Abstract
With the availability of whole-genome sequences for an increasing number of species, we are now faced with the challenge of decoding the information contained within these DNA sequences. Comparative analysis of DNA sequences from multiple species at varying evolutionary distances is a powerful approach for identifying coding and functional noncoding sequences, as well as sequences that are unique for a given organism. In this review, we outline the strategy for choosing DNA sequences from different species for comparative analyses and describe the methods used and the resources publicly available for these studies.
The remarkable accomplishments of the past decade in genomic biology were achieved in large part by significant technological advances in DNA sequencing as well as data and information processing systems. The biosciences community currently has access to whole-genome sequences for over 85 microbial organisms (see http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/micr.html) as well as a handful of eukaryotic species, including a yeast (Saccharomyces cerevisiae), a nematode (Caenorhabditis elegans), a fruitfly (Drosophila melanogaster), thale cress (Arabidopsis thanliana), a variety of rice (Oryza sativa japonica), a pufferfish (Fugu ribripes) and humans (Homo sapiens). Other eukaryotic genomic sequences, including those of the mouse and rat, will be completed in the near future, and the genomes of additional eukaryotes, including the chimpanzee, zebrafish, and bumblebee, are slated for sequencing in the next few years.
The present ability to sequence almost entire genomes outpaces in some aspects current computational and experimental methods to decode the information contained within these sequences. The existing high-throughput sequence annotation pipelines combine the results of database similarity searches and gene-predicting algorithms to identify coding sequences with good but not complete accuracy. In some cases only a fraction of the sequences comprising a gene are identified and some genes are missed entirely. Moreover, current annotation methods are largely unable to identify reliably other types of functional elements, such as transcriptional regulatory regions, noncoding RNA genes, and elements involved in chromosome structure and function.
Comparing the DNA sequences of different species is a powerful method for decoding genomic information, because functional sequences tend to evolve at a slower rate than nonfunctional sequences. By comparing the genomic sequences of species at different evolutionary distances, one can identify coding sequences and conserved noncoding sequences with regulatory functions, and determine which sequences are unique for a given species. Here we review the basis for selecting DNA sequences of species at appropriate evolutionary distances for comparative analysis depending on the biological question being addressed, and describe the algorithms commonly used in these studies. We also compare a small interval of human chromosome 7q31 with DNA sequences of four species at different evolutionary distances to demonstrate the multistep process of comparative sequence analysis, and discuss several of the public resources available for these studies.
Selection, Annotation, and Alignment of DNA Sequences for Cross-Species Comparisons
Comparative genomics requires at least two DNA sequences that are evolutionarily related (Fig. 1). The biological question being addressed by the comparative analysis will determine the appropriate level of nucleotide similarity (evolutionary distance) of the sequences and the alignment method used.
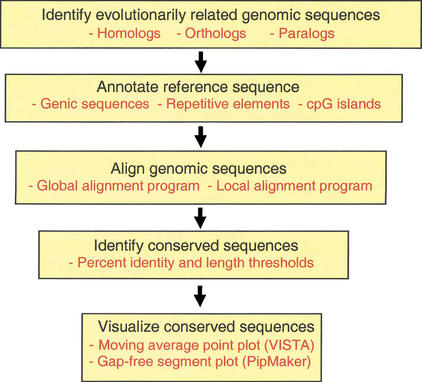
Figure 1.
Multistep process of comparative sequence analysis. Evolutionarily related sequences must be identified. The reference sequence should be annotated for known functional elements. Sequences to be compared must be aligned. Evolutionarily conserved sequences are identified based on specified thresholds (such as percent identity and length) of conservation. Visualization tools allow the individual to view the annotations, sequence alignments, and conserved elements simultaneously.
Evolutionarily Related Gene Sequences
Previous comparative DNA sequence studies have revealed that genes similar to each other at the nucleotide level can be related to one another by different evolutionary histories. Although a few examples of convergent evolution (two previously unrelated genes that became similar as they acquired new, related functions) have been reported, most comparative genomic approaches investigate sequences that are related by divergent evolution from a common ancestor. Genes derived from a common ancestral gene are homologs, and the level of similarity in their sequences often reflects the time since they diverged. Homologous genes can be generated by speciation, which produces pairs of orthologs (genes in different species that are derived from the same gene in the last common ancestral species, and thus usually have similar functions). Homologous genes can also result from the duplication of a chromosomal segment, which produces paralogs (duplicate gene pairs that have diverged and typically have different functions).
When performing cross-species DNA sequence comparisons to identify functional elements, it is important to distinguish between orthologous sequences and paralogous sequences. Comparisons between paralogs that are descended from last common ancestral species do not reveal as many evolutionarily conserved sequences as comparisons between orthologs, simply because those paralogous sequences have been apart longer, and thus are more divergent.
Evolutionarily Related Chromosomal Segments
Numerous studies comparing the relative order of gene orthologs in the human and mouse genomes revealed that long-range sequence organization to a large extent has been preserved from their last common ancestor. These observations have led to the following definition of terms to describe similarity between evolutionarily related chromosomal segments in different species (Gregory et al. 2002): (1) Synteny (literally “same thread”) refers to two or more genes that are located on the same chromosome, and hence is only relevant within a species. (2) When the orthologs of genes that are syntenic in one species are also located on a single chromosome in a second species (without regard to their order), the chromosomal segments in the two species have conserved synteny (Fig. 2). (3) When the order of multiple orthologous genes is the same in two species, the genomic intervals are referred to as conserved segments (also called “conserved linkages”). As gene orthology and map information has been obtained for a larger number of species, it has become clear that conserved segments can be observed between all mammals. Although conserved synteny has been observed between organisms as evolutionarily distant as humans and pufferfish, which diverged approximately 450 million years ago (Aparicio et al. 2002), conserved long-range sequence organization has not been reported for more distantly related species.
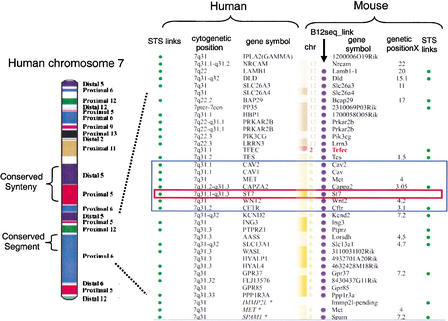
Figure 2.
