Abstract
We seek to define sequence-based predictive criteria to identify polymorphic and hypermutable minisatellites in the human genome. Polymorphism of a representative pool of minisatellites, selected from human chromosomes 21 and 22, was experimentally measured by PCR typing in a population of unrelated individuals. Two predictive approaches were tested. One uses simple repeat characteristics (e.g., unit length, copy number, nucleotide bias) and a more complex measure, termed HistoryR, based on the presence of variant motifs in the tandem array. We find that HistoryR and percentage of GC are strongly correlated with polymorphism and, as predictive criteria, reduce by half the number of repeats to type while enriching the proportion with heterozygosity ≥0.5, from a background level of 43% to 59%. The second approach uses length differences between minisatellites in the two releases of the human genome sequence (from the public consortium and Celera). As a predictor, this similarly enriches the number of polymorphic minisatellites, but fails to identify an unexpectedly large number of these. Finally, typing of the highly polymorphic minisatellites in large families identified one new hypermutable minisatellite, located in a predicted coding sequence. This may represent the first coding human hypermutable minisatellite.
[Supplemental material is available online at www.genome.org.]
Tandem repeats represent a significant fraction of vertebrate genomes and have been classified as satellites, minisatellites, and microsatellites according to the length of the repeated unit and the overall length of the array. Minisatellites are usually defined as the tandem repeats of a short (10- to 100-bp) motif spanning several hundred to several thousand base pairs and are associated with interesting features of genome biology (for review, see Vergnaud and Denoeud 2000).
Minisatellites frequently exhibit length polymorphism, which results from variation in the number of internal copies, making them valuable genomic markers. They provided the first highly polymorphic, multiallelic markers for linkage studies (Bell et al. 1982; Nakamura et al. 1987) and were used in the early stages of human genome mapping (NIH/CEPH Collaborative Mapping Group, 1992). Chromosomal distribution of minisatellites in the human genome is highly skewed toward telomeres and ancestrally telomeric regions (Amarger et al. 1998). Highly polymorphic minisatellites are thus a good tool for detection of microdeletions in the ends of chromosomes, associated with human pathologies such as mental retardation (Giraudeau et al. 2001). Polymorphic minisatellites are also found in bacterial genomes (Le Fleche et al. 2001), in which they have proven to be a powerful tool for bacterial strain identification.
Although the abundance of polymorphic minisatellites suggests that they are fast-evolving sequences, most of them are, in fact, quite stable. New alleles that display changes in the number of tandem copies have been observed at only a few loci, called hypermutable minisatellites. Changes at these loci in the germline can be observed in the next generation, and in humans, one locus, D2S90 (CEB1), has been found to change in as many as 13% of the gametes (Vergnaud et al. 1991; Vergnaud and Denoeud, 2000). Hypermutable minisatellites may provide a potent source of information on the mechanism of minisatellite instability. In humans, this instability apparently arises at least in part through gene conversion events, during or shortly after meiosis, many of which involve interallelic transfers of information (Buard and Vergnaud 1994; Jeffreys et al. 1994; May et al. 1996; Buard et al. 1998). Similar intraallelic and interallelic recombination events are found in MS32 and CEB1 minisatellite sequences, when they are placed close to a meiotic hotspot in Saccharomyces cerevisiae (Appelgren et al. 1997, 1999; Debrauwère et al. 1999). Most likely, these events result from the gene conversion repair of double-strand breaks, as recent evidence indicates that meiotic recombination in mammals and yeast is initiated by the Spo11p endonuclease (Bergerat et al. 1997; Keeney et al. 1997; Baudat et al. 2000; Romanienko and Camerini-Otero 2000), which is also essential to the meiotic instability of the minisatellites introduced in yeast (Debrauwère et al. 1999). In agreement with these observations, it has been proposed that the meiotic hypermutability of some minisatellite structures is the byproduct of the coincidence of an ordinary minisatellite with a double-strand break hotspot (Vergnaud and Denoeud 2000).
Interestingly, hypermutable minisatellites might additionally provide biomarkers for low-dose exposure of the human germline to ionizing radiation (Dubrova et al. 1993, 1997; Dubrova and Plumb 2002). Unfortunately, <10 human hypermutable loci have been characterized so far, using approaches developed >10 years ago, whereas the population studies conducted to evaluate the effect of low-dose irradiation would greatly benefit from the availability of a larger panel of probes.
Given the multifaceted utility of minisatellites, determining which are polymorphic/hypermutable would seem a valuable task. Efficient tandem repeat detection software enables the identification of tandem repeats across entire genomes (Benson 1999; Vergnaud and Denoeud 2000), so that testing for polymorphism is all that is required. But although the polymorphism of the few dozen minisatellites usually present in a small genome can be systematically assayed at a reasonable cost (Le Fleche et al. 2001), this is not a realistic option for the human genome. There, the number of minisatellite loci is estimated in the thousands (based on the sequence of chromosome 22; Vergnaud and Denoeud 2000), the proportion of highly polymorphic minisatellites among these is not known, and previous efforts to identify hypermutable loci among minisatellites have produced only very low yields (∼1% to 3% of those examined). Furthermore, sequence analysis of a few hypermutable loci has not yet revealed specific features that might facilitate their identification (Murray et al. 1999). Of need are predictive criteria that can be applied before the expensive and labor-intensive step of polymorphism typing.
Earlier attempts at polymorphism prediction for tandem repeats focused on microsatellites. Fondon III et al. (1998) identified polymorphic loci by selecting microsatellites in which the individual copies were at least 90% identical to a core pattern, but that study did not include a control group to test whether selection yielded higher polymorphism values than the background rate. Wren et al. (2000) improved polymorphic microsatellite identification by requiring perfect homogeneity of the repetitive unit. Such results are in accordance with the mutation process of microsatellites (replication slippage): They are stabilized by variant repeats (Weber 1990), the presence of which facilitates detection of slipped-strand DNA by the mismatch repair system (Strand et al. 1993; Heale and Petes 1995). In the case of minisatellites, in which internal conservation is not the rule at currently known hypermutable loci (Murray et al. 1999; Vergnaud and Denoeud 2000), such a high conservation requirement imposes too great a restriction on the set of potentially useful repeats and, as we report below, would preclude finding both highly polymorphic and hypermutable repeats.
The purpose of this report is to define inexpensive strategies to accelerate the search for highly polymorphic minisatellites. The goal has been the development of sequence-based predictive criteria for polymorphism. Results are based on the study of a representative pool of minisatellites selected from human chromosomes 21 and 22. Polymorphism for these loci was experimentally measured by typing in a population of unrelated individuals. This was followed by typing the most polymorphic loci across a number of large families to test for hypermutability. Two predictive approaches were tested. The most straightforward takes advantage of the availability of two different releases of the human genome sequence: one from the public genome sequencing project and the other from the private Celera project. The second approach uses sequence-based characteristics of the repeats—including such simple measures as unit length, copy number, degree of conservation, percentage of GC (%GC)— and a more complex measure based on the internal organization of variant motifs in the tandem array. A repeat that contains several distinct sets of nearly identical mutations exhibits prima facie evidence of multiple rounds of expansion and may be more likely to exist as multiple alleles than a repeat that contains mostly unique mutations (Fig. 1). This later measure is analyzed by using history reconstruction (Benson and Dong 1999), a type of parsimony analysis that infers how the present day sequence could have evolved from a single ancestral copy while undergoing a minimum number of point mutations interspersed with duplications.
