Abstract
Hauling and anchoring the nucleus within immobile or motile cells, tissues and/or syncytia represents a major challenge. In the past 15 years, Linkers of the Nucleoskeleton to the Cytoskeleton (LINC complexes) have emerged as evolutionary-conserved molecular devices that span the nuclear envelope and provide interacting interfaces for cytoskeletal networks and molecular motors to the nuclear envelope. Here, we will review the molecular composition of LINC complexes and focus on how their genetic alteration in vivo has provided a wealth of information related to the relevance of nuclear positioning during tissue development and homeostasis with a special emphasis on the central nervous system. As it may be relevant for metastasis in a range of cancers, the involvement of LINC complexes in migration of non-neuronal cells via its interaction with the perinuclear actin cap will also be developed.
Keywords: LINC complexes, Sun protein, Nesprin, KASH domain, SUN domain, nuclear lamina, interkinetic nuclear migration, neuronal migration, nuclear anchorage, actin cap, skeletal muscle, retina, cell motility
Introduction
The nuclear envelope (NE) physically separates the genome from the cytoplasm (Fig. 1). It is composed of an inner and outer nuclear membrane (INM and ONM, respectively) that connect at nuclear pores and delineate the luminal compartment that is continuous with the lumen of the endoplasmic reticulum (ER). Whereas the ONM is an extension of the rough ER, the INM is devoid of ribosomes and displays a unique set of resident proteins that are immobilized within the INM by virtue of the interaction of their nucleoplasmic domains with the nuclear lamina and/or chromatin [1]. The nuclear lamina is a meshwork of nuclear type-V intermediate filaments represented by A- and B-type lamins [2–4]. Whereas B-type lamins appear to be ubiquitously expressed both within progenitors and differentiated tissues, A-type lamins expression is restricted to subsets of differentiated cells and tissues [5–7].
Figure 1. Linkers of the Nucleoskeleton to the Cytoskeleton (LINC complexes) organization at the nuclear envelope in mammals (see text for more details).
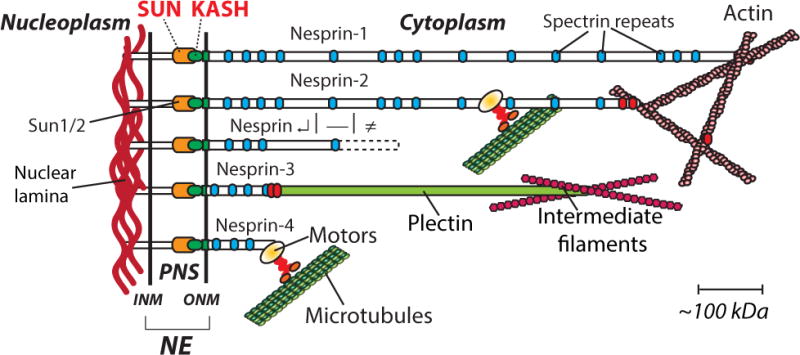
Nesprin α, β, γ refers to shorter spliced isoforms of Nesprin 1 and 2. INM, ONM: inner and outer nuclear membrane, respectively; NE: nuclear envelope; SUN: Sad1 and Unc84 domain; KASH: Klarsicht/ANC1/Syne homology domain.
LINC complexes: macromolecular assemblies that span the nuclear envelope and mediate nuclear migration and anchorage
The diversity of physiological functions exerted by resident proteins of the INM has completely redefined the functionality of the NE [8]. In particular, major progress has been made in the identification and physiological roles of macromolecular complexes that span the NE and mediate nuclear movements during development as well as nuclear anchorage in differentiated tissues. The term LINC (LInkers of the Nucleoskeleton to the Cytoskeleton) was coined to describe these molecular assemblies [9] (Fig. 1). They consist of interactions, within the lumen, between evolutionarily conserved motifs that characterize two families of integral transmembrane proteins of the NE: Sun (Sad1/Unc84) proteins and Nesprins (Nuclear Envelope SPectrin Repeat containing proteINS). In the following sections, we will review the molecular nature of Sun proteins and Nesprins as well as their role as LINC complexes in nuclear dynamics and single cell migration.
Sun proteins
Fifteen years ago, a seminal study by Malone et al identified unc84, a gene whose mutation prevented both the migration and subsequent anchorage of nuclei populating the developing hypodermal syncytium of C. elegans [10]. UNC84 was identified as a transmembrane protein residing at the NE. One of its remarkable features was a C-terminal region of about 200 amino acids that displayed a strong homology with the C-terminal region of Sad1, a spindle pole body-associated protein in S. pombe [11]. This region, called the SUN domain (Sad1-Unc84 homology, PFAM family PF03856), was also identified in two human genes called SUN1 and SUN2. Mammalian Sun1 and Sun2 proteins were later identified as type II integral membrane proteins of the INM with their conserved C-terminal SUN domain protruding within the luminal region between the INM and ONM of the NE [12–14]. Since then, orthologs have been identified in many phyla as well as in the plant kingdom [15] confirming the strong evolutionary conservation of SUN domains [16]. Recent crystallography studies have shown that SUN domains form homotrimeric structures displaying a cloverleaf-like arrangement [17,18].
The N-terminal nucleoplasmic region of Sun proteins is less well characterized. This region interacts with nuclear lamins [9,13] but lamins are not required for human Sun proteins localization at the NE [19]. An emerging feature of Sun1 is the versatility of its primary sequence [20] due to the alternative splicing of exons 5 to 10 that encode the central nucleoplasmic region (Fig. 2). The nucleoplasmic region of Sun2, by contrast, does not display any comparable alternative splicing (data not shown). Interestingly, the extensively spliced region encodes a region reported to bind to nucleoplasmic binding partners of Sun1 [21] suggesting that specific Sun1 isoforms interact with distinct nucleoplasmic proteins. To date, the best-characterized nucleoplasmic variant of Sun1 was identified in testes and functionally defined by Gob et al. as being involved in mammalian sperm head formation [22]. Another intriguing feature of the nucleoplasmic region of Sun1 is the presence of a Mitochondrial RNA binding Protein domain (MRP, PFAM family PF09387) whose motif is highly conserved in mammalian Sun1 but not Sun2 (Fig. 2). In Trypanosoma, MRP1 and MRP2 belong to a complex machinery involved in mitochondrial RNA editing, a hallmark of kinetoplastids [23]. Whether the synthesis of MRP-like proteins by Sun1 genes actually occurs in mammals and their functional roles remains to be established.
Figure 2. The nucleoplasmic region of Sun1 is alternatively spliced and contains an MRP-homology region.