Comparative map between human chromosome 7 and the mouse genome. On the left is a representation of human chromosome 7 with blocks of conserved synteny color-coded based on their chromosomal position in the mouse genome (indicated on right-hand side; diagram from Thomas et al. 2000). In the middle of human chromosome 7 is one block of conserved synteny subdivided into two conserved segments, one from distal murine 5 and one from proximal murine 5. At the resolution of this diagram, intrachromosomal rearrangements present in the mouse genome cannot be visualized. On the right is a detailed comparative map of the human 7q31 interval with conserved synteny intervals in the mouse (http://www.ncbi.nlm.nih.gov/Homology). The black rectangle shows the position of the ∼1.8-Mb CFTR interval, and the red rectangle highlights the position of ST7 in this region. This map was constructed using NCBI build 28 and the MGI composite map. The order of the loci presented is based on the human reference sequence. The indicated mouse loci are not consecutive based on their MGI cM positions (Tes 1.5 cM, Met 4.0 cM, Cappa2 3.05 cM, Wnt2 4.2 cM, Cftr 3.1 cM, and Kend2 7.2 cM). The curvy line between Tcfec and Tes represents a conserved synteny breakpoint between humans and mice. The following NCBI links are provided: STS links are linked to the dbSTS pages, human cytogenetic positions are linked to NCBI's MapViewer, gene symbols are linked to LocusLink, the Bl2_seq links provide an alignment of two representative transcripts using Blast2Seq, and genetic positions (cM) are linked to the NCBI's Mapviewer.
Selection of Species for DNA Sequence Comparisons
The comparison of DNA sequences between pairs of species that diverged ∼40–80 million years ago from a common ancestor, such as humans with mice, or two species of fruitflies (Drosophila melanogaster with Drosophila pseudoobscura), or two species of nematodes (Caenorhabditis elegans with Caenorhabditis briggsae; Kent and Zahler 2000), or Escherichia coli with Salmonella species (McClelland et al. 2000) reveals conservation in both coding sequences and a significant number of noncoding sequences. To date, only a limited number of conserved noncoding sequences identified by comparing sequences from species at this evolutionary distance have been characterized functionally. Those whose functions have been assigned have been transcriptional regulatory elements of nearby genes (Gumucio et al. 1996; Hardison et al. 1997) or genes as much as 200 kb away (Loots et al. 2000). A question remains as to whether most of the conserved noncoding sequences are present because of functional constraints or are the result of a lack of divergence time. Sequence comparisons between multiple species that diverged approximately 40–80 million years ago, such as humans with mice and humans with cows, allow one to determine which noncoding sequences have been conserved in several species and thus are more likely due to active conservation than shared ancestry. A second issue that is necessary to be aware of when sifting through conserved elements identified by comparative analysis of species at this evolutionary distance is that it is difficult to definitively distinguish between yet undiscovered coding sequences and functional non-coding sequences.
The comparison of orthologous DNA sequences between evolutionary distantly related species, such as humans and pufferfish, which diverged approximately 450 million years ago, primarily reveals coding sequences as conserved (Aparicio et al. 2002). This is due to the fact that protein coding sequences are tightly constrained to retain function and thus evolve slowly, resulting in readily detectable sequence homology even over this large phylogenetic distance. Therefore, the addition of distantly related organisms (∼450 million years) to a multi-species sequence comparison improves the ability to classify conserved elements into coding sequences and non-coding sequences.
Comparative analyses of genomic DNA from closely related species, such as humans with chimpanzees or other nonhuman primates, identify sequences that have changed and genomic rearrangements that have occurred in recent evolutionary history (Frazer et al. 2003). Some of these recent genomic events identified by comparison of orthologous sequences between closely related species may be responsible for gene differences between the organisms, and thus are of interest because of their potential functional consequences. Thus, by adding a closely related organism to a multispecies comparative sequence analysis, one can identify not only coding sequences and functional noncoding sequences but also those genomic sequences which may be responsible for traits that are unique to the reference species.
Obtaining Genomic Sequences for Comparative Analyses
Genomic DNA sequences for both completely sequenced organisms and those for which sequencing is in progress can be obtained from public Web sites (see Table 1). From some of these Web servers, such as the National Center for Biotechnology (NCBI), the majority of publicly available DNA sequences for all species can be obtained. Species-specific sequence databases also exist, but primarily for those organisms with whole-genome sequences currently available.
Table 1.
List of Resources for Obtaining and Analyzing Genomic Sequences
This is not meant to be a comprehensive list, but to the reader an idea of the multitude of choices available.
Annotation of Reference DNA Sequence
After obtaining a set of DNA sequences for comparative analysis, the first step is to annotate the reference sequence for known sequence features. The patterns of sequence conservation between species can be interpreted only if the locations of known coding sequences and repetitive elements are indicated in the reference sequence. For species with whole-genome sequences currently available, such as human, high-throughput sequence annotation pipelines have already determined the locations of known coding sequences and other types of functional information (Table 1). In the event that annotations are not precomputed for the sequence of interest, the position of known genes can be deduced by using the genomic sequence to search databases of genes, proteins, or expressed sequence tags (ESTs; see http://www.ncbi.nlm.nih.gov). There are also several programs available (Burge and Karlin 1997; Kulp et al. 1997; Solovyev and Salamov 1997) which predict the locations of putative genes by searching the DNA sequence for common features of coding sequences (evolutionary conservation, open reading frames, GC content, etc.
Repetitive sequence elements account for a large fraction of most mammalian genomes. There are several classes of repetitive elements (Lander at el. 2001); the members of each class are highly similar to each other at the nucleotide level. If these repetitive elements are not masked (marked so that they are ignored by the alignment program) in a DNA sequence prior to a comparative analysis, they will generate very large numbers of alignments that do not reflect biologically significant similarities. RepeatMasker is the most commonly used program for screening DNA sequences for repetitive elements (Smit and Green 1999).
After the location of genes and repetitive elements are known in the reference DNA sequence, the remaining conserved elements identified in a comparative analysis are of interest as putative regulatory elements, novel structural features, or uncharacterized genes.
Alignments of Orthologous Sequences
An alignment is a mapping of one DNA sequence onto another evolutionarily related DNA sequence in order to identify regions that have been conserved. There are two basic types of alignment programs, local and global (Table 1). Local alignments are computed to produce optimal similarity scores between subregions of the sequences. The rationale behind local alignments is that the two sequences being compared may differ in ways that preclude an accurate end-to-end alignment. For example, when long continuous sequences (encoding multiple genes) with conserved synteny but scrambled gene order are compared, some of the orthologous subregions of the two sequences will have changed order and/or orientation, and some of the subregions may not be orthologous because of insertions or deletions. Thus, the various aligning orthologous subregions may be more accurately viewed as separate segments, that is, by local alignments.
Global alignments are computed to produce optimal similarity scores over the entire length of the two sequences being compared. Global alignments may be better than local alignments for detecting highly diverged but orthologous subregions in the comparison of two long contiguous sequences. However, if the DNA sequences of only two species are being compared, it is difficult to assess whether a subregion matching in a global alignment but not in a local alignment is highly divergent but orthologous, or present due to a deletion or insertion and thus, nonorthologous sequence. The global alignment technique is appropriate for comparing portions of genomic sequences from two species that are expected to share similarity over their entire length, such as conserved segments.
Critical, objective comparisons of the two approaches for sensitivity and specificity have not yet been performed, and many users will likely find both approaches to be informative. In later sections, we will describe two servers for producing and visualizing alignments of genomic DNA, one that generates local alignments (PipMaker) and one that generates global alignments (VISTA).
