylation in the developing endosperm (Gehring et al., 2009; Hsieh et al., 2009). Surprisingly, maize does not show these global patterns of DNA hypomethylation in the endosperm (Zhang et al., 2011, 2014). Instead, allele-specific bisulfite sequencing of endosperm DNA revealed a pattern of DNA hypomethylation at maternal alleles with corresponding hypermethylation at paternal alleles for specific sites within the genome (Zhang et al., 2014). These maize results are consistent with DNA demethylation specifically occurring in the central cell.
The differential loss of DNA methylation sets-up contrasting chromatin marks in Arabidopsis repeat sequences near the paternal and maternal alleles. Methylation marks can then be interpreted by the genome with a variety of molecular mechanisms. For example, methylation of the paternal allele can lead to a transcriptionally silent state, while the demethylated maternal allele would become transcriptionally active (Kinoshita et al., 2004; Jullien et al., 2006a; Hermon et al., 2007; Tiwari et al., 2008). There are also a few examples where RNA-directed DNA methylation (RdDM) is critical in the male parent to ensure silencing of the paternal allele at MEG loci, suggesting that small RNAs can have a significant role in setting MEG expression patterns (Bratzel et al., 2012; Vu et al., 2013). Although these models can explain MEG patterns of expression, PEGs can also be hypermethylated at the paternal allele and hypomethylated at the maternal allele (Gehring et al., 2009; Hsieh et al., 2009; Zhang et al., 2014). This maternal hypomethylation is essential for silencing of the maternal allele for many PEGs (Hsieh et al., 2011; Wolff et al., 2011).
How can the same epigenetic mark of reduced DNA methylation in the maternal allele result in opposite MEG and PEG expression patterns? Trimethylation of lysine 27 on histone H3 (H3K27me3) is another chromatin mark that is required for imprinted gene expression (Schuettengruber and Cavalli, 2009; Köhler et al., 2012). H3K27me3 marks are catalyzed by the Polycomb Repressive Complex2 (PRC2). In the Arabidopsis endosperm, the PRC2 complex is referred to as the FERTILIZATION INDEPENDENT SEED (FIS) complex and is composed of four core subunits: the MEDEA (MEA) Enhancer of zeste homolog, the FERTILIZATION INDEPENDENT SEED2 (FIS2) Suppressor of zeste homolog, the FERTILIZATION INDEPENDENT ENDOSPERM (FIE) Extra sex combs homolog, and the MULTICOPY SUPPRESSOR OF IRA1 (MSI1), which is a WD-40 repeat protein that is homologus to Drosophila p55 (Grossniklaus et al., 1998; Kiyosue et al., 1999; Luo et al., 1999; Köhler et al., 2003). The H3K27me3 post-translational modification is a repressive chromatin mark, and FIS-PRC2 is known to be required to repress paternal alleles of MEGs as well as maternal alleles of PEGs (Köhler et al., 2003, 2005; Baroux et al., 2006; Makarevich et al., 2006; Fitz Gerald et al., 2009; Weinhofer et al., 2010).
It is not inherently obvious how PRC2 would differentially target hypo- or hypermethylated DNA. PRC2 is recruited to cis-elements at repressed loci. These PRC2 recruitment elements have been identified in multiple organisms and can include repeat sequences (Kinoshita et al., 2007; Makarevich et al., 2008), small segments of CG-rich sequence (Jermann et al., 2014), or transcription factor binding sites (Berger et al., 2011; Liu et al., 2011; Lodha et al., 2013). In addition, non-coding RNA has been shown to interact with PRC2 and target it to specific loci in plants (Heo and Sung, 2011). DNA methylation interferes with PRC2 function and prevents H3K27me3 modification (Weinhofer et al., 2010; Deleris et al., 2012; Jermann et al., 2014). Thus, hypermethylation of paternal alleles can interfere with PRC2 recruitment sites allowing expression of the paternal allele, while PRC2 activity at the maternal, hypomethylated allele would result in transcriptional silencing to give a PEG pattern of expression. Global analysis of H3K27me3 sites in maize supports this model (Makarevitch et al., 2013). Indeed, Zhang et al. (2014) found that PEGs showed enrichment for maternal H3K27me3 marks concomitant with hypomethylation at the maternal allele and hypermethylation at the paternal allele.
PRC2 is also required to repress the paternal allele of some MEGs (Baroux et al., 2006; Gehring et al., 2006; Jullien et al., 2006b). However, there is no molecular mechanism proposed for how PRC2 would preferentially target the hypermethylated, paternal allele at a MEG locus. Maternal specific expression is also observed for the Arabidopsis ZIX locus, but the MEG pattern of expression does not dependent upon DME or FIS-PRC2 (Ngo et al., 2012). Moreover, imprinted gene expression is documented within the embryo of both maize and Arabidopsis (Jahnke and Scholten, 2009; Raissig et al., 2013). The DME DNA glycosylase is not expressed significantly in the Arabidopsis egg cell (Choi et al., 2002), and there is no evidence for global DNA demethylation in the embryo (Hsieh et al., 2009). The FIS-PRC2 complex is required for some embryo imprinted gene expression suggesting that H3K27me3 does have a functional role in setting-up MEG and PEG expression in the embryo (Raissig et al., 2013). These observations indicate that we are far from completely understanding the molecular mechanisms guiding imprinted expression patterns during seed development.
IS IMPRINTING NECESSARY FOR SEED DEVELOPMENT?
Genetic analysis of imprinted genes suggests a similar spectrum of developmental functions as for biallelic-expressed genes. As mentioned earlier, imprinting in maize can affect non-essential genes regulating anthocyanin biosynthesis or storage protein accumulation. There are also numerous imprinted genes that have been shown to have critical roles in seed development. Superficially, it is simple to conclude that imprinted expression patterns are therefore critical to seed development. However, we argue that most of these examples fail to provide conclusive evidence that the imprinted pattern is indispensable as opposed to a minimum expression level of the gene being critical.
