Abstract
The pharmacokinetic and pharmacodynamic disciplines address pharmacological traits, including efficacy and adverse events. Pharmacogenomics studies have identified pervasive genetic effects on treatment outcomes, resulting in the development of genetic biomarkers for optimization of drug therapy. Pharmacogenomics-based tests are already being applied in clinical decision making. However, despite substantial progress in identifying the genetic etiology of pharmacological response, current biomarker panels still largely rely on single gene tests with a large portion of the genetic effects remaining to be discovered. Future research must account for the combined effects of multiple genetic variants, incorporate pathway-based approaches, explore gene-gene interactions and nonprotein coding functional genetic variants, extend studies across ancestral populations, and prioritize laboratory characterization of molecular mechanisms. Because genetic factors can play a key role in drug response, accurate biomarker tests capturing the main genetic factors determining treatment outcomes have substantial potential for improving individual clinical care.
Keywords: genetic architecture, genomics, heritability, linear mixed modeling, pharmacogenomics, polygenic architecture, polygenic modeling
The NIH has envisioned translation of the detailed information collected about the human genome into improvements in human health and well being [1]. Pharmacogenomics (PGx), the study of how genetic architecture influences pharmacological traits and outcomes, provides immediate applications for directing clinical decision making. Pharmacogenomic information can aid choices about selecting pharmacological treatments, optimal time courses and drug dosage on the basis of a patients' genetic architecture.
Interpatient variability manifests itself in different ways for pharmacological traits, described quantitatively by pharmacokinetics (PK) and pharmacodynamics (PD). Pharmacological traits include efficacy, adverse events and the balance between efficacy and toxicity, defining the therapeutic ‘window.’ Further definitions of these terms are presented in Box 1. By connecting genetic variation to measurable interpatient variability, a course of action can be defined on an individual basis.
Box 1. Definitions.
Pharmacological trait: Measurable variation in response to pharmacological treatments.
Pharmacokinetics (PK): The degree or rate of absorption, metabolism, distribution and elimination of a drug within a living system.
Pharmacodynamics (PD): The relationship between drug concentration and effect on a living system, or the microorganisms affected within a living system by a drug.
ADME: Absorption, distribution, metabolism and elimination.
Efficacy: The quality of the effect of a pharmacological treatment on a living system in relation to the quantity of the drug.
Adverse event: A detrimental response to a drug within a living system.
Idiosyncratic adverse event: An unexpected response to a drug within a living system.
Therapeutic window: The range of drug concentrations efficacious with minimal toxicity.
Efficacy/toxicity balance: Drug levels have an impact on drug efficacy, but also toxicity.
Pharmacogenomics (PGx): Study of the relationship between genetic variation and drug response, including but not limited to pharmacological traits, PK, PD, efficacy, toxicity and/or adverse events.
NIH Pharmacogenomics Research Network (PGRN): A collaboration between an ever growing number of study sites, all investigating the pharmacogenomics of a variety of traits [2,3].
The Pharmacogenomics Knowledge base (PharmGKB): A database of comprehensively collected information about the relationships between genes and PT, PK, PD, efficacy, adverse events and/or disease [4,5].
Polygenic genetic architecture: The contribution of multiple common SNPs to phenotypic variance in aggregate.
Polygenic modeling: Method that develops an additive polygenic risk score based on SNPs that pass a p-value threshold in a discovery set of samples, tested in an independent set of samples.
Mixed Linear Modeling (MLM): Estimation of an additive genetic variance under a mixed linear model with a random effect representing the polygenic component of underlying trait variation.
Investigating the genetic architecture of PGx traits can be pursued in multiple ways, and each of the approaches taken has advantages and limitations, discussed further in this review. PGx studies typically reveal SNP biomarkers that link genetic variation to treatment outcomes. Elucidating the underlying molecular genetic mechanism underlying the association between genetic variants and pharmacological traits is a major goal in the PGx field. In contrast, genome-wide association studies (GWAS) of complex diseases have yielded numerous variants with significant associations, but for a vast majority of these results, the causative variants and mechanisms remain unknown [6]. In addition, pharmacogenomic variants tend to exert stronger effects on drug response phenotypes than those discovered for complex disorders – perhaps because a relatively limited number of genes influence PK and PD traits. Moreover, drugs are targeted to specific pathways thought to be involved in complex disease phenotypes, thereby narrowing the number of candidate genes, with each displaying a proportionally larger effect size. We assume here that complex disorders such as cardiovascular diseases and cancer represent a collection of disease subtypes each with similar symptoms – where drug therapy would typically target a specific subtype.
A series of limitations and challenges confront the field of PGx. One current limitation is the wide use of single common-variant/outcome trait association testing. Alternative modeling methodologies and strategies that incorporate multiple genetic markers, as well as the inclusion of lower-frequency variants, may prove effective for enhancing PGx trait prediction. A challenge for PGx studies includes the difficulty of estimating the heritability of PGx traits, since it is untenable to administer medications to unaffected individuals. Attaining adequate statistical power presents a challenge when faced with the frequently small sample size in PGx studies, especially for adverse drug reaction studies. Effective biomarker identification has also remained a challenge, with potential biomarkers that have not shown clinical efficacy [7,8], and important work yet to be done understanding the biology underlying effective biomarkers [8]. In addition, the PGx field needs to broaden investigations of PGx traits across samples of diverse ancestry/race/ethnicity, to derive relevant actionable information for clinicians. Finally, ‘risk’ is difficult to define for PGx traits, as metrics for drug efficacy can be difficult to ascertain when compared with the probability (rate) of risk of a common complex disease.
Herein, we describe the current understanding of the genetic architecture of PGx traits. We discuss in detail some of the aforementioned challenges and limitations while also pointing out opportunities and future directions for the field of PGx. These include new methods development such as polygenic modeling, pathway analyses, and systems biology approaches for the development of further robust biomarkers, all with the goal of discovering the etiology underlying interindividual variations in drug response, and designing robust biomarker panels predictive of treatment outcomes.
What have we learned?
Heritability of PGx traits: challenges & successes
The rationale for PGx is the underlying assumption that genetic variation plays a substantial role in pharmacological outcome. The heritability of a PGx trait should be measurable if variant transmission from parent to offspring is the basis of the genetic architecture influencing PGx traits. While determining the heritability provides the rationale for a PGx study, estimating the heritability of PGx traits is nontrivial.
Defining the heritability of PGx traits encounters hurdles distinct from those found in the analysis of complex disorders. By definition, any drug response trait represents a gene–environment interaction, where the drug is only one component of multiple environmental exposures, and multiple genes may contribute to the PGx response. Moreover, each drug, even closely related ones such as the statins, has different degrees of heritability and must be studied individually. Also, drug effects are highly dependent on the dosage, and hence, genetic factors differ with drug dose, as shown for the impact of SLCO1B1 variants on simvastatin's muscle toxicity, only detectable when high doses are needed to control cholesterol levels [9]. Lastly, drug therapy commonly involves multiple drugs. As a result, predictive biomarker tests of the future will have to evolve to consider complex gene–gene–environment interactions.
Few drugs can be used in a familial setting to monitor the variability of drug response as most family members will not have a condition warranting treatment. Drug treatment of a group of individuals without a need for treatment is limited for safety and ethical reasons. In addition, sample size is often low for PGx studies, especially when investigating adverse events. Access to large numbers of related individuals taking a specific drug is limited to communities that have a common need for a particular drug, such as lipid lowering treatments [10]. This is distinct from estimating heritability for complex diseases or outcomes where twin and family studies are facilitated, even when trait complexity can present challenges for heritability estimation.
A few examples of familial studies of drug response clearly indicate the heritability of some PGx traits, underscoring considerable influence of genetic factors; these are presented in Table 1. For example, dicumarol was monitored in the plasma for a group of identical and fraternal twins [11], revealing little difference in the variability of dicumarol half-life in plasma response between identical twins, minor differences between fraternal twins and wide differences between nonrelated subjects demonstrating a significant heritable component. Investigating heritability of PGx traits across communities of related individuals where a specific drug is commonly administered is another way to determine heritability of certain PGx traits. For instance, a study of the heritability of platelet response, measured by ex vivo platelet aggregometry, involved the administration of clopidogrel to 429 Amish persons and revealed the platelet response to be highly heritable [10].
Table 1.
Heritability of pharmacogenomics traits.
| Study type | Trait | Drug | Heritability measure | Heritability estimate | SE | Ref. |
|---|---|---|---|---|---|---|
| Pedigree | Response (platelet aggregation) | Clopidogrel | h2 | 0.73 | ±0.12 | [10] |
|
| ||||||
| Twins | Half-life | Dicumarol | h2 | 0.97 | NA | [11] |
|
| ||||||
| Twins | Half-life | Anti purine | h2 | 0.98 | NA | [11] |
|
| ||||||
| Pedigree | Platelet response measured by phenotypes indirectly related to inhibition of COX-1 | Aspirin (acetylsalicylic acid) | h2 | 0.266–0.762 | SE | [12] |
| PEP aggregation 2 μg/ml collagen | 0.451 | ±0.080 | [12] | |||
| PEP lag time 2 μg/ml collagen | 0.309 | ±0.095 | [12] | |||
| Whole blood aggregation 1 μg/ml collagen | 0.365 | ±0.073 | [12] | |||
| PEP aggregation 10 μmol/l ADP | 0.475 | ±0.083 | [12] | |||
| Whole blood aggregation 10 μmol/l ADP | 0.434 | ±0.070 | [12] | |||
| PEP aggregation 10 μmol/l epinephrine | 0.535 | ±0.077 | [12] | |||
| β-thromboglobulin release | 0.65 | ±0.086 | [12] | |||
NA: Not applicable; PEP: Platelet rich plasma; SE: Standard error.
To circumvent limitations of familial studies for PGx traits, estimation of heritability in an ex vivo manner has been successful. This approach has been applied for measuring drug cytotoxicity within familial-derived lymphoblastoid cell lines (LCL) [13]. Further work with the LCL approach has lead to a detailed understanding of effective LCL study design, enabling identification of loci related to variability of PGx traits which in turn guide studies in humans and model organisms. For example, heritability of chemotherapeutic cisplatin-induced cytotoxicity has been estimated at approximately 57% through the LCL approach, with evidence for multiple causative variants [14]. Table 2 presents a series of heritability results from cell line experiments. Detailed work has characterized factors that confound interpretation of these experiments, such as the portion of the genetic variation of drug-induced cytotoxicity accounted for by heritability of variation in cellular growth rate [13]. In addition, cellular assays are amenable to high-throughput testing of multiple drugs. For example, one study investigated the cytotoxic effect of 29 chemotherapeutic agents on 125 LCL from 14 extended families, and found a range of heritabilities from <15% (gemcitabine) to >60% (epirubicin) [15].
Table 2.
