Abstract
CD38 encodes a ligand in the oxytocin signaling pathway. Some single nucleotide polymorphisms in this gene have been associated with low serum oxytocin levels in autism spectrum disorder (ASD) patients. Oxytocin disruption has been hypothesized to account for features of ASD, including impaired communication and social behavior, based on animal studies. Recent human studies have shown administration of oxytocin improving emotion recognition, promoting social behavior, and improving auditory processing of social stimuli in ASD patients. In addition to its role in oxytocin signaling, CD38 is involved in the regulation of calcium concentration in airway smooth muscle with impairment of CD38 being implicated in airway diseases like asthma. While a number of studies have implicated rare chromosomal deletions and duplications in helping determine genetic risk for autism, there are to our knowledge no reports describing rearrangements involving CD38 or deletions in patients with ASD. Here, we present two sisters diagnosed with autism and with features of regression—previously acquired speech lost in the second year of life. The younger sister, who also had asthma, inherited a maternal deletion of 4p15.32 that results in a BST1-CD38 fusion transcript. Their mother's deletion was mosaic and she was not affected. Although further work is required to assess functional consequences of the fusion transcript, we hypothesize that the proband's deletion may have served as a risk factor for autism that, when combined with other susceptibility variants, resulted in a more severe presentation than her sister.
Keywords: autism, CD38, oxytocin, CNV, fusion transcript
Introduction
Autism spectrum disorders (ASD) are characterized by deficits in several domains: impairment in social interaction, communication, and the presence of restricted, repetitive, and stereotyped patterns of behavior [American Psychiatric Association, 2000]. Estimates of autism heritability range from very high [Bailey et al., 1995] to moderate [Hallmayer et al., 2011]. The estimated prevalence of ASD is 1 in 88 according to a recent Centers for Disease Control and Prevention report (“Prevalence of autism spectrum disorders—Autism and Developmental Disabilities Monitoring Network, 14 sites, United States, 2008”, 2012); autism has a 4:1 male to female ratio [Chakrabarti & Fombonne, 2005].
Copy number variants (CNV) appear to play a signifi-cant role in determining genetic susceptibility to autism. For instance, when focusing on sporadic cases, there appears to be a higher rate of de novo CNV when compared with controls [Sebat et al., 2007]. There is also a higher genic burden of rare CNV in cases compared with controls [Pinto et al., 2010]. In most cases, it is thought that CNV mediate their effects by influencing gene expression. However, a previous study showed that ~6% of rare CNV may have potential to result in fusion transcripts, which could lead to novel proteins with gain-of-function effects [Holt et al., 2012].
Modahl and colleagues found that children with autism had lower plasma oxytocin levels compared with a control group [Modahl et al., 1998]. Studies of oxytocin and vasopressin in animal models indicate that the disruption of these neuropeptide pathways may result in some of the core symptoms seen in ASD, such as impaired social attachments, stereotyped behaviors, and impaired communication [Insel, O'Brien, & Leckman, 1999].
CD38, a transmembrane protein involved in oxytocin secretion (OMIM #107270), has been found to be critical for maternal nurturing and social behavior in mice [Jin et al., 2007]. Cd38 knockout mice were found to grow normally and to be viable; however, they had lower plasma oxytocin concentrations when compared with wild-type mice; additionally, they had impaired social memory/recognition and impaired maternal nurturing behavior, both of which were rescued by exogenous oxytocin administration [Jin et al., 2007]. In these mice, Cd38 was found to play an important role in the process of oxytocin release in the hypothalamus; Cd38 knockout mice could produce and package oxytocin into vesicles, but they could not release oxytocin into the blood [Higashida et al., 2011]. Munesue et al. [2010] found the single nucleotide polymorphism (SNP) rs3796863 of CD38 to be associated with high-functioning autism in some populations. There is also data suggesting that the oxytocin pathway, and in particular CD38 function, may be important because of lower CD38 expression in lymphoblastoid cells from ASD subjects [Lerer et al., 2010]. Recently, a pharmacological challenge study with oxytocin reported that CD38 variants of rs3796863 were related to the activation of the fusiform brain regions in healthy males, and authors suggest this may be connected to hypoactivation of amygdala and fusiform brain regions observed in other imaging studies of ASD patients [Sauer, Montag, Worner, Kirsch, & Reuter, 2012].
In the current study, we describe a rare deletion of 4p15.32 that involves CD38—which we believe is the first report of a disruption of this gene in a patient with autism. We provide evidence that this deletion was inherited from the mother in whom the deletion was mosaic, suggesting that it arose during her early development. Fine mapping indicated that the deletion removed the first exon of CD38 and the last two exons of the gene BST1 (bone marrow stromal cell antigen 1), and this leads to the creation of an in-frame fusion transcript. Although the microdeletion is present in a mosaic form in the clinically normal mother and absent from an affected sibling, we hypothesize that it may represent a risk factor that, when combined with other susceptibility variants, shared or unshared with the affected sibling, results in a more severe presentation of clinical symptoms in the proband.
Clinical Description
The family is part of the International Molecular Genetics Study of Autism Consortium (IMGSAC) multiplex family collection that was initially described in 1998 [IMGSAC, 1998]. The proband (3388_4) is the younger of two sisters. Both sisters were clinically diagnosed with autism, as well as meeting autism classification on both the Autism Diagnostic Observation Schedule [Lord et al., 2000] and the Autism Diagnostic Interview-Revised [Lord, Rutter, & Le Couteur, 1994]. However, as detailed below, the proband has continued to need one-to-one assistance in social and unfamiliar settings, and has lower language ability and IQ than her sister, who in contrast has been able to function well in the mainstream educational system. The family is of African American ancestry and so had been excluded from our previous scan for potential fusion-gene CNV [Holt et al., 2012].