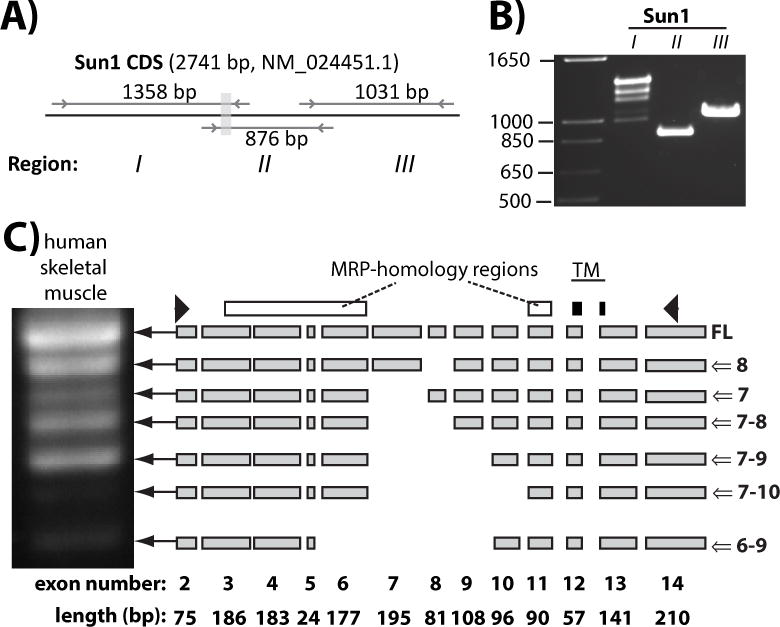
A) Primers location (arrows) and size of each amplicon predicted from indicated mRNA reference sequence of Sun1. Gray shaded rectangle corresponds to the transmembrane-encoding region. B) RT-PCR amplification of Region I, II and III of Sun1 transcripts carried out on total RNA of C2C12 mouse myoblasts. Note the amplification of multiple bands from Region I of Sun1 that encodes the nucleoplasmic domain. C) PCR amplification of Sun1 transcripts from human skeletal muscle first strand cDNAs with depiction of alternatively spliced isoforms. White rectangles: exonic region displaying MRP homology. Black rectangles: exonic region encoding the transmembrane domain of Sun1. Arrowheads: position of primers used in PCR amplification.
Nesprins
Within the lumen, the SUN domain interacts directly with the evolutionary-conserved KASH (Klarsicht/Anc-1, Syne Homology, PFAM family PF10541) domain, a stretch of ~60 amino acids made up of a transmembrane domain followed by a short stretch of ~30 luminal C-terminal amino acids (Fig. 1). To date, KASH domain-containing proteins have been functionally identified in S. pombe, D. melanogaster, C. elegans and mammals [16]. In the latter, it is the typical molecular signature of a family of mammalian NE proteins called Nesprins that are encoded by five distinct genes (Nesprin-1 to -5) [24–28]. Whereas Nesprin-1, -2 and -3 are expressed in a wide variety of tissues, Nesprin-4 expression is more restricted and Nesprin-5 is a meiosis-specific KASH protein [28]. Nesprins harbor variable numbers of spectrin repeats along their cytoplasmic region that extend from ~50 kDa (Nesprin-4) to an astounding 1,000 kDa (giant isoform of Nesprin-1). Importantly, due to their gene size, a challenging plethora of Nesprin-1 and -2 isoforms (with and without KASH domains), are expressed to different degrees in different tissues and at different development times [29–31]. Next to these common structural features, giant isoforms of Nesprin-1 and -2 directly interact with actin through N-terminal actin binding domains [32,33] and Nesprin-3 with plectin [26] (Fig. 1). Nesprins also associate with molecular motors. In C. elegans, nuclear migration is mediated through direct interactions of the cytoplasmic region of UNC-83 with both kinesin-1 and dynein and their regulators [34,35]. In mammals, Nesprin-2 coimmunoprecipitates with the dynein complex and Nesprin-4 with kinesin-1 [27,36]. Together, these finding strongly support a model whereby LINC complexes connect the nucleus to the cytoskeleton and molecular motors.
LINC complexes: hubs for force transduction across the NE
Interactions between SUN and KASH domain-containing proteins across the NE are direct and essential for the recruitment of KASH domain-containing proteins at the NE. Indeed, studies in different biological systems clearly demonstrate that the presence of SUN domains is strictly required for the ONM localization of KASH domain-containing proteins [9,36,37]. In addition, the KASH domain, by itself, is both sufficient to localize at the NE and strictly required to specify the NE localization of KASH proteins [38]. Accordingly, either the targeted expression of SUN domains within the perinuclear space or the overexpression of recombinant tagged KASH domains act in a dominant-negative manner by dislodging endogenous KASH domain-containing proteins from the ONM to the ER [39]. As we will see below, this property of recombinant KASH domain, such as EGFP-KASH, has multiple experimental applications to examine the role of LINC complexes in different species.
Several lines of evidence indicate that LINC complexes transduce forces across the NE. Physical coupling between the nucleus and the cytoplasm was directly demonstrated in harpooning experiments of the cytoplasm using microneedles. In this experimental setting, the NE clearly protrudes in the direction of an outward cytoplasmic pull whereas it invaginates when the nucleoplasm itself is harpooned [40]. The NE is also distorted by manipulating microbeads attached to integrins, thereby suggesting that mechanical forces can be directly applied at the NE from the cytoplasmic membrane. As described above, the structural analysis and domain composition of Sun proteins and Nesprins strongly suggested a central role for LINC complexes in establishing such physical connections. Accordingly, disruption of SUN/KASH interactions drastically reduces nuclear deformation in microneedle manipulations [41] and further disrupts stretch-induced nuclear rotation [39]. Beside nuclear mechanics and dynamics, disruption of LINC complexes induces an overall loss of mechanical stiffness across the cytoskeleton [39], a phenotype that most likely reflects reported alterations of the perinuclear cytoskeleton [41] as well as decreased cellular migration and loss of polarization ensuing from LINC complex disruption [39].
Human SUN/KASH complexes have recently been crystallized [17]. Solved structures and biochemical approaches indicate that SUN domains physiologically assemble as trimers whose interacting interfaces provide large grooves for KASH domain binding. Because these interactions consist of an extensive network of non-covalent interactions between SUN and KASH triads and SUN and KASH domains interact covalently through disulfide bonds [17], SUN/KASH interactions appear well adapted as force-resistant coupling devices to move or still anchor nuclei within cells or syncytia [42]. Accumulating data indicate that forces transduced by LINC complexes are used in two important biological phenomena: nuclear positioning and chromosome movements. For more information on the role of SUN/KASH interaction in chromosome movements, readers are referred to a recent and thorough review by Kracklauer et al. [43]. Here, we will further focus on the involvement of LINC complexes in nuclear positioning.
