Abstract
Patient-derived human-in-mouse xenograft models of breast cancer (PDX models) that exhibit spontaneous lung metastases offer a potentially powerful model of cancer metastasis. In this study, we evaluated the malignant character of lung micro-metastases that emerge in such models after orthotopic implantation of human breast tumor cells into the mouse mammary fat pad. Interestingly, relative to the parental primary breast tumors, the lung metastasis (met)-derived mammary tumors exhibited a slower growth rate and a reduced metastatic potential with a more differentiated epithelial status. Epigenetic correlates were determined by gene array analyses. Lung met-derived tumors displayed differential expression of negative regulators of cell proliferation and metabolism and positive regulators of mammary epithelial differentiation. Clinically, this signature correlated with breast tumor subtypes. We identified microRNA-138 as a novel regulator of invasion and epithelial-mesenchymal transition in breast cancer cells, acting by directly targeting the polycomb epigenetic regulator EZH2. Mechanistic investigations showed that GATA3 transcriptionally controlled miR-138 levels in lung metastases. Notably, the miR-138 activity signature served as a novel independent prognostic marker for patient survival beyond traditional pathologic variables, intrinsic subtypes or a proliferation gene signature. Our results highlight the loss of malignant character in some lung micro-metastatic lesions and the epigenetic regulation of this phenotype.
Keywords: breast cancer, pulmonary metastasis, microRNA, EZH2, malignancy
Introduction
Metastasis can occur at the very early stages of cancer disease, especially in the form of micro-metastatic lesions (1,2). The multi-step process of metastasis comprises tumor cell invasion to surrounding tissue, intravasation to vascular or lymphatic structures, survival in circulation, homing and extravasation to distant organs, and colonization with distant tumor growth (3). An open debate in cancer research focuses on whether the metastatic tumor cells in distant organs, such as the lungs, possess a more metastatic potential than the primary tumor cells. Another important related question is what morphological and epigenetic features may be dynamically acquired or selected within spontaneous metastases.
By tail vein injection and in vivo selection of MDA-MB-231 breast cancer cells in the lungs, Minn et al. isolated lung-tropic LM1 and LM2 cells with increased lung metastatic capacity upon tail vein injection (4). However, this blood stream-inoculation approach bypasses the first two steps of metastasis: invasion and intravasation. In contrast, we wanted to determine whether the lung metastases spontaneously developed from breast tumors carry more malignant features compared to the parent tumor cells.
To generate spontaneous metastasis models representing human disease, we have previously employed an orthotopic implantation approach to xenograft human breast tumor specimens into NOD/SCID or NSG mouse mammary fat pads, called patient-derived human-in-mouse breast tumor xenograft (PDX) models (5). Most of our established metastatic PDX models with spontaneous pulmonary micro-metastases are triple-negative for estrogen receptor (ER), progesterone receptor (PR), and HER 2 expression (ER-PR-HER2-) (5). Based on gene expression profiles, breast tumors are also divided into six intrinsic molecular subtypes: normal-like, Luminal A, Luminal B, Claudin-low, Basal-like, and HER2-enriched (6,7). The advantages of classifying six molecular subtypes include distinct clinical implications and prognosis correlations. Basal-like tumors are one of the most aggressive subtypes with the worst patient prognosis (6,7). A majority, but not all, of the triple-negative breast tumors belong to the Basal-like subtype (8), which can be determined by prediction analysis of microarray 50 (PAM50) (9).
We focused on a few triple negative, metastatic breast tumor models (“parental” primary tumors) with Basal-like characteristics and set out to determine whether their spontaneous lung metastases remain malignant after being re-implanted back into mouse mammary fat pads to form “met-derived” mammary tumors. We originally expected that the met-derived mammary tumor models upon orthotopic re-implantation would be more aggressive and metastatic than the parental primary tumors. Surprisingly, we found that certain lung met-derived mammary tumors were less metastatic than routinely passaged parental tumors in NOD/SCID mice. Since these two models originated from the same patient with minimal, if any, genetic alterations, we hypothesized that epigenetic regulations contributed significantly to the differential metastatic capacity of these models.
MicroRNAs (miRNAs, miRs) have emerged as critical epigenetic regulators of stem cell functions, epithelial-mesenchymal transition (EMT), and metastasis (10–21). With the combined approaches of gene expression arrays and global miRNA analyses, we identified genes, miRNAs, and their pathways that had altered expression levels in the lung-met derived tumor model and also regulated the growth and invasion of breast cancer cells. Further clinical data analyses based on multiple independent datasets confirmed the clinical relevance of our discoveries and the prognostic value of the candidate miR-138 activity signature.
Materials and Methods
Animal studies
Human-in-mouse breast tumor models with spontaneous metastases were generated using patient tumors (5). Experiments were performed under the approval of the Institutional Biosafety Committee, Institutional Review Board and the Administrative Panel on Laboratory Animal Care of Case Western Reserve University and the University of Chicago.
Tumor dissociation and transplantation in mice
As previously described, tumors were dissociated with collagenase III (Worthington) and DNase I (Sigma) (5). Red blood cells were lysed with ACK lysis buffer (Invitrogen). Dissociated tumor cells were stained with CD44-APC, H2Kd-biotin and strepavidin-PE-cy7 (BD), and DAPI in HBSS/2% FBS for further flow analysis or sorting on FacsAriaII (BD). Tumor cells were mixed 1:1 with Matrigel (BD) and transplanted into NOD/SCID mouse mammary fad pads.
Bioluminescence imaging
Mice were injected IP with D-luciferin (30 mg/ml, Biosynth AG) and anesthetized with Isoflurane (2% in 1 L/min oxygen) (5). Bioluminescence images (0.1 sec to 5 min) were acquired ten-fifteen minutes after D-luciferin injection using the IVIS Spectrum® system (Caliper). For ex vivo BLI of dissected lungs and tumors, D-luciferin (300 μg/ml in PBS) was used to soak freshly isolated lungs or tumors for 2–3 min before imaging. Data was analyzed using LivingImage 2.5 Software (Caliper) and expressed as total flux (photons/second).
Gene expression microarray analyses
Samples were profiled as described previously using oligo microarrays (22) (Agilent Technologies, Santa Clara, CA, USA). All microarray data is available in the University of North Carolina (UNC) Microarray Database and have been deposited in the Gene Expression Omnibus (GEO) under the accession number GEO-GSE 59750. Each gene signature (i.e. Met-derived and miR-138) was derived by comparing two groups using an unpaired two-class Significance Analysis of Microarrays (SAM). The miR-138 signature of up- and down-regulated genes was then summarized as a single “activity score” by multiplying the SAM score of each gene by its expression value in the tested sample and then summing all of the values of each sample.