The MEA, FIE, and FIS2 genes encode subunits of PRC2 and are MEGs in Arabidopsis (Luo et al., 2000). Loss-of-function mutations in these genes have profound effects on seed development with 50% seed abortion, delays in endosperm and embryo development, and increased cell proliferation in the developmentally delayed endosperm and embryo (Ohad et al., 1996; Chaudhury et al., 1997; Grossniklaus et al., 1998; Kiyosue et al., 1999). Moreover, these mutants can begin central cell divisions even when the ovule is not fertilized. The FIS-PRC2 phenotypes have been interpreted as imprinted expression of PRC2 being a key repressor of seed growth. However, very similar phenotypes are observed in mutants of the non-imprinted subunit of the FIS-PRC2 complex, MSI1, suggesting that imprinted gene expression is not directly responsible for the seed phenotypes (Köhler et al., 2003; Guitton and Berger, 2005; Leroy et al., 2007). This conclusion is further supported by the maize MEA/Enhancer of zeste (Mez1) gene, which is an endosperm MEG (Haun et al., 2007). A transposon insertion in the promoter region of the locus causes biallelic expression of Mez1 but no change in seed phenotype (Haun et al., 2009). These data show that FIS-PRC2 function needs to be expressed at sufficient levels in both the female gametophyte and developing seed. However, direct evidence is lacking to support the hypothesis that imprinted expression of the PRC2 subunits is required.
The PHERES1 (PHE1) gene is consistently up-regulated in Arabidopsis mea, fie, and fis2 mutants (Köhler et al., 2003). The PHE1 locus was the first PEG to be identified and encodes the AGAMOUS-LIKE37 (AGL37) MADS-domain protein that is a predicted transcription factor (Köhler et al., 2003, 2005). Knocking-down expression of PHE1 in a mea mutant can partially rescue mea defective seed phenotypes suggesting that part of the FIS-PRC2 mutant phenotypes are due to increased PHE1 expression (Köhler et al., 2003). However, insertion mutants in the 3′ regulatory region of PHE1 can cause a loss of imprinting, switching to a biallelic pattern, with no effect on seed phenotype reported (Makarevich et al., 2008). PHE1 is one of several AGL genes, including AGL28, AGL36, AGL40, AGL62, and AGL90, which are up-regulated when endosperm cellularization is delayed either by PRC2 mutants or genome dosage imbalances (Kradolfer et al., 2013a). Although AGL28 and AGL36 are MEGs (Shirzadi et al., 2011; Wolff et al., 2011), the other AGL genes are expressed from both parental alleles. Mutations in AGL62 cause recessive seed defects illustrating that FIS-PRC2 complex influences biallelic genes as well as imprinted genes (Kang et al., 2008). Total expression levels of the AGL co-expression network correlates well with the timing of endosperm cellularization and embryo development (Walia et al., 2009; Tiwari et al., 2010; Kradolfer et al., 2013a). The divergent mechanisms of epigenetic control for these AGL genes and the lack of a requirement for paternal expression of PHE1 suggest that imprinting per se is not likely the primary regulator of this developmental node.
An additional Arabidopsis PEG, ADMETOS (ADM), has been implicated in regulating the AGL gene node (Kradolfer et al., 2013b). ADM encodes a recently evolved J-domain protein that is only found in a few genera of the Brassicaceae. Consistent with its PEG expression, the adm locus was identified as a paternal-specific suppressor of seed abortion due to paternal genome excess (Kradolfer et al., 2013b). When mutated, adm reduces the overexpression of PHE1 and other AGL genes toward normal both in interploidy crosses and in mea mutants. Although ADM is a PEG in multiple Arabidopsis accessions (Hsieh et al., 2011; Wolff et al., 2011), natural variation reducing ADM expression level in the Ler accession is correlated with improved seed development and viability in paternal genome excess crosses (Kradolfer et al., 2013b). When adm is mutant in both maternal and paternal gametes, adm more effectively reduces AGL expression as well as more effectively suppresses seed abortion due to either paternal genome excess or mea, suggesting the maternal allele expresses at a developmentally significant level (Kradolfer et al., 2013b). Interestingly, homozygous adm/adm plants, overexpression of ADM, and biallelic expression of ADM have no seed phenotype in diploid crosses, suggesting that imprinting of this gene is not necessary for normal diploid seed development. The wild-type function of ADM is primarily to block interploidy and interspecific hybridizations.
The Arabidopsis FORMIN HOMOLOGUE5 (AtFH5) gene is a MEG in which the paternal allele is repressed by PRC2 (Fitz Gerald et al., 2009). ATFH5 is an actin nucleator and is critical for cell plate formation and endosperm cellularization (Ingouff et al., 2005). Ectopic expression of the paternal allele of AtFH5 does not impact mea mutant phenotypes (Fitz Gerald et al., 2009), suggesting paternal silencing of AtFH5 may not be required for normal endosperm development. Moreover, double mutants of mea and atfh5 show additive endosperm cellularization and morphogenic defects (Fitz Gerald et al., 2009). These genetic results typically would be interpreted as indicating mea and atfh5 act in different genetic pathways. Although AtFH5 is clearly an imprinted gene with a critical endosperm development function, it is unclear whether the imprinted gene expression pattern has a significant role in endosperm development.
The role of imprinting for the Arabidopsis MATERNALLY EXPRESSED PAB C-TERMINAL (MPC) gene is even less clear than for AtFH5. MPC encodes the C-terminal domain of poly(A) binding proteins (PABP) and is hypothesized to have a role in regulating translation of mRNA (Tiwari et al., 2008). The MPC gene is a MEG, and homozygous mpc RNAi lines show abnormal embryo and endosperm development. However, the role of imprinted gene expression for MPC function is difficult to address, since the gene body sequence is necessary to confer maternal specific expression (Tiwari et al., 2008).
