Abstract
Drastic biodiversity declines have raised concerns about the deterioration of ecosystem functions and have motivated much recent research on the relationship between species diversity and ecosystem functioning. A functional trait framework has been proposed to improve the mechanistic understanding of this relationship, but this has rarely been tested for organisms other than plants. We analysed eight datasets, including five animal groups, to examine how well a trait-based approach, compared with a more traditional taxonomic approach, predicts seven ecosystem functions below- and above-ground. Trait-based indices consistently provided greater explanatory power than species richness or abundance. The frequency distributions of single or multiple traits in the community were the best predictors of ecosystem functioning. This implies that the ecosystem functions we investigated were underpinned by the combination of trait identities (i.e. single-trait indices) and trait complementarity (i.e. multi-trait indices) in the communities. Our study provides new insights into the general mechanisms that link biodiversity to ecosystem functioning in natural animal communities and suggests that the observed responses were due to the identity and dominance patterns of the trait composition rather than the number or abundance of species per se.
Keywords: functional traits, biodiversity, pollination, biocontrol, nutrient cycling, dung removal
1. Introduction
Unprecedented species extinctions during the past decades have raised concerns about the consequences of biodiversity loss for the functioning of ecosystems and associated ecosystem services that are fundamental for human well-being [1]. Ample evidence shows that species richness and diversity can enhance ecosystem functioning [2,3]. However, much variation in the relationship between biodiversity and functioning (BEF) remains to be explained. To improve predictions and mechanistic understanding of BEF, it has been increasingly accepted that instead of focusing on the taxonomic identity of organisms, the diversity of functional traits of species within a community should be studied [2–5]. However, the usefulness of such trait-based compared with species-based approaches, as well as the relative importance of single versus multiple traits for ecosystem functioning remained largely unexplored in organisms other than plants.
In early attempts to link species traits to ecosystem functioning, species were sorted into functional groups based on the similarity of their traits often according to experts’ opinion (e.g. [6]). Although this was a step forward and a useful exercise, the approach was criticized because functional groups failed to consider within-group variation in traits, and they rarely explained more variation in ecosystem functioning compared with randomly assembled groups of species [7]. Recently, quantitative measures have been developed that use multivariate techniques to integrate multiple traits into a single continuous trait diversity index. These measures capture value, range or distribution of functional traits in a community (hereafter ‘functional diversity’). They are promising tools that could increase our understanding of the mechanisms that drive ecosystem functioning [8–11]. However, most studies have used functional diversity merely as a proxy for ecosystem functioning, but without actually measuring the function and explicitly linking it to the functional diversity measure. For functional diversity measures to be useful for explaining ecosystem functioning, their predictive ability needs to be tested, and they should provide information beyond that given by measures based exclusively on species richness and abundances [5]. Here, we intend to fill this gap in BEF research by examining the relationship between trait- or species-based indices and a number of animal provided ecosystems functions measured below- and above-ground.
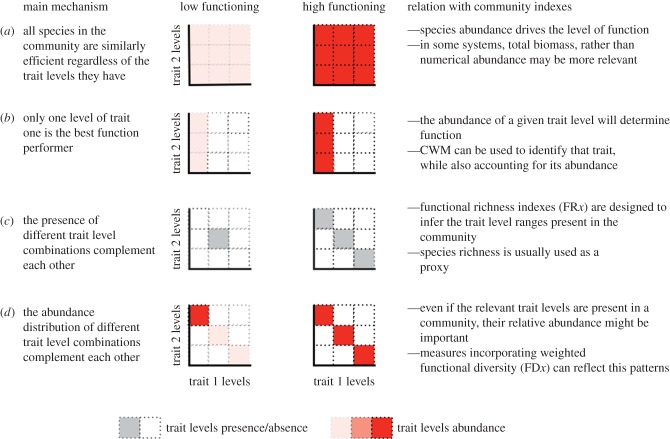
There is an ongoing debate about which of the many functional diversity measures should best predict ecosystem functioning, and which mechanisms these relationships reflect [5]. We summarize the main mechanisms of ecosystem functioning that different trait-based indices emphasize (figure 1). First, if differences among species are unimportant, the overall numerical or biomass abundance of organisms in a community might be better predictors than any of the measures that incorporate functional traits (figure 1a). Thus, overall abundance provides a null model in which all species in the community are equally efficient regardless of the trait levels they have. Note that trait-based indices consider both, which traits are assumed to be important, but also their trait values (continuous traits, e.g. different values of body size) or levels (discrete traits, e.g. diet ‘specialist’ or ‘generalist’). For simplicity, we refer only to trait levels throughout. Second, if a single trait level is strongly linked to an ecosystem function, abundance of this trait level may best predict the functioning (the functional identity hypothesis) ([12–14]; figure 1b). Alternatively, the complementarity of different traits in the community may be important for the functioning in the ecosystem (the functional complementarity hypothesis) [4,15]. In this case, indices that measure presence or absence of certain trait levels (i.e. functional richness, figure 1c), or those that consider abundance of different trait levels in the community (figure 1d) will explain most of the functioning. In the latter case, weighted functional diversity indices will best predict ecosystem functioning. It should be noted that only positive functional diversity–ecosystem functioning relationships indicate functional complementarity. Negative relationships reflect components of both functional identity and complementarity with only a few dominant trait levels being important. Hence, the functional identity and complementarity hypotheses are not mutually exclusive and several studies have found that a combination of the two explained most of the variation for several ecosystem functions [16–20]. Analysing which functional diversity indices can best explain a set of ecosystem functions may provide clues to the main drivers of these functions and increase our mechanistic understanding of the BEF relationship.
Figure 1.