Cell lines and transfections
MDA-MB-231, BT-20 and Hek293Ts were obtained from ATCC and cultured for less than 6 months. ATCC the verified cell lines using STR profiling. Cells were maintained at 10% fetal bovine serum + 1% Penicillin-Streptomycin in DMEM (MDA-MB-231, Hek293T with G418) and EMEM (BT-20). MiRNAs (Dharmacon, negative control #4) were transfected at 100nM (MDA-MB-231) or 50nM (BT-20, Hek293T) and repeated on subsequent days. 3′UTR luciferase vectors and cDNA vectors were transfected using Fugene (Roche-Promega). For target gene rescue experiments, cells were transfected twice with cDNA vectors after two miRNA transfections.
Quantitative invasion assays in vitro
MDA-MB-231 cells were plated in serum-free DMEM to the top of 8 μm transwell inserts (BD) precoated 1:100 with growth factor-reduced Matrigel (BD) in 24-well companion plates (BD) containing DMEM + 10% FBS (no gradient for controls). After 24 hours, cells were stained for one hour at 37°C in HBSS+ 4 μg/μl calcein AM (BD). Cells on the top of inserts were removed using cotton swabs, and invaded cells were dissociated using dissociation buffer (Trevigen) (shaken for an hour at 37°C), and read at 485/535nM.
Cell growth assays
MDA-MB-231 cells were plated in 24- well plates in DMEM +10% FBS +1% Penicillin-Streptomycin one day prior to starting analysis. Cells were trypsinized and counted using tryphan blue and hemacytometer every day for four days. Each time point was an average of three wells.
Western blot
MDA-MB-231 cells were plated in 10cm dishes and harvested 48 hours after transfections. Cells were lysed and sonicated in RIPA buffer and 100 μg of total protein was loaded onto 4–20% gradient gels for immunoblots with antibodies to EZH2 (BD biosciences) and β-Actin (Sigma-Aldrich, AC-15). Fluorescence-conjugated secondary antibody (red for β-Actin and green for others) was used and blots were read using Odyssey (Li-Cor).
RNA extraction and miRNA real-time PCR
Total RNAs for array analysis were extracted from culture cells or frozen tumor samples using Trizol (Invitrogen) and cleaned using the RNAeasy mini kit (Qiagen). RNA from sorted cell populations were precipitated with isopropanol and glycogen (Invitrogen). Reverse transcription reactions and real-time PCR for miRNAs were performed using the TaqmanR microRNA reverse transcription kit and individual miRNA primers (Applied Biosystems). Real-time PCR was performed in Onestep Real time PCR system (Applied Biosystems).
Apoptosis and sub-G1 population
MDA-MB-231 cells were plated in 12-well plates and transfected as above. Both adherent and floated cells were collected at 24hrs or 72hrs and fixed in 70% ethanol and then stained with propidium iodide (Sigma). DNA content was analyzed using BD LSRII flow cytometer.
Immunofluorescence, Immunohistochemistry, and phase contrast imaging
MDA-MB-231 cells were plated on microscope slide wells (Millipore) and transfected. For F-actin staining, cells were fixed in 4% paraformaldehyde/PBS and stained with fluorescent phalloidin (Invitrogen F432) and SlowFade Gold antifade reagent with DAPI (Invitrogen). Cell images were taken under Axiovert 100tv (fluorescent) (Zeiss). Tumor and lung samples were fixed in 10% neutral buffered formalin, embedded and sectioned into 5um-thin tissue sections. Immunohistochemical staining was performed using antibodies to hEZH2 (BD Biosciences) at Histology core facility. Images were taken by Pannoramic Scan Whole Slide Scanner (CRi, Cambrige Research and Instrumentation).
Statistical analysis
For all assays and analyses in vitro, a student T test was used to evaluate the significance. For animal studies, tumor growth curves and lung metastases were analyzed using a linear mixed model or Wilcoxon rank sum test in R software.
Additional method information including detailed statistical analysis methods can be found in the supplemental data.
Results
Generation of a lung met-derived breast tumor model
Based on the previously developed human-in-mouse triple-negative breast tumor models that exhibited spontaneous pulmonary micro-metastases (5), we term these primary tumor models as “parental” models, such as Metastatic-1 (M1-parental) which belongs to the Basal-like subtype. To further examine the metastatic potential of lung metastases (mets), we dissociated tumor cells from M1 lung mets and then engrafted the cells back into the mammary fat pads of new recipient mice. Consequently, a second breast tumor xenograft model was established as M1 lung-metastasis-derived mammary tumor model, termed as “M1-met-derived”, in contrast to the normally passaged M1-parental breast tumor model (Fig. 1A), both grown in the mouse mammary fat pads to exclude the effects of different microenvironments between the primary and distant sites.
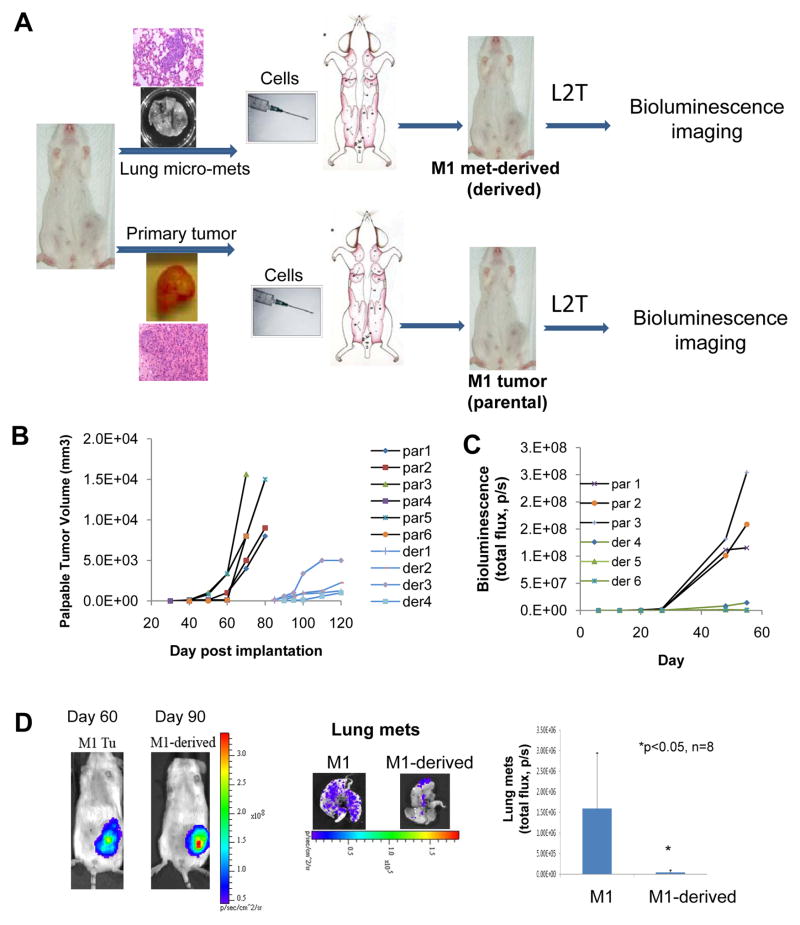
Figure 1. Tumor growth and metastatic phenotype of the lung met-derived model.