Resources and Software Tools for Cross-Species Sequence Comparisons
As a means for describing some of the public resources available for comparative sequence studies, we focus on an ∼1.8-Mb interval of human chromosome 7q31 commonly referred to as the cystic fibrosis transmembrane conductance regulator (CFTR) region (Fig. 3). Although the resources described here are primarily aimed at analyzing human genomic sequences by comparative analyses, the software tools for sequence annotation, alignment, and visualization are applicable regardless of the species being compared.
Figure 3.
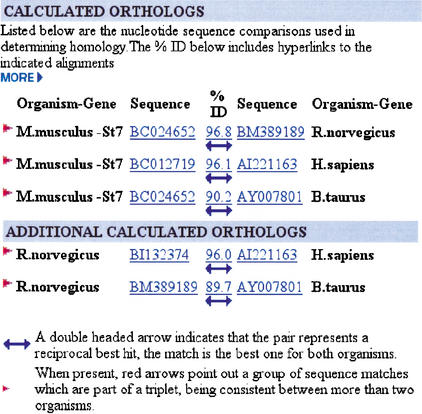
HomoloGene calculated reciprocal best match analysis between the mouse (M. musculus), human (H. sapiens), rat (R. norvegicus), and cow (B. taurus) ST7 genes. In the HomoloGene Calculated Orthologs section, a double-headed arrow indicates that the pairwise alignment represents a reciprocal best match between the indicated species. The red arrow indicates that the sequence matches are part of a triplet, being consistent between more than two species. When a pair of genes is part of a triplet relationship, the other members of the triplet are shown in the Additional Calculated Orthologs section. The accession numbers are hyperlinked to the GenBank entry, and the arrow and identity score are linked to a BLAST alignment of the two sequences.
The NIH Intramural Sequencing Center
As a complement to ongoing efforts to sequence completely a handful of vertebrate genomes, the U.S. National Institutes of Health (NIH) Intramural Sequencing Center (NISC) has established a program for sequencing targeted genomic regions in many evolutionarily diverse species (see http://www.nisc.nih.gov). To date, more than 40 genomic regions that together encompass more than 30 Mb are being sequenced in multiple vertebrates, including chimpanzee, baboon, dog, cat, cow, pig, mouse, rat, chicken, zebrafish, and pufferfish. Some of these intervals, such as the ∼1.8 Mb CFTR region, are being sequenced by the NISC Comparative Sequencing Program in over 10 additional vertebrates. Among its many goals, this program aims to create multispecies sequence data sets that might help guide decisions about which vertebrate genomes to sequence more completely in the future.
In this review, to demonstrate the strategy as well as some of the commonly used software tools for comparative sequence analysis (Fig. 1), we focus on a small interval in the CFTR region on human 7q31 for which the orthologous regions have been sequenced in multiple species by the NISC Comparative Sequencing Program. Specifically, we compare an ∼308-kb segment that contains the ST7 (suppression of tumorigenicity 7) gene, which is believed to play an important role in some types of human cancer (Zenklusen et al. 2001), with the conserved baboon, cow, mouse, and pufferfish DNA segments.
HomoloGene
HomoloGene is a relatively new resource at the NCBI of both curated and calculated gene orthology information between species. Though curated orthologs are a valuable resource, they are limited in number due to the laborious nature of hand curation and the fact that most of the pairs are based on known genes. Thus, the majority of orthologies in HomoloGene are calculated in an automated high-throughput fashion.
Orthology is calculated by comparing sequences from different species in a pairwise, reciprocal fashion. When transcript sequences from two species are each other's best match (reciprocal best hits), the corresponding genes are considered as being putative orthologs. The calculated orthologous relationships for ST7 human, cow, mouse, and rat gene sequences are shown in Figure 3. For these four species, all six pairwise transcript sequence comparisons identified the ST7 gene as the reciprocal best match, suggesting that this is a true set of orthologs.
For some gene sequences it is not possible to determine a unique orthologous relationship across species; for example, several transcript sequences in one species can have the same best match in the second species. This can result from biological events, such as lineage-specific gene duplications generating multiple paralogs in one species (all of which have the same ortholog in a second species), or a deletion event in one species resulting in the loss of the “true ortholog” of a gene(s) in the second species. In some cases, this will be due to the accuracy of the present data set; the gene sequence may not yet be deposited in one of the species-specific databases, or a UniGene error may result in either the artificial splitting of a gene into two clusters or the combining of two homologous genes into a single cluster.
In the future, the HomoloGene resource will include more species-specific sequence databases to calculate orthologous relationships. In addition, the algorithms used to automatically calculate gene orthologs are constantly being refined, and the sequence data quality is continually being improved.
NCBI Human–Mouse Conserved Synteny Maps
The National Center for Biotechnology (NCBI) currently constructs homology maps of the human and mouse genomes based on the locations of known genes (see http://www.ncbi.nlm.nih.gov/Homology/). The chromosomal locations and order of human genes are based on the most recent NCBI build of the human genome (see http://www.ncbi.nlm.nih.gov/genome/guide/human/), and the MGI composite map is used to determine the location and order of murine genes (see http://www.informatics.jax.org/ and http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/map_srchdb?chr=mouse_chr.inf). The most difficult task in constructing these homology maps is determining the locations where intrachromosomal inversions and interchromosomal rearrangements have occurred during the evolution of the present-day human and mouse chromosome structures from a common ancestor.
A recent comparative analysis study between human chromosome 7 and the mouse genome using a high density of markers identified 20 conserved segments in 16 regions of conserved synteny (Fig. 2; Thomas et al. 2000). The ∼1.8-Mb CFTR region on human 7q31 is contained within a single conserved segment on mouse chromosome 6 but is relatively close to a conserved synteny breakpoint (Fig. 2). The relative genetic positions (cM) of the mouse loci in the MGI composite map in Figure 2 suggest that intrachromosomal inversions have occurred between humans and mice in the ∼1.8-Mb CFTR region. However, the recent assembly of the mouse genome sequence (MGSCv3) and prior bacterial artificial chromosome (BAC) fingerprint mapping studies (Thomas et al. 2000) indicate that the genes in this region are in the same linear order in humans and mice. These contradictory data result from the fact that the MGI composite map is low-resolution compared with sequence data (the MGSCv3 sequence map). At the resolution of the MGI composite map, conserved synteny blocks can be identified reasonably well, but subdividing these blocks into conserved segments is difficult.
In the near future, the human–mouse homology maps will be constructed using computational annotation of the MGSCv3 sequence map of the mouse genome (Waterston et al. 2002) as the reference for chromosomal location and order of mouse loci. The current plan is to continue constructing the homology maps based on orthologous human and mouse transcripts.
Annotation of Human Sequence Encoding ST7
In the next two sections, we describe the use of two different Web servers, PipMaker and VISTA, for comparative sequence analysis. Although PipMaker and VISTA differ in many aspects (Table 2), the “gene annotation files” that they use are similarly formatted. The gene annotation file provides information about coordinates of coding sequences in the reference sequence, including gene name(s), exon positions, and direction of transcription.
Table 2.
