Abstract
DNA mismatch repair involves is a widely conserved set of proteins that is essential to limit genetic drift in all organisms. The same system of proteins plays key roles in many cancer related cellular transactions in humans. Although the basic process has been reconstituted in vitro using purified components, many fundamental aspects of DNA mismatch repair remain hidden due in part to the complexity and transient nature of the interactions between the mismatch repair proteins and DNA substrates. Single molecule methods offer the capability to uncover these transient but complex interactions and allow novel insights into mechanisms that underlie DNA mismatch repair. In this review, we discuss applications of single molecule methodology including electron microscopy, atomic force microscopy, particle tracking, FRET, and optical trapping to studies of DNA mismatch repair. These studies have led to formulation of mechanistic models of how proteins identify single base mismatches in the vast background of matched DNA and signal for their repair.
Introduction
All organisms require a stable genome. Continuous assault by endogenous and exogenous chemicals and the imperfect fidelities of DNA polymerases tend to degrade the genome. Consequently, multiple DNA repair pathways have evolved to maintain genomic integrity. If these repair pathways fall short, cell cycle checkpoints result that cause cell-cycle arrest and/or apoptosis. These pathways include DNA mismatch repair (MMR), which is responsible for correcting errors made during DNA replication. The MMR proteins also are involved in several other DNA transactions. Inactivation of the MMR genes not only dramatically increases the frequency of mutations, it also decreases apoptosis, increases cell survival, and results in resistance to many chemotherapeutic agents [1–3]. In humans, mutations in the genes responsible for the initiation of MMR are associated with > 80% of hereditary non-polyposis colorectal cancers (HNPCC) and certain sporadic cancers [4]. Patients with cancers associated with defects in MMR genes may be at particular risk because the loss of MMR results in resistance to the cytotoxic effects of several DNA damaging agents, such as cisplatin and alkylating agents, as well as increased mutagenesis due to the inability to repair replication errors generated from copying both normal and damaged bases. Together, these effects are thought to contribute to selective growth advantages for MMR defective cells during multistage carcinogenesis [5]. Understanding the molecular mechanisms that underlie these different activities will be essential for developing effective treatments, with minimal side effects.
MMR is initiated by MutS and MutL homologs, which are highly conserved throughout prokaryotes and eukaryotes. MutS and MutL homologs are dimers and contain DNA binding and ATPase activities that are essential for MMR in vivo [6, 7]. Eukaryotes have multiple heterodimeric MutS and MutL homologs [8–14]. MSH2-MSH6 (MutSα) is primarily responsible for repairing single base-base mismatches and one and two base insertions or deletions (IDLs), whereas MSH2-MSH3 (MutSβ) is primarily responsible for repairing larger IDLs [11, 15–18]. MutLα (MLH1-PMS2 in humans, Mlh1-Pms1 in yeast) is the major MutL homolog involved in MMR. The MMR proteins must both locate mismatches in a vast excess of correctly paired DNA and direct repair to the daughter strand.
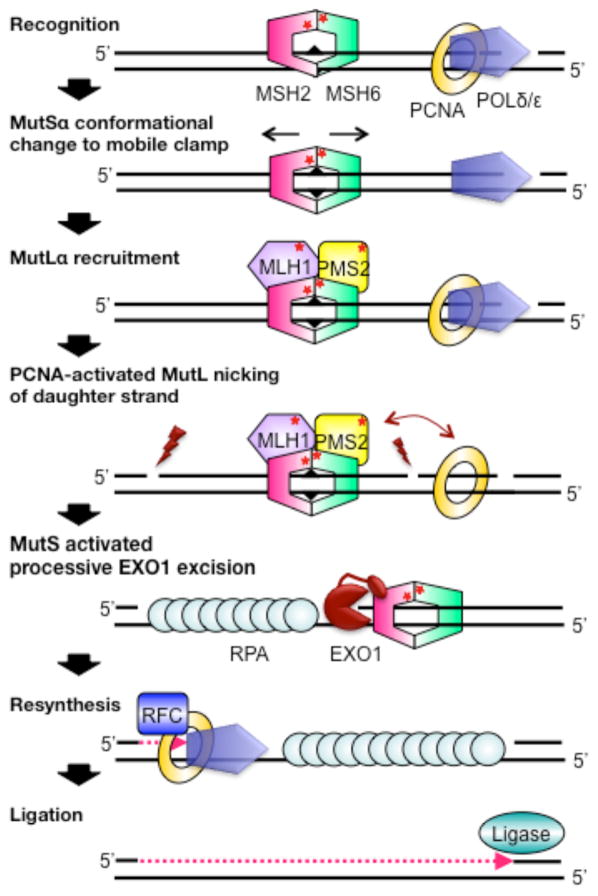
MMR has been reconstituted in vitro with E. coli and human proteins using plasmid DNA containing a mismatch and a nick (or a hemimethylated GATC site for E. coli) either 5′ or 3′ to the mismatch [19–26]. In eukaryotes, MMR is initiated by MutSα (or MutSβ) first recognizing a mismatch or IDL (Figure 1). ATP and mismatch binding induce a conformational change in MutSα, such that it forms a mobile clamp state that can move along the DNA. This activated state of MutSα, in turn, promotes its interaction with one or more MutLα proteins [27–35]. Subsequently, PCNA, which is a component of the replication apparatus, activates MutLα to incise the daughter strand hundreds of base pairs from a mismatch both distally (preferential) and proximally. [36, 37]. Once MutLα nicks the DNA, MutSα activates the exonuclease EXO1 to processively excise the DNA containing the incorrect nucleotide [20, 38–40]. Alternatively, POLδ/ε can initiate strand-displacement synthesis from the nick[41]. Finally, DNA resynthesis is catalyzed by DNA polymerases δ (lagging strand) or ε (leading strand), and DNA ligase seals the nick [42–44]. Because PCNA is loaded asymmetrically on DNA at the replication fork (or at a nick), the PCNA-activated nicking of the DNA by MutLα may serve as a strand-discrimination signal in MMR [45]. For leading strand synthesis, this nicking is thought to be essential to provide a nick 5′ to the mismatch in the daughter strand; however, for lagging strand synthesis, the 5′-end of the Okazaki fragment could also provide a transient strand-discrimination signal [46–48]. Consistent with this latter suggestion, MutLα is not required for 5′-nick directed repair in vitro [49, 50].
Figure 1.
Mechanism of human mismatch repair. Triangles, stars and lightning bolts represent the mismatch, ATPase sites, and MutLα nicking, respectively.
The early steps of MMR are similar in prokaryotes; however, processing of the DNA after mismatch recognition differs in prokaryotes and eukaryotes, which puts different constraints on signaling repair. In prokaryotes, the MutS-MutL initiation complex “directs” UvrD-catalyzed unwinding towards the mismatch, followed by excision by the appropriate exonuclease [6, 7, 51]. In contrast, no helicases are known to be involved in eukaryotic MMR, and EXO1, a double-stranded 5′ to 3′ exonuclease, is the only exonuclease that is clearly involved in repair [7]. Consequently, it is not necessary for MutSα (or MutLα) to confer directionally on EXO1, only to activate it.
Here, we review applications of single molecule experimentation to investigate these MMR phenomena.
MutS searching homoduplex DNA for mismatches
In the 1970s and 1980s, it was proposed that DNA binding proteins underwent 1D diffusion along DNA to explain the observation that the rate of association of proteins with their specific sites on long DNA molecules is faster than theoretically possible based on 3D diffusion alone [52–54]. Many subsequent biochemical studies, which investigated the DNA length dependence of proteins binding to their specific sites, supported 1D diffusion, but it was the onset of single-molecule tracking studies that allowed the direct observation of such behavior. Early tracking and AFM studies with RNA polymerase demonstrated 1D diffusion. [55–58]
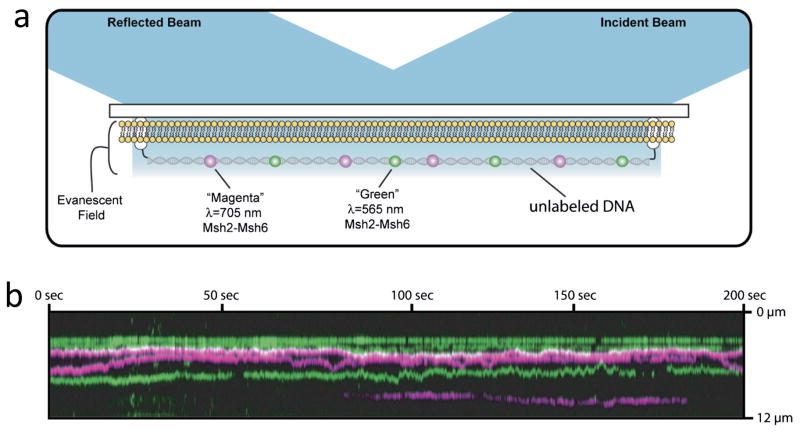
Single particle tracking of proteins on DNA with fluorescence microscopy requires long, extended DNA substrates along with fluorescent labeling of the proteins. Experiments typically use λ DNA that is tethered to a microscope slide and stretched with a fluid flow (Figure 2). After the DNA is stretched, a distinct tether on the opposite end of the DNA can be linked to the surface to maintain the DNA in an elongated configuration upon termination of the flow (Figure 2). Fluorescent labeling without perturbing molecular function is a challenge in all fluorescence studies. In MutS studies, homologs have been labeled with both organic fluorophores and quantum dots (QDs). Specific labeling sites of MutS used to date have been tested to preserve DNA binding properties and ATPase behaviors [59–62]. While small organic dyes have minimal impact of protein diffusivity, QD labeling slows the observed diffusivity of other proteins sliding on DNA in comparison to smaller probes [63, 64 2012].
Figure 2.