A. Diagram showing the generation of the M1 parental and met-derived model with L2T transduced tumor cells.
B. Tumor growth curves of caliper-measured tumor volumes for both models as measured over 3–4 months. Representative examples for six parental (par 1–6) and four met-derived models (der 1–4).
C. Tumor growth curves of L2T-labeled parental and met-derived tumors as measured by bioluminescence imaging (BLI) in total flux (photons/second, p/s) over two months. Three representative examples are shown for both parental (par 1–3) and met-derived models (der 4–6).
D. Representative images of mammary tumors and lung metastases for M1 parental and met-derived model and quantification of BLI of dissected lungs from tumor-bearing mice.
The tumor cells from both the parental tumor model and the met-derived model were labeled with a luciferase 2-tdTomato fusion vector pFU-L2T (5) to allow for the monitoring of growth and metastasis using bioluminescence imaging (BLI) (Fig. 1A). Measured by a caliper as well as BLI, the mammary tumors of the met-derived model had a slower growth rate compared to that of the parental tumors, replicated with multiple lung metastases isolated from seven different mice of the M1 parental model (Fig. 1B and C). Specifically, it took met-derived mammary tumors 90 days on average to form palpable tumors that were comparable in size to the parental tumors formed on average in 60 days (Fig. 1D). In addition to tumor growth, BLI of the dissected lungs from breast tumor-bearing mice showed that the met-derived tumors exhibited reduced lung micro-metastases than the parental tumors (Fig. 1D). These data suggest that the lung met-derived tumor models do not necessarily remain aggressive but instead become less malignant compared to the parental breast tumor models.
To further understand wthat could be responsible for the altered tumor growth and lung metastasis in the met-derived model, we performed both mRNA microarrays and global miRNA real-time PCR analyses.
Global gene expression alterations in the met-derived breast tumors
By gene expression microarray analyses, the met-derived tumors showed 934 up-regulated probes (corresponding to 686 genes) and 297 down-regulated probes (corresponding to 229 genes), compared to the Basal-like parental tumors (False Discovery Rate [FDR] < 5%; Fig. 2A and Supplementary Table S1). Many of the up-regulated genes in the met-derived tumors were related to cell adhesion and differentiation as well as tumor suppressors and cell cycle inhibitors, such as KRT19 (CK19), CEACAM1 (CD66a), and CDKN1A (Fig. 2A and Supplementary Table S1). Many of the down-regulated genes were promoters of self-renewal, cell motility, and cell cycle, such as EZH2, SMAD2, and CCNB1(Fig. 2A and Supplementary Table S1). We then utilized a Differentiation Predictor that measures each sample’s position along a normal mammary developmental axis from mammary stem cell, luminal progenitor, and mature luminal cell states (23). Consistently, the differentiation scores of the lung met-derived tumors were significantly higher than those of the primary tumors (p=0.007) (Supplementary Fig. S1).
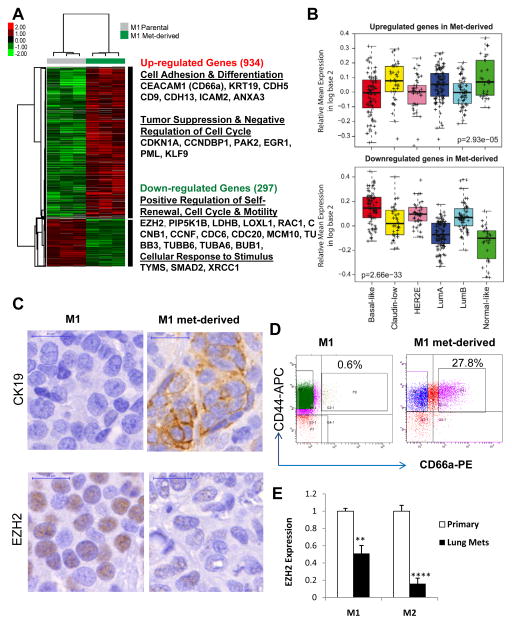
Figure 2. Gene expression differences between the M1 parental and met-derived tumor models.
A. Heat-map and a list of representative genes differentially expressed between the M1 parental and met-derived tumors as measured by microarrays.
B. Mean expression of the up (upper panel) and down (lower panel) gene signatures obtained by comparing met-derived tumors versus the parental across the intrinsic subtypes of breast cancer (UNC337 dataset).
C. Immunochemical staining of CK19 (upper panel) and EZH2 (lower panel) in the M1 parental and met-derived tumor sections.
D. Flow profiles of M1 parental and met-derived tumor cells for CEACAM1-coded CD66a expression.
E. Decreased EZH2 expression in tumor cells (tdTomato+) directly sorted from the lung metastases compared to paired primary tumor cells from both M1 and M2 xenograft models generated from two triple negative patient tumors. Expression levels were measured by real-time PCR with sorted 10,000 –40,000 tumor cells labeled by pFU-L2T.
To further link the met-derived gene signatures to breast cancer intrinsic subtypes, we examined the mean expression pattern of the up-regulated and down-regulated genes across the UNC337 database (23). The up- and down-regulated genes in the met-derived model were found highly expressed in the subset of normal-like, Luminal A, and Claudin-low subtypes and relatively lowly expressed in the other subset of three subtypes: Luminal B, HER2-enriched, and Basal-like (p<0.0001, Student’s t-test; Fig. 2B). Thus, the met-derived signature was found more enriched in Luminal A and Normal-like with good outcomes than in poor outcome and undifferentiated tumor subtypes (i.e. Luminal B, HER2-enriched, and Basal-like) (Fig. 2B). However, it is surprising that the met-derived signature with a list of cell adhesion and differentiation genes is highly expressed in Claudin-low, which shows low expression of Claudins 3,4,7 and not necessarily good outcomes (23).
By immunohistochemical (IHC) staining and flow cytometry analyses, we further validated the differential expression of representative genes at the protein level. CK19 is a luminal epithelial cell marker with a favorable prognosis in breast cancer patients (24). EZH2 regulates self-renewal and it promotes tumor growth and metastasis (25–27). CK19 was up regulated and EZH2 was down regulated in met-derived tumor sections (Fig. 2C). CD66a is a mammary epithelial cell differentiation marker (28) and a suppressor of cell proliferation and tumorigenesis (29,30). By flow analyses we observed a 27.2% increase of CD66a positive cells in the met-derived model compared to that of the parental tumor (Fig. 2D). All these markers confirmed a more differentiated status of met-derived breast tumors versus parental breast tumors.
