Abstract
Accumulating evidence suggests that fatty livers are particularly more susceptible to several pathological conditions, including hepatic inflammation, cirrhosis and liver cancer. However the exact mechanism of such susceptibility is still largely obscure. The current study aimed to elucidate the effect of hepatocytes lipid accumulation on the nuclear electrophilic stress. Accumulation of intracellular lipids was significantly increased in HepG2 cells incubated with fatty acid (FA) complex (1 mM, 2:1 oleic and palmitic acids). In FA-treated cells, lipid droplets were localized around the nucleus and seemed to induce mechanical force, leading to the disruption of the nucleus morphology. Level of reactive oxygen species (ROS) was significantly increased in FA-loaded cells and was further augmented by treatment with moderate stressor (CoCl2). Increased ROS resulted in formation of reactive carbonyls (aldehydes and ketones, derived from lipid peroxidation) with a strong perinuclear accumulation. Mass-spectroscopy analysis indicated that lipid accumulation per-se can results in modification of nuclear protein by reactive lipid peroxidation products (oxoLPP). 235 Modified proteins involved in transcription regulation, splicing, protein synthesis and degradation, DNA repair and lipid metabolism were identified uniquely in FA-treated cells. These findings suggest that steatosis can affect nuclear redox state, and induce modifications of nuclear proteins by reactive oxoLPP accumulated in the perinuclear space upon FA-treatment.
Keywords: Fatty liver diseases, Lipid peroxidation, Electrophilic stress, Reactive carbonyls, Protein modifications
Abbreviations: BSA, bovine serum albumin; CoCl2, cobalt chloride; FA, fatty acids; hnRNPs, heterogeneous nuclear ribonucleoproteins; H2AX, histone 2AX; HHE, hydroxy-hexenal; HNE, hydroxy-nonenal; LPP, lipid peroxidation products; MCL, Markov cluster algorithm; MS, mass-spectroscopy; NAFLD, non-alcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; OHE, oxo-hexenal; ONE, oxo-nonenal; PPARα, peroxisome proliferator-activated receptor α; PGC-1α, peroxisome-proliferator-activated receptor-γ coactivator 1α; PUFA, poly unsaturated fatty acids; ROS, reactive oxygen species; VEGF, vascular endothelial growth factor
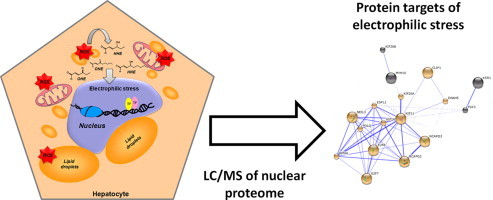
Graphical abstract
Highlights
-
•
Effects of fatty acids on modification of nuclear proteins in hepatocytes was studied.
-
•
Lipid accumulation was associated with abnormal nuclear morphology.
-
•
Lipid accumulation promoted mitochondrial activity and enhanced ROS generation.
-
•
Nuclear proteins were modified by lipid peroxidation products.
-
•
Data suggest nuclear stress as a mechanism for fatty liver disease progression.
Introduction
Non-alcoholic fatty liver disease (NAFLD) is currently one of the most common chronic liver diseases for both adults and children. NAFLD includes a spectrum of liver pathologies encompassed by the initial early stage of steatosis to the more pathological conditions of steatohepatitis (NASH) and fibrosis, which are recognized as a potential precursors of cirrhosis and hepatocellular carcinoma. Emerging evidence indicates that oxidative stress and dysregulation of redox-sensitive signaling pathways are central to the pathobiology of fatty liver diseases [1]. The role of liver steatosis, which affects around 30% of the general population, in development of liver deteriorations still remains unclear. Reactive oxygen species (ROS) are by-products of normal cellular metabolic processes. When ROS exceed certain level and not adequately removed, the balance between pro- and antioxidants is impaired, resulting in oxidative stress [2]. Oxidative stress can lead to the modifications of biological molecules, including DNA, lipids and proteins and has been implicated in many pathological conditions [3,4]. Recent data emphasize the importance of the local sites of ROS generation [2,3,5,6]. Some analysis have shown that the relative redox states from the most reducing to the most oxidizing can be assigned as mitochondria>nuclei>cytoplasm>endoplasmic reticulum>extracellular space [2]. However, it is logical to speculate that at specific conditions, when ROS production has been augmented in one of the cellular compartments, this equation will no longer be accurate.
The nucleus is one of the most prominent cellular organelles and it is continuously exposed to ROS derived from numerous endogenous and exogenous sources. The nuclei of most of the cells are either round or oval. However, this define morphology can be altered under several conditions. For example, several diseases, as well as aging, are associated with alterations in nuclear shape [7]. Nuclear structure is mainly determined by the nuclear matrix, which consists of the peripheral lamins, protein complexes, internal ribonucleic protein network, and residual nucleoli [7,8]. Nuclear shape is usually thought to be modified in response to the changes in nuclear lamina, it can also be altered by forces that act from the cytoplasm [7]. Although it is still not entirely clear how changes in nuclear shape affects its function, it was suggested that it can result in chromatin reorganization and/or affect DNA transcription or replication, and thereby gene expression [7–9]. Beside the effects on a nuclear shape, oxidative stress and modifications of nuclear proteins can also have a great impact on signaling pathways as many transcription factors are redox sensitive, including AP-1, HIF-1, NF-κB, Nrf2, p53, glucocorticoid receptor, and others [3,5,6].
In this study we used saturated and monounsaturated fatty acids (FA; palmitic and oleic) to induce steatosis in hepatocytes and elucidate the effects of FA treatment on the nuclear redox state under basal (only FA) and moderate oxidative stress (FA+CoCl2) conditions. Specifically, we aimed to evaluate the association between alterations in nuclear morphology and oxidative stress by identifying protein adducts with reactive lipid peroxidation products (oxoLPP). Defining these protein targets may provide a novel insights into the pathogenesis of NAFLD.
Material and methods
Cell culture and treatments
HepG2 cells were maintained at 37 °C in 5% CO2 in DMEM medium supplemented with 10% FCS, 2 mM l-glutamine, 1 mM sodium pyruvate, 100 U/mL penicillin and 100 mg/mL streptomycin. FA-loading was performed according to experimental model of in-vitro hepatocellular steatosis [10,11]. Briefly, free FA (2:1, molar ratio, oleic and palmitic acids) were mixed with bovine serum albumin (BSA). Cells were incubated with FA–BSA complex in FCS-free culture medium at 1 mmol/L final concentration of FA and 1% of BSA. Control cell cultures were incubated with medium containing the vehicle. After 24 h of incubation, the medium was replaced with a fresh medium with or without 100 µM of CoCl2 and cells were incubated for 4 h.
Cell viability
Cell viability was measured using Neutral red lysosomal uptake assay as previously described [12].
Evaluation of intracellular fat accumulation and nuclear staining
Cells were stained with DMSO-dissolved Nile-red (1 µg/mL). The fluorescence was measured with excitation at 488 nm and emission at 575 nm by flow cytometry (FACSCalibur, Becton, Dickinson, CA, USA). Intracellular lipid distribution was monitored by fluorescence microscopy following Nile-red staining [13]. Nuclei were stained with Hoechst. The fluorescence intensity for each image was adjusted differently to maximize the quality. Image processing was performed equally and simultaneously to all groups and was carried out using GIMP 1.2 software and/or ImageJ (background subtraction, Fig. 1B).
Fig. 1.