Cell-line based estimates of heritability for drug cytotoxicity.
| Drug | h2 heritability estimate | Ref. |
|---|---|---|
| 5-fluorouracil | 0.26–0.65 (dose dependent) | [16] |
| Dovetail | 0.21–0.70 (dose dependent) | [16] |
| Isolating | 0.47 | [14] |
| Daunorubicin | 0.18–0.63 (dose dependent) | [17] |
| 5-Fluorouracil | 29.2 | [15] |
| Arsenic trioxide | 24.4 | [15] |
| Azacitidine | 20.7 | [15] |
| Neomycin | 17.3 | [15] |
| Bu sulfa | 14.2 | [15] |
| Carbonating | 43.2 | [15] |
| Cladribine | 27.3 | [15] |
| Cytarabine | 41.7 | [15] |
| Daunorubicin | 37.1 | [15] |
| Dovetail | 30.1 | [15] |
| Doxorubicin | 35.3 | [15] |
| Epirubicin | 59.5 | [15] |
| Topside | 41.3 | [15] |
| Floxuridine | 27 | [15] |
| Fludarabine | 13.5 | [15] |
| Gemcitabine | 8.1 | [15] |
| Hydroxy urea | 43.2 | [15] |
| Ida rubicon | 45.8 | [15] |
| Neomycin | 26.7 | [15] |
| Mitoxantrone | 46.5 | [15] |
| Oxaliplatin | 50 | [15] |
| Paclitaxel | 45.9 | [15] |
| Rapamycin | 15.1 | [15] |
| Temozolomide | 63.5 | [15] |
| Teniposide | 36.4 | [15] |
| Topotecan | 46.1 | [15] |
| Vin blasting | 31.2 | [15] |
| Vin pristine | 23.1 | [15] |
| Vino reline | 34.1 | [15] |
Furthermore, HapMap cell lines from multiple ancestries can be used in these cellular assay based studies to represent multiple ancestries, allowing for characterization of the relationship between PGx traits across ancestry groups. For example, an exploratory analysis used HapMap cells to investigate genetic variants and their functional consequences for the enzyme deoxycytidine kinase (DCK) in two ancestries, European and African (Yoruba) [18]. DCK is a rate-limiting enzyme in the activation of nucleoside analogs. Cytarabine (ara-C), a chemotherapeutic agent commonly used in in acute myeloid leukemia, is one such nucleoside analog. DCK activity was lower for subjects heterozygous for coding changes compared with homozygous subjects, and DCK activity in general was higher in the African cell lines when compared with the European cell lines.
Another approach available for determining the genetic component influencing PGx traits involves Repeated Drug Administration (RDA). In this method, a drug is administered multiple times to unrelated individuals, and the variability in the PGx trait of interest between and within individuals is compared [19]. RDA information can be used to calculate the Relative Genetic Component (rGC), an estimate on a scale of 0 to 1 of the genetic component of a PK or PD parameter. This measurement has also been referred to as ‘intraclass correlation’ or ICC [20]. The rGC measurement is calculated through the following formula: (variability between individuals - variability within individuals)/variability between individuals. The measurement can be interpreted as a rough estimate of heritability, where a trait with high rGC will likely have high heritability. The rGC measurement can also be calculated from monozygotic twin pairs, when dizygotic twins are not available [19]. For example, the genetic component of variation in renal clearance of amoxicillin, ampicillin, metformin, terodiline, digoxin and iohexol, was investigated using this approach [21]. Results from these rGC based studies are summarized in Table 3. Limitations of this approach include the high variability of PGx traits over limited time periods even in the absence of genetic factors, potentially leading to large error estimates.
Table 3.
Repeated drug administration and resultant relative genetic component measurements.
| Study type | Trait | Drug | Relative genetic component estimate | 95% CI | Ref. |
|---|---|---|---|---|---|
| RDA | Renal clearance | Metformin | 0.94 | NA | [22] |
| RDA | Renal clearance | Amoxicillin | 0.91 | NA | [23] |
| RDA | Renal clearance | Ampicillin | 0.64 | NA | [23] |
| RDA | Renal clearance | Terodiline | 0.37 | NA | [24] |
| RDA | Renal clearance | Iohexol | 0.2 | NA | [25] |
| RDA | Renal clearance | Digoxin | 0.12 | NA | [26] |
| RDA | Metabolism by CYP1A2 | Caffeine | 0.69 | NA | [19,27] |
| RDA | Metabolism by NAT2 | Caffeine | 0.95 | NA | [19,27] |
| RDA | Metabolism by Xanthippe oxidate | Caffeine | 0.24 | NA | [19,27] |
| RDA | Metabolism by UGT2B7 | Oxazepam | 0.98 | NA | [19,27] |
| RDA | Metabolism by ADJ | Ethanol | 0.57 | NA | [19,27] |
| RDA | Metabolism by CYP2D6 | Dextromethorphan | 0.97 | NA | [19,27] |
| RDA | Metabolism by CYP3A4 (adriamycinol-AUC[0-Inf] to adriamycin-AUC[0-Inf]) | Aureomycin | 0.55 | 0.00–0.86 | [28] |
| RDA | Metabolism by CYP3A4 (terminal elimination half-life (unbound drug)) | Cyclosporine | 0.83 | 0.12–0.97 | [28] |
| RDA | Metabolism by CYP3A4 (erythromycin N-demethylation rate) | Erythromycin | 0.89 | 0.65–0.98 | [28] |
| RDA | Metabolism by CYP3A4 (serum AUX[0–24]) | Ethinylestradiol | 0.79 | 0.48–0.94 | [28] |
| RDA | Metabolism by CYP3A4 (serum AUX[0–24]) | Ethinylestradiol | 0.94 | 0.83–0.98 | [28] |
| RDA | Metabolism by CYP3A4 (serum AUX[0–24]) | Ethinylestradiol | 0.86 | 0.48–0.96 | [28] |
| RDA | Metabolism by CYP3A4 (ethyl morphine N-demethylation metabolic ratios) | Ethyl morphine | 0.98 | 0.87–1.00 | [28] |
| RDA | Metabolism by CYP3A4 (plasma clearance) | Midazolam | 0.96 | 0.92–0.98 | [28] |
| RDA | Metabolism by CYP3A4 (plasma AUX[0–24]) | Nifedipine | 0.82 | 0.55–0.94 | [28] |
| RDA | Metabolism by CYP3A4 (plasma AUX[0-Inf]) | Nifedipine | 0.98 | 0.95–0.99 | [28] |
| RDA | Metabolism by CYP3A4 (plasma AUX[0–24]) | Nitrendipine | 0.66 | 0.00–0.92 | [28] |
| RDA | Pharmacokinetics of nevi rapine (plasma AUX[0–6]) | Nevi rapine | European Americans: 0.904 African–Americans: 0.902 | European Americans: 0.64–0.97 African–Americans: 0.42–0.98 | [29] |
| Twins | AUC [0–12] | d0-digoxin | 0.89 | NA | [30] |
| Twins | AUC [0–12] | d3-digoxin | 0.79 | NA | [30] |
| Twins | Oral clearance | Digoxin (oral) | 0.36 | NA | [31] |
| Twins | Renal clearance | Digoxin (oral) | 0.19 | NA | [31] |
| Twins | Renal clearance | Metformin | 0.95 | NA | [32] |
AUC: Area under the curve; NA: Not applicable; RDA: Repeated drug administration.
The PGx landscape: drug efficacy & adverse events
Among the many PGx success stories is the use of genetic information to facilitate prescription of optimal warfarin dosage levels to prevent cardioembolic stroke, myocardial infarction and venous thrombosis as well as prevent adverse events. Warfarin is widely prescribed after placing arterial stents or after myocardial infarction. However, warfarin causes serious side effects including hemorrhage, especially during drug initiation when patients are titrated to the optimal dosage level [33,34]. Variants in CYP2C9 influence the PK [35] and VKORC1 variants influence the PD of warfarin [36–39]. It is noteworthy that the two-gene biomarker test for warfarin dosing still represents an exception; most other genetic biomarker PGx tests only include one gene, and the identification of multigene robust biomarkers remains an important area for expansion within PGx research. An algorithm for estimating individualized warfarin dosages was defined using clinical and PGx data [40]. As a result, the FDA updated the label for warfarin, detailing the use of pharmacogenetic testing for clinical decision making [33]. Recent studies have evaluated genotypic bio markers for warfarin dosing with different conclusions; one study indicated genotype-guided dosing of warfarin was ineffective when compared with dosing without genotypic information [41]. A separate study indicated genotype-guided dosing was associated with a patients being within the therapeutic range for a greater period of time when compared with the standard initiation of warfarin [42].
In another example, clopidogrel is prescribed to prevent atherothrombotic events after myocardial infarction but exhibits notable variability in successfully preventing further cardiovascular events. This has at least in part been attributed to genetically determined variation in the drug metabolizing enzyme CYP2C19, largely responsible for converting clopidogrel to its active metabolite. The most common loss-of-function allele is CYP2C19*2 (rs4244285), associated with increased risk of cardiovascular events [43]. Indeed the CYP2C19*2 variant is considered a major determinant of prognosis for patients <45 years of age on clopidogrel treatment after myocardial infarction [44].
While the list of PGx traits continues to grow, translating the complex and sometimes conflicting research results from PGx to clinical action requires accessible information that is updated as new findings come to light. FDA labels are already being modified in response to emerging PGx findings, listed here [45]. However, FDA label changes are only one way to provide information to clinicians for implementing PGx results in treatment decisions. The Clinical Pharmacogenetics Implementation Consortium (CPIC) [46], the Royal Dutch Association for the Advancement of Pharmacy (DPWG) [47] and other professional medical societies, have also been publishing pharmacogenetic dosing guidelines for an increasing number of drugs [48].
CPIC in particular has carefully reviewed the criteria for translation of PGx traits, and as a result, has developed a framework for identifying key evidence justifying clinical implementation. Published CPIC guidelines target specific gene/drug pairs (Table 4), reviewing the existing research for each gene/drug pair [49]. In addition, CPIC provides a standardized web-interface of gene/drug pair summary information, including outcome phenotype based on genotype, dosing recommendations and allele frequency differences and impact of specific variants across distinct ancestry. CPIC continues to review ongoing research on gene/drug pairs to determine whether the existing information needs updating or new gene–drug pairs can be recommended for clinical use.
Table 4.
Reviewed Clinical Pharmacogenetics Implementation Consortium drug/gene pairs.