To validate that the altered gene expression accumulated in the pulmonary metastases, we sorted tumor cells directly from the lung metastases of the parental models, M1 and another independent model, Metastatic-2 (M2) which is derived from a second triple negative patient tumor (5). Measured by real-time PCR, EZH2 expression decreased in the pulmonary metastatic cells compared to the paired primary tumor cells in both M1 and M2 models (Fig. 2E). These data suggest that the altered gene expression in the met-derived model occurs in the pulmonary metastatic site prior to re-engraftment.
Importing the 492 genes into DAVID (The Database for Annotation, Visualization and Integrated Discovery, http://david.abcc.ncifcrf.gov/) revealed they were enriched in pathways related to cell proliferation, cytoskeleton, and cell cycle processes (Supplementary Table S2).
Differentially expressed miRNAs in the met-derived breast tumors
To identify candidate miRNAs that may regulate cell cycle, apoptosis, and metastasis (10–21), we performed real time PCR analyses for 384 miRNAs in both parental and met-derived tumors. Thirty-two miRNAs were differentially expressed with twenty-seven miRNAs being up-regulated in the met-derived tumors, including miR-200 family members, miR-138, and others (Supplementary Table S3). The global up-regulation of miRNA expression might suggest a relatively benign status for the met-derived breast tumors, as aggressive tumors have a greater majority of down-regulated miRNAs in their expression profiles (31).
The up-regulated miRNAs contained a few luminal subtype miRNAs, such as known miR-200, miR-29, and miR-30 family members (22,32,33). Based on the miRNA expression profiles of the human breast tumors we collected at the University of Chicago (GSE39543) (22), a few miRNAs displayed differential expression profiles across the subtypes of clinical breast tumors, including miR-138-1*, miR-143, miR-26b, and miR-331-3p with down-regulated levels in triple negative tumors versus non-triple negative tumors, and miR-576-3p up-regulated in tumors versus normal breast tissue adjacent to tumors (Supplementary Fig. S2). However, miR-138-1* is not the candidate miR-138 but the 3′ passenger strand produced from the miR-138-1 precursor during miRNA maturation. From the analyses of GSE37407 (34) with paired primary tumors with metastatic tumors (both lymph nodes and distant organs) from 14 breast tumor patients, we found that miR-138, along with miR-138-1* and miR-138-2*, were upregulated in the metastases compared to the paired primary tumors (n=23, p<0.05 or <0.01) (Supplementary Fig. S3 A–B).
We subsequently characterized the functions as well as regulation of candidate miRNAs, and then identified their direct target genes in regulating breast cancer phenotypes.
MiR-138 regulates breast cancer cell invasion, growth, and EMT
From the selected miRNA candidates with potential clinical relevance (Supplementary Fig. S2–S3), we further characterized their role in regulating breast cancer cell invasion and proliferation. While most of the chosen miRNAs, such as miR-143, miR-26b, and miR-331-5p, did not show effects on invasion in the screening assays, enforced expression of miR-138 suppressed invasion in both MDA-MB-231 and BT-20 cells (Fig. 3A). The level of miR-138 was vastly up regulated in the met-derived model versus parental tumor model (Fig. 3A and Supplementary Table S3).
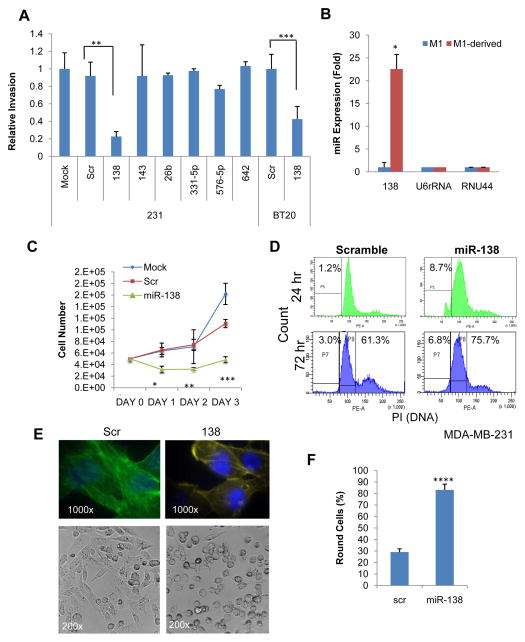
Figure 3. miR-138 regulates growth, invasion, and cell cycle.
A. Cell invasion of MDA-MB-231 and BT-20 breast cancer cells upon transfection with candidate miRNAs, scramble and mock transfected controls as measured by a transwell assay over a 24hr period. **p<0.01, ***p<0.001
B. Expression level of miR-138 in parental and met-derived models as compared to U6 or RNU44 controls. *p<0.05
C. Proliferation curves of MDA-MB-231 cells post 36hr-transfections of miR-138 (Day 0–3), mock and scrambled controls, measured by cell counting. *p<0.05, **p<0.01, ***p<0.001 comparing the miR-138 group to other controls.
D. Histograms of PI-stained DNA content of MDA-MB231 cells, 24hr and 72 hr post transfection of miR-138 and scrambled control. The populations of P5 at 24hr and P7 at 72hr represent the sub-G1 apoptotic cells, and P8 gates the cells at G1 phase.
E. F-actin staining (upper panels) and phase contrast (bottom panels) of MDA-MB-231 cells 72hrs post transfection with miR-138 and scrambled control (Scr).
F. Quantitative counting (percentage) of round cells for scrambled control and miR-1138-transfected MDA-MB-231 cells, as shown in bright field images. **** p<0.0001 (n=4 sets).
Furthermore, we examined whether upregulated miR-138 contributed to the slower growth rate of the met-derived tumor model. In the cell counting assay, enforced expression of miR-138 inhibited the proliferation of breast tumor cells (Fig. 3C). Cell cycle and apoptosis analyses using propidium iodide staining showed that miR-138 slightly increased the percentage of sub-G1 apoptotic cells at the early (24 hr) and late (72 hr) time points and induced G1 arrest at 72 hr post transfection (Fig. 3D). Phalloidin staining revealed that increased levels of miR-138 reduced F-actin associated stress fibers (22,33) and promoted a rounder and more epithelial like cell phenotype in MDA-MB-231 cells (Fig. 3E–F), suggesting that miR-138 inhibits EMT which is important for invasion at the early stage of metastasis.
GATA3 regulates miR-138 in breast cancer metastasis
To examine what mechanisms caused the increased level of miR-138 in the lung-derived model, we performed fluorescence in situ hybridization (FISH) staining to detect the copy number of the miR-138 gene in both tumor models. There were equal numbers (three per cell on average) of the miR-138 gene present in both parental and met-derived tumor models (Supplementary Fig. S4), indicating that the copy number change was not a factor involved in the increased level in the lung-derived model.