Single molecule tracking experiments. a) Schematic of the experimental approach. DNA is tethered to a surface at the ends. The intervening surface is passivated in this example by a lipid bilayer. Fluorescent dye-labeled MMR proteins are shown diffusing on the DNA. The surface is illuminated by total internal reflection. b) Kymograph representation of single particle tracking of protein diffusion on DNA. The vertical direction represents position along one molecule of DNA that is extended and tethered to the surface. The horizontal direction is time during an experimental imaging experiment. The individual colored lines are the location of diffraction limited fluorescence spots from single proteins. Horizontal lines represent position vs. time of proteins along the DNA. Motion of a protein along the DNA appears as vertical changes in the line as time advances from left to right. Note, some molecules bind or unbind the DNA during the observation, indicated by colored lines that begin or end abruptly as they are tracked horizontally across the image. Adapted from [60].
Single particle tracking of Cy3-labeled Thermus aquaticus (Taq) MutS and QD-labeled MutSα on λ DNA [65] yielded diffusivities of 0.036±0.018 μm2/s (0.31 M(bp)2/s) [61, 65] and 0.012±0.018 (0.1 M(bp)2/s) [60], respectively, in the presence of ADP during the searching phase. These diffusivities are similar to many other DNA binding proteins [66]. In addition, using two-color single particle tracking of MutSα labeled with two different color QDs, it was demonstrated that two QD-MutSα proteins could not pass one another while scanning the DNA [60]. QD-MutSα also cannot diffuse past nucleosomes [67]. As expected, the binding lifetime of MutS and QD-MutSα on λ DNA decreased with increasing salt concentration. In 100 mM salt, it was ~ 3 s for Taq MutS [61, 65] and ~ 30 s for QD-MutSα [60], which is longer than most DNA binding proteins [66].
The mobility of MMR proteins on DNA substrates also has been examined by single-molecule Förster Resonance Energy Transfer (smFRET) using a single donor fluorophore attached to MutS and an acceptor fluorophore attached to the DNA (Figure 3a). Donor fluorescence is excited with an external light source. When donor-MutS is far from the acceptor fluorophore on the DNA, there is no FRET coupling and only donor emission is seen. FRET will occur only when donor-MutS is within ~5 nm of the acceptor attached to DNA. Consequently, these FRET approaches do not directly image the motion of the protein on the DNA substrate but deduce the moments when MutS is within ~5 nm from the fluorophore attached on the DNA. Note, that single particle tracking studies work in the opposite limit where the precise location of QD-MutSα on long DNAs can be determined only with resolution of ~1000 base pairs (see Figure 4c of [68]), although relative locations can be measured more accurately. This smFRET approach using acceptor-labeled, mismatch-containing DNAs (50–500 bp) was used to examine mobility of Taq MutS on DNA in the searching phase [62]. When donor-labeled MutS bound at the mismatch a characteristic, high FRET efficiency signal was observed. Because, the affinity of the MutS for the mismatch is substantially higher than for matched DNA, in most observations, MutS bound at the mismatch faster than the temporal resolution of the experiments. By analyzing relatively rare events where low FRET was observed briefly before high FRET associated with mismatch binding, a limit on the mobility was determined [69], and this mobility is consistent with values observed by single particle tracking [61].
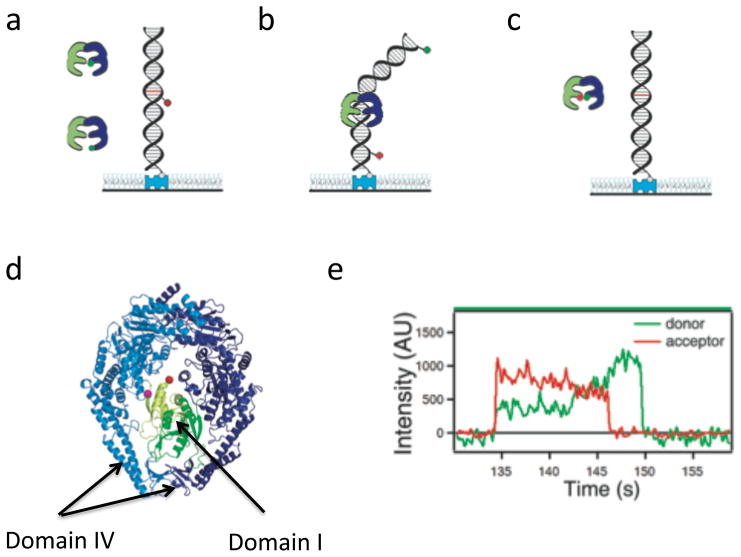
Figure 3.
Single molecule FRET studies. Duplex DNA is tethered to a surface by a biotin/streptavidin interaction and MutS is added in solution. FRET pairs used include a) donor-labeled MutS and acceptor-labeled DNA (used for reporting MutS position on DNA), b) donor and acceptor both on DNA spanning a mismatch (used for reporting DNA bending at the mismatch) and c) donor and acceptor both on MutS with unlabeled DNA (used for reporting internal conformation of MutS). d) A crystal structure of the Taq MutS dimer with DNA not shown (PDB-1EWR) with domains I colored green and domains IV indicated by arrows. e) Intensity vs. time graphs for donor and acceptor emission from a single acceptor-labeled Taq MutS interacting with T-bulge mismatch DNA with the acceptor dye 9 bp from the mismatch (adapted [69]). Note FRET efficiency ~0.7 upon initial binding converts to FRET~0.5 during an intermediate state before a transition to FRET~0 indicates MutS sliding away from the mismatch. The disappearance of emission finally indicates MutS sliding off the end of the DNA.
MutS tracks double helix
Single particle tracking studies not only allow the direct observation of proteins diffusing along DNA, but also permit the determination of the relative importance of diffusion with rotation in register with the DNA helix compared to weaker DNA interactions allowing hopping or jumping. [60, 63, 64, 66, 70]. By examining the salt dependence of the diffusion rate and/or comparing the observed diffusion coefficients with theoretical predictions of the diffusion coefficients based on purely translational motion or tracking of the protein along the helix of DNA, it is possible to discern the mechanism by which proteins move along the DNA [60, 63, 64, 66, 70]. Groove tracking by proteins predicts a slower rate of diffusion than diffusion without tracking, especially with a QD attached to the protein. Greene and co-workers estimated that attaching a QD to MutSα will slow the rate of diffusion by a factor of 2 if the protein is not tracking the DNA helix and a factor of 10 if the protein tracks the DNA double helix as it searches [60]. Desbiolles and coworkers refined this modeling to predict dramatically different diffusivities depending on whether the QD is attached by rigid or flexible linker [63, 64]. These differences result from partial uncoupling of the Brownian motion of the QD and protein when they are connected with a flexible linker.
The diffusivities of both Taq MutS and QD-MutSα are independent of salt concentration [60, 61], consistent with a sliding mechanism over a hopping mechanism, but this salt-dependence does not distinguish between sliding with free rotation or with the protein tracking the groove of the DNA. Comparison of the measured searching phase diffusivities for QD-MutSα with theoretical predictions supports a diffusion model in which the QD-MutSα rotates in register with the DNA [60], as has been found for many other proteins [66].
Both smFRET and sm-fluorescence polarization have been used to examine if Taq MutS tracks the DNA groove during searching [61, 65]. These studies compared observations to theoretical predictions of models where MutS rotates around the DNA following the groove or where MutS only translates along the DNA with no rotation at all. In 4–30 ms averaged smFRET experiments, FRET between donor-MutS and 74 bp or 100 bp acceptor-labeled homoduplex DNA with blocked ends (Figure 3a) was lower on the longer DNA [61]. Because the DNA is longer than the FRET range of their dye pair, the lower FRET on the longer DNA is consistent with MutS scanning the whole DNA by diffusion and thus spending on average less time near the acceptor on the DNA during a camera integration period. Comparing the breadth of the distributions of FRET efficiencies measured at 4 ms and 30 ms acquisition times with predictions based on models of diffusion with rotation in register with the DNA or diffusion without rotation supported a model of diffusion with helix tracking. Specifically, the rotating model led to a smaller range of distances between the donor and acceptor as MutS passed near the labeled DNA location and thus a tighter distribution of averaged FRET values [61]; however, comparing the helix tracking to a freely rotating model would be more realistic than one in which MutS cannot rotate around the helix.
In the sm-fluorescence polarization experiments, it was first confirmed that the Cy3 label on MutS was sufficiently restricted to allow for polarization to report MutS orientation, and then the polarization of Cy3-labeled MutS was monitored on homoduplex DNA [65] (Figure 3a). No polarization signal was observed for Cy3-MutS searching homoduplex DNA. Zero polarization is consistent with DNA helix coupled rotation while diffusing because the known diffusion rate predicts a rotation rate that will result in zero average polarization at the experimental resolution, but it does not distinguish between the rapid rotation either coupled or uncoupled to the DNA helix. To distinguish between these 2 possibilities, the authors used 26 to 100 base DNA substrates that are short enough to limit the number of full turns MutS can make if remaining in sync with the DNA helix. The effect of DNA length on polarization averaging was confirmed in the searching phase where short substrates have broad polarization distributions that trend toward narrow distributions on longer substrates where more complete averaging is allowed [65].
These MutS diffusion experiments complement a flurry of analogous studies of a variety of DNA binding proteins. All sequence specific DNA binding proteins examined thus far (as well as some non-specific binders) are determined to rotate in register with the DNA helix during linear diffusive motion [60, 63, 64, 66, 70]. Only two proteins studied to date have exhibited diffusive properties that are consistent with a hopping mechanism (herpes simplex virus UL42 processivity factor [71] and MutLα (see below)), and neither exhibit sequence specific recognition. These generalizations have broad reaching implications for the mechanisms of specific site recognition in DNA binding proteins
MutS bends DNA during its search
Crystal structures of Taq and E. coli MutS and human MutSα bound to several different mismatched DNA bases and base insertion/deletions [72–75] reveal only two specific amino acid contacts between MutS or MutSα and the mismatched base: a phenylalanine which stacks with the mismatched base and a glutamate, which forms a hydrogen bond with the N3 of the mismatched thymine or N7 of the mismatched purine [72–75]. All other interactions between MutS and the DNA are nonspecific backnone contacts. These specific and non-specific interactions induce significant DNA bending at the mismatch and rotation of one of the mismatched bases toward the minor groove, with this rotation being stabilized by stacking with the phenylalanine in the conserved Phe-X-Glu motif.