LINC complexes and nuclear positioning in CNS development
As described above, pioneering studies in C. elegans and D. melanogaster [44] clearly pointed out the role of SUN and KASH domain-containing proteins in nuclear migration and anchorage. More importantly, they paved the way to more recent studies aimed at understanding the physiological relevance of different types of nuclear movement observed during central nervous system (CNS) development [16]. The latter proceeds through the transformation of a pseudostratified layer of precursor cells into laminated layers of differentiated neurons whose interconnections establish the CNS circuitry. This transformation and its accompanying nuclear movements are well illustrated by the development of the mammalian retina. In the latter, the pseudostratified neuroepithelium, called the neuroblast layer, morphs into three distinct laminae of differentiated neurons. This process can be divided in different steps: 1) exit of retinal progenitor cells, which populate the neuroblast layer, from the cell cycle, 2) migration of post-mitotic newborn neurons towards their final laminar position, 3) anchorage of differentiated neurons to their specific laminar position. Below, we describe the various types of nuclear movements as well as what is known about the involvement of LINC complexes in these different developmental steps.
Interkinetic nuclear migration in neuronal progenitors
Interkinetic nuclear migration (IKNM, Fig. 3) consists of cell cycle-dependent oscillations of neuronal progenitors nuclei within pseudostratified neuroepithelia (recently reviewed in [45,46]). Importantly, IKNM appears to be a universal property of pseudostratified epithelia. It has mostly been studied in developing neural tissues but, importantly, it is neither restricted to developing CNS tissues nor confined to vertebrates [46–48]. During IKNM, nuclei migrate towards the basal side of neuroepithelia during the G1 phase of the cell cycle and move back to the apical side during G2. As a result, mitoses take place exclusively at the apical side (Fig. 3A) while S-phase proceeds at the basal side of neuroepithelia. Anti-phospho-Histone H3, which specifically labels mitotic cells, and BrdU labeling, which labels S-phase cells, is commonly used to track IKNM in fixed samples.
Figure 3. Nuclear movements during embryonic retinal development.
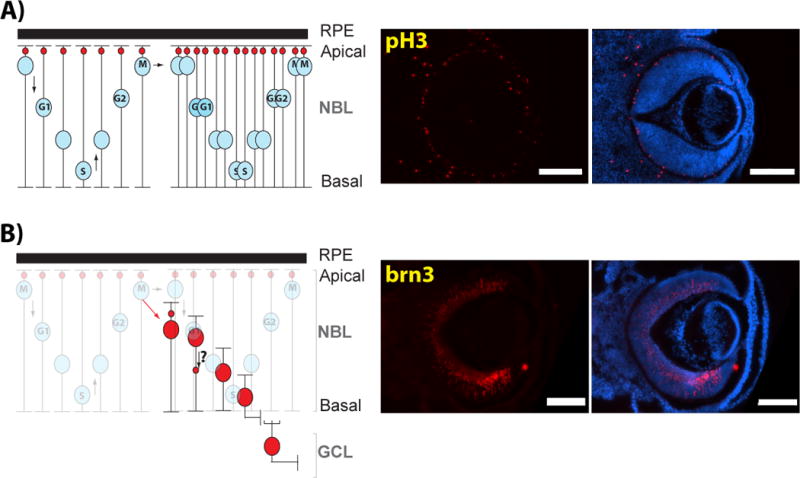
A) Interkinetic nuclear migration consists of the basoapical migration of retinal progenitor cell nuclei (RPC, blue nuclei) in phase with the cell cycle. A symmetric mitotic division generating two progenitor cells is also illustrated. Small red circles depict the position of the centrosome. Right panel: Immunofluorescence of an E14.5 mouse ocular globe showing the apical localization of mitotic progenitors (labeled with anti-phosphoHistone3, pH3) on the apical side of the neuroretina. B) In the case of an asymmetric division, the post-mitotic cells (in this case a retinal ganglion cell, red nuclei) migrate towards its final laminar position within the ganglion cell layer. Right panel: Immunofluorescence of an E14.5 mouse ocular globe showing newborn retinal ganglion cells (labeled with brn3) migrating towards the GCL. M: M-phase nuclei; S: S-phase nuclei; RPE: Retinal Pigment Epithelium; NBL: Neuroblast layer; GCL: Ganglion Cell Layer. Scale bar: 200mm.
Blocking nuclear oscillations during IKNM does not alter cell cycle progression of progenitors [49,50] whereas cell cycle arrest blocks nuclear oscillations [51,52]. These nuclear oscillations require an intact cytoskeleton (microtubule, actin and centrosome) and molecular motors (actomyosin, kinesins and the dynein complex). The identification of these molecular actors and their respective roles in apicobasal migration within neuroepithelial cells has been recently reviewed [45,46]. By comparison to the wide amplitude of apicobasal migration of neuroblast nuclei, the centrosome is relatively stationary on the apical side of neuroepithelia [53] (Fig. 3A). However, recent time-lapse video microscopy experiments on chicken neural tube indicate that the centrosome migrates basally to “meet” apically migrating nuclei at late G2 [54].
To date, two reports indicate that LINC complexes directly mediate IKNM in mammals. Using time-lapse video microscopy, Zhang et al. showed that nuclear migration towards the apical side of the ventricular zone was significantly slower in brain slices of Nesprin-2 KO and Sun1/Sun2 double knockout (DKO) mouse embryos by comparison to wild-type brains [36]. Accordingly, mitotic cells labeled with phospho-Histone H3 antibodies mislocalized across the length of the ventricular zone rather than on its apical side. IKNM is also significantly altered in Sun1 and Nesprin2 KO mouse retina [55]. In zebrafish retina, either the downregulation of Nesprin-2 or the expression of a recombinant dominant-negative EGFP-KASH protein during IKNM correlates with the accelerated genesis of earlier born neurons such as retinal ganglion cells at the expense of later-born photoreceptors [56]. These experiments did not directly examine nuclear positioning during IKNM. However, a similar phenotype of accelerated neurogenesis is also observed upon nonsense mutations of dynactin that directly affects nuclear positioning during IKNM. Because Nesprin2 interacts with dynactin in mouse retina and brain lysates [36,55] and these interactions are evolutionary conserved, these experimental results predict that LINC complex disruption should affect IKNM in Zebrafish retina. Furthermore they would suggest that intact LINC complexes are required to maintain an appropriate pool of neural progenitors while generating post-mitotic neurons in a timely fashion [50,51,56].
Together, these results strongly suggest that LINC complexes provide essential nucleocytoskeletal connections that directly mediate nuclear positioning during IKNM. Molecular motors-generated forces that move nuclei across the neuroepithelial cell length are most likely transduced to the NE through LINC complexes. The identification of unc-83 binding domains to dyneins, kinesins and their accessory proteins further suggest that the Nesprin interaction with molecular motors may be direct [35]. According to a model whereby basalmost migration of progenitor nuclei favors a subsequent asymmetric division into a newborn neuron and another retinal progenitor cells [49,51,56], faulty IKNM within retinal progenitor cells may affect the balance of asymmetric vs. symmetric division thereby affecting the relative abundance of retinal cell types in adult retinas. By contrast to the alteration of molecular motors that are involved in a diverse array of cellular functions, disruption of LINC complexes may provide a more specific mean to disrupt nuclear positioning during IKNM. Hence, the development of genetic tools to disrupt the SUN/KASH interactions in different species may provide additional insight into the possible physiological function(s) of IKNM.