We then hypothesized that miR-138 might be transcriptionally regulated in the lung metastases. Within the putative promoter regions (−3kb) of miR-138-1 and miR-138-2 genes, JASPAR online analyses (35) revealed multiple GATA3 binding sites located upstream of both genes (Supplementary Table S4). We then analyzed the expression of miR-138 and GATA3 by real-time PCR in paired lung metastases and primary tumor cells sorted from M1 and M2 models. Both miR-138 and GATA3 were upregulated in the lung metastases compared to paired primary tumor cells (Fig. 4A–B). In addition, forced expression of wild-type GATA3 induced a significant increase (8–10 fold) of miR-138 levels in MDA-MB-231 cells (Fig. 4C).
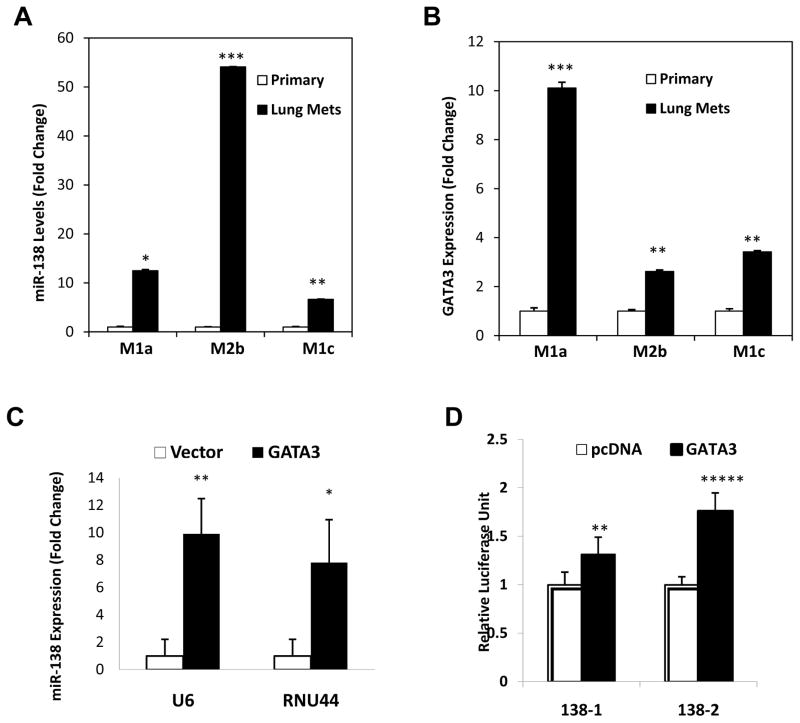
Figure 4. GATA3 regulates miR-138 expression in breast tumor metastasis.
A. Increased miR-138 expression in sorted tumor cells from the lung metastases compared to the paired primary tumor cells of patient-derived xenograft models M1 parental and M2 parental, including M1a (M1, mouse a), M2a (M2, mouse b), and M1c (M1, mouse c), measured by real-time PCR an normalized based on U6. T Test p values *<0.05, **<0.01, ***<0.001. n=3.
B. Elevated GATA3 mRNA levels in sorted lung met cells compared to paired primary tumor cells in patient-derived xenograft models M1 parental and M2 parental, including M1a (M1, mouse a), M2a (M2, mouse b), and M1c (M1, mouse c), measured by real-time PCR and normalized on GAPDH. **p<0.01, ***p<0.001. n=3.
C. Relative miR-138 expression levels (fold change) in MDA-MB-231 cells transfected by the vector control and GATA3 cDNA, measured by real-time PCR and normalized on U6 and RNU44 controls. *p<0.05, **p<0.01. n=3.
D. Luciferase activity of MDA-MB-231 cell lysates 48 hours post co-transfection of GATA3/vector control and the luciferase reporter pGL4 with the promoter regions (−3kb) of miR-138-1 and miR-138-2 genes. **p<0.01, ***p<0.001. n=6.
To further determine whether GATA3 functionally interacts with the miR-138 promoters (138-1 and 138-2), we cloned the putative promoter regions (−3kb) into the luciferase reporter vector pGL4. We detected a positive transcriptional regulation of the luciferase reporter gene by GATA3 through inserted promoter regions of miR-138-1 or miR-138-2 genes (Fig. 4D), which harbor two or three putative GATA3 binding sites respectively (Supplementary Table S4). These data indicate that GATA3 transcriptionally promotes miR-138 expression in the lung metastases.
Identification of miR-138 activity gene signature
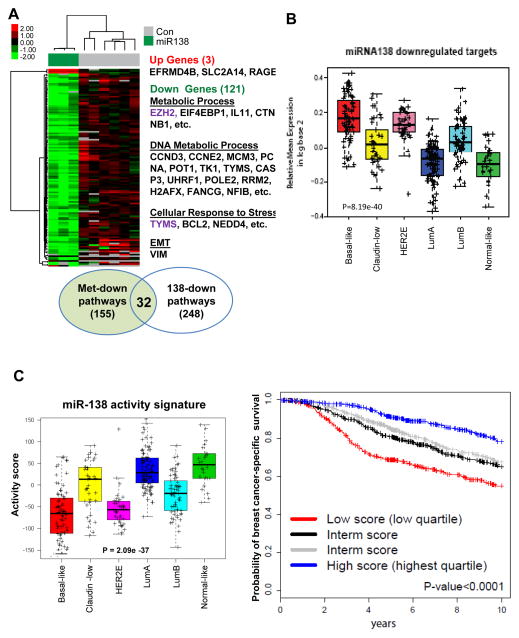
Individual miRNAs regulate mRNAs of up to hundreds of genes, which renders miRNAs as powerful regulators (36). To test the hypothesis that miRNAs contribute to the altered gene expression profile of the met-derived tumor models, we examined the effects of miR-138 on global gene expression. The microarray analysis of MDA-MB-231 cells upon miR-138 transfection revealed a miR-138 gene signature (FDR < 8.5%), with 121 down-regulated probes (corresponding to 96 genes) and 3 up-regulated probes (corresponding to 3 genes) (Fig. 5A and Supplementary Table S5).
Figure 5. Clinical relevance of miR-138 activity.
A. Microarray analyses of global gene expression changes induced by miR-138. Left panels: a heat-map of up-regulated genes (3) and down-regulated genes (121) by miR-138 (n=3), compared to mock and scramble controls (n=6). Representative genes regulate metabolic process, DNA replication, cell cycle, and EMT.
B. Mean expression of miR-138 down-regulated genes across intrinsic molecular subtypes of UNC337 breast tumor set (n=337, p=8.19e-40).