In maize, only one imprinted gene has been functionally characterized in seed development. The meg1 locus encodes a small, secreted peptide that is expressed specifically in the basal endosperm transfer cell layer of the developing endosperm (Gutierrez-Marcos et al., 2004). Meg1 is initially expressed from the maternal allele and becomes biallelic around 12 days after pollination. RNAi of meg1 results in reduced transfer cell differentiation and smaller seeds than non-transgenic controls (Costa et al., 2012). Ectopic expression of Meg1 results in patchy, ectopic transfer cell differentiation throughout the epidermal endosperm, indicating that MEG1 protein is a positive regulator of transfer cell fate.
Meg1 is part of a gene family with six members expressing at significant levels in transfer cells during seed development (Xiong et al., 2014). Only Meg1 shows imprinted expression with all other Meg family members expressing similarly when inherited through either parent. The developmental function of these other Meg family members has not yet been experimentally tested. However, the expression patterns of these genes are very similar to Meg1 in both developmental timing and location, suggesting that these genes are also likely to be regulators of transfer cell differentiation (Xiong et al., 2014). To directly test the role of imprinting on Meg1 function, Costa et al. (2012) developed a non-imprinted, synthetic Meg1 gene with two different transfer cell specific promoters. These transgenics show a dosage-sensitive increase both in the number of transfer cells and in seed size suggesting that imprinting of Meg1 serves to limit nutrient uptake and seed size. Thus, among more than a dozen seed developmental genes studied in detail, only Meg1 has strong evidence indicating that imprinted gene expression has a significant impact on development and growth of the seed.
WHY DOES IMPRINTING EXIST IN ANGIOSPERMS?
Parent-of-origin specific gene expression is a fascinating pattern of molecular regulation of the genome, and its evolution has been the subject of extensive theoretical debate (Patten et al., 2014). The most accredited explanation for imprinting in plants is provided by the parental-conflict hypothesis, also known as the kinship theory of selection (Haig and Westoby, 1989; Haig and Westoby, 1991; Haig, 2013). This hypothesis argues that imprinting evolves when the maternal parent provides resources during offspring development. In angiosperms, seeds require nutrition from the maternal parent from fertilization until seed maturation. The parental-conflict hypothesis states that the paternal genome expression is selected to increase support for individual progeny, while the maternal genome expression is selected to limit resources to maximize seed set.
The parental-conflict hypothesis predicts that MEGs should reduce seed size and potentially reduce seed set in unfavorable conditions. Conversely, PEGs would increase seed size and promote seed set. Loss-of-function phenotypes of the FIS-PRC2 mutants mea, fis2, fie, and msi1 have been interpreted to support parental-conflict theory, because these mutants extend cell proliferation in the endosperm and embryo at the cost of failing to complete development (Grossniklaus et al., 1998; Kiyosue et al., 1999; Guitton and Berger, 2005; Ingouff et al., 2005). However, parental-conflict predicts stable networks of imprinted genes with MEGs and PEGs balancing each other for normal seed development (Patten et al., 2014). As discussed above, most imprinted genes, except for Meg1, that have been studied in detail can lose imprinted gene expression without significant consequence to seed development, suggesting MEG or PEG expression does not undergo significant selection pressure. The case of the Meg1 gene also argues against the parental-conflict hypothesis. Meg1 is normally maternally expressed, and a non-imprinted Meg1 transgene shows a positive dosage effect for increasing seed size (Costa et al., 2012). The parental-conflict hypothesis would predict that a maternally expressed peptide like MEG1 should be a repressor of transfer cell development or that Meg1 should be a PEG.
An alternate hypothesis to explain imprinting is the maternal-offspring coadaptation model of gene expression, in which maternal alleles may be selected for imprinted expression to provide the greatest combined fitness for the mother and offspring (Wolf and Hager, 2006). This model is meant primarily to explain the larger number of MEGs over PEGs that have been identified in both mammals and angiosperms. For Meg1, the model correctly predicts maternal specific expression, but maternal-offspring coadaptation does not explain the relatively extensive number of PEGs or the apparent mutability of most angiosperm imprinted genes to switch between biallelic and imprinted states (Patten et al., 2014).
More in-depth evolutionary analysis of the identified imprinted genes in Arabidopsis suggests that imprinting correlates with rapid evolution of gene duplicates. More than two-thirds of Arabidopsis imprinted genes derive from recent gene duplication events (Qiu et al., 2014). Arabidopsis imprinted genes also show reduced domains of expression and increased evolutionary rates over non-imprinted paralogs. This analysis argues strongly that imprinted genes are undergoing neofunctionalization. Neither the parental-conflict nor the coadaptation models predict that recent gene duplication events would be favored for imprinted expression, although the bias toward gene duplicates does not specifically argue against these evolutionary models (Patten et al., 2014).
The current understanding of cis-elements targeting genes for imprinting further suggests that imprinting is primarily a rapid form of evolution. Transposons and short repeats appear to be the targets of differential demethylation in the central cell versus sperm cells (Gehring et al., 2009; Hsieh et al., 2009). Transposon movement allows random conversion of genes to an imprinted pattern of expression. For example, differences in transposon insertions near genes are associated with allele-specific imprinting in Arabidopsis (Pignatta et al., 2014). Transposon and other insertions can also convert imprinted genes to biallelic expression patterns, providing a fast mechanism to revert alleles into Mendelian, diploid expression (Haun et al., 2009). Importantly, transposition is known to increase in plants exposed to abiotic and biotic stress, suggesting imprinted gene expression is expected to change more rapidly when plants are poorly adapted to an environment (reviewed in Chénais et al., 2012). Although transposon insertions are generally thought to reduce gene expression, genome-wide analysis of gene expression and DNA methylation of 140 Arabidopsis accessions suggests transposon insertion within genes is associated with increased expression levels specifically during seed and pollen development (Schmitz et al., 2013). This more permissive epigenetic state allows genes silenced in other tissues to be expressed during seed development.