Nuclear translocation in post-mitotic neurons
Exit of progenitor cells from the cell cycle is followed by the migration of post-mitotic neurons from the apical side of the neuroepithelium toward their final laminar destination (Fig. 3B). In the developing neocortex, trekking of these newborn neurons towards the pial surface requires the active translocation of their nuclei [57,58]. This actin- and microtubule-dependent process is initiated by the rapid extension and subsequent retraction of the leading neurite. The centrosome, which is invariably positioned ahead of the nucleus in these cells, moves away from the nucleus towards the leading process. The nucleus then translocates closer to the centrosome [59]. Hence, by contrast to IKNM where the centrosome remains relatively stationary, nuclear translocation within migrating post-mitotic neurons requires the coupling of the nucleus to the centrosome via a “fork-like” structure of microtubules wrapping the nucleus. The dynein/dynactin complex, LIS1 and other proteins that bind to microtubules and regulate dynein activity closely regulate nuclear translocation in these cells [57,59–61]. Abnormal nuclear translocation underlies the erroneous lamination of cortical layers, a phenotype that characterizes a wide range of human pathologies of the CNS called lissencephalies [62]. Similar cortical lamination defects, caused by hypoglycosylation of basement membrane components, are also observed in a subgroup of congenital muscular dystrophies called dystroglycanopathies. These include Muscle-Eye-Brain disease, Fukuyama congenital muscular dystrophy and the Walker-Warburg syndrome [63,64]. Severe retinal development abnormalities such as microphthalmia, optic nerve hypoplasia and retinal dysgenetic stratification are also prominent in patients affected by lissencephalies [65–70] and ocular defects are also observed in mouse models of human dystroglycanopathies [71,72]. It is therefore likely that similar molecular mechanisms underlying nuclear migration in the neocortex are at play during retinogenesis. For these reasons, a better understanding of the role of nuclear movements during retinogenesis will most likely provide essential information about molecular mechanisms underlying both congenital retinopathies and congenital brain development disorders.
LINC complexes are required for nuclear translocation during the migration of newborn cortical neurons. Indeed, radial migration of newborn cortical neurons is severely hampered in the developing cerebral cortex of SUN1/2 DKO and Nesprin2 KO embryos [36]. These defects are clearly associated to the failure of newborn cortical neurons to translocate their nuclei due to a loss of physical coupling between the nucleus and the centrosome. Indeed, whereas nuclei undergo robust translocation towards the pial surface in close association with the centrosome in wild-type brain slices, newborn cortical neuron nuclei remain at the ventricular side of SUN1/2 DKO or Nesprin2 KO brain slices even though centrosomes keep moving towards the pial surface. These observations strongly suggest that transduction of apical forces on the migrating neuron nuclei are abolished upon LINC complex disruption. Whereas this mode of nuclear translocation has been clearly demonstrated for tangentially migrating cortical interneurons and radial cortical neurons [73,74], other modes of neuronal nuclear translocation may exist within the CNS. Indeed, during radial migration of granule cells in cerebellar slices, the centrosome most often localizes between the anterior and posterior poles of the migrating nucleus rather than leading the nucleus [75]. Another example of “unconventional” nuclear translocation is provided by newborn retinal ganglion cells migrating towards their final laminar position at the beginning of zebrafish retinogenesis. In that case, the centrosome localizes at the trailing edge of migrating retinal ganglion cells [53]. The postnatal development of cone photoreceptors may provide another example of unconventional nuclear translocation within post-mitotic neurons. Whereas cone photoreceptors are specified early during embryonic retinal development [76], their nuclei remain on the apical surface of the neuroblast layer. However, during postnatal retinal development, mouse cone nuclei localize within the upper 2/3 of the developing outer plexiform layer before regaining their apical position [77]. Through the spatiotemporal control of EGFP-KASH2 expression during mouse cone photoreceptor maturation, we recently showed that LINC complexes mediate the apical migration leg of that oscillation whereas basal migration appears unaffected. As a result, cone photoreceptor nuclei mislocalized at the basal edge of the outer nuclear layer as well as within the outer plexiform layer in adult retina [78]. Though less severe, a mislocalization phenotype of cone photoreceptor nuclei was also reported in adult retinas from Sun1 KO mice [55]. Even though the localization of the centrosome has not been formally examined during these oscillations, their apical location is required to elaborate the connecting cilium of presumptive photoreceptors. Hence, nuclear oscillations within maturing cone photoreceptors most likely take place without any significant movement of the centrosome thereby representing another case of unconventional nuclear translocation within a post-mitotic neuron.
Molecular mechanisms mediating photoreceptor nuclei positioning appear to be evolutionary conserved. Indeed, in D. melanogaster and zebrafish, genetic alterations of either the dynein/dynactin complex or SUN/KASH interactions clearly affect nuclear positioning during photoreceptor maturation [79–81].
As mentioned above, the nucleoplasmic region of Sun proteins interacts with nuclear lamins. Therefore, forces generated by molecular motors and transduced through LINC complexes should act on the lamina meshwork to pull the nucleus in the direction of cellular migration. This essential contribution of lamins is best exemplified by nuclear movements associated with the development of the compound eye of D. melanogaster. Indeed, genetic alteration of lamin Dm0 prevents the apical migration of photoreceptor precursors nuclei [82]. Importantly, that same failure of apical migration is phenocopied by genetic alteration of klaroid (SUN protein), Klarsicht (KASH protein) or glued (Dynactin) mutants [80–83]. In mammals, A- and B-type lamins make up the nuclear lamina of most differentiated cells. However, mostly B-type lamins are expressed during CNS development. Accordingly, B-type lamins KO mice display severe neurodevelopmental defects and die at birth whereas A-type lamin KO mice develop normally but die by 4–6 weeks and present muscular dystrophy phenotypes [84,85].
Nuclear anchorage
Mammalian Nesprin-1 was initially discovered as a direct binding partner of MuSK, a tyrosine kinase enriched at the post-synaptic membrane of the neuromuscular junction (NMJ), and further identified as a NE protein whose abundance was significantly higher in specialized nuclei that cluster beneath the NMJ (synaptic nuclei) by comparison to extrasynaptic nuclei [24]. Constitutive transgenic overexpression of the dominant negative KASH domain of Nesprin-1 induced a phenotype where synaptic nuclei mislocalize away from the NMJ [86]. The development of Nesprin-1 KO mice confirmed that phenotype and further indicated that Nesprin1 also mediate the spacing of extrasynaptic nuclei along skeletal muscle fibers. Because Nesprin2 is dispensable for synaptic nuclei to localize at the NMJ [87], anchoring of synaptic nuclei at the NMJ is one of the few cellular functions of Nesprin-1 that does not overlap with Nesprin-2. Similar synaptic nuclei mispositioning phenotypes have been reported in different models of genetic alterations of the Nesprin1 gene in mice [88,89], and, in agreement with a central role of SUN/KASH interactions in synaptic nuclei anchoring, Sun1 and Sun2 function redundantly in myonuclear anchorage [90]. Synaptic nuclei mislocalization has also been reported in skeletal muscle biopsies from patients affected by autosomal recessive cerebellar ataxia type 1 (ARCA1) associated to mutations of Nesprin-1 [91]. In agreement with mouse models, NMJ structural organization is not affected by nuclear mislocalization. Whereas these results clearly indicate that genetic alteration of Nesprin1 alters nuclear positioning in skeletal muscle, the phenotypical outcome in these different mouse models greatly varies from the lack of any overt phenotype [87] to Emery-Dreifuss muscular dystrophy-like [89] or growth retardation [88] phenotypes. The reader is referred to [88] for further discussion about the potential reasons underlying this phenotypical variability. Taken together, these results clearly indicate that LINC complexes, mostly made of Nesprin1 and Sun1 in this case, mediate the anchorage of myonuclei within skeletal muscle fibers. Furthermore, results from different mouse models and ARCA1 patients suggest that whereas genetic alterations of Nesprin1 are clearly associated with mispositioning of synaptic nuclei away from the NMJ, this mispositioning is not a good indicator of the clinical presentation.