C. Left panel: miR-138 activity signature across the intrinsic subtypes of breast cancer based on the UNC337 data set (n=320, p=2.09e-37). Right panel: Kaplan-Meier curves of survival outcomes based on miR-138 activity score signature to predict breast cancer-specific survival in the METABRIC dataset (n=1848, p<0.0001). Samples have been rank ordered based on their scores and grouped based on quartiles. Multivariate Analyses (MVA) show adjusted p value <0.05 based on the classical pathological variables, intrinsic subtypes, and proliferation signature (Supplementary Table S6).
We compared the genes down-regulated by miR-138 with the ones down-regulated in met-derived models and found two common genes, EZH2 and TYMS (Fig. 2A, Fig. 5A, Supplementary Tables S1 and S5). However, GO analyses of the down-regulated genes suggested a total of 32 common biological processes altered in both miR-138 expressing cells and in the met-derived models (Fig. 5A, Supplementary Tables S1 and S5). The pathways regulated by miR-138 included metabolic processes (EZH2, IL11, etc), DNA metabolic processes (CCND3, CCNE2, MCM3, PCNA, RRM2, H2AFX, etc), cellular response to stress (TYMS), and EMT (VIM) (Fig. 5A and Supplementary Table S5). We identified the similar expression pattern of miR-138 downregulated genes (Fig. 5B), as that of downregulated genes in the met-derived gene signature across breast tumor subtypes in UNC337 (Fig. 2B), suggesting that miR-138 may contribute to the altered gene expression in the met-derived model.
To complement the limited sample size of patient tumors with both miRNA and clinical-pathological analyses, we employed a global gene signature regulated by miR-138 to mirror the expression and activity status of miR-138 in tumor samples, termed as “miR-138 activity gene signature” which is distinct and opposite to miR-138 downregulated genes. We evaluated the activity of the miR-138 gene signature across the intrinsic subtypes in UNC337. Mirroring the pattern of up-regulated genes in the met-derived gene signature (Fig. 2B), a high activity of miR-138 was found in normal-like, Luminal A, and Claudin-low subtypes, and in other words, miR-138 activity was down-regulated in poor outcome and undifferentiated tumor subtypes (i.e. Basal-like, HER2 enriched, and Luminal B) (Fig. 5C, left panel).
Furthermore, we determined the prognostic value of the miR-138 activity signature using two independent breast tumor data sets, METABRIC (n=1848) (37) and MDACC588 (n=588) (38). Kaplan-Meier (KM) plots showed that patient tumors with high miR-138 activity gene expression correlated with an increased probability of breast cancer specific-survival or distant relapse-free survival (Fig. 5C right panel and Supplementary Fig. S5). Multivariate analyses (MVA) further demonstrated the independent prognostic role of the miR-138 activity gene signature in predicting survival outcome beyond classical clinical-pathological variables (age, tumor size, grade, nodal status, and systemic treatment), intrinsic subtype, and the proliferation gene signature (39,40) in the METABRIC data set (n=1848, adjusted P<0.05) (Supplementary Table S6).
EZH2 is an essential direct target of miR-138
MiRNAs regulate gene expression by targeting the 3′UTR of the relevant mRNAs. To strategically identify the important direct targets of miR-138, we performed the GeneSet2miR prediction (41) with the list of down regulated genes (Supplementary Table S5) and validated the genes using experimental analyses, such as real-time PCR, immunoblots, luciferase assays, and functional rescue studies.
Four out of 13 algorithms of the GeneSet2miR analyses (41) predicted 9 out of the 121 down-regulated genes as direct target genes of miR-138, including EZH2 and PPIP5K1 (Supplementary Fig. S6A). We also found the EMT marker VIM predicted as a miR-138 target by one algorithm. Real-time PCR analyses of four chosen genes validated microarray data in which miR-138 inhibited mRNA expression levels of EZH2, PPIP5K1, VIM, and IL-11 in MDA-MB-231 cells (Supplementary Fig. S6B–C). Immunoblotting and ELISA analyses also confirmed miR-138-mediated inhibition of target genes EZH2 and IL-11 at protein levels respectively (Fig. 6A and Supplementary Fig. S6D). Luciferase assays with the 3′UTR of the candidate genes cloned upstream of the luciferase gene demonstrated that miR-138 interacts with the 3′UTR of EZH2, PPIP5K1, and VIM and induces an inhibitory effect on the expression of these target genes (Supplementary Fig. S6E–F).
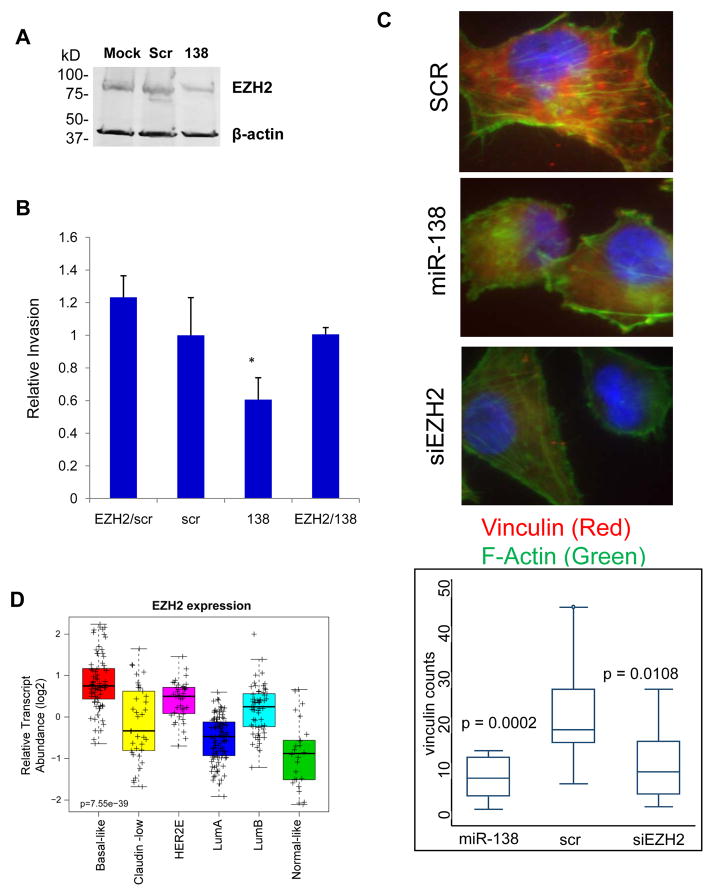
Figure 6. EZH2 is an essential target of miR-138.
A. Immunoblots of EZH2 and the loading control β-actin with lysates of MDA-MB-231 cells transfected with miR-138, mock control and scrambled (Scr) control.