Main mechanisms linking traits to ecosystem function. The x- and y-axes represent different trait levels (e.g. ‘large body size’, ‘medium body size’ and ‘small body size’). For simplicity, only two traits are presented. Darker colours indicate higher trait level abundance in the community. Different mechanisms predict that high functioning levels can be achieved by having (a) high abundance of any trait present in the community, (b) high abundance of the efficient trait level of the relevant trait, (c) the presence of complementary trait levels combinations or (d) an even distribution of complementary trait level combinations. Figures should be seen as simplified examples and other trait combinations are possible. See text for explanation for the calculation of indices.
Most tests of how well multivariate functional diversity is linked to ecosystem functioning (figure 1 c,d) have been conducted in small-scale, highly controlled plant communities. In addition, we have not been able to find any investigations of this relationship for terrestrial animals (see the literature summary in electronic supplementary material, table S1). Hence, we analysed eight datasets collected from the field along land-use gradients, and covering five terrestrial animal groups and seven ecosystem functions above- and below-ground: bees (pollination), carabid beetles (biocontrol of crop pests, biocontrol of weeds), earthworms (bioturbation), soil nematodes (nutrient cycling) and dung beetles (dung removal and seed burial). Increased understanding of the BEF relationship in these systems is important because both species and functional diversity are under great threat from land-use intensification [21,22]. Furthermore, sustainable development of human society in the face of rapidly increasing human populations will depend on the ways we manage these ecosystems and the services they provide. However, we do not attempt to describe direct effects of land-use on biodiversity or ecosystem functioning as this is done in numerous previous studies (e.g. [22–24]). Instead, we use the land-use gradients in order to assure we capture variability in different aspects of the community composition such that we can detect and assess its impacts on functioning.
We tested which of the four groups of indices in figure 1 best predicted ecosystem functioning in our datasets. More precisely, we explored (i) whether trait-based approaches offer greater explanatory power of ecosystem functioning than indices based only on species presence and abundance; (ii) whether single-trait measures calculated as community weighted trait means (CWM, reflecting the functional identity hypothesis) explain ecosystem functioning better or worse than multivariate functional diversity measures (reflecting the functional complementarity hypothesis) and (iii) whether the predictive power of multivariate functional diversity measures increases when the traits are weighted by numerical or biomass abundance of the species in the communities.
2. Material and methods
(a). Data description
We analysed eight field studies that included five animal groups (bees, carabid beetles, earthworms, soil nematodes and dung beetles) which deliver seven key ecosystem functions (pollination, biocontrol of crop pests, biocontrol of weeds, bioturbation, nutrient cycling, dung removal and seed burial). We focus on field studies because knowledge gained by them is an important complement to the numerous experimental studies in BEF research. Despite difficulties in demonstrating direct causal links [25], field studies better reflect the relative importance of mechanisms in real-world situations that are governed by processes acting at other scales than the commonly investigated small-scale BEF experiments. The data we used had not been analysed in this context previously. For each animal group, we collected species’ trait information from identification keys and from a number of published research papers and databases. We included traits that are often measured for a specific animal group and shown to be key traits in affecting the organisms' response to environmental change, and/or to have functional significance (see electronic supplementary material, S1 for the discussion about the trait choice and list of traits and references). Adult specimens were identified to species, except for pollinators and nematodes where similar species not identifiable in the laboratory were assigned to the same morphspecies. Analyses were done independently for each separate dataset and ecosystem function.
(i). Pollination
We analysed three separate datasets conducted in three crop systems (field beans, strawberries and spring oilseed rape) in UK, Germany and Sweden, respectively [26]. Bees were sampled in 10 fields in each crop type by hand-netting along a fixed transect. Fields were located along a gradient of landscape complexity measured as percentage arable land. Functioning was measured as total weight of fruits on five to 10 plants (depending on the crop) in four plots per field.
(ii). Biocontrol of pests
We analysed data from studies replicated in six European regions: Ireland, West Germany, East Germany, Poland and two provinces in Sweden: Uppsala and Scania [27]. In each country, eight cereal fields were located in contrasting landscapes with low versus high levels of agricultural intensification. Carabid beetles were collected with five pitfall traps per field. To measure function delivery by ground-dwelling predators, exclosure experiments were used to calculate the difference between aphid population growth in full exclosure (excluding ground-dwelling and flying predators and parasitoids using cages and barriers), and aphid population growth when ground-dwelling predators (mainly carabids) had access to aphids (excluding flying predators and parasitoids using cages).
(iii). Biocontrol of weeds
We used data from a study conducted in Germany in 22 winter wheat fields selected along a gradient in landscape complexity measured as percentage arable land (11 paired fields [28]). Carabids were sampled using four pitfall traps per field. Biocontrol of weeds was calculated for four common species: goosegrass (Galium aparine L.; seed consumption Ga), creeping thistle (Cirsium arvense L. Scop.; seed consumption Ca), rough-stalked meadow-grass (Poa trivialis L.; seed consumption Pt) and loose silky bentgrass (Apera spica-venti L; seed consumption As) separately. To measure percentage of seed loss due to ground-dwelling invertebrates, exclosure experiments were used to calculate the difference between percentage of remaining seeds from the initial seed number or seed weight in full exclosure (vertebrates and invertebrates excluded using cages with a small mesh size) and when only the vertebrates were excluded (using cages with a large mesh size) so that carabids had access to seeds.
(iv). Bioturbation
Earthworm communities were studied in cereal fields in the Swedish provinces of Uppland and Scania. In each province, earthworm communities were assessed in six sets of three farms that differed in farm management in close proximity to one another (see [29] for design of the study). Earthworm communities were estimated from four soil samples (30 × 30 × 30 cm) per field, taken at least 20 m from the field edges and with a 20 m distance between each sample. Earthworms were carefully hand sorted. Bioturbation was measured as above-ground cast production estimated by measuring in situ cast production over time on four observation squares at each field (dry matter soil per unit area and time). Bioturbation is an important ecosystem function as it affects soil formation, water supply and flood and erosion control through its influence on paedogenesis and infiltration and storage of water in soil [30]. Earthworms actively participate to the process of bioturbation as they may ingest large amounts of soil and litter, and hence become major regulators of the dynamics of litter and SOM in the ecosystem [31].