Figure 1.
Multiple alignments of tandem repeats CEB252 and CEB233. In each alignment, the upper darker line is a consensus pattern for the basic unit, shown for reference, and the lighter lines are the individual copies, ordered from top to bottom as they occur in the repeat. Only differences with the consensus are shown. Heterozygosity for CEB252 is 0.6. Note several redundant patterns of mutation resulting in a high HistoryR score. Heterozygosity for CEB233 is zero. No clear organization of mutations resulting in a low HistoryR score.
RESULTS
Characterization of Chromosome 21 and 22 Minisatellites
Human chromosomes 21 and 22 contain ∼15,000 tandem repeats each (as detected by tandem repeats finder [TRF] in the publicly available sequences that exclude heterochromatin; Benson 1999). For this study, the empirical definition of minisatellites follows the suggestion made in Vergnaud and Denoeud (2000), which is more stringent than the usual definition of minisatellites mentioned in the introduction: (1) unit length ≥17 bp, (2) copy number ≥10, (3) total length ≥350 bp, (4) percent matches ≥70%, and (5) GC bias (i.e., strand asymmetry for G and C; see Methods section) ≥0.35. This definition includes repeats clearly classified as minisatellites, not microsatellites, allows minisatellites shorter than the ≥800 bp usually identified by Southern blotting (Vergnaud 1989; Amarger et al. 1998) and removes repeats with highly diverged copies. On chromosomes 21 and 22, 127 tandem repeats fulfill these criteria. Table 1 indicates their position on the chromosomes. As described before for minisatellites derived by classical approaches (Amarger et al. 1998), they are mainly located toward chromosome ends (both chromosomes are acrocentric). Analysis shows no statistically significant differences between the minisatellites from chromosome 21 and 22 for any of the characteristics listed in Supplementary Table 1. The two chromosomes will subsequently be considered together.
Table 1.
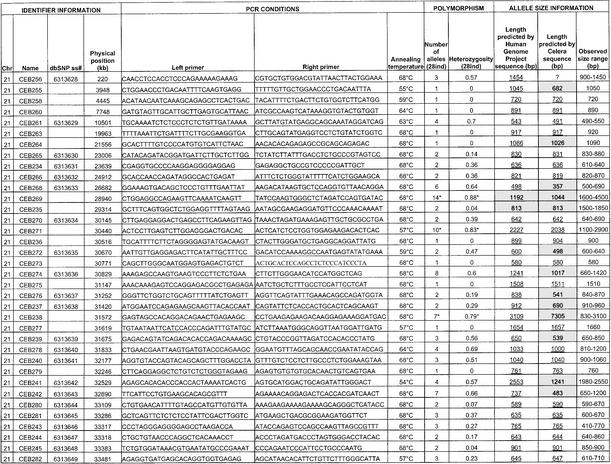
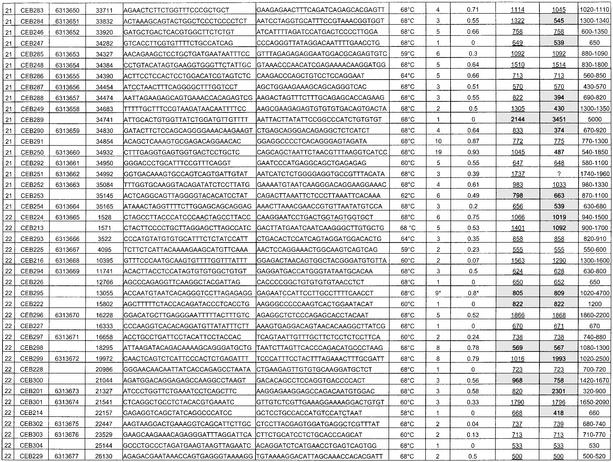
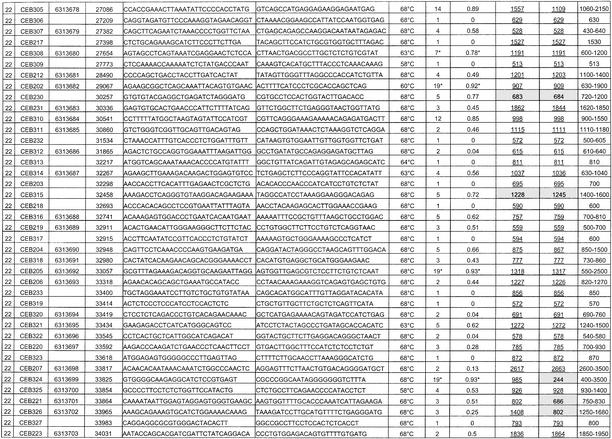
List of the 118 Minisatellites That Were Typed: PCR Conditions, Polymorphism Results, and Allele Size Information
PCR Typing Results
Polymorphism results, that is, number of alleles observed and heterozygosity, are given in Table 1, as well as dbSNP accession numbers for the polymorphic minisatellites that were submitted to the SNP database (http://www.ncbi.nlm.nih.gov/SNP/index.html). Supplementary data about polymorphism is also available at http://minisatellites.u-psud.fr. For the minisatellites that were typed first (“training set”), the study was made on a population of 76 unrelated individuals. Results were comparable to those obtained with a subset of 28 unrelated individuals from the set of 76. Subsequent PCR typings (minisatellites from the “test set”) were performed only on the 28 individuals, except for the most polymorphic loci that were typed in all 76 individuals in order to evaluate their polymorphism more accurately.
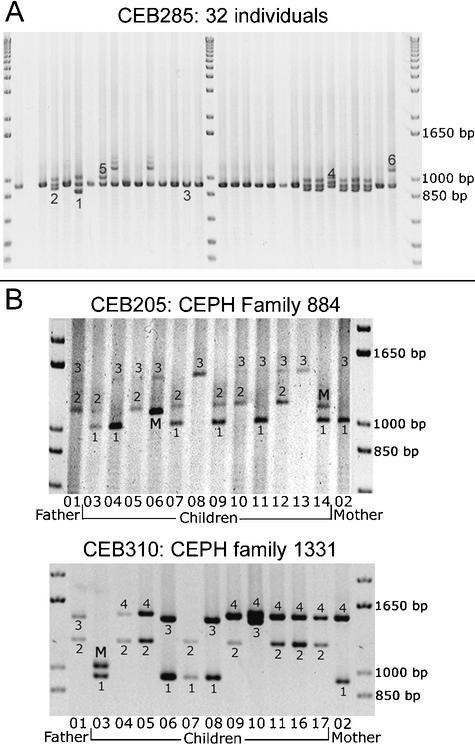
Among the 127 minisatellites, 118 were successfully amplified (55 on chromosome 21 and 63 on chromosome 22) by using the selected primer pair (Table 1). Not surprisingly, long minisatellites (>2 kb) are the most difficult to amplify: Only five among eight were successfully amplified under the conditions used. Figure 2A shows the image of the gel obtained for minisatellite CEB285 on 32 individuals (including 28 unrelated individuals): Six different alleles can be assigned. About 75% of the minisatellites successfully amplified are polymorphic (i.e., two alleles or more), and 42% have a heterozygosity value ≥0.5.