The proband required speech/language and developmental therapy. During her pregnancy with the proband, the mother had spotting/bleeding and was seen by a physician regularly. There were no complications during the delivery. At the age of 9 months, the proband began using words. Around 15–18 months, her parents realized her development was not typical—she stopped babbling and using words to communicate, and was less responsive to her parents. The family's concern with the proband's development prompted them to see a professional when she was 20 months old. She received speech therapy at 23 months for expressive and receptive delays, and was diagnosed with autism and global developmental delay at 27 months. She did not begin speaking again until the age of 3 years. By age 3 and a half years, she used occasional phrases.
At age 3 years and 5 months, she was enrolled in the IMGSAC project at the University of Chicago according to a protocol approved by the University of Chicago Institutional Review Board (IRB), and subsequently the University of Illinois at Chicago IRB. Adaptive skills were evaluated at age 3 years 7 months via the Vineland Adaptive Behavior Scales, Second Edition (VABS-II) [Sparrow, Cicchetti, & Balla, 2005], and her adaptive behavior composite standard score was 65. Mullen Scales of Early Learning [Mullen, 1995] was administered at age 3 years 8 months and demonstrated nonverbal ratio IQ (NVRIQ) = 59 and a verbal ratio IQ (VRIQ) = 30. At age 4 years and 10 months, she was readministered the VABS-II and her standard score was 66, revealing a mild deficit. When the Mullen Scales of Early Learning was readministered, her NVRIQ = 55 and her VRIQ = 31.
The proband attended a special education program and had limited communication skills. This was consistent with her minimal spontaneous verbalizations, reduced range of affect, and lack of imaginative play; she used about 25 words when she was 4 years 10 months old. Her mother described her language as severely impaired. She was described as a loving, kind, tenacious child. Her parents were concerned that they may never know how much she understands and will always have to watch her because she does not know when something might hurt her.
At age 11, she needed help finding the bathroom and required guidance throughout the day. She sporadically and unpredictably used about 100 words. She still showed limited cognitive, communicative, and adaptive skills; these difficulties became more pronounced with age. Her parents again expressed concern that the proband was not aware of danger. Temper tantrums were occurring approximately once a week up until the age of 10. She took guanfacine, lisdexamfetamine, and risperidone to manage several behavioral symptoms, and her temper tantrums became less frequent, although spontaneous, loud utterances are still an occasional concern. Overeating became a problem around the age of 8, and she was overweight at age 11.
Aside from her autism diagnosis, the proband was diagnosed with asthma at age 18 months. Her parents reported that her breathing had been inconsistent from age 16 months, and she was put on oxygen briefly around 18 months of age. Symptoms thereafter were successfully treated with a nebulizer and inhalers with levalbuterol (bronchodilator) and budesonide (corticosteroid). The proband has had no asthma episodes since the age of 5, and the use of inhalers had subsequently stopped. Her sister (3388_3) was never diagnosed with asthma, although both of them experience seasonal allergies with congestion and runny noses, successfully managed with the antihistamine loratadine. The father of the girls also reports having asthma, the mother does not.
The proband's sister (3388_3) was 20 months older. She was born prematurely at 33 weeks of gestation weighing 3 lbs and 13 oz, and was found to have congenital anal stenosis and poor sucking that required IV fluids. She was jaundiced during her first month and was placed in an incubator. At the age of 1, she had surgery to repair the anal stenosis. 3388_3 was later to start talking than her sister and began to use single words at age 15 months. She also showed signs of regression, and at 18 months began to lose the use of words, not speaking again until 24 months. She began to use phrases at 30 months. 3388_3 was enrolled as the “affected sibling” in the IMGSAC study. At age 5 years 7 months, her adaptive behavior composite standard score on the VABS-II was 60. Her mother described her language impairment as mild. Her nonverbal deviation IQ was 110 on the Raven's Colored Progressive Matrices [Raven, 1947, 2000]. On the Peabody Picture Vocabulary Test Form IIIA [Dunn, 1997], her standard score was 92. Her Clinical Evaluation of Language Fundamentals-Preschool [Wiig, Secord, & Semel, 1992] total language standard score was 66.
The sister has attended mainstream public school at an age-appropriate grade level with only slight modifications to the standard curriculum. She continued to do well, with a grade average of A−. The use of pragmatic language was age-appropriate. She assists her sister (the proband) with daily activities like finding the bathroom. This older sister's cognitive and adaptive skills followed a more typical developmental trajectory after the delays of early childhood. This is consistent with her more creative play, and early ability to describe events and people who were not present. Although described to have a cautious temperament, the parents said she learned quickly and they are not concerned about her ability to determine if a situation is dangerous. She has been described as helpful and cooperative, particularly with her younger sister, the proband. This older sister took guanfacine to manage her attention, but otherwise her behavior is reported to be within normal limits.
Methods
Genotyping and CNV Analysis
Genotyping was performed for three family members (mother, father, proband) but not individual 3388_3. Genotyping was carried out using the Infinium 1 M single sample SNP microarray (Illumina, San Diego, CA) that contains around one million genome-wide SNP as well as some copy number-specific probes, according to the manufacturer's guidelines. For the parents, DNA had been extracted from lymphoblastoid cell lines (LCLs) that had previously been generated from peripheral blood leukocytes. In contrast, for the proband, DNA had been extracted directly from blood. Extractions were performed using the Nucleon BACC2 kit (Tepnel Life Sciences, Manchester, UK). In the parent–proband trio, the parent–parent–child Mendelian error rate was 0.0005. Three different algorithms were used to call CNV from the SNP data—QuantiSNP, iPattern, and PennCNV—as previously described [Pinto et al., 2010].