(v). Nutrient cycling
Soil surveys from 44 agricultural sites in The Netherlands were analysed [32]. In each field, 320 soil cores were randomly collected and mixed. Nematodes were extracted from 100 g sub-subsamples. One hundred and fifty randomly chosen individual nematodes were identified per site. As a measure of ecosystem function, we used total amount of phosphorous (P total) in soil as a proxy for nutrient cycling. Nematode abundance is strongly correlated to soil P and through their micro-bioturbation activity, high nematode abundances might contribute to high P retention [33].
(vi). Dung removal and seed burial
We used data collected from six forest sites in Sabah, Malaysian Borneo (two old-growth forest, two low-intensity selectively logged forest and two high-intensity logged forest) [34]. Dung beetles were sampled using 10 dung-baited pitfall traps per site. Dung removal was measured by placing a pile of cattle dung at each of the 10 points one month after the trapping and collecting the remaining dung after 24 h. Plastic beads of three sizes (small, medium and large) were used as seed mimics and placed in the dung to measure seed removal rates.
(b). Diversity indices
For each community, we calculated several biodiversity indices (table 1) divided into the four groups shown in figure 1. For indices that were weighted by numerical or biomass abundance, we used the subscripts ‘/n’ and ‘/b’, respectively. Biomass abundance of each species in a community was obtained by multiplying the number of individuals of each species by its average body mass. For bees and carabid beetles, average body masses were estimated from a measure of body size using allometric relationships (based on intertegular distance for bees [37]; total body length for carabids [38]). For earthworms, nematodes and dung beetles, we used body mass measurements; dry body mass measured directly or fresh weight converted to dry body mass. For earthworms and nematodes, body mass was estimated separately for field populations of adults and juveniles, and then weighted by their proportional numerical abundances.
Table 1.
Explanation of the indices used in the analyses. Groups of the indices (a–d) correspond to the groups in figure 1. Note that Ssh, Seve, Stot, as well as all indices in the groups (b,d) can be weighted by numerical or biomass abundances.
| name | index | refs | |
|---|---|---|---|
| (a) | Srich | species richness | |
| Seve | Pielous' J species evenness | ||
| Ssh | Shannon diversity | ||
| Stot | total abundances | ||
| (b) | CWM | community weighted means | [35] |
| (c) | FRdendr | functional richness | [8,9] |
| FRminvol | functional richness | [11] | |
| (d) | FDdendr.wc | weighted FRdendr | our adjusted index |
| FDdendr.ac | weighted FRdendr | our adjusted index | |
| FDdis | functional dispersion | [10] | |
| FDeve | functional evenness | [11] | |
| FDdiv | functional divergence | [11] | |
| FDRao | Rao's quadratic entropy | [36] |
First, we calculated species-based indices from species presence, and numerical or biomass abundance (Sx, where x is the diversity index used): species richness (Srich), Pielou's evenness based on species numerical or biomass abundance (Seve/n and Seve/b), Shannon diversity index based on numerical or biomass abundance (Ssh/n and Ssh/b) and total abundance or biomass of the community (Stot/n and Stot/b).
Second, we calculated single-trait-based indices, i.e. community weighted means for each trait in a community (figure 1b), weighted by their relative numerical (CWMx/n, [35]) or biomass abundances (our adjusted index, CWMx/b), where x is the name of the trait or a trait dominant level for categorical traits. If a trait was categorical, we used the frequency of the most abundant trait level in the community.
Multi-trait indices are often described by three independent groups of measures [39]—functional richness, functional evenness and functional diversity [11,40], which capture different aspects of the functional diversity [11]. Each group of measures can be calculated in several different ways, but there is no consensus on which index within each group performs best. To test our question about relative importance of weighted versus non-weighted FD indices, we calculated 14 commonly used multivariate functional diversity measures, which we divided into two groups. The first group considers only the presence or absence of trait levels (two functional richness indices FRx, figure 1c). The second group comprises 12 functional diversity indices weighted by numerical and biomass abundance (FDx/n and FDx/b, figure 1d), therefore including both functional divergence and functional evenness measures. All indices are based on a species per species trait–distance matrix. Given that all datasets contain traits coded as categorical variables, all distance matrices based on species traits were calculated using Gower distance with Podani's extension to ordinal variables [11,41,42].
For the two functional richness measures (FRx), we first calculated a measure based on dendrograms (FRdendr, [8]). The dendrogram was constructed using the UPGMA clustering algorithm, as it yielded a dendrogram with the highest cophenetic correlation with our original distance matrices and has also been identified to perform best in most cases [42]. The cophenetic correlation measures how faithfully a dendrogram preserves the original pairwise distances. Second, we estimated the minimum volume required to contain a set of points in trait space (FRminvol, [11]). A Cailliez correction was applied when the species-by-species distance matrix could not be represented in a Euclidean space [43]. However, the quality of the reduced space was not as high as the quality measured as cophenetic correlation for the dendogram-based approach (quality FRminvol = 0.51 ± 0.11, quality FRdendr = 0.8 ± 0.04).