Figure 2.
(A) Ethidium bromide–stained agarose gel showing PCR products for minisatellite CEB285. Six different alleles are scored among 32 individuals (in some cases, three bands are seen for one individual [the upper one is a PCR artifact as shown by segregation patterns in families]; this artifact occurs only in heterozygotes [data not shown], indicating a mechanism involving an interaction between the two alleles). (B) Image of the gels obtained for minisatellites CEB205 and CEB310 on CEPH families 884 and 1331, respectively. Two children inherit mutant alleles for CEB205, and one child inherits a mutant allele for CEB310. For CEB205, larger alleles are missed in the procedure used: The results were confirmed by Southern blot.
Polymorphism Prediction: Sequence Characteristics and History Reconstruction
Training Set
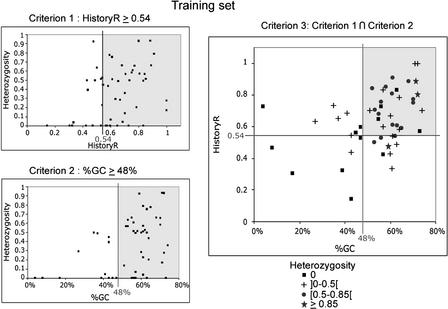
Twenty-five out of 60 and 32 out of 67 minisatellites were picked randomly, from chromosomes 21 and 22 respectively, to be typed first: They form the training set. PCR amplification was successful on 51 out of 57. A comparison of the sequence and polymorphism characteristics between the training set and the remaining minisatellites showed that the two sets have comparable distributions except for percentage of matches, purine/pyrimidine bias, and GC bias. To determine if some sequence characteristics are associated with high polymorphism, correlations between sequence characteristics and allele number or heterozygosity were calculated for the training set. The greatest correlations were obtained for HistoryR (a measure derived from the tandem repeats history reconstruction algorithm [Benson and Dong 1999]; see Methods section) and %GC (Fig. 3). Weaker correlations were also found for average entropy (strongly correlated with HistoryR), and unit length (data not shown). Based on these observations, we chose to test three predictive criteria: criterion 1, minisatellites with HistoryR ≥ 0.54; criterion 2, minisatellites with %GC ≥ 48%; and criterion 3, minisatellites with HistoryR ≥ 0.54 and %GC ≥ 48%.
Figure 3.
Criteria 1, 2, and 3 applied to the training set. For criteria 1 and 2, heterozygosity (28 individuals) versus HistoryR (criterion 1) or percentage of GC (%GC; criterion 2) are plotted. Correlations are significant at the 0.01 level. For criterion 3, HistoryR versus %GC is plotted, with different symbols representing the polymorphism. Lines represent the selected thresholds, and shaded areas contain the minisatellites selected by the criteria (criterion 1, HistoryR ≥0.54; criterion 2, %GC ≥48%; criterion 3, criteria 1 and 2 combined). Plots show that criteria select most of the polymorphic minisatellites and eliminate a majority of monomorphs or slightly polymorphic ones.
Test Set
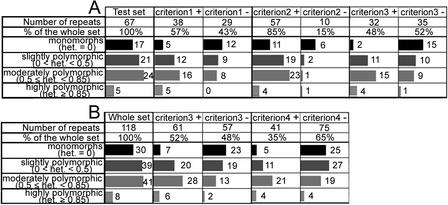
Of the remaining 70 minisatellites, 67 were successfully amplified and used as a test set in order to confirm the predictive criteria deduced from the training set. For each of the three criteria, the test set was partitioned into two groups: a positive group fitting the predictive criterion and a negative group. Figure 4A illustrates the results: All three criteria are predictive, that is, heterozygosity and allele number are significantly higher in the positive group compared with the negative group. The best polymorphism prediction was obtained with criterion 3 (HistoryR and %GC combined). It produces an enrichment of repeats having heterozygosity ≥0.5 from 43% (29 of 67) in the test set to 59% (19 of 32) in the positive group and a diminishment of monomorphic repeats from 25% (17 of 67) in the test set to 6% (two of 32) in the positive group. Criterion 3 thus reduces by half (67 to 32) the number of minisatellites to type while eliminating most monomorphic minisatellites and keeping most polymorphic ones (Fig. 4A). One among five highly polymorphic minisatellites (heterozygosity ≥0.85%) would have been missed using criterion 3.
Figure 4.
(A) Application of criteria 1 (HistoryR ≥0.54), 2 (%GC ≥48%), and 3 (HistoryR ≥0.54 and %GC ≥48%) to the test set. For each criterion, the distributions of minisatellites (from monomorphs to highly polymorphic) between positive (retained by the criterion) and negative (excluded by the criterion) sets are compared. All differences between sets + and − are statistically significant at the 0.01 level. (B) On the whole set, comparison of the results obtained with criterion 4 and criterion 3.
Polymorphism Prediction: Direct Sequence Comparison
The experimental polymorphism values measured here indicate that greatly enhanced efficiency of polymorphic loci identification is possible if the sequences of two independent alleles for each locus are available. The reasoning is that two random samples of a moderately or highly polymorphic locus will, with high probability, yield different alleles, whereas for a monomorphic or only slightly polymorphic locus, the alleles will likely be identical. Thus, selection based on observed allele difference in the two samples should enhance the proportion of loci obtained that are polymorphic. The applicability of this approach was directly tested by comparing sequences from the Human International Genome Sequencing Consortium (HGP) and Celera genomics. We establish selection criterion 4 to be different reported lengths in these two sequences. For the 127 minisatellites previously identified in the HGP sequence, repeat sizes in the sequence provided by Celera (Venter et al. 2001) were obtained by BLAST with the PCR primers. Three tandem repeats were not found in the Celera sequence, including two that were typed (CEB230, CEB256) and one long repeat (CEB215; length expected from HGP = 2834 bp) that could not be typed. Of the remainder, 51% (29 of 57) have a different length in the two sequences for chromosome 21 and 22% (15 of 67) for chromosome 22. From the measured heterozygosity values, we would expect 37% (43 of 116) to have different lengths between the two sequences, essentially the same as found. None of these should be monomorphic, and ∼75% (32 of 43) should have a heterozygosity value ≥0.5.
Heterozygosity and allele number are significantly higher in the positive group for criterion 4 (over the entire set of typed repeats) compared with the negative group (Fig. 4B). Criterion 4 produces an enrichment of repeats having heterozygosity ≥0.5 from 42% (49 of 118) in the whole set to 61% (25 of 41) in the positive group and a diminishment of monomorphic repeats from 25% (30 of 118) in the whole set to 12% (5 of 41) in the positive group. Criterion 4 thus reduces to nearly one third (116 to 41) the number of minisatellites to type while eliminating most monomorphic minisatellites and retaining 50% of the most polymorphic ones. By comparison, criterion 3, if applied to the entire set of typed repeats, (Fig. 4B) would reduce their number by roughly half (118 to 61), eliminating just two fewer monomorphs while retaining 69% (34 of 49) of the most polymorphic repeats. Additionally, criterion 4 eliminates half (four of eight) of the highly polymorphic (heterozygosity ≥0.85) minisatellites, whereas criterion 3 retains 75% (six of eight) of these.