| Drug class | Drugs | Drug use in the clinic | Concern | Gene | Gene function | Genotype information and SNPs |
Other notes | Ref. | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Thiopurines | Azathioprine, mercaptopurine and thioguanine | Critical anticancer agents; imunosuppressants in inflammatory bowel disease, rheumatoid arthritis and other immune conditions | Individuals with two nonfunctional TPMT alleles are at 100% risk for life-threatening myelosupression | TPMT | Thiopurine methyltransferase activity | Multiple SNPs in TPMT are detailed here [50] CPIC defines specific haplotypes and SNPs for TPMT [51] | Substantial ethnic differences in the frequencies of low-activity variant alleles | [52] | ||
| Clopidogrel | Antiplatelet therapy, particularly for those undergoing percutaneous coronary intervention | Dependent on CYP2C19 haplotype, patients have a variation in clopidogrel response By haplotype, patients can be grouped as follows: ultrarapid metabolizer, extensive metabolizer, intermediate metabolizer or poor metabolizer | CYP2C19 | Inhibition of platelet aggregation | The key polymorphism is CYP2C19*2 (c.681G>A; rs4244285), specifically studied with respect to clinical outcomes However, multiple SNPs are under investigation and detailed here [53] | The frequencies of CYP2C19 poor metabolizers are ∼2–5% among Caucasians and Africans and ∼15% in Asians | [54] | |||
| Warfarin | Treatment and prevention of thrombotic disorders | The variant alleles CYP2C9*2 and *3 impair metabolism of S-warfarin by ∼30–40% and ∼80–90%, respectively. If a patient receives too much warfarin, or is a fast metabolizer, they may be at risk of a bleeding event. If a patient receives too little warfarin, or is a slow metabolizer, they may be at risk of a thrombotic event | CYP2C9 | CYP2C9 is a hepatic drug-metabolizing enzyme in the CYP450 superfamily, and is the primary metabolizer of S-warfarin | CYP2C9 has more than 30 known variant alleles CYP2C9*2 (rs1799853) and CYP2C9*3 (rs1057910) are the two most common variants with reduced enzyme activity | The frequencies of the CYP2C9 variant alleles differ between racial/ethnic groups. In addition, the CYP2C9 variant alleles with reduced activity (CYP2C9*5, *6, *8 and *11) contribute to dose variability among African-Americans | [55] | |||
| Warfarin | Treatment and prevention of thrombotic disorders | Warfarin has a narrow therapeutic index and wide interindividual variability If a patient receives too much warfarin, or is a fast metabolizer, they may be at risk of a bleeding event If a patient receives too little warfarin, or is a slow metabolizer, they may be at risk of a thrombotic event | VKORC1 | VKORC1 encodes the vitamin K-epoxide reductase protein, the target enzyme of warfarin | Variation in rs9923231 is significantly associated with warfarin sensitivity and reduced dose requirements. The polymorphism alters a VKORC1 transcription factor binding site, leading to lower protein expression There are rare nonsynonymous VKORC1 variants that confer warfarin resistance resulting in high warfarin requirements | The variation in allele frequency of rs9923231 across ancestry largely explains the average dose requirement differences across white subjects, black subjects and Asians | [55] | |||
| Codeine | Treatment for pain management | Poor metabolizers may lack any effect of codeine, warranting the use of an alternative analgesic Ultrarapid metabolizers of codeine are at risk of severe toxicity associated with a ‘normal’ dose of codeine | CYP2D6 | Hepatic CYP2D6 bioactivates codeine to morphine | More than 80 CYP2D6 alleles have been defined, and are detailed here [56] Based on diplotype, a likely phenotypic outcome can be defined: ultrarapid metabolizer, extensive metabolizer, intermediate metabolizer and poor metabolizer | In some cases, patients have more than two copies of the CYP2D6 gene CYP2D6 allele frequencies differ substantially between racial and ethnic groups | [57] | |||
| Abacavir | Anti-HIV treatment which suppresses HIV's ability to convert its RNA genome into DNA before insertion into a host cell's genome | Variants in HLA-B can result in an increased risk in hypersensitivity, a multiorgan illness | HLA-B | HLA-B encodes a cell surface protein for presenting intracellular antigens to the immune system | Variation in HLA-B:HLA-B*57:01 | HLA-B*57:01 has also been shown to be over-represented in HIV long-term nonprogressors | [58] | |||
| Simvastatin | Used for lipid lowering | Muscle toxicity (myopathy) | SLC01B1 | TheOATP1B1 protein that is encoded by SLCOIB1 facilitates the hepatic uptake of statins, and variation in SNP rs4149056can have a damaging effect on OATP1B1 | A minor allele C for SNP rs4149056 in SLC01B1 increases systemic exposure to simvastatin and the risk of muscle toxicity | Drug among the most commonly used prescription medications | [9] | |||
| Allopurinol | Treatment of hyperuricemia and gout | Cutaneous adverse reactions including: drug hypersensitivity syndrome, Stevens-Johnson syndrome, toxic epidermal necrolysis | HLA-B | HLA-B encodes a cell surface protein for presenting intracellular antigens to the immune system | Variant allele of the HLA-B, HLA-B*58:01 | [59] | ||||
| Tricyclics | Amitriptyline, clomipramine, doxepin, imipramine and trimipramine, desipramine and nortriptyline | Treatment for pain management and psychiatric disorders | Variation in drug metabolism based on polymorphism profile | CYP2D6 | These drugs undergo hydroxylation by CYP2D6toless active metabolites | Although likely there is a combined effect of polymorphisms in both CYP2D6 and CYP2C19, further research needs to be carried out to characterize the combined effect of polymorphisms in both genes | Pharmacogenomics guidelines are specific to treatment of psychiatric disorders versus pain management Clomipramine, doxepin, imipramine and trimipramine | [60] | ||
| Amitriptyline, clomipramine, doxepin, imipramine and trimipramin | Treatment for pain management and psychiatric disorders | Variation in drug metabolism based on polymorphism profile | CYP2C19 | These drugs are demethylated by CYP2C19 to pharmacologically active metabolites | Although likely there is a combined effect of polymorphisms in both CYP2D6 and CYP2C19, little information is available on how to adjust initial doses based on combined genotype information | Pharmacogenomics guidelines are specific to treatment of psychiatric disorders vs pain management | [60] | |||
| Carbamazepine | Treatment for seizures, nerve pain and bipolar disorder | Increased risk of serious blistering cutaneous adverse drug reactions: Stevens-Johnson syndrome and toxic epidermal necrolysis | HLA-B | HLA-B encodes a cell surface protein for presenting intracellular antigens to the immune system | Considered ‘positive’ if one or two copies of HLA-*15:02 are present or as ‘negative’ if no copies of HLA-B*15:02 are present | HLA-B*15:02 is most prevalent in Oceanian, east Asian and south/central Asian populations | [61] | |||
| Fluoro-pyrimidines | Capecitabine | Fluoropyrimidines are chemotherapeutic agents for the treatment of many types of cancers | Deficiency in DPD can result in profound toxicity, such as myelosuppression, mucositis, neurotoxicity, hand-foot syndrome and diarrhea, when taking 5-fluorouracil | DPYD-5FU | This gene encodes for an enzyme critical for detoxifying metabolism of fluoropyrimidines | Homozygotes of *2A, *13 and rs67376798 are considered deficient in DPD Heterozygotes for any combination of *2A, *13 and rs67376798 have intermediate or partial DPD activity Individuals with none of these alleles are likely to have normal activity | Weak or conflicting evidence for other alleles of DPYD | [62] | ||
| Pegylated IFN-α | PEG-IFN-α, PEG-IFN 2a and 2b, ribavirin | Treatment of hepatitis C virus genotype 1 | IL28B genotype strongest baseline predictor of response to PEG-IFN-α and RBV therapy | IFNL3I IL28B | The gene encodes IFN-λ 3, a member of the type 3 IFN- λ family with antiviral, antiproliferative and immune-modulatory activities | The two most commonly tested SNPs are rs12979860 and rs8099917 | The rs12979860 allele frequency varies among different ethnic groups, explaining the differences in treatment response rates among east Asians, Caucasians, African–Americans and Hispanics with chronic hepatitis C virus infection | [63] | ||
| Ivacaftor | Treatment for cystic fibrosis | CFTR | Mutations within CFTR cause cystic fibrosis | G551D-CFTR variant in at least one allele, guidelines indicate ivacaftor therapy if no contraindications Ivacaftor therapy is not recommended for cystic fibrosis patients with other CFTR mutations | While 1900 disease-causing CFTR variants have been identified, they can be grouped by shared features Growing understanding of how various mutations disrupt CFTR protein function is beginning to help clinicians categorize cystic fibrosis patients for viable therapeutic strategies | [64] | ||||
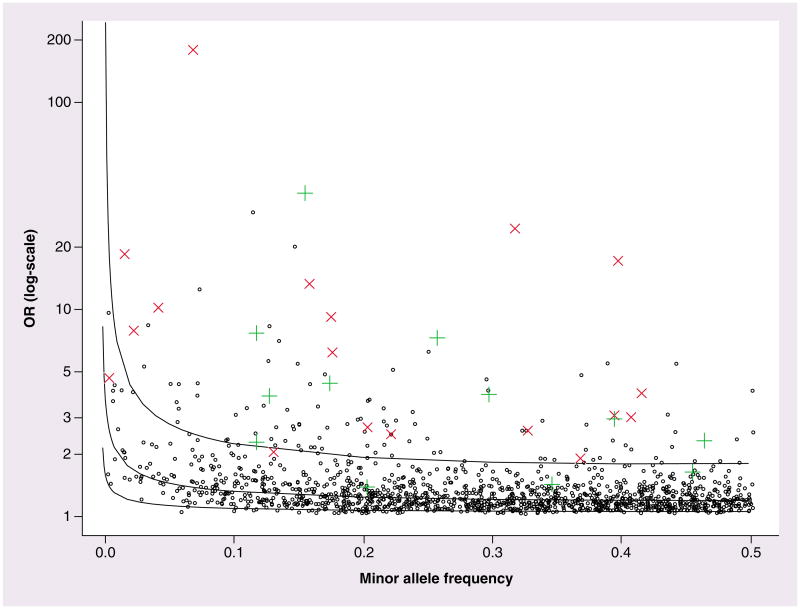
The effect size of genetic variants affecting PGx traits tends to exceed that of SNPs derived from GWAS of complex human disorders. In Figure 1, we illustrate this trend by comparing the odds ratios for efficacy and toxicity related PGx results with those from the NHGRI GWAS catalog, where the phenotypic outcome covers a range of non-PGx complex traits [65]. Figure 1 displays larger relative strength of effect size for PGx traits compared with the range of effect sizes observed with complex-trait GWAS. This observation is consistent with our expectation that PGx variants affect targeted subsets of genes and pathways; however, ascertainment bias cannot be excluded resulting from the different methods used for discovery of the genetic variants.
Figure 1.
Contrasting the effect size and minor allele frequency range of pharmacogenomic variants versus variants from the NHGRI GWAS catalog. Black circles are the NHGRI GWAS catalog results, plotted by OR results in logarithmic (base10) scale versus minor allele frequency. Each green cross represents a replicated efficacy result for a pharmacogenomics study. Each red X represents a replicated toxicity result for pharmacogenomics. The solid lines represent the 80% power equivalent curves across minor allele frequency, from top to bottom for n = 1 × 103, 10 × 103 and 100 × 103, respectively (assuming n/2 cases and n/2 controls). OR: Odds ratio.
Functional role of genomic architecture in PGx traits
The PGx field faces challenges in understanding the mechanistic role of genomic variation in PGx traits. Much of the focus of interpretation of the relationship between genetic variability and outcome for both complex disease and PGx has been centered on protein-coding regions. In PGx studies there has been a particular focus on candidate gene approaches targeting Absorption, Distribution, Metabolism and Execretion (ADME) genes in addition to GWAS. This makes particular sense for PGx traits, as genetic variation can have an impact on the protein structure of drug-metabolizing enzymes resulting in changes in enzymatic activity. Furthermore, there are known important PK pathways where the impact of genetic variation has been demonstrated on a protein-coding modification level. One example is CYP2D6, an enzyme involved in the metabolism of up to 25% of clinical drugs, where nonsynonomous variants can result in enzymatic changes and subsequent changes in catalytic activity [66]. Many of these very important pharmacogenes (VIP) are summarized in the Pharmacogenomics Knowledge Base (PharmGKB) [67]. However, GWAS and now full-genome sequencing have identified many biomarkers that do not cause functional protein-coding modification, or are located in genes whose role in drug disposition, response or toxicity was previously not well characterized. For example, a distal enhancer variant >100 kb downstream of the coding region of CYP2D6 strongly increases gene expression in the liver, accounting for cases of ultra rapid metabolism [68]. Likewise, in a genome wide association study of flucloxacillin-induced liver injury, a novel association between ST6GAL1, an enzyme with a possible role in B-cell immune response, and the drug induced liver toxicity was identified [69]. In some cases, there are genetic variants that account for substantial outcome variability, and are already used as a clinical biomarker, but we have a more limited understanding of the impact of that genetic factor, such as the functional mechanisms of HLA variants and drug response variability [70,71].
Resolving the mechanistic etiology of the impact of genetic variation on PGx traits is a critical focus for future PGx studies. Exploration of the function of nonprotein coding genetic regions will be essential, including regulatory regions and noncoding RNA [72]. Regulatory variants may account for a large portion of genetic variability, and should be incorporated into analyses as knowledge of the functional impact of genetic variation on genetic enhancers, promoters and gene expression is accrued and shared through projects such as ENCODE and related databases [73–76].
Beyond PGx GWAS: polygenic analyses
New methodologies will continue to drive advances in the field of PGx. The majority of PGx results have arisen from investigation of the association between single, common, genetic variants and pharmacological outcome. As found with GWAS for common complex traits [6], this approach may have varied or limited success in future studies, as the genetic architecture of any trait can be complex. On a biological level, a variety of potential genetic mechanisms influencing PGx traits fail to be captured when investigating only the relationship between single, common, genetic-variants and outcomes. Thus, we need to diversify the methodologies being used to better define polygenic traits.