Quantitative Polymerase Chain Reaction (qPCR)
To confirm the presence of a deletion and test the sibling, qPCR was performed using SYBR-green (Applied Biosystems, Foster City, CA), and primer pairs specific for the deleted region of CD38 (exon1-F 5’-AGC CTC TCT CTT GCT GCC TA-3’, exon1-R 5’-GAC AGA GTT GGG CTC TCC TA-3’) and the nondeleted region (exon2-F 5’-ATG CTT TCA AGG GTG CAT TTA T-3’, exon2-R 5’-ATT GGC TTT TAT CCC CAG TTT T-3’). For each sample, qPCR was performed twice in triplicate, with mean values used for final data points.
Long-Range PCR and Sequencing
To determine the genomic breakpoints of the deletion, long-range polymerase chain reaction (LR-PCR) was carried out using the Bio-X-ACT long DNA polymerase (Bioline, London, UK) and the manufacturer's suggested protocol. The following primers were used to amplify across the 4p15.32 deletion: BST1-F 5’-GCT GAG TCA AGG ACA GAA GAC AT-3’ and CD38-R 5’-TAG GCA GAA GGA ATA AGC GTC AC-3’. PCR products were purified using exonuclease I (NEB, Ipswich, MA) and shrimp alkaline phosphatase (USB, Cleveland, OH). Bidirectional Sanger sequencing was then performed using BigDye chemistry (Applied Biosystems) and run on a 3730 × l DNA Analyzer (Applied Biosystems).
RNA Extraction and cDNA Synthesis
Epstein–Barr virus transformed LCLs were available for all members of family 3388 from the European Collection of Cell Cultures (https://www.phe-culturecollections.org.uk/). The cells were grown in RPMI-1640 media supplemented with 10% fetal bovine serum (PAA Laboratories, Pasching, Austria), L-glutamine (final concentration 2 mM), penicillin (500 U/mL), and streptomycin (5 μg/ mL). When the cells reached an amount of approximately 1 × 107, RNA was extracted using the RNeasy Mini kit (Qiagen, Crawley, UK), according to the manufacturer's suggested protocol. cDNA was synthesized using the QuantiTect Reverse Transcription kit (Qiagen) using approximately 1 μg of RNA as template.
RT-PCR and Sequencing
To confirm the transcription of a BST1-CD38 fusion gene, PCR primers were first designed in the exons closest to the CNV breakpoints: the forward primer was in exon 7 of BST1 (BST1x7F; 5’-AAG GCA GCA TGA AAG TCC TG-3’) and the reverse primer was in exon 2 of CD38 (CD38x2R; 5’-TTG TTG CAA GGT ACG GTC TG-3’). Forty cycles of PCR amplification were performed in a 25 μL volume, with 10× NH4 Reaction Buffer, a final Mg2+ concentration of 2.5 mM, each primer at final concentration of 0.2 μm, dNTPs at final concentration of 0.2 mM, and BIOTAQ DNA Polymerase (Bioline) and Pfu DNA Polymerase (Thermo Scientific, Loughborough, UK) used in a ratio 9:1 and 5 μL of cDNA diluted 1:10 as template. PCR products were visualized by UV illumination of 2% agarose gel stained with SYBR Safe (Invitrogen, Paisley, UK). To ensure integrity of cDNA, we also used primer pairs for IMMP2L and ZNF277, and obtained bands of the expected size for all family members. To confirm and further characterize the fusion transcript, RT-PCR was repeated using primers in exon 3 of BST1 (5’-GTA TGG CAG GGT TGC AGA TT-3’) and in exon 7 of CD38 (5’-TCG ATT CCA GCT CTT TTA TGG-3’). The BST1-CD38 RT-PCR products were purified and sequenced using a bidirectional Sanger protocol, as described above.
Expression of Canonical Transcripts and Fusion Transcript
RT-PCR primers were designed that would amplify the canonical transcripts but not the fusion transcript: CD38x1F (5’-CGA TGC GTC AAG TAC ACT GAA A-3’), CD38x3R (5’-ATG GGC CAG ATC TTT TAT TCT G-3’), and BST1x7F and BST1x9R (5’-ACT TGG GGC TTT TCT TTG AGT A-3’). The same primers used to amplify and sequence the BST1-CD38 transcript (BST1x7F and CD38x2R) were also used in qPCR experiments to evaluate the expression of the fusion transcript compared with the expression of the canonical BST1 transcript. qPCR was performed using SYBR-green (Applied Biosystems), and the results were normalized against the housekeeping gene beta glucuronidase (GUSB), which was amplified using primers in exon 8 (5’-CAC CTA GAA TCT GCT GGC TAC T-3’) and exon 9 (5’-AGA GTT GCT CAC AAA GGT CAC A-3’).
Western Blot Analysis
LCLs for the family were grown as described above (RNA extraction and cDNA synthesis), as well as HeLa cells which were used as a positive control for each antibody. LCLs were harvested by centrifugation at 900 rpm for 5 min and cell pellets washed in phosphate-buffered saline. All cells were lysed in RIPA buffer (50 mM Tris-HCl pH 7.4, 1% NP-40, 150 mM NaCl, 1 mM EDTA, Roche complete protease inhibitor). Post-nuclear supernatants were heat-denatured in reducing Lithium dodecyl sulfate loading dye and run on a 4–12% Bis-Tris gel (Invitrogen) under reducing conditions (30 μg total protein loaded per lane). Proteins were transferred onto Polyvinylidene fluo-ride membrane, and Western blotting was performed according to standard protocols using a monoclonal antibody raised against the C-terminus of anti-CD38 (ab108403, Abcam, Cambridge, UK) or a polyclonal antibody raised against amino acids 1–146 of anti-bone marrow stromal cell antigen 1 (ab137718, Abcam), followed by appropriate secondary antibodies (Bio-Rad, Hemel Hempstead, UK). Immunoreactive bands were visualized using ECL Plus Western blot detection reagent (GE Healthcare, Little Chalfont, UK).