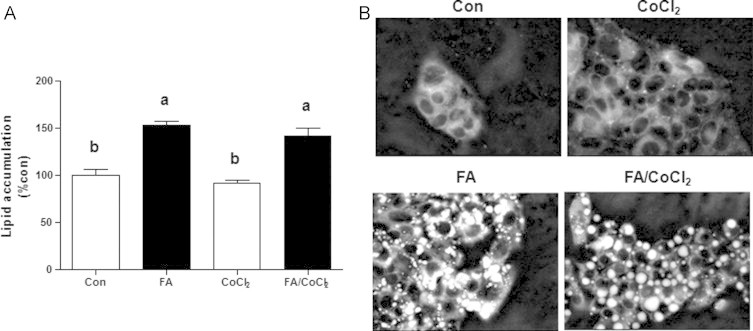
Effects of fatty acids treatment and/or CoCl2 on cellular lipids accumulation in HepG2 cells. Cells were incubated with 1 mM fatty acid mixture (oleate/palmitate, 2/1; black bars) or vehicle in which fatty acids were dissolved (white bars) for 24 h. Afterwards, cells were treated with or without 100 µM CoCl2 for 4 h. (A) Intracellular lipid levels were evaluated quantitatively and qualitatively using Nile red staining and flow cytometry analysis or (B) fluorescence microscopy. Results are means±SE. Means with different letters in each group differ at p<0.05. Con, control; FA, fatty acids.
Fluorescence microscopy of reactive lipid peroxidation products (oxoLPP)
Cells were grown on cover slips and treated as described above. Cells were washed with PBS, fixed (4% paraformaldehyde, 15 min, 37 °C), washed again (PBS), blocked in blocking solution (5% FBS, 0.1% Tween-20 in PBS, 1 h, RT) and incubated with 7-(diethylamino)-coumarin-3-carbohydrazide (CHH; 0.2 mM, 2 h, RT). Cells were washed (PBS; 3 times), nuclei were counterstained with propidium-iodide, washed (PBS), and cover slips were mounted on cover slides using Immunoselect Antifading mounting medium (Dianova GmbH, Hamburg, Germany). Images processing was performed equally and simultaneously to all groups and was carried out using GIMP 1.2 software.
Determination of mitochondrial activity by MTT assay
The MTT assay was used to measure mitochondrial activity as previously described with minor modifications [14]. After exposure to the treatments, the cell culture medium was discarded, and the cells were incubated with 100 µL of MTT solution (0.5 mg/mL in DMEM, 2 h at 37 °C, 5% CO2). The MTT solution was discarded, and 100 µL of DMSO was added to each well. Optical density was read on a microplate reader at 550 nm.
Evaluation ROS
Cells were grown on a flat bottom black 96-well plates (Greiner CELLSTAR®). Following FA-loading, medium was replaced with transparent DMEM containing DCFDA (Sigma Aldrich GmbH, Germany; 10 µmol/L) and incubated (37 °C, 1 h). Cells were washed and incubated in DMEM. After 30 min the medium was exchanged to DMEM with or without CoCl2 and the fluorescence (485/535 nm) was recorded for 5 h on a Paradigm™ Detection Platform (Molecular devices, Salzburg, Austria).
Protein extraction and Western blot analyses
Whole-cell lysates were prepared in RIPA buffer and nuclear extracts were prepared as previously described [15] with slight modifications [16]. In brief, nuclear pellets were resuspended in 3 mL of 0.25 M sucrose containing 10 mM MgCl2, layered over with 3 mL cushion of 0.35 M sucrose containing 0.5 mM MgCl2 and centrifuged at 1400g for 5 min at 4 °C. The resulting nuclear pellets were resuspended in ice-cold buffer containing 20 mM HEPES pH 7.9, 400 mM NaCl, 1 mM each of DTT, EDTA, EGTA and PMSF, placed on a rotatory shaker for 15 min followed by centrifugation at 14,000 rpm for 10 min.
Aliquots of the protein extracts were subjected to the western blot analysis (whole cell extracts in RIPA buffer) or separated by SDS-PAGE (10% T; BioRad mini protean III cell; BioRad Laboratories GmbH, München, Germany) for further in-gel digestion (nuclear-enriched extracts). Western blot analysis was performed using following primary antibodies: phospho-specific (Ser139) and total histone H2AX (Cell signaling Technology) and β-actin (BD Biosciences). Secondary antibodies were obtained from Jackson ImmunoResearch (West Grove, PA). Total and phosphorylated histone H2AX levels were normalized to β-actin. Western blots were developed using chemiluminescence detection and analyzed by densitometry.
CHH labeling and mass spectrometry analysis of carbonylated lipids
For the derivatization of lipid bound carbonyls, cell pellets were mixed with 50 µL of 0.1% aqueous ammonium-acetate and derivatized with 7-(diethylamino)coumarin-3-carbohydrazide (CHH; 3.5 µL of 100 mmol/L, 1 h, 37 °C) [17]. Lipids were extracted using methyl-tert-butyl ether (MTBE) as described previously [18]. Samples were diluted in a mixture of methanol and chloroform (2:1, v/v) containing ammonium formate (5 mmol/L) and analyzed using robotic nanoflow ion source (TriVersaNanoMate; AdvionBioSciences, Ithaca, NY) equipped with nanoelectrospray chip (1.5 kV ionization voltage, 0.4 psi backpressure) coupled to an LTQ Orbitrap XL ETD mass spectrometer (Thermo Fischer Scientific GmbH, Bremen, Germany). The temperature of the transfer capillary was set to 200 °C and the tube lens voltage to 110 V. Mass spectra were acquired with a target mass resolution of 100,000 at m/z 400 in a data-dependent acquisition (DDA) mode using FT-MS survey scan followed by consecutive CID fragmentations of the five most abundant ions in the LTQ using gas phase fractionation. Acquired data were analyzed by using Xcalibur software (version 2.0.7).
nUPLC–ESI–MS of modified proteins
For LC–MS analysis protein bands were cut out from the corresponding gels and digested with trypsin [19]. A nanoACQUITY UPLC (Waters GmbH, Eschborn, Germany) was coupled on-line to an LTQ Orbitrap XL ETD mass spectrometer equipped with a nano-ESI source (Thermo Fischer Scientific, Bremen, Germany). Eluent A was aqueous formic acid (0.1%, v/v) and eluent B was formic acid (0.1%, v/v) in acetonitrile. Tryptic peptides were loaded onto the trap column (nanoACQUITY symmetry C18, internal diameter 180 µm, length 20 mm, particle diameter 5 µm) at a flow rate of 10 µL/min. Peptides were separated on BEH 130 column (C18-phase, internal diameter 75 µm, length 100 mm, particle diameter 1.7 µm) with a flow rate of 0.4 µL/min using two step gradient: up to 30% B in 18 min and then to 85% B in 1 min. Together with an equilibration time of 12 min the samples were injected every 33 min. The transfer capillary temperature was set to 200 °C and tube lens voltage to 120 V. An ion spray voltage of 1.5 kV was applied to a PicoTip™ on-line nano-ESI emitter (New Objective, Berlin, Germany). The CID-tandem mass spectra (isolation width 2, activation Q=0.25, normalized collision energy 35%, activation time 30 ms) were recorded by data dependent acquisition (DDA) for the top six most abundant ions in each survey scan with dynamic exclusion for 60 s using Xcalibur software (Version 2.0.7).
Database search and STRING analysis
The acquired tandem mass spectra were searched against the human proteins database using Sequest search engine (Proteome Discoverer 1.1, Thermo Fischer). The setting allowed up to two missed cleavage sites and a mass tolerance of 10 ppm for precursor and 0.8 u for product ion scans. Each sample was analyzed using a set of variable modifications including propionamide on Cys, oxidation of Met, Cys, His, Trp and four Michael adducts: HNE (delta mass 156,115), HHE (delta mass 114,068), ONE (delta mass 154,099), and OHE (delta mass 112,052) on Cys, His and Lys. In this study only peptides with high and medium confidence, Xcorr value≥1.5, ranked in position 1 in the database search were considered, if the corresponding protein was identified in three technical replicates within each experimental group. Functional protein interaction analysis was performed with Search Tool for the Retrieval of Interacting Genes (STRING), i.e. a database of known and predicted protein interactions [20]. Resulted networks were clustered based on the Markov cluster algorithm (MCL), disconnected nodes were removed and resulted clusters were manually annotated. It is important to note that STRING protein–protein interaction analysis, performed in the current study, relay on the knowledge databases which are summing up reported physical interactions (proteins binding) as well as neighborhood in the genome, gene fusions, co-occurrence across genomes, co-expression, experimental/biochemical data, association in curated databases as well as co-mentioning in PubMed abstracts thus creating a final score which summarize our current knowledge on certain proteins. Clustering of the networks relay on these global STRING scores and bringing together proteins with highest scores related to direct and functional interactions.