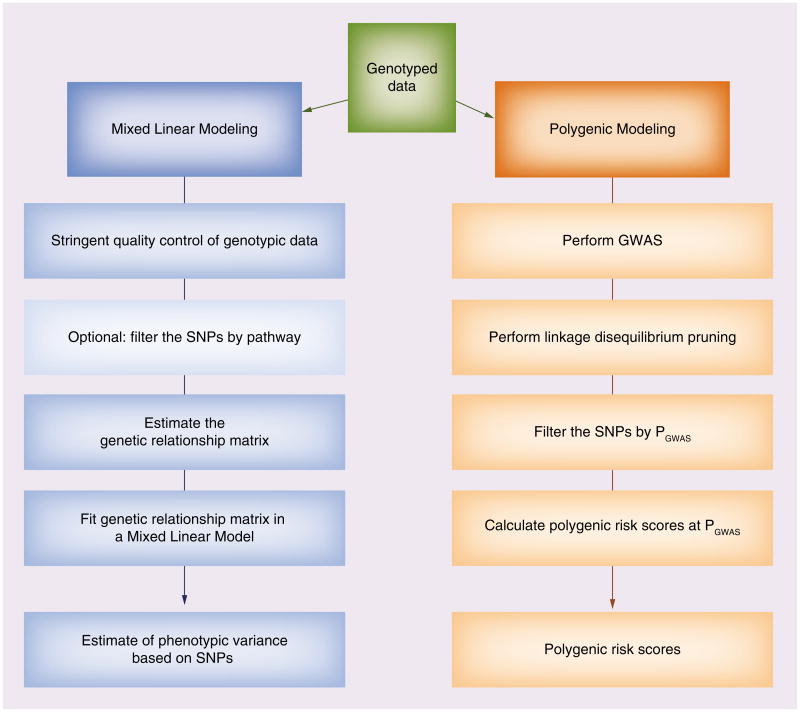
One alternate approach considers polygenic genetic architecture, or the contribution of multiple common SNPs to phenotypic variance in aggregate. Two methods have already been used for a variety of complex outcomes for non-PGx traits: mixed linear modeling (MLM) and polygenic modeling. Both methods test a polygenic model for the relationship between multiple SNPs and outcome, as illustrated in Figure 2. MLM estimates the additive genetic variance under a mixed linear model with a random effect representing the polygenic component of trait variation. The software tool GCTA (Genome-wide Complex Trait Analysis) has been developed for use of MLM in estimation of the proportion of phenotypic variance accounted for by genome-wide association genotypic data [77]. The MLM/GCTA approach has been used successfully for identifying the collective contribution of GWAS-polymorphisms to traits including height [78], Crohn's disease, bipolar disorder and Type 1 diabetes [79] and other complex outcomes [80,81].
Figure 2.
Overview of polygenic analysis methods. On the left, the general work flow of using Mixed Linear Modeling pursued using the software genome-wide complex trait analysis. On the right, the general workflow of Polygenic Modeling. Both methodologies allow the user to identify multiple SNPs related to pharmacogenomics outcome, with different information resulting from each approach. GWAS: Genome-wide association study.
Polygenic modeling develops an additive polygenic risk score for a given trait based on a group of SNPs filtered by a GWAS-based p-value threshold in a discovery sample set. The polygenic risk score is then tested in an independent set of samples. This approach has been successfully used to detect the contribution of multiple variants with small effects to the heritability of diseases/traits/outcomes such as schizophrenia [82], multiple sclerosis [83], height [84], body mass index [85] and rheumatoid arthritis [86]. Polygenic modeling analyses for complex traits yield results consistent with simulated genetic models in which hundreds of associated loci harbor common causal variants and a smaller number of loci harbor multiple rare causal variants [86]. The heritability estimates derived from these polygenic approaches have been consistent with previously reported estimates for these complex traits. MLM and polygenic modeling methods are now being applied to PGx data. MLM/GCTA analyses have been used to investigate asthma PGx traits [87], and paclitaxel-induced sensory peripheral neuropathy [88].
Paclitaxel is a chemotherapeutic agent commonly prescribed to treat carcinomas of the breast, ovaries, lung, head and neck. Peripheral neuropathy is one of the most common toxicities with paclitaxel treatment, and occurs in a substantial subset of patients. Known causes of peripheral neuropathy do not completely explain the incidence of toxicity amongst patients treated with paclitaxel, suggesting a genetic basis for susceptibility to the toxicity. Small candidate gene studies have had mixed results identifying variants related to variability paclitaxel-induced peripheral neuropathy [4,89]. One study reports a high risk odds ratio (OR: 19.1) for paclitaxel neurotoxicity associated with CYP3A4*22 [90], as a result of reduced metabolic activity of the *22 allele [91], but this result requires replication (CYP3A4*22 is not on earlier GWAS panels and cannot be readily imputed). GWAS for this PGx trait have identified some candidate SNPs, but replication has been inconsistent [2,92].
Chhibber et al. (2014) [88] investigated a polygenic etiology of paclitaxel-induced neuropathy. They estimated the variance explained by common SNPs (MAF >1%) for two outcomes: the maximum grade of sensory peripheral neuropathy, and the dose at first instance of peripheral neuropathy. They investigated the variance explained by all autosomal SNPs, SNPs selected based on genomic location, and SNPs in gene sets selected based on prior knowledge regarding possible mechanisms of the pathogenesis of paclitaxel-induced peripheral neuropathy using the GCTA software tool. They found whole genome estimates of heritability were not significant; however, using a pathway-based approach for filtering SNPs yielded significant results. Specifically, the Axonogenesis GO Term set (GO: 0007409) had significant estimates of heritability close to 20%, suggesting a portion of the heritability of paclitaxel-induced neuropathy is driven by genes involved in the regulation of axon extension. These results show both the utility of polygenic approaches for PGx traits, as well as the utility of exploring pathway-based expert knowledge filtering of SNPs before investigating polygenic architecture.
McGeachie et al. (2013) estimated the heritability of bronchodilator response (∼30%), airway hyper-responsiveness (∼50%) and asthma liability (∼61%) due to SNPs in aggregate using the MLM/GCTA approach [87]. Linkage studies have yielded comparable heritability estimates for both bronchodilator response (∼12–40%) and airway responsiveness (∼67%) [20,93,94], supporting the validity of the polygenic modeling approach. In addition, the estimate obtained for the heritability of asthma corresponds to published asthma heritability from twin studies ranging from 70 to 90% [95]. With polygenic approaches, the total variance explained by a series of alleles should approach the heritability estimates by other methods, unless there are nonadditive mechanisms or causal alleles are not well tagged in the GWAS SNP panels. This study indicates polygenic modeling can provide heritability estimates within the range of heritability measured in familial studies. Therefore, MLM/GCTA are suitable for providing narrow-sense heritability estimates for PGx traits where family or other approaches are not possible for estimating heritability.
MLM/GCTA methods were largely developed for GWAS data of complex traits, and at this point, most GWAS for non-PGx traits have very large sample size. With PGx traits, low sample size is common and this can limit the utility of polygenic approaches unless strategies are implemented to increase sample size, such as multi-institution collaborations to combine datasets. In addition, all methods have expectations of the type of phenotype that will be used, implicit in the development of the method. Pharmacological measurements and outcome measures can be complex, such as ordinal variables or survival times subject to censoring, to which current polygenic models can be difficult to apply. Despite these limitations, polygenic analyses are showing utility in providing an additional tool for seeking information about the relationship between genetic architecture and PGx traits and estimates of heritability of these traits.
Further limitations of MLM/GCTA methods include the underlying assumptions that genotype effects are predominantly additive, thereby limiting assessment of the ‘mutational burden’ as a measure of genetic influence on a trait, including response to therapy. For example, this approach ignores the pervasive influence of epistatic gene–gene interactions, where the effect of one variant is contingent on the presence of another variant. Also, while this approach may yield an estimate of the trait's heritability, it remains to be determined whether mutational load of many variants can serve as clinical biomarker panels to guide therapeutic decision.
Future methods
In addition to polygenic analyses, other approaches may provide keys to elucidating the etiology of PGx traits. Different predictive models based on genetic architecture may be necessary to explain many PGx traits that remain to be elucidated. These models may not include loci of large effect, and some of these models may not be additive and fail assumptions of linear regression. Novel approaches are being introduced and refined at a fast rate, and these may emerge as key tools for exposing further the genetic architecture under lying PGx traits. New technologies for characterizing the genome are also emerging. These technologies include large-scale high-throughput sequencing to detect comprehensive genetic variation data including low-frequency variants [96], genetic and genomic variation such as copy-number variants, new gene expression technologies and methods to detect the complex epigenetic landscape of the human genome. One can argue that drug therapy ranks among those environmental stimuli that alter the epigenetic chromatin landscape, thereby adding another dimension to PGx. Novel analysis methods include pathway [97] and prior knowledge based approaches [98], rare variant analyses [99,100] and interaction studies [101]. Integration of these diverse large-scale datasets has the potential for driving PGx discovery and clinical applications.
Pathway approaches are becoming more common, taking advantage of prior knowledge previously obtained in molecular and cellular biology studies. Diverse databases cataloging the results of countless PGx studies include the PharmGKB database mentioned earlier. In addition, newly developed tools allow users to tap simultaneously into multiple database sources for pathway based analyses, such as Biofilter [102–104] and PARIS [102]. Furthermore, as mentioned, pathway based approaches can serve as ‘input’ for polygenic analyses, reducing the search space for variables by collapsing multiple genes into groups [88].
Methods are now emerging that enable exploring rare-variant data, usually defined as SNPs with allele frequencies <0.01, data of greater abundance with comprehensive sequencing data becoming available. The impact of rare variants on PGx traits are just beginning to be explored. Examples of already discovered rare-variants for PGx traits include rare variants found within the SLCO1B1 gene, where haplotypes have been associated with reduced methotrexate clearance during treatment of childhood acute lymphoblastic leukemia. SLC01B1 variants accounted for 10.7% of the population variability in clearance. Of those variants, common nonsynonymous variants contributed the most to variability, but rare nonsynonymous variants contributed to 1.9% of total variation in clearance [105]. This example illustrates the promise of searching for rare variants but also cautions against optimistic expectations regarding clinical utility. In this case, the rare variants contribute a relatively small portion to the variability attributable to SLCO1B1, and for clinical utility in and an individual patient, the SLCO1B1 phasing is typically unknown, adding uncertainty to any clinical recommendations.
Rare-variant collapsing strategies have now been developed for assessing their influence on traits, as the power for detecting the relationship between single low-frequency variants is limited. Collapsing approaches provide a way to identify specific patterns of genetic variation predictive of outcome variation. Several collapsing methods have been published in the past 5 years [106–114]. An example of a novel collapsing strategy is BioBin [115–117], a low-frequency variant collapsing method that considers the cumulative effect of rare variants within genetic features chosen by the users. These features can include genes but can also be pathways, or other biologically based criteria such as evolutionarily conserved regions.
The role of epistasis in PGx traits
One of the reasons for the popularity of the GWAS and candidate gene approach is the simplicity of the regressive model for interpretation, and clear guidelines for ascertainment of significance and multiple hypothesis testing corrections. However, a variety of tools exist for the development of more complex predictive models beyond single-variant/outcome association testing for common variants. For example, step-wise regression can be used to develop models with additional terms, instead of using single variant data. More complex models may show better outcome prediction, such as gene-by-gene (GxG) interaction models.
The overall role of dynamic GxG interactions remains a matter of debate. One can argue that a substantial portion of the ‘missing heritability’ of complex traits is accounted for by epistasis [118], but few studies document this in PGx. It may require identifying the interplay of more than one genetic variant to adequately predict the outcome of drug administration [119]. An example of an interaction has been found between the dopamine D2 receptor and the dopamine transporter, encoded by DRD2 and DAT, respectively. Both genes harbor several common regulatory variants but only DRD2 is associated with lethal risk resulting from cocaine abuse when each gene is studied separately. Yet, a combination of a single variants from both DRD2 and DAT convey a seven- to eight-fold increased risk in a highly significant interaction model [120]. Such cases of gene–gene–environment interaction may be more prevalent than currently anticipated and need to be explored on a broader basis.