Summary of PipMaker and VISTA Server Features
| PipMaker | Both | VISTA | |
| Input files | DNA sequences | ||
| Annotation of the base sequence | |||
| Base sequence mask file | |||
| Underlay files (for any sequence) | |||
| Embedded hyperlink file | |||
| Output files | Alignments in different formats (nucleotide level) | ||
| Ordered and oriented sequence relative to first sequence | |||
| The percent identity plot | VISTA plot | ||
| Dot plot | Conserved sequences | ||
| Analysis of exons: splice junctions, predicted coding sequence | |||
| Limit on the length | ∼2mb, time limited | 4 mb | |
| Implementation | Web server and stand alone programs, finished and draft sequences | ||
| Underlying alignment | Local | Global | |
| Features to be visualized | CpG islands | Genes, exons, repeats, CNSs, order and orientation of aligned sequences | Gaps in both sequences |
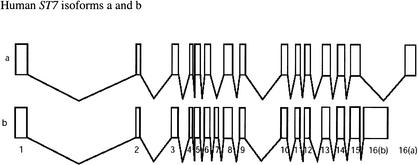
The sequence of the ∼308-kb interval on human 7q31 encoding ST7 was obtained from the NISC, and the gene annotation file was generated by using the program sim4 (Florea et al. 1998) to align the ST7 cDNA (RefSeq file: http://www.ncbi.nlm.nih.gov/LocusLink/LocRpt.cgi?l=7982) to the genomic DNA. The ST7 locus produces two transcripts, known as isoforms a and b, which result from alternative splicing (Fig. 4). The mRNA transcripts of the human ST7 isoforms a and b contain distinctly different 3′-end exons, and isoform a is missing the alternatively spliced exon 7.
Figure 4.
The human ST7 locus on 7q31 produces two transcripts. Isoform a is shorter in length than isoform b due to the fact that it is missing the alternatively spliced exon 7 and has a shorter length 3′-end exon. The two isoforms, a and b, each have their own NCBI accession numbers, NM_018412 and NM_021908, respectively.
PipMaker Alignment and Visualization Tool
PipMaker (http://bio.cse.psu.edu) is a WorldWide Web server used to compare two long genomic sequences and identify conserved segments between them (Schwartz et al. 2000). A companion server at the same site, MultiPipMaker, will align three or more genomic DNA sequences. The underlying alignment software, BLASTZ (Schwartz et al. 2002), used by both of these servers computes local alignments, following the general design of the “gapped BLAST” family of programs, which start by finding short, exact matches, then extend those matches to alignments that include gaps (Altschul et al. 1997). BLASTZ uses an empirically determined scoring matrix for matches and mismatches (Chiaromonte et al. 2002) plus an affine gap penalty (gap open and gap extension penalties).
To use PipMaker and MultiPipMaker for comparative sequence analysis, two or more sequences in FastA format (plain text only) along with files to mask repeats, annotate genes, and provide highlighting of other functional sequence features, are submitted to the Web servers (Fig. 5). The repeat file documents the position and type of repeats in the reference sequence and is usually generated with output from the program RepeatMasker (Smit and Green 1999). The PipMaker server will mask the positions of repeats with lowercase letters, allowing the BLASTZ alignment program to skip these positions during the first step of the alignment. In the subsequent step extending the primary alignment seeds, these “soft-masked” regions can be included in the alignment if doing so increases the similarity score. Thus, repeats that predate the separation of the species being compared can be aligned. The exon text file (gene annotation file) provides positional information about coding sequences in the reference sequence. This information can be obtained from a number of resources, including GenBank entries (Wheeler et al. 2002) and genome browsers (Table 1; Hubbard et al. 2002; Kent et al. 2002). Regions of interest in the first sequence, such as the locations of exons or regulatory elements, can be shaded in the background to help distinguish them from other genomic features, with an underlay file (Fig. 5). Detailed instructions on using PipMaker and MulitiPipMaker, as well as a suite of software tools (PipTools) to aid users in constructing the input files for these Web servers, are available (Elnitski et al. 2002a,b).
Figure 5.
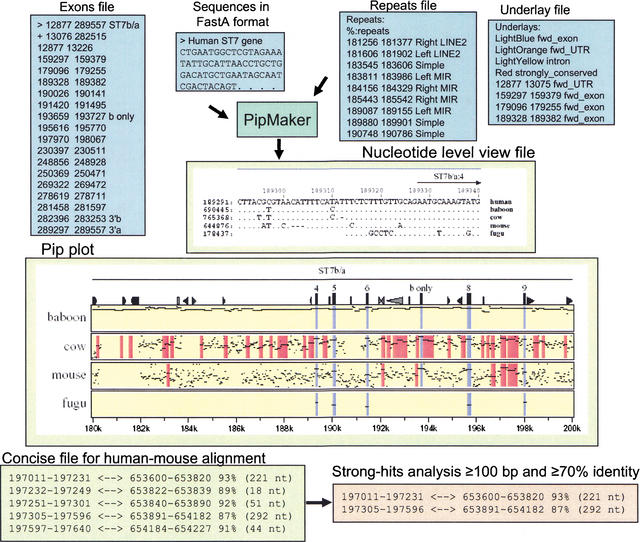
PipMaker: input and output files. Files for submission to PipMaker include Sequences (required), Repeats (recommended), Underlay (optional), and Exon annotations (optional). The Repeats file is made by simplifying RepeatMasker output using the program rmask2repeats (from the PipTools program package). The simplified version is shown. The coordinates in the Repeats file and Underlay file correspond to the coordinates in the Pip plot. PipMaker generates three multiple output files. The Pip plot shown is a subregion of the human ST7 interval compared with the orthologous baboon, cow, mouse, or fugu sequences. Each panel represents a pairwise comparison between human sequence and that of the indicated species. Each alignment consists of a series of horizontal lines that represent the gap-free aligning segments that are graphed on a vertical scale of 50%–100% and relative to the coordinates in the human sequence on the horizontal axis. Icons across the top panel represent annotations for the reference human sequence and include triangles for various repeats and rectangles for exons and CpG islands. The names of gene (ST7) and direction of transcription are indicated above the alignments. The Nucleotide-Level View shows the multiple alignment of the multispecies comparison at the nucleotide level. Dots represent nucleotides that are the same. Dashes represent gaps. The interval shown includes ST7 exon sequences, and thus some nucleotides are conserved between humans and fugu. The Concise output gives a coordinate-based format of the aligning segments. The gap-free interval in the first sequence and the corresponding interval in the second sequence are listed, along with the percent identity and length. The program strong-hits reads the concise file format and returns alignments that fulfill user-specified thresholds (length of the alignment and the percent identity).
We submitted to MultiPipMaker the ∼308-kb region on human 7q31 encompassing ST7 for comparison with the homologous baboon, cow, mouse, and fugu DNA segments. The server returned a percent identity plot (Pip) displaying the position, length, and percent identity (from 50%–100%) of each gap-free segment in the pairwise BLASTZ alignments of the human sequence with DNA from each of the four vertebrate species (Fig. 5, Suppl. Fig. 1). The coordinates (lower horizontal axis) are the nucleotide positions in the human reference sequence. The icons across the top of the Pip represent features in the human sequence such as repeats (from the repeats file), gene names, exons, and direction of transcription (from the exons file) and CpG islands (computed by the server). The portions of the Pip corresponding to specific genomic features are color-coded: Coding exons are colored blue, light orange corresponds to untranslated regions (UTRs), light yellow indicates intronic sequences, and red is used for highlighting regions of strong conservation but no known function.