Statistical analysis
Data were expressed as mean±SE. For multiple groups, data were analyzed by analysis of variance (ANOVA). Differences were considered significant at probability levels of p<0.05 with the Tukey–Kramer HSD method. Statistical analysis was performed using JMP version 7 (SAS Institute, Cary, NC).
Results
Effects of FA and/or CoCl2 on cellular lipid accumulation
Intracellular lipid accumulation was evaluated quantitatively and qualitatively using Nile red staining. Lipid content increased in HepG2 cells incubated with fatty acids (FA) compared to controls (Fig. 1). Cobalt chloride (CoCl2; 100 µM ) had no lipid accumulative effect in control and FA-loaded hepatocytes.
Effects of FA and/or CoCl2 on cell viability and mitochondrial activity
Mitochondrial activity was evaluated by MTT assay. Although MTT assay is usually used to asses cell viability, it can also serve as a rough estimation of mitochondrial function by providing the information on activity of mitochondrial oxidoreductases. FA treatment significantly enhanced the mitochondrial activity (Fig. 2A). Conversely, CoCl2 had no significant effect on the mitochondrial function both in control and FA-treated cells. Although cell viability was unaffected (Fig. 2B), increased mitochondrial activity in FA-loaded cells was associated with higher ROS levels (Fig. 2C). As expected, exposing cells to CoCl2 for 4 h significantly promoted ROS production both in FA-treated and untreated cells (Fig. 2C).
Fig. 2.
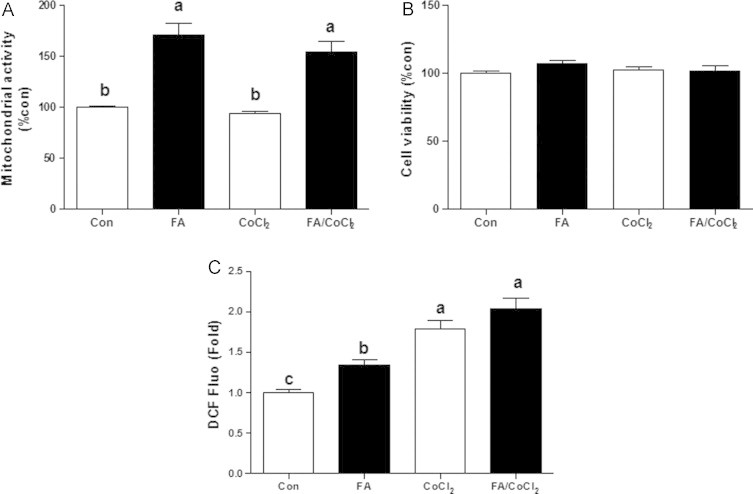
Effects of fatty acids treatment and/or CoCl2 on mitochondrial activity and ROS production. Cells were incubated with 1 mM fatty acid mixture (oleate/palmitate, 2/1; black bars) or vehicle in which fatty acids were dissolved (white bars) for 24 h. Afterwards, cells were treated with or without 100 µM CoCl2 for 4 h. (A) Mitochondrial activity was evaluated using MTT assay and (B) cell viability was assessed using neutral-red staining. (D) ROS levels following 3.5 h of incubation with or without CoCl2 in normal and FA-loaded cells measured by DCFDA assay. Results are means±SE. Means with different letters in each group differ at p<0.05. Con, control; FA, fatty acids.
Effects of FA and/or CoCl2 on nuclear morphology
Nuclear morphology was significantly disrupted in FA-treated cells (Fig. 3A). For some but not all cells the nuclei size was reduced. This reduction might be attributed to an early stage of apoptosis (as cell viability was still unaffected) or, alternatively, the alterations in nuclear matrix. Moreover, some of the cells demonstrated altered nuclear shapes both in FA and FA/CoCl2 treatment groups (Fig. 3A).
Fig. 3.
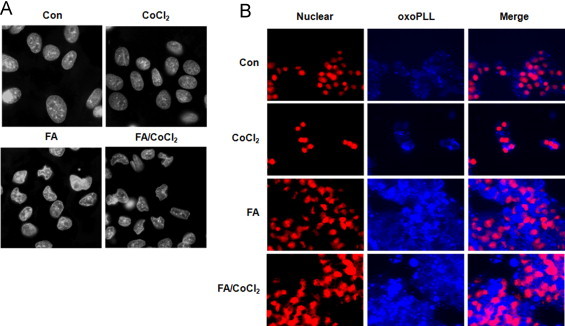
Effects of fatty acids treatment and/or CoCl2 on nuclear morphology and lipid peroxidation products (oxoLPP) formation. Cells were incubated with 1 mM fatty acid mixture (oleate/palmitate; 2/1) or vehicle in which fatty acids were dissolved for 24 h. Afterwards, cells were treated with or without 100 µM CoCl2 for 4 h. (A) Nuclear morphology and (B) oxoLPP spatial distribution were observed using fluorescence microscopy. Con, control; FA, fatty acids.
Given the increase in ROS upon FA treatment and vast accumulation of lipid droplets in cytoplasmic region, the formation of reactive lipid peroxidation products (oxoLPP) was further evaluated using carbonyl-specific chemical probe (CHH) and fluorescence microscopy (Fig. 3B). OxoLPP-specific fluorescence intensity was significantly higher in both FA and FA/CoCl2 treated cells, whereas CoCl2 alone only slightly increased the signal in comparison to control. OxoLPP-specific fluorescence signals were localized in the cytoplasm, including the perinuclear space. Interestingly, intra-nuclear space did not show any significant increase in oxoLPP.
Mass spectrometry (MS) based identification of carbonylated LPP
To further confirm the presence of carbonylated LPP observed by microscopy studies (Fig. 3B), cells were collected, derivatized with CHH and lipids were analyzed by direct infusion ESI-LTQ-Orbitrap MS [17]. Low molecular weight carbonylated LPP were identified by manual data analysis, based on a high mass accuracy of MS survey scan in Orbitrap and CID MS/MS experiments performed in LTQ. 47 Different low molecular weight oxoLPP were identified (Supplementary file S1). Among them were (hydroxy)alkanals, (hydroxy)alkenals, (oxo/hydroxy)alkadienals, (oxo)alkatrienals and corresponding oxo-carboxylic acids. 29, 37, 37 and 31 oxoLPP were identified in Control, FA, CoCl2 and FA/CoCl2 groups, respectively. Fig. 4A exemplify CID tandem mass spectrum of CHH-derivatized 4-hydroxy-nonenal (HNE) which was identified in all four experimental groups.
Fig. 4.