One challenge for seeking more complex models is the number of options to investigate, when investigating pairwise GxG and SNP-by-SNP (SNP×SNP) interaction models, as the number of potential interactions skyrockets as the number of variants grows. Tools exist for generating pair-wise GxG interaction models that address this. For example, Biofilter [104] is a tool that allows users to filter and annotate genetic data, as well as generate pairwise SNP×SNP models prioritized by the biological evidence supporting the genetic interaction. Multifactor dimensionality reduction (MDR) performs an exhaustive analysis of all n-wise interacting loci to generate models [101]. The Analysis Tool for Heritable and Environmental Network Associations (ATHENA) is a software tool that combines advanced filtering and machine learning analytical techniques to generate multi-variable models that can predict categorical or quantitative outcomes [121,122]. ATHENA can be used for both G×G/SNP×SNP interaction models that move beyond pairwise interactions, as well as for metadimensional analysis, where different data types of high-throughput genetic predictor variables are incorporated. However, all these methodologies require large sample cohorts, which are rarely encountered in PGx studies.
Clinical & regulatory decision making: moving from ‘bench to bedside’
The list of drugs for which genetic information has potential utility in guiding individualized therapy is growing. Clinical implementation is lagging behind our current knowledge in part because of multiple challenges faced in clinical practice. Recognizing the mandate to optimize drug therapy, the FDA maintains a website with current assessments of how clinicians should utilize PGx information (see Table of Pharmaco genomic Biomarkers in Drug Labels [45]). As more of the complex genetic architecture of PGx traits is uncovered, substantial challenges remain for translating PGx findings to the clinic. Key questions include: Is this an effective biomarker with clinical utility? How many individuals will be helped by geno-typing a specific PGx variant? Will there be an impact on survival, recovery and/or prevention of a major adverse reaction and how much of an impact? Will a genetic variant manifest only in one population or is there evidence of consistency across multiple ancestral populations? Will the cost of genotyping for a PGx variant confer sufficient benefit to offset the cost?
A major concern for moving PGx findings to the clinic is the impact of ancestry on genetic variation. Highly significant associations between variants and PGx traits may differ considerably across ancestries, which has a direct impact on dosing decisions. For example, a significant association was found between the HLA-B*1502 variant and carbamazepine-induced (CBZ-induced) Stevens–Johnson syndrome in Han Chinese and Thai individuals [123–126]. However, separate studies have indicated that HLA-B*1502 is not a marker for all forms of CBZ-induced hypersensitivity in individuals of European decent [127,128]. In one study, the only four individuals out of 12 cases with CBZ-induced hypersensitivity had the HLA-B*1502 marker; these four individuals also had Asian ancestry [128].
Ancestry specific PGx differences in association of the arginine (Arg) 16 allele in the beta2-anderegenic receptor (beta2-AR) with asthma severity and broncho-dilator response [129] have been found. Two admixed populations, Puerto Ricans and Mexicans, have different proportions of European, African and Native American ancestry. These two populations have the highest and lowest asthma prevalence, morbidity and mortality respectively. In the study by Choudhury et al. (2005), associations between bronchodilator response, asthma severity and the beta2-AR (Arg) 16 allele were found in Puerto Ricans, but not in Mexicans. These results are likely accounted for by the presence of more than one causative variant in the same gene, or in interaction genes, with distinct population distribution.
We have mentioned already CYP2C9 variation and warfarin dosing. Polymorphisms in CYP2C9 account for 18% of the variance in warfarin dose, and polymorphisms in VKORC1 account for 30%, in European Americans; however, these variants account for a smaller portion of variability in patients with Asian or African Ancestry [40,130–137]. Additional CYP2C9 variant alleles with reduced activity (CYP2C9*5, *6, *8 and *11) have been found to contribute to dose variability among African–Americans [55,138].
The field of PGx already has a record of investigations in groups beyond European Americans, when contrasted with much of the initial work of GWAS that was focused on European American ancestry. CPIC guidelines usually contain statements about existing knowledge of gene/drug pair information across ancestry. Work is being done to determine repeated drug administration rGC values across ancestry [29]. These analyses incorporating multiple ancestries should continue to be an important pursuit for the field of PGx moving forward. FDA labeling should also consistently reflect what populations PGx discoveries were made in, as that may impact the utility of a biomarker for a given patient.
Finally, substantial inconsistencies exist in study design, dosing regimens, study population and analysis methods for the field of PGx. For example, three studies, with differences in ethnic background and disease state of patients, study size and methods used to measure response to treatment, have reported contradictory results on the association of the FcγRIIIA 158V/F polymorphism and response to etanercept or infliximab in patients with Rheumatoid Arthritis [139–141]. Such inconsistencies are common between PGx studies, making interpretation of results across studies challenging, even for the same PGx trait. Further more, huge variability exists in the information that is reported when a PGx study is published. A standardized way of reporting PGx results and more consistency in study design could assist in developing clear guidelines for what constitutes a validated and actionable PGx result, and provide the means for comparing results across studies. The CPIC-authored studies have made recommendations for evaluating PGx results and reporting information accessible to clinicians [46].
Conclusion
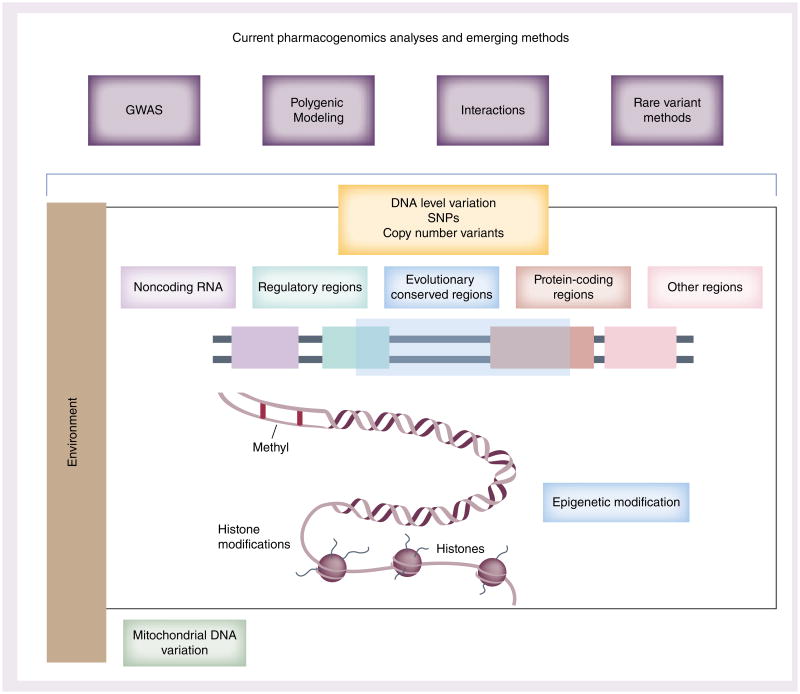
Current understanding of the genetic architecture of PGx traits presents a picture of a substantial impact of genetic variation on PK, PD, adverse events and other pharmacological outcomes. We have detailed here several examples of successes for the field of PGx. This information is being moved into the clinic for aiding decision making and results of these studies guide future drug development. While this knowledge is proving useful, we have outlined here key considerations for future PGx research and use of clinical biomarkers. Figure 3 provides an overview of important aspects that should be integrated in PGx studies to identify more robust markers for PGx traits and advance a more comprehensive understanding of the relationship between genetic architecture and drug response.
Figure 3.
Uncovering the genetic etiology of pharmacogenomic traits: methodologies and data. Along the top of the figure: pharmacogenomics studies should incorporate multiple types of analyses, beyond GWAS moving forward. Lower part of the figure: pharmacogenomics methods need to incorporate multiple types of genomic data, and consider the importance of environment as a modifier. Combining these elements may to yield improved predictions of pharmacogenomics outcomes. Furthermore, detailed molecular genetics studies following up on genomic association discovery will be important for identifying robust biomarkers for clinical decision-making.
GWAS: Genome-wide association study.
DNA/histone lower part of figure adapted with permission from [142].
Improving the way the PGx field has been sharing PGx association results, and expanding what is considered ‘validation’ and ‘replication’ for PGx association results, has broad potential for improving the utility of PGx findings as robust clinical biomarkers. Standardized reporting of PGx results will assist in compiling evidence and subsequent interpretation of multiple study results. Furthermore, molecular evaluation and validation of the mechanism by which polymorphisms have an effect on outcome needs to be an important step after association studies have identified variants of interest. While seeking replication of association results over multiple studies can provide evidence for a bio-marker, establishing the biological mechanistic role of a genetic variant on outcome can identify robust markers for clinical trial [8,72]. Multiple polymorphisms have known effects on protein coding genes, such as the well understood ADME genes. However, GWAS have identified numerous genetic variants outside of protein coding genes. As nearly 80% of the human genome is transcribed while only 1.2% encodes proteins, and as countless genomic regions carry epigenetic regulatory marks, our emerging understanding of the dynamic nature of nonprotein-coding regions of the genome must be leveraged for studying functionality of SNPs identified in association studies.
The field of PGx and GWAS of complex traits have focused almost exclusively on SNPs of common frequency. Rare variants, as well as other genetic variation such as copy number variation and mitochondrial variants may also prove important moving forward. Much of the original GWAS for complex traits was limited to individuals of European descent. The PGx field has stronger track record of studies across ancestry, accruing information about variation in drug response and genetic variation across multiple ancestries. Such ancestry information has clinical relevance and is being incorporated with FDA drug labeling.
An emphasis on cross-disciplinary work has become increasingly critical, with involvement of clinicians, genetic epidemiologists, statisticians, bioinformaticists and molecular and cellular biologists. The Pharmaco-genomics Research Network (PGRN), and the related Pharmacogenomics Statistical Analysis Resource (P-STAR), exemplify cross-disciplinary collaborations supporting PGx discovery. Furthermore, novel computational and statistical methods will prove critical, given the explosion of data generated in recent years. Simulations will be useful for exploring models for PGx traits.
Future work utilizing electronic health records will provide new dimensions for the successful development of PGx phenotypes, cohorts, studies and hypotheses, made available on a large scale through consortia such as the eMERGE network [143]. For analytical methods development, future efforts should include extension of MLM/GCTA/polygenic methods for better handling non-normal PGx traits, investigating sensitivity of heritability estimates with model specification and covariate selection and the development of integrated analysis methods for simultaneously incorporating different types of genomic features (genetic and epigenetic) and prior knowledge (pathways, gene sets, etc.) in each PGx study. Going beyond additive models, future studies should also focus on dynamic (epistatic) gene–gene interactions, and the impact of environmental influence.
Future perspective
PGx research has already yielded numerous examples of the pervasive effect of genetic factors on drug response. These advances demonstrate that clinical applications of pharmacogenomic biomarker tests have outstanding potential to enhance efficacy and reduce adverse effects, considered a main cause of morbidity and mortality – thereby showing promise for advancing the NIH mandate for the future of genomics. However, much of the genetic influence on treatment outcomes remains hidden, leaving uncertain how many genes and genetic variants contribute to pharmacological traits, how common and rare variants affect response and whether gene–gene interactions play a role. These relationships form the ‘pharmacogenomics architecture’ that still needs to be elucidated, presaging a profound evolution of the field of PGx, as is occurring in genomics studies of complex disorders. New approaches and studies across multiple human populations will prove critical for the characterization of the genetic architecture of pharmacogenomic traits required for realizing the full potential of PGx in guiding the development of optimal individualized therapies. With these advances realized over the next 5–10 years, the findings of PGx will dramatically increase use of genotypic data by clinicians in decisions on individual therapies, with substantially improved health outcomes.
Executive summary.
