A novel functional role for WAVE1 is found that is lacking in N-WASP and WAVE2. Through its unique WH2 domain, WAVE1 dramatically reduces the rate of actin filament elongation independently of its interactions with the Arp2/3 complex. These findings help explain how cells build actin networks with distinct geometries and growth rates.
Abstract
Branched actin filament networks in cells are assembled through the combined activities of Arp2/3 complex and different WASP/WAVE proteins. Here we used TIRF and electron microscopy to directly compare for the first time the assembly kinetics and architectures of actin filament networks produced by Arp2/3 complex and dimerized VCA regions of WAVE1, WAVE2, or N-WASP. WAVE1 produced strikingly different networks from WAVE2 or N-WASP, which comprised unexpectedly short filaments. Further analysis showed that the WAVE1-specific activity stemmed from an inhibitory effect on filament elongation both in the presence and absence of Arp2/3 complex, which was observed even at low stoichiometries of WAVE1 to actin monomers, precluding an effect from monomer sequestration. Using a series of VCA chimeras, we mapped the elongation inhibitory effects of WAVE1 to its WH2 (“V”) domain. Further, mutating a single conserved lysine residue potently disrupted WAVE1's inhibitory effects. Taken together, our results show that WAVE1 has unique activities independent of Arp2/3 complex that can govern both the growth rates and architectures of actin filament networks. Such activities may underlie previously observed differences between the cellular functions of WAVE1 and WAVE2.
INTRODUCTION
Precise spatial and temporal control of actin filament network assembly is critical for a range of cellular processes, including cell division, cell migration, and neuronal process formation (Pollard and Cooper, 2009). Electron microscopy and superresolution imaging studies of actin networks have revealed a diversity of branched and unbranched actin architectures in cells, which appear to be uniquely tailored to their different roles (Svitkina and Borisy, 1999; Korobova and Svitkina, 2010; Bear et al., 2002; Collins et al., 2011; Xu et al., 2012a, 2013; Blanchoin et al., 2014). Even among branched networks or within a single branched network, there can be considerable variation in branch density and filament length (Svitkina and Borisy, 1999; Korobova and Svitkina, 2010; Bear et al., 2002; Vinzenz et al., 2012; Mueller et al., 2014). However, there is still relatively little understanding of how this diversity in actin network architecture is specified.
Branched actin networks are assembled by the actin-related protein 2/3 (Arp2/3) complex, which contains seven evolutionarily conserved subunits, including two actin-related proteins, Arp2 and Arp3. The nucleation activities of Arp2/3 complex are stimulated by direct interactions with nucleation-promoting factors (NPFs), the best characterized of which are members of the Wiskott–Aldrich syndrome protein (WASP)/WASP and verprolin homologue (WAVE) family (Pollard, 2007; Campellone and Welch, 2010; Padrick and Rosen, 2010). All WASP/WAVE proteins have a C-terminal verprolin homology/central/acidic (VCA) domain, in which the V (WH2) binds monomeric actin and the C and A bind Arp2/3 complex (Boczkowska et al., 2014). Through these interactions, the VCA directly stimulates Arp2/3 complex–mediated nucleation by 1) inducing conformational changes in Arp2/3 complex to align the Arp2 and Arp3 subunits, 2) increasing association of Arp2/3 complex with the sides of preexisting (mother) filaments, and 3) recruiting the first actin monomers to nucleate the nascent daughter branch (Goley et al., 2004; Rodal et al., 2005; Rouiller et al., 2008; Xu et al., 2012b; Smith et al., 2013a). WASP/WAVE-family proteins are believed to function as dimers and/or clusters on cell surfaces, and indeed there are two VCA binding sites on the Arp2/3 complex (Westphal et al., 2000; Padrick et al., 2008, 2011; Padrick and Rosen, 2010; Gohl et al., 2010). Recent single-molecule studies show that VCA-Arp2/3 complex associates with sides of mother filaments, but daughter filament elongation does not proceed until VCA dissociates from the nascent branch (Smith et al., 2013b). These observations, together with structural studies on VCA-Arp2/3 complex-actin interactions, suggest that the WH2 domain of VCA transiently obstructs further actin subunit addition to the Arp2/3-VCA-actin complex.
In cells, a variety of signaling pathways lead to NPF recruitment and activation at membranes. This in turn stimulates Arp2/3 complex–dependent assembly of actin networks in which mother and daughter filament barbed ends are oriented toward the membrane. Thus branched networks grow through continuous rounds of NPF-Arp2/3 complex interaction at the membrane, triggering nucleation, followed by rapid dissociation of membrane-bound NPF from Arp2/3. In addition to their interactions with Arp2/3 complex, NPFs interact transiently with barbed ends of filaments to maintain attachment of the growing network to the NPF-bound surface (Co et al., 2007; Hu and Kuhn, 2012; Khanduja and Kuhn, 2014). Arp2/3 complex stimulation and barbed-end attachment are considered to be the two general features of VCA-containing NPFs. However, other than some noted differences in potencies of Arp2/3 complex activation (Zalevsky et al., 2001; Yarar et al., 2002; Kang et al., 2010), there has been little investigation into whether VCA domains of different NPFs have unique effects on actin network growth rate and/or architecture.
The eight members of the mammalian WASP/WAVE superfamily (WASP, neuronal WASP [N-WASP], WAVE1, WAVE2, WAVE3, WASH, WHAMM, and JMY) have a wide range of expression and localization patterns, as well as distinct cellular and physiological roles, for example, in endocytosis, phagocytosis, intracellular traffic, cell motility, cell adhesion, and cell and tissue morphogenesis (Rottner et al., 2010; Veltman and Insall, 2010). These functional differences are attributed primarily to the domains located outside of the VCA region, which mediate interactions with a variety of cellular factors and link the NPFs to different signaling pathways. However, it has been more perplexing why some cell types express multiple WASP/WAVE family members with similar localization patterns. For instance, in neurons, WAVE1, WAVE2, and N-WASP all localize to growth cones and dendritic spines and make important contributions to their formation (Irie and Yamaguchi, 2002; Dahl et al., 2003; Nozumi et al., 2003; Kawano et al., 2005; Kim et al., 2006; Soderling et al., 2007; Sanchez et al., 2009; Ito et al., 2010; Spillane et al., 2012), but it is not yet known what mechanistic roles each one plays. In nonneuronal cells, whereas N-WASP functions in endocytosis and possibly filopodia formation, WAVE1 and WAVE2 both localize to the leading edge and regulate cell motility (Miki et al., 1998; Nozumi et al., 2003; Yamazaki et al., 2003, 2005; Kim et al., 2006). However, WAVE2 knockout causes complete loss of lamellipodial protrusion in mouse embryonic fibroblasts, whereas WAVE1 knockout causes faster lamellipodial protrusion and increased ruffling (Yamazaki et al., 2003, 2005). The high degree of similarity between WAVE1 and WAVE2 in their domain layouts suggests that differences in their cellular activities may be specified by relatively minor sequence differences. Here we used total internal reflection fluorescence (TIRF) and electron microscopy to compare quantitatively the assembly kinetics and architectures of branched actin filament networks produced by Arp2/3 complex in combination with the VCA regions of WAVE1, WAVE2, or N-WASP. We observed novel and striking differences in their activities, which may underlie some of the aforementioned differences in their cellular roles.