By contrast, there is a clear clinical correlation between genetic alteration of Nesprin-4 and deafness. Indeed, a study by Horn et al. recently reported that patients affected by progressive high frequency hearing loss carry genetic alterations of the gene encoding nesprin4 [92], a phenotype that can be reproduced in Nesprin-4 KO mice. In agreement for a direct role of LINC complexes in hearing, Sun1KO mice also present with severe hearing loss. Within outer hair cells from both KO models, nuclei mislocalized on the apical side whereas wild-type outer hair cells maintain their nuclei at significantly more basal positions. This mispositioning is accompanied by a severe degeneration of outer hair cells that occurs in a basal to apical gradient across the cochlea. Disruption of LINC complexes upon expression of dominant-negative KASH proteins (under the control of a heat shock promoter) induced after the full differentiation of zebrafish photoreceptors also induce nuclear mislocalization, a phenotype that is also accompanied by severe photoreceptor degeneration. A challenging question regarding the consequences of LINC complex disruption in sensory neurons is the molecular mechanisms leading to their degeneration. Indeed, disruption of LINC complexes in cultured cells is accompanied by significant cytoskeletal and mechanical cellular defects. Hence, one can wonder whether either nuclear positioning per se or the alteration of cytoskeletal organization ensuing from LINC complex disruption underlies sensory neurons degeneration.
Role of the nucleus and LINC complexes in single cell migration
Since the nucleus is topologically enclosed within the intracellular space of migrating eukaryotic cells, nuclear movements and overall cell movements are highly correlated. But whether the interphase nucleus plays an active role in cell migration – and is not simply dragged along by the cell – is only beginning to be investigated. Recent discovery and characterization of LINC complexes suggests a much more tightly regulated functional connection between nuclear movements and positioning in the cell and cell migration than previously thought.
During planar cell migration in vitro, the nucleus is typically positioned near the myosin-rich contractile tail of the cell, while the microtubule-associated organizing center (MTOC) polarizes the cell and is positioned between the nucleus and the cell’s leading edge, where active actin filament assembly and turnover occur [93]. Mesenchymal migrating cells dynamically switch back and forth between persistent moves and periods of migratory arrest, generating trajectories that are well approximated by a so-called persistent random walk. A persistent random walk is completely defined by just two parameters: cell speed and persistence time, the time it takes for the cell to deviate from a straight line, straightforwardly computed from fits of the mean squared displacements of cells. A long persistence time signifies a directionally persistent migration of the cell. During such migratory patterns, the cell dynamically changes its shape, from a mostly elongated morphology during persistent moves to a mostly rounded morphology during transient arrest and re-polarization of the MTOC for the cell’s next persistent move in a direction uncorrelated to the previous persistent move.
Recent work has revealed the existence of a highly contractile acto-myosin filamentous structure, the perinuclear actin cap, which tightly wraps around the nucleus and regulates its shape [94]. The perinuclear actin cap is composed of thick and mostly parallel fibers that are rich in phosphorylated myosin II and F-actin crosslinker α-actinin and are dynamically anchored to the apical surface and lateral sides of the interphase nucleus and its lamina through LINC complexes [94,95] (Fig. 4A). Hence, nucleus-anchored actin-cap fibers are topologically different from conventional stress fibers, which lie entirely at the bottom of adherent cells and are anchored to the plasma membrane. In interphase cells, the actin cap is not permanently affixed to the nucleus; rather it forms and dissolves dynamically due to both rapid turnover dynamics of F-actin fibers in the cap and the cap’s sliding motion above the apical surface of the nucleus, which causes the actin-cap fibers to take on characteristics of basal stress fibers [95].
Figure 4. The architecture and function of the LINC-anchored actin cap in 3D cell migration.
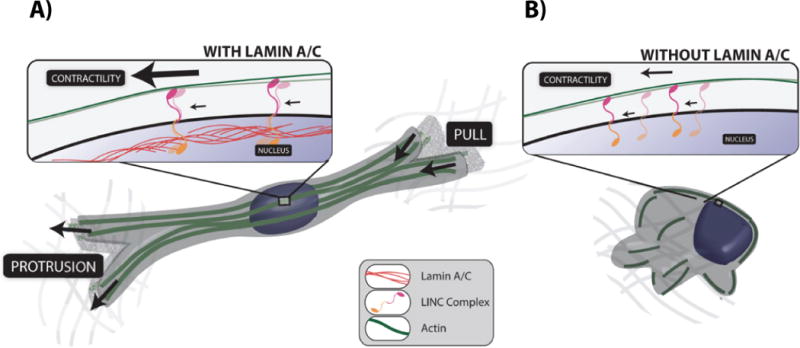
A. Cells with an intact actin cap, form thick pseudopodial protrusions that can pull on the surrounding matrix for net translocation. Actin cap fibers (green) provide mechanical support to the protrusion thanks to LINC-mediated anchorage to the nuclear lamina. B. Cells lacking an actin cap (caused by lamin A/C deficiency) cannot generate protrusions and, therefore, cannot translocate efficiently.
Cells forced to elongate on narrow adhesive (fibronectin- or collagen-coated) patterns flanked with non-adhesive PEG surfaces display an elongated nuclear morphology, while cells on round adhesive micropatterns display a round nucleus [94]. Accordingly, elongated cells show a prominent perinuclear actin cap, while rounded cells show no actin cap, suggesting that an important function of the actin cap is nuclear shaping and relating nuclear shape to cell shape. Cells on narrow patterns, which typically show an actin cap oriented along the long cell axis, are forced to undergo highly persistent migration, and cells on round patterns, which lack an actin cap, do not undergo net translocation. This global functional correlation between cell shape, nuclear shape, actin-cap status and mode of migration predict that cells migrating on flat (unpatterned) substrates, which undergo persistent migration, will dynamically show an actin cap during directionally persistent moves and no or disrupted actin cap during transient migratory arrest and re-polarization events [96,97]. These results also predict that disruption of the actin cap and LINC complexes, either through genetic depletion of LINC complex molecules or pharmacological inhibition of myosin activity or actin filament assembly, should affect cell persistence and speed. Cells harvested from mouse models of laminopathies typically show a disrupted or missing actin cap [94] (Fig. 4B). Therefore they should undergo reduced migration, which is indeed experimentally verified [98–100].