What have we learned?
- Heritability of pharmacogenomic traits: challenges & successes
- Defining the heritability of pharmacogenomic (PGx) traits encounters hurdles distinct from those found in the analysis of complex disorders.
- Heritability estimates in PGx: methods and current estimates.
- The PGx landscape: drug efficacy & adverse events
- PGx in the clinic: warfarin, clopidogrel.
- Dosing guidelines: US FDA Labels and Clinical Pharmacogenetics Implementation Consortium (CPIC).
- Larger relative strength of effect size for PGx traits compared with the range of effect sizes observed with complex-trait genome-wide association studies.
- Functional role of genomic architecture in PGx traits
- Focus of interpretation of the relationship between genetic variability and outcome for PGx has been centered on protein-coding regions.
- Much more to discover.
- Mechanistic etiology of genetic variation on PGx traits is a critical focus for future PGx studies.
Beyond PGx genome-wide association studies: polygenic analyses
- Polygenic genetic architecture
- Contribution of multiple common SNPs to phenotypic variance in aggregate.
- Two methods have already been used for a variety of complex outcomes for non-PGx traits:
- Mixed linear modeling.
- Polygenic modeling.
Overview of current method use, discovery and limitations.
Future methods
Summary of other methods of utility for future PGx research.
The role of epistasis in PGx traits.
Clinical & regulatory decision-making: moving from ‘bench to bedside’
As more of the complex genetic architecture of PGx traits is uncovered, substantial challenges remain for translating PGx findings to the clinic.
Highly significant associations between variants and PGx traits may differ considerably across ancestries, which has a direct impact on dosing decisions.
Substantial inconsistencies exist in study design, dosing regimens, study population and analysis methods for the field of PGx that should be addressed.
Conclusion
Much has been learned in the PGx field about the genetic architecture of PGx; however, there is more to be understood.
A variety of additional methods and approaches should be included in future PGx research.
There are many challenges still faced by the field, but ultimately promise for incorporating understanding of individuals genetic architecture for personalized/precision medicine.
Acknowledgments
The authors thank the Pharmacogenomics Research Network (PGRN) and its trans-network project on ‘Polygenic Modeling': P-STAR (PGRN Statistical Analysis Resource (funded as part of PAT – HL065962, XGEN (Expression Genetics in Drug Therapy) PMT GM092655, PAAR4Kids (Pharmacogenom-cs of Anticancer Agents Research in Children) GM092666, PEAR (Pharmacogenomic Evaluation of Antihypertensive Responses) GM074492, PARC (Pharmacogenomics and Risk of Cardiovascular Disease) HL069757, PHAT (Pharmacogenetics of Asthma Treatment) HL065899, PAPI (Pharmacogenomics of Anti-Platlet Intervention) HL105198, PHRAT (Pharmacogenomics of Rheumatoid Arthritis Therapy) GM092691.
Footnotes
Financial & competing interests disclosure: The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest;
•• of considerable interest
- 1.Green ED, Guyer MS. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470(7333):204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 2.Bergmann TK, Vach W, Feddersen S, et al. GWAS-based association between RWDD3 and TECTA variants and paclitaxel induced neuropathy could not be confirmed in Scandinavian ovarian cancer patients. Acta Oncol. 2013;52(4):871–874. doi: 10.3109/0284186X.2012.707787. [DOI] [PubMed] [Google Scholar]
- 3.PGRN. www.pgrn.org/display/pgrnwebsite/PGRN+Home.
- 4.Ofverholm A, Einbeigi Z, Manouchehrpour S, Albertsson P, Skrtic S, Enerbäck C. The ABCB1 3435 T allele does not increase the risk of paclitaxel-induced neurotoxicity. Oncol Lett. 2010;1(1):151–154. doi: 10.3892/ol_00000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.PharmGKB. www.pharmgkb.org/
- ••6.Maher B. Personal genomes: the case of the missing heritability. Nature. 2008;456(7218):18–21. doi: 10.1038/456018a. After many genome-wide association studies for common traits, very little of the heritability of traits has been explained. This commentary outlines several ways more can be elucidated about the genomic architecture underlying traits and outcomes. These ideas can also be applied to future work in pharmacogenomics (PGx), as the PGx field moves forward. [DOI] [PubMed] [Google Scholar]
- 7.Ioannidis JPA. Biomarker Failures. Clin Chem. 2013;59(1):202–204. doi: 10.1373/clinchem.2012.185801. [DOI] [PubMed] [Google Scholar]
- ••8.Sadee W. Pharmacogenomic biomarkers: validation needed for both the molecular genetic mechanism and clinical effect. Pharmacogenomics. 2011;12(5):675–680. doi: 10.2217/pgs.11.23. Discussion of the importance of laboratory research into the molecular mechanisms of PGx findings. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilke RA, Ramsey LB, Johnson SG, et al. The clinical pharmacogenomics implementation consortium: CPIC guideline for SLCO1B1 and simvastatin-induced myopathy. Clin Pharmacol Ther. 2012;92(1):112–117. doi: 10.1038/clpt.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shuldiner AR, O'Connell JR, Bliden KP, et al. Association of cytochrome P450 2C19 genotype with the anti platelet effect and clinical efficacy of clopidogrel therapy. JAMS. 2009;302(8):849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vessel ES, Page JG. Genetic control of dicumarol levels in man. J Clin Invest. 1968;47(12):2657–2663. doi: 10.1172/JCI105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faraday N, Yank LR, Mathias R, et al. Heritability of platelet responsiveness to aspirin in activation pathways directly and indirectly related to cyclooxygenase-1. Circulation. 2007;115(19):2490–2496. doi: 10.1161/CIRCULATIONAHA.106.667584. [DOI] [PubMed] [Google Scholar]
- •13.Stark AL, Zheng W, Mi S, et al. Heritable and non-genetic factors as variables of pharmacology phenotypes in lymphoblastoid cell lines. Pharmacogenomics J. 2010;10(6):505–512. doi: 10.1038/tpj.2010.3. Research into considerations for the use of lymphoblastoid cell lines for PGx research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dilan ME, New bold KG, Nagasubramanian R, et al. Heritability and linkage analysis of sensitivity to cisplatin-induced cytotoxicity. Cancer Res. 2004;64(12):4353–4356. doi: 10.1158/0008-5472.CAN-04-0340. [DOI] [PubMed] [Google Scholar]
- •15.Peters EH, Motsinger-Reif A, Havened TM, et al. Pharmacogenomic characterization of US FDA-approved cytotoxic drugs. Pharmacogenomics. 2011;12(10):1407–1415. doi: 10.2217/pgs.11.92. High-throughput evaluation of the cytotoxic effect of 29 commonly prescribed chemotherapeutics across 125 lymphoblastoid cell lines derived from 14 extended CUFF families. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waters J, Kaja A, Merci MA, Province MA, Cleo JL. Genome-wide discovery of loci influencing chemotherapy cytotoxicity. Proc Natl Acad Sci USA. 2004;101(32):11809–11814. doi: 10.1073/pnas.0404580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dean S, Bluebell WK, Huang RS, et al. Mapping genes that contribute to daunorubicin-induced cytotoxicity. Cancer Res. 2007;67(11):5425–5433. doi: 10.1158/0008-5472.CAN-06-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambs JK, Crews K, Pounds S, et al. Pharmacogenetics of deoxycytidine kinase: identification and characterization of novel genetic variants. J Pharmacol Exp Ther. 2007;323(3):935–945. doi: 10.1124/jpet.107.128595. [DOI] [PubMed] [Google Scholar]
- •19.Allow W, Tang BK, Endrenyi L. Hypothesis: comparisons of inter- and intra-individual variations can substitute for twin studies in drug research. Pharmacogenetics. 1998;8(4):283–289. doi: 10.1097/00008571-199808000-00001. Introduction and explanation of the use of repeated drug administration for estimation of heritability. [DOI] [PubMed] [Google Scholar]
- 20.We AC, Tantisira K, Li L, Schumann B, Weirs S Childhood Asthma Management Program Research Group. Repeatability of response to asthma medications. J Allergy Clin Immune. 2009;123(2):385–390. doi: 10.1016/j.jaci.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Legman MK, Giacomini KM. Estimating the contribution of genes and environment to variation in renal drug clearance. Pharmacogenetics. 2003;13(9):581–584. doi: 10.1097/00008571-200309000-00007. [DOI] [PubMed] [Google Scholar]
- 22.Pencilälinen PH, Neurone PH, Pencilä A. Pharmacokinetics of metformin after intravenous and oral administration to man. Eur J Clin Pharmacol. 1979;16(3):195–202. doi: 10.1007/BF00562061. [DOI] [PubMed] [Google Scholar]
- 23.Sjöval J, Albán G, Heartfelt B. Intra- and inter-individual variation in pharmacokinetics of intravenously infused amoxicillin and ampicillin to elderly volunteers. Br J Clin Pharmacol. 1986;21(2):171–181. doi: 10.1111/j.1365-2125.1986.tb05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hallén B, Gail baud O, Stöberg S, Lind eke B. Single-dose pharmacokinetics of terodiline, including a stable isotope technique for improvement of statistical evaluations. Bop harm Drug Dispose. 1988;9(3):229–250. doi: 10.1002/bod.2510090302. [DOI] [PubMed] [Google Scholar]
- 25.Davidson A, Headman A. Plasma and renal clearance of iohexol – a study on the reproducibility of a method for the glomerular filtration rate. Sand J Clin Lab Invest. 1990;50(7):757–761. doi: 10.3109/00365519009091069. [DOI] [PubMed] [Google Scholar]
- 26.Wagner JG. Inter- and intra subject variation of digoxin renal clearance in normal adult males. Drug Intel Clin Farm. 1988;22(7–8):562–567. doi: 10.1177/106002808802200708. [DOI] [PubMed] [Google Scholar]
- 27.Allow W, Endrenyi L, Tang B. Repeat administration of drugs as a means to assess the genetic component in pharmacological variability. Pharmacology. 1999;58(6):281–284. doi: 10.1159/000028292. [DOI] [PubMed] [Google Scholar]
- 28.Oz demur V, Allow W, Tang BK, et al. Evaluation of the genetic component of variability in CYP3A4 activity: a repeated drug administration method. Pharmacogenetics. 2000;10(5):373–388. doi: 10.1097/00008571-200007000-00001. [DOI] [PubMed] [Google Scholar]
- 29.Michele J, Chin LW, Sugars SB, et al. Measuring the overall genetic component of nevi rapine pharmacokinetics and the role of selected polymorphisms: towards addressing the missing heritability in pharmacogenetic phenotypes? Pharmacogenes Genomics. 2013;23(11):591–596. doi: 10.1097/FPC.0b013e32836533a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Birkenfield AL, Jordan J, Human U, et al. Genetic influences on the pharmacokinetics of orally and intravenously administered digoxin as exhibited by monozygotic twins. Clin Pharmacol Ther. 2009;86(6):605–608. doi: 10.1038/clpt.2009.170. [DOI] [PubMed] [Google Scholar]
- 31.Kroetz FL, Guyed T, Gang T, et al. Heritability of digoxin pharmacokinetics. Clin Pharmacol Ther. 2005;77(2):P21–P21. [Google Scholar]
- 32.Legman M, Brown C, Cheng J, et al. Heritability of metformin renal clearance. Clin Pharmacol Ther. 2005;77(2):P61–P61. [Google Scholar]
- 33.Gage BF, Lesli J. Pharmacogenetics of warfarin: regulatory, scientific, and clinical issues. J Thrombi Thrombosis. 2008;25(1):45–51. doi: 10.1007/s11239-007-0104-y. [DOI] [PubMed] [Google Scholar]
- 34.Schwas AI, Ritchie MD, Bradford Y, et al. Genetic determinants of response to warfarin during initial anticoagulation. N Eng J Med. 2008;358(10):999–1008. doi: 10.1056/NEJMoa0708078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tobe J, Halal D, Bugling T. Influence of cytochrome P-450 CYP2C9 polymorphisms on warfarin sensitivity and risk of over-anticoagulation in patients on long-term treatment. Blood. 2000;96(5):1816–1819. [PubMed] [Google Scholar]
- 36.Roast S, Foreign A, Ivaskevicius V, et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427(6974):537–541. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- 37.Reeder MK, Reine AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcription regulation and warfarin dose. N Eng J Med. 2005;352(22):2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 38.Scott SA, Telemann L, Lorn reich R, De snick RH. Warfarin pharmacogenetics: CYP2C9 and VKORC1 genotypes predict different sensitivity and resistance frequencies in the Ashkenazim and Sephardi Jewish populations. Am J Hum Gene. 2008;82(2):495–500. doi: 10.1016/j.ajhg.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sinatra FL, You HS, Reeder MK, et al. Association of Vitamin K epoxied reduct as complex 1 (VKORC1) variants with warfarin dose in a Hong King Chinese patient population. Pharmacogenes Genomics. 2005;15(10):687–691. doi: 10.1097/01.fpc.0000174789.77614.68. [DOI] [PubMed] [Google Scholar]
- 40.International Warfarin Pharmacogenetics Consortium. Klein TE, Atman RB, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Eng J Med. 2009;360(8):753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kimbell SE, French B, Kasper SE, et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Eng J Med. 2013;369(24):2283–2293. doi: 10.1056/NEJMoa1310669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pirmohamed M, Burnside G, Erik son N, et al. A randomized trial of genotype-guided dosing of warfarin. N Eng J Med. 2013;369(24):2294–2303. doi: 10.1056/NEJMoa1311386. [DOI] [PubMed] [Google Scholar]
- 43.Scott SA, Sang kohl K, Gardner E, et al. Clinical pharmacogenetics implementation consortium guidelines for cytochrome P450–2C19 (CYP2C19) genotype and clopidogrel therapy. Clin Pharmacol Ther. 2011;90(2):328–332. doi: 10.1038/clpt.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colet JP, Helot JS, Penna A, et al. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet. 2009;373(9660):309–317. doi: 10.1016/S0140-6736(08)61845-0. [DOI] [PubMed] [Google Scholar]
- 45.Table of Pharmacogenomic Biomarkers in Drug Labeling. www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm.