AFM studies demonstrated that E. coli and Taq MutS induce DNA bending when bound to homoduplex DNA, as well as when bound to mismatches, which led to the suggestions that MutS induces DNA bending during its diffusive search for a mismatch and that this bending facilitates mismatch recognition [76, 77]. The idea that MutS propagates the DNA bend as it scans DNA is consistent with the conclusion from the tracking studies that MutS rotates along the DNA during its search phase and suggests that MutS, as well as many (or most) other specific DNA binding proteins closely track the DNA double helix. In many ways such a scenario should not be surprising because the majority of the binding energy for proteins that recognize specific DNA sequences results from nonspecific protein-DNA interactions, with specific interactions providing sequence preference. For these nonspecific interactions to be maximized, the DNA needs to adopt a conformation that results in optimal geometry of the protein’s DNA binding pocket, which in turn facilitates sampling of contacts required for specific site recognition and maximizes specificity by paying the cost of bending the DNA with nonspecific protein-DNA interactions [77, 78]
Mismatch recognition by MutS
Conformational properties of MutS-mismatch complexes
Several single-molecule studies have investigated conformational changes in DNA associated with binding of E. coli and Taq MutS to different mismatches. In an early study, AFM was used to directly visualize MutS bound to mismatched and to homoduplex DNA [76]. The capability of AFM to distinguish binding of MutS at homoduplex (nonspecific) DNA sites from binding on mismatches allows identification of conformations unique to the mismatch (or specific site). In addition, analysis of the occupancies of MutS on DNA as a function of position along the DNA in AFM images allowed the determination of the specificities and binding affinities of MutS for mismatches, and the binding affinities determined by AFM agreed with those determined by bulk fluorescence anisotropy studies [79]. Notably, AFM studies found only bent DNA in MutS-DNA complexes at homoduplex sites, but both bent and unbent conformations were observed at mismatch sites. The unbent state is thus the result of unique interactions between the mismatch base and MutS, which suggests that the bent conformation is an intermediate when forming the unbent state [6, 76, 77]. Notably, the DNA is bent in the crystal structure [72–75], suggesting that the crystal structure may represent an initial recognition state. Together with the crystal structures, these data led to the proposal that MutS binds to DNA nonspecifically and bends it in search of a mismatch; upon specific recognition of a mismatch, MutS undergoes a conformational change to an initial recognition complex (IRC) in which the DNA is kinked, with interactions similar to those in the crystal structures; and MutS then undergoes further conformational changes to the ultimate recognition complex (URC) in which the DNA is unbent (or slightly bent) with the mismatched base possibly being flipped out [6, 76, 77]. In this model, bending facilitates the initial recognition of the mismatched base and provides the opportunity for the specific interaction of a conserved phenylalanine with the mispaired base. After recognition, MutS uses the energy stored in the bent DNA to help drive a conformational change in MutS to the unbent state, providing a “double check” before repair is initiated. In this model, whether or not the downstream events that lead to repair occur will be determined, in part, by the ability of MutS to form the unbent URC, and therefore, the efficiency of repair will be governed by the relative stability of the bent (or kinked) and unbent state.
Further support for this model came from additional AFM studies of two mutants of Taq MutS, with alanine substitutions in the conserved Phe-Xaa-Glu mismatch recognition motif (MutS-F39A and MutS-E41A) [77]. These studies showed a direct correlation between the ability of MutS to signal repair and its ability to induce an unbent conformation at the mismatch. In addition, these studies found that the conserved Glu, but not the Phe, facilitates MutS-induced DNA bending on homoduplex DNA, whereas at mismatches, both Phe and Glu promote the formation of the unbent conformation. The data reveal an unusual role for the Phe residue in that it promotes the unbending, not bending, of DNA at mismatch sites and provided a structural explanation for the mechanism by which MutS searches for and recognizes mismatches. A significant limitation of these studies is that AFM provides only static snapshots of MutS-DNA complexes and thus cannot provide proof of the proposed pathway; however, subsequent smFRET studies monitoring DNA bending support the proposed pathway.
Sass et al. used surface-tethered DNA substrates containing a GT mismatch halfway between a FRET donor (TAMRA) and acceptor (Cy5) located 19 bp apart, to measure DNA bending-induced FRET efficiency changes in real time (Figure 3b) [80]. These studies revealed that Taq MutS-GT mismatch complexes are highly dynamic, adopting as many as six different conformations with different degrees of DNA bending. The authors developed a method (called FRET TACKLE) to analyze the dynamic data in which they combined direct analysis of smFRET transitions with examination of kinetic lifetimes to identify all of the conformational states and characterize the kinetics of the binding and conformational equilibria. They found lifetimes differing by as much as 20-fold and rates of interconversion varying by 2 orders of magnitude. In a more recent study, they examined the dynamics of MutS and MutS-E41A bound to different mismatches, including a T-bulge and GT and CC mismatches [81]. These studies lead to the suggestion that the dynamics of the MutS-mismatch complex could govern the efficiency of repair of different DNA mismatches [80, 81]. Although, these studies revealed additional conformational states of MutS-mismatch complexes relative to the AFM studies, they confirmed the suggestion from the AFM studies that MutS first induces DNA bending upon binding and then undergoes a conformational change to a state in which the DNA is not bent (or minimally bent). One caveat to both the AFM and smFRET studies is that they were conducted in the absence of nucleotide or in the presence of ADP but not in the presence of ATP. Consequently, it is possible that the conformational properties are different in the presence of ATP.
Single-molecule multiparameter fluorescence spectroscopy has also been used to examine E. coli MutS-induced DNA bending as well as the orientation of binding of MutS to a mismatch [82]. In these studies, the authors used substrates similar to the previous study, in which different mismatches are placed between a donor and acceptor fluorophores; however, their measurements were conducted on freely diffusing molecules instead of surface immobilized DNA molecules. They located the dyes such that MutS would interact with the donor in one binding orientation (e.g., the phenylalanine stacking on the G vs. the T in a GT mismatch) and measured both FRET and the anisotropy of the donor. These measurements reported both DNA bending and the orientation of MutS binding to the mismatch. In addition, they used fluorescence correlation spectroscopy to measure the apparent diffusion times, which allowed them to determine whether or not MutS was bound to the DNA. Consistent with the studies discussed above, they observed both bent and unbent conformations of MutS-mismatch complexes; however, because their observation time is very short (~1 ms), they could not observe transitions between different conformations. In addition, their work confirmed the suggestion based on MutS-mismatch crystal structures that MutS exhibits a strong orientational preference on asymmetric mismatches (e.g., GT vs. TG and AC vs. CA) but not on symmetric mismatches such as GG. Coupling these data with bulk measurements, they suggest that this orientational binding may be important for the efficiency of repair [82].
smFRET is difficult to apply to DNA bending by MutS when the FRET from protein-bound unbent states overlaps with the FRET from free DNA states. DeRocco et al. labeled yMutSα using tris-NTA linked fluorophores [83, 84] bound to a His6 tag on Msh6 [85]. Cysteine engineering approaches are not feasible for labeling MutSα because it contains more than 30 cysteines. They used 2-color excitation and three-color sm-fluorescence detection to simultaneously measure DNA bending and colocalization of yMutSα. These studies on CC mismatched DNA resolved multiple bent DNA states of MutSα-DNA complexes, some of which overlapped with free DNA.
smFRET studies also have been conducted on human MutSβ [86], which is responsible for signaling the repair of IDLs of 2–10 bp and also can promote triplet repeat expansion [87]. They monitored the DNA conformational dynamics in the presence and absence of MutSβ of three DNA substrates: a (CA)4 insertion which is well repaired via MutSβ signaling and a (CAG)13 or (CAG)11-(CTG)2 insertion, which can form hairpins containing AA mismatches that are resistant to repair and can lead to mutations. MutSβ induced a single conformation on the (CA)4 substrate in which the DNA was more bent than the unbound conformation in the presence and absence of ATP, and ATP increased dissociation. In contrast, on (CAG)13, MutSβ induced two conformations that could interconvert, one less bent and one more bent than the unbound substrate, and on (CAG)11-(CTG)2, a single less bent state was observed in the absence of ATP. Interestingly, in the presence of ATP, the more bent population of MutSβ-(CAG)13 complexes shifted to the less bent population from which Mutβ dissociated slowly, consistent with its expected inability to convert to a sliding clamp. These studies coupled with biochemical studies led the authors to suggest that the conformational dynamics of the different DNA substrates may control the ability of MutSβ to signal repair [86].
In addition to monitoring DNA bending, smFRET has also been used to report conformational changes of Taq MutS in several contexts: the absence of DNA, during scanning of homoduplex DNA, upon mismatch recognition, and following ATP activation [69]. These studies employed a single cysteine mutant of Taq MutS (C42A/M88C) that allowed donor and acceptor fluorophore attachment in domains I of the homodimer, which contains the mismatch recognition motif (Figure 3c, 3d). smFRET revealed that domains I interconvert between conformations in which domains I are open (low FRET) and in which domains I are closed (high FRET) while scanning homoduplex DNA. Upon mismatch recognition, MutS adopts a conformation in which domains I are closed (high FRET), consistent with the position of these domains in the crystal structure of MutS (Figure 3d).