Actin cap fibers are terminated by focal adhesions that are significantly larger, more elongated, and longer-lived than conventional focal adhesions terminating stress fibers at the basal surface of the cell [95]. These actin-cap-associated focal adhesions are localized at the leading edge of migrating cells, while conventional focal adhesions are smaller and localized further away from the leading edge. The location and long lifetime of actin-cap-associated focal adhesions may maintain a productive lamellipodium in a given direction before retraction and cell repolarization. During random migration, upon spontaneous dissolution of the actin cap and of actin-cap-associated focal adhesions, the lamellipodium retracts, slowing down cell translocation. While the actin cap is present, the LINC interconnections between actin cap fibers on the nuclear surface and nuclear lamina would prevent nuclear rotation and only allows for nuclear translocation, inducing persistent migration [95,101]. When the actin cap (transiently) disappears, nuclear translocation stops, the brakes to nuclear rotation are released, and dynein-mediated nuclear rotation can occur. Accordingly, the MTOC can re-orient as it is physically connected to the NE and the cell repolarizes to prepare for the next persistent move of the cell in a new direction.
Fibroblasts and post-epidermal-mesenchymal transition cancer cells migrate within a mostly three-dimensional (3D) collagen I-rich matrix. In 3D cell migration, the interphase nucleus is typically smaller and more elongated than the same cells on 2D flat collagen-coated surfaces and located in the middle of the cell [102–105]. The 3D equivalent of the actin cap for cells inside 3D collagen matrices is constituted of acto-myosin fibers that are now isotropically located all around the nucleus and prolong the nuclear region into long and thick protrusions [103]. These pseudopodial protrusions mediate traction forces on the collagen fibers surrounding the cell, inducing net cell translocation. Cell migration in 3D matrices is much more persistent than in 2D migration presumably because the 3D actin cap is longer-lived compared to its 2D counterpart. Moreover, activated fibroblasts and cancer cells will locally digest the matrix through the expression of membrane-bound matrix metalloproteinases (i.e. MT1-MMP), forming in their wake thin open channels of cross-sectional size smaller than the nuclear size [96,97,102,106]. Disruption of LINC complexes significantly reduces the ability of cells to protrude and in turn reduces cell-induced matrix traction and therefore cell migration. Hence, the LINC-anchored contractile actin cap plays a central role in 3D cell migration by promoting the formation of protrusions and by actively compressing the nucleus [96].
Lateral confinement of cells by microfabricated channels of cross-sectional size smaller than nuclear size typically induces actin cap formation, which may help migration of these highly confined cells even for microchannel sizes that are multiple-fold smaller than the natural size of the nucleus [105,107]. How these in vitro systems mimic the in vivo context and whether actin cap formation is necessary for efficient cancer cell migration in the stromal matrix remains to be experimentally tested.
Concluding remarks
For the past fifteen years, our view of the NE has been radically transformed from a mere physical barrier between the nucleoplasm and the cytoplasm to a multifunctional compartment with “a life on its own”. In particular, the identification of LINC complexes and their roles as force transmission hubs involved in the physical communication between cytoplasmic and nucleoplasmic networks across the NE is currently fueling new research avenues. Whereas the “core” of LINC complexes is now well defined, we are starting to have an appreciation of the variability of their cytoplasmic and nucleoplasmic interfaces. For these reasons, appropriate tools need to be developed to address the variable composition of Nesprins in different tissues at different development stages (Which nesprin? Which isoform thereof? When?). By the same token, the putative variability of LINC complexes nucleoplasmic interface(s) needs to be further addressed. Finally, whereas current knockout animal models have been central in our understanding of how the LINC complex affects CNS development, conditional models of LINC complex disruption need to be developed in order to examine their physiological roles in vivo in a cell-autonomous manner and at specific developmental stages. Recent advances in methods allowing for the expression of exogenous proteins in either organotypic mammalian tissue slices or primary cell cultures combined to time-lapse video microscopy also provide very attractive experimental options [108,109]. Taken together, these approaches will most likely further fuel our expanding understanding of the role of the NE in normal and pathological biological processes. Given the central involvement of both LINC complexes and actin cap in cellular migration, examining new hypotheses about the relevance of these NE components in metastasis will be of particular interest.
Acknowledgments
The authors are supported by the National Institute of Health (R01GM084204 to D.W. and D.H., R01CA174388 and U54CA143868 to D.W., R01EY022632 to D.H.), and by NIH training grant T32EY013360, core grant P30EY002687 and Unrestricted Grants from Research to Prevent Blindness, Inc. awarded to the Department of Ophthalmology and Visual Sciences at Washington University School of Medicine.
Abbreviations
- CNS
central nervous system
- DKO
double knock-out
- ER
endoplasmic reticulum
- IKNM
interkinetic nuclear migration
- INM
inner nuclear membrane
- KO
knockout
- LINC
linkers of the nucleoskeleton to the cytoskeleton
- MTOC
microtubule organizing center
- NE
nuclear envelope
- NMJ
neuromuscular junction
- ONM
outer nuclear membrane
Contributor Information
David Razafsky, Email: Razafsky@vision.wustl.edu.
Denis Wirtz, Email: wirtz@jhu.edu.
Didier Hodzic, Email: hodzicd@vision.wustl.edu.