We note that for some highly polymorphic minisatellites, (CEB202, CEB205, CEB310, CEB291), predicted lengths are identical in the two sequences. In addition, the results for criterion 4 are not uniform for the two chromosomes, owing to the much greater agreement on predicted loci length in chromosome 22. We presume that this reflects the fact that the Celera sequence was assembled by using both public and Celera sequence reads (Venter et al. 2001). More surprisingly, for five minisatellites, which we found to be monomorphic, predicted lengths differ (CEB214, CEB255, CEB264, CEB247, CEB289). These findings raise unresolved questions about the accuracy of the HGP and Celera sequences with regard to minisatellites. Tandem arrays can present significant sequence assembly problems, in particular when the internal array contains regions of high homology and, potentially more seriously, when the repeat exhibits length polymorphism and data are drawn from more than one individual, as was done for the Celera sequence (Venter et al. 2001).
To examine this further, we compared the HGP and Celera predictions to the alleles we detected (in Table 1, predicted lengths are underlined and not shaded when they correspond to an observed allele). In 65% (75 of 116) of the repeats, HGP and Celera predict an identical allele length, which corresponds to an observed allele length with five exceptions (Table 2) and is the most common allele in 81% of these cases. In 35% of the repeats (41 of 116), HGP and Celera predict different length alleles (Table 2). The length predicted by the HGP sequence fits with an observed allele size in 36 cases (most common allele length in 20 of these), whereas the Celera prediction fits with an observed size in 10 cases (and was once the most common allele).
Table 2.
Success of the Public Human Genome Project (HGP) and Celera Sequences in Predicting Alleles That Were Actually Found to Occur
| 116 tandem repeats (predicted by both sequences) | |||||
| Predicted lengths match: n = 75 | Predicted lengths differ: n = 41 | ||||
| Allele found with predicted length: | Allele found with predicted length: | ||||
| Yes | No | Yes, both predictions | HGP prediction only | Celera prediction only | Neither prediction |
| 70 | 5 | 10 | 26 | 0 | 5 |
Among the tandem repeats that provide PCR products unmatched by the HGP sequence, six sufficiently informative ones (CEB230, CEB253, CEB295, CEB298, CEB315, CEB269), with at least three different alleles among the four parental chromosomes, were typed in large CEPH families to check their chromosomal origin. All map to the expected area of chromosome 21 or 22, indicating that the discrepancy between sequence data and PCR product size probably results from a sequencing error (or the sequencing of a very rare allele) and not from a PCR specificity problem.
χ2 tests were used to examine whether the similarities in prediction of the HGP and Celera findings could be explained by chance (see Methods). Differences identified by the tests had, in all cases, less than one one-thousandth probability of occurring by chance. Specifically, cases in which predictions disagreed and both allele sizes were detected were underrepresented (compared to expected frequency) in all tests, and cases in which only one or neither predicted size was detected were overrepresented in all but one test.
Identifying Hypermutable Loci
Hypermutable minisatellites are expected to belong to the class of highly polymorphic loci because they are, by definition, subject to frequent rearrangements that generate new alleles. For practical reasons linked to the size of available pedigrees, a minisatellite will usually be classified as hypermutable if its average mutation rate in the germline is >0.5%, that is, if an average of at least one or two mutant alleles is observed among 100 children.
We typed the eight most polymorphic minisatellites (i.e., with heterozygosity ≥0.85) in the eight largest CEPH families (102 children) to search for mutant alleles. Comparing the results obtained by PCR and Southern blotting shows that even when some larger alleles are missing in the PCR products, the estimated heterozygosity rate (see Methods) is close to the heterozygosity rate obtained with Southern blots. This helps validate the simplified PCR-based polymorphism measurement. Among the eight minisatellites (CEB202, CEB205, CEB250, CEB310, CEB269, CEB291, CEB305, CEB324), two showed mutant alleles (CEB205 and CEB310; Fig. 2B). Both yielded two mutant alleles among 204 meioses, that is, 102 children (mutation rate, 0.12% to 3.5%; 95% confidence interval). For minisatellite CEB205, one mutation event occurred in the mother and the other in the father, whereas for CEB310, both mutations occurred in the father. The remaining six minisatellites yielded no mutant allele among 102 children (mutation rate, 0 to 1.79%; 95% confidence interval). They were not investigated further but can not be strictly excluded from being hypermutable. The two minisatellites that appeared hypermutable among 102 children were then typed in more families (32 other reference CEPH families). For CEB205, one new mutant allele was found among 352 meioses (mutation rate, 0.54%; 95% confidence interval, 0.11% to 1.57%), but no other mutant allele was detected for CEB310 among 476 additional meioses (mutation rate, 0.29%; 95% confidence interval, 0.04% to 1.06%). Based on these results, CEB205 appears to be hypermutable. It is a GC-rich minisatellite with a unit length of 33 bp repeated 10 to 70 times, located at 1.5 Mb from the end of the chromosome 22 sequence. It seems to be part of a predicted coding region (gene LOC129238; see http://www.ncbi.nlm.nih.gov/LocusLink/LocRpt.cgi?l=129238, 31 July 2002 update).
DISCUSSION
This study, performed on the scale of entire human chromosomes, provides a first global evaluation of minisatellite polymorphism based on genome sequence data. The repeats studied here, chosen by using a detailed definition that is more stringent than the broad definition mentioned in the Introduction, are, in majority (75%), polymorphic in the population investigated, and 42% have a heterozygosity value ≥0.5. Minisatellites from chromosomes 21 and 22 are similar in physical distribution (higher frequency toward chromosome ends), sequence features, and polymorphism. Assuming that chromosomes 21 and 22 are representative of all human chromosomes and given that the two chromosomes represent ∼2% of the genome, we speculate that the entire human genome contains ∼6,000 minisatellites that match our definition, including 4,800 polymorphic and 2,500 very polymorphic ones. A few 10s of these might be expected to qualify as hypermutable loci. Because our definition precluded many other potentially polymorphic minisatellites, future research should seek to expand the category of minisatellites that are tested against our polymorphism prediction criteria.
Predicting Polymorphism
We showed that using the sequence properties %GC and HistoryR effectively improves polymorphic minisatellite selection. With them, we reduce the number of minisatellites for typing by about half while increasing the frequency of repeats with heterozygosity ≥0.5% from the background rate of 43% to 59%. Internal conservation, used as a polymorphism predictor for microsatellites, is not applicable to minisatellites, presumably owing to the greater complexity of their mutation processes.