Monitoring mismatch recognition and ATP activation of MutS
The earliest single molecule study of MMR was the electron microscopy (EM) investigation of the properties of E. coli MutS-DNA and MutS-MutL-DNA complexes bound to 6 kbp DNA containing a single GT mismatch in the presence of ATP [88]. This study observed ATP-dependent formation of large α-loop structures. MutS (or MutS and MutL) was at the base of the loop, with the mismatch in the middle. The size of the loop increased linearly with the incubation time and was dependent on ATP hydrolysis. MutL increased the rate of loop formation. These findings led to the authors to suggest that ATP induces MutS to leave the mismatch and translocate bidirectionally along the DNA away from the mismatch, forming loops in an ATP hydrolysis dependent manner. Subsequent bulk studies, which monitored the rate of dissociation of MutS from DNA with blocked or unblocked ends upon the addition of ATP or ATP-γS, have indicated that MutS can form a mobile clamp in the absence of ATP hydrolysis; however, these studies used shorter substrates [18, 29, 89]. Although it appears clear that ATP hydrolysis is not necessary for activation of MutS to a mobile clamp state, it is possible that ATP hydrolysis could be necessary for activated MutS to travel long distances on the DNA, which may be important in E. coli where MutS-MutL complexes must interact with MutH at hemi-methylated GATC sites, which could be up to 1000 bp away.
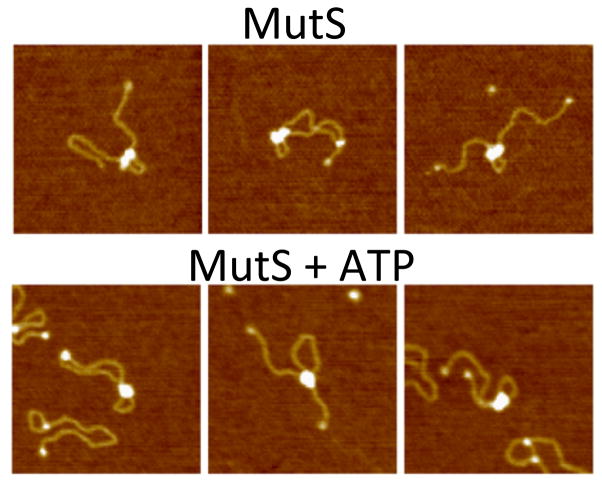
A recent AFM study [90] of E. coli MutS bound to mismatch-containing DNA used significantly shorter DNA. ATP-dependent loop formation was observed in this study; however, loops were also seen in the absence of nucleotide and the presence of ADP with a similar frequency but smaller size than in ATP. These latter results differ from the EM study and previous AFM studies, in which very few loops were observed with E. coli or Taq MutS in the presence or absence of ADP [76, 77, 88]. By analyzing the AFM volume [91, 92] of MutS bound to DNA, Jiang and Marszalek demonstrated that two MutS dimers mediate loop formation. The conformations of the two MutS dimers in the complex stabilizing the loop were different in the presence and absence of ATP (Figure 4). In the absence of ATP, two individual MutS dimers were often discernable; whereas, in the presence of ATP, the two dimers were generally indistinguishable. A notable difference between this study and the previous EM study [88] is that in the EM study, MutS rarely remained in the vicinity of the mismatch upon addition of ATP, while in this AFM study, one of the MutS dimers often remained in the vicinity of the mismatch even in the presence of ATP. This latter result led to the suggestion that the two MutS dimers are nonequivalent, with one marking the mismatch and one searching the DNA. The apparent differences between these two studies may result from the differences in the lengths of the substrates (1.1 kbp vs. 6.4 kbp) and size of the loops. In fact, in the EM study, those loops in which one MutS remains near the mismatch are predominantly seen for complexes with shorter loops [88].
Figure 4.
AFM images of E coli MutS induced DNA looping of 1.1 kbp DNA containing a single GT-mismatch in the absence (top) and presence of ATP (bottom). Adapted from [90]
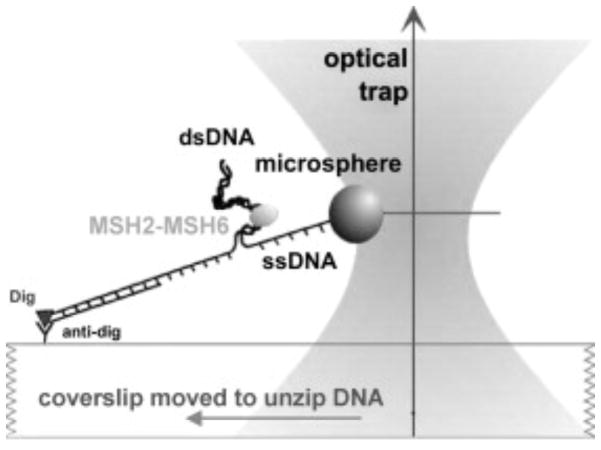
The earliest single molecule experiment detecting mismatch recognition and subsequent sliding clamp formation employed optical trapping to monitor the force required to unwind end-blocked DNA as a function of the position of a mismatch (Figure 5) [59]. Addition of yMutSα increased the required unwinding force at the location of the mismatch in the presence of ADP. In ATP or ATP-γS, the position of the increased force (presumed to be the location of MutSα) no longer coincided with the mismatch, suggesting a sliding clamp mode. These behaviors required a mismatch in the DNA, further supporting the presence of a sliding clamp. This experimental method, however, could not ascertain the dynamics of the process.
Figure 5.
Schematic of DNA unwinding assay used to detect mismatch recognition and subsequent sliding clamp formation. A bead is attached to one DNA strand and the other strand is attached to the surface. An optical trap is used to trap the bead and pull on the DNA to promote unwinding, and the force required to unwind the DNA is monitored. Adapted from [59].
Single molecule fluorescence methods provide a window into the dynamics of the transition from recognition to ATP activation. Gorman et al. constructed a λ DNA substrate containing 3 mismatches spaced by 38 base pairs for use in their surface tethered tracking assay [68]. They observed QD-MutSα searching homoduplex DNA and binding specifically to the mismatches in the presence of ADP. They found the dwell time at positions consistent with the position of the mismatches to be 9.6 minutes, which is in stark contrast to ensemble studies that find 25 second lifetimes for yMutSα bound to a GT mismatch in the presence of ADP [93]. These single molecule tracking and ensemble stopped flow assays may be sensitive to different subpopulations, or the differences may result from the different substrates employed in the single molecule and bulk studies. Alternately, the differences may derive from the presence of three closely spaced mismatches in the single molecule studies and/or the presence of a QD on MutSα. Examination of the diffusivity for QD-MutSα in the presence of ATP revealed that QDMutS diffused modestly faster after sliding clamp formation relative to during the searching phase (D= 0.057±0.064 μm2/s for the activated clamp vs. 0.009±0.011 μm2/s for the searching phase). Studies of Taq MutS also revealed a slightly faster (1.6 fold) diffusing sliding clamp relative to the searching phase. These results led to the suggestion that MutS no longer tracks the double helix after activation [61, 68]. The response of the mobility of Taq MutS to altering both the salt concentration and buffer flow rate (which puts force on the MutS bound to DNA) further supports this conclusion [61]. The diffusivity for Taq MutS has also been estimated from studies measuring smFRET between the protein and DNA [69]. In these studies, a slower diffusivity was found for the clamp than the searching phase. The source of these differences remains unknown, but may be related to differences in the details of the salt and buffer conditions or the dye-labeling of MutS.
The most significant change in the ATP/ATPγS-mismatch activated state compared to the searching state is that dwell time on the DNA is dramatically increased (hundreds vs. a few seconds), and this activated state no longer interacts with the mismatch [60, 61, 68]. In addition, smFRET studies on Taq MutS revealed that multiple MutS proteins could load onto mismatch DNA with blocked DNA ends in the presence of ATP, consistent with a sliding clamp leaving the mismatch, [61, 69].
smFRET provides detailed information about the position of MutS at a mismatch. Two studies have monitored mismatch recognition by monitoring FRET between donor-labeled MutS and an acceptor fluorophore attached 9 bp from a mismatch. In one study, MutS was labeled on domain IV [61] and in the other, it was labeled on domain I [69] (Figure 3d). Both of these studies detected binding of MutS to the mismatch (one in the absence of nucleotide and the other in the presence of ADP), and the dwell time at the mismatch was consistent with bulk biochemical studies. In addition both of these studies monitored recognition in the presence of ATP (see below).
Experiments of this sort are complicated by the fact that fluorescent dyes attached to DNA are flaws that MutS can recognize. In the FRET studies of MutS, the 9 bp offset of the acceptor from the mismatch generates FRET less than 1.0 upon mismatch binding, but direct MutS binding to the dye results in FRET nearly 1. Using this difference in FRET values, these studies have observed direct dye binding of 10% [69] and 30% [61]. The single molecule approach allows those direct dye-binding events to be rejected from further analysis.
smFRET assays have also characterized conformational changes within MutS during the ATP-dependent conversion to the post-recognition sliding clamp. The domain I label site used by Qiu et al. allowed detection of two sequential conformational changes as the mismatch recognition state converts to a sliding clamp [69]. These two conformations were detected both by monitoring intra-protein FRET between dual-labeled domains I (Figure 3c) and by monitoring FRET between one domain I and an acceptor dye 9 bases away from the mismatch (Figure 3a). In the latter experiments, MutS bound a mismatch yielding FRET ~0.7. About 20% of such mismatch binding events eventually converted to sliding clamps in the presence of ATP. Before sliding commenced, MutS converted to an intermediate conformation that generated FRET 0.5 (Figure 3e). This intermediate state, which lasts about 1.5 seconds before MutS leaves the mismatch, is consistent a substantial movement of the DNA interacting domain I. Measurement of intraprotein FRET (Figure 3c) also revealed an intermediated state with the same kinetic properties as the protein-DNA FRET measurements, further confirming this two-step transition [69]. Another FRET study labeling MutS in domain IV did not observe an intermediate FRET level before mismatch release. [61] The different FRET results when labeling domain I vs. IV may indicate the domains move in distinct ways or alternately could reflect the differences in the DNA substrates used in the studies. Future measurements with different label attachment sites on MutS will be useful to further clarify the precise conformation of this intermediate state. This intermediate state could provide new surfaces that interact with downstream signaling proteins, and in particular MutL, to regulate or determine the continuation of the mismatch repair cascade.