- ••46.Telling MB, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89(3):464–467. doi: 10.1038/clpt.2010.279. Explanation of the development and goals of Clinical Pharmacogenetics Implementation Consortium, and discussion of important considerations for the clinical community for moving PGx findings from ‘bench to bedside.’. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swen JJ, Nijenhuis M, de Boer A, et al. Pharmacogenetics: from bench to byte - an update of guidelines. Clin Pharmacol Ther. 2011;89(5):662–673. doi: 10.1038/clpt.2011.34. [DOI] [PubMed] [Google Scholar]
- 48.PharmGKB: Dosing Guidelines – CPIC. www.pharmgkb.org/view/dosing-guidelines.do?source=CPIC.
- 49.PharmGKB: CPIC pairs. www.pharmgkb.org/page/cpicGeneDrugPairs.
- 50.PMT nomenclature committee. www.imh.liu.se/tpmtalleles.
- 51.PharmGKB: PMT. www.pharmgkb.org/gene/PA356#Tabbie=tab4&suntan=31.
- 52.Telling MB, Gardner E, Sand born WK, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for theo purine methyltransferase genotype and theo purine dosing. Clin Pharmacol Ther. 2011;89(3):387–391. doi: 10.1038/clpt.2010.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.CYP2C19 allele nomenclature. www.cypalleles.ki.se/cyp2c19.htm.
- 54.Scott SA, Sang kohl K, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013;94(3):317–323. doi: 10.1038/clpt.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson JAE, Gong L, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin Pharmacol Ther. 2011;90(4):625–629. doi: 10.1038/clpt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.The Human Cytochrome P450 (CUP) Allele Nomenclature Database. www.cypalleles.ki.se.
- 57.Crews KR, Gae disk A, Dunnenberger HM, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for Cytochrome P450 2D6 Genotype and Codeine Therapy: 2014 Update. Clin Pharmacol Ther. 2014;95(4):376–382. doi: 10.1038/clpt.2013.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin MA, Housman JIM, Freud RR, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for HLA-B Genotype and Abacavir Dosing: 2014 Update. Clin Pharmacol Ther. 2014;95(5):499–500. doi: 10.1038/clpt.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hersh field MS, Callaghan HT, Tassaneeyakul W, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for human leukocyte antigen-B genotype and allopurinol dosing. Clin Pharmacol Ther. 2013;93(2):153–158. doi: 10.1038/clpt.2012.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hicks JK, Swen JJ, Thorn CF, et al. Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6 and CYP2C19 genotypes and dosing of tricycle antidepressants. Clin Pharmacol Ther. 2013;93(5):402–408. doi: 10.1038/clpt.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neckband SG, Kellsie JR, Dunnenberger HM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for HLA-B genotype and carbamazepine dosing. Clin Pharmacol Ther. 2013;94(3):324–328. doi: 10.1038/clpt.2013.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Candle K, Thorn CF, Klein TE, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for dihydropyrimidine dehydrogenate genotype and Fluoropyrimidines dosing. Clin Pharmacol Ther. 2013;94(6):640–645. doi: 10.1038/clpt.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muir AH, Gong L, Johnson SG, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for IFNL3 (IL28B) genotype and PEG interferon-α-based regimens. Clin Pharmacol Ther. 2014;95(2):141–146. doi: 10.1038/clpt.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chancy JP, Johnson SG, Ye SW, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for ivacaftor therapy in the context of CTR genotype. Clin Pharmacol Ther. 2014 doi: 10.1038/clpt.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hind off LA, Seth apathy P, Jenkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chou SF. Polymorphism of human cytochrome P450 2D6 and its clinical significance: Part I. Clin Pharmacokinetic. 2009;48(11):689–723. doi: 10.2165/11318030-000000000-00000. [DOI] [PubMed] [Google Scholar]
- ••67.Thorn CF, Klein TE, Atman RB. PharmGKB: The Pharmacogenomics Knowledge Base. Methods Moll Biol. 2013;1015:311–320. doi: 10.1007/978-1-62703-435-7_20. The Pharmacogenomics Knowledge Base, through the Pharmacogenomics Research Network (PGRN). They describe the PharmGKB website ( www.pharmgkb.org), a knowledge base including genotypic, molecular and clinical PGx knowledge with links to additional external resources for PGx research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang D, Poi MK, Sun X, Gae disk A, Leader JS, Sadee W. Common CYP2D6 polymorphisms affecting alternative splicing and transcription: long-range haplotypes with two regulatory variants modulate CYP2D6 activity. Hum Moll Gene. 2014;23(1):268–278. doi: 10.1093/hmg/ddt417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dally AK. Genome-wide association studies in pharmacogenomics. Nat Rev Gene. 2010;11(4):241–246. doi: 10.1038/nrg2751. [DOI] [PubMed] [Google Scholar]
- 70.Dally AK. Pharmacogenomics of adverse drug reactions. Genome Med. 2013;5(1):5. doi: 10.1186/gm409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pompey YA, Stewart JD, Molal S, Phillips E, Peters B, Strove DA. The structural basis of HLA-associated drug hypersensitivity syndromes. Immune Rev. 2012;250(1):158–166. doi: 10.1111/j.1600-065X.2012.01163.x. [DOI] [PubMed] [Google Scholar]
- 72.Sadee W, Wang D, Pap AC, et al. Pharmacogenomics of the RNA world: structural RNA polymorphisms in drug therapy. Clin Pharmacol Ther. 2011;89(3):355–365. doi: 10.1038/clpt.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.ENCODE Project Consortium. A user's guide to the encyclopedia of DNA elements (ENCODE) Los Biol. 2011;9(4):e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ward LED, Kellia M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Null Acids Res. 2011;40(Database issue):D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Amazon ER, Zheng W, Konkashbaev A, et al. SCAN: SNP and copy number annotation. Bioinformation. 2010;26(2):259–262. doi: 10.1093/bioinformatics/btp644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang J, Lee SH, Goddard ME, Fischer PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Gene. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang J, Benyamin B, McCoy BP, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Gene. 2010;42(7):565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee SH, Wary N, Goddard ME, Fischer PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Gene. 2011;88(3):294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang J, Manolo TA, Pasquale LR, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Gene. 2011;43(6):519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang J, Furriers T, Morris AP, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Gene. 2012;44(4):369–375. S1–S3. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Purcell SM, Wary N, Stone JL, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bush WSW, Saucer SJ, de Ager PL, et al. Evidence for polygenic susceptibility to multiple sclerosis – the shape of things to come. Am J Hum Gene. 2010;86(4):621–625. doi: 10.1016/j.ajhg.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lang Allen H, Strata K, Letter G, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Peyotes UK, Willer CJ, Brent SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Gene. 2010;42(11):937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stahl EA, Legman D, Trunks G, et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat Gene. 2012;44(5):483–489. doi: 10.1038/ng.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McGeachie MK, Stahl EA, Hemes BE, et al. Polygenic heritability estimates in pharmacogenetics: focus on asthma and related phenotypes Pharmacogenes. Genomics. 2013;23(6):324–328. doi: 10.1097/FPC.0b013e3283607acf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chhibber A, Mumford J, Stahl EA, et al. Polygenic inheritance of paclitaxel-induced sensory peripheral neuropathy driven by axon outgrowth gene sets in CALEB 40101 (Alliance) Pharmacogenomics J. 2014;142(4):336–34. doi: 10.1038/tpj.2014.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bergmann TK, Gréen H, Brasch-Andersen C, et al. Retrospective study of the impact of pharmacogenetic variants on paclitaxel toxicity and survival in patients with ovarian cancer. Eur J Clin Pharmacol. 2011;67(7):693–700. doi: 10.1007/s00228-011-1007-6. [DOI] [PubMed] [Google Scholar]
- 90.De Groan SJM, Elans L, Sprawl JAE, et al. CYP3A4*22 genotype and systemic exposure affect paclitaxel-induced net toxicity. Clin Cancer Res. 2013;19(12):3316–3324. doi: 10.1158/1078-0432.CCR-12-3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang D, GAO Y, Brighton SA, Coke GE, Sadee W. Intrinsic polymorphism in CYP3A4 affects hepatic expression and response to stating drugs. Pharmacogenomics J. 2011;11(4):274–286. doi: 10.1038/tpj.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Baldwin RM, War K, Zembutsu H, et al. A genome-wide association study identifies novel loci for paclitaxel-induced sensory peripheral neuropathy in CALEB 40101. Clin Cancer Res. 2012;18(18):5099–5109. doi: 10.1158/1078-0432.CCR-12-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Richen B, Shorten JP, Weirs ST, Rosier B, De Cries K, Van der Lend R. Long-term variability of bronchial responsiveness to histamine in a random population sample of adults. Am Rev Respire Dis. 1993;148(4 Pt 1):944–949. doi: 10.1164/ajrccm/148.4_Pt_1.944. [DOI] [PubMed] [Google Scholar]
- 94.Nonfat MFR, Gut EEG, Dementias F, et al. A large-scale, consortium-based genome wide association study of asthma. N Eng J Med. 2010;363(13):1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thomson SF, van der Slues S, Kivim KO, Scythe A, Backer V. Estimates of asthma heritability in a large twin sample. Clin Exp Allergy. 2010;40(7):1054–1061. doi: 10.1111/j.1365-2222.2010.03525.x. [DOI] [PubMed] [Google Scholar]
- 96.Van Disk EL, Auger H, Jaszczyszyn Y, Hermes C. Ten years of next-generation sequencing technology. Trends Gene. 2014;30(9):418–426. doi: 10.1016/j.tig.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 97.Harri P, Scrota M, Butte AH. Ten Years of Pathway Analysis: Current Approaches and Outstanding Challenges. Los Compute Biol. 2012;8(2):e1002375. doi: 10.1371/journal.pcbi.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Peterson A, Sprat J, Title BL. Incorporating prior knowledge to increase the power of genome-wide association studies. In: Hondo C, van der Were J, Hayes B, editors. Genome-Wide Association Studies and Genomic Prediction. Humans Press; NJ, USA: 2013. pp. 519–541. [DOI] [PubMed] [Google Scholar]
- 99.Worthy EA. Analysis and annotation of whole-genome or whole-exome sequencing-derived variants for clinical diagnosis. Curr Proton Hum Gene. 2013;79 doi: 10.1002/0471142905.hg0924s79. Unit 9.24. [DOI] [PubMed] [Google Scholar]