RESULTS
WAVE1 dimeric VCA stimulates formation of branched actin networks with short daughter filaments
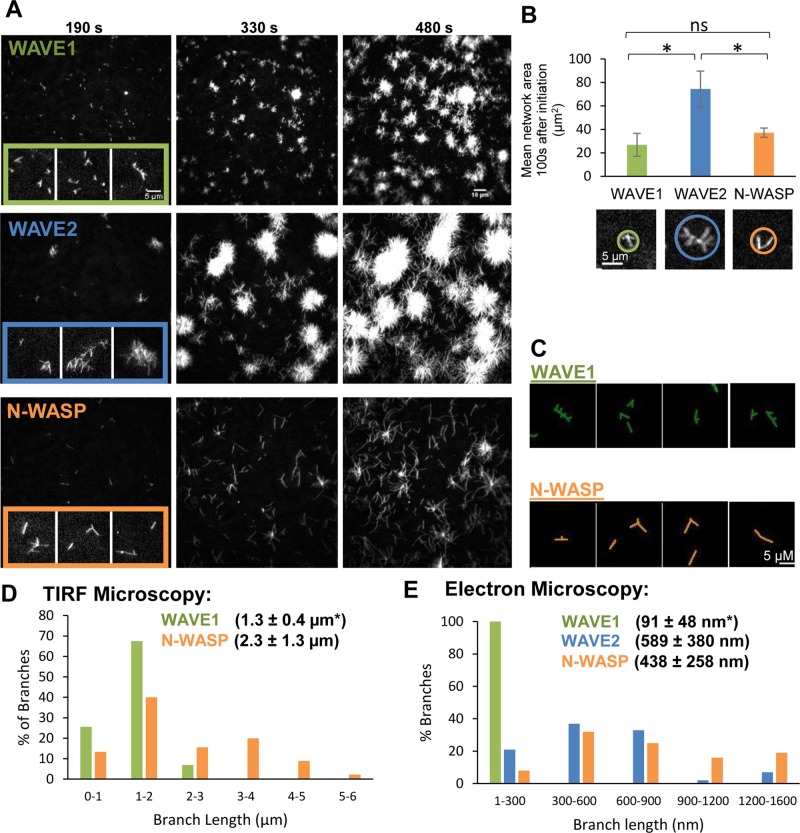
Previous studies used bulk assays to compare the VCA regions of different WASP/WAVE- family proteins for their potencies in stimulating Arp2/3 complex–dependent actin polymerization (Yamaguchi et al., 2000; Zalevsky et al., 2001; Yarar et al., 2002; Kang et al., 2010). However, there have been no studies comparing VCA activities by TIRF microscopy, by which filament growth rates and spatial organization can be monitored. Using TIRF microscopy, we directly compared branched actin network formation by Arp2/3 complex in combination with dimeric VCA regions (glutathione S-transferase [GST]-VCAs) of WAVE1, WAVE2, and N-WASP (Figure 1A and Supplemental Movie S1). WAVE2 had the most robust effects in stimulating actin network assembly. However, we also noticed key differences in the appearance of the networks. WAVE1 generated numerous small, tight nodes of actin, WAVE2 produced relatively dense networks of larger dimensions, and N-WASP produced fewer, less dense asters with longer filaments extending from them. Further, the two-dimensional areas of WAVE1 networks were smaller (by [3.6 ± 1.4]-fold) than WAVE2 networks measured 190 s after the network nucleation (Figure 1B). A closer examination of filaments at earlier time points in the TIRF reactions (insets in Figure 1A, traces in Figure 1C, quantification in Figure 1D) revealed that WAVE1 produced much shorter filaments than with N-WASP, consistent with the smaller dimensions of WAVE1 networks. Although branches formed by WAVE2 appeared to be long, similar to those formed by N-WASP, individual branches could not be traced since strong NPF activity of WAVE2 led to very dense networks whose branches appeared to overlap at the resolution of TIRF microscopy.
FIGURE 1:
TIRF microscopy analysis of branched actin networks produced by different WASP/WAVE family members in the presence of Arp2/3 complex. (A) Branched actin filament networks visualized by TIRF microscopy 190, 330, and 480 s after the initiation of assembly. Reactions contained 1 μM actin (10% Oregon green labeled), 10 nM bovine Arp2/3 complex, and 100 nM GST-VCA (human WAVE1, human WAVE2, or bovine N-WASP). Insets, higher- magnification views of branched filaments at 190 s; in the insets only, each image was individually autocontrasted to optimize the visualization of filaments. (B) Mean branched network surface area 100 s after initiation of assembly. For each condition, 10 networks were measured from each of three independent trials. Error bars, SEM. *p < 0.01; ns, not significant. Below the graph are representative images of the branched networks with circles defining the measured areas. (C) Skeletalized images from the 190-s time point highlighting differences in architecture between filaments assembled by WAVE1-Arp2/3 and N-WASP-Arp2/3. Owing to filament crowding, WAVE2 filaments could not be accurately traced. (D) Branch length distributions from the 190-s time point from TIRF reactions described in A. From 43 to 45 branches were pooled from three replicates of each condition. Mean branch lengths ± SD are given in parentheses. *p < 0.00001 relative to N-WASP by Student's t test. Owing to branch crowding, WAVE2 filaments could not be accurately measured. (E) Electron microscopy analysis of branch length distributions from actin filament networks produced by different WASP/WAVE family members with Arp2/3 complex. Actin networks were assembled by Arp2/3 and GST-VCA of WAVE1, WAVE2, or N-WASP. Representative images are shown in Supplemental Figure S1. Branch lengths (n = 39–45 for each condition) measured 5 min after initiation of assembly. Mean lengths for each condition ± SD are given in parentheses. *p < 0.01 relative to WAVE2 and N-WASP samples determined by ANOVA followed by Tukey's honestly significant difference (HSD) test.
As a complementary assay for comparing the lengths of actin filament branches, we used electron microscopy (EM; Figure 1E; example micrographs in Supplemental Figure S1). Although filaments can break during specimen preparation and thus reduce mean filament length (Xu et al., 1999), EM can nonetheless be a useful approach for assessing relative differences in filament length. Indeed, our EM analysis yielded mean daughter branch lengths of 91 (± 48), 589 (± 380), and 438 (± 258) nm for WAVE1, WAVE2, and N-WASP, respectively, confirming that WAVE1 produces shorter daughter branches.
WAVE1 VCA inhibits actin filament elongation independently of Arp2/3 complex
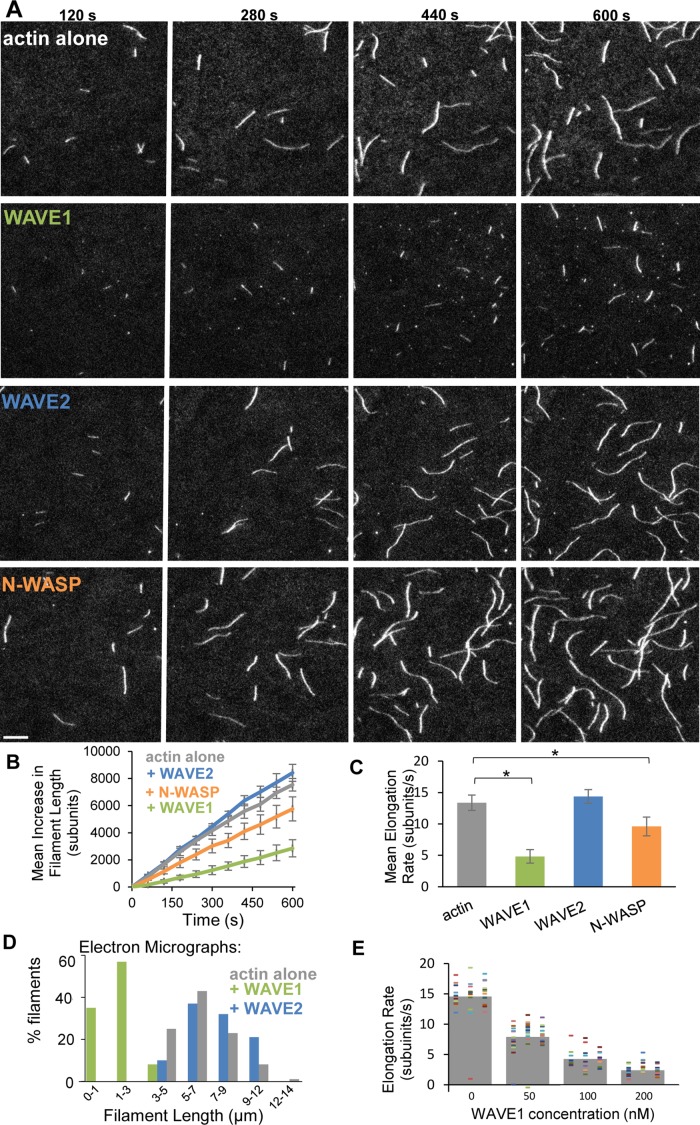
The foregoing observations led us to ask whether the effects of WAVE1 on filament length are dependent on Arp2/3 complex. To address this, we used TIRF microscopy to compare the effects of each dimeric VCA domain on the growth rates of individual filaments in the absence of Arp2/3 complex. With WAVE1, filaments grew ∼30% as fast and consequently were much shorter than filaments in reactions containing WAVE2 or N-WASP or control reactions (Figure 2, A–C, Supplemental Figure S2, A and B, and Supplemental Movie S2). These results were confirmed by bulk seeded polymerization assays (Supplemental Figure S2C) and by EM analysis of filament lengths (Figure 2D and Supplemental Figure S2D). Further, the inhibitory effect of WAVE1 was concentration dependent and displayed a unimodal distribution of elongation rates at each concentration of WAVE1 tested (Figure 2E), indicating that all filaments, and not merely a subset of filaments, were elongating at the reduced rate in the presence of WAVE1. Elongation inhibition by WAVE1 also was observed at different salt concentrations, including 150 mM KCl (Supplemental Figure S2E), and occurred in both the presence and absence of profilin (Supplemental Figure S2F). Inhibition required dimerization of the WAVE1 VCA region, as monomeric WAVE1 (MBP-VCA) failed to inhibit elongation in TIRF assays (Supplemental Figure S2G). Together these results indicate that the shorter filaments in reactions containing dimerized WAVE1 (Figures 1 and 2) result from inhibition of elongation, which occurs independently of Arp2/3 complex.