B. Rescue invasion assays using transwell inserts with MDA-MB-231 cells transfected with miR-138, scrambled (Scr) control, and EZH2 cDNA. *p<0.05
C. Top three panels: representative images of vinculin (red) and F-actin (green) staining in MDA-MB-231 cells transfected with the scramble control, miR-138, and siEZH2. Bottom panel: quantification of the amount of stained vinculin-mediated focal adhesions (red dots) per cell in each of the three samples mentioned above.
D. EZH2 transcript abundance across the intrinsic subtypes of breast cancer in the UNC337 data set (n=320, p=7.55e-39).
To determine the importance of the target genes, we performed rescue experiments in which candidate cDNA was overexpressed in MDA-MB-231 cells to compensate for the increased levels of miR-138. EZH2 was able to restore the invasiveness of breast tumor cells back to the control levels in the presence of transfected miR-138 (Fig. 6B). Knockdown of EZH2 by siRNAs mimicked the inhibitory effect of miR-138 on two cytoskeleton markers of EMT (22,33), F-actin and focal adhesion (vinculin) (Fig. 6C). These data demonstrate that EZH2 is an essential target of miR-138 in regulating both breast tumor invasion and EMT.
We further compared the expression pattern of EZH2 across the intrinsic breast cancer subtypes. EZH2 showed similar expression patterns across subtypes as those genes downregulated in met-derived tumors and those targeted by miR-138, namely higher expression in poor prognostic and undifferentiated subtypes such as Basal-like, HER2-enriched, and Luminal B (Fig. 2B, 5B and 6D). Concordant with these findings, higher EZH2 expression was associated with estrogen receptor negative status by IHC in 50 tumor samples from the University of Chicago tumor dataset (Supplementary Fig. S6G). These data suggest that EZH2 is a relevant regulator of breast cancer.
Discussion
In this study we report that in contrast to the malignant primary tumors, the spontaneous pulmonary metastases developed in PDX models can lose their metastatic potential and mesenchymal status and gain a more epithelial and differentiated status. This feature might be generalized to a group of breast cancer patients with tumor metastases in the brain, liver, and lymph nodes (34). This phenomenon had initially been unexpected and was against the EMT dogma proposed in metastasis (42). However, more and more studies support that MET along with an epithelial state is required for colonization at the late stage of metastasis (43–45). While there is still debate in the field, our studies indicate that metastasis is a complex procedure and the tumor cells that make it to the last step of colonization may not necessarily be very proliferative and mobile again. Similarly, an acquired luminal and non-metastatic feature had been previously observed in certain breast cancer cell lines derived from the metastatic sites of breast tumors, such as MCF-7 (46) which is derived from pulmonary metastases (pleural effusions) of an invasive breast cancer patient, but has lost the invasive and mesenchymal features of primary tumors and gained an epithelial phenotype with CK19 and ER expression (47). The open question is whether selection or adaptation plays a more important role in this phenomenon.
Consistent with our findings of an altered expression of the miR200 family members in the epithelial lung met-derived models, the recent study on paired metastases and primary tumors of breast cancer patients also demonstrates an increased epithelial feature (E-cadherin expression) along with elevated expression of miR-200 family members in almost all of the distant metastasis and lymph nodes compared to corresponding primary tumors (34). Furthermore, certain ER− primary tumors might gain ER expression in their brain metastases although other ER+ primary tumors might still maintain or lose ER expression in distant metastases and lymph nodes (34). A future comprehensive analysis will be necessary to examine the global gene expression of paired metastases and primary tumors of breast cancer.
Nevertheless, the distinct properties of the met-derived breast tumor models versus parental tumors have served as a great model system for us to examine the epigenetic regulation (and possibly genetic selection) process of tumor cells during metastasis. Gene and miRNA expression profile comparisons have led us to identify new pairs of miRNA/gene regulators, such as miR-138 and EZH2. Interestingly, EZH2 is not only a representative gene down regulated in the met-derived model, but also a direct and important target of miR-138 which is oppositely up regulated in that model and regulates EMT. EZH2 is a chromatin-bound component of PRC complex II and plays a critical and powerful role in self-renewal and tumor progression in lymphoma, breast cancer, and prostate cancer (48–52). Previous data has reported that EZH2 directly binds to the promoter of vinculin, a marker of focal adhesion and EMT (24). The clinical relevance of both EZH2 and miR-138 activity gene signature implies innovative biomarkers and targets in human breast cancer development and progression.
Although our previous data suggests that IL-11 plays a role in breast tumor invasion and EMT (22,33), IL-11 is not on the list of direct targets and is likely an indirect target of miR-138. Because IL-11 is a target of NF-kB (53), there is a possible regulation of IL-11 by EZH2. While EZH2 binds to RelA/RelB directly in Basal-like breast cancer cells to promote expression of NF-kB targets (54), EZH2 may also positively regulate IL-11 in the Basal-like breast tumors.
Supplementary Material
Acknowledgments
Grant Support
This study was supported in part by Case Western Reserve University start-up fund, National Cancer Institute Paul Calabresi K12 Scholar 1K12CA139160-02, K99/R00 CA160638-02, Case Comprehensive Cancer Center Pilot Project P30 CA043703-23, and Northern Ohio Golf Charities & Foundation (H.L.).
We thank Dr. Michael Clarke and Dr. Geoffrey Greene for suggestions and comments. We appreciate the technical support from Ravand Samaeekia, Kathy Yee, Rachel Dalton, Yuhao Wang, Grigoriy Moskalenk, Junyuan Wu, Dalong Qian and Yohei Shimono. We specifically acknowledge the animal facilities, optical imaging core facilities, integrated microscopy core facilities, DNA sequencing facility and functional genomics facilities, flow cytometer cores at Stanford University, the University of Chicago and Case Western Reserve University.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
Authors’ Contributions
J.B., A.P., Y-F.C., X.L., S.H., M.S., C.N., M.J.G-V., C.H., and H.L. designed and performed experiments, and analyzed data. A.P. performed microRNA arrays and analyzed the arrays and other tumor datasets. O.I.O. and C.M.P. advised on the project. H.L. designed the project. J.B. and H.L. wrote the manuscript. A.P. and S.H. edited the manuscript.