Results
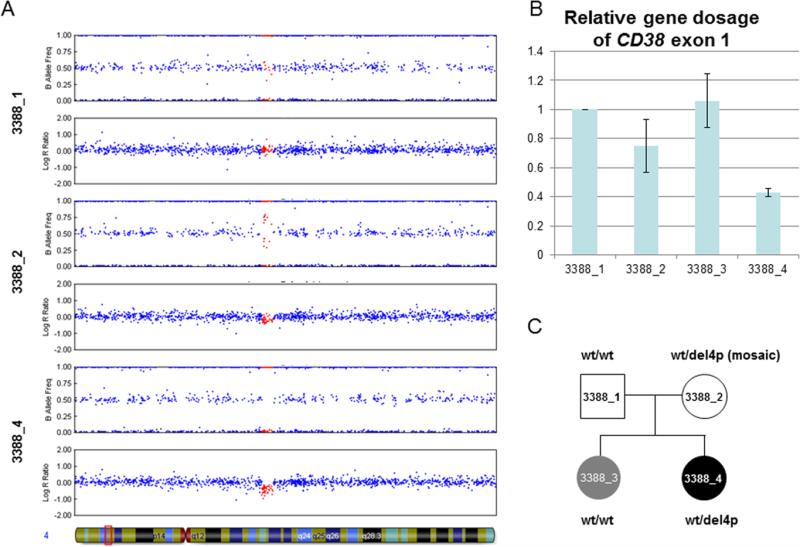
In a previous study, the Autism Genome Project genotyped 1275 ASD cases using the 1 M SNP microarray and used these data in the first instance for a genome-wide association study [Anney et al., 2010] and an analysis of rare CNV [Pinto et al., 2010]. Noting a strong link in the literature between oxytocin and the regulation of social behavior (reviewed recently in [Neumann & Landgraf, 2012], we interrogated these data for rare CNV that disrupts genes related to oxytocin. These included OXT (20p13), OXTR (3p25), AVPR1A (12q14–15), AVPR1B (1q32), AVP (20p13), and CD38 (4p15). The only rare CNV identified was a ~80 kb deletion of 4p15.32 identified in individual 3388_4. This event involved 46 of the genotyped SNP and was called with high confidence by all three algorithms (with a log Bayes factor = 204 using QuantiSNP). This deletion involved the 5’ end of CD38 and the 3’ end of its neighboring gene, BST1. Analyses of the parental data revealed that although the father carried no CNV at 4p15.32, the mother was predicted to carry a CNV event in the same region as the child's deletion. Inspection of the B-allele frequency (BAF) plot showed a split in the intermediate heterozygote cluster, suggesting that she was mosaic for the deletion (Fig. 1A).
Figure 1.
Detection of a 4p15.32 deletion inherited from mosaic mother. (A) Illumina 1 M-single SNP data viewed with Genome Studio V2011.1. The father (3388_1) shows normal copy number across the region, whereas the proband (3388_4) shows a heterozygous deletion. In contrast, the pattern shown for the mother (3388_2) suggests she is mosaic for the deletion. The 46 SNP inside the deletion are highlighted in red. Region shown is chr4:14 000 000–17 000 000 (NCBI36/hg18 build). (B) Quantitative PCR showing the relative gene dosage of CD38 exon 1 (deleted) relative to exon 2 (not deleted). Results are normalized using the father (3388_1) who has a normal copy number at this locus. Two independent qPCR experiments were performed and the results combined (error bars: +/− SD). The results help validate the deletion in 3388_4, and the intermediate result seen for the mother is consistent with her being a mosaic for this CNV. Although there is some variability between the two runs, the results suggest that the older sister (3388_3) has not inherited the deletion. (C) Pedigree drawing summarizing the CD38deletion status. Gray shading indicates autism, while black shading indicates autism and asthma.
To validate the CNV predicted by the algorithms, qPCR was performed. This confirmed that the dosage of CD38 exon 1 was reduced for the proband compared with exon 2 (Fig. 1B). While a smaller decrease was confirmed in the mother (consistent with her being a genetic mosaic), the gene dosage for the sibling 3388_3 was not decreased, suggesting that she had not inherited the deletion (Fig. 1B).
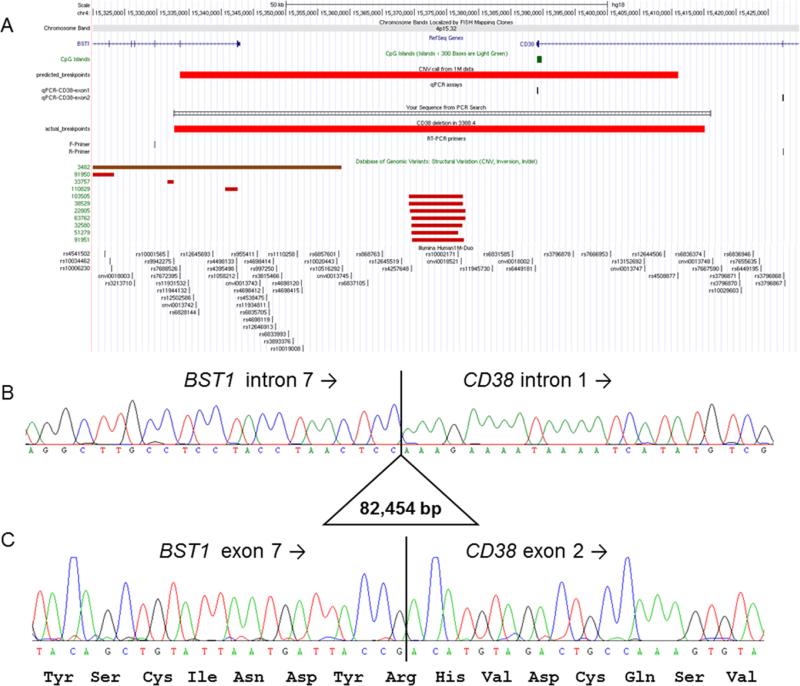
To characterize the deletion breakpoints, LR-PCR was carried out across the predicted deletion region. The size of the genomic region prohibited amplification of the wild-type allele but created a 1 kb product for the deleted allele. PCR products were obtained for the proband and mother, but not for the father or the sibling, thus confirming the segregation of this deletion (Fig. 1C). By Sanger sequencing this junction fragment, the precise breakpoints of the deletion were resolved to be chr4:15 332 700–15 415 153, showing the deletion was slightly larger than the original CNV call (Fig. 2A) and confirming that the distal breakpoints lie in intron 7 and not intron 8 of BST1 (Fig. 2B). Similar deletions are not reported in the Database of Genomic Variants. Three base pairs of microhomology were seen at either end of the deletion, which may have helped mediate the deletion through a mechanism such as fork stalling and template switching (FoSTeS) [Lee, Carvalho, & Lupski, 2007].