Based on the recent genome-wide analyses of imprinting and epigenetic regulation, we suggest that imprinting is a form of epigenetic regulation that allows more rapid selection on recent gene duplicates. Imprinting uncovers individual alleles by converting genes into a pseudohaploid mode of expression during seed development. There is no evidence for prolonged, imprinted expression of genes after germination, and many imprinted genes are expressed later in plant development (Pignatta et al., 2014). Thus, imprinting of one copy of a gene duplicate enables the imprinted gene to accumulate mutations without compromising whole plant fitness. Monoallelic expression in the seed exposes an imprinted allele to more rapid selection acting primarily upon the seed phenotype. Imprinting of recessive, advantageous alleles can confer greater fitness if only expressed from one parent. By contrast, selection against deleterious imprinted alleles is not as strong as in true haploid inheritance. Deleterious imprinted alleles would only be selected against when inherited from the parent conferring expression. For example, a deleterious PEG would be neutral when inherited from the mother. If a deleterious PEG is linked to an advantageous MEG allele, it could be maintained in a population for a significant period, potentially allowing time for additional compensatory mutations. Thus, imprinted expression may allow plant genomes to explore a larger space of allelic and phenotypic variation in the seed while avoiding deleterious plant phenotypes. The mature seed phenotype is expected to be a major driver of species fitness, and we suggest imprinting is a form of gene expression that allows for more efficient diversifying selection on the seed phenotype.
An important consequence of hypothesizing imprinting as a form of diversifying selection is that most imprinted expression patterns would be expected to have neutral effects on the fitness of the seed. Gene networks that appear to fit parental-conflict or coadaptation models are expected to evolve under diversifying selection. However, the prediction is that the bulk of imprinted expression patterns could revert to biallelic expression with no consequence on seed phenotype. Similarly, allele specific imprinting and novel imprinted loci would be expected to evolve at high frequency. Additional functional data of imprinted genes in outcrossing species such as maize, would help resolve whether parental-conflict or other types of selection is the primary driving force for the evolution of imprinted genes.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mary Daliberti for assistance in preparing Figure 1 and the reviewers for their helpful comments on the manuscript. We apologize to the authors of relevant research articles that were not highlighted in this review due to space constraints. The authors’ research on seed development is supported by grants from the National Science Foundation (awards IOS-1031416 and MCB-1412218), the National Institute of Food and Agriculture (awards 2010-04228 and 2011-67013-30032), and the Vasil-Monsanto Endowment.
REFERENCES
- Arnaud P., Feil R. (2006). MEDEA takes control of its own imprinting. Cell 124 468–470 10.1016/j.cell.2006.01.020 [DOI] [PubMed] [Google Scholar]
- Baroux C., Gagliardini V., Page D. R., Grossniklaus U. (2006). Dynamic regulatory interactions of polycomb group genes: MEDEA autoregulation is required for imprinted gene expression in Arabidopsis. Genes Dev. 20 1081–1086 10.1101/gad.378106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger N., Dubreucq B., Roudier F., Dubos C., Lepiniec L. (2011). Transcriptional regulation of Arabidopsis LEAFY COTYLEDON2 involves RLE, a cis-element that regulates trimethylation of histone H3 at lysine-27. Plant Cell 23 4065–4078 10.1105/tpc.111.087866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi J., Lanubile A., Li Q. B., Kumar D., Kladnik A., Cook S. D., et al. (2012). Impaired auxin biosynthesis in the defective endosperm18 mutant is due to mutational loss of expression in the ZmYuc1 gene encoding endosperm-specific YUCCA1 protein in maize. Plant Physiol. 160 1318–1328 10.1104/pp.112.204743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratzel F., Yang C., Angelova A., López-Torrejón G., Koch M., del Pozo J. C., et al. (2012). Regulation of the new Arabidopsis imprinted gene AtBMI1C requires the interplay of different epigenetic mechanisms. Mol. Plant 5 260–269 10.1093/mp/ssr078 [DOI] [PubMed] [Google Scholar]
- Brown R., Lemmon B. E., Nguyen H., Olsen O.-A. (1999). Development of endosperm in Arabidopsis thaliana. Sex. Plant Reprod. 12 32–42 10.1007/s004970050169 [DOI] [Google Scholar]
- Brown R. C., Lemmon B. E., Olsen O.-A. (1994). Endosperm development in barley: microtubule involvement in the morphogenetic pathway. Plant Cell 6 1241–1252 10.1105/tpc.6.9.1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R. C., Lemmon B. E., Olsen O.-A. (1996). Development of the endosperm in rice (Oryza sativa L.): cellularization. J. Plant Res. 109 301–313 10.1007/BF02344477 [DOI] [Google Scholar]
- Chandler J., Nardmann J., Werr W. (2008). Plant development revolves around axes. Trends Plant Sci. 13 78–84 10.1016/j.tplants.2007.11.010 [DOI] [PubMed] [Google Scholar]
- Chaudhuri S., Messing J. (1994). Allele-specific parental imprinting of dzr1, a posttranscriptional regulator of zein accumulation. Proc. Natl. Acad. Sci. U.S.A. 91 4867–4871 10.1073/pnas.91.11.4867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury A. M., Ming L., Miller C., Craig S., Dennis E. S., Peacock W. J. (1997). Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 94 4223–4228 10.1073/pnas.94.8.4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chénais B., Caruso A., Hiard S., Casse N. (2012). The impact of transposable elements on eukaryotic genomes: from genome size increase to genetic adaptation to stressful environments. Gene 509 7–15 10.1016/j.gene.2012.07.042 [DOI] [PubMed] [Google Scholar]