Properties of MutL homologs
MutL and MutL homologs are members of the GHL ATPase family [94–98], which includes DNA Gyrase, Hsp90, Grp94 and the type II topoisomerases. ATP binding and/or hydrolysis induces large conformational changes in GHL proteins, which are thought to be involved in the signaling of cellular processes [94, 96, 99–104]. Both homodimeric bacterial MutL and heterodimeric eukaryotic MutLα dimerize via their C-terminal domains, which are connected to an N-terminal ATPase domain via a linker region that is predicted to be unstructured [105–108].
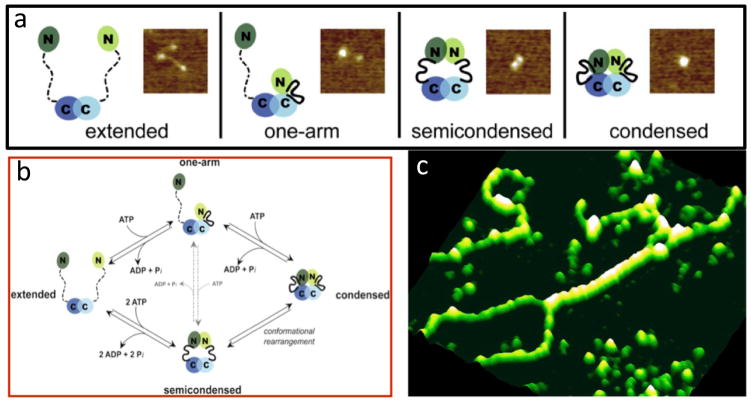
Sacho et al. used AFM coupled with biochemical techniques to examine conformational properties of MutLα [109]. They demonstrated that adenine nucleotides induce large asymmetric conformational changes that are associated with significant increases in secondary structure in yeast and human MutLα (Figure 6a). By examining the conformations as a function of nucleotides, they identified the changes in conformations associated with nucleotide binding, hydrolysis, and release (Figure 6b). They suggested these nucleotide-dependent conformational changes mediate the interactions of MutLα with other proteins in the MMR pathway, coordinating the recognition of DNA mismatches by MutSα and the activation of MutLα with the downstream events that lead to repair.
Figure 6.
Single molecule AFM imaging of MutLα and MutLα-DNA complexes. a) AFM image and cartoons showing different conformational states of MutLα. In the cartoons, domains are indicated by ovals, connected by a flexible linker, with the disordered linker shown as a dashed line and ordered linker shown as a solid heavy line. Mlh1 is depicted in light green and blue, and Pms2 (yPms1) is depicted in dark green and blue. (Adapted from [109]) b) Cartoon of the ATPase cycle deduced from the nucleotide dependence of MutLα conformations (Adapted from [109]). c) AFM image showing yMutLα forming long protein tracts on DNA and bring two double-stranded DNAs together (adapted from [112]).
Prokaryotic MutL and eukaryotic MutLα exhibit different DNA binding preferences, with MutL preferentially binding single-stranded DNA and MutLα preferentially binding double-stranded DNA [95, 110]. smFRET has been used to monitor MutL interactions with a donor/acceptor labeled DNA containing a single strand overhang [111]. Multiple MutL proteins bound the single strand region at low salt, but there was little binding at high salt. AFM was used in investigate the DNA binding properties of MutLα. [112] These studies revealed that MutLα can bind cooperatively to form long, continuous tracts of protein along duplex DNA in low salt. The AFM images also showed that MutLα can interact simultaneously with two different strands of duplex DNA (Figure 6c). This latter result is consistent with experiments demonstrating that the N-terminal domains of both yMlh1 and yPms1 bind double-stranded DNA [110]. Because these behaviors emerged only at low salt concentrations, with only weak DNA binding by MutLα at physiological salt concentrations, the physiological relevance of these findings is questionable. Notably, however, MutLα must interact directly with DNA to carry out its essential MMR function of nicking the daughter strand [36, 37]. Perhaps the interaction of MutLα with MutSα/β promotes MutLα-DNA interactions under physiological conditions.
Single molecule tracking using QD-labeled MutLα and λ DNA tethered to the surface at one or both ends has also been used to examine the interaction of MutLα with DNA [67]. In these studies, the observed diffusivity of MutLα, unlike MutSα, increased with increasing NaCl concentration, with MutLα exhibiting a similar diffusivity to MutSα at 50 mM NaCl but an ~5-fold higher diffusivity than MutSα at 150 mM NaCl. This salt dependence suggests that diffusion of MutLα along DNA involves hopping or stepping of MutLα along the DNA [67], at least at the higher salt concentrations. An increase in diffusivity as a function of salt concentration is expected for 1D diffusion involving hopping because increased electrostatic screening from higher salt concentration reduces the DNA binding affinity, increasing the probability of micro-dissociations from the DNA [53]. The authors also found that MutLα, unlike MutSα, could bypass nucleosomes, further supporting the idea that MutLα may move along the DNA through a stepping mechanism. Stepping is also consistent with the biochemical observation that the N-terminal domains of both Mlh1 and Pms1 can bind DNA and with the AFM observation that MutLα can simultaneously bind two DNA double strands [110, 112].
In addition to this stepping mechanism, Gorman et al. also suggested that MutLα can form a ring-like structure that encircles the DNA loosely, giving a greater diffusivity than expected if it is required to rotate in register with the DNA helix. This latter suggestion is based on the observation that QD-MutLα preferentially dissociates from free DNA ends relative to blocked DNA ends when hydrodynamic force (fluid flow) was used to push QD-MutLα along the DNA; whereas, QD-Mlh1 alone dissociated from both blocked and unblocked ends. Such a model seems inconsistent with their proposed stepping/hopping mechanism of movement. It also seems inconsistent with the DNA binding and electrostatic properties of MutLα. Specifically, the N-terminal domains contain MutLα’s DNA binding activity [110], and the linker region (59 negative and 39 positive residues) and C-terminal domain of Pms1 are highly negatively charged [113]. The differences in end dissociation that Gorman et al. observed between MutLα (Mhl1-Pms1) and Mlh1 may simply result from the significantly lower DNA binding affinity of Mlh1 relative to MutLα [110] and to the weak homo-dimerization constant of Mlh1 (3 μM [114]). One concern with this study is that the authors were able to analyze the diffusion properties of only 10% of the total QD-MutLα proteins that were bound to DNA, making it possible that they are monitoring a population that may not represent the bulk of MutLα proteins.
Downstream mismatch repair signaling involving MutL
In a recent study, Gorman et al. examined the interaction of QD-MutSα and QD-MutLα on λ-DNA containing three GT mismatches spaced by 38 bp, using MutSα and MutLα labeled with QDs with distinct emission wavelengths. Surprisingly, they found that QD-MutLα colocalized with QD-MutSα bound to a mismatch in the presence of ADP with a lifetime of 7.8 minutes, which is similar to the dwell time of MutSα alone in this assay. This result is in stark contrast to bulk studies, which do not detect any interaction between MutLα and MutSα bound to DNA containing mismatches in the presence of ADP [93, 115]. They subsequently examined the fate of a subset of these complexes (n=35) upon the addition of ATP. They found that 14% remain stationary, 23% dissociate directly, and the remaining 63% of QD-MutSα and QD-MutLα diffuse together as a complex, with properties that are nearly identical ATP-activated MutSα alone. One significant limitation of these studies is that the analysis is limited to only a single QD-MutSα and a single QD-MutLα, resulting in only a small number of the total bound proteins being analyzed. If MMR signaling involves multiple MutSα and/or MutLα proteins, as suggested by in vivo studies of fluorescently tagged MutS and MutL homologs [35, 116], studies that focus on single proteins may miss important multi-protein complexes. Perhaps in the future, it will be possible to expand these studies to more broadly analyze the entire population of protein-DNA complexes.
The future of single molecule methods in studies of DNA mismatch repair
DNA mismatch repair proceeds by a series of transient, multi-protein interactions that present significant challenges to experimentation using traditional biochemical methods. Single molecule approaches are well suited to overcome these challenges and uncover the key signaling events that lead to repair of flawed DNA. In this review, we have described the existing state of the art in applying single molecule investigation to DNA MMR. To date, these studies have mainly focused on the binary interaction of MutS with mispaired DNA while only a few have included MutL. Sometime ago, DNA MMR was successfully reconstituted in vitro with 8–10 distinct, purified proteins depending on the particular system studied [7, 117, 118]. As our understanding of these proteins increases and as single molecule methods continue to mature, we can envision eventually reconstituting the entire DNA MMR reaction in a single molecule assay similar to the recent progress in studies of replication [119–122]. Such an assay would be an exceptional tool to tease apart the multiple interactions that both promote and regulate DNA repair.
DNA MMR operates within the complex environment of living cells. In vitro studies, however sophisticated, may not capture the full complement of interactions of the MMR system with other cellular processes. Ultimately, studies in live cells will be the most useful. Kolodner and co-workers recently took exciting steps toward visualizing DNA MMR in live yeast cells by quantitatively imaging individual MMR foci [35]; however, these experiments were not sensitive to assemblies of fewer than ~6 labeled molecules. Single molecule fluorescence methods including fluorescence stoichiometry, super-resolution imaging, and single molecule FRET have been recently applied within live cells for increasingly complex phenomena. [123–127]. The possibility of in vivo MMR studies using smFRET is particularly exciting because of its sensitivity to transient multimolecular complexes, which are difficult to observe by other methods in living cells. The use of single molecule methods has the potential to resolve longstanding questions and controversies in the MMR field. Furthermore, the ability to measure molecular mechanisms of DNA MMR in vivo will enable breakthroughs in understanding connections between aberrant repair and disease.