Figure 2.
Molecular characterization of CD38 deletion. (A) Figure from UCSC genome browser showing the position of BST1 and CD38, predicted deletion breakpoints based on 1 M SNP data, position of qPCR assays used to validate the deletion, and position of long-range PCR primers used to determine the true breakpoint positions. There are no similar CNV listed in the Database of Genomic Variants v10 (February 2011). Region shown is chr4:15 320 000–15 430 000 (NCBI36/hg18 build). (B) Sanger sequencing of the long-range PCR product from the BST1-F primer spanned the deletion breakpoints and shows the true deletion coordinates to be chr4:15 332 700– 15 415 153 (NCBI36/hg19). TCC microhomology was seen at each end of the deleted region. (C) Sequencing of a 201 bp RT-PCR product confirmed that the reading frame of the fusion transcript was maintained. Similar results were obtained using a second set of primers.
As both disrupted genes are encoded on the same (+) strand of chromosome 4, we hypothesized that the deletion would lead to the presence of a fusion transcript between exons 1–7 of BST1 and exons 2–8 of CD38. Accordingly, RT-PCR was carried out using primers designed to bind to the exons adjacent to the deletion breakpoints. RT-PCR products of the expected size were obtained for 3388_4 and 3388_2 but not for the other two family members. Sequencing confirmed that these bands corresponded to the predicted fusion transcript and showed that the reading frame of CD38 is maintained (Fig. 2C). RT-PCR was repeated using primers in BST1 exon 3 and CD38 exon 7. PCR products of the expected size were again only seen in the proband and the mother, consistent with the inheritance pattern. Sanger sequencing confirmed not only the fusion breakpoints but also additional exon–exon junctions, demonstrating that the fusion transcript involves at least exons 3–7 of BST1 and exons 2–7 of CD38.
As well as confirming the presence of a fusion transcript, the relative levels of the canonical transcripts were also measured. Using qPCR, we showed that there was an apparent reduction of both BST1 and CD38 in cells from the proband when compared with cell lines derived from other family members (Fig. S1A and B). Moreover, since the regulatory regions of BST1 should be driving the expression of the fusion transcript, we tested by qPCR the expression of the BST1-CD38 fusion transcript in the proband cell line and observed that the fusion transcript is expressed at a lower level compared with the canonical BST1 transcript (Fig. S1C).
To determine whether this fusion transcript was translated into a stable protein, Western blot analyses were performed using antibodies raised against both the N-terminus of BST1 and the C-terminus of CD38. There were no additional bands detected in the proband (3388_4) or her mother (3388_2) (Fig. S2).
Discussion
Several studies have shown that CNV plays a significant role in determining genetic susceptibility to autism [Pinto et al., 2010; Sebat et al., 2007]. In most cases, it is likely that CNV mediates its effects either by influencing gene expression, or else by unmasking a recessive allele on the homologous chromosome. However, in previous studies, we have identified cases where rare CNV found in ASD patients can lead to the formation of novel fusion transcripts that are easily detectable by RT-PCR [Holt et al., 2012]. In this study, we scanned for CNV disrupting genes involved in oxytocin function and identified a novel 82 kb deletion that fuses the BST1 and CD38 genes. Unlike the previously reported IMMP2L-DOCK4 [Pagnamenta et al., 2010] and MAPKAPK5-ACAD10 fusion transcripts [Holt et al., 2012], the reading frame of the BST1-CD38 fusion transcript is maintained and so may lead to a novel 486 amino acid fusion protein involving amino acids 1–264 (N-terminal) of BST1 and 79–300 (C-terminal) of CD38. However, in light of the reduced expression of both canonical transcripts (Fig. S1A and B), we cannot discount that haploinsufficiency may also modulate the phenotype seen in 3388_4.
Expression of a gene is largely determined by various promoter sequences that mostly lie immediately 5’ of the gene. Therefore, the tissue-specific expression pattern of the hypothetical BST1-CD38 fusion protein would be likely to match that of BST1 rather than CD38. Quantification of the fusion transcript in the proband's cell line by qPCR showed a lower level of expression in relation to the wild-type BST1 transcript. A preliminary Western analysis was unable to detect an aberrant protein in cells from 3388_2 or 3388_4. However, further proteomic studies would be needed to determine whether this is because (a) the aberrant protein is simply present at low levels, (b) it is unstable, or (c) the fusion of BST1 and CD38 changes the proteins’ structure such that the epitopes are no longer present/accessible.