- Chia J. M., Song C., Bradbury P. J., Costich D., De Leon N., Doebley J., et al. (2012). Maize HapMap2 identifies extant variation from a genome in flux. Nat. Genet. 44 803–807 10.1038/ng.2313 [DOI] [PubMed] [Google Scholar]
- Choi Y., Gehring M., Johnson L., Hannon M., Harada J. J., Goldberg R. B., et al. (2002). DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110 33–42 10.1016/S0092-8674(02)00807-3 [DOI] [PubMed] [Google Scholar]
- Costa L. M., Marshall E., Tesfaye M., Silverstein K. A., Mori M., Umetsu Y., et al. (2014). Central cell-derived peptides regulate early embryo patterning in flowering plants. Science 344 168–172 10.1126/science.1243005 [DOI] [PubMed] [Google Scholar]
- Costa L. M., Yuan J., Rouster J., Paul W., Dickinson H., Gutierrez-Marcos J. F. (2012). Maternal control of nutrient allocation in plant seeds by genomic imprinting. Curr. Biol. 22 160–165 10.1016/j.cub.2011.11.059 [DOI] [PubMed] [Google Scholar]
- Day R. C., Herridge R. P., Ambrose B. A., Macknight R. C. (2008). Transcriptome analysis of proliferating Arabidopsis endosperm reveals biological implications for the control of syncytial division, cytokinin signaling, and gene expression regulation. Plant Physiol. 148 1964–1984 10.1104/pp.108.128108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris A., Stroud H., Bernatavichute Y., Johnson E., Klein G., Schubert D., et al. (2012). Loss of the DNA methyltransferase MET1 induces H3K9 hypermethylation at PcG target genes and redistribution of H3K27 trimethylation to transposons in Arabidopsis thaliana. PLoS Genet. 8:e1003062 10.1371/journal.pgen.1003062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I., Lau S., Mayer U., Jürgens G. (2010). Embryogenesis – the humble beginnings of plant life. Plant J. 61 959–970 10.1111/j.1365-313X.2010.04143.x [DOI] [PubMed] [Google Scholar]
- Fatihi A., Zbierzak A. M., Dormann P. (2013). Alterations in seed development gene expression affect size and oil content of Arabidopsis seeds. Plant Physiol. 163 973–985 10.1104/pp.113.226761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitz Gerald J. N., Hui P. S., Berger F. (2009). Polycomb group-dependent imprinting of the actin regulator AtFH5 regulates morphogenesis in Arabidopsis thaliana. Development 136 3399–3404 10.1242/dev.036921 [DOI] [PubMed] [Google Scholar]
- Fouquet R., Martin F., Fajardo D. S., Gault C. M., Gomez E., Tseung C. W., et al. (2011). Maize rough endosperm3 encodes an RNA splicing factor required for endosperm cell differentiation and has a nonautonomous effect on embryo development. Plant Cell 23 4280–4297 10.1105/tpc.111.092163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M. (2013). Genomic imprinting: insights from plants. Annu. Rev. Genet. 47 187–208 10.1146/annurev-genet-110711-155527 [DOI] [PubMed] [Google Scholar]
- Gehring M., Bubb K. L., Henikoff S. (2009). Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324 1447–1451 10.1126/science.1171609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M., Huh J. H., Hsieh T. F., Penterman J., Choi Y., Harada J. J., et al. (2006). DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124 495–506 10.1016/j.cell.2005.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M., Missirian V., Henikoff S. (2011). Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS ONE 6:e23687 10.1371/journal.pone.0023687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U., Vielle-Calzada J. P., Hoeppner M. A., Gagliano W. B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280 446–450 10.1126/science.280.5362.446 [DOI] [PubMed] [Google Scholar]
- Guitton A. E., Berger F. (2005). Loss of function of MULTICOPY SUPPRESSOR OF IRA 1 produces nonviable parthenogenetic embryos in Arabidopsis. Curr. Biol. 15 750–754 10.1016/j.cub.2005.02.066 [DOI] [PubMed] [Google Scholar]
- Gutierrez-Marcos J. F., Costa L. M., Biderre-Petit C., Khbaya B., O’sullivan D. M., Wormald M., et al. (2004). Maternally expressed gene1 is a novel maize endosperm transfer cell-specific gene with a maternal parent-of-origin pattern of expression. Plant Cell 16 1288–1301 10.1105/tpc.019778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D. (2013). Kin conflict in seed development: an interdependent but fractious collective. Annu. Rev. Cell Dev. Biol. 29 189–211 10.1146/Annurev-Cellbio-101512-122324 [DOI] [PubMed] [Google Scholar]
- Haig D., Westoby M. (1989). Parent-specific gene-expression and the triploid endosperm. Am. Nat. 134 147–155 10.1086/284971 [DOI] [Google Scholar]
- Haig D., Westoby M. (1991). Genomic imprinting in endosperm - its effect on seed development in crosses between species, and between different ploidies of the same species, and its implications for the evolution of apomixis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 333 1–13 10.1098/Rstb.1991.0057 [DOI] [Google Scholar]
- Haun W. J., Danilevskaya O. N., Meeley R. B., Springer N. M. (2009). Disruption of imprinting by mutator transposon insertions in the 5’ proximal regions of the Zea mays Mez1 locus. Genetics 181 1229–1237 10.1534/genetics.108.093666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haun W. J., Laoueille-Duprat S., O’Connell M. J., Spillane C., Grossniklaus U., Phillips A. R., et al. (2007). Genomic imprinting, methylation and molecular evolution of maize enhancer of zeste (Mez) homologs. Plant J. 49 325–337 10.1111/j.1365-313X.2006.02965.x [DOI] [PubMed] [Google Scholar]