Acknowledgments
This work was supported by a research scholar grant from the American Cancer Society RSG-10-048 (K.R.W.) and by National Institutes of Health grant GM079480 (D.A.E.) and GM080294 (D.A.E.)
References
- 1.Karran P, Offman J, Bignami M. Human mismatch repair, drug-induced DNA damage, and secondary cancer. Biochimie. 2003;85:1149–1160. doi: 10.1016/j.biochi.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Bignami M, Casorelli I, Karran P. Mismatch repair and response to DNA-damaging antitumour therapies. Eur J Cancer. 2003;39:2142–2149. doi: 10.1016/s0959-8049(03)00569-0. [DOI] [PubMed] [Google Scholar]
- 3.Fedier A, Fink D. Mutations in DNA mismatch repair genes: implications for DNA damage signaling and drug sensitivity (review) Int J Oncol. 2004;24:1039–1047. [PubMed] [Google Scholar]
- 4.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 5.Fishel R. The selection for mismatch repair defects in hereditary nonpolyposis colorectal cancer: revising the mutator hypothesis. Cancer Res. 2001;61:7369–7374. [PubMed] [Google Scholar]
- 6.Kunkel TA, Erie DA. DNA mismatch reapir. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 7.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 8.Santucci-Darmanin S, Paquis-Flucklinger V. Homologs of MutS and MutL during mammalian meiosis. Med Sci (Paris) 2003;19:85–91. doi: 10.1051/medsci/200319185. [DOI] [PubMed] [Google Scholar]
- 9.Svetlanov A, Cohen PE. Mismatch repair proteins, meiosis, and mice: understanding the complexities of mammalian meiosis. Exp Cell Res. 2004;296:71–79. doi: 10.1016/j.yexcr.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 10.Flores-Rozas H, Kolodner RD. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc Natl Acad Sci U S A. 1998;95:12404–12409. doi: 10.1073/pnas.95.21.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harfe BD, Jinks-Robertson S. DNA mismatch repair and genetic instability. Annu Rev Genet. 2000;34:359–399. doi: 10.1146/annurev.genet.34.1.359. [DOI] [PubMed] [Google Scholar]
- 12.Lipkin SM, Wang V, Jacoby R, Banerjee-Basu S, Baxevanis AD, Lynch HT, Elliott RM, Collins FS. MLH3: a DNA mismatch repair gene associated with mammalian microsatellite instability. Nat Genet. 2000;24:27–35. doi: 10.1038/71643. [DOI] [PubMed] [Google Scholar]
- 13.Harfe BD, Minesinger BK, Jinks-Robertson S. Discrete in vivo roles for the MutL homologs Mlh2p and Mlh3p in the removal of frameshift intermediates in budding yeast. Curr Biol. 2000;10:145–148. doi: 10.1016/s0960-9822(00)00314-6. [DOI] [PubMed] [Google Scholar]
- 14.Lipkin SM, Moens PB, Wang V, Lenzi M, Shanmugarajah D, Gilgeous A, Thomas J, Cheng J, Touchman JW, Green ED, et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat Genet. 2002;31:385–390. doi: 10.1038/ng931. [DOI] [PubMed] [Google Scholar]
- 15.Schofield MJ, Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu Rev Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- 16.Li GM. DNA mismatch repair and cancer. Front Biosci. 2003;8:d997–1017. doi: 10.2741/1121. [DOI] [PubMed] [Google Scholar]
- 17.McCulloch SD, Gu L, Li GM. Bi-directional processing of DNA loops by mismatch repair-dependent and -independent pathways in human cells. J Biol Chem. 2003;278:3891–3896. doi: 10.1074/jbc.M210687200. [DOI] [PubMed] [Google Scholar]
- 18.Gradia S, Acharya S, Fishel R. The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell. 1997;91:995–1005. doi: 10.1016/s0092-8674(00)80490-0. [DOI] [PubMed] [Google Scholar]
- 19.Modrich P. Strand-specific mismatch repair in mammalian cells. J Biol Chem. 1997;272:24727–24730. doi: 10.1074/jbc.272.40.24727. [DOI] [PubMed] [Google Scholar]
- 20.Genschel J, Bazemore LR, Modrich P. Human exonuclease I is required for 5′ and 3′ mismatch repair. J Biol Chem. 2002;277:13302–13311. doi: 10.1074/jbc.M111854200. [DOI] [PubMed] [Google Scholar]
- 21.Ramilo C, Gu L, Guo S, Zhang X, Patrick SM, Turchi JJ, Li GM. Partial reconstitution of human DNA mismatch repair in vitro: characterization of the role of human replication protein A. Mol Cell Biol. 2002;22:2037–2046. doi: 10.1128/MCB.22.7.2037-2046.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iams K, Larson ED, Drummond JT. DNA template requirements for human mismatch repair in vitro. J Biol Chem. 2002;277:30805–30814. doi: 10.1074/jbc.M200846200. [DOI] [PubMed] [Google Scholar]
- 23.Wang H, Hays JB. Mismatch repair in human nuclear extracts. Time courses and ATP requirements for kinetically distinguishable steps leading to tightly controlled 5′ to 3′ and aphidicolin-sensitive 3′ to 5′ mispair-provoked excision. J Biol Chem. 2002;277:26143–26148. doi: 10.1074/jbc.M200358200. [DOI] [PubMed] [Google Scholar]
- 24.Raschle M, Dufner P, Marra G, Jiricny J. Mutations within the hMLH1 and hPMS2 subunits of the human MutLalpha mismatch repair factor affect its ATPase activity, but not its ability to interact with hMutSalpha. J Biol Chem. 2002;277:21810–21820. doi: 10.1074/jbc.M108787200. [DOI] [PubMed] [Google Scholar]
- 25.Wei K, Clark AB, Wong E, Kane MF, Mazur DJ, Parris T, Kolas NK, Russell R, Hou H, Jr, Kneitz B, et al. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 2003;17:603–614. doi: 10.1101/gad.1060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo S, Presnell SR, Yuan F, Zhang Y, Gu L, Li GM. Differential requirement for proliferating cell nuclear antigen in 5′ and 3′ nick-directed excision in human mismatch repair. J Biol Chem. 2004;279:16912–16917. doi: 10.1074/jbc.M313213200. [DOI] [PubMed] [Google Scholar]
- 27.Palombo F, Gallinari P, Iaccarino I, Lettieri T, Hughes M, D’Arrigo A, Truong O, Hsuan JJ, Jiricny J. GTBP, a 160-kilodalton protein essential for mismatchbinding activity in human cells. Science. 1995;268:1912–1914. doi: 10.1126/science.7604265. [DOI] [PubMed] [Google Scholar]
- 28.Drummond JT, Li GM, Longley MJ, Modrich P. Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 29.Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci U S A. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iaccarino I, Marra G, Palombo F, Jiricny J. hMSH2 and hMSH6 play distinct roles in mismatch binding and contribute differently to the ATPase activity of hMutSalpha. EMBO J. 1998;17:2677–2686. doi: 10.1093/emboj/17.9.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Risinger JI, Umar A, Boyd J, Berchuck A, Kunkel TA, Barrett JC. Mutation of MSH3 in endometrial cancer and evidence for its functional role in heteroduplex repair. Nat Genet. 1996;14:102–105. doi: 10.1038/ng0996-102. [DOI] [PubMed] [Google Scholar]
- 32.Iaccarino I, Palombo F, Drummond J, Totty NF, Hsuan JJ, Modrich P, Jiricny J. MSH6, a Saccharomyces cerevisiae protein that binds to mismatches as a heterodimer with MSH2. Curr Biol. 1996;6:484–486. doi: 10.1016/s0960-9822(02)00516-x. [DOI] [PubMed] [Google Scholar]
- 33.Alani E. The Saccharomyces cerevisiae Msh2 and Msh6 proteins form a complex that specifically binds to duplex oligonucleotides containing mismatched DNA base pairs. Mol Cell Biol. 1996;16:5604–5615. doi: 10.1128/mcb.16.10.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark AB, Cook ME, Tran HT, Gordenin DA, Resnick MA, Kunkel TA. Functional analysis of human MutSalpha and MutSbeta complexes in yeast. Nucleic Acids Res. 1999;27:736–742. doi: 10.1093/nar/27.3.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hombauer H, Campbell CS, Smith CE, Desai A, Kolodner RD. Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell. 2011;147:1040–1053. doi: 10.1016/j.cell.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 37.Kadyrov FA, Holmes SF, Arana ME, Lukianova OA, O’Donnell M, Kunkel TA, Modrich P. Saccharomyces cerevisiae MutLa is a mismatch repair endonuclease. J Biol Chem. 2007 doi: 10.1074/jbc.M707617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szankasi P, Smith GR. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science. 1995;267:1166–1169. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- 39.Tishkoff DX, Boerger AL, Bertrand P, Filosi N, Gaida GM, Kane MF, Kolodner RD. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci U S A. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tran PT, Simon JA, Liskay RM. Interactions of Exo1p with components of MutLalpha in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2001;98:9760–9765. doi: 10.1073/pnas.161175998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kadyrov FA, Genschel J, Fang Y, Penland E, Edelmann W, Modrich P. A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8495–8500. doi: 10.1073/pnas.0903654106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Longley MJ, Pierce AJ, Modrich P. DNA polymerase delta is required for human mismatch repair in vitro. J Biol Chem. 1997;272:10917–10921. doi: 10.1074/jbc.272.16.10917. [DOI] [PubMed] [Google Scholar]
- 43.Constantin N, Dzantiev L, Kadyrov FA, Modrich P. Human mismatch repair: reconstitution of a nick-directed bidirectional reaction. J Biol Chem. 2005;280:39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li GM. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 45.Pluciennik A, Burdett V, Lukianova O, O’Donnell M, Modrich P. Involvement of the beta clamp in methyl-directed mismatch repair in vitro. The Journal of biological chemistry. 2009;284:32782–32791. doi: 10.1074/jbc.M109.054528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liberti SE, Larrea AA, Kunkel TA. Exonuclease 1 preferentially repairs mismatches generated by DNA polymerase alpha. DNA Repair. 2013;12:92–96. doi: 10.1016/j.dnarep.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pavlov YI, Mian IM, Kunkel TA. Evidence for preferential mismatch repair of lagging strand DNA replication errors in yeast. Curr Biol. 2003;13:744–748. doi: 10.1016/s0960-9822(03)00284-7. [DOI] [PubMed] [Google Scholar]