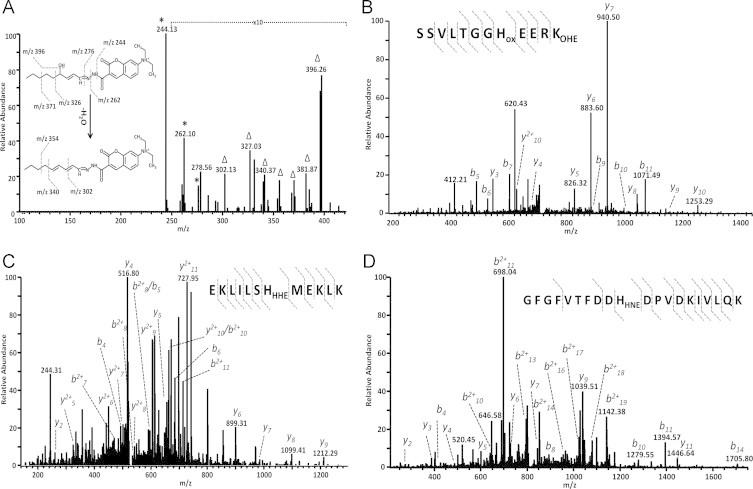
LTQ CID spectra of carbonylated LPP and oxoLPP modified peptides. For detection of oxoLPP cell pellets were collected, derivatized with 7-(diethylamino)coumarin-3-carbohydrazide (CHH), lipids were extracted and analyzed by direct infusion ESI-LTQ-Orbitrap. (A) LTQ CID spectrum of CHH-derivatized 4-hydroxy-nonenal. For identification of oxoLPP-modified proteins, nuclear proteins were separated by SDS-PAGE, in-gel digested with trypsin and analyzed by nUPLC-ESI-LTQ-Orbitrap MS. LTQ CID spectra of peroxisome-proliferator-activated receptor-γ coactivator 1α OHE-modified peptide 361SSVLTGGHoxEERKOHE371 (B), histone acetyltransferases KAT6B HHE-modified peptide 961EKLILSHHHEMEKLK972 (C), and hnRNP A/B HHE-modified peptide 154GFGFVTFDDHHNEDPVDKIVLQK173 (D).
Four new oxoLPP, namely octadienal and oxo-pentanoic, oxo-hexanoic and oxo-octenoic acids, were identified in all three treatment conditions. Interestingly, FA and FA/CoCl2 showed high degree of similarity in terms of new oxoLPP formed. Thus undecenal, undecatrienal, hydroxy-octenal, hydroxy-decadienal, oxo-hexenoic and oxo-nonadienoic acids were identified in both FA and FA/CoCl2 groups but not in control and CoCl2. Other six oxoLPP were unique for CoCl2 group (decatrienal, hydroxy-butanal, hydroxy-nonanal, oxo-nonadienal, oxo-decatrienal, oxo-octatrienoic acid). It is important to note that analytical method used here did not provide any quantitative information and was directed only to identification of low abundant carbonylated LPP.
Effects of FA and/or CoCl2 on histones proteins
Separation of nuclear proteins by SDS-PAGE identified three clearly visible bands with increased intensities in samples obtained from FA-treated cells (with or without CoCl2; Fig. 5A). Analysis of these bands using MS allowed to identify core histone species H2A.1, H2B.1, H3.3 and H4. Although it was not identified by MS, we investigated the levels of histone 2AX (H2AX) protein and its phosphorylated form by Western blot analysis. H2AX is H2A isoform which undergoes phosphorylation upon formation of double strand DNA break, telomere dysfunction, interrupted replication/transcription, virus infection, and apoptosis. Phosphorylated H2AX foci serve as platforms for the recruitment of DNA repair and chromatin remodeling factors as well as factors involved in the cell-cycle checkpoint. Therefore, phosphorylated H2AX is a useful marker of DNA damage [21]. Both the total as well as the phosphorylated H2AX protein levels were enhance by FA treatment (Fig. 5B and C), and the ratio between phosphorylated H2AX to total H2AX was considerably higher in these cells (Fig. 5D). By contrast, CoCl2 had no significant effect on total or H2AX phosphorylation.
Fig. 5.
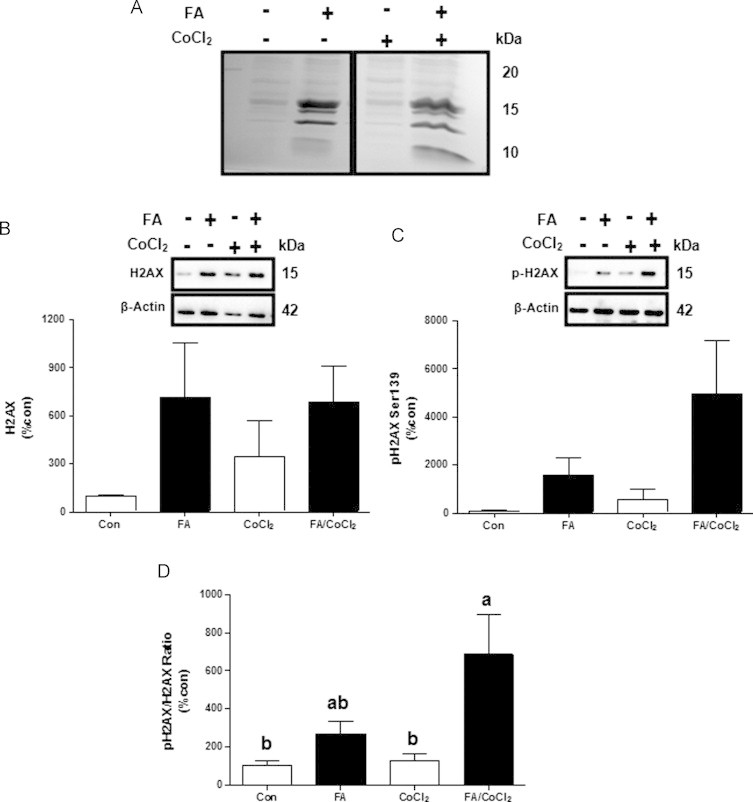
Effects of fatty acids treatment and/or CoCl2 on histone H2AX. Cells were incubated with 1 mM fatty acid mixture (oleate/palmitate, 2/1; black bars) or vehicle in which fatty acids were dissolved (white bars) for 24 h. Afterwards, cells were treated with or without 100 µM CoCl2 for 4 h. (A) Representative SDS PAGE gels and (B) total and (C) phosphorylation status of histone 2AX (H2AX) at Ser139. (D) H2AX total to phosphorylated ratio. Results are means±SE. Means with different letters in each group differ at p<0.05. Con, control; FA, fatty acids.
Mass spectrometry based identification of modified proteins
To identify nuclear proteins modified by reactive oxoLPP, untreated cells and cells treated with FA, CoCl2 or combination of both were collected and nuclear proteins were enriched. To reduce sample complexity, proteins were separated by SDS-PAGE, each line was divided into several equal parts, cut out, subjected to in-gel digestion and analyzed by LC–MS/MS. It allowed identification of modified proteins without additional enrichment. To evaluate protein modifications, four α,β-unsaturated LPP, namely hydroxy- and oxo-nonenal (HNE and ONE) and hydroxy- and oxo-hexenal (HHE and OHE), were used as variable modifications for database search. These oxoLPP are generally known as representative markers of n-6 (HNE and ONE) and n-3 (HHE and OHE) polyunsaturated fatty acids (PUFA) oxidation and were shown to play a role in NAFLD and alcoholic liver disease pathogenesis [22–25].
Overall 751 unique modified proteins present in three experimental replicates were identified (Fig. S1; Supplementary file S2). FA-treated group showed the highest number of total LPP-modified (332) as well as unique modified proteins (235). Number of modified protein in CoCl2 and FA/CoCl2 treated cells were similar both in terms of total and unique protein identities (227 and 229 total, 140 and 146 unique, respectively). Finally, 182 in total including 99 unique modified proteins were detected in untreated hepatocytes.
11 Proteins were found to be modified by oxoLPP in all four experimental groups (Supplementary file S2). Among them actin, chromodomain-helicase-DNA-binding protein 4, heterogeneous nuclear ribonucleoprotein L, neuroblastoma differentiation-associated protein AHNAK were also shown to be modified by HNE in several other studies [23,26]. Interestingly, several isoforms of heterogeneous nuclear ribonucleoproteins (hnRNPs), a component of spliceosome complex, were identified in the current study including hnRNP L in all four groups, hnRNP D0, H2 and U in FA, hnRNP H2 in CoCl2 and hnRNP A/B in FA/CoCl2 group. Modified hnRNPs A/B and D0, identified upon FA and FA/CoCl2 treatment, were shown previously as LPP targets in murine model of early alcoholic liver disease [23]. Overall, different isoforms of hnRNPs were detected among targets of protein carbonylation in several models of oxidative stress using different cell types and conditions [27,28]. The mechanism of such high susceptibility of hnRNPs to LPP modifications is not clear and needs to be addressed in more details. However, such ubiquitous identification of carbonylated hnRNPs demonstrates non-random nature of protein carbonylation.