That %GC correlates with polymorphism is in agreement with earlier observations. Some of the first minisatellites to be characterized were detected via a shared 10- to 15-bp “core” sequence similar to the generalized recombination signal (χ) of Escherichia coli (GCTGTGG; Jeffreys et al. 1985). The majority of classical minisatellites (mostly polymorphic and/or hypermutable ones) are GC-rich, with a strong purine/pyrimidine strand asymmetry (Vergnaud and Denoeud 2000). In other genomes, though, (for instance bacterial genomes), %GC does not seem to be associated with minisatellites polymorphism (Le Fleche et al. 2001). Such a criterion may therefore not be universal, especially because GC content varies significantly across genomes.
The HistoryR criterion is based on the hypothesis that tandem repeats expand through multiple rounds of duplication, with the new copies sharing the mutations that occur before duplication, whereas unique mutations accumulate once the repeat is no longer evolving. For example (Fig. 1), minisatellite CEB252 shows several redundant patterns of mutation, resulting in a high HistoryR score, whereas CEB233 shows no clear organization of mutations, resulting in a low HistoryR score.
This polymorphism criterion is likely to be applicable to any genome, even though the history reconstruction algorithm makes simplifying assumptions about the possible biological mechanisms involved in array expansion. These mechanisms, which include mutational events during mitotic replication and meiotic recombination, comprising both intraallelic and interallelic events, might occur independently or jointly. At present, there are no rules to predict which mechanism will occur preferentially at which locus (Maleki et al. 2002). Moreover, the individual mechanisms themselves are still poorly understood and, thus, impossible to model. Meiotic events, for instance, have been shown to result from the activity of nearby meiosis-specific double-strand break hot-spots. The nature of these sites, better known in yeast, is still unknown in the human genome (Debrauwère et al. 1999; Tamaki et al. 1999; Vergnaud and Denoeud 2000). In view of the current state of knowledge, it may be premature to hope for a perfect polymorphism predictor based on apparent array expansion.
Use of Two Human Sequences to Select for Polymorphic Loci Is Problematic
The availability of two versions of the human genome sequence provides an additional avenue to improve polymorphic minisatellite identification. However, in the repeats studied here, selection based on reported length differences discarded half the highly polymorphic minisatellites and, in particular, the hypermutable one from chromosome 22. In both chromosomes, the number of loci with different predicted lengths in the HGP and Celera sequences that were nonetheless both found was significantly underrepresented. This is apparently owing to the lack of independence resulting from sharing of data during assembly of the Celera sequence. In addition, in both chromosomes, the number of loci in which only one or no predicted allele was found is overrepresented, apparently owing to assembly errors. Because the Celera sequence—which when not in agreement with the HGP data—usually provides copy numbers unobserved in any allele, it appears that the Celera sequence, at least with respect to minisatellites, is more prone to assembly error. As a result of the lack of independence/assembly errors in the Celera/HGP data, polymorphism prediction based on sequence comparison did not perform as well as anticipated.
One New Hypermutable Locus in a Coding Region
This study revealed one hypermutable minisatellite, CEB205, showing three mutant alleles among 278 children (mutation rate, 0.54%; 95% confidence interval, 0.11% to 1.57%). Interestingly, CEB205, with a 33-bp pattern, may be part of a coding region. The corresponding putative protein is 614 amino acids long, half of which are derived from the tandem repeat (11 codon repetition) at the N terminus. Of the minisatellites studied here, 26 among 60 (43%) on chromosome 21, and 22 among 67 (33%) on chromosome 22 belong to genes (i.e., exons, introns, or UTRs), as determined by sequence similarity analyses in the human genome sequence (using BLAST and http://www.ncbi.nlm.nih.gov/genome/seq/, release of November 2001). None except CEB205 appear to contribute to the coding sequence itself. Although the proportion of tandem repeats that contribute to coding regions is important in bacterial genomes, it is relatively low in the human genome, and CEB205 might represent the first known, coding hypermutable minisatellite.
CEB310—which exhibited meiotic mutation events, but which we do not here classify as hypermutable—is unusual in that its sequence is 80% AT. It is reminiscent of the tandem repeats studied in Giraudeau et al. (1999), that is, minisatellites made up of degenerated microsatellite-like repeated units (in this case, [AC]m[AT]n). Although most hypermutable minisatellites known to date are GC-rich, some have been described as having a very high AT content, for instance, the one constituting the chromosomal fragile site FRA16B (Yu et al. 1997; Yamauchi et al. 2000). The highly polymorphic minisatellite MSY1, from human chromosome Y, is also very AT-rich (75% to 80%; Jobling et al. 1998).
Future research will expand the systematic exploration of human tandem repeat polymorphism by testing the %GC, HistoryR, and HGP/Celera criteria on other human chromosomes as the sequences are progressively finished and released (Deloukas et al. 2001).
METHODS
Constructing the Tandem Repeats Database
Tandem repeats were identified from chromosome 21 (Hattori et al. 2000) and chromosome 22 (Dunham et al. 1999) sequences by using the TRF software (Benson 1999) with the following options: alignment parameters of (2,3,5), minimum alignment score to report repeat of 50, maximum period size of 500. When the program reported redundant (overlapping) repeats, the redundancy was eliminated in the following way. For each group of overlapping repeats, two values were determined: Lmax, the maximal total length among the redundant alignments, and Mmax, the maximal percent matches among the redundant alignments with total length ≥80% of Lmax. Then, of all the alignments in the group with total length ≥80% of Lmax and percentage of matches ≥Mmax − 0.1, the one with smallest unit length was stored in the database. The nominal length of the stored repeat is the total length of the overlapping region, that is, from the first position of the first overlapping repeat to the last position of the last overlapping repeat. Twenty-two tandem repeats showed differences of >5% between the nominal length and the length of the stored repeat, (the difference exceeded 10% in 14 cases, and 30% in three cases: CEB311, 33%; CEB320, 46%; and CEB327, 50%). For these latter three, TRF cut the repeats into two parts, which were combined for further analysis. Variation between nominal and stored size of repeats does not affect allele size prediction, which is based on length of sequence between primers. The database, publicly available at http://minisatellites.u-psud.fr, can be queried according to a number of simple features (e.g., total length, unit length, copy number, %GC) and provides links to repeat alignments and flanking sequence data as described previously (Le Fleche et al. 2001).
PCR Typing of Minisatellites
DNA was provided by Centre d'Etudes du Polymorphisme Humain (CEPH; http://www.cephb.fr/). PCRs were performed in 15 μL reactions, using 50 ng of genomic DNA, Roche long template PCR buffer (1.75 mM MgCl2, 50 mM Tris-HCl at pH 9.2 and 25°C, 16 mM [NH4]2[SO4]), 0.033 U/μL Taq polymerase (Roche), 0.003 U/μL Pwo (Roche), 200 μM of each dNTP (Amersham-Pharmacia biotech), and 0.6 μM of each flanking primer (Table 1; primers were selected within the flanking sequences provided by TRF using Primer3 software: http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). PCRs were cycled for 5 min at 96°C, then for 15 sec at 96°C: for 20 sec at annealing temperature (Table 1; this temperature was optimized for each primer pair by using the temperature gradient provided by MJResearch PTC200), for 5 min at 68°C for 30 cycles, and for 10 min at 68°C, on Perkin Elmer 9600 thermocycler or MJResearch PTC200. Samples were run through a 13-cm-long 1% standard agarose (Qbiogen) gel in 0.5× TBE buffer at 10 V/cm for 1.5 h and visualized by ethidium bromide staining using UV (1× TBE buffer is 89 mM Tris, 89 mM boric acid, 2 mM EDTA at pH 8).