References
- 1.Schirmer EC, Florens L, Guan T, Yates JR, 3rd, Gerace L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 2003;301:1380–1382. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]
- 2.Stuurman N, Heins S, Aebi U. Nuclear lamins: their structure, assembly, and interactions. J Struct Biol. 1998;122:42–66. doi: 10.1006/jsbi.1998.3987. [DOI] [PubMed] [Google Scholar]
- 3.Hutchison CJ. Lamins: building blocks or regulators of gene expression? Nat Rev Mol Cell Biol. 2002;3:848–858. doi: 10.1038/nrm950. [DOI] [PubMed] [Google Scholar]
- 4.Burke B, Stewart CL. The nuclear lamins: flexibility in function. Nat Rev Mol Cell Biol. 2013;14:13–24. doi: 10.1038/nrm3488. [DOI] [PubMed] [Google Scholar]
- 5.Stewart C, Burke B. Teratocarcinoma stem cells and early mouse embryos contain only a single major lamin polypeptide closely resembling lamin B. Cell. 1987;51:383–392. doi: 10.1016/0092-8674(87)90634-9. [DOI] [PubMed] [Google Scholar]
- 6.Rober RA, Weber K, Osborn M. Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: a developmental study. Development. 1989;105:365–378. doi: 10.1242/dev.105.2.365. [DOI] [PubMed] [Google Scholar]
- 7.Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013;152:584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Stewart CL, Roux KJ, Burke B. Blurring the boundary: the nuclear envelope extends its reach. Science. 2007;318:1408–1412. doi: 10.1126/science.1142034. [DOI] [PubMed] [Google Scholar]
- 9.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, et al. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malone CJ, Fixsen WD, Horvitz HR, Han M. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development. 1999;126:3171–3181. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- 11.Hagan I, Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodzic DM, Yeater DB, Bengtsson L, Otto H, Stahl PD. Sun2 is a novel mammalian inner nuclear membrane protein. J Biol Chem. 2004;279:25805–25812. doi: 10.1074/jbc.M313157200. [DOI] [PubMed] [Google Scholar]
- 13.Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, et al. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q, Du X, Cai Z, Greene MI. Characterization of the structures involved in localization of the SUN proteins to the nuclear envelope and the centrosome. DNA Cell Biol. 2006;25:554–562. doi: 10.1089/dna.2006.25.554. [DOI] [PubMed] [Google Scholar]
- 15.Graumann K, Runions J, Evans DE. Characterization of SUN-domain proteins at the higher plant nuclear envelope. Plant J. 2010;61:134–144. doi: 10.1111/j.1365-313X.2009.04038.x. [DOI] [PubMed] [Google Scholar]
- 16.Razafsky D, Hodzic D. Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J Cell Biol. 2009;186:461–472. doi: 10.1083/jcb.200906068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sosa BA, Rothballer A, Kutay U, Schwartz TU. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell. 2012;149:1035–1047. doi: 10.1016/j.cell.2012.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Z, Du X, Cai Z, Song X, Zhang H, et al. Structure of Sad1-UNC84 homology (SUN) domain defines features of molecular bridge in nuclear envelope. J Biol Chem. 2012;287:5317–5326. doi: 10.1074/jbc.M111.304543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasan S, Guttinger S, Muhlhausser P, Anderegg F, Burgler S, et al. Nuclear envelope localization of human UNC84A does not require nuclear lamins. FEBS Lett. 2006;580:1263–1268. doi: 10.1016/j.febslet.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 20.Gob E, Meyer-Natus E, Benavente R, Alsheimer M. Expression of individual mammalian Sun1 isoforms depends on the cell type. Commun Integr Biol. 2011;4:440–442. doi: 10.4161/cib.4.4.15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haque F, Mazzeo D, Patel JT, Smallwood DT, Ellis JA, et al. Mammalian SUN protein interaction networks at the inner nuclear membrane and their role in laminopathy disease processes. J Biol Chem. 2009;285:3487–3498. doi: 10.1074/jbc.M109.071910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gob E, Schmitt J, Benavente R, Alsheimer M. Mammalian sperm head formation involves different polarization of two novel LINC complexes. PLoS One. 2010;5:e12072. doi: 10.1371/journal.pone.0012072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schumacher MA, Karamooz E, Zikova A, Trantirek L, Lukes J. Crystal structures of T. brucei MRP1/MRP2 guide-RNA binding complex reveal RNA matchmaking mechanism. Cell. 2006;126:701–711. doi: 10.1016/j.cell.2006.06.047. [DOI] [PubMed] [Google Scholar]
- 24.Apel ED, Lewis RM, Grady RM, Sanes JR. Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J Biol Chem. 2000;275:31986–31995. doi: 10.1074/jbc.M004775200. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Q, Ragnauth C, Greener MJ, Shanahan CM, Roberts RG. The nesprins are giant actin-binding proteins, orthologous to Drosophila melanogaster muscle protein MSP-300. Genomics. 2002;80:473–481. [PubMed] [Google Scholar]
- 26.Wilhelmsen K, Litjens SH, Kuikman I, Tshimbalanga N, Janssen H, et al. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J Cell Biol. 2005;171:799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, et al. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc Natl Acad Sci U S A. 2009;106:2194–2199. doi: 10.1073/pnas.0808602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morimoto A, Shibuya H, Zhu X, Kim J, Ishiguro K, et al. A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. J Cell Biol. 2012;198:165–172. doi: 10.1083/jcb.201204085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajgor D, Mellad JA, Autore F, Zhang Q, Shanahan CM. Multiple novel nesprin-1 and nesprin-2 variants act as versatile tissue-specific intracellular scaffolds. PLoS One. 2012;7:e40098. doi: 10.1371/journal.pone.0040098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cottrell JR, Borok E, Horvath TL, Nedivi E. CPG2: a brain- and synapse-specific protein that regulates the endocytosis of glutamate receptors. Neuron. 2004;44:677–690. doi: 10.1016/j.neuron.2004.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Randles KN, Lam le T, Sewry CA, Puckelwartz M, Furling D, et al. Nesprins, but not sun proteins, switch isoforms at the nuclear envelope during muscle development. Dev Dyn. 2010;239:998–1009. doi: 10.1002/dvdy.22229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhen YY, Libotte T, Munck M, Noegel AA, Korenbaum E. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J Cell Sci. 2002;115:3207–3222. doi: 10.1242/jcs.115.15.3207. [DOI] [PubMed] [Google Scholar]
- 33.Padmakumar VC, Abraham S, Braune S, Noegel AA, Tunggal B, et al. Enaptin, a giant actin-binding protein, is an element of the nuclear membrane and the actin cytoskeleton. Exp Cell Res. 2004;295:330–339. doi: 10.1016/j.yexcr.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 34.Meyerzon M, Fridolfsson HN, Ly N, McNally FJ, Starr DA. UNC-83 is a nuclear-specific cargo adaptor for kinesin-1 mediated nuclear migration. Development. 2009 doi: 10.1242/dev.038596. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fridolfsson HN, Ly N, Meyerzon M, Starr DA. UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Dev Biol. 338:237–250. doi: 10.1016/j.ydbio.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, et al. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGee MD, Rillo R, Anderson AS, Starr DA. UNC-83 IS a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol Biol Cell. 2006;17:1790–1801. doi: 10.1091/mbc.E05-09-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, et al. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J Cell Sci. 2005;118:3419–3430. doi: 10.1242/jcs.02471. [DOI] [PubMed] [Google Scholar]