In the mother, the mosaic status of the deletion meant that it was originally detected as a loss by iPattern and as a gain by QuantiSNP. This difference reflects the relative weights placed upon log R ratio and BAF by different CNV calling algorithms. Detection of the same event as both a loss and gain by different algorithms may, therefore, represent a signature of mosaic deletions. As the deletion was transmitted to the youngest daughter, we know the deletion must be present in the mother's germline. Because oogenesis in humans is completed at around the time of birth, and given the mosaic nature of the deletion seen in DNA derived from blood, we hypothesize that the deletion represents a post-zygotic de novo event occurring very early in the mother's development. As the DNA for the mother's parents was not available, we are unable to confirm this. Although we note that the maternal DNA sample used for the SNP array was obtained from cell line-derived material, and thus cannot totally rule out the apparent mosaic nature of the deletion in the maternal sample being a cell line artifact, a precise reversion event is unlikely.
CD38 regulates oxytocin secretion in mice [Jin et al., 2007], a function that is believed to be conserved in humans [Munesue et al., 2010]. The highest levels of CD38 are found in the hypothalamus of both mice and humans [Jin et al., 2007; Munesue et al., 2010]. Higashida and colleagues demonstrated reduced plasma oxytocin levels in Cd38 knockout mice and elevated levels of oxytocin in the hypothalamus and pituitary tissues, suggesting that oxytocin is produced and packaged but is not released into the brain and blood stream in mice that were Cd38-/- [Higashida et al., 2011]. In mice, Cd38 was found to be critical for social behavior [Jin et al., 2007]. Higashida and colleagues showed that Cd38 knockout mice have impairment of maternal behavior and male social recognition [Higashida et al., 2011].
In human studies, children with ASD had lower plasma oxytocin levels and different patterns of association between oxytocin level and age than a control group [Modahl et al., 1998]. There is evidence that polymorphisms in the oxytocin receptor gene (OXTR) may be associated with diagnosis of ASD or with specific symptom domains related to social behavior [Campbell et al., 2011; Gregory et al., 2009; Jacob et al., 2007; Lerer et al., 2008; Liu et al., 2010; Wermter et al., 2010; Wu et al., 2005; Yrigollen et al., 2008]. In addition, the OXTR is susceptible to epigenetic regulation, possibly by a number of factors or developmental experiences. Currently, there are several trials underway with intranasal oxytocin in autism to target social behavioral deficits. Understanding the pathophysiology of the oxytocin system in ASD and its relationship to receptor, peptide, and regulator pathways like CD38 will be helpful in identifying individuals who may or may not respond to these methods of treatment interventions. Unfortunately, we were unable to test serum oxytocin levels in this family as they refused further blood draws.
In CD38, the SNP rs1800561, a single point mutation where tryptophan replaced arginine at amino acid residue 140 (R140W), has been associated with lower oxytocin levels in patients with ASD [Munesue et al., 2010]. Furthermore, administration of intranasal oxytocin helped with symptoms of ASD in a patient with autism and the R140W allele [Higashida et al., 2012]. The family and clinicians found improvement in eye contact and communication, as well as smiling [Higashida et al., 2012; Munesue et al., 2010]. Of note, not all patients with ASD who were administered oxytocin showed improvement, and not all individuals carrying this variant develop autism, indicating that the CD38 R140W allele alone is not sufficient to cause ASD [Higashida et al., 2012].
Also of interest in this case is the evidence that CD38 may play a role in autoimmune diseases, such as diabetes and asthma. In addition to the regulation of oxytocin levels, CD38 plays a second messenger role in the regulation of calcium concentration in smooth muscle, including airway smooth muscle. CD38 is involved in the synthesis and degradation of cyclic ADP ribose (cADPR)—an endogenous ligand for ryanodine receptors in airway smooth muscle [Matsumoto et al., 2012]. When ryanodine receptors are bound to cADPR, mobilization of intracellular calcium stores from the airway smooth muscle cells occurs [Deshpande et al., 2005], which results in contraction of the airway smooth muscles.
Less is known regarding the function of the second gene in the detected fusion transcript. BST1 encodes a glycosylphosphatidylinositol-anchored protein that facilitates the stroma cell-dependent growth of pre-B cell line [Kaisho et al., 1994]. BST1 is a candidate gene for Parkinson's disorder, spurring ongoing investigation of brain function [Peeraully & Tan, 2012]. Interestingly, BST1 and CD38 share around ~30% identity [Kaisho et al., 1994]. Both have ADP ribosyl cyclase activity and cADPR hydrolase activity, and are known as bifunctional enzymes [Deshpande et al., 2005].
In summary, in this study, we report a deletion of CD38 and BST1 in a patient with autism and asthma, inherited from her mosaic mother. While this event does not cosegregate with autism in the pedigree, the absence of similar CNV in screened controls or in the Database of Genomic Variants, the detection of a novel fusion tran script and the documented links among CD38, oxytocin levels, and social behavior lead us to hypothesize that the 82 kb deletion and its disruption of CD38 may contribute to the phenotype of autism and asthma in the proband. Known autism risk factors, such as deletions of 16p11.2, do not always cosegregate with autism within pedigrees, suggesting that genetic heterogeneity often exists within families [Weiss et al., 2008]. Under the second hit hypothesis, shared environmental factors, or as yet undiscovered genetic or epigenetic factors, may modify the penetrance of this event. It has been shown that for many genomic disorders, a higher rate of genetic second hit is seen in individuals with developmental delay when compared with controls [Girirajan et al., 2012]. Further studies should include functional studies to assess the potential consequences of the predicted BST1-CD38 protein and further interrogation of rare variants in oxytocin pathway genes in data from recent exome studies.
Supplementary Material
Figure S1. Relative expression levels of both canonical transcripts and of the BST1-CD38 fusion transcript measured in lymphoblastoid cell line derived from the proband (3388_4) and other family members.