Next, we calculated the 12 functional diversity measures weighted by numerical or biomass abundance (FDx). The first four indices (FDdendr.wc/n, FDdendr.wc/b, FDdendr.ac/n and FDdendr.ac/b) are weighted versions of FRdendr implemented specifically for this paper. In order to construct the weighted indexes, before summing the branches of a dendrogram, each branch is weighted by the relative numerical or biomass abundance of each species within the community (FDdendr.wc/n, FDdendr.wc/b). Hence, for each terminal branch, the weighting is done according to the abundance of the terminal species in this branch, but for each internal branch, the weighting is done by the average of the abundances of all the species descending form this internal branch. This index is highly correlated with the weighting procedure proposed in [44], but has the advantage that instead of building a different dendogram for each community, it builds a single dendrogram for all communities, which is the recommended approach [42]. The next two indices are constructed in the same way, but weighted by the mean relative proportion of numerical or biomass abundance of each species with respect to the species with highest numerical or biomass abundance across all communities (FDdendr.ac/n, FDdendr.ac/b). While the first index relates to the evenness of species in a community, the second one takes into account the relative numerical or biomass abundances in a community with respect to all the other analysed communities. The remaining eight indices are based on the convex hull space: functional divergence (FDdiv/n, FDdiv/b, [11]), functional dispersion (FDdis/n, FDdis/b, [10]) and Rao's quadratic entropy (FDRao/n, FDRao/b [36]). Functional dispersion and Rao's quadratic entropy are highly correlated, but we included both to enable comparison with other studies that have used these indices. Finally, we calculated two measures of functional evenness (FDeve/n, FDeve/b; [11]).
(c). Statistical analysis
For each dataset, we ranked the indices according to their relative performance in explaining functioning. For that, we focus only on the explanatory power (measured as R2) of different indices. First, we used linear mixed-effect models and calculated their marginal R2 [45]. For each ecosystem function (response variable), we built one single-variable model for each of the diversity indices (explanatory variable). For datasets that included observations that were collected at multiple times within a region or a field, we included these (Field or Region) as random factors. The residuals from all models were plotted and visually inspected. When necessary, data were transformed by log10(x + 1) or arcsine square root to meet model assumptions of normality. To meet the assumptions of homoscedasticity, we used a constant variance function when necessary. We only provide p-values in the appendix for completeness, and we do not interpret them as indicators of statistical significance due to the risk of type I errors from multiple testing on the same data. Indices were ranked according to the R2 value obtained and a relative rank bounded between 0 and 1 was calculated for each dataset, with 0 being the best ranked index.
To compare the relative performance among groups of indices, we used linear mixed-effects model to regress the arcsine square-root transformed relative rank of the indices within each of the 14 datasets (response variable) against its category (factor with four levels: species-based indices (Sx), functional richness (FRx), functional diversity (FDx), community-weighted means (CWMx), and weighting method (factor with two levels: biomass or numerical abundance). Given that FDRao and FDdis are mathematically correlated, we excluded FDdis from this comparison. We used ‘dataset’ in the random structure to control for multiple calculations of the indices belonging to the same group in each dataset. We used general linear hypothesis testing (‘glht’ function) with two-tailed test and Hochberg correction for multiple testing [46] for post hoc comparisons among groups of indices. Note that studies are conducted at different scales (within versus across regions) with a consequence of having more confidence in the results for highly replicated designs (i.e. biocontrol of pests and nutrient cycling). However, we do not correct for this as each dataset contributes with only one set of values to the linear model.
(d). Influence of traits on functional diversity–ecosystem functioning relationship
All included traits were chosen a priori based on the authors’ ecological knowledge. To test whether our choice of traits had a large influence on the observed effect of functional diversity measures on ecosystem functioning, we used a jackknife approach for the functional diversity predictors (FRx or FDx) that explained most variance.
We built models with all traits included, and we then removed one trait at a time from the full model. We calculated the difference in explanatory power (ΔR2) between the full model and the model without a given trait. Negative ΔR2's reflect traits that are important in explaining the relationship between diversity and function, whereas positive values indicate traits that, when excluded, improved the model. All calculations of diversity indices and statistical analyses where performed in R (v. 2.15.1, [47]) using packages ‘nlme’ [48], ‘MuMIn’ [49], ‘FD’ [10,50], ‘multcomp’ [51] and our own R script. The R function to calculate all indices used in our analysis is available at https://github.com/ibartomeus/fundiv.
All relevant data including all indices calculated for each dataset can be found in electronic supplementary material, dataset S1.
3. Results
(a). Performance of functional diversity in predicting ecosystem functioning
We compared explanatory power between groups of biodiversity indices: species-based versus trait-based, single trait versus multiple traits, community weighted versus non-weighted indices and indices weighted by numerical versus biomass abundance. We found large differences in the average performance between the index groups (F3,478 = 11.16, p < 0.0001). Post hoc comparison among relative ranks of indices groups revealed that weighted trait-based indices, both community weighted means (CWMx) and functional diversity (FDx), performed consistently better than species-based indices (Sx) across all datasets studies (difference in means: Sx − CWMx = 0.23 ± 0.04, p < 0.0001; Sx − FDx = 0.13 ± 0.05, p = 0.02; figure 2; electronic supplementary material, table S2). However, non-weighted functional richness (FRx) did not perform better than species-based indices (Sx − FRx = 0.10 ± 0.07, p = 0.36), while single-trait measures (CWMx) were on average better ranked than functional diversity measures (CWMx − FDx = −0.10 ± 0.04, p = 0.02). Multi-trait functional diversity measures weighted by numerical abundance (FD/n) performed equally good as measures weighted by biomass abundance (FD/b; F1,478 = 0.078, p = 0.93). Note that the lower the relative rank, the better the performance of the index.
Figure 2.