Polymorphism Measures
A population of 96 CEPH individuals (from the 40 reference families) were typed for minisatellite polymorphism. This population includes 13 mother/father/child trios and altogether comprises 76 unrelated individuals. The 76 unrelated individuals form subpopulation 1. A subset of 28 unrelated individuals forms subpopulation 2. The exact list of the 96 individuals typed is provided in Supplementary Table 2.
In this study, we examined only length polymorphism, not internal sequence variation. Two values, calculated on unrelated individuals, were used to quantify polymorphism: the number of alleles observed and the heterozygosity, calculated as 1 − Σf2, where f are the allelic frequencies observed in the population of unrelated individuals. Heterozygosity represents the probability of having two different alleles. The simple PCR and ethidium bromide staining assay used here will usually detect only the smallest allele in individuals showing large length differences between alleles (as is often the case for highly polymorphic loci). The shorter allele often masks the longer one because it is easier to amplify. Such PCR artifacts are indicated with an asterisk in Table 1. They were detected because of the mother/father/child segregation controls and also because they do not satisfy the Hardy-Weinberg equilibrium, as tested with the HWE program, from the publicly available Linkage Utilities package (Ott 1999). For these loci, the heterozygosity value calculated from allelic frequencies was obtained by counting only one allele for individuals showing a single band (i.e., by assuming that the individual is heterozygous with one allele masked) instead of counting the same allele twice, as was done for loci in which homozygosity was not in question. The resulting heterozygosity value could be underestimated (if too many alleles are not seen), but it is sufficient to roughly evaluate the polymorphism.
Mutation Rate Estimation
Mutation rate of the most polymorphic (i.e., potentially hypermutable) minisatellites was evaluated by a combination of Southern blot hybridization and PCR typings, in recognition of the “masking” phenomenon described above. Typings were performed by using DNA from the eight largest CEPH families (F102, F884, F1331, F1332, F1347, F1362, F1413, F1416). Five μg of DNA were digested with AluI (CEB202, CEB250, CEB269, CEB291) or HinfI (CEB205, CEB324, CEB305; Boehringer Mannheim), electrophoresed through a 1% agarose gel and transferred to nylon membranes (Nytran+, Schleicher and Schuell) under vacuum (Pharmacia Biotech). Probes were obtained from PCR products and recovered from agarose using QIAquick gel extraction kit (Qiagen). Probes were labeled with a-[32P]dCTP (Amersham Pharmacia Biotech) by the random priming procedure (Feinberg and Vogelstein 1984). Hybridization was conducted as described in Vergnaud (1989) in an hybridization oven at 65°C. After hybridization, the filters were washed in 1× SSC/0.1% SDS or 0.1× SSC/0.1% SDS at 65°C. Membranes were revealed by using a phosphoimager (Storm 860 Molecular Dynamics).
Sequence Characteristics of Repeats
The following sequence characteristics (calculated from the HGP sequence) were tested for correlation with either allele number or heterozygosity. Characteristics did not differ markedly when evaluated in the Celera sequence (in which differences with HGP typically involved deletion of adjacent copies reported in the HGP sequence):
Unit length: the length of the repetitive unit (consensus pattern).
Copy number: the number of copies of the repetitive unit.
Total length: the length of the entire tandem array.
Percent matches: the frequency at which a nucleotide at a position in one unit matches the corresponding nucleotide in the next unit (reading from left to right).
%GC: the percentage of nucleotides that are either G or C.
GC bias: strand asymmetry for G and C, |%G − %C|/(%G + %C).
Purine/Pyrimidine bias: strand asymmetry for purines and pyrimidines, |%Pur − %Pyr|/(%Pur + %Pyr).
Average entropy: from the columns of a multiple alignment of the repeat copies, the average, over all columns, of the entropy calculated from nucleotide frequencies.
HistoryR: described below.
HistoryR is derived from the tandem repeats history reconstruction algorithm (Benson and Dong 1999), a greedy algorithm that chooses a series of least-cost contractions to convert a multicopy tandem array into a single putative ancestral copy. Greedy algorithms are not guaranteed to find the overall least-cost solution, but testing has shown this approach to work very well on simulated sequences. Input is a multiple alignment, M, of the individual copies in the repeat, with n rows (number of copies) and k columns (length of alignment). Mi,j represents the ith row and jth column of M, and each Mi,j contains one of the alphabet symbols (A,C,G,T,–). In a contraction, two or more consecutive, equal-length subsequences (the contraction copies) are replaced by a single subsequence (the merged copy) of the same length (all subsequences selected have length equal to a multiple of k). Each contraction reduces the number of rows in M. If the contraction copies are identical, then one becomes the merged copy. Otherwise at every position at which the contraction copies differ, the merged copy contains the character that occurs most often, with ties being represented by an ambiguous character, that is, a set of all the most frequently occurring characters at that position. An ambiguous character created in one contraction may be converted to a single character in a subsequent contraction. This method is analogous to that used by Sankoff (1975; Sankoff and Rousseau 1975). The cost of a contraction is a ratio. The numerator is the cost of obtaining the contraction copies from the merged copy; that is, at each position of the merged copy, subtract the number of times the most frequent character occurs in the contraction copies from the total number of contraction copies, then sum all these differences. The denominator is the combined length of rows by which M is reduced, that is, the length of all contraction copies minus the length of the merged copy.
History reconstruction yields four numerical values: (1) Max, the maximum possible history cost; (2) Min, the minimum possible history cost; (3) BinaryActual, the calculated history cost when the number of contraction copies in every contraction is restricted to exactly two; and (4) ManyActual, the calculated cost when the number of contraction copies is unrestricted. Max and Min are sums of column values from the original alignment M. In the case of Max, the value of a single column is the number of characters that are not the most frequent character. Max is therefore the cost if the most frequent character is ancestral and if every character different from the ancestral character was produced by its own mutation. For Min, the value is one less than the number of distinct characters in a column, that is, at most four. Min is the history cost if every distinct character different from the ancestral character arose by a single mutation (with identical characters produced by duplication).
Combinations of the four numerical values were tested for polymorphism prediction in the training set and HistoryR, which produced the highest correlation with heterozygosity, was used for the remainder of the study. It is defined as
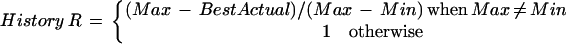 |
where BestActual is the minimum of BinaryActual and ManyActual. Usually, this was BinaryActual. The HistoryR value can be thought of as the proportion of mutations that could be accounted for by duplication that actually are. When Max ≠ Min, HistoryR ≤1, with a higher ratio indicating more mutations accounted for by duplications (Fig. 1). When Max = Min, each mutation is unique, and we arbitrarily set the ratio to one. This occurred in only one repeat with a total of four mutations. The history reconstruction program is freely available for interactive use at http://tandem.biomath.mssm.edu/cgi-bin/history/history.exe.