- 48.Nick McElhinny SA, Kissling GE, Kunkel TA. Differential correction of lagging-strand replication errors made by DNA polymerases {alpha} and {delta} Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21070–21075. doi: 10.1073/pnas.1013048107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol Cell. 2003;12:1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- 50.Genschel J, Modrich P. Functions of MutLalpha, replication protein A (RPA), and HMGB1 in 5′-directed mismatch repair. The Journal of biological chemistry. 2009;284:21536–21544. doi: 10.1074/jbc.M109.021287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morita R, Nakane S, Shimada A, Inoue M, Iino H, Wakamatsu T, Fukui K, Nakagawa N, Masui R, Kuramitsu S. Molecular mechanisms of the whole DNA repair system: a comparison of bacterial and eukaryotic systems. Journal of nucleic acids. 2010;2010:179594. doi: 10.4061/2010/179594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riggs AD, Bourgeois S, Cohn M. The lac repressor-operator interaction. 3 Kinetic studies. J Mol Biol. 1970;53:401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- 53.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1 Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 54.von Hippel PH, Berg OG. Facilitated target location in biological systems. The Journal of biological chemistry. 1989;264:675–678. [PubMed] [Google Scholar]
- 55.Kabata H, Kurosawa O, Arai I, Washizu M, Margarson SA, Glass RE, Shimamoto N. Visualization of single molecules of RNA polymerase sliding along DNA. Science. 1993;262:1561–1563. doi: 10.1126/science.8248804. [DOI] [PubMed] [Google Scholar]
- 56.Bustamante C, Guthold M, Zhu X, Yang G. Facilitated target location on DNA by individual Escherichia coli RNA polymerase molecules observed with the scanning force microscope operating in liquid. J Biol Chem. 1999;274:16665–16668. doi: 10.1074/jbc.274.24.16665. [DOI] [PubMed] [Google Scholar]
- 57.Guthold M, Zhu X, Rivetti C, Yang G, Thomson NH, Kasas S, Hansma HG, Smith B, Hansma PK, Bustamante C. Direct observation of one-dimensional diffusion and transcription by Escherichia coli RNA polymerase. Biophys J. 1999;77:2284–2294. doi: 10.1016/S0006-3495(99)77067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harada Y, Funatsu T, Murakami K, Nonoyama Y, Ishihama A, Yanagida T. Single-molecule imaging of RNA polymerase-DNA interactions in real time. Biophys J. 1999;76:709–715. doi: 10.1016/S0006-3495(99)77237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang J, Bai L, Surtees JA, Gemici Z, Wang MD, Alani E. Detection of high-affinity and sliding clamp modes for MSH2-MSH6 by single-molecule unzipping force analysis. Mol Cell. 2005;20:771–781. doi: 10.1016/j.molcel.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 60.Gorman J, Chowdhury A, Surtees JA, Shimada J, Reichman DR, Alani E, Greene EC. Dynamic basis for one-dimensional DNA scanning by the mismatch repair complex Msh2-Msh6. Mol Cell. 2007;28:359–370. doi: 10.1016/j.molcel.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jeong C, Cho WK, Song KM, Cook C, Yoon TY, Ban C, Fishel R, Lee JB. MutS switches between two fundamentally distinct clamps during mismatch repair. Nat Struct Mol Biol. 2011;18:379–385. doi: 10.1038/nsmb.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schopf B, Bregenhorn S, Quivy JP, Kadyrov FA, Almouzni G, Jiricny J. Interplay between mismatch repair and chromatin assembly. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1895–1900. doi: 10.1073/pnas.1106696109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bonnet I, Desbiolles P. The diffusion constant of a labeled protein sliding along DNA. The European physical journal E, Soft matter. 2011;34:1–10. doi: 10.1140/epje/i2011-11025-8. [DOI] [PubMed] [Google Scholar]
- 64.Dikic J, Menges C, Clarke S, Kokkinidis M, Pingoud A, Wende W, Desbiolles P. The rotation-coupled sliding of EcoRV. Nucleic Acids Res. 2012;40:4064–4070. doi: 10.1093/nar/gkr1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cho W-K, Jeong C, Kim D, Chang M, Song K-M, Hanne J, Ban C, Fishel R, Lee J-B. ATP Alters the Diffusion Mechanics of MutS on Mismatched DNA. Structure/Folding and Design. 2012:1–11. doi: 10.1016/j.str.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blainey PC, Luo G, Kou SC, Mangel WF, Verdine GL, Bagchi B, Xie XS. Nonspecifically bound proteins spin while diffusing along DNA. Nature Structural & Molecular Biology. 2009;16:1224–1229. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gorman J, Plys AJ, Visnapuu ML, Alani E, Greene EC. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat Struct Mol Biol. 2010;17:932–938. doi: 10.1038/nsmb.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gorman J, Wang F, Redding S, Plys AJ, Fazio T, Wind S, Alani EE, Greene EC. Single-molecule imaging reveals target-search mechanisms during DNA mismatch repair. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3074–3083. doi: 10.1073/pnas.1211364109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qiu R, DeRocco VC, Harris C, Sharma A, Hingorani MM, Erie DA, Weninger KR. Large conformational changes in MutS during DNA scanning, mismatch recognition and repair signalling. The EMBO Journal. 2012:1–13. doi: 10.1038/emboj.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Komazin-Meredith G, Mirchev R, Golan DE, van Oijen AM, Coen DM. Hopping of a processivity factor on DNA revealed by single-molecule assays of diffusion. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10721–10726. doi: 10.1073/pnas.0802676105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 73.Natrajan G, Lamers MH, Enzlin JH, Winterwerp HH, Perrakis A, Sixma TK. Structures of Escherichia coli DNA mismatch repair enzyme MutS in complex with different mismatches: a common recognition mode for diverse substrates. Nucleic Acids Res. 2003;31:4814–4821. doi: 10.1093/nar/gkg677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 75.Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, Beese LS. Structure of the human MutSalpha DNA lesion recognition complex. Mol Cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 76.Wang H, Yang Y, Schofield MJ, Du C, Fridman Y, Lee SD, Larson ED, Drummond JT, Alani E, Hsieh P, et al. DNA bending and unbending by MutS govern mismatch recognition and specificity. Proc Natl Acad Sci U S A. 2003;100:14822–14827. doi: 10.1073/pnas.2433654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tessmer I, Yang Y, Zhai J, Du C, Hsieh P, Hingorani MM, Erie DA. Mechanism of MutS searching for DNA mismatches and signaling repair. J Biol Chem. 2008;283:36646–36654. doi: 10.1074/jbc.M805712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Erie DA, Yang G, Schultz HC, Bustamante C. DNA bending by Cro protein in specific and nonspecific complexes: implications for protein site recognition and specificity [see comments] Science. 1994;266:1562–1566. doi: 10.1126/science.7985026. [DOI] [PubMed] [Google Scholar]
- 79.Yang Y, Sass L, Du C, Hsieh P, Erie DA. Determination of protein-DNA binding constants and specificities from statistical analyses of single molecules: MutS-DNA interactions. Nucleic Acids Res. 2005 doi: 10.1093/nar/gki708. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sass LE, Lanyi C, Weninger K, Erie DA. Single-molecule FRET TACKLE reveals highly dynamic mismatched DNA-MutS complexes. Biochemistry. 2010;49:3174–3190. doi: 10.1021/bi901871u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Derocco VC, Sass LE, Qiu RR, Weninger KR, Erie DA. Dynamics of MutS-mismatched DNA complexes are predictive of their repair phenotypes. Biochemistry. 2014 doi: 10.1021/bi401429b. http://dx.doi.org/10.1021/bi401429b. [DOI] [PMC free article] [PubMed]
- 82.Cristovao M, Sisamakis E, Hingorani MM, Marx AD, Jung CP, Rothwell PJ, Seidel CA, Friedhoff P. Single-molecule multiparameter fluorescence spectroscopy reveals directional MutS binding to mismatched bases in DNA. Nucleic Acids Res. 2012;40:5448–5464. doi: 10.1093/nar/gks138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lata S, Piehler J. Stable and functional immobilization of histidine-tagged proteins via multivalent chelator headgroups on a molecular poly(ethylene glycol) brush. Anal Chem. 2005;77:1096–1105. doi: 10.1021/ac048813j. [DOI] [PubMed] [Google Scholar]
- 84.Lata S, Gavutis M, Tampe R, Piehler J. Specific and stable fluorescence labeling of histidine-tagged proteins for dissecting multi-protein complex formation. J Am Chem Soc. 2006;128:2365–2372. doi: 10.1021/ja0563105. [DOI] [PubMed] [Google Scholar]