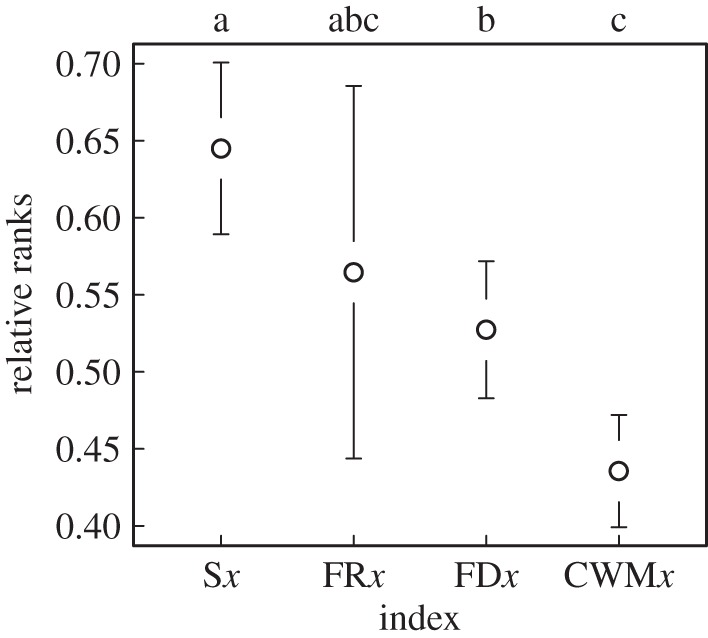
Performance of different groups of diversity indices across ecosystem functions and groups of organisms investigated. The mean and standard error of the relative ranking of species-based indices (Sx, n = 94), functional richness (FRx, n = 28), functional diversity (FDx, n = 168) and community weighted means (CWMx, n = 194). Different letters indicate post hoc significant differences after correcting for multiple comparisons. Lower rank values indicate better explanatory power. See the text and table 1 for description of the ecosystem functions and codes for biodiversity indices, and electronic supplementary material, table S2 for the results for all predictors.
Interestingly, species richness and abundance did not only obtain low rankings, but their explanatory power was on average less than half that of FD indices (electronic supplementary material, table S2). Shannon diversity and species evenness tended to explain most functions better than species richness and abundance. Within the weighted multi-trait functional diversity measures, FDeve and FDdiv were the best performers. In fact, in nine of 14 cases, they ranked as the overall best predictors. Notably, the direction of the effects of biodiversity indices on ecosystem functioning was positive in the majority of cases, the exception being a few FD indices (electronic supplementary material, table S2).
(b). Influence of traits on functional diversity–ecosystem functioning relationship
Jackknife analysis showed that our results are relatively robust with respect to the choice of traits included (see electronic supplementary material, figure S1). Changes in R2 after excluding any trait were small and mainly negative. The few exceptions were ‘dung manipulation strategy’ for large seed burial by dung beetles, ‘light preference’ for consumption of A. spica-venti seeds by carabid beetles, and ‘body length’ and ‘trophic level’ for nutrient cycling by nematodes. Traits with high negative values are highly influential because they increase the explanatory power. By contrast, we only found one trait, ‘hibernation’, which induced large positive R2-changes in the consumption of A. spica-venti seeds and G. aparine seeds by carabid beetles, indicating that this trait reduces the model performance (electronic supplementary material, figure S1).
4. Discussion
Indices solely based on the numbers and abundances of species were consistently poor at predicting ecosystem functioning across the seven ecosystem functions investigated here. Moreover, they performed worse than indices using a trait-based approach, both in previous studies of plants (electronic supplementary material, table S1) and in our current analysis of animals. As in many plant studies, single-trait indices (CWMx) were often ranked as the best predictors of ecosystem functioning in our analyses on animals. Hence, functioning is in the majority of cases maximized by a single trait. However, we also found that multi-trait functional diversity measures (e.g. FDeve, FDdiv) can best predict functions provided by some animal groups. Thus, it appears that the distribution of functionally dissimilar traits is also relevant for several functions.
Despite the diversity of ecosystems and of organisms and ecosystem functions provided by animals investigated here, and by plants in previous studies, some general conclusions can be made. First, species numerical and biomass abundance appear to be poor sole predictors of the functions investigated, although they are often positively correlated with ecosystem functions (figure 1a, e.g. [52]). Second, non-weighted indices that have commonly been used as proxies of functional diversity were also poor predictors of ecosystem functioning. These include species richness, but also newly developed multi-trait indices of functional richness (FRx) that have been useful for analysing community assembly [40]. This suggests that the number of species in a community, or the trait ranges they encompass, are insufficient to fully explain ecosystem functioning.
Current knowledge of the role of species richness for ecosystem functioning is mainly based on small-scale experiments [3]. There is increasing evidence that results from such studies do not always agree with findings from more realistic and species-rich assemblages where skewed species abundance distributions have been suggested to play a key role [53,54]. Our findings indicate that we need to integrate the abundance and distribution not only of species, but also of their trait levels within the community to better understand BEF relationships in terrestrial animal communities (figure 1b,d). On one hand, we show that weighted functional diversity indices (especially functional evenness and divergence) in many cases were the best predictors of ecosystem functioning provided by animals, and this relationship was most often positive. This means that communities with a more even distribution of species across the trait space, will deliver higher levels of ecosystem functioning; a result that supports the functional complementarity hypothesis. On the other hand, we also found negative relationships between functional evenness and functioning in some cases, as well as single traits being consistently good predictors of functioning. This exemplifies that a dominant trait level of a single or just a few traits are needed to maximize functioning in some communities.
The functions studied here were performed by different taxa with different traits, and hence the mechanisms driving high functioning levels vary among functions. Given the exploratory nature of our analyses, we restrain from discussing specific traits and mechanisms for different organisms, but rather propose that our findings provide a starting point for future research in these communities. On a more general level, there are some interesting questions emerging from our study that future BEF research should focus on. First, why does functional identity often appear as the best mechanism and under which scenarios does it interplay with functional complementarity? For example, a reason for the better support of the functional identity rather than functional complementarity hypothesis for some functions may be that ecosystem functions, such as predation of just one pest species, provide a narrow niche with less opportunity for niche partitioning than the predation of different species. Second, how can increasing the spatial and temporal scales, or the number of functions performed by the same animal group, increase the importance of functional diversity? For example, it appears that even when the same animal group (e.g. bees) is performing a given function (e.g. pollination), the key traits explaining functioning for a particular crop are specific for each plant. Hence, for pollination to be maximized at the landscape level and simultaneously for several crops, the functional diversity of the pollinator community would have to be increased. In this case, functional diversity will be more important than single-trait values as it provides insurance across varying conditions across space and time. However, the situation may be different when there are trade-offs between functions provided by the same community [15].
The choices we make in BEF research, such as which traits and indices to use, can strongly affect the observed relationship between functional diversity and ecosystem functioning [5]. First, the trait selection is extremely important for characterizing trait-based indices, especially for single-trait measures, such as CWM. Preferably, we should use a priori knowledge based on experimental manipulations investigating which traits are likely to drive different functions, but this information is rarely available for animals. However, we found that most multi-trait functional diversity indices were weakly affected by trait choice (see also [55]), and while excluding traits worsens explanatory power in some cases, it rarely increased it. We propose that the jackknife approach can be used to exclude or weight traits that contribute little to predicting functioning. Second, we show that the choice of weighted versus non-weighted indices is important. Weighted indices always explained ecosystem functions better, demonstrating the importance of considering the abundance distribution of traits in communities. Weighting by biomass should be superior to weighting by numerical abundance in cases where the process is size-based, often by being related to metabolic rate of individuals (i.e. individual's performance increase with body size). However, we found no clear preference for indices scaled by biomass versus numerical abundances in the communities we investigated.
Several new avenues have been proposed to better quantify functional diversity and increase the predictive power of biodiversity–functioning relationships: taking into account single and multi-trait indices simultaneously, phylogenetic diversity [56], within-species trait variability [57], abiotic factors [58] and nonlinearities in the response [3]. We show that the power to predict ecosystem functions using trait distributions in natural communities is relatively low (less than 50%). This is not surprising given that most ecosystem functions, such as crop pollination and thereby yield production, depend on multiple abiotic and biotic processes including several organism groups [59,60]. Direct links between organisms and functions, such as between aphid predation and predators, are stronger than indirect links, such as between P retention and nematodes. However, we show that for predicting ecosystem functioning, trait-based measures are substantially better than measures of species richness and abundances, commonly used by researchers and policymakers. Our study thus provides new insights into general mechanisms that link biodiversity to ecosystem functioning in natural animal communities and suggests that the observed responses were due to the identity and dominance patterns of the trait composition rather than to the number or abundance of species per se. Hence, using a trait-based approach in BEF research is a promising step forward and may greatly increase our understanding and aid management of multiple ecosystem functions.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Piotr Ceryngier, Michael Kuhlmann and Stuart Roberts for providing data. We also thank two anonymous reviewers for their insightful comments on the manuscript. I.B. and V.G. designed the research; A.T., C.W., C.F., E.S., S.P., I.S-D., M.E., T.T., W.W. and R.B. collected and provided data; V.G. and I.B. analysed the data; V.G. and I.B. wrote the first draft of the manuscript, and all authors contributed substantially to revisions.
Data accessibility
Data available from the Dryad Digital Repository: http://doi.org/10.5061/dryad.177g0.
Funding statement
Funding was provided by the Swedish Research Council FORMAS to the project ‘SAPES—Multifunctional agriculture: harnessing biodiversity for sustaining agricultural production and ecosystem services'. S.P. and I.S-D. were supported by EC FP7 grant no. 244090, STEP Project (Status and Trends of European Pollinators, www.step-project.net), and T.T. by the DFG and BMBF. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Millennium Ecosystem Assessment. 2005. Ecosystems and human well-being: synthesis. Washington, DC: Island Press. [Google Scholar]
- 2.Hooper DU, et al. 2005. Effects of biodiversity on ecosystem functioning: a consensus of current knowledge. Ecol. Monogr. 75, 3–35. ( 10.1890/04-0922) [DOI] [Google Scholar]
- 3.Cardinale BJ, et al. 2012. Biodiversity loss and its impact on humanity. Nature 486, 59–67. ( 10.1038/nature11148) [DOI] [PubMed] [Google Scholar]
- 4.Díaz S, Cabido M. 2001. Vive la différence: plant functional diversity matters to ecosystem processes. Trends Ecol. Evol. 16, 646–655. ( 10.1016/S0169-5347(01)02283-2) [DOI] [Google Scholar]
- 5.Cadotte MW, Carscadden K, Mirotchnick N. 2011. Beyond species: functional diversity and the maintenance of ecological processes and services. J. Appl. Ecol. 48, 1079–1087. ( 10.1111/j.1365-2664.2011.02048.x) [DOI] [Google Scholar]
- 6.Tilman D, et al. 1997. The Influence of functional diversity and composition on ecosystem processes. Science 277, 1300–1302. ( 10.1126/science.277.5330.1300) [DOI] [Google Scholar]
- 7.Wright JP, Naeem S, Hector A, Lehman C, Reich PB, Schmid B, Tilman D. 2006. Conventional functional classification schemes underestimate the relationship with ecosystem functioning. Ecol. Lett. 9, 111–120. ( 10.1111/j.1461-0248.2005.00850.x) [DOI] [PubMed] [Google Scholar]