STING analysis of LPP-modified proteins
To evaluate possible enrichment of modified proteins in certain biological pathways, systems biology analysis was performed using STRING knowledge database of protein–protein interactions (Figs. 6 and S2–S4). Despite a similar size of imported protein sets (182, 332, 227 and 229 protein entries for control, FA, CoCl2 and FA/CoCl2 groups, respectively), network clustering indicated significant differences in functional protein enrichment. Thus, the highest cluster number and density were observed for modified proteins from FA group (Fig. 6). Based on the manual revision of obtained network, several distinct functional groups were defined (Fig. 6, Table 1), including cytoskeletal–plasma membrane–extracellular matrix proteins (40 proteins), nuclear pore organization (3), cytoskeletal proteins involved in cell division (22), regulation of transcription (18), splicing (12), protein synthesis and degradation (25), DNA repair (9) and regulation of lipid metabolism (5). For instance, among modified proteins involved in regulation of lipid metabolism acyl-CoA thioreductase 2, peroxisome proliferator-activated receptor α (PPARα), and peroxisome-proliferator-activated receptor-γ coactivator 1α (PGC-1α; tandem mass spectrum of OHE-modified peptide 361SSVLTGGHoxEERKOHE371 exemplified in Fig. 4B) were identified. Interestingly, it was previously shown by us that PGC-1α mRNA level increased in animal model of NAFLD, however the protein level and thus the activity of PGC-1α were reduced [29]. Thus, PGC-1α modification by LPP might suggest an additional regulatory mechanism of its activity.
Fig. 6.
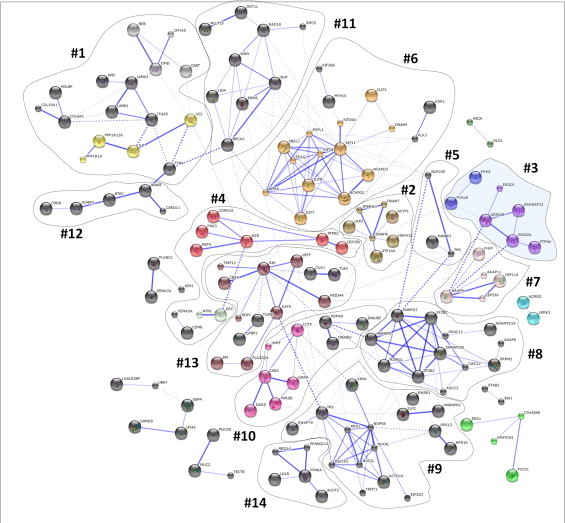
STRIG protein–protein interaction and clustering analysis of oxoLPP modified proteins identified in fatty acids treated HepG2 cells. 332 oxoLPP modified proteins identified in FA group by LC–MS analysis were uploaded to STRIG database of protein–protein interaction. Resulted network was clustered based on the Markov cluster algorithm (MCL), disconnected nodes were removed and resulted clusters were manually annotated. Corresponding cluster numbering legend is provided in Table 1.
Table 1.
Functional protein–protein interaction clusters of oxoLPP-modified proteins, identified in untreated cells and cells treated with FA, CoCl2 and a combination of both, assigned by STRING analysis and manual annotation. Cluster numbers corresponds to the one provided in Figs. 6 and S2–S4. Number of proteins involved in a cluster shown in [brackets]. PM—plasma membrane; C—cytoskeleton; ECM—extracellular matrix.
| Control | FA | CoCl2 | FA/CoCl2 |
|---|---|---|---|
| I. Cytoskeleton–plasma membrane–ECM (C–PM–ECM) | |||
| 1. C–PM–ECM architecture [16] | 1. C–PM architecture [7] | 2. C–PM architecture [7] | |
| 2. PM associated ATPases [7] | 2. C–PM architecture [11] | ||
| 3. C–PM receptors signaling [7] | |||
| 4. C–PM signaling [10] | |||
| II. Plasma membrane and secreted proteins | |||
| 5. PM and secreted proteins [5] | 3. PM and secreted proteins [14] | ||
| 4. Proteoglycan synthesis [3] | |||
| III. Cytoskeleton related transport and motility | |||
| 7. Cytoskeletal transport [8] | 9. Dynein related motility [3] | ||
| 8. Cytoskeletal transport [6] | |||
| IV. Nuclear pore organization | |||
| 5. Nuclear pore and transport [3] | |||
| V. Cytoskeletal proteins involved in cell division | |||
| 6. Cell division, chromosome segregation [17] | |||
| 7. Centrosome organization [5] | |||
| VI. Signaling | |||
| 3. Cytoskeleton mediated signaling [6] | 3. Inositol-phosphate related [3] | 1. PM-cytoskeleton signal transduction [18] | |
| #4. Signaling [3] | |||
| VII. Regulation of transcription | |||
| 12. NFκB related transcription [5] | |||
| 13. Regulation of transcription [13] | |||
| VIII. Chromatin remodeling | |||
| 1. Chromatin remodeling [4] | 6. Chromatin remodeling and DNA replication [15] | ||
| IX. Splicing | |||
| 8. Regulation of transcription and splicing [12] | 6. Splicing [3] | 7. Splicing [7] | |
| X. Protein synthesis and degradation | |||
| 9. Ribosome biogenesis and regulation [16] | |||
| 10. Protein synthesis and folding [9] | 5. Protein synthesis and folding [7] | ||
| 2. Proteosome [3] | |||
| XI. DNA repair | |||
| 11. DNA repair [9] | |||
| XII. Regulation of lipid metabolism | |||
| 14. Regulation of lipid metabolism [5] | 8. Regulation of lipid metabolism [5] | ||
30 Different proteins involved in regulation of transcription and splicing were shown to be modified. Among modified proteins involved in regulation of transcription were IκB kinase β, tumor suppressor Rb1 (retinoblastoma-associated protein), two members of E2F family of transcriptional factors (E2F7 and E2F8, acting as transcription suppressors), and serine–threonine kinase ATM. Within proteins involved in splicing events four hnRNPs proteins mentioned above, namely hnRNPs L, D, H2 and U, were shown to be modified as well as two subunits of splicing factor 3b. Although proteins involved in regulation of splicing were found to be modified in CoCl2 (3 proteins) and FA/CoCl2 groups (7 proteins) as well, the number of modified proteins was significantly higher in FA group.
Interestingly, modified proteins from several function groups were identified exclusively in FA group. Those included three proteins of nuclear pore organization, namely two nucleoporins (Nup250 and Nup358) and nucleoprotein TPR. Furthermore, cytoskeletal proteins involved in cell division were show only for FA group, including several members of kinesin family and subunits of condensin complex. Additionally, modified proteins involved in DNA repair, such as DNA mismatch repair protein Msh6, post-replication repair protein RAD18 and DNA ligase IV, were exclusively identified. Interestingly, DNA damage and activation of DNA repair upon FA and FA/CoCl2 treatment was confirmed by western blot analysis of H2AX phosphorylation (Fig. 5). However, among LPP modified proteins in FA group only linker histone H1.3 was found to be modified by HHE in three experimental replicates. In FA/CoCl2 group several other histones were identified to form adducts with LPP, including core histones H2A.2 and H2B.1, and linker histones H1.5 and H1x.