Figure S2. Western blot showing (A) CD38 and (B) BST1 expression from Hela (lane 1) and lymphoblastoid cell lines (lanes 2–5).
Acknowledgments
We thank the patients and their family for their cooperation, assistance, and support in this project. This work was supported in part by 5T32MH067631-07 Training in the Neuroscience of Mental Health (A.S.), NIH Autism Center of Excellence P50 HD055751 (E.H.C.), K23MH082121 (S.J.), the MRC [G1000569/1] (D.N.), the Wellcome Trust (090532/Z/09/Z), and the NIHR Biomedical Research Centre Oxford, with funding from the Department of Health's NIHR Biomedical Research Centres funding scheme. The views expressed in this publication are those of the authors and not necessarily those of the Department of Health. Dianne Newbury is an MRC Career Development Fellow and a Junior Research Fellow at St John's College. We would like to acknowledge Zoe Holloway for technical assistance with the preliminary Western analysis, and Steve Guter for assistance with the clinical data.
Footnotes
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
The authors have no conflicts of interest to disclose.
References
- American Psychiatric Association . Diagnostic and statistical manual of mental disorders. 4th ed., text revision American Psychiatric Association; Washington, DC: 2000. [Google Scholar]
- Anney R, Klei L, Pinto D, Regan R, Conroy J, et al. A genome-wide scan for common alleles affecting risk for autism. Human Molecular Genetics. 2010;19:4072–4082. doi: 10.1093/hmg/ddq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autism and Developmental Disabilities Monitoring Network Surveillance Year 2008 Principal Investigators; Centers for Disease Control and Prevention Prevalence of autism spectrum disorders—Autism and Developmental Disabilities Monitoring Network, 14 sites, United States, 2008. MMWR Surveillance Summaries. 2012;61:1–19. [PubMed] [Google Scholar]
- Bailey A, Lecouteur A, Gottesman I, Bolton P, Simonoff E, et al. Autism as a strongly genetic disorder—Evidence from a British twin study. Psychological Medicine. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- Campbell DB, Datta D, Jones ST, Batey Lee E, Sutcliffe JS, et al. Association of oxytocin receptor (OXTR) gene variants with multiple phenotype domains of autism spectrum disorder. Journal of Neurodevelopmental Disorders. 2011;3:101–112. doi: 10.1007/s11689-010-9071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Fombonne E. Pervasive developmental disorders in preschool children: Confirmation of high prevalence. The American Journal of Psychiatry. 2005;162:1133–1141. doi: 10.1176/appi.ajp.162.6.1133. [DOI] [PubMed] [Google Scholar]
- Deshpande DA, White TA, Dogan S, Walseth TF, Panettieri RA, et al. CD38/cyclic ADP-ribose signaling: Role in the regulation of calcium homeostasis in airway smooth muscle. American Journal of Physiology. Lung Cellular and Molecular Physiology. 2005;288:L773–L788. doi: 10.1152/ajplung.00217.2004. [DOI] [PubMed] [Google Scholar]
- Dunn LM. Peabody picture vocabulary test-III. American Guidance Service; Circle Pines, MN: 1997. [Google Scholar]
- Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. The New England Journal of Medicine. 2012;367:1321–1331. doi: 10.1056/NEJMoa1200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory SG, Connelly JJ, Towers AJ, Johnson J, Biscocho D, et al. Genomic and epigenetic evidence for oxytocin receptor deficiency in autism. BMC Medicine. 2009;7:62. doi: 10.1186/1741-7015-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmayer J, Cleveland S, Torres A, Phillips J, Cohen B, et al. Genetic heritability and shared environmental factors among twin pairs with autism. Archives of General Psychiatry. 2011;68:1095–102. doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashida H, Yokoyama S, Huang JJ, Liu L, Ma WJ, et al. Social memory, amnesia, and autism: Brain oxytocin secretion is regulated by NAD(+) metabolites and single nucleotide polymorphisms of CD38. Neurochemistry International. 2012;61:828–938. doi: 10.1016/j.neuint.2012.01.030. [DOI] [PubMed] [Google Scholar]
- Higashida H, Yokoyama S, Munesue T, Kikuchi M, Minabe Y, et al. CD38 gene knockout juvenile mice: A model of oxytocin signal defects in autism. Biological and Pharmaceutical Bulletin. 2011;34:1369–1372. doi: 10.1248/bpb.34.1369. [DOI] [PubMed] [Google Scholar]
- Holt R, Sykes NH, Conceicao IC, Cazier JB, Anney RJ, et al. CNVs leading to fusion transcripts in individuals with autism spectrum disorder. European Journal of Human Genetics. 2012;20:1141–1147. doi: 10.1038/ejhg.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IMGSAC A full genome screen for autism with evidence for linkage to a region on chromosome 7q. International Molecular Genetic Study of Autism Consortium. Human Molecular Genetics. 1998;7:571–578. doi: 10.1093/hmg/7.3.571. [DOI] [PubMed] [Google Scholar]
- Insel TR, O'Brien DJ, Leckman JF. Oxytocin, vasopressin, and autism: Is there a connection? Biological Psychiatry. 1999;45:145–157. doi: 10.1016/s0006-3223(98)00142-5. [DOI] [PubMed] [Google Scholar]
- Jacob S, Brune CW, Carter CS, Leventhal BL, Lord C, et al. Association of the oxytocin receptor gene (OXTR) in Caucasian children and adolescents with autism. Neuroscience Letters. 2007;417:6–9. doi: 10.1016/j.neulet.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D, Liu HX, Hirai H, Torashima T, Nagai T, et al. CD38 is critical for social behaviour by regulating oxytocin secretion. Nature. 2007;446:41–45. doi: 10.1038/nature05526. [DOI] [PubMed] [Google Scholar]