Statistical Analysis
All statistical analysis was done with the SPSS program except for χ2 tests which were done with StatXact 4. Correlations were determined by three methods: Pearson correlation, and nonparametric Kendall's τ–b and Spearman's ρ. Correlations are considered significant at the 0.01 level (two-tailed) of the test statistics. Group comparisons were determined by first conducting two tests of normality, Kolmogorov-Smirnov and Shapiro-Wilkinson, on the values within each group. Values were assumed to be normally distributed unless the test statistic fell within the 0.05 level of significance. If the values were normally distributed in the two groups, then a t test was used to compare the means, which were judged significantly different at the 0.01 level of the statistic (two-tailed). If the values were not normally distributed in either of the two groups, then a nonparametric Mann-Whitney test was used to compare the distributions, which were judged significantly different at the 0.01 level of the statistic (two-tailed).
χ2 tests were used to analyze HGP/Celera prediction data for chromosomes 21 and 22 separately. The data were divided into three categories: (1) identical predictions/allele size detected, (2) different predictions/both alleles sizes detected, and (3) one or neither predicted allele size detected. Two estimates for frequency of unobserved alleles were used (in order to calculate the probability of alleles being detected): 10% which corresponds to the largest frequency in the population for which the chance of not appearing in our sample of 28 individuals is ≥0.05, and an arbitrary low estimate of 1%. The probability of identical predictions in the HGP and Celera sequences was obtained by summing the estimated heterozygosity values calculated separately for each locus based on observed frequencies in our sample (equivalent to using the average observed heterozygosity over all loci).
WEB SITE REFERENCES
http://minisatellites.u-psud.fr; the tandem repeats database.
http://tandem.biomath.mssm.edu/cgi-bin/history/history.exe; history reconstruction program
http://www.ncbi.nlm.nih.gov/genome/seq/; Human Genome Sequencing at NCBI.
http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi; Primer3 primer picking software.
http://www.cephb.fr; Centre d'Etudes du Polymorphisme Humain.
http://www.ncbi.nlm.nih.gov/LocusLink/LocRpt.cgi?l=129238; locuslink at NCBI, predicted gene LOC129238.
http://www.ncbi.nlm.nih.gov/SNP/index.html; dbSNP home page.
Acknowledgments
We would like to thank Carol Bodian for extensive discussions and help with design of the χ2 analysis. G.B. is supported in part by NSF grants CCR-0073081 and DBI-0090789. F.D. and G.V. are supported by grants from Délégation Générale de l'Armement.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL France.Denoeud@igmors.u-psud.fr; FAX 33-1-69-15-66-78.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.574403. Article published online before print in April 2003.
REFERENCES
- 1.Amarger V., Gauguier, D., Yerle, M., Apiou, F., Pinton, P., Giraudeau, F., Monfouilloux, S., Lathrop, M., Dutrillaux, B., Buard, J., et al. 1998. Analysis of the human, pig, and rat genomes supports a universal telomeric origin of minisatellite sequences. Genomics 52: 62-71. [DOI] [PubMed] [Google Scholar]
- 2.Appelgren H., Cederberg, H., and Rannug, U. 1997. Mutations at the human minisatellite MS32 integrated in yeast occur with high frequency in meiosis and involve complex recombination events. Mol. Gen. Genet. 256: 7-17. [DOI] [PubMed] [Google Scholar]
- 3.___, 1999. Meiotic interallelic conversion at the human minisatellite MS32 in yeast triggers recombination in several chromatids. Gene 239: 29-38. [DOI] [PubMed] [Google Scholar]
- 4.Baudat F., Manova, K., Yuen, J.P., Jasin, M., and Keeney, S. 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6: 989-998. [DOI] [PubMed] [Google Scholar]
- 5.Bell G.I., Serby, M.J., and Rutter, W.J. 1982. The highly polymorphic region near the human insulin gene is composed of simple tandemly repeating sequences. Nature 295: 31-35. [DOI] [PubMed] [Google Scholar]
- 6.Benson G. 1999. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 27: 573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson, G. and Dong, L. 1999. Reconstructing the duplication history of a tandem repeat. Proc. Int. Conf. Intell. Syst. Mol. Biol. 44–53. [PubMed]
- 8.Bergerat A., de Massy, B., Gadelle, D., Varoutas, P.C., Nicolas, A., and Forterre, P. 1997. An atypical topoisomerase II from archaea with implication for meiotic recombination. Nature 386: 414-417. [DOI] [PubMed] [Google Scholar]
- 9.Buard J. and Vergnaud, G. 1994. Complex recombination events at the hypermutable minisatellite CEB1 (D2S90). EMBO J. 13: 3203-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buard J., Bourdet, A., Yardley, J., Dubrova, Y., and Jeffreys, A.J. 1998. Influences of array size and homogeneity on minisatellite mutation. EMBO J. 17: 3495-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Debrauwère H., Buard, J., Tessier, J., Aubert, D., Vergnaud, G., and Nicolas, A. 1999. Meiotic instability of human minisatellite CEB1 in yeast requires DNA double-strand breaks. Nat. Genet. 23: 367-371. [DOI] [PubMed] [Google Scholar]
- 12.Deloukas P., Matthews, L.H., Ashurst, J., Burton, J., Gilbert, J.G., Jones, M., Stavrides, G., Almeida, J.P., Babbage, A.K., Bagguley, C.L., et al. 2001. The DNA sequence and comparative analysis of human chromosome 20. Nature 414: 865-871. [DOI] [PubMed] [Google Scholar]
- 13.Dubrova Y.E. and Plumb, M.A. 2002. Ionising radiation and mutation induction at mouse minisatellite loci: The story of the two generations. Mutat. Res. 499: 143-150. [DOI] [PubMed] [Google Scholar]
- 14.Dubrova Y.E., Jeffreys, A.J., and Malashenko, A.M. 1993. Mouse minisatellite mutations induced by ionizing radiation. Nat. Genet. 5: 92-94. [DOI] [PubMed] [Google Scholar]
- 15.Dubrova Y.E., Nesterov, V.N., Krouchinsky, N.G., Ostapenko, V.A., Vergnaud, G., Giraudeau, F., Buard, J., and Jeffreys, A.J. 1997. Further evidence for elevated human minisatellite mutation rate in Belarus eight years after the Chernobyl accident. Mut. Res. 381: 267-278. [DOI] [PubMed] [Google Scholar]
- 16.Dunham I., Shimizu, N., Roe, B.A., Chissoe, S., Hunt, A.R., Collins, J.E., Bruskiewich, R., Beare, D.M., Clamp, M., Smink, L.J., et al. 1999. The DNA sequence of human chromosome 22. Nature 402: 489-495. [DOI] [PubMed] [Google Scholar]