FIGURE 2:
WAVE1 inhibits actin filament elongation in the absence of Arp2/3 complex. (A) Representative TIRF images of filaments at different time points after initiation of actin assembly. Reactions contained 1 μM G-actin (10% Oregon green labeled) and 100 nM GST-VCA (human WAVE1, human WAVE2, or bovine N-WASP). (B) Mean increase in filament length over time from reactions as shown in A. From 9 to15 filaments per reaction were analyzed from three independent trials. Error bars, SEM. (C) Mean filament elongation rates from reactions in A. Error bars, SEM. *p < 0.01 by one-way ANOVA and Tukey's HSD tests. (D) Electron microscopy analysis of filament length distributions (n = 18–78 for each condition). Actin was assembled in the presence or absence of different WASP/WAVE GST-VCAs (all human) and negatively stained 15 min after initiation of assembly. (E) Concentration-dependent effects of WAVE1 on actin filament elongation. TIRF microscopy reactions contained 1 μM G-actin (10% Oregon green labeled), 3 μM human profilin, and indicated concentrations of human WAVE1 GST-VCA. Dashes represent elongation rates for individual filaments; owing to overlap, not all dashes may be visible. From 30 to 45 filaments were pooled from three independent trials.
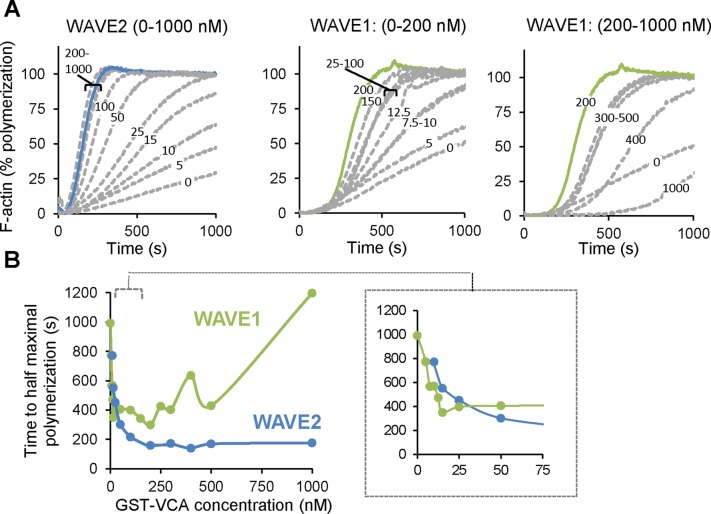
These results suggested that WAVE1 VCA domain has dual functions in network formation, serving both to stimulate actin nucleation by Arp2/3 complex and decrease rate of filament elongation independent of Arp2/3 complex. To further dissect these two roles of WAVE1, we compared WAVE1 and WAVE2 for stimulation of Arp2/3 complex–mediated actin assembly over a wide concentration range in bulk assays (Figure 3, A and B). For WAVE2, the time to half-maximal polymerization decreased with increasing concentration of WAVE2 until the effects plateaued at ∼200 nM. For WAVE1, it was only at lower concentrations that we observed faster assembly rates correlating with increasing concentrations of WAVE1 (Figure 3, A, middle, and B, inset), and then at higher concentrations of WAVE1 (>200 nM) the assembly rate instead decreased with increasing concentration (Figure 3, A, right, and B). An important point made by these results is that the nucleation efficiency of WAVE1 can be masked in bulk assays due to its inhibitory effects on filament elongation.
FIGURE 3:
Concentration-dependent effects of WAVE1 and WAVE2 on Arp2/3-mediated actin assembly. (A) Assembly kinetics for reactions containing 2 μM G-actin (5% pyrene labeled), 20 nM Arp2/3, and 0–1000 nM WAVE1 or WAVE2 GST-VCA as indicated. Curves for 200 nM GST-VCA are depicted as solid blue (WAVE2) or green (WAVE1) lines for easier comparison. (B) Time to half-maximal actin polymerization measured for reactions in A; inset highlights the effects of low concentrations of GST-VCA.
The elongation-inhibitory activity in WAVE1 maps to its WH2 domain
To determine what portion(s) of the WAVE1 VCA region mediate the inhibitory effects on elongation, we generated a series of chimeras between WAVE1 and N-WASP, or WAVE1 and WAVE2, and compared their effects on filament growth rate in TIRF assays (Figure 4, A and B, and Supplemental Figure S3A). All chimeras containing the V or VC portions of WAVE1 reduced elongation rates similar to WAVE1 VCA, and, conversely, all chimeras containing the V or VC portions of N-WASP produced elongation rates similar to N-WASP VCA (Figure 4A). A longer construct of N-WASP that included both of its tandem WH2 domains (GST-VVCA) had activity similar to the shorter N-WASP (GST-VCA; Supplemental Figure S3D). Further, we found that WAVE1-WAVE2 chimeras containing the V domain of WAVE1 had reduced rates of elongation similar to WAVE1 VCA, and those containing the V domain of WAVE2 did not (Figure 4B). Collectively these data show that the effects of WAVE1 in slowing elongation are derived from its V (WH2) domain.
FIGURE 4:
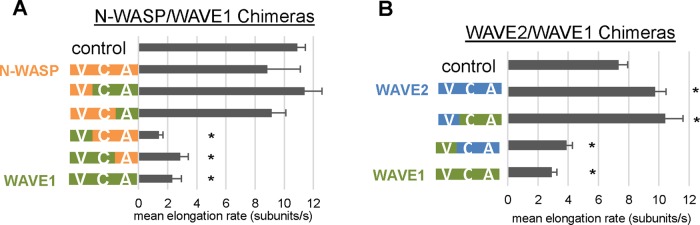
Chimera analysis reveals that WAVE1 inhibitory effects on filament elongation stem from its WH2 (V) domain. (A) TIRF analysis of mean elongation rates for filaments polymerized in the presence of GST-VCA chimeras between bovine N-WASP and human WAVE1. Reactions contained 1 μM G-actin (10% Oregon green labeled), 3 μM profilin, and 0 or 100 nM GST-VCA. From 11 to 15 filaments per reaction were analyzed for each of three independent trials. Error bars, SEM. *p <0.01 by one-way ANOVA and Tukey's HSD tests. (B) TIRF analysis of mean elongation rates for filaments polymerized in the presence of GST-VCA chimeras between human WAVE1 and human WAVE2. Reactions were performed and analyzed as in A.