- Kaisho T, Ishikawa J, Oritani K, Inazawa J, Tomizawa H, et al. BST-1, a surface molecule of bone marrow stromal cell lines that facilitates pre-B-cell growth. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:5325–5329. doi: 10.1073/pnas.91.12.5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Lerer E, Levi S, Israel S, Yaari M, Nemanov L, et al. Low CD38 expression in lymphoblastoid cells and haplotypes are both associated with autism in a family-based study. Autism Research. 2010;3:293–302. doi: 10.1002/aur.156. [DOI] [PubMed] [Google Scholar]
- Lerer E, Levi S, Salomon S, Darvasi A, Yirmiya N, et al. Association between the oxytocin receptor (OXTR) gene and autism: Relationship to Vineland Adaptive Behavior Scales and cognition. Molecular Psychiatry. 2008;13:980–988. doi: 10.1038/sj.mp.4002087. [DOI] [PubMed] [Google Scholar]
- Liu X, Kawamura Y, Shimada T, Otowa T, Koishi S, et al. Association of the oxytocin receptor (OXTR) gene polymorphisms with autism spectrum disorder (ASD) in the Japanese population. Journal of Human Genetics. 2010;55:137–141. doi: 10.1038/jhg.2009.140. [DOI] [PubMed] [Google Scholar]
- Lord C, Risi S, Lambrecht L, Cook EH, Jr., Leventhal BL, et al. The Autism Diagnostic Observation Schedule-Generic: A standard measure of social and communication deficits associated with the spectrum of autism. Journal of Autism and Developmental Disorders. 2000;30:205–223. [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview-Revised: A revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. Journal of Autism and Developmental Disorders. 1994;24:659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Hirata Y, Otsuka K, Iwata T, Inazumi A, et al. Interleukin-13 enhanced Ca2 + oscillations in airway smooth muscle cells. Cytokine. 2012;57:19–24. doi: 10.1016/j.cyto.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Modahl C, Green L, Fein D, Morris M, Waterhouse L, et al. Plasma oxytocin levels in autistic children. Biological Psychiatry. 1998;43:270–277. doi: 10.1016/s0006-3223(97)00439-3. [DOI] [PubMed] [Google Scholar]
- Mullen EM. Mullen Scales of Early Learning manual. AGS ed. American Guidance Service; Circle Pines, MN: 1995. [Google Scholar]
- Munesue T, Yokoyama S, Nakamura K, Anitha A, Yamada K, et al. Two genetic variants of CD38 in subjects with autism spectrum disorder and controls. Neuroscience Research. 2010;67:181–191. doi: 10.1016/j.neures.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Neumann ID, Landgraf R. Balance of brain oxytocin and vasopressin: Implications for anxiety, depression, and social behaviors. Trends in Neurosciences. 2012;35:649–659. doi: 10.1016/j.tins.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Pagnamenta AT, Bacchelli E, de Jonge MV, Mirza G, Scerri TS, et al. Characterization of a family with rare deletions in CNTNAP5 and DOCK4 suggests novel risks loci for autism and dyslexia. Biological Psychiatry. 2010;68:320–328. doi: 10.1016/j.biopsych.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeraully T, Tan EK. Genetic variants in sporadic Parkinson's disease: East vs west. Parkinsonism and Related Disorders. 2012;18(Suppl. 1):S63–S65. doi: 10.1016/S1353-8020(11)70021-9. [DOI] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven J. The Raven's Progressive Matrices: Change and stability over culture and time. Cognitive Psychology. 2000;41:1–48. doi: 10.1006/cogp.1999.0735. [DOI] [PubMed] [Google Scholar]
- Raven JC. Sets A, AB, B: Board and book forms. Lewis; London: 1947. Progressive matrices. [Google Scholar]
- Sauer C, Montag C, Worner C, Kirsch P, Reuter M. Effects of a common variant in the CD38 gene on social processing in an oxytocin challenge study: possible links to autism. Neuropsychopharmacology. 2012;37:1474–1482. doi: 10.1038/npp.2011.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow SS, Cicchetti DV, Balla DA. Vineland Adaptive Behavior Scales. 2nd ed. American Guidance Service; Circle Pines, MN: 2005. [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, et al. Association between Microdeletion and Micro-duplication at 16p11.2 and Autism. New England Journal of Medicine. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- Wermter AK, Kamp-Becker I, Hesse P, Schulte-Korne G, Strauch K, et al. Evidence for the involvement of genetic variation in the oxytocin receptor gene (OXTR) in the etiology of autistic disorders on high-functioning level. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics. 2010;153B:629–639. doi: 10.1002/ajmg.b.31032. [DOI] [PubMed] [Google Scholar]
- Wiig EH, Secord WA, Semel E. Clinical evaluation of language fundamentals—Preschool. Psychological Corporation; San Antonio: 1992. [Google Scholar]
- Wu S, Jia M, Ruan Y, Liu J, Guo Y, et al. Positive association of the oxytocin receptor gene (OXTR) with autism in the Chinese Han population. Biological Psychiatry. 2005;58:74–77. doi: 10.1016/j.biopsych.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Yrigollen CM, Han SS, Kochetkova A, Babitz T, Chang JT, et al. Genes controlling affiliative behavior as candidate genes for autism. Biological Psychiatry. 2008;63:911–916. doi: 10.1016/j.biopsych.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Relative expression levels of both canonical transcripts and of the BST1-CD38 fusion transcript measured in lymphoblastoid cell line derived from the proband (3388_4) and other family members.
Figure S2. Western blot showing (A) CD38 and (B) BST1 expression from Hela (lane 1) and lymphoblastoid cell lines (lanes 2–5).