References
- 1.Husemann Y, Geigl JB, Schubert F, Musiani P, Meyer M, Burghart E, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13(1):58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148(1–2):349–61. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nature reviews. 2002;2(8):563–72. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 4.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436(7050):518–24. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu H, Patel MR, Prescher JA, Patsialou A, Qian D, Lin J, et al. Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc Natl Acad Sci U S A. 2010;107(42):18115–20. doi: 10.1073/pnas.1006732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 7.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Molecular oncology. 2011;5(1):5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prat A, Adamo B, Cheang MC, Anders CK, Carey LA, Perou CM. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. The oncologist. 2013;18(2):123–33. doi: 10.1634/theoncologist.2012-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prat A, Parker JS, Fan C, Perou CM. PAM50 assay and the three-gene model for identifying the major and clinically relevant molecular subtypes of breast cancer. Breast Cancer Res Treat. 2012;135(1):301–6. doi: 10.1007/s10549-012-2143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27(15):2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 11.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10(2):202–10. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 12.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 13.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451(7175):147–52. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 15.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9(6):582–9. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes & development. 2008;22(7):894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283(22):14910–4. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138(3):592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131(6):1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 20.Hurst DR, Edmonds MD, Welch DR. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer research. 2009;69(19):7495–8. doi: 10.1158/0008-5472.CAN-09-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo X, Wu Y, Hartley RS. MicroRNA-125a represses cell growth by targeting HuR in breast cancer. RNA Biol. 2009;6(5):575–83. doi: 10.4161/rna.6.5.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bockhorn J, Dalton R, Nwachukwu C, Huang S, Prat A, Yee K, et al. MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat Commun. 2013;4:1393. doi: 10.1038/ncomms2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortenson ED, Park S, Jiang Z, Wang S, Fu YX. Effective Anti-Neu-Initiated Antitumor Responses Require the Complex Role of CD4+ T Cells. Clin Cancer Res. 2013;19(6):1476–86. doi: 10.1158/1078-0432.CCR-12-2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer Cell. 2011;19(1):86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren G, Baritaki S, Marathe H, Feng J, Park S, Beach S, et al. Polycomb Protein EZH2 Regulates Tumor Invasion via the Transcriptional Repression of the Metastasis Suppressor RKIP in Breast and Prostate Cancer. Cancer research. 2012;72(12):3091–104. doi: 10.1158/0008-5472.CAN-11-3546. [DOI] [PubMed] [Google Scholar]
- 27.Min J, Zaslavsky A, Fedele G, McLaughlin SK, Reczek EE, De Raedt T, et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat Med. 2010;16(3):286–94. doi: 10.1038/nm.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang J, Hardy JD, Sun Y, Shively JE. Essential role of biliary glycoprotein (CD66a) in morphogenesis of the human mammary epithelial cell line MCF10F. J Cell Sci. 1999;112 (Pt 23):4193–205. doi: 10.1242/jcs.112.23.4193. [DOI] [PubMed] [Google Scholar]
- 29.Leung N, Turbide C, Balachandra B, Marcus V, Beauchemin N. Intestinal tumor progression is promoted by decreased apoptosis and dysregulated Wnt signaling in Ceacam1−/− mice. Oncogene. 2008;27(36):4943–53. doi: 10.1038/onc.2008.136. [DOI] [PubMed] [Google Scholar]
- 30.Leung N, Turbide C, Olson M, Marcus V, Jothy S, Beauchemin N. Deletion of the carcinoembryonic antigen-related cell adhesion molecule 1 (Ceacam1) gene contributes to colon tumor progression in a murine model of carcinogenesis. Oncogene. 2006;25(40):5527–36. doi: 10.1038/sj.onc.1209541. [DOI] [PubMed] [Google Scholar]
- 31.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 32.Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8(10):R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bockhorn J, Yee K, Chang YF, Prat A, Huo D, Nwachukwu C, et al. MicroRNA-30c targets cytoskeleton genes involved in breast cancer cell invasion. Breast Cancer Res Treat. 2013;137(2):373–82. doi: 10.1007/s10549-012-2346-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gravgaard KH, Lyng MB, Laenkholm AV, Sokilde R, Nielsen BS, Litman T, et al. The miRNA-200 family and miRNA-9 exhibit differential expression in primary versus corresponding metastatic tissue in breast cancer. Breast Cancer Res Treat. 2012;134(1):207–17. doi: 10.1007/s10549-012-1969-9. [DOI] [PubMed] [Google Scholar]
- 35.Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nature reviews. 2004;5(4):276–87. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 36.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nature reviews Genetics. 2004;5(7):522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 37.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hatzis C, Pusztai L, Valero V, Booser DJ, Esserman L, Lluch A, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA: the journal of the American Medical Association. 2011;305(18):1873–81. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res. 2010;16(21):5222–32. doi: 10.1158/1078-0432.CCR-10-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Antonov AV, Dietmann S, Wong P, Lutter D, Mewes HW. GeneSet2miRNA: finding the signature of cooperative miRNA activities in the gene lists. Nucleic Acids Res. 2009;37(Web Server issue):W323–8. doi: 10.1093/nar/gkp313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science (New York, NY) 2011;331(6024):1559–64. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 43.Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22(6):725–36. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ocana OH, Corcoles R, Fabra A, Moreno-Bueno G, Acloque H, Vega S, et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell. 2012;22(6):709–24. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 45.Stankic M, Pavlovic S, Chin Y, Brogi E, Padua D, Norton L, et al. TGF-beta-Id1 signaling opposes Twist1 and promotes metastatic colonization via a mesenchymal-to-epithelial transition. Cell reports. 2013;5(5):1228–42. doi: 10.1016/j.celrep.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soule HD, Vazguez J, Long A, Albert S, Brennan M. A human cell line from a pleural effusion derived from a breast carcinoma. J Natl Cancer Inst. 1973;51(5):1409–16. doi: 10.1093/jnci/51.5.1409. [DOI] [PubMed] [Google Scholar]
- 47.Harrell JC, Pfefferle AD, Zalles N, Prat A, Fan C, Khramtsov A, et al. Endothelial-like properties of claudin-low breast cancer cells promote tumor vascular permeability and metastasis. Clinical & experimental metastasis. 2014;31(1):33–45. doi: 10.1007/s10585-013-9607-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100(20):11606–11. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren G, Baritaki S, Marathe H, Feng J, Park S, Beach S, et al. Polycomb protein EZH2 regulates tumor invasion via the transcriptional repression of the metastasis suppressor RKIP in breast and prostate cancer. Cancer research. 2012;72(12):3091–104. doi: 10.1158/0008-5472.CAN-11-3546. [DOI] [PubMed] [Google Scholar]
- 50.Di Meglio T, Kratochwil CF, Vilain N, Loche A, Vitobello A, Yonehara K, et al. Ezh2 orchestrates topographic migration and connectivity of mouse precerebellar neurons. Science (New York, NY) 2013;339(6116):204–7. doi: 10.1126/science.1229326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science (New York, NY) 2012;338(6113):1465–9. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 53.Steinbrecher KA, Harmel-Laws E, Sitcheran R, Baldwin AS. Loss of epithelial RelA results in deregulated intestinal proliferative/apoptotic homeostasis and susceptibility to inflammation. Journal of immunology. 2008;180(4):2588–99. doi: 10.4049/jimmunol.180.4.2588. [DOI] [PubMed] [Google Scholar]
- 54.Lee ST, Li Z, Wu Z, Aau M, Guan P, Karuturi RK, et al. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Mol Cell. 2011;43(5):798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.