We also examined the effects of chimeric VCA regions in combination with Arp2/3 complex in TIRF microscopy assays (Supplemental Movie S3 and Supplemental Figure S3, B and C). Chimeras containing the V or VC domain of N-WASP with the A domain of WAVE1 gave rise to large, densely branched actin networks, similar to WAVE2-Arp2/3 complex, and an accelerated rate of total polymer mass assembly. Conversely, chimeras containing the V or VC domain of WAVE1 and the A domain of N-WASP produced smaller networks with slower rates of total polymer assembly. These observations support the view that VCA regions have a modular design and that the CA region specifies the potency of Arp2/3 complex nucleation-promoting activity, whereas the WH2 (V) domain independently regulates the rate of elongation.
Lys-507 in the WH2 domain of WAVE1 is critical for the effects on elongation
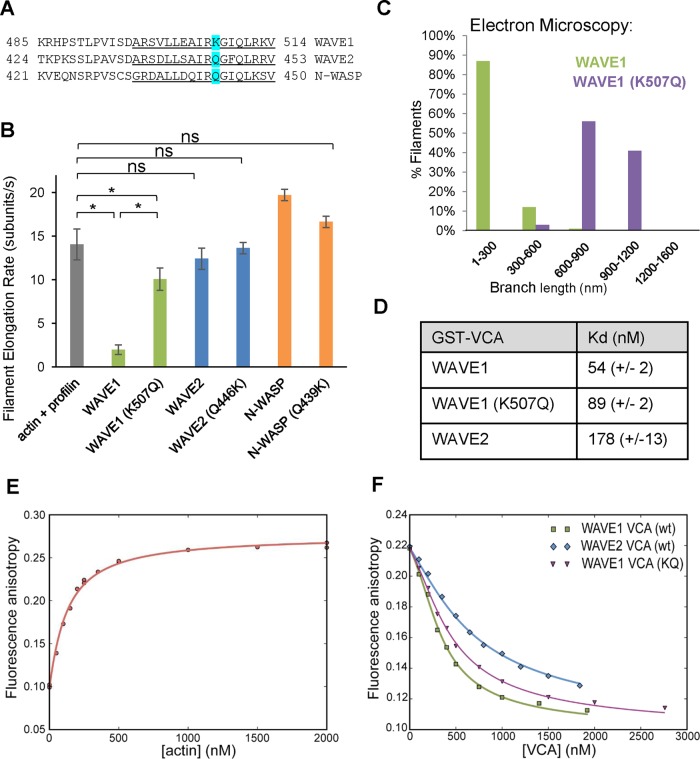
We noted that just one residue in the WH2 domain was the same in WAVE2 (Q446) and N-WASP (Q443) but different in WAVE1 (K507), making it a candidate to explain their different effects on actin filament elongation (Figure 5A). A K507Q substitution in WAVE1 greatly reduced its inhibitory effects on elongation, although Q-to-K substitutions in WAVE2 and N-WASP did not convert them into inhibitors of elongation (Figure 5, B and C). Thus K507 is required but not sufficient for VCA to inhibit elongation. Lysine 507 is conserved in WAVE1s of diverse vertebrate species with multiple WAVE genes yet is absent from nonvertebrate species with single SCAR/WAVE genes (Supplemental Figure S4A). Further, Saccharomyces cerevisiae Las17 (the only WASP/WAVE protein in budding yeast) lacks this lysine, and its GST-VCA domain did not inhibit elongation (Supplemental Figure S4B). Fluorescence anisotropy assays showed that the affinity of WAVE1 for actin monomers was threefold higher than for WAVE2 and that the K507Q mutation had only a small effect on the affinity of WAVE1 for actin monomers (Figure 5, D–F). This small difference in affinity is unlikely to account for the large effect of the K507Q mutation on elongation (discussed later).
FIGURE 5:
Identification of a residue (Lys-507) in WAVE1 that is necessary for inhibition of filament elongation. (A) Sequence alignment of “V” regions from WAVE1, WAVE2, and N-WASP (all human) constructs used. The WH2 domains in each sequence are underlined. Lys-507 in WAVE1, and the corresponding residues in WAVE2 and N-WASP are highlighted in cyan. (B) Mean elongation rates for filaments assembled in the presence of wild-type or mutant GST-VCAs measured by TIRF microscopy. Reactions contained 1 μM G-actin (10% Oregon green labeled) and 3 μm profilin with or without 100 nM GST-VCAs. From 8 to 15 filaments per reaction were analyzed from each of three independent trials. Error bars, SEM. *p < 0.01 using one-way ANOVA and Tukey's HSD tests. (C) Electron microscopy analysis of branch length distributions (n = 34–62 for each condition) measured 15 min after initiation of actin assembly in reactions containing Arp2/3 complex and wild-type or mutant (K507Q) WAVE1 GST-VCA. (D) Apparent binding affinities (Kd) of GST-VCAs for G-actin determined by anisotropy competition experiments, as in F. Indicated errors are 1σ confidence interval on the fits of the data. (E) Fluorescence anisotropy measured for 40 nM Alexa 594–N-WASP VCA and different concentrations of G-actin, which gives a binding isotherm of Kd = 106 nM. (F) Concentration-dependent competitive effects of different (unlabeled) GST-VCAs on the fluorescence anisotropy of 40 nM Alexa 594–N-WASP VCA and 250 nM G-actin.
DISCUSSION
WASP/WAVE proteins function downstream of multiple signaling pathways, linking Arp2/3 complex–mediated actin assembly to various cellular functions (Padrick and Rosen, 2010; Rotty et al., 2013). Until now, functional differences among WASP/WAVE proteins have primarily been ascribed to domains residing outside of their VCA regions, since these non-VCA domains interact with a range of different cellular factors to govern protein localization and release from transinhibited or autoinhibited states (Padrick and Rosen, 2010). The only reported differences in the activities of WASP/WAVE VCA regions have been the relative potencies in stimulating actin nucleation by Arp2/3 complex (Yamaguchi et al., 2000; Suetsugu et al., 2001; Zalevsky et al., 2001; Yarar et al., 2002; Kang et al., 2010). In this study, we uncovered a novel actin-filament elongation-inhibitory activity that specifically resides in the VCA region of WAVE1 but not WAVE2 or N-WASP and is independent of Arp2/3 complex.
Although interactions with filament barbed ends have been reported for N-WASP VCA, the functional effects of those interactions are distinct from the effects we observed for WAVE1 VCA (Co et al., 2007; Hu and Kuhn, 2012; Khanduja and Kuhn, 2014; Carlier et al., 2013). For example, the WH2 domain of N-WASP helped link actin filament comet tails to N-WASP–coated beads (Co et al., 2007), and nanoparticle-tethered N-WASP VVCA accelerated actin filament elongation more than threefold compared with untethered filaments (Khanduja and Kuhn, 2014). However, the latter effects were primarily observed for bundled filaments, whereas we did not observe strong effects of N-WASP on elongation for nonbundled filaments. Thus the ability of N-WASP to accelerate barbed-end elongation may depend on higher-order clustering of N-WASP at a surface and/or a specific filamentous architecture (e.g., bundles instead of branched or single filaments). Nonetheless, our study is the first to compare directly the effects of different WASP/WAVE proteins on filament growth rates. In doing so, we identified WAVE1 as an inhibitor rather than an accelerator of elongation, thus expanding the functional roles of the WASP/WAVE family.