- Heo J. B., Sung S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331 76–79 10.1126/science.1197349 [DOI] [PubMed] [Google Scholar]
- Hermon P., Srilunchang K. O., Zou J., Dresselhaus T., Danilevskaya O. N. (2007). Activation of the imprinted polycomb group fie1 gene in maize endosperm requires demethylation of the maternal allele. Plant Mol. Biol. 64 387–395 10.1007/s11103-007-9160-0 [DOI] [PubMed] [Google Scholar]
- Hsieh T. F., Ibarra C. A., Silva P., Zemach A., Eshed-Williams L., Fischer R. L., et al. (2009). Genome-wide demethylation of Arabidopsis endosperm. Science 324 1451–1454 10.1126/science.1172417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T. F., Shin J., Uzawa R., Silva P., Cohen S., Bauer M. J., et al. (2011). Regulation of imprinted gene expression in Arabidopsis endosperm. Proc. Natl. Acad. Sci. U.S.A. 108 1755–1762 10.1073/pnas.1019273108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M., Fitz Gerald J. N., Guerin C., Robert H., Sorensen M. B., Van Damme D., et al. (2005). Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nat. Cell Biol. 7 374–380 10.1038/ncb1238 [DOI] [PubMed] [Google Scholar]
- Jahnke S., Scholten S. (2009). Epigenetic resetting of a gene imprinted in plant embryos. Curr. Biol. 19 1677–1681 10.1016/j.cub.2009.08.053 [DOI] [PubMed] [Google Scholar]
- Jermann P., Hoerner L., Burger L., Schübeler D. (2014). Short sequences can efficiently recruit histone H3 lysine 27 trimethylation in the absence of enhancer activity and DNA methylation. Proc. Natl. Acad. Sci. U.S.A. 111 E3415–E3421 10.1073/pnas.1400672111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Zhao H., Ren L., Song W., Zeng B., Guo J., et al. (2012). Genome-wide genetic changes during modern breeding of maize. Nat. Genet. 44 812–815 10.1038/ng.2312 [DOI] [PubMed] [Google Scholar]
- Jullien P. E., Kinoshita T., Ohad N., Berger F. (2006a). Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell 18 1360–1372 10.1105/tpc.106.041178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien P. E., Katz A., Oliva M., Ohad N., Berger F. (2006b). Polycomb group complexes self-regulate imprinting of the polycomb group gene MEDEA in Arabidopsis. Curr. Biol. 16 486–492 10.1016/j.cub.2006.01.020 [DOI] [PubMed] [Google Scholar]
- Kang I. H., Steffen J. G., Portereiko M. F., Lloyd A., Drews G. N. (2008). The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20 635–647 10.1105/tpc.107.055137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle J. L. (1969). Androgenesis conditioned by a mutation in maize. Science 166 1422–1424 10.1126/science.166.3911.1422 [DOI] [PubMed] [Google Scholar]
- Kermicle J. L. (1970). Dependence of the R-mottled aleurone phenotype in maize on mode of sexual transmission. Genetics 66 69–85 10.3410/f.718486711.793497084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesselbach T. A. (1949). The Structure and Reproduction of Corn. Research Bulletin 161. Lincoln: Agricultural Experiment Station, University of Nebraska College of Agriculture. [Google Scholar]
- Kinoshita T., Miura A., Choi Y., Kinoshita Y., Cao X., Jacobsen S. E., et al. (2004). One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303 521–523 10.1126/science.1089835 [DOI] [PubMed] [Google Scholar]
- Kinoshita Y., Saze H., Kinoshita T., Miura A., Soppe W. J., Koornneef M., et al. (2007). Control of FWA gene silencing in Arabidopsis thaliana by SINE-related direct repeats. Plant J. 49 38–45 10.1111/j.1365-313X.2006.02936.x [DOI] [PubMed] [Google Scholar]
- Kiyosue T., Ohad N., Yadegari R., Hannon M., Dinneny J., Wells D., et al. (1999). Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 96 4186–4191 10.1073/pnas.96.7.4186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C., Hennig L., Bouveret R., Gheyselinck J., Grossniklaus U., Gruissem W. (2003). Arabidopsis MSI1 is a component of the MEA/FIE polycomb group complex and required for seed development. EMBO J. 22 4804–4814 10.1093/emboj/cdg444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C., Page D. R., Gagliardini V., Grossniklaus U. (2005). The Arabidopsis thaliana MEDEA polycomb group protein controls expression of PHERES1 by parental imprinting. Nat. Genet. 37 28–30 10.1038/ng1495 [DOI] [PubMed] [Google Scholar]
- Köhler C., Wolff P., Spillane C. (2012). Epigenetic mechanisms underlying genomic imprinting in plants. Annu. Rev. Plant Biol. 63 331–352 10.1146/annurev-arplant-042811-105514 [DOI] [PubMed] [Google Scholar]
- Kradolfer D., Hennig L., Köhler C. (2013a). Increased maternal genome dosage bypasses the requirement of the FIS polycomb repressive complex 2 in Arabidopsis seed development. PLoS Genet. 9:e1003163 10.1371/journal.pgen.1003163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kradolfer D., Wolff P., Jiang H., Siretskiy A., Köhler C. (2013b). An imprinted gene underlies postzygotic reproductive isolation in Arabidopsis thaliana. Dev. Cell 26 525–535 10.1016/j.devcel.2013.08.006 [DOI] [PubMed] [Google Scholar]
- Leroy O., Hennig L., Breuninger H., Laux T., Köhler C. (2007). Polycomb group proteins function in the female gametophyte to determine seed development in plants. Development 134 3639–3648 10.1242/dev.009027 [DOI] [PubMed] [Google Scholar]
- Li J., Berger F. (2012). Endosperm: food for humankind and fodder for scientific discoveries. New Phytol. 195 290–305 10.1111/j.1469-8137.2012.04182.x [DOI] [PubMed] [Google Scholar]