- 85.DeRocco V, Anderson T, Piehler J, Erie DA, Weninger K. Four-color single-molecule fluorescence with noncovalent dye labeling to monitor dynamic multimolecular complexes. BioTechniques. 2010;49:807–816. doi: 10.2144/000113551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lang WH, Coats JE, Majka J, Hura GL, Lin Y, Rasnik I, McMurray CT. Conformational trapping of mismatch recognition complex MSH2/MSH3 on repair-resistant DNA loops. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E837–844. doi: 10.1073/pnas.1105461108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brouwer JR, Willemsen R, Oostra BA. Microsatellite repeat instability and neurological disease. Bioessays. 2009;31:71–83. doi: 10.1002/bies.080122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Allen DJ, Makhov A, Grilley M, Taylor J, Thresher R, Modrich P, Griffith JD. MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 1997;16:4467–4476. doi: 10.1093/emboj/16.14.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schofield MJ, Nayak S, Scott TH, Du C, Hsieh P. Interaction of Escherichia coli MutS and MutL at a DNA mismatch. J Biol Chem. 2001;276:28291–28299. doi: 10.1074/jbc.M103148200. [DOI] [PubMed] [Google Scholar]
- 90.Jiang Y, Marszalek PE. Atomic force microscopy captures MutS tetramers initiating DNA mismatch repair. The EMBO journal. 2011;30:2881–2893. doi: 10.1038/emboj.2011.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ratcliff GC, Erie DA. A novel single-molecule study to determine protein--protein association constants. JACS. 2001;123:5632–5635. doi: 10.1021/ja005750n. [DOI] [PubMed] [Google Scholar]
- 92.Yang Y, Wang H, Erie DA. Quantitative characterization of biomolecular assemblies and interactions using atomic force microscopy. Methods. 2003;29:175–187. doi: 10.1016/s1046-2023(02)00308-0. [DOI] [PubMed] [Google Scholar]
- 93.Mendillo ML, Mazur DJ, Kolodner RD. Analysis of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 and MLH1-PMS1 complexes with DNA using a reversible DNA end-blocking system. J Biol Chem. 2005;280:22245–22257. doi: 10.1074/jbc.M407545200. [DOI] [PubMed] [Google Scholar]
- 94.Ban C, Junop M, Yang W. Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell. 1999;97:85–97. doi: 10.1016/s0092-8674(00)80717-5. [DOI] [PubMed] [Google Scholar]
- 95.Ban C, Yang W. Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell. 1998;95:541–552. doi: 10.1016/s0092-8674(00)81621-9. [DOI] [PubMed] [Google Scholar]
- 96.Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 97.Guarne A, Junop MS, Yang W. Structure and function of the N-terminal 40 kDa fragment of human PMS2: a monomeric GHL ATPase. EMBO J. 2001;20:5521–5531. doi: 10.1093/emboj/20.19.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hu X, Machius M, Yang W. Monovalent cation dependence and preference of GHKL ATPases and kinases. FEBS Lett. 2003;544:268–273. doi: 10.1016/s0014-5793(03)00519-2. [DOI] [PubMed] [Google Scholar]
- 99.Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yu J, Ha T, Schulten K. Structure-based Model of the Stepping Motor of PcrA Helicase. Biophys J. 2006 doi: 10.1529/biophysj.106.088203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Corbett KD, Berger JM. Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI. Structure. 2005;13:873–882. doi: 10.1016/j.str.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 102.Dollins DE, Immormino RM, Gewirth DT. Structure of unliganded GRP94, the endoplasmic reticulum Hsp90. Basis for nucleotide-induced conformational change. The Journal of biological chemistry. 2005;280:30438–30447. doi: 10.1074/jbc.M503761200. [DOI] [PubMed] [Google Scholar]
- 103.Immormino RM, Dollins DE, Shaffer PL, Soldano KL, Walker MA, Gewirth DT. Ligand-induced conformational shift in the N-terminal domain of GRP94, an Hsp90 chaperone. The Journal of biological chemistry. 2004;279:46162–46171. doi: 10.1074/jbc.M405253200. [DOI] [PubMed] [Google Scholar]
- 104.Shiau AK, Harris SF, Southworth DR, Agard DA. Structural Analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell. 2006;127:329–340. doi: 10.1016/j.cell.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 105.Guarne A, Ramon-Maiques S, Wolff EM, Ghirlando R, Hu X, Miller JH, Yang W. Structure of the MutL C-terminal domain: a model of intact MutL and its roles in mismatch repair. EMBO J. 2004;23:4134–4145. doi: 10.1038/sj.emboj.7600412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kosinski J, Steindorf I, Bujnicki JM, Giron-Monzon L, Friedhoff P. Analysis of the quaternary structure of the MutL C-terminal domain. J Mol Biol. 2005;351:895–909. doi: 10.1016/j.jmb.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 107.Pang Q, Prolla TA, Liskay RM. Functional domains of the Saccharomyces cerevisiae Mlh1p and Pms1p DNA mismatch repair proteins and their relevance to human hereditary nonpolyposis colorectal cancer-associated mutations. Mol Cell Biol. 1997;17:4465–4473. doi: 10.1128/mcb.17.8.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tran PT, Liskay RM. Functional studies on the candidate ATPase domains of Saccharomyces cerevisiae MutLalpha. Mol Cell Biol. 2000;20:6390–6398. doi: 10.1128/mcb.20.17.6390-6398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sacho EJ, Kadyrov FA, Modrich P, Kunkel TA, Erie DA. Direct visualization of asymmetric adenine-nucleotide-induced conformational changes in MutL alpha. Mol Cell. 2008;29:112–121. doi: 10.1016/j.molcel.2007.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hall MC, Shcherbakova PV, Fortune JM, Borchers CH, Dial JM, Tomer KB, Kunkel TA. DNA binding by yeast Mlh1 and Pms1: implications for DNA mismatch repair. Nucleic Acids Res. 2003;31:2025–2034. doi: 10.1093/nar/gkg324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Park J, Jeon Y, In D, Fishel R, Ban C, Lee JB. Single-molecule analysis reveals the kinetics and physiological relevance of MutL-ssDNA binding. PloS one. 2010;5:e15496. doi: 10.1371/journal.pone.0015496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hall MC, Wang H, Erie DA, Kunkel TA. High affinity cooperative DNA binding by the yeast Mlh1-Pms1 heterodimer. J Mol Biol. 2001;312:637–647. doi: 10.1006/jmbi.2001.4958. [DOI] [PubMed] [Google Scholar]
- 113.Pillon MC, Lorenowicz JJ, Uckelmann M, Klocko AD, Mitchell RR, Chung YS, Modrich P, Walker GC, Simmons LA, Friedhoff P, et al. Structure of the endonuclease domain of MutL: unlicensed to cut. Mol Cell. 2010;39:145–151. doi: 10.1016/j.molcel.2010.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shcherbakova PV, Hall MC, Lewis MS, Bennett SE, Martin KJ, Bushel PR, Afshari CA, Kunkel TA. Inactivation of DNA mismatch repair by increased expression of yeast MLH1. Mol Cell Biol. 2001;21:940–951. doi: 10.1128/MCB.21.3.940-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Blackwell LJ, Wang S, Modrich P. DNA chain length dependence of formation and dynamics of hMutSalpha. hMutLalpha.heteroduplex complexes. J Biol Chem. 2001;276:33233–33240. doi: 10.1074/jbc.M105076200. [DOI] [PubMed] [Google Scholar]
- 116.Elez M, Radman M, Matic I. Stoichiometry of MutS and MutL at unrepaired mismatches in vivo suggests a mechanism of repair. Nucleic Acids Res. 2012;40:3929–3938. doi: 10.1093/nar/gkr1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Grilley M, Griffith J, Modrich P. Bidirectional excision in methyl-directed mismatch repair. J Biol Chem. 1993;268:11830–11837. [PubMed] [Google Scholar]
- 118.Dzantiev L, Constantin N, Genschel J, Iyer RR, Burgers PM, Modrich P. A defined human system that supports bidirectional mismatch-provoked excision. Mol Cell. 2004;15:31–41. doi: 10.1016/j.molcel.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 119.Yardimci H, Loveland AB, van Oijen AM, Walter JC. Single-molecule analysis of DNA replication in Xenopus egg extracts. Methods (San Diego, Calif) 2012:1–8. doi: 10.1016/j.ymeth.2012.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tanner NA, van Oijen AM. Visualizing DNA replication at the single-molecule level. Methods Enzymol. 2010;475:259–278. doi: 10.1016/S0076-6879(10)75011-4. [DOI] [PubMed] [Google Scholar]
- 121.Kulczyk AW, Tanner NA, Loparo JJ, Richardson CC, van Oijen AM. Direct observation of enzymes replicating DNA using a single-molecule DNA stretching assay. Journal of visualized experiments : JoVE. 2010 doi: 10.3791/1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tanner NA, Loparo JJ, Hamdan SM, Jergic S, Dixon NE, van Oijen AM. Real-time single-molecule observation of rolling-circle DNA replication. Nucleic Acids Res. 2009;37:e27. doi: 10.1093/nar/gkp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Uphoff S, Reyes-Lamothe R, Garza de Leon F, Sherratt DJ, Kapanidis AN. Single-molecule DNA repair in live bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8063–8068. doi: 10.1073/pnas.1301804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sakon JJ, Weninger KR. Detecting the conformation of individual proteins in live cells. Nat Methods. 2010;7:203–U256. doi: 10.1038/nmeth.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li GW, Xie XS. Central dogma at the single-molecule level in living cells. Nature. 2011;475:308–315. doi: 10.1038/nature10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Huang J, Nagy SS, Koide A, Rock RS, Koide S. A Peptide Tag System for Facile Purification and Single-Molecule Immobilization. Biochemistry. 2009;48:11834–11836. doi: 10.1021/bi901756n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Reyes-Lamothe R, Sherratt DJ, Leake MC. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science. 2010;328:498–501. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]