How the unique activities of WAVE1 may contribute to its cellular functions
The activities we observed for WAVE1 in slowing actin filament elongation may be uniquely tailored to its cellular roles and could help explain why vertebrates express multiple WAVE proteins. The strong NPF activity of WAVE1 on Arp2/3 complex combined with its inhibitory effects on elongation produce dense actin networks that expand slowly, whereas WAVE2 produces fast-growing networks. In mouse embryonic fibroblasts, which express both WAVE1 and WAVE2, deletion of WAVE2 greatly diminishes lamellipodia formation, whereas deletion of WAVE1 has opposite effects, resulting in rapidly protruding but shorter-lived lamellipodia (Yamazaki et al., 2005). WAVE1−/− cells also formed a deeper leading-edge actin network with reduced intensity of F-actin staining (Yamazaki et al., 2005), consistent with the idea that WAVE1 generates more densely branched but slower-growing actin networks. Our observations also may help explain why loss of WAVE1 in neuronal growth cones increases rather than decreases lamellipodial extension velocity (Yamazaki et al., 2005; Soderling et al., 2007). Further, WAVE1 plays important roles in dendritic spine formation and in learning and memory (Soderling et al., 2003; Kim et al., 2006; Hazai et al., 2013); our results suggest that WAVE1 could influence these processes by governing the assembly of both the unbranched and branched actin filaments found in spines (Korobova and Svitkina, 2010).
In addition, cells may tune membrane protrusion dynamics and steer migration by expressing and locally activating different WAVE proteins. We found that WAVE1 still inhibits filament elongation in vitro in the presence of excess WAVE2 (Supplemental Figure S5, A and B, and Supplemental Movie S4), suggesting that activation of WAVE1 may be sufficient to slow membrane protrusion even if WAVE2 were present and active. Thus local control of WAVE1 activity could transiently accelerate or decelerate protrusions and help steer cell migration, similar to the recently described effects of Arpin, an inhibitor of Arp2/3 complex (Dang et al., 2013).
Mechanism by which WAVE1 inhibits actin filament elongation and relationship to other WH2 domain-containing proteins
The mechanism of WAVE1's inhibitory effects on actin filament elongation has yet to be resolved but seems likely to involve WAVE1 forming either a “transient cap” or a “processive cap” at the growing barbed end. In the transient-cap model, WAVE1 VCA dimers would first deliver actin monomers to the barbed end but then dissociate slowly, creating a temporary obstruction to elongation. In the processive-cap model, WAVE1 VCA dimers would ride the growing barbed end (similar to formins), allowing insertional assembly but at a reduced rate compared with free barbed ends. The processive-cap model predicts that at low concentrations of WAVE1, only a subset of filaments in TIRF reactions should be bound by WAVE1 and thus elongate at the slower rate while the remaining filaments grow at the uninhibited rate. This would produce a bimodal distribution of elongation rates. The transient-cap model instead predicts that all of the filaments in the population experience similar rates of elongation, which scale with WAVE1 concentration. This produces a unimodal distribution of elongation rates at any concentration. Because we observed a unimodal distribution of elongation rates at different concentrations of WAVE1 (Figure 2E), our results seem to support a transient cap model. This might involve the WH2 domain of WAVE1 blocking actin subunit addition at the barbed end by binding to the cleft between subdomains 1 and 3 in the ultimate and penultimate subunits of the filament (modeled in Figure 6); this would partly occlude engagement of the D loop of the next actin subunit, thus hindering monomer addition.
FIGURE 6:
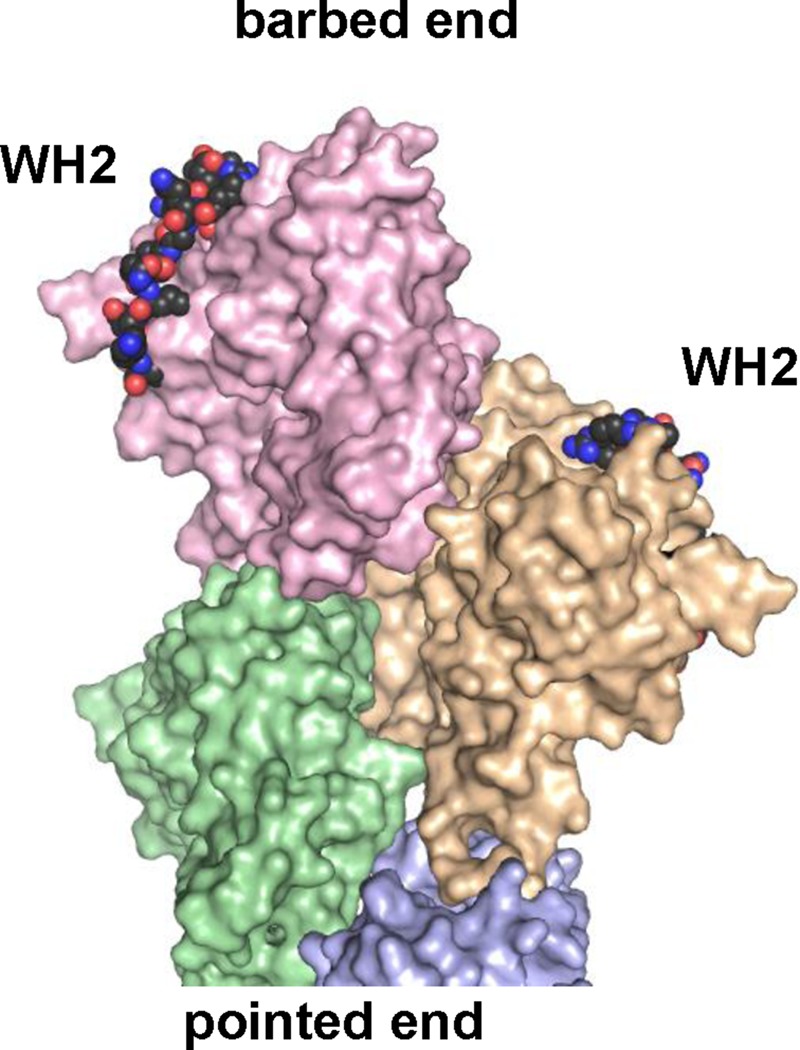
Two WH2 domains modeled on the barbed end of an actin filament. Four actins in a filament conformation (Oda et al., 2009) are shown as pale-colored surfaces. Actin-bound WH2s (from WAVE2; Chereau et al., 2005) were docked onto the barbed end of the four-actin cluster using the actin to guide alignment. Guide actin is hidden, and WH2s are rendered as colored spheres with black carbons.
Another open question is how Lys-507 in WAVE1 contributes critically to the inhibitory mechanism. Based on the WAVE2 WH2-actin monomer cocrystal structure (Chereau et al., 2005), Gln-446 (which corresponds to WAVE1 Lys-507) is not predicted to contact actin directly (Supplemental Figure S3E; Chereau et al., 2005). Alternatively, Lys-507 in WAVE1 might stabilize dynamics within the WH2 α-helix to strengthen WH2 interactions with the barbed end. Given that mutating Lys-507 in WAVE1 (K507Q) virtually abolished barbed-end inhibitory effects but only reduced actin monomer affinity less than twofold (Figure 5, D and F), this suggests that K507 may be particularly important for WAVE1 interactions with filament ends compared with monomers.
Beyond their roles in WASP/WAVE-family proteins, WH2 domains are found in diverse actin regulators and perform a range of critical functions, including nucleation, capping, and severing (Carlier et al., 2013). These activity differences can be specified by the precise sequences of the WH2 domains in these proteins and the spatial arrangement of the WH2 domains. Because the inhibitory effects of WAVE1 depend on Lys-507, we aligned WH2 sequences from a number of actin-regulatory proteins (Supplemental Figure S4C) and found a lysine at the same position in the WH2 domains of four other proteins (SPIRE, WIP, WIRE, and INF2). Further, in each of these cases, there was an aspartate residue located three or four residues N-terminal to the lysine, introducing the potential for a helix-stabilizing salt bridge. It will be intriguing to learn whether any of these other WH2 domain–containing proteins have activities and/or mechanisms related to those of WAVE1.