- 17.Feinberg A.P. and Vogelstein, B. 1984. Addendum: a technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 137: 266-267. [DOI] [PubMed] [Google Scholar]
- 18.Fondon J.W., Mele, G.M., Brezinschek, R.I., Cummings, D., Pande, A., Wren, J., O'Brien, K.M., Kupfer, K.C., Wei, M.H., Lerman, M., et al. 1998. Computerized polymorphic marker identification: Experimental validation and a predicted human polymorphism catalog. Proc. Natl. Acad. Sci. 95: 7514-7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giraudeau F., Petit, E., Avet-Loiseau, H., Hauck, Y., Vergnaud, G., and Amarger, V. 1999. Finding new human minisatellite sequences in the vicinity of long CA-rich sequences. Genome Res. 9: 647-653. [PMC free article] [PubMed] [Google Scholar]
- 20.Giraudeau F., Taine, L., Biancalana, V., Delobel, B., Journel, H., Moncla, A., Bonneau, D., Lacombe, D., Moraine, C., Croquette, M.F., et al. 2001. Use of a set of highly polymorphic minisatellite probes for the identification of cryptic 1p36.3 deletions in a large collection of patients with idiopathic mental retardation: Three new cases. J. Med. Genet. 38: 121-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hattori M., Fujiyama, A., Taylor, T.D., Watanabe, H., Yada, T., Park, H.S., Toyoda, A., Ishii, K., Totoki, Y., Choi, D.K., et al. 2000. The DNA sequence of human chromosome 21: The chromosome 21 mapping and sequencing consortium. Nature 405: 311-319. [DOI] [PubMed] [Google Scholar]
- 22.Heale S.M. and Petes, T.D. 1995. The stabilization of repetitive tracts of DNA by variant repeats requires a functional mismatch repair system. Cell 83: 539-545. [DOI] [PubMed] [Google Scholar]
- 23.Jeffreys A.J., Wilson, V., and Thein, S.L. 1985. Hypervariable “minisatellite” regions in human DNA. Nature 314: 67-73. [DOI] [PubMed] [Google Scholar]
- 24.Jeffreys A.J., Tamaki, K., MacLeod, A., Monckton, D.G., Neil, D.L., and Armour, J.A.L. 1994. Complex gene conversion events in germline mutation at human minisatellites. Nat. Genet. 6: 136-145. [DOI] [PubMed] [Google Scholar]
- 25.Jobling M.A., Bouzekri, N., and Taylor, P.G. 1998. Hypervariable digital DNA codes for human paternal lineages: MVR-PCR at the Y-specific minisatellite, MSY1 (DYF155S1). Hum. Mol. Genet. 7: 643-653. [DOI] [PubMed] [Google Scholar]
- 26.Keeney S., Giroux, C.N., and Kleckner, N. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375-384. [DOI] [PubMed] [Google Scholar]
- 27.Le Fleche P., Hauck, Y., Onteniente, L., Prieur, A., Denoeud, F., Ramisse, V., Sylvestre, P., Benson, G., Ramisse, F., and Vergnaud, G. 2001. A tandem repeats database for bacterial genomes: application to the genotyping of Yersinia pestis and Bacillus anthracis. BMC Microbiol. 1: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maleki S., Cederberg, H., and Rannug, U. 2002. The human minisatellites MS1, MS32, MS205 and CEB1 integrated into the yeast genome exhibit different degrees of mitotic instability but are all stabilised by RAD27. Curr. Genet. 41: 333-341. [DOI] [PubMed] [Google Scholar]
- 29.May C.A., Jeffreys, A.J., and Armour, J.A.L. 1996. Mutation rate heterogeneity and the generation of allele diversity at the human minisatellite MS205 (D16S309). Hum. Mol. Genet. 5: 1823-1833. [DOI] [PubMed] [Google Scholar]
- 30.Murray J., Buard, J., Neil, D.L., Yeramian, E., Tamaki, K., Hollies, C.R., and Jeffreys, A.J. 1999. Comparative sequence analysis of human minisatellites showing meiotic repeat instability. Genome Res. 9: 130-136. [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura Y., Leppert, M., O'Connell, P., Wolff, T., Holm, T., Culver, M., Martin, C., Fujimoto, E., Hoff, M., Kumlin, E., et al. 1987. Variable number of tandem repeat (VNTR) markers for human gene mapping. Science 235: 1616-1622. [DOI] [PubMed] [Google Scholar]
- 32.NIH/CEPH collaborative mapping group 1992. A comprehensive genetic linkage map of the human genome. Science 258: 67-83. [PubMed] [Google Scholar]
- 33.Ott J., 1999. Analysis of human genetic linkage, 3d ed. Johns Hopkins University Press, Baltimore, MD.
- 34.Romanienko P.J. and Camerini-Otero, R.D. 2000. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6: 975-987. [DOI] [PubMed] [Google Scholar]
- 35.Sankoff D. 1975. Minimal mutation trees of sequences. J. Appl. Math. 28: 35-42. [Google Scholar]
- 36.Sankoff D. and Rousseau, P. 1975. Locating the vertices of a Steiner tree in an arbitrary metric space. Math. Programming 9: 240-246. [Google Scholar]
- 37.Strand M., Prolla, T.A., Liskay, R.M., and Petes, T.D. 1993. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 365: 274-276. [DOI] [PubMed] [Google Scholar]
- 38.Tamaki K., May, C.A., Dubrova, Y.E., and Jeffreys, A.J. 1999. Extremely complex repeat shuffling during germline mutation at human minisatellite B6.7. Hum. Mol. Genet. 8: 879-888. [DOI] [PubMed] [Google Scholar]
- 39.Venter J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- 40.Vergnaud G. 1989. Polymers of random short oligonucleotides detect polymorphic loci in the human genome. Nucleic Acids Res. 17: 7623-7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vergnaud G. and Denoeud, F. 2000. Minisatellites: Mutability and genome architecture. Genome Res. 10: 899-907. [DOI] [PubMed] [Google Scholar]
- 42.Vergnaud G., Mariat, D., Apiou, F., Aurias, A., Lathrop, M., and Lauthier, V. 1991. The use of synthetic tandem repeats to isolate new VNTR loci: Cloning of a human hypermutable sequence. Genomics 11: 135-144. [DOI] [PubMed] [Google Scholar]
- 43.Weber J.L. 1990. Informativeness of human (dC-dA)n (dG-dT)n polymorphisms. Genomics 7: 524-530. [DOI] [PubMed] [Google Scholar]
- 44.Wren J.D., Forgacs, E., Fondon, J.W., III, Pertsemlidis, A., Cheng, S.Y., Gallardo, T., Williams, R.S., Shohet, R.V., Minna, J.D., and Garner, H.R. 2000. Repeat polymorphisms within gene regions: phenotypic and evolutionary implications. Am. J. Hum. Genet. 67: 345-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamauchi M., Tsuji, S., Mita, K., Saito, T., and Morimyo, M. 2000. A novel minisatellite repeat expansion identified at FRA16B in a Japanese carrier. Genes Genet. Syst. 75: 149-154. [DOI] [PubMed] [Google Scholar]
- 46.Yu S., Mangelsdorf, M., Hewett, D., Hobson, L., Baker, E., Eyre, H.J., Lapsys, N., Le Paslier, D., Doggett, N.A., Sutherland, G.R., et al. 1997. Human chromosomal fragile site FRA16B is an amplified AT-rich minisatellite repeat. Cell 88: 367-374. [DOI] [PubMed] [Google Scholar]