- Liu X., Kim Y. J., Müller R., Yumul R. E., Liu C., Pan Y., et al. (2011). AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of polycomb group proteins. Plant Cell 23 3654–3670 10.1105/tpc.111.091538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodha M., Marco C. F., Timmermans M. C. (2013). The ASYMMETRIC LEAVES complex maintains repression of KNOX homeobox genes via direct recruitment of polycomb-repressive complex2. Genes Dev. 27 596–601 10.1101/gad.211425.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig S. R., Habera L. F., Dellaporta S. L., Wessler S. R. (1989). Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc. Natl. Acad. Sci. U.S.A. 86 7092–7096 10.1073/pnas.86.18.7092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Bilodeau P., Dennis E. S., Peacock W. J., Chaudhury A. (2000). Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. U.S.A. 97 10637–10642 10.1073/pnas.170292997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Bilodeau P., Koltunow A., Dennis E. S., Peacock W. J., Chaudhury A. M. (1999). Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 96 296–301 10.1073/pnas.96.1.296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevich G., Leroy O., Akinci U., Schubert D., Clarenz O., Goodrich J., et al. (2006). Different polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 7 947–952 10.1038/sj.embor.7400760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevich G., Villar C. B., Erilova A., Köhler C. (2008). Mechanism of PHERES1 imprinting in Arabidopsis. J. Cell Sci. 121 906–912 10.1242/jcs.023077 [DOI] [PubMed] [Google Scholar]
- Makarevitch I., Eichten S. R., Briskine R., Waters A. J., Danilevskaya O. N., Meeley R. B., et al. (2013). Genomic distribution of maize facultative heterochromatin marked by trimethylation of H3K27. Plant Cell 25 780–793 10.1105/tpc.112.106427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo Q. A., Baroux C., Guthörl D., Mozerov P., Collinge M. A., Sundaresan V., et al. (2012). The Armadillo repeat gene ZAK IXIK promotes Arabidopsis early embryo and endosperm development through a distinctive gametophytic maternal effect. Plant Cell 24 4026–4043 10.1105/tpc.112.102384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad N., Margossian L., Hsu Y. C., Williams C., Repetti P., Fischer R. L. (1996). A mutation that allows endosperm development without fertilization. Proc. Natl. Acad. Sci. U.S.A. 93 5319–5324 10.1073/pnas.93.11.5319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen O.-A. (2004). Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell 16 S214–S227 10.1105/tpc.017111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly C., Shepherd N. S., Pereira A., Schwarz-Sommer Z., Bertram I., Donald S., et al. (1985). Molecular cloning of the a1 locus of Zea mays using the transposable elements En and Mu1. EMBO J. 4 877–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten M. M., Ross L., Curley J. P., Queller D. C., Bonduriansky R., Wolf J. B. (2014). The evolution of genomic imprinting: theories, predictions and empirical tests. Heredity (Edinb.) 113 119–128 10.1038/hdy.2014.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris C. I., Rademacher E. H., Weijers D. (2010). Green beginnings - pattern formation in the early plant embryo. Curr. Top. Dev. Biol. 91 1–27 10.1016/S0070-2153(10)91001-6 [DOI] [PubMed] [Google Scholar]
- Perrot G. H., Cone K. C. (1989). Nucleotide sequence of the maize R-S gene. Nucleic Acids Res. 17:8003 10.1093/nar/17.19.8003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatta D., Erdmann R. M., Scheer E., Picard C. L., Bell G. W., Gehring M. (2014). Natural epigenetic polymorphisms lead to intraspecific variation in Arabidopsis gene imprinting. Elife 3:e03198 10.7554/eLife.03198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires N. D., Grossniklaus U. (2014). Different yet similar: evolution of imprinting in flowering plants and mammals. F1000Prime Rep. 6 63 10.12703/P6-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Liu S. L., Adams K. L. (2014). Frequent changes in expression profile and accelerated sequence evolution of duplicated imprinted genes in Arabidopsis. Genome Biol. Evol. 6 1830–1842 10.1093/gbe/evu144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig M. T., Bemer M., Baroux C., Grossniklaus U. (2013). Genomic imprinting in the Arabidopsis embryo is partly regulated by PRC2. PLoS Genet. 9:e1003862 10.1371/journal.pgen.1003862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli P. A., Larkins B. A. (2009a). The contribution of cell cycle regulation to endosperm development. Sex. Plant Reprod. 22 207–219 10.1007/s00497-009-0105-4 [DOI] [PubMed] [Google Scholar]
- Sabelli P. A., Larkins B. A. (2009b). The development of endosperm in grasses. Plant Physiol. 149 14–26 10.1104/pp.108.129437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R. J., Schultz M. D., Urich M. A., Nery J. R., Pelizzola M., Libiger O., et al. (2013). Patterns of population epigenomic diversity. Nature 495 193–198 10.1038/nature11968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoft V. K., Chumak N., Choi Y., Hannon M., Garcia-Aguilar M., Machlicova A., et al. (2011). Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc. Natl. Acad. Sci. U.S.A. 108 8042–8047 10.1073/pnas.1105117108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B., Cavalli G. (2009). Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136 3531–3542 10.1242/dev.033902 [DOI] [PubMed] [Google Scholar]
- Selinger D. A., Chandler V. L. (2001). B-Bolivia, an allele of the maize b1 gene with variable expression, contains a high copy retrotransposon-related sequence immediately upstream. Plant Physiol. 125 1363–1379 10.1104/pp.125.3.1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirzadi R., Andersen E. D., Bjerkan K. N., Gloeckle B. M., Heese M., Ungru A., et al. (2011). Genome-wide transcript profiling of endosperm without paternal contribution identifies parent-of-origin-dependent regulation of AGAMOUS-LIKE36. PLoS Genet. 7:e1001303 10.1371/journal.pgen.1001303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangeland B., Salehian Z., Aalen R., Mandal A., Olsen O.-A. (2003). Isolation of GUS marker lines for genes expressed in Arabidopsis endosperm, embryo and maternal tissues. J. Exp. Bot. 54 279–290 10.1093/jxb/erg031 [DOI] [PubMed] [Google Scholar]