FA/CoCl2 group showed the second highest functional enrichment of LPP modified proteins (Fig. S2). In total STRING analysis and MCL clustering allowed to define eight functional clusters which were further manually annotated. These clusters included cytoskeletal–plasma membrane proteins (7 proteins), secreted proteins (14) and proteins involved in proteoglycan synthesis (3 proteins), dynein related motility (3), signal transduction (18), lipid metabolism (5), chromatin remodeling and DNA replication (15), splicing (7) and protein synthesis and folding (7). In addition to histone proteins discussed above, among modified proteins involved in chromatin remodeling and DNA replication were N-lysine methyltransferase SETD8 and histone acetyltransferases KAT6B (tandem mass spectrum of HHE-modified peptide 961EKLILSHHHEMEKLK972 exemplified in Fig. 4C) and KAT8, which might indicate interference of LPP protein adducts with histone methylation and acetylation pathways of epigenetic regulation of gene transcription. Among lipid metabolism associated proteins PGC-1α, carnitine palmitoyltransferase, retinoid X receptor and phospholipase A2 were identified. Seven modified proteins within FA/CoCl2 group were related to mRNA splicing including hnRNP A/B (tandem mass spectrum of HHE-modified peptide 154GFGFVTFDDHHNEDPVDKIVLQK173 exemplified on Fig. 4D) and L, small nuclear ribonucleoprotein snRNP D2, and splicing factor Prp8. In contrast to the FA group, in which most of the annotated clusters were related to nuclear proteins (8 clusters out of 14), in FA/CoCl2 group only two clusters (chromatin remodeling/DNA replication, and splicing) corresponded to the nuclear proteome. Many of modified proteins were associated with cytoskeleton. Thus, proteins involved in cytoskeleton interaction with plasma membrane, dynein mobility and signal transduction from plasma membrane to cytoskeleton were enriched. Additionally, 14 plasma membrane associated and secreted proteins were clustered together as well as three proteins involved in proteoglycan synthesis.
For CoCl2-treated cells protein–protein interaction analysis revealed only few defined clusters with low number of proteins involved (Fig. S3) despite similar to FA/CoCl2 group number of entries subjected to STRING analysis (227 and 229 protein entries for CoCl2 and Fat/CoCl2 groups, respectively). Among them only three nuclear proteins involved in splicing were enriched by functional interaction analysis, whereas the rest contained proteins involved in organization of cytoskeleton–plasma membrane architecture (18 proteins), cytoskeleton transport (14 proteins), signaling (6 proteins) or plasma membrane associated and secreted proteins (5 proteins).
Even lower number of functional interaction clusters was shown for control group (Fig. S4). It was possible to define only three distinct clusters involved in chromatin remodeling (4 proteins), related to proteasomal protein degradation (3 proteins) and cytoskeleton mediated signaling (6 proteins).
Discussion
Excessive accumulation of lipid droplets in hepatocytes is a common feature of several diseases, including NAFLD [25]. Although liver steatosis is currently considered benign, fatty livers are particularly more susceptible to variety of insults. Indeed, excessive lipid accumulation was found to predispose hepatocytes to hepatocellular injury, which may lead to the progression of simple steatosis to NASH and fibrosis [30]. Hepatocellular injury can be caused by external as well as internal factors, including cytotoxic effects of free FA excess, oxidative stress and lipid peroxidation [31]. By using an in-vitro model of hepatic steatosis, this study illustrates that steatosis per-se, without the addition of external stimuli, induces nuclear alterations while not affecting cell viability. Data presented here suggests FA-induced nuclear oxidative/electrophilic stress as an important contributor to NAFLD progression.
The exact mechanism by which lipid accumulation induces nuclear stress is not entirely comprehended. Nevertheless, it seems that lipid accumulation within hepatocytes might promote mitochondrial FA oxidation as a compensatory mechanism which subsequently enhances ROS generation. Furthermore, it was recently demonstrated that palmitic acid overload can induce mitophagy deficiency resulting in increased ROS production by damaged mitochondria [32]. Taking into account unchanged cell viability, enhanced mitochondrial activity accompanied by increase in ROS production, we can speculate that FA treatment had a similar effect on hepatocytes. It is important to note, that mild oxidative stressor CoCl2 itself did not induce mitochondrial activity, although significantly increased ROS production.
Increased ROS production resulted in the vast lipid peroxidation and the production of biogenic reactive oxoLPP, as was monitored by CHH labeling and fluorescence microscopy. OxoLPP, formed in close proximity to nuclear membrane, can further affect the nuclear compartment and induce electrophilic stress in this organelle. Additionally, formation of a large number of low molecular weight oxoLPP was confirmed by MS using derivatization technique specific for carbonylated lipids [17]. Interestingly, high similarity of generated oxoLPP was shown for both FA-treated cells (FA and FA/CoCl2 groups), whereas CoCl2 treated group showed specific set of new oxoLPP formation. Lipid peroxidation is one of the main events induced by oxidative stress and it can be particularly damaging to the liver nuclear membrane rich in PUFA [33]. Despite the fact that liver can metabolize reactive oxoLPP faster than most of the other tissues [34], numerous publications indicated high level of modifications of liver and hepatocyte proteins by reactive oxoLPP, and HNE in particular [35–37].
Indeed, electrophilic stress resulted in a high number of proteins modified by reactive oxoLPP in FA-treated cells. Results of this work are in agreement with earlier studies demonstrating the reactivity of oxoLPP towards nuclear biomolecules in hepatocytes [33,38,39] and thus further suggest the importance of these alterations in NAFLD progression. LC–MS allowed to identify over 300 proteins modified by oxoLPP in FA-treated cells and 751 protein modified in four experimental groups. By comparing the list of modified proteins with published data, several dozen of proteins were already shown as carbonylation targets. Thus, 30 proteins identified here were identical to the LPP-modified proteins extracted from the liver of mice with early alcoholic liver disease [23]. 59 and 23 identical proteins were present in the study on RKO cells treated with LPP [26] and HeLa cells treated with hydrogen peroxide [40]. Interestingly, 34 carbonylated proteins were already identified in the human plasma samples from obese patients with and without type II diabetes [41]. These similarities might illustrate the specificity of certain protein as carbonylation targets. However, in the current study enriched nuclear protein fractions were used which allowed to get deeper coverage of oxoLPP modified nuclear proteome in hepatocytes for the first time, to the best of our knowledge.
Our study demonstrated specificity of protein targets in FA-treated cells. Proteins involved in nuclear pore organization, chromosome segregation, cell cycle control, centrosome organization, regulation of transcription and splicing, ribosomal biogenesis and DNA repair were specifically modified by oxoLPP. Clear enrichment of modified proteins involved in nuclear riboprotein complexes, chromatin remodeling, including histones and histone methyl- and actyltransferases, as well as transcription and splicing can provide the link between lipid accumulation, oxidative stress and changes in protein expression levels as well as protein functional activities. Our study indicated significant increase in phosphorylation level of histone H2AX in FA-treated cells indicating possible malfunction of replication/transcription and/or activation of DNA repair mechanisms. It was recently demonstrated that treatment of RKO cells with HNE and ONE resulted in multiple adducts of oxoLPP with histones H2B, H2, H3 and H4. Furthermore, modifications of H3 and H4 resulted in disruption of nucleosome formation, which may challenge chromatin dynamics and histone turnover [42]. Furthermore, modifications of lysines residues by LPP might hamper epigenetic regulation via acetylation and methylation on this residues. Interestingly, several key players of epigenetic regulation such as N-lysine methyltransferase SETD8 and histone acetyltransferases KAT6B and KAT8 were shown among modified proteins.
Additionally we demonstrated that nuclear shape seems to be modified due to a mechanical pressure of the lipid droplets in FA-treated cells. However, it is also possible that oxidative changes in structural proteins resulted in this outcome. Changes in nuclear shape were suggested to influence chromatin organization and gene expression and are associated with the development and progression of several pathological conditions [7–9]. We demonstrated that several main proteins involved in a nuclear membrane architecture were modified by reaction with oxoLPP, including prelamin A/B and nesprin-1 and 2. These proteins directly determine the shape, mechanical stiffness and extensibility of nuclear membrane [43]. Additionally, several components of nuclear pore complex, such as nucleoporins, were found to be modified.