We examined the highly conserved elements in the ST7 locus for each of the four pairwise sequence comparisons: human–baboon, human–cow, human–mouse, and human–fugu (Suppl. Fig. 1). For this analysis, gap-free segments (≥100 bp and ≥70% identity) were identified by analyzing the concise output from a PipMaker alignment using the PipTools program, strong-hits (Fig. 5; Elnitski et al. 2002a). As expected, the amount of sequence in these highly conserved, gap-free segments decreases with increasing phylogenetic distance. The human and baboon sequences align along almost their entire lengths, including intronic and intragenic regions, at a level of ∼90% identity. Only repetitive elements that inserted since the two species diverged do not align. For both the human–cow and human–mouse comparisons, the majority of highly conserved elements identified is located outside known exonic sequences of ST7, and thus are potential noncoding functional sequences, such as regulatory elements. In contrast, the human–fugu comparison reveals only exonic sequences as conserved, reinforcing previous conclusions that many coding sequences but few noncoding functional sequences can be aligned at this large phylogenetic distance. The ST7 coding sequences are highly similar in humans, baboons, and cows, but isoform b is completely absent in fugu (both the terminal 3′ exon and the alternatively spliced exon 7) and is only partially present in the mouse (the terminal 3′ coding exon of b is absent; Suppl. Fig. 1).
The multiple sequence alignment in the Nucleotide Level View output file of MultiPipMaker (Fig. 5) is constructed by merging the pairwise alignments, pruning them so that each position in the reference sequence aligns with at most one position in the secondary sequence, and repeating the process to improve the alignment according to rigorously defined multiple alignment scores. These multiple alignments can be evaluated in a number of ways, including column matching scores, amount of evolutionary change, information content, and more complex analyses that examine contiguous matches in each row, which mimics the expected behavior of transcription factor binding sites (Stojanovic et al. 1999). Regions in the alignment that have multiple sequences similar at the nucleotide level have been conserved in several species, and thus are more likely present due to selection.
With the available high-quality draft genomic sequences of human (Lander et al. 2001) and mouse (Waterston et al. 2002), the two genomes have been aligned using BLASTZ in an all-versus-all comparison (Waterston et al. 2002). Results of this computation can be accessed in several ways. Pips have been computed across these genomes and are available from the PipDispenser (http://bio.cse.psu.edu/genome/hummus/). The human–mouse whole-genome Pip plots are annotated with the names of RefSeq genes (Pruitt and Maglott 2001) and will be updated as new annotations are added. Tracks showing the positions of BLASTZ alignments and links to nucleotide-level alignments of conserved human–mouse sequences are at the interactive Human Genome Browser at the University of California Santa Cruz (UCSC; Fig. 6; Kent et al. 2002). In addition, the alignments have been analyzed for the likelihood that they reflect selection (i.e., functional constraint) relative to nearby neutrally evolving DNA (Waterston et al. 2002). Plots of this function are also at the Human Genome Browser (Table 1).
Figure 6.
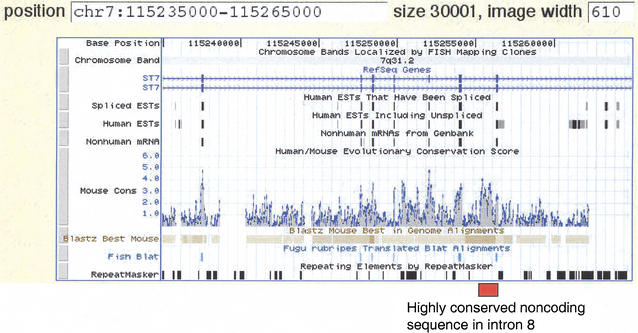
UCSC Genome Browser view of the human–mouse sequence alignment in the ST7 region. The Browser tracks provide different types of information on the ST7 gene, in this case showing the region from exon 3 through exon 9, as thin rectangles on the track beneath “RefSeq Genes“. The two diagrams for ST7 represent the two isoforms, a and b. The direction of transcription is indicated by the light blue arrows. The next three tracks (Spliced ESTs, Human ESTs, Nonhuman mRNAs) show evidence of transcription into stable mRNA. The Mouse Cons track plots a log-likelihood score that gives the probability that an aligned segment is under selection, adjusted for the neutral substitution rate measured in nearby ancestral repeats. The positions of blastz alignments are plotted on the Blastz Best Mouse track, and nucleotide-level alignments can be obtained by clicking on this track. Regions that align with Fugu rubripes are shown on the Fish Blat track, followed by repeats as identified by RepeatMasker. The position of the highly conserved noncoding sequence in intron 8 of ST7 is indicated by the red box. Note that it has a score on the MouseCons track of 4.0, meaning that it is 10,000 times more likely to be under selection than to be under evolutionary drift.
VISTA Alignment and Visualization Tool
The VISTA Web server (http://www-gsd.lbl.gov/vista) is an integrated set of software tools for comparing two or more genomic sequences. The server consists of two autonomous modules—one for alignment of long genomic sequences, and one for the visualization and identification of conserved elements (Dubchak et al. 2000; Mayor et al. 2000). The VISTA server currently uses AVID, a global alignment program (Bray et al. 2003) that works by first finding maximal exact matches between two sequences using a suffix tree, and then recursively identifies the best anchor points based on the length of the exact matches and the similarity in their flanking regions. The VISTA visualization module is also configured to use global alignments produced by (GLASS [Batzoglou et al. 2000] and) the limited-area global alignment of nucleotides (LAGAN) algorithm (Brudno et al. in prep.; Table 1).
To use VISTA for comparative sequence analysis, two or more sequences in FastA format (plain text only) along with a gene annotation file are submitted to the Web server (Fig. 7). The server automatically uses RepeatMasker (Smit and Green 1999) to mask repetitive elements in the reference sequence. Detailed instructions on using the VISTA server and a stand-alone program are available (see http://www-gsd.lbl.gov/vista, Mayor et al. 2000).
Figure 7.
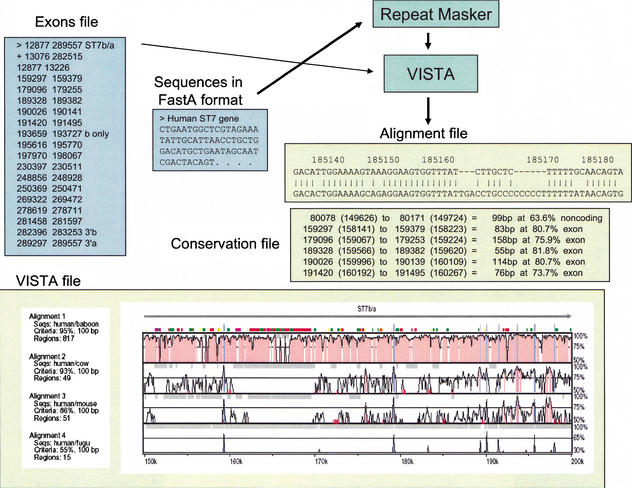
VISTA: input and output files. Files for submission to VISTA include Sequences (required) and Exons (optional). Repeats are masked in the reference sequence using RepeatMasker upon its submission to VISTA. VISTA generates three output files. The VISTA plot shown here is a subregion of the human ST7 interval compared with the orthologous baboon, cow, mouse, or fugu sequences. Conserved sequences represented as peaks [noncoding (red) and coding (blue)] are shown relative to their positions in the human genome (horizontal axes), and their percent identities (50%–100%) are indicated on the vertical axes. The locations of ST7 exons are indicated by tall blue rectangles, and the direction of transcription is indicated by a horizontal arrow. The locations of repetitive elements are indicated by color rectangles (see Suppl. Fig. 2). The Alignment file shows the alignment between the human reference sequence and the orthologous mouse DNA; coordinates correspond to the positions in the human sequence shown in the VISTA plot. The Conservation file gives the coordinates of the conserved sequences at predefined cutoffs.
We submitted to VISTA the ∼308-kb region on human 7q31 encompassing ST7 as the reference sequence for comparison with the baboon, cow, mouse, and fugu conserved DNA segments. The server returned a VISTA plot visualizing the pairwise global alignments between the human reference sequence and the DNA of other species (Fig. 7, Suppl. Fig. 2). The VISTA plot was generated by sliding the default window (100 bp long) along each pairwise sequence alignment and calculating the percent identity at each base pair position. The VISTA plot coordinates are the nucleotide positions in the human 7q31 reference sequence (horizontal axis) and the percent identities of the conserved elements (vertical axis). We also submitted a gene annotation file to the VISTA Web server, and thus the gene name (ST7), exon positions, and direction of transcription are marked above the VISTA plot (Fig. 7, Suppl. Fig. 2). Conserved sequences are highlighted under the curve, with blue indicating a conserved exon, turquoise indicating a conserved UTR, and red indicating a conserved noncoding region.
We examined each of the four pairwise sequence comparisons between the human sequence and the orthologous baboon, cow, mouse, and fugu DNA for conserved elements. For this analysis, conserved sequences ≥100 bp (including gaps) and ≥70% identity were identified by analyzing the VISTA conservation file (Fig. 7); the amount of highly conserved sequence decreased with increasing phylogenetic distance, as expected.
To analyze conserved elements identified in the multispecies comparison, we also used an algorithm available at the VISTA server that simultaneously compares two sequence alignments (in this case human–cow and human–mouse) to determine percent identity and length thresholds that primarily identify sequences that have been conserved in three species (Dubchak et al. 2000). This algorithm is based on the assumption that conserved sequences present in three species (humans/cows/mice) are more likely due to active conservation than shared ancestry. For this analysis, the thresholds calculated by the program for the human–cow comparisons (≥93% identity/100 bp) identified 46 conserved elements, and for the human–mouse comparison (≥86% identity/100 bp) identified 48 conserved elements. The human and baboon ST7 orthologous sequences are too similar and the human and fugu orthologous sequences are too dissimilar for this type of analysis; we therefore arbitrarily chose 95% identity/100 bp as the human–baboon thresholds (791 conserved sequences), and 55% identity/100 bp for the human–fugu thresholds (16 conserved sequences). Using these stringent criteria for defining sequences as evolutionarily conserved results in the identification of fewer conserved noncoding sequences, but those identified are more likely to be present due to active conservation and thus have biological function.
High-quality draft human (Lander et al. 2001) and mouse (Waterston et al. 2002) genomic sequences have been aligned using a computational strategy where mouse sequence contigs are anchored on the human genome by local alignment matches (Kent 2002) and then globally extended by AVID (Couronne et al. 2003). Alignments on the whole-genome scale can be visualized using an interactive tool, Vista Genome Browser, accessible at the gateway Web site http://pipeline.lbl.gov. Vista Genome Browser is an applet that allows for displaying results of comparative sequence analysis in a VISTA format on the scale of whole chromosomes. It displays the RefSeq gene annotations (Pruitt and Maglott 2001) and has a number of interactive options, including the ability to extract the sequence and conserved elements of a displayed region, define thresholds of sequence conservation, and determine zoom level, as well as other options.
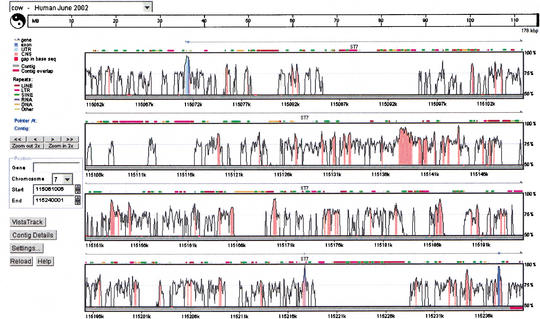
The computational strategy of anchoring sequence contigs from one species onto a base genome sequence assembly of a second species by local alignment matches and then globally aligning these contigs to candidate regions is also implemented for user-submitted sequences at another VISTA server, http://pipeline.lbl.gov/cgi-bin/GenomeVista. This server assists in finding candidate orthologous regions for a submitted sequence from any species on either the human or mouse genome sequence assembly, and provides detailed comparative analysis. We submitted the ST7 baboon, mouse, and cow genomic sequences to the server, and for each sequence the orthologous human ST7 region on 7q31 was unambiguously identified in the whole-human-genome assembly (June 2002). The GenomeVista server also generates corresponding pairwise alignments accompanied by the analysis of conservation. A VISTA Genome Browser display generated by the submission of the cow ST7 genomic sequence to the server is shown in Figure 8.
Figure 8.
VISTA Genome Browser display generated by the submission of the cow ST7 genomic sequence to the GenomeVista server. The cow ST7 sequence is automatically aligned against the orthologous human region (June 2002 assembly). Details of the display including chromosome and nucleotide position of the sequence alignment are given in the legend on the left-hand side of the plot. The ‘Contig Details’ button opens another window that provides access to files of nucleotide-level alignment, individual sequences in the alignment, corresponding RefSeq annotation, conserved regions, and other results. The ‘Settings’ button brings up a window where a user can customize a cutoff for calculating conservation level (percent identity and the window size) and many display options, such as image format.
A Candidate Regulatory Element
The Pip and VISTA plots were examined to find candidate regulatory elements. The aim is to find noncoding sequences that are evolutionarily conserved at a higher level than surrounding sequences, based on the assumption that these sequences are likely to be under purifying selection; that is, they are likely to be functional. Both the Pip and VISTA plots (Figs. 5, 7) show that the human and baboon DNA sequences are much too similar to distinguish functional from nonfunctional noncoding sequences, and indeed even the human–cow comparison shows much more noncoding conservation than can be studied intensively. In contrast, in the human–mouse comparisons there are many fewer conserved noncoding sequences observed, and a small number stand out dramatically. In particular, a 500-bp noncoding region between exons 8 and 9 (beginning at position 197 k) aligns with a high percent identity and only one gap in the Pip plot (Fig. 5). This region is also one of the more striking noncoding conserved sequences in the VISTA plot (Fig. 7). Furthermore, comparison with nearby neutrally evolving DNA suggests that it is about 10,000 times more likely to be under selection than to be under evolutionary drift (Fig. 6; Waterston et al. 2002).
Based on the analyses of the two different sequence alignments [BLASTZ (local) and LAGAN (global)], the 500 bp conserved noncoding sequence between exons 8 and 9 of the ST7 gene is probably under purifying selection, and thus is likely functional. Two possibilities immediately come to mind: It could be an undiscovered exon or it could be an intronic regulatory element. The first possibility can be tested in silico by searching for ESTs that include the conserved sequence; none are present in the current databases (Fig. 5). High-sensitivity assays such as RT-PCR can be done to look for expression in numerous tissues. The possibility that this is an intronic regulatory sequence could be tested by gain-of-function assays, e.g. by adding the putative regulator to a reporter gene with the ST7 promoter and transfecting appropriate cell lines. Now that the results of the whole-genome human–mouse alignments are readily available, we expect such interspecies alignments to become a routine part of the strategic planning for studies of regulation of mammalian genes.
Final Remarks
The annotation of whole-genome sequences for functional elements is clearly one of the most important and difficult challenges facing the biosciences community. The strategy of using cross-species DNA comparisons for identifying functionally important sequences is a powerful approach, but some factors complicate its application genome-wide. One caveat of using this approach to identify functional elements is the fact that the neutral rate of evolution varies across the genome (Wolfe et al. 1989; Waterston et al. 2002). Thus whole-genome sequence comparisons between two species that diverged ∼40–80 million years ago from a common ancestor have background levels of sequence conservation that vary from one genomic region to the next. For example, there are many regions in the human–mouse whole-genome sequence comparison for which the rate of divergence is slow enough to allow the alignment of orthologous sequences that are undergoing neutral drift (such as in ancient repeats; Waterston et al. 2002). For this reason, it is impossible to pick universal thresholds (length and percent identity) of conservation for the purpose of identifying sequences that are under selection. Based on the assumption that sequences which have been actively conserved due to purifying selection are more likely to be present in multiple species than sequences which are conserved due to a lack of divergence time, previous studies have used multispecies sequence comparisons as a means for assigning increased likelihoods that a conserved element is present because of functional constraints (Dubchak et al. 2000; Frazer et al. 2001). Because of the importance of this issue, additional efforts are underway to develop other approaches for distinguishing between these two types of evolutionarily conserved sequences, including analyzing sequence alignments for statistically significant conservation scores while allowing the basal rate of evolution to vary (Li and Miller 2002; Waterston et al. 2002).
Even the apparently straightforward task of annotating a whole-genome sequence for all coding sequences by cross-species DNA comparisons has some caveats. For instance, distantly related species, for which DNA sequence comparisons readily identify coding sequences, share only a subset of their genes. In contrast, species that are more closely related, such that they share the majority of their genes, also share a significant amount of highly conserved noncoding sequences, and thus it is not possible to identify new genes by merely looking for sequence conservation. Furthermore, coding sequences of genes under positive selection evolve faster than surrounding intronic and intergenic noncoding sequences, and thus, identifying genes of this nature by cross-species DNA comparisons requires looking for depressions of conservation between closely related species (Johnson et al. 2001).
So, what information can be deduced from cross-species sequence comparisons? They provide a window through which to look at the mechanisms of genome evolution. Only a few short years ago it would have been hard to imagine that a large fraction of the sequences conserved between humans and mice would be noncoding (Frazer et al. 2001; Mural et al. 2002; Waterston et al. 2002). The important role that small scale deletions and insertions play in genome evolution is only beginning to be understood through cross-species sequence comparisons (Frazer et al. 2003). Thus, cross-species sequence comparisons provide us with knowledge about the underlying mechanisms of genome evolution and point us in the direction of functionally important sequences. In this sense, comparative genomics provides key pieces of information necessary to develop new experimental and computational methods of sequence analysis which will eventually allow the complete annotation of whole-genome sequences, including not only the identification but also classification of functional elements.
WEB SITE REFERENCES
http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/micr.html; access to microbial genome sequences.
http://www.ncbi.nlm.nih.gov; National Center for Biotechnology Information Web site.
http://www.ncbi.nlm.nih.gov/Homology/; Human–Mouse homology map.
http://www.ncbi.nlm.nih.gov/genome/guide/human/; Human genome resources.
www.informatics.jax.org/; Mouse Genome Informatics Web site.
http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/map_srchdb?chr=mouse_chr.inf; mouse genome view.
http://www.ncbi.nlm.nih.gov/LocusLink/LocRpt.cgi?l=7982; Locus Link report of human ST7 gene.
http://bio.cse.psu.edu; Penn State Bioinformatics Group.
http://www-gsd.lbl.gov/vista; VISTA server.
http://pipeline.lbl.gov; comparative analysis pipeline gateway at Lawrence Berkeley National Laboratory.
Acknowledgments
We thank Ryan Weber, David Haussler, and the UCSC browser development team for developing the “Mouse Cons” track at the Genome Browser, and Pamela Jacques Thomas at NISC for help annotating the nonhuman species ST7 sequences. R.H. and L.E. were supported by PHS grants HG02238, HG02325, and DK27635. K.F. was supported in part by NIH grant GM-5748202. I.D. was supported by Programs for Genomic Applications grant from NHLBI/NIH.
E-MAIL kelly_frazer@perlegen.com; FAX (650) 625-4510.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.222003.
REFERENCES
- 1.Altschul S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aparicio S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1283-1285. [DOI] [PubMed] [Google Scholar]
- 3.Batzoglou S., Pachter, L., Mesirov, J.P., Berger, B., and Lander, E.S. 2000. Human and mouse gene structure: Comparative analysis and application to exon prediction. Genome Res. 10: 950-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bray, N., Dubchak, I., and Pachter, L. 2003. AVID: A global alignment program for large genomic sequences. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- 5.Burge C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78-94. [DOI] [PubMed] [Google Scholar]
- 6.Chiaromonte, F., Yap, V.B., and Miller, W. 2002. Scoring pair-wise genomic sequence alignments. Pac. Symp. Biocomput. 115–126. [DOI] [PubMed]
- 7.Couronne, O., Poliakov, A., Bray, N., Ishkhanov, T., Ryaboy, D., Rubin, E., Pachter, L., and Dubchak, I. 2003. Strategies and tools for whole genome alignments. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- 8.Dubchak I., Mayor, C., Brudno, M, Pachter, L.S., Rubin, E.M., and Frazer, K.A. 2000. Active conservation of noncoding sequences revealed by 3-way species comparisons. Genome Res. 10: 1304-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elnitski, L., Riemer, C., Petrykowska, H., Schwartz, S., Florea, L., Miller, W., and Hardison, R. 2002a. PipTools, a toolkit for annotating genomic sequence alignments: Predictions of DNase I hypersensitive sites. Genomics (in press).