- 100.Wagner MK. Rare-variant genome-wide association studies: a new frontier in genetic analysis of complex traits. Pharmacogenomics. 2013;14(4):413–424. doi: 10.2217/pgs.13.36. [DOI] [PubMed] [Google Scholar]
- 101.Cordell JJ. Genome-wide association studies: detecting gene-gene interactions that underlie human diseases. Nat Rev Gene. 2009;10(6):392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ya span BL, Bush WSW, Tors tendon ES, et al. Genetic analysis of biological pathway data through genomic randomization. Hum Gene. 2011;129(5):563–571. doi: 10.1007/s00439-011-0956-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pendergrass SA, Fraser A, Wallace J, et al. Genomic analyses with biofilter 2.0: knowledge driven filtering, annotation, and model development. iodate Min. 2013;6(1):25. doi: 10.1186/1756-0381-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bush WSW, Dude SM, Ritchie MD. Biofilter: a knowledge-integration system for the multi-locus analysis of genome-wide association studies. Pac Sump Biocomput. 2009:368–379. [PMC free article] [PubMed] [Google Scholar]
- 105.Ramsey LB, Braun E, Yang W, et al. Rare versus common variants in pharmacogenetics: SLCO1B1 variation and methotrexate disposition. Genome Res. 2012;22(1):1–8. doi: 10.1101/gr.129668.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Basia G, Banal V, Harris mendy O, et al. A covering method for detecting genetic associations between rare variants and common phenotypes. Los Compute Biol. 2010;6(10):e1000954. doi: 10.1371/journal.pcbi.1000954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Morgen haler S, Dilly G. A strategy to discover genes that carry multi-allelic or mono-allelic risk for common diseases: a cohort allele sums test (CAST) Mutate Res. 2007;615(1–2):28–56. doi: 10.1016/j.mrfmmm.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 108.Li B, Lil SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Gene. 2008;83(3):311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Madden BE, Browning SR. A group wise association test for rare mutations using a weighted sum statistic. Los Gene. 2009;5(2):e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Han F, Pan W. A data-adaptive sum test for disease association with multiple common or rare variants. Hum Herd. 2010;70(1):42–54. doi: 10.1159/000288704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Price AL, Kr yukon GOV, de Balker PI, et al. Pooled association tests for rare variants in exon-resequencing studies. Am J Hum Gene. 2010;86(6):832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Housman TH, Marine NJ, Witt JS. Comprehensive approach to analyzing rare genetic variants. Los ONE. 2010;5(11):e13584. doi: 10.1371/journal.pone.0013584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.We MC, Lee S, Cai T, Li Y, Boone M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Gene. 2011;89(1):82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Randell M, Huff C, He H, et al. A probabilistic disease-gene finder for personal genomes. Genome Res. 2011;21(9):1529–1542. doi: 10.1101/gr.123158.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Moore CB, Wallace JR, Fraser AT, Pendergrass SA, Ritchie MD. BioBin: a Bioinformation tool for automating the binning of rare variants using publicly available biological knowledge. BMC Med Genomics. 2013;6(Suppl. 2):S6. doi: 10.1186/1755-8794-6-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Moore CB, Wallace JR, Wolfe DJ, et al. Low frequency variants, collapsed based on biological knowledge, uncover complexity of population stratification in 1000 genomes project data. Los Gene. 2013;9(12):e1003959. doi: 10.1371/journal.pgen.1003959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Moore CB, Wallace JR, Fraser AT, Pendergrass SA, Ritchie MD. Using BioBin to explore rare variant population stratification. Pac Sump Biocomput. 2013:332–343. [PMC free article] [PubMed] [Google Scholar]
- ••118.Sadee W. The relevance of “missing heritability” Pharmacogenomics. Clin Pharmacol Ther. 2012;92(4):428–430. doi: 10.1038/clpt.2012.116. Discussion of moving beyond single SNP associations for PGx research to gene–gene and gene–environment explorations, as well as considering regulatory variants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••119.Evans WE, Telling MB. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286(5439):487–491. doi: 10.1126/science.286.5439.487. Discussion of how the effect of medications are not monomeric in nature, describing how they are actually determined by the interplay of several genes encoding proteins involved in multiple pathways of drug metabolism, disposition and effects. Discussion of other important considerations for PGx research. [DOI] [PubMed] [Google Scholar]
- 120.Sullivan D, Pinsonneault JK, Pap AC, et al. Dopamine transporter DAT and receptor DRD2 variants affect risk of lethal cocaine abuse: a gene–gene–environment interaction. Transl Psychiatry. 2013;3:e222. doi: 10.1038/tp.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Haul zinger ER, Dude SM, Fraser AT, Krauts RM, Medina MW, Ritchie MD. Athena: a tool for meta-dimensional analysis applied to genotypes and gene expression data to predict Hal cholesterol levels. Pac Sump Biocomput. 2013:385–396. [PMC free article] [PubMed] [Google Scholar]
- 122.Haul zinger ER, Buchanan CC, Dude SM, Tors tendon EC, Turner SD, Ritchie MD. Initialization parameter sweep in ATHENA: optimizing neural networks for detecting gene-gene interactions in the presence of small main effects. Gene Evil Compute Cong. 2010;12:203–210. doi: 10.1145/1830483.1830519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen P, Lin JJ, Lu CS, et al. Carbamazepine-induced toxic effects and HLA-B*1502 screening in Taiwan. N Eng J Med. 2011;364(12):1126–1133. doi: 10.1056/NEJMoa1009717. [DOI] [PubMed] [Google Scholar]
- 124.Man BL, Kean P, Boom L, et al. Association between HLA-B*1502 allele and anti epileptic drug-induced cutaneous reactions in Han Chinese. Epilepsy. 2007;48(5):1015–1018. doi: 10.1111/j.1528-1167.2007.01022.x. [DOI] [PubMed] [Google Scholar]
- 125.Hung SI, Cheng W, Liu SS, et al. Common risk allele in aromatic antiepileptic-drug induced Stevens-Johnson syndrome and toxic epidermal necrosis in Han Chinese. Pharmacogenomics. 2010;11(3):349–356. doi: 10.2217/pgs.09.162. [DOI] [PubMed] [Google Scholar]
- 126.Locharernkul C, Loplumlert J, Limo tao C, et al. Carbamazepine and phenytoin induced Stevens-Johnson syndrome is associated with HLA-B*1502 allele in Thai population. Epilepsy. 2008;49(12):2087–2091. doi: 10.1111/j.1528-1167.2008.01719.x. [DOI] [PubMed] [Google Scholar]
- 127.Allergic A, Morgen sen AL, Williamson PR, Chadwick D, Park BK, Pirmohamed M. HLA-B locus in Caucasian patients with carbamazepine hypersensitivity. Pharmacogenomics. 2006;7(6):813–818. doi: 10.2217/14622416.7.6.813. [DOI] [PubMed] [Google Scholar]
- 128.Bonjour C, Thomas L, Boot N, et al. A marker for Stevens-Johnson syndrome …: ethnicity matters. Pharmacogenomics J. 2006;6(4):265–268. doi: 10.1038/sj.tpj.6500356. [DOI] [PubMed] [Google Scholar]
- 129.Choudhury S, Eng N, Aila PC, et al. Pharmacogenetic differences in response to albuterol between Puerto Ricans and Mexicans with asthma. Am J Respire CRT Care Med. 2005;171(6):563–570. doi: 10.1164/rccm.200409-1286OC. [DOI] [PubMed] [Google Scholar]
- 130.Lindi N, Gold stein J, Blair dell J, Basely T, Rivers C, Acton R. Influence of CYP2C9 genotype on warfarin dose among African American and European Americans. Per Med. 2007;4(2):157–169. doi: 10.2217/17410541.4.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Scott SA, Kameko M, Kibitz SA, Lorn reich R, Haltering JL, De snick RH. CYP2C9*8 is prevalent among African-Americans: implications for pharmacogenetic dosing. Pharmacogenomics. 2009;10(8):1243–1255. doi: 10.2217/pgs.09.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tai G, Faring F, Reeder MK, et al. In-vitro and in-vivo effects of the CYP2C9*11 polymorphism on warfarin metabolism and dose. Pharmacogenes Genomics. 2005;15(7):475–481. doi: 10.1097/01.fpc.0000162005.80857.98. [DOI] [PubMed] [Google Scholar]
- 133.Tedman AR, Dick man J, Kidd RS, Gold stein JAE, Ritchie FM, Hon TY. CYP2C9 genetic polymorphisms and warfarin. Clin Apple Thrombi He most. 2004;10(2):149–154. doi: 10.1177/107602960401000205. [DOI] [PubMed] [Google Scholar]
- 134.Cavalry LG, Lanae TY, Mammary KM, et al. Genetic and clinical predictors of warfarin dose requirements in African Americans. Clin Pharmacol Ther. 2010;87(4):459–464. doi: 10.1038/clpt.2009.223. [DOI] [PubMed] [Google Scholar]
- 135.Delius M, Chen LY, Lind JD, et al. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2009;113(4):784–792. doi: 10.1182/blood-2008-04-149070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gage BF, Evy C, Johnson JAE, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84(3):326–331. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lindi NA, Delius M, Cavalry L, et al. Warfarin pharmacogenetics: a single VKORC1 polymorphism is predictive of dose across 3 racial groups. Blood. 2010;115(18):3827–3834. doi: 10.1182/blood-2009-12-255992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Suarez-Kurtz G, Bottom MR. Pharmacogenomics of warfarin in populations of African descent. Br J Clin Pharmacol. 2013;75(2):334–346. doi: 10.1111/j.1365-2125.2012.04354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kastbom A, Brat J, Ernestus S, et al. Gamma receptor type RIIIA genotype and response to tumor necrosis factor alpha-blocking agents in patients with rheumatoid arthritis. Arthritis Rheum. 2007;56(2):448–452. doi: 10.1002/art.22390. [DOI] [PubMed] [Google Scholar]
- 140.Tut uncut Z, Havana A, Vainer N, Corr M, Deutsche R, Boyle D. Gamma receptor type RIIIA polymorphisms influence treatment outcomes in patients with inflammatory arthritis treated with tumor necrosis factor alpha-blocking agents. Arthritis Rheum. 2005;52(9):2693–2696. doi: 10.1002/art.21266. [DOI] [PubMed] [Google Scholar]
- 141.CañET JD, SeaáRex B, HerbáAndes MB, et al. Influence of variants of Fc gamma receptors ILIA and RIIIA on the American College of Rheumatology and European League Against Rheumatism responses to anti-tumour necrosis factor alpha therapy in rheumatoid arthritis. Ann Rheum Dis. 2009;68(10):1547–1552. doi: 10.1136/ard.2008.096982. [DOI] [PubMed] [Google Scholar]
- 142.Walker FM, Gore AC. Trans generational neuron endocrine disruption of reproduction. Nat Rev Endocrine. 2011;7(4):197–207. doi: 10.1038/nrendo.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hotelman O, Kuivaniemi H, Tromp G, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Gene Med. 2013;15(10):761–771. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]