The effect of moderate external stressor in control or FA-treated cells was evaluated by using CoCl2 at relatively low concentrations. Surprisingly, the addition of CoCl2 did not significantly influence level of nuclear protein modifications despite significant increase in ROS generation. Thus, the current study demonstrated that FA-treatment alone without additional inducers of oxidative stress, can result in ROS generation, most probably via defective mitophagy. Enhanced generation of ROS resulted in the vast lipid peroxidation and production of reactive oxoLPP capable to modify nuclear proteins. Such electrophilic stress resulted in the modifications of numerous proteins involved in nuclear pore organization, chromosome segregation, cell cycle control, centrosome organization, regulation of transcription and splicing, ribosomal biogenesis and DNA repair. The loss of functional activity and structural integrity of nuclear proteome may provide the link between lipid accumulation and NAFLD progression.
Conflict of interest
None.
Acknowledgment
Financial support from the European Fund for Regional Structure Development (EFRE, European Union and Free State Saxony; 100146238 and 100121468 to M.F), and COST Action CM1001 are gratefully acknowledged. We thank Prof. Ralf Hoffmann (Institute of Bioanalytical Chemistry, University of Leipzig) for providing access to laboratory space and ESI-LTQ-Orbitrap mass spectrometer.
Contributor Information
Oren Tirosh, Email: oren.tirosh@mail.huji.ac.il.
Maria Fedorova, Email: maria.fedorova@bbz.uni-leipzig.de.
Appendix. Supplementary material
The Supplementary data associated with this article can be found online version at 10.1016/j.redox.2014.12.009.
Appendix. Supplementary material
Supplementary material
Supplementary file S1
Supplementary file S2
References
- 1.Feldstein A.E., Bailey S.M. Emerging role of redox dysregulation in alcoholic and nonalcoholic fatty liver disease. Antioxidants & Redox Signaling. 2011;15(2):421–424. doi: 10.1089/ars.2011.3897. 21254858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hansen J.M., Go Y.M., Jones D.P. Nuclear and mitochondrial compartmentation of oxidative stress and redox signaling. Annual Review of Pharmacology and Toxicology. 2006;46:215–234. doi: 10.1146/annurev.pharmtox.46.120604.141122. 16402904 [DOI] [PubMed] [Google Scholar]
- 3.Lukosz M., Jakob S., Büchner N., Zschauer T.C., Altschmied J., Haendeler J. Nuclear redox signaling. Antioxidants & Redox Signaling. 2010;12(6):713–742. doi: 10.1089/ars.2009.2609. 19737086 [DOI] [PubMed] [Google Scholar]
- 4.Strand J.M., Scheffler K., Bjørås M., Eide L. The distribution of DNA damage is defined by region-specific susceptibility to DNA damage formation rather than repair differences. DNA Repair. 2014;18:44–51. doi: 10.1016/j.dnarep.2014.03.003. 24685126 [DOI] [PubMed] [Google Scholar]
- 5.Shlomai J. Redox control of protein–DNA interactions: from molecular mechanisms to significance in signal transduction, gene expression, and DNA replication. Antioxidants & Redox Signaling. 2010;13(9):1429–1476. doi: 10.1089/ars.2009.3029. 20446770 [DOI] [PubMed] [Google Scholar]
- 6.Go Y.M., Jones D.P. Redox control systems in the nucleus: mechanisms and functions. Antioxidants & Redox Signaling. 2010;13(4):489–509. doi: 10.1089/ars.2009.3021. 20210649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Webster M., Witkin K.L., Cohen-Fix O. Sizing up the nucleus: nuclear shape, size and nuclear-envelope assembly. Journal of Cell Science. 2009;122(10):1477–1486. doi: 10.1242/jcs.037333. 19420234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brünagel G., Schoen R.E., Bauer A.J., Vietmeier B.N., Getzenberg R.H. Nuclear matrix protein alterations associated with colon cancer metastasis to the liver. Clinical Cancer Research. 2002;8(10):3039–3045. 12374670 [PubMed] [Google Scholar]
- 9.Rowat A.C., Lammerding J., Herrmann H., Aebi U. Towards an integrated understanding of the structure and mechanics of the cell nucleus. BioEssays News and Reviews in Molecular, Cellular and Developmental Biology. 2008;30(3):226–236. doi: 10.1002/bies.20720. 18293361 [DOI] [PubMed] [Google Scholar]
- 10.Gómez-Lechón M.J., Donato M.T., Martínez-Romero A., Jiménez N., Castell J.V., O’Connor J.E. A human hepatocellular in vitro model to investigate steatosis. Chemico-Biological Interactions. 2007;165(2):106–116. doi: 10.1016/j.cbi.2006.11.004. 17188672 [DOI] [PubMed] [Google Scholar]
- 11.Gunduz F., Aboulnasr F.M., Chandra P.K., Hazari S., Poat B., Baker D.P., Balart L.A., Dash S. Free fatty acids induce ER stress and block antiviral activity of interferon alpha against hepatitis C virus in cell culture. Virology Journal. 2012;9:143. doi: 10.1186/1743-422X-9-143. 22863531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Repetto G., del Peso A., Zurita J.L. Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nature Protocols. 2008;3(7):1125–1131. doi: 10.1038/nprot.2008.75. 18600217 [DOI] [PubMed] [Google Scholar]
- 13.Anavi S., Harmelin N.B., Madar Z., Tirosh O. Oxidative stress impairs HIF1alpha activation: a novel mechanism for increased vulnerability of steatotic hepatocytes to hypoxic stress. Free Radical Biology and Medicine. 2012;52(9):1531–1542. doi: 10.1016/j.freeradbiomed.2012.02.014. 22343340 [DOI] [PubMed] [Google Scholar]
- 14.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. Journal of Immunological Methods. 1983;65(1–2):55–63. doi: 10.1016/0022-1759(83)90303-4. 6606682 [DOI] [PubMed] [Google Scholar]
- 15.Lahiri D.K., Ge Y. Electrophoretic mobility shift assay for the detection of specific DNA–protein complex in nuclear extracts from the cultured cells and frozen autopsy human brain tissue. Brain Research Brain Research Protocols. 2000;5(3):257–265. doi: 10.1016/s1385-299x(00)00021-0. 10906491 [DOI] [PubMed] [Google Scholar]
- 16.Vascotto C., Fantini D., Romanello M., Cesaratto L., Deganuto M., Leonardi A., Radicella J.P., Kelley M.R., D’Ambrosio C., Scaloni A., Quadrifoglio F., Tell G. APE1/Ref-1 interacts with NPM1 within nucleoli and plays a role in the rRNA quality control process. Molecular and Cellular Biology. 2009;29(7):1834–1854. doi: 10.1128/MCB.01337-08. 19188445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milic I., Hoffmann R., Fedorova M. Simultaneous detection of low and high molecular weight carbonylated compounds derived from lipid peroxidation by electrospray ionization-tandem mass spectrometry. Analytical Chemistry. 2013;85(1):156–162. doi: 10.1021/ac302356z. 23186270 [DOI] [PubMed] [Google Scholar]
- 18.Matyash V., Liebisch G., Kurzchalia T.V., Shevchenko A., Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. Journal of Lipid Research. 2008;49(5):1137–1146. doi: 10.1194/jlr.D700041-JLR200. 18281723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shevchenko A., Jensen O.N., Podtelejnikov A.V., Sagliocco F., Wilm M., Vorm O., Mortensen P., Shevchenko A., Boucherie H., Mann M. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proceedings of the National Academy of Sciences of the United States of America A. 1996;93(25):14440–14445. doi: 10.1073/pnas.93.25.14440. 8962070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snel B., Lehmann G., Bork P., Huynen M.A. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Research. 2000;28(18):3442–3444. doi: 10.1093/nar/28.18.3442. 10982861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma A., Singh K., Almasan A. Histone H2AX phosphorylation: a marker for DNA damage. Methods in Molecular Biology. 2012;920:613–626. doi: 10.1007/978-1-61779-998-3_40. 22941631 [DOI] [PubMed] [Google Scholar]