- 10.Elnitski, L., Riemer, R., Schwartz, S., Hardison, R., and Miller, W. 2002b. PipMaker: A world wide web server for genomic sequence alignments. Current Protocols (in press). [DOI] [PubMed]
- 11.Florea L., Hartzell, G., Zhang, Z., Rubin, G.M., and Miller, W. 1998. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 8: 967-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frazer K.A., Sheehan, J.B., Stokowski, R.P., Chen, X., Hosseini, R., Cheng, J.F., Fodor, S.P., Cox, D.R., and Patil, N. 2001. Evolutionarily conserved sequences on human chromosome 21. Genome Res. 11: 1651-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frazer, K.A., Chen, X., Hinds, D.A., Pant, P.V.K., Patil, N., and Cox, D.R. Genomic DNA insertions and deletions occur frequently between humans and nonhuman primates. Genome Res. (in press). [DOI] [PMC free article] [PubMed]
- 14.Gregory S.G., Sekhon, M., Schein, J., Zhao, S., Osoegawa, K., Scott, C.E., Evans, R.S., Burridge, P.W., Cox, T.V., Fox, C.A., et al. 2002. A physical map of the mouse genome. Nature 418: 743-750. [DOI] [PubMed] [Google Scholar]
- 15.Gumucio D., Shelton, D., Zhu, W., Millinoff, D., Gray, T., Bock, J., Slightom, J., and Goodman, M. 1996. Evolutionary strategies for the elucidation of cis and trans factors that regulate the developmental switching programs of the β-like globin genes. Mol. Phylogenet. and Evol. 5: 18-32. [DOI] [PubMed] [Google Scholar]
- 16.Hardison R., Slightom, J.L., Gumucio, D.L., Goodman, M., Stojanovic, N., and Miller, W. 1997. Locus control regions of mammalian β-globin gene clusters: Combining phylogenetic analyses and experimental results to gain functional insights. Gene 205: 73-94. [DOI] [PubMed] [Google Scholar]
- 17.Hubbard T., Barker, D., Birney, E., Cameron, G., Chen, Y., Clark, L., Cox, T., Cuff, J., Curwen, V., Down, T., et al. 2002. The Ensembl genome database project. Nucleic Acids Res. 30: 38-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson M.E., Viggiano, L., Bailey, J.A., Abdul-Rauf, M., Goodwin, G., Rocchi, M., and Eichler, E.E. 2001. Positive selection of a gene family during the emergence of humans and African apes. Nature 413: 514-519. [DOI] [PubMed] [Google Scholar]
- 19.Kent J. 2002. BLAT—The BLAST-like alignment tool. Genome Res. 12: 656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kent W.J. and Zahler, A.M. 2000. Conservation, regulation, synteny, and introns in a large-scale C. briggsae-C. elegans genomic alignment. Genome Res. 10: 1115-1125. [DOI] [PubMed] [Google Scholar]
- 21.Kent W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, D. 2002. The human genome browser at UCSC. Genome Res. 12: 996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulp, D., Haussler, D., Reese, M.G., and Eeckman, F.H. 1997. Integrating database homology in a probabilistic gene structure model. Pac. Symp. Biocomput. 232–244. [PubMed]
- 23.Lander E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- 24.Li, J. and Miller, W. 2002. Significance of interspecies matches when evolutionary rate varies. RECOMB 2002. 216–224. [DOI] [PubMed]
- 25.Loots G., Ovcharenko, I., Pachter, L., Dubchak, I., and Rubin, E. 2002. rVISTA for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 12: 832-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayor C., Brudno, M., Schwartz, J.R., Poliakov, A., Rubin, E.M., Frazer, K.A., Pachter, L.S., and Dubchak, I. 2000. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16: 1046-1047. [DOI] [PubMed] [Google Scholar]
- 27.McClelland M., Florea, L., Sanderson, K., Clifton, S.W., Parkhill, J., Churcher, C., Dougan, G., Wilson, R.K., and Miller, W. 2000. Comparison of the Escherichia coli K-12 genome with sampled genomes of a Klebsiella pneumoniae and three salmonella enterica serovars, Typhimurium, Typhi and Paratyphi. Nucleic Acids Res. 28: 4974-4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mural R.J., Adams, M.D., Myers, E.W., Smith, H.O., Miklos, G.L., Wides, R., Halpern, A., Li, P.W., Sutton, G.G., Nadeau, J., et al. 2002. A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome. Science 296: 1661-1671. [DOI] [PubMed] [Google Scholar]
- 29.Pruitt K.D. and Maglott, D.R. 2001. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 29: 137-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwartz S., Zhang, Z., Frazer, K.A., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R., and Miller, W. 2000. PipMaker-A web server for aligning two genomic DNA sequences. Genome Res. 10: 577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartz, S., Kent, W.J., Smit, A., Zhang, Z., Baertsch, R., Hardison, R., Haussler, D. and Miller, W. 2003. Human–mouse alignments with BLASTZ. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- 32.Smit, A. and Green, P. 1999. RepeatMasker. http://ftp.genome.washington.edu/RM/RepeatMasker.html.
- 33.Solovyev V. and Salamov, A. 1997. The Gene-Finder computer tools for analysis of human and model organisms genome sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 5: 294-302. [PubMed] [Google Scholar]
- 34.Stojanovic N., Florea, L., Riemer, C., Gumucio, D., Slightom, J., Goodman, M., Miller, W., and Hardison, R. 1999. Comparison of five methods for finding conserved sequences in multiple alignments of gene regulatory regions. Nucleic Acids Res. 27: 3899-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas J.W., Summers, T.J., Lee-Lin, S.Q., Maduro, V.V., Idol, J.R., Mastrian, S.D., Ryan, J.F., Jamison, D.C., and Green, E.D. 2000. Comparative genome mapping in the sequence-based era: Early experience with human chromosome 7. Genome Res. 10: 624-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waterston R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- 37.Wheeler D.L., Church, D.M., Lash, A.E., Leipe, D.D., Madden, T.L., Pontius, J.U., Schuler, G.D., Schriml, L.M., Tatusova, T.A., Wagner, L., et al. 2002. Database resources of the National Center for Biotechnology Information: 2002 update. Nucleic Acids Res. 30: 13-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolfe K.H., Sharp, P.M., and Li, W.H. 1989. Mutation rates differ among regions of the mammalian genome. Nature 337: 283-285. [DOI] [PubMed] [Google Scholar]
- 39.Zenklusen J.C., Conti, C.J., and Green, E.D. 2001. Mutational and functional analyses reveal that ST7 is a highly conserved tumor-suppressor gene on human chromosome 7q31. Nat. Genet. 27: 392-398. [DOI] [PubMed] [Google Scholar]