MATERIALS AND METHODS
Plasmids
Human VCA sequences were PCR amplified from pET11a-WAVE1-VCA (Panchal et al., 2003), pET11-WAVE2-VCA, and pGEX2T-N-WASP-VVCA (Panchal et al., 2003) parent plasmids. Inserts were ligated into a pGAT2 vector at the BamHI and NsiI sites as indicated in Supplemental Table S1. Bovine N-WASP VCA domain was purified using pGEX6-N-WASP-VCA (amino acids 422–505), a gift from David Kovar (University of Chicago, Chicago, IL). Point mutations in VCA domains were generated by primer-based site-directed mutagenesis, and chimeras were generated by PCR-amplifying VCA subdomains using primers with long 5′ tails overlapping the “new” neighboring domain and then combining the subdomains in a second PCR and religating the new chimeric VCA into pGAT2.
Protein purification
Unlabeled rabbit muscle actin (RMA) was purified and gel filtered, and RMA was labeled with N-(1-pyrene) iodoacetamide or Oregon green as described in detail (Graziano et al., 2013). Bovine Arp2/3 complex was generously provided by Brad Nolen (University of Oregon, Eugene, OR) and purified as previously described (Higgs et al., 1999; Higgs and Pollard, 2000). GST-VCA polypeptides were expressed in Escherichia coli, and lysates were cleared at 18,000 rpm for 15 min. Cleared lysates were either incubated with glutathione-agarose beads or loaded onto a Profinia Protein Purification System (Bio-Rad, Hercules, CA) equipped with a 1-ml GST cartridge and 10-ml desalting cartridge. Aliquots of GST-VCA in 20 mM Tris, pH 7.5, and 50 mM KCl were snap frozen and stored at −80°C. Freshly gel-filtered WAVE1 GST-VCA showed the same effects on actin filaments, both in the presence and absence of Arp2/3 complex, as non–gel-filtered WAVE1 GST-VCA (Supplemental Figure S6), demonstrating that the effects were not due to WAVE1 GST-VCA aggregation. Untagged human profilin was expressed from plasmid pBG942 in E. coli. E. coli pellets resuspended in 50 mM Tris-HCl, pH 8.0, with DNase and lysozyme were sonicated and centrifuged at 80,000 rpm for 25 min. Profilin was then purified from cell lysates by sequential chromatography on a Q-High trap column, followed by a Superdex5 gel filtration column. For anisotropy experiments, proteins were purified independently as follows. Plasmids were transformed into BL21(DE3) T1R bacteria, which were grown at 37°C in Luria broth in shaker flasks to an OD600 nm of 0.6–1.0 and then induced using 1 mM isopropyl-β-d-thiogalactoside for 3 h at 37°C. Cells were harvested by centrifugation, resuspended in 200 mM NaCl, 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.5, 2 mM ethylenediaminetetraacetic acid, 2 mM dithiothreitol (DTT), and 1 mM phenylmethylsulfonyl fluoride, and snap frozen in liquid nitrogen. Cells were thawed in cool water, lysed by extrusion (Emulsiflex C5; Avestin, Ottawa, Canada) with inline chilling, and clarified by centrifugation at 19,500 rpm in a JA25.5 rotor (Beckman Coulter, Brea, CA). Clarified lysates from 4.5 l of culture were purified using glutathione Sepharose (GE Biosciences, Uppsala, Sweden) affinity chromatography, and eluates were snap froze in liquid nitrogen. Glutathione eluates were thawed and further purified by Mono Q anion exchange chromatography and Superdex 200 gel filtration chromatography. Concentrations were determined by ultraviolet absorption at 280 nm, and proteins were used without concentration or freezing. Alexa 594–labeled N-WASP VCA (S430C) was prepared as described (Doolittle et al., 2013; Smith et al., 2013b).
Pyrene-actin assembly assays
Pyrene-actin assembly assays were performed in a fluorescence spectrophotometer (Photon Technologies International, Lawrenceville, NJ) as described (Ydenberg et al., 2013). In seeded elongation assays, 5 μl if preformed F-actin (1 μM final) was mixed with 20 μl of 20/50 buffer (20 mM Tris, pH 7.5; 50 mM KCl) alone or with proteins. Filaments were sheared by six passages through a 27-gauge needle and syringe. We added 20 μl of sheared filaments/protein mixture (final 333 nM F-actin) to 40 μl of monomeric actin (0.5 or 1 μM final, 1% pyrene labeled) in F-buffer (3 mM Tris, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2, 2 mM MgCl2, 50 mM KCl, 0.5 mM DTT) to initiate filament elongation. Pyrene fluorescence was monitored as described.
TIRF microscopy
TIRF microscopy was performed as described (Graziano et al., 2013). TIRF reactions contained 1 μM actin monomers (10% Oregon green labeled) and different concentrations as indicated of Arp2/3 complex and GST-VCA in TIRF buffer (10 mM imidazole, 50 mM KCl, 1 mM MgCl2, 1 mM ethylene glycol tetraacetic acid [EGTA], 0.2 mM ATP, 10 mM DTT, 15 mM glucose, 20 μg/ml glucose oxidase, and 0.5% methylcellulose [4000 cP], pH 7.4). Movies were recorded for 5–10 min on a Nikon Ti2000 microscope (Nikon Corporation, Tokyo, Japan), acquiring 200 ms exposures every 10 s. Three fields of view were analyzed per reaction, and all reactions were performed in triplicate. In some reactions, 3 μM human profilin was included to suppress spontaneous nucleation and avoid filament crowding in fields. TIRF analysis was performed in ImageJ (National Institutes of Health, Bethesda, MD). For quantification of branch length, 13–15 branches were measured for each of three replicates for WAVE1 and N-WASP conditions 190 s after reaction initiation. Only filaments that could be clearly distinguished as stable branches (as opposed to mother filaments or overlapping filaments) were measured. Actin filament elongation rates were calculated by measuring lengths for 8–15 filaments per reaction every 60s over 520–600 s in ImageJ. Significance groups were determined using a one-way analysis of variance (ANOVA; www.danielsoper.com/statcalc3/calc.aspx?id=43) and a Tukey Test (faculty.vassar.edu/lowry/hsd.html). Total fluorescence per field of view was measured using the Raw Integrated Density measurement feature with or without background subtraction with a rolling ball radius of 50 pixels. Network surface areas were determined in ImageJ by selecting the smallest circular region of interest that enclosed the network 100 s after the first filament in the network could be observed.
Electron microscopy
RMA (10 μM) was polymerized by addition of 2 mM MgCl2 and 50 mM KCl in the presence or absence of Arp2/3 complex (0.1 μM) and different GST-VCAs (1 μM). Either 5 or 15 min after initiation of polymerization, samples were diluted 10-fold in F-buffer (5 mM Tris, pH 8.0, 50 mM KCl, 2 mM MgCl2, 0.2 mM EGTA, 1 mM DTT), applied for 30 s to freshly glow discharged formvar-carbon–coated 200-mesh copper grids, blotted to remove excess solution, negatively stained with 1% (wt/vol) uranyl acetate for 1 min, blotted, and allowed to air-dry. Images were recorded on a charge-could device with a FEI Morgani 268 transmission electron microscope (FEI, Hillsboro, OR) at an acceleration voltage of 80 kV and magnifications of 3500, 5600, and 11,000. For measurement of daughter-filament branch lengths, pictures were taken at a magnification of 5600 or 11,000. Branches were counted only if they met three criteria: 1) they had a visible Arp2/3 complex at the junction, 2) the branch angle was close to 70°, and 3) the daughter filament was oriented in the same direction as the mother filament. Because filaments polymerized in the absence of Arp2/3 complex were too long to be captured on a single micrograph, the micrographs of several adjacent areas on the grid (10–20 images taken at magnification of 3500) were combined into one picture using Photoshop (Adobe, San Jose, CA). The contour length of each filament was measured using ruler tool in Photoshop. The number-average length (Ln) was defined as Ln = (1/n) Σ li, where n is the number of filaments and li is the contour length of each filament.