- 39.Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res. 2008;314:1892–1905. doi: 10.1016/j.yexcr.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci U S A. 1997;94:849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, et al. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J Biol Chem. 2011;286:26743–26753. doi: 10.1074/jbc.M111.233700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rothballer A, Schwartz TU, Kutay U. LINCing complex functions at the nuclear envelope: What the molecular architecture of the LINC complex can reveal about its function. Nucleus. 2013;4:29–36. doi: 10.4161/nucl.23387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kracklauer MP, Link J, Alsheimer M. LINCing the nuclear envelope to gametogenesis. Curr Top Dev Biol. 2013;102:127–157. doi: 10.1016/B978-0-12-416024-8.00005-2. [DOI] [PubMed] [Google Scholar]
- 44.Starr DA, Fischer JA. KASH ‘n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. Bioessays. 2005;27:1136–1146. doi: 10.1002/bies.20312. [DOI] [PubMed] [Google Scholar]
- 45.Taverna E, Huttner WB. Neural progenitor nuclei IN motion. Neuron. 2010;67:906–914. doi: 10.1016/j.neuron.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 46.Kosodo Y. Interkinetic nuclear migration: beyond a hallmark of neurogenesis. Cell Mol Life Sci. 2012;69:2727–2738. doi: 10.1007/s00018-012-0952-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spear PC, Erickson CA. Interkinetic nuclear migration: a mysterious process in search of a function. Dev Growth Differ. 2012;54:306–316. doi: 10.1111/j.1440-169X.2012.01342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee HO, Norden C. Mechanisms controlling arrangements and movements of nuclei in pseudostratified epithelia. Trends Cell Biol. 2013;23:141–150. doi: 10.1016/j.tcb.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 49.Murciano A, Zamora J, Lopez-Sanchez J, Frade JM. Interkinetic nuclear movement may provide spatial clues to the regulation of neurogenesis. Mol Cell Neurosci. 2002;21:285–300. doi: 10.1006/mcne.2002.1174. [DOI] [PubMed] [Google Scholar]
- 50.Schenk J, Wilsch-Brauninger M, Calegari F, Huttner WB. Myosin II is required for interkinetic nuclear migration of neural progenitors. Proc Natl Acad Sci U S A. 2009;106:16487–16492. doi: 10.1073/pnas.0908928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baye LM, Link BA. Interkinetic nuclear migration and the selection of neurogenic cell divisions during vertebrate retinogenesis. J Neurosci. 2007;27:10143–10152. doi: 10.1523/JNEUROSCI.2754-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ueno M, Katayama K, Yamauchi H, Nakayama H, Doi K. Cell cycle progression is required for nuclear migration of neural progenitor cells. Brain Res. 2006;1088:57–67. doi: 10.1016/j.brainres.2006.03.042. [DOI] [PubMed] [Google Scholar]
- 53.Zolessi FR, Poggi L, Wilkinson CJ, Chien CB, Harris WA. Polarization and orientation of retinal ganglion cells in vivo. Neural Dev. 2006;1:2. doi: 10.1186/1749-8104-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spear PC, Erickson CA. Apical movement during interkinetic nuclear migration is a two-step process. Dev Biol. 2012;370:33–41. doi: 10.1016/j.ydbio.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu J, Lei K, Zhou M, Craft CM, Xu G, et al. KASH protein Syne-2/Nesprin-2 and SUN proteins SUN1/2 mediate nuclear migration during mammalian retinal development. Hum Mol Genet. 2011;20:1061–1073. doi: 10.1093/hmg/ddq549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Del Bene F, Wehman AM, Link BA, Baier H. Regulation of neurogenesis by interkinetic nuclear migration through an apical-basal notch gradient. Cell. 2008;134:1055–1065. doi: 10.1016/j.cell.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghashghaei HT, Lai C, Anton ES. Neuronal migration in the adult brain: are we there yet? Nat Rev Neurosci. 2007;8:141–151. doi: 10.1038/nrn2074. [DOI] [PubMed] [Google Scholar]
- 58.Ayala R, Shu T, Tsai LH. Trekking across the brain: the journey of neuronal migration. Cell. 2007;128:29–43. doi: 10.1016/j.cell.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 59.Tsai LH, Gleeson JG. Nucleokinesis in neuronal migration. Neuron. 2005;46:383–388. doi: 10.1016/j.neuron.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 60.Hatten ME. New directions in neuronal migration. Science. 2002;297:1660–1663. doi: 10.1126/science.1074572. [DOI] [PubMed] [Google Scholar]
- 61.Solecki DJ, Trivedi N, Govek EE, Kerekes RA, Gleason SS, et al. Myosin II motors and F-actin dynamics drive the coordinated movement of the centrosome and soma during CNS glial-guided neuronal migration. Neuron. 2009;63:63–80. doi: 10.1016/j.neuron.2009.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kato M, Dobyns WB. Lissencephaly and the molecular basis of neuronal migration. Hum Mol Genet 12 Spec No. 2003;1:R89–96. doi: 10.1093/hmg/ddg086. [DOI] [PubMed] [Google Scholar]
- 63.Labelle-Dumais C, Dilworth DJ, Harrington EP, de Leau M, Lyons D, et al. COL4A1 mutations cause ocular dysgenesis, neuronal localization defects, and myopathy in mice and Walker-Warburg syndrome in humans. PLoS Genet. 7:e1002062. doi: 10.1371/journal.pgen.1002062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Godfrey C, Clement E, Mein R, Brockington M, Smith J, et al. Refining genotype phenotype correlations in muscular dystrophies with defective glycosylation of dystroglycan. Brain. 2007;130:2725–2735. doi: 10.1093/brain/awm212. [DOI] [PubMed] [Google Scholar]
- 65.Nabi NU, Mezer E, Blaser SI, Levin AA, Buncic JR. Ocular findings in lissencephaly. J AAPOS. 2003;7:178–184. doi: 10.1016/s1091-8531(02)42005-8. [DOI] [PubMed] [Google Scholar]
- 66.Dobyns WB, Pagon RA, Armstrong D, Curry CJ, Greenberg F, et al. Diagnostic criteria for Walker-Warburg syndrome. Am J Med Genet. 1989;32:195–210. doi: 10.1002/ajmg.1320320213. [DOI] [PubMed] [Google Scholar]
- 67.Cormand B, Pihko H, Bayes M, Valanne L, Santavuori P, et al. Clinical and genetic distinction between Walker-Warburg syndrome and muscle-eye-brain disease. Neurology. 2001;56:1059–1069. doi: 10.1212/wnl.56.8.1059. [DOI] [PubMed] [Google Scholar]
- 68.Levine RA, Gray DL, Gould N, Pergament E, Stillerman ML. Warburg syndrome. Ophthalmology. 1983;90:1600–1603. doi: 10.1016/s0161-6420(83)34345-1. [DOI] [PubMed] [Google Scholar]
- 69.Dobyns WB, Truwit CL. Lissencephaly and other malformations of cortical development: 1995 update. Neuropediatrics. 1995;26:132–147. doi: 10.1055/s-2007-979744. [DOI] [PubMed] [Google Scholar]
- 70.Santavuori P, Somer H, Sainio K, Rapola J, Kruus S, et al. Muscle-eye-brain disease (MEB) Brain Dev. 1989;11:147–153. doi: 10.1016/s0387-7604(89)80088-9. [DOI] [PubMed] [Google Scholar]
- 71.Gupta V, Kawahara G, Gundry SR, Chen AT, Lencer WI, et al. The zebrafish dag1 mutant: a novel genetic model for dystroglycanopathies. Hum Mol Genet. 2011;20:1712–1725. doi: 10.1093/hmg/ddr047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chan YM, Keramaris-Vrantsis E, Lidov HG, Norton JH, