Abstract
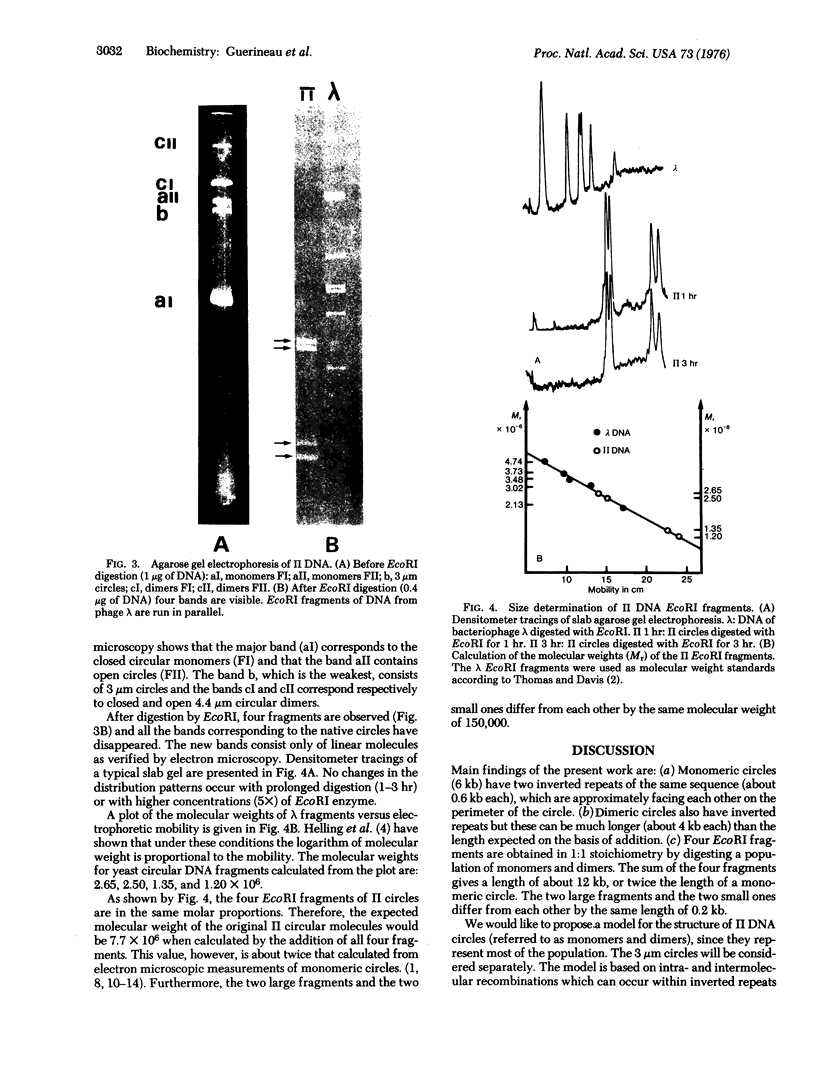
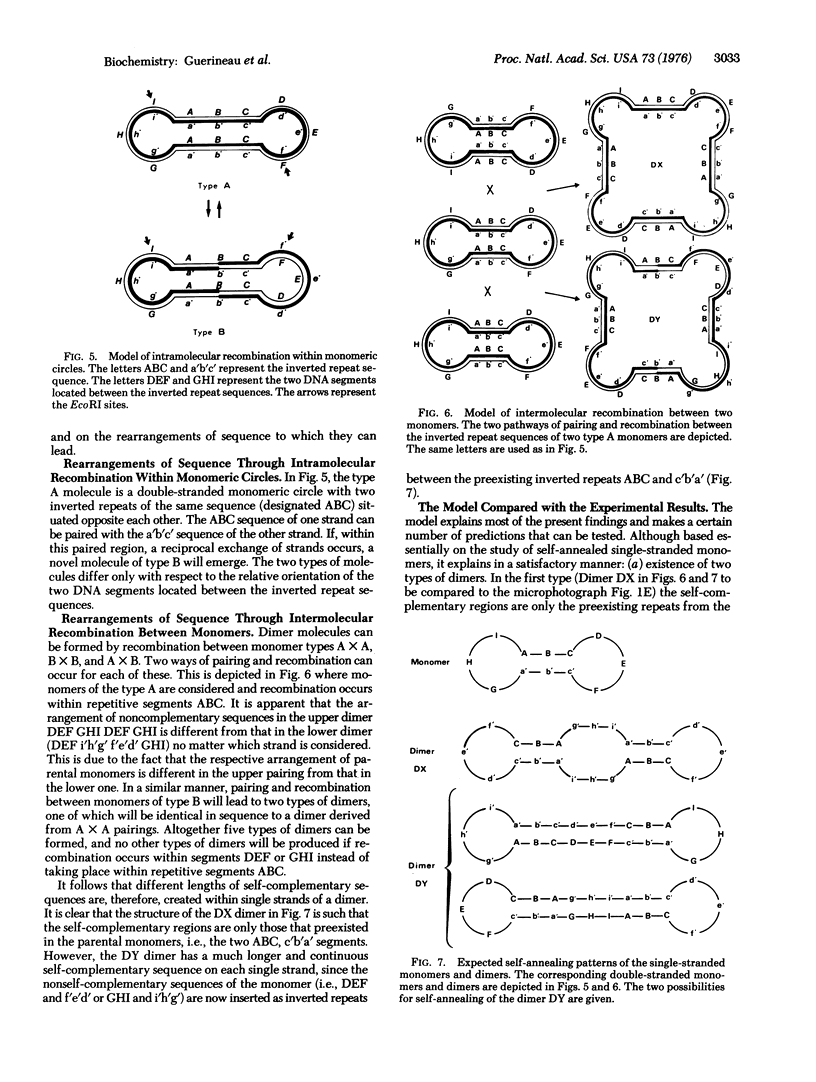
Small circular DNA molecules from genetically characterized clones of Saccharomyces cerevisiae have been studied by restriction endonuclease analysis and electron microscopy. The circular monomers (6000 bases) are shown to contain two inverted repeats of the same sequence (600 bases) situated opposite each other along the perimeter. Four endonuclease EcoRI fragments are obtained in 1:1:1:1 stoichiometry, and their sum gives a length of about 12,000 bases. The two large fragments and the two small ones differ from each other by 200 bases. We propose a model for the structure of the monomer molecule. Two classes of monomers can be generated by intramolecular recombinations within inverted repeats; they differ by the relative orientation of nonrepeated segments. The structure of dimers as predicted by the model is verified by self-renaturation of single-stranded circles. Inverted repeats in circular molecules may be related to the insertion release faculty of II episome in the chromosomes.
Full text
PDF
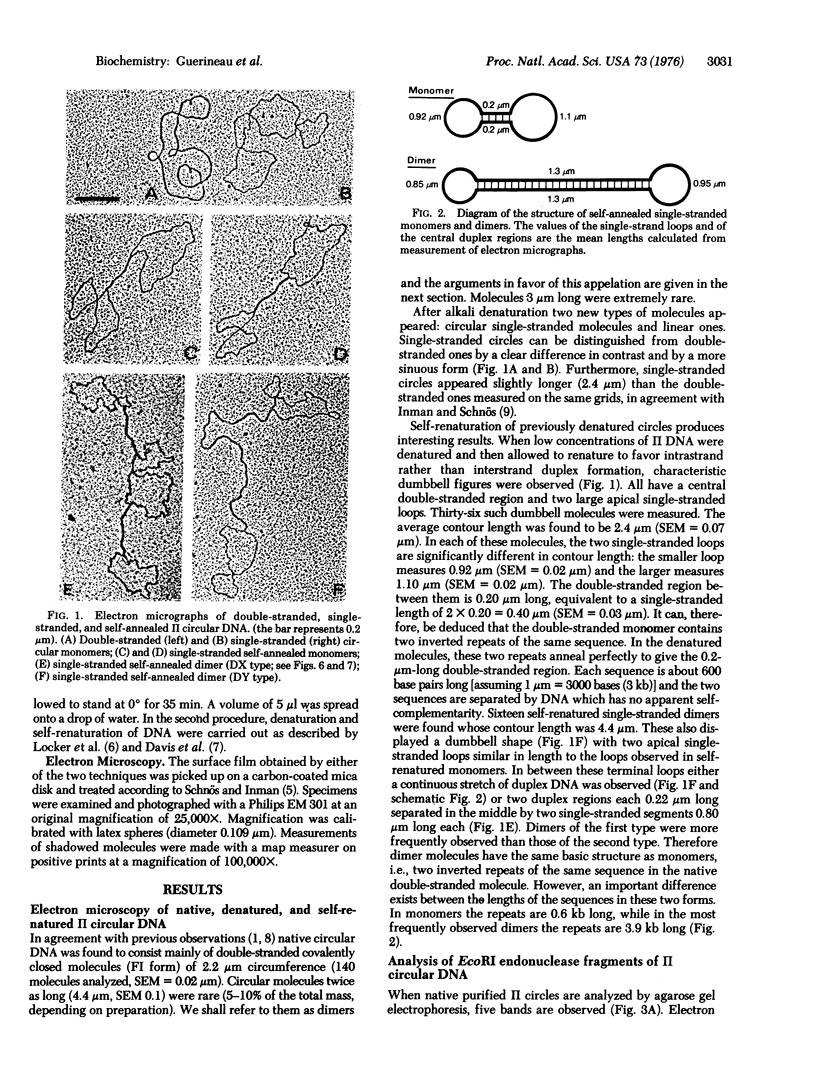

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bak A. L., Christiansen C., Christiansen G. Circular, repetitive DNA in yeast. Biochim Biophys Acta. 1972 May 29;269(3):527–530. doi: 10.1016/0005-2787(72)90144-x. [DOI] [PubMed] [Google Scholar]
- Clark-Walker G. D. Size distribution of circular DNA from petite-mutant yeast lacking rho DNA. Eur J Biochem. 1973 Jan 15;32(2):263–267. doi: 10.1111/j.1432-1033.1973.tb02606.x. [DOI] [PubMed] [Google Scholar]
- Guerineau M., Grandchamp C., Paoletti C., Slonimski P. Characterization of a new class of circular DNA molecules in yeast. Biochem Biophys Res Commun. 1971 Feb 5;42(3):550–557. doi: 10.1016/0006-291x(71)90406-2. [DOI] [PubMed] [Google Scholar]
- Guerineau M., Slonimski P. P., Avner P. R. Yeast episome: oligomycin resistance associated with a small covalently closed non-mitochondrial circular DNA. Biochem Biophys Res Commun. 1974 Nov 27;61(2):462–469. doi: 10.1016/0006-291x(74)90979-6. [DOI] [PubMed] [Google Scholar]
- Heffron F., Rubens C., Falkow S. Translocation of a plasmid DNA sequence which mediates ampicillin resistance: molecular nature and specificity of insertion. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3623–3627. doi: 10.1073/pnas.72.9.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling R. B., Goodman H. M., Boyer H. W. Analysis of endonuclease R-EcoRI fragments of DNA from lambdoid bacteriophages and other viruses by agarose-gel electrophoresis. J Virol. 1974 Nov;14(5):1235–1244. doi: 10.1128/jvi.14.5.1235-1244.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg C. P., Borst P., van Bruggen E. F. Mitochondrial DNA. V. A 25 micron closed circular duplex DNA molecule in wild-type yeast mitochondria. Stucture and genetic complexity. Biochim Biophys Acta. 1970 May 21;209(1):1–15. [PubMed] [Google Scholar]
- Hsu M. T., Davidson N. Electron microscope heteroduplex study of the heterogeneity of Mu phage and prophage DNA. Virology. 1974 Mar;58(1):229–239. doi: 10.1016/0042-6822(74)90157-3. [DOI] [PubMed] [Google Scholar]
- Inman R. B., Schnös M. Partial denaturation of thymine- and 5-bromouracil-containing lambda DNA in alkali. J Mol Biol. 1970 Apr 14;49(1):93–98. doi: 10.1016/0022-2836(70)90378-5. [DOI] [PubMed] [Google Scholar]
- Kopecko D. J., Cohen S. N. Site specific recA--independent recombination between bacterial plasmids: involvement of palindromes at the recombinational loci. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1373–1377. doi: 10.1073/pnas.72.4.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker J., Rabinowitz M., Getz G. S. Tandem inverted repeats in mitochondrial DNA of petite mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1366–1370. doi: 10.1073/pnas.71.4.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saedler H., Reif H. J., Hu S., Davidson N. IS2, a genetic element for turn-off and turn-on of gene activity in E. coli. Mol Gen Genet. 1974;132(4):265–289. doi: 10.1007/BF00268569. [DOI] [PubMed] [Google Scholar]
- Schnös M., Inman R. B. Position of branch points in replicating lambda DNA. J Mol Biol. 1970 Jul 14;51(1):61–73. doi: 10.1016/0022-2836(70)90270-6. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Cohen S. N., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. II. Structure of drug resistance (R) factors and F factors. J Mol Biol. 1973 Apr 5;75(2):235–255. doi: 10.1016/0022-2836(73)90018-1. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Sinclair J. H., Stevens B. J., Sanghavi P., Rabinowitz M. Mitochondrial-satellite and circular DNA filaments in yeast. Science. 1967 Jun 2;156(3779):1234–1237. doi: 10.1126/science.156.3779.1234. [DOI] [PubMed] [Google Scholar]
- Stevens B. J., Moustacchi E. ADN satellite gamma et molécules circulaires torsadées de petite taille chez la levure Saccharomyces cerevisiae. Exp Cell Res. 1971 Feb;64(2):259–266. doi: 10.1016/0014-4827(71)90075-9. [DOI] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]