- Tiwari S., Schulz R., Ikeda Y., Dytham L., Bravo J., Mathers L., et al. (2008). MATERNALLY EXPRESSED PAB C-TERMINAL, a novel imprinted gene in Arabidopsis, encodes the conserved C-terminal domain of polyadenylate binding proteins. Plant Cell 20 2387–2398 10.1105/tpc.108.061929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S., Spielman M., Schulz R., Oakey R. J., Kelsey G., Salazar A., et al. (2010). Transcriptional profiles underlying parent-of-origin effects in seeds of Arabidopsis thaliana. BMC Plant Biol. 10:72 10.1186/1471-2229-10-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu T. M., Nakamura M., Calarco J. P., Susaki D., Lim P. Q., Kinoshita T., et al. (2013). RNA-directed DNA methylation regulates parental genomic imprinting at several loci in Arabidopsis. Development 140 2953–2960 10.1242/dev.092981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia H., Josefsson C., Dilkes B., Kirkbride R., Harada J., Comai L. (2009). Dosage-dependent deregulation of an AGAMOUS-LIKE gene cluster contributes to interspecific incompatibility. Curr. Biol. 19 1128–1132 10.1016/j.cub.2009.05.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters A. J., Bilinski P., Eichten S. R., Vaughn M. W., Ross-Ibarra J., Gehring M., et al. (2013). Comprehensive analysis of imprinted genes in maize reveals allelic variation for imprinting and limited conservation with other species. Proc. Natl. Acad. Sci. U.S.A. 110 19639–19644 10.1073/pnas.1309182110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters A. J., Makarevitch I., Eichten S. R., Swanson-Wagner R. A., Yeh C. T., Xu W., et al. (2011). Parent-of-origin effects on gene expression and DNA methylation in the maize endosperm. Plant Cell 23 4221–4233 10.1105/tpc.111.092668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhofer I., Hehenberger E., Roszak P., Hennig L., Köhler C. (2010). H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet. 6:e1001152 10.1371/journal.pgen.1001152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendrich J. R., Weijers D. (2013). The Arabidopsis embryo as a miniature morphogenesis model. New Phytol. 199 14–25 10.1111/nph.12267 [DOI] [PubMed] [Google Scholar]
- Wolf J. B., Hager R. (2006). A maternal-offspring coadaptation theory for the evolution of genomic imprinting. PLoS Biol. 4:e380 10.1371/journal.pbio.0040380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff P., Weinhofer I., Seguin J., Roszak P., Beisel C., Donoghue M. T., et al. (2011). High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genet. 7:e1002126 10.1371/journal.pgen.1002126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Brown R. C., Lemmon B. E., Harada J. J., Goldberg R. B., Fischer R. L. (2006). Regulation of seed size by hypomethylation of maternal and paternal genomes. Plant Physiol. 142 1160–1168 10.1104/pp.106.088849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M., Yang R., Li G., Chen H., Laurie J., Ma C., et al. (2013). Dynamic expression of imprinted genes associates with maternally controlled nutrient allocation during maize endosperm development. Plant Cell 25 3212–3227 10.1105/tpc.113.115592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Q., Creff A., Waters A., Tanaka H., Goodrich J., Ingram G. C. (2013). ZHOUPI controls embryonic cuticle formation via a signaling pathway involving the subtilisin protease ABNORMAL LEAF-SHAPE1 and the receptor kinases GASSHO1 and GASSHO2. Development 140 770–779 10.1242/dev.088898 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Mei W., Kim E. D., Mukherjee K., Hassanein H., Barbazuk W. B., et al. (2014). Adaptive expansion of the maize maternally expressed gene (Meg) family involves changes in expression patterns and protein secondary structures of its members. BMC Plant Biol. 14:204 10.1186/s12870-014-0204-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Johnston N., Talideh E., Mitchell S., Jeffree C., Goodrich J., et al. (2008). The endosperm-specific ZHOUPI gene of Arabidopsis thaliana regulates endosperm breakdown and embryonic epidermal development. Development 135 3501–3509 10.1242/dev.026708 [DOI] [PubMed] [Google Scholar]
- Young T. E., Gallie D. R. (2000). Programmed cell death during endosperm development. Plant Mol. Biol. 44 283–301 10.1023/A:1026588408152 [DOI] [PubMed] [Google Scholar]
- Young T. E., Gallie D. R., DeMason D. A. (1997). Ethylene-mediated programmed cell death during maize endosperm development of wild type and shrunken2 genotypes. Plant Physiol. 115 737–751 10.1104/pp.115.2.737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Chaudhury A., Wu X. (2013). Imprinting in plants and its underlying mechanisms. J. Genet. Genomics 40 239–247 10.1016/j.jgg.2013.04.003 [DOI] [PubMed] [Google Scholar]
- Zhang M., Xie S., Dong X., Zhao X., Zeng B., Chen J., et al. (2014). Genome-wide high resolution parental-specific DNA and histone methylation maps uncover patterns of imprinting regulation in maize. Genome Res. 24 167–176 10.1101/gr.155879.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Zhao H., Xie S., Chen J., Xu Y., Wang K., et al. (2011). Extensive, clustered parental imprinting of protein-coding and noncoding RNAs in developing maize endosperm. Proc. Natl. Acad. Sci. U.S.A. 108 20042–20047 10.1073/pnas.1112186108 [DOI] [PMC free article] [PubMed] [Google Scholar]