- 8.Petchey OL, Gaston KJ. 2002. Functional diversity (FD), species richness and community composition. Ecol. Lett. 5, 402–411. ( 10.1046/j.1461-0248.2002.00339.x) [DOI] [Google Scholar]
- 9.Petchey OL, Gaston KJ. 2006. Functional diversity: back to basics and looking forward. Ecol. Lett. 9, 741–758. ( 10.1111/j.1461-0248.2006.00924.x) [DOI] [PubMed] [Google Scholar]
- 10.Laliberté E, Legendre P. 2010. A distance-based framework for measuring functional diversity from multiple traits. Ecology 91, 299–305. ( 10.1890/08-2244.1) [DOI] [PubMed] [Google Scholar]
- 11.Villéger S, Mason NWH, Mouillot D. 2008. New multidimensional functional diversity indices for a multifaceted framework in functional ecology. Ecology 89, 2290–2301. ( 10.1890/07-1206.1) [DOI] [PubMed] [Google Scholar]
- 12.Grime JP. 1998. Benefits of plant diversity to ecosystems: immediate, filter and founder effects. J. Ecol. 86, 901–910. ( 10.1046/j.1365-2745.1998.00306.x) [DOI] [Google Scholar]
- 13.Garnier E, et al. 2004. Plant functional markers capture ecosystem properties during secondary succession. Ecology 85, 2630–2637. ( 10.1890/03-0799) [DOI] [Google Scholar]
- 14.Vile D, Shipley B, Garnier E. 2006. Ecosystem productivity can be predicted from potential relative growth rate and species abundance. Ecol. Lett. 9, 1061–1067. ( 10.1111/j.1461-0248.2006.00958.x) [DOI] [PubMed] [Google Scholar]
- 15.Hillebrand H, Matthiesen B. 2009. Biodiversity in a complex world: consolidation and progress in functional biodiversity research. Ecol. Lett. 12, 1405–1419. ( 10.1111/j.1461-0248.2009.01388.x) [DOI] [PubMed] [Google Scholar]
- 16.Schumacher J, Roscher C. 2009. Differential effects of functional traits on aboveground biomass in semi-natural grasslands. Oikos 118, 1659–1668. ( 10.1111/j.1600-0706.2009.17711.x) [DOI] [Google Scholar]
- 17.Mouillot D, Villéger S, Scherer-Lorenzen M, Mason NWH. 2011. Functional structure of biological communities predicts ecosystem multifunctionality. PLoS ONE 6, e17476 ( 10.1371/journal.pone.0017476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butterfield BJ, Suding KN. 2013. Single-trait functional indices outperform multi-trait indices in linking environmental gradients and ecosystem services in a complex landscape. J. Ecol. 101, 9–17. ( 10.1111/1365-2745.12013) [DOI] [Google Scholar]
- 19.Conti G, Díaz S. 2013. Plant functional diversity and carbon storage—an empirical test in semiarid forest ecosystems. J. Ecol. 101, 18–28. ( 10.1111/1365-2745.12012) [DOI] [Google Scholar]
- 20.Lavorel S. 2013. Plant functional effects on ecosystem services. J. Ecol. 101, 4–8. ( 10.1111/1365-2745.12031) [DOI] [Google Scholar]
- 21.Sala OE, et al. 2000. Global biodiversity scenarios for the year 2100. Science 287, 1770–1774. ( 10.1126/science.287.5459.1770) [DOI] [PubMed] [Google Scholar]
- 22.Flynn DFB, et al. 2009. Loss of functional diversity under land use intensification across multiple taxa. Ecol. Lett. 12, 22–33. ( 10.1111/j.1461-0248.2008.01255.x) [DOI] [PubMed] [Google Scholar]
- 23.Rader R, Bartomeus I, Tylianakis JM, Laliberté E. 2014. The winners and losers of land use intensification: pollinator community disassembly is non-random and alters functional diversity. Divers. Distrib. 20, 908–917. ( 10.1111/ddi.12221) [DOI] [Google Scholar]
- 24.Vandewalle M, et al. 2010. Functional traits as indicators of biodiversity response to land use changes across ecosystems and organisms. Biodivers. Conserv. 19, 2921–2947. ( 10.1007/s10531-010-9798-9) [DOI] [Google Scholar]
- 25.Scherer-Lorenzen M. 2005. Biodiversity and ecosystem functioning: basic principles. In Biodiversity: structure and function. Encyclopedia of life support systems (eds Barthlott W, Linsenmair KE, Porembski S.). Oxford, UK: EOLSS. [Google Scholar]
- 26.Bartomeus I, et al. 2014. Contribution of insect pollinators to crop yield and quality varies with agricultural intensification. PeerJ 2, e328 ( 10.7717/peerj.328) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thies C, et al. 2011. The relationship between agricultural intensification and biological control: experimental tests across Europe. Ecol. Appl. 21, 2187–2196. ( 10.1890/10-0929.1) [DOI] [PubMed] [Google Scholar]
- 28.Fischer C, Thies C, Tscharntke T. 2011. Mixed effects of landscape complexity and farming practice on weed seed removal. Perspect. Plant Ecol. Evol. Syst. 13, 297–303. ( 10.1016/j.ppees.2011.08.001) [DOI] [Google Scholar]
- 29.Jonason D, Andersson GKS, Öckinger E, Rundlöf M, Smith HG, Bengtsson J. 2011. Assessing the effect of the time since transition to organic farming on plants and butterflies. J. Appl. Ecol. 48, 543–550. ( 10.1111/j.1365-2664.2011.01989.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavelle P, et al. 2006. Soil invertebrates and ecosystem services. Eur. J. Soil. Biol. 42, 3–15. ( 10.1016/j.ejsobi.2006.10.002) [DOI] [Google Scholar]
- 31.Lavelle P. 1997. Faunal activities and soil processes: adaptive strategies that determine ecosystem function. Adv. Ecol. Res. 27, 93–132. ( 10.1016/S0065-2504(08)60007-0) [DOI] [Google Scholar]
- 32.Mulder C, Vonk JA. 2011. Nematode traits and environmental constraints in 200 soil systems: scaling within the 60–6,000 mm body size range. Ecology 92, 2004 ( 10.1890/11-0546.1) [DOI] [Google Scholar]
- 33.Vonk JA, Breure AM, Mulder C. 2013. Environmentally-driven dissimilarity of trait-based indices of nematodes under different agricultural management and soil types. Agric. Ecosyst. Environ. 179, 133–138. ( 10.1016/j.agee.2013.08.007) [DOI] [Google Scholar]