Fluorescence anisotropy–detected actin binding
N-WASP VCA (S430C) was labeled with an Alexa Fluor 594 maleimide dye. Fluorescence anisotropy was performed at 20°C using a T-form PTI QuantaMaster spectrofluorimeter (Photon Technology International, Edison, NJ) equipped with temperature control and Glan–Thompson linear polarizers. Alexa 594 was excited using 590-nm light (10-nm bandpass) and detected at 612 nm (10-nm bandpass), averaging for 4 min. Actin polymerization was avoided by keeping actin monomer concentration low (final 250 nM) and by exchanging actin from buffer G immediately before the sample was mixed. Alexa 594 VCA and competitor (unlabeled) GST-VCAs were added to actin and incubated at 20°C for 5–10 min. Final buffer conditions were 10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM EGTA, 1 mM MgCl2, 20 μM ATP, and 0.5 mM DTT. Binding isotherms were fitted to a complete competition-binding solution for a single-site receptor, using Levenberg–Marquardt nonlinear least squares methods. Direct-binding isotherms were fitted with the concentration of competitor ligand set to zero. Both the bound and free fluorescence anisotropy values were optimized parameters, and competition-binding isotherm fitting used the observed Kd from the optimized direct-binding isotherm fit. Optimized values of free Alexa 594–VCA fluorescence anisotropy values were similar to the actual free anisotropy, and the values of free and bound fluorescence anisotropy determined from competition-binding experiments was similar to those obtained in fitting the direct-binding system.
Supplementary Material
Acknowledgments
We are extremely grateful to Brad Nolen for providing purified bovine Arp2/3 complex. We also thank Jeff Gelles, Adam Johnston, Ben Smith, Casey Ydenberg, and Richa Jaiswal for many helpful discussions, technical advice, and editing of the manuscript. S.B.P. was supported by grants to Michael K. Rosen from the Howard Hughes Medical Institute, National Institutes of Health (R01-GM56322), and the Welch Foundation (I-1544). This work was supported primarily by a grant from the National Institutes of Health (GM063691) to B.G.
Abbreviations used:
- Arp2/3
actin-related protein 2/3
- NPF
nucleation-promoting factor
- N-WASP
neuronal Wiskott–Aldrich syndrome protein
- WAVE
WASP and verprolin homologue
- TIRF
total internal reflection fluorescence
- VCA
verprolin homology/central/acidic.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-10-1477) on December 3, 2014.
REFERENCES
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, et al. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev. 2014;94:235–263. doi: 10.1152/physrev.00018.2013. [DOI] [PubMed] [Google Scholar]
- Boczkowska M, Rebowski G, Kast DJ, Dominguez R. Structural analysis of the transitional state of Arp2/3 complex activation by two actin-bound WCAs. Nat Commun. 2014;5:3308. doi: 10.1038/ncomms4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Pernier J, Avvaru BS. Control of actin filament dynamics at barbed ends by WH2 domains: From capping to permissive and processive assembly. Cytoskeleton (Hoboken) 2013;70:540–549. doi: 10.1002/cm.21124. [DOI] [PubMed] [Google Scholar]
- Chereau D, Kerff F, Graceffa P, Grabarek Z, Langsetmo K, Dominguez R. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci USA. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Co C, Wong DT, Gierke S, Chang V, Taunton J. Mechanism of actin network attachment to moving membranes: barbed end capture by N-WASP WH2 domains. Cell. 2007;128:901–913. doi: 10.1016/j.cell.2006.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A, Warrington A, Taylor KA, Svitkina T. Structural organization of the actin cytoskeleton at sites of clathrin-mediated endocytosis. Curr Biol. 2011;21:1167–1175. doi: 10.1016/j.cub.2011.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JP, Wang-Dunlop J, Gonzales C, Goad ME, Mark RJ, Kwak SP. Characterization of the WAVE1 knock-out mouse: implications for CNS development. J Neurosci. 2003;23:3343–3352. doi: 10.1523/JNEUROSCI.23-08-03343.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang I, Gorelik R, Blin C, Derivery E, Guerin C, Linkner J, Nemethova M, Dumortier JG, Giger FA, Chipysheva TA, et al. Inhibitory signalling to the Arp2/3 complex steers cell migration. Nature. 2013;503:281–284. doi: 10.1038/nature12611. [DOI] [PubMed] [Google Scholar]
- Doolittle LK, Rosen MK, Padrick SB. Measurement and analysis of in vitro actin polymerization. Methods Mol Biol. 2013;1046:273–293. doi: 10.1007/978-1-62703-538-5_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohl C, Banovic D, Grevelhorster A, Bogdan S. WAVE forms hetero- and homo-oligomeric complexes at integrin junctions in Drosophila visualized by bimolecular fluorescence complementation. J Biol Chem. 2010;285:40171–40179. doi: 10.1074/jbc.M110.139337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley ED, Rodenbusch SE, Martin AC, Welch MD. Critical conformational changes in the Arp2/3 complex are induced by nucleotide and nucleation promoting factor. Mol Cell. 2004;16:269–279. doi: 10.1016/j.molcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Graziano BR, Jonasson EM, Pullen JG, Gould CJ, Goode BL. Ligand-induced activation of a formin-NPF pair leads to collaborative actin nucleation. J Cell Biol. 2013;201:595–611. doi: 10.1083/jcb.201212059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazai D, Szudoczki R, Ding J, Soderling SH, Weinberg RJ, Sotonyi P, Racz B. Ultrastructural abnormalities in CA1 hippocampus caused by deletion of the actin regulator WAVE-1. PLoS One. 2013;8:e75248. doi: 10.1371/journal.pone.0075248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs HN, Blanchoin L, Pollard TD. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry. 1999;38:15212–15222. doi: 10.1021/bi991843+. [DOI] [PubMed] [Google Scholar]
- Higgs HN, Pollard TD. Activation by Cdc42 and PIP(2) of Wiskott-Aldrich syndrome protein (WASp) stimulates actin nucleation by Arp2/3 complex. J Cell Biol. 2000;150:1311–1320. doi: 10.1083/jcb.150.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Kuhn JR. Actin filament attachments for sustained motility in vitro are maintained by filament bundling. PLoS One. 2012;7:e31385. doi: 10.1371/journal.pone.0031385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie F, Yamaguchi Y. EphB receptors regulate dendritic spine development via intersectin, Cdc42 and N-WASP. Nat Neurosci. 2002;5:1117–1118. doi: 10.1038/nn964. [DOI] [PubMed] [Google Scholar]
- Ito H, Morishita R, Shinoda T, Iwamoto I, Sudo K, Okamoto K, Nagata K. Dysbindin-1, WAVE2 and Abi-1 form a complex that regulates dendritic spine formation. Mol Psychiatry. 2010;15:976–986. doi: 10.1038/mp.2010.69. [DOI] [PubMed] [Google Scholar]
- Kang H, Wang J, Longley SJ, Tang JX, Shaw SK. Relative actin nucleation promotion efficiency by WASP and WAVE proteins in endothelial cells. Biochem Biophys Res Commun. 2010;400:661–666. doi: 10.1016/j.bbrc.2010.08.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano Y, Yoshimura T, Tsuboi D, Kawabata S, Kaneko-Kawano T, Shirataki H, Takenawa T, Kaibuchi K. CRMP-2 is involved in kinesin-1-dependent transport of the Sra-1/WAVE1 complex and axon formation. Mol Cell Biol. 2005;25:9920–9935. doi: 10.1128/MCB.25.22.9920-9935.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanduja N, Kuhn JR. Processive acceleration of actin barbed end assembly by N-WASP. Mol Biol Cell. 2014;25:55–65. doi: 10.1091/mbc.E12-11-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Sung JY, Ceglia I, Lee KW, Ahn JH, Halford JM, Kim AM, Kwak SP, Park JB, Ho Ryu S, et al. Phosphorylation of WAVE1 regulates actin polymerization and dendritic spine morphology. Nature. 2006;442:814–817. doi: 10.1038/nature04976. [DOI] [PubMed] [Google Scholar]