- 22.Alkhouri N., Berk M., Yerian L., Lopez R., Chung Y.M., Zhang R., McIntyre T.M., Feldstein A.E., Hazen S.L. OxNASH score correlates with histologic features and severity of nonalcoholic fatty liver disease. Digestive Diseases and Sciences. 2014;59(7):1617–1624. doi: 10.1007/s10620-014-3031-8. 24464211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galligan J.J., Smathers R.L., Fritz K.S., Epperson L.E., Hunter L.E., Petersen D.R. Protein carbonylation in a murine model for early alcoholic liver disease. Chemical Research in Toxicology. 2012;25(5):1012–1021. doi: 10.1021/tx300002q. 22502949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dou X., Li S., Wang Z., Gu D., Shen C., Yao T., Song Z. Inhibition of NF-κB activation by 4-hydroxynonenal contributes to liver injury in a mouse model of alcoholic liver disease. American Journal of Pathology. 2012;181(5):1702–1710. doi: 10.1016/j.ajpath.2012.08.004. 22982442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart B.J., Roede J.R., Doorn J.A., Petersen D.R. Lipid aldehyde-mediated cross-linking of apolipoprotein B-100 inhibits secretion from HepG2 cells. Biochimica et Biophysica Acta. 2009;1791(8):772–780. doi: 10.1016/j.bbalip.2009.04.004. 19393338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vila A., Tallman K.A., Jacobs A.T., Liebler D.C., Porter N.A., Marnett L.J. Identification of protein targets of 4-hydroxynonenal using click chemistry for ex vivo biotinylation of azido and alkynyl derivatives. Chemical Research in Toxicology. 2008;21(2):432–444. doi: 10.1021/tx700347w. 18232660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanito M., Haniu H., Elliott M.H., Singh A.K., Matsumoto H., Anderson R.E. Identification of 4-hydroxynonenal-modified retinal proteins induced by photooxidative stress prior to retinal degeneration. Free Radical Biology and Medicine. 2006;41(12):1847–1859. doi: 10.1016/j.freeradbiomed.2006.09.012. 17157187 [DOI] [PubMed] [Google Scholar]
- 28.Baglo Y., Sousa M.M., Slupphaug G., Hagen L., Håvåg S., Helander L., Zub K.A., Krokan H.E., Gederaas O.A. Photodynamic therapy with hexyl aminolevulinate induces carbonylation, posttranslational modifications and changed expression of proteins in cell survival and cell death pathways. Photochemical & Photobiological Sciences. 2011;10(7):1137–1145. doi: 10.1039/c0pp00369g. 21448498 [DOI] [PubMed] [Google Scholar]
- 29.Aharoni-Simon M., Hann-Obercyger M., Pen S., Madar Z., Tirosh O. Fatty liver is associated with impaired activity of PPARgamma-coactivator 1alpha (PGC1alpha) and mitochondrial biogenesis in mice. Laboratory Investigation. 2011;91(7):1018–1028. doi: 10.1038/labinvest.2011.55. 21464822 [DOI] [PubMed] [Google Scholar]
- 30.Yilmaz Y. Review article: is non-alcoholic fatty liver disease a spectrum, or are steatosis and non-alcoholic steatohepatitis distinct conditions? Alimentary Pharmacology & Therapeutics. 2012;36(9):815–823. doi: 10.1111/apt.12046. 22966992 [DOI] [PubMed] [Google Scholar]
- 31.El-Zayadi A.R. Hepatic steatosis: a benign disease or a silent killer. World Journal of Gastroenterology. 2008;14(26):4120–4126. doi: 10.3748/wjg.14.4120. 18636654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang S., Xia C., Li S., Du L., Zhang L., Zhou R. Defective mitophagy driven by dysregulation of rheb and KIF5B contributes to mitochondrial reactive oxygen species (ROS)-induced nod-like receptor 3 (NLRP3) dependent proinflammatory response and aggravates lipotoxicity. Redox Biology. 2014;3:63–71. doi: 10.1016/j.redox.2014.04.001. 25462067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marmunti M., Gavazza M., Catalá A. Non-enzymatic and enzymatic lipid peroxidation of microsomes and nuclei obtained from rat liver. Molecular and Cellular Biochemistry. 2004;265(1–2):1–9. doi: 10.1023/b:mcbi.0000044302.59193.00. 15543928 [DOI] [PubMed] [Google Scholar]
- 34.Siems W.G., Zollner H., Grune T., Esterbauer H. Metabolic fate of 4-hydroxynonenal in hepatocytes: 1,4-dihydroxynonene is not the main product. Journal of Lipid Research. 1997;38(3):612–622. 9101442 [PubMed] [Google Scholar]
- 35.Shearn C.T., Backos D.S., Orlicky D.J., Smathers-McCullough R.L., Petersen D.R. Identification of 5′ AMP-activated kinase as a target of reactive aldehydes during chronic ingestion of high concentrations of ethanol. Journal of Biological Chemistry. 2014;289(22):15449–15462. doi: 10.1074/jbc.M113.543942. 24722988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaudhary P., Sharma R., Sahu M., Vishwanatha J.K., Awasthi S., Awasthi Y.C. 4-hydroxynonenal induces G2/M phase cell cycle arrest by activation of the ataxia telangiectasia mutated and Rad3-related protein (ATR)/checkpoint kinase 1 (Chk1) signaling pathway. Journal of Biological Chemistry. 2013;288(28):20532–20546. doi: 10.1074/jbc.M113.467662. 23733185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shearn C.T., Fritz K.S., Reigan P., Petersen D.R. Modification of Akt2 by 4-hydroxynonenal inhibits insulin-dependent Akt signaling in HepG2 cells. Biochemistry. 2011;50(19):3984–3996. doi: 10.1021/bi200029w. 21438592 [DOI] [PubMed] [Google Scholar]
- 38.Vaca C.E., Harms-Ringdahl M. Interaction of lipid peroxidation products with nuclear macromolecules. Biochimica et Biophysica Acta. 1989;1001(1):35–43. doi: 10.1016/0005-2760(89)90304-4. 2912492 [DOI] [PubMed] [Google Scholar]
- 39.Cao E.H., Liu X.Q., Wang L.G., Wang J.J., Xu N.F. Evidence that lipid peroxidation products bind to DNA in liver cells. Biochimica et Biophysica Acta. 1995;1259(2):187–191. doi: 10.1016/0005-2760(95)00162-6. 7488640 [DOI] [PubMed] [Google Scholar]
- 40.Bollineni R.C., Hoffmann R., Fedorova M. Proteome-wide profiling of carbonylated proteins and carbonylation sites in HeLa cells under mild oxidative stress conditions. Free Radical Biology and Medicine. 2014;68:186–195. doi: 10.1016/j.freeradbiomed.2013.11.030. 24321318 [DOI] [PubMed] [Google Scholar]
- 41.Bollineni R.C., Fedorova M., Blüher M., Hoffmann R. Carbonylated plasma proteins as potential biomarkers of obesity induced type 2 diabetes mellitus. Journal of Proteome Research. 2014;13:5081–5093. doi: 10.1021/pr500324y. 25010493 [DOI] [PubMed] [Google Scholar]
- 42.Galligan J.J., Rose K.L., Beavers W.N., Hill S., Tallman K.A., Tansey W.P., Marnett L.J. Stable histone adduction by 4-oxo-2-nonenal: a potential link between oxidative stress and epigenetics. Journal of the American Chemical Society. 2014;136(34):11864–11866. doi: 10.1021/ja503604t. 25099620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson K.L., Berk J.M. The nuclear envelope at a glance. Journal of Cell Science. 2010;123(12):1973–1978. doi: 10.1242/jcs.019042. 20519579 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary file S1
Supplementary file S2