- 34.Slade EM, Mann DJ, Lewis OT. 2011. Biodiversity and ecosystem function of tropical forest dung beetles under contrasting logging regimes. Biol. Conserv. 144, 166–174. ( 10.1016/j.biocon.2010.08.011) [DOI] [Google Scholar]
- 35.Lavorel S, et al. 2008. Assessing functional diversity in the field—methodology matters! Funct. Ecol. 22, 134–147. [Google Scholar]
- 36.Botta-Dukát Z. 2005. Rao's quadratic entropy as a measure of functional diversity based on multiple traits. J. Vegetation Sci. 16, 533–540. ( 10.1111/j.1654-1103.2005.tb02393.x) [DOI] [Google Scholar]
- 37.Cane JH. 1987. Estimation of bee size using intertegular span (Apoidea). J. Kans. Entomol. Soc. 60, 145–147. [Google Scholar]
- 38.Jelaska LŠ, Dumbović V, Kučinić M. 2011. Carabid beetle diversity and mean individual biomass in beech forests of various ages. Zookeys 405, 393–405. ( 10.3897/zookeys.100.1536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mason NWH, Mouillot D, Lee WG, Wilson JB. 2005. Functional richness, functional evenness and functional divergence: the primary components of functional diversity. Oikos 111, 112–118. ( 10.1111/j.0030-1299.2005.13886.x) [DOI] [Google Scholar]
- 40.Mouchet MA, Villeger S, Mason NWH, Mouillot D. 2010. Functional diversity measures: an overview of their redundancy and their ability to discriminate community assembly rules. Funct. Ecol. 24, 867–876. ( 10.1111/j.1365-2435.2010.01695.x) [DOI] [Google Scholar]
- 41.Podani J. 1999. Extending Gower's general coefficient of similarity to ordinal characters. Taxon 48, 331–340. ( 10.2307/1224438) [DOI] [Google Scholar]
- 42.Podani J, Schmera D. 2006. On dendrogram-based measures of functional diversity. Oikos 115, 179–185. ( 10.1111/j.2006.0030-1299.15048.x) [DOI] [Google Scholar]
- 43.Cailliez F. 1983. The analytical solution of the additive constant problem. Psychometrika 48, 305–310. ( 10.1007/BF02294026) [DOI] [Google Scholar]
- 44.Clark CM, Flynn DFB, Butterfield BJ, Reich PB. 2012. Testing the link between functional diversity and ecosystem functioning in a Minnesota grassland experiment. PLoS ONE 7, e52821 ( 10.1371/journal.pone.0052821) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakagawa S, Schielzeth H. 2013. A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol. Evol. 4, 133–142. ( 10.1111/j.2041-210x.2012.00261.x) [DOI] [Google Scholar]
- 46.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300. [Google Scholar]
- 47.R Development Core Team. 2012. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 48.Pinheiro J, Bates D, DebRoy S, Sarkar D, the R Development Core Team. 2012. nlme: linear and nonlinear mixed effects models. R package v. 3.1–104 See http://CRAN.R-project.org/package=nlme. [Google Scholar]
- 49.Barton K. 2013. MuMIn: multi-model inference. R package v. 1.9.13 See http://CRAN.R-project.org/package=MuMIn.
- 50.Laliberté E, Shipley B. 2011. FD: measuring functional diversity from multiple traits, and other tools for functional ecology. R package v. 1.0–11 See http://CRAN.R-project.org/package=FD. [DOI] [PubMed]
- 51.Hothorn T, Bretz F, Westfall P. 2008. Simultaneous Inference in general parametric models. Biom. J. 50, 346–363. ( 10.1002/bimj.200810425) [DOI] [PubMed] [Google Scholar]
- 52.Garibaldi LA, et al. 2013. Wild pollinators enhance fruit set of crops regardless of honey bee abundance. Science 339, 1608–1611. ( 10.1126/science.1230200) [DOI] [PubMed] [Google Scholar]
- 53.Smith MD, Knapp AK. 2003. Dominant species maintain ecosystem function with non-random species loss. Ecol. Lett. 6, 509–517. ( 10.1046/j.1461-0248.2003.00454.x) [DOI] [Google Scholar]
- 54.Hillebrand H, Benett DM, Cadotte MW. 2008. Consequences of dominance: a review of evenness effects on local and regional ecosystem processes. Ecology 89, 1510–1520. ( 10.1890/07-1053.1) [DOI] [PubMed] [Google Scholar]
- 55.Sasaki T, Katabuchi M, Kamiyama C, Shimazaki M, Nakashizuka T, Hikosaka K. 2014. Vulnerability of moorland plant communities to environmental change: consequences of realistic species loss on functional diversity. J. Appl. Ecol. 51, 299–308. ( 10.1111/1365-2664.12192) [DOI] [Google Scholar]
- 56.Flynn DFB, Mirotchnick N, Jain M, Palmer MI, Naeem S. 2011. Functional and phylogenetic diversity as predictors of biodiversity—ecosystem–function relationships. Ecology 92, 1573–1581. ( 10.1890/10-1245.1) [DOI] [PubMed] [Google Scholar]
- 57.Albert CH, de Bello F, Boulangeat I, Pelle G, Lavorel S, Thuiller W. 2012. On the importance of intraspecific variability for the quantification of functional diversity. Oikos 121, 116–126. ( 10.1111/j.1600-0706.2011.19672.x) [DOI] [Google Scholar]
- 58.Díaz S, Lavorel S, de Bello F, Quétier F, Grigulis K, Robson TM. 2007. Incorporating plant functional diversity effects in ecosystem service assessments. Proc. Natl Acad. Sci. USA 104, 20 684–20 689. ( 10.1073/pnas.0704716104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Bello F, et al. 2010. Towards an assessment of multiple ecosystem processes and services via functional traits. Biodivers. Conserv. 19, 2873–2893. ( 10.1007/s10531-010-9850-9) [DOI] [Google Scholar]
- 60.Lundin O, Smith HG, Rundlöf M, Bommarco R. 2013. When ecosystem services interact: crop pollination benefits depend on the level of pest control. Proc. R. Soc. B 280, 20122243 ( 10.1098/rspb.2012.2243) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available from the Dryad Digital Repository: http://doi.org/10.5061/dryad.177g0.