- Korobova F, Svitkina T. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol Biol Cell. 2010;21:165–176. doi: 10.1091/mbc.E09-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Suetsugu S, Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 1998;17:6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller J, Pfanzelter J, Winkler C, Narita A, Le Clainche C, Nemethova M, Carlier MF, Maeda Y, Welch MD, Ohkawa T, et al. Electron tomography and simulation of baculovirus actin comet tails support a tethered filament model of pathogen propulsion. PLoS Biol. 2014;12:e1001765. doi: 10.1371/journal.pbio.1001765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozumi M, Nakagawa H, Miki H, Takenawa T, Miyamoto S. Differential localization of WAVE isoforms in filopodia and lamellipodia of the neuronal growth cone. J Cell Sci. 2003;116:239–246. doi: 10.1242/jcs.00233. [DOI] [PubMed] [Google Scholar]
- Oda T, Iwasa M, Aihara T, Maeda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- Padrick SB, Cheng HC, Ismail AM, Panchal SC, Doolittle LK, Kim S, Skehan BM, Umetani J, Brautigam CA, et al. Hierarchical regulation of WASP/WAVE proteins. Mol Cell. 2008;32:426–438. doi: 10.1016/j.molcel.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. Arp2/3 complex is bound and activated by two WASP proteins. Proc Natl Acad Sci USA. 2011;108:E472–E479. doi: 10.1073/pnas.1100236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, Rosen MK. Physical mechanisms of signal integration by WASP family proteins. Annu Rev Biochem. 2010;79:707–735. doi: 10.1146/annurev.biochem.77.060407.135452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchal SC, Kaiser DA, Torres E, Pollard TD, Rosen MK. A conserved amphipathic helix in WASP/Scar proteins is essential for activation of Arp2/3 complex. Nat Struct Biol. 2003;10:591–598. doi: 10.1038/nsb952. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal AA, Sokolova O, Robins DB, Daugherty KM, Hippenmeyer S, Riezman H, Grigorieff N, Goode BL. Conformational changes in the Arp2/3 complex leading to actin nucleation. Nat Struct Mol Biol. 2005;12:26–31. doi: 10.1038/nsmb870. [DOI] [PubMed] [Google Scholar]
- Rottner K, Hanisch J, Campellone KG. WASH, WHAMM and JMY: regulation of Arp2/3 complex and beyond. Trends Cell Biol. 2010;20:650–661. doi: 10.1016/j.tcb.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol. 2013;14:7–12. doi: 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- Rouiller I, Xu XP, Amann KJ, Egile C, Nickell S, Nicastro D, Li R, Pollard TD, Volkmann N, Hanein D. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol. 2008;180:887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez AM, Flamini MI, Fu XD, Mannella P, Giretti MS, Goglia L, Genazzani AR, Simoncini T. Rapid signaling of estrogen to WAVE1 and moesin controls neuronal spine formation via the actin cytoskeleton. Mol Endocrinol. 2009;23:1193–1202. doi: 10.1210/me.2008-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BA, Daugherty-Clarke K, Goode BL, Gelles J. Pathway of actin filament branch formation by Arp2/3 complex revealed by single-molecule imaging. Proc Natl Acad Sci USA. 2013a;110:1285–1290. doi: 10.1073/pnas.1211164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BA, Padrick SB, Doolittle LK, Daugherty-Clarke K, Correa IR, Jr, Xu MQ, Goode BL, Rosen MK, Gelles J. Three-color single molecule imaging shows WASP detachment from Arp2/3 complex triggers actin filament branch formation. Elife. 2013b;2:e01008. doi: 10.7554/eLife.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderling SH, Guire ES, Kaech S, White J, Zhang F, Schutz K, Langeberg LK, Banker G, Raber J, Scott JD. A WAVE-1 and WRP signaling complex regulates spine density, synaptic plasticity, and memory. J Neurosci. 2007;27:355–365. doi: 10.1523/JNEUROSCI.3209-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderling SH, Langeberg LK, Soderling JA, Davee SM, Simerly R, Raber J, Scott JD. Loss of WAVE-1 causes sensorimotor retardation and reduced learning and memory in mice. Proc Natl Acad Sci USA. 2003;100:1723–1728. doi: 10.1073/pnas.0438033100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillane M, Ketschek A, Donnelly CJ, Pacheco A, Twiss JL, Gallo G. Nerve growth factor-induced formation of axonal filopodia and collateral branches involves the intra-axonal synthesis of regulators of the actin-nucleating Arp2/3 complex. J Neurosci. 2012;32:17671–17689. doi: 10.1523/JNEUROSCI.1079-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S, Miki H, Yamaguchi H, Obinata T, Takenawa T. Enhancement of branching efficiency by the actin filament-binding activity of N-WASP/WAVE2. J Cell Sci. 2001;114:4533–4542. doi: 10.1242/jcs.114.24.4533. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman DM, Insall RH. WASP family proteins: their evolution and its physiological implications. Mol Biol Cell. 2010;21:2880–2893. doi: 10.1091/mbc.E10-04-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinzenz M, Nemethova M, Schur F, Mueller J, Narita A, Urban E, Winkler C, Schmeiser C, Koestler SA, Rottner K, et al. Actin branching in the initiation and maintenance of lamellipodia. J Cell Sci. 2012;125:2775–2785. doi: 10.1242/jcs.107623. [DOI] [PubMed] [Google Scholar]
- Westphal RS, Soderling SH, Alto NM, Langeberg LK, Scott JD. Scar/WAVE-1, a Wiskott-Aldrich syndrome protein, assembles an actin-associated multi-kinase scaffold. EMBO J. 2000;19:4589–4600. doi: 10.1093/emboj/19.17.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Casella JF, Pollard TD. Effect of capping protein, CapZ, on the length of actin filaments and mechanical properties of actin filament networks. Cell Motil Cytoskeleton. 1999;42:73–81. doi: 10.1002/(SICI)1097-0169(1999)42:1<73::AID-CM7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Xu K, Babcock HP, Zhuang X. Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton. Nat Methods. 2012a;9:185–188. doi: 10.1038/nmeth.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Zhong G, Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 2013;339:452–456. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XP, Rouiller I, Slaughter BD, Egile C, Kim E, Unruh JR, Fan X, Pollard TD, Li R, Hanein D, et al. Three-dimensional reconstructions of Arp2/3 complex with bound nucleation promoting factors. EMBO J. 2012b;31:236–247. doi: 10.1038/emboj.2011.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Miki H, Suetsugu S, Ma L, Kirschner MW, Takenawa T. Two tandem verprolin homology domains are necessary for a strong activation of Arp2/3 complex-induced actin polymerization and induction of microspike formation by N-WASP. Proc Natl Acad Sci USA. 2000;97:12631–12636. doi: 10.1073/pnas.190351397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki D, Fujiwara T, Suetsugu S, Takenawa T. A novel function of WAVE in lamellipodia: WAVE1 is required for stabilization of lamellipodial protrusions during cell spreading. Genes Cells. 2005;10:381–392. doi: 10.1111/j.1365-2443.2005.00845.x. [DOI] [PubMed] [Google Scholar]
- Yamazaki D, Suetsugu S, Miki H, Kataoka Y, Nishikawa S, Fujiwara T, Yoshida N, Takenawa T. WAVE2 is required for directed cell migration and cardiovascular development. Nature. 2003;424:452–456. doi: 10.1038/nature01770. [DOI] [PubMed] [Google Scholar]
- Yarar D, D’Alessio JA, Jeng RL, Welch MD. Motility determinants in WASP family proteins. Mol Biol Cell. 2002;13:4045–4059. doi: 10.1091/mbc.E02-05-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ydenberg CA, Padrick SB, Sweeney MO, Gandhi M, Sokolova O, Goode BL. GMF severs actin-Arp2/3 complex branch junctions by a cofilin-like mechanism. Curr Biol. 2013;23:1037–1045. doi: 10.1016/j.cub.2013.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky J, Lempert L, Kranitz H, Mullins RD. Different WASP family proteins stimulate different Arp2/3 complex-dependent actin-nucleating activities. Curr Biol. 2001;11:1903–1913. doi: 10.1016/s0960-9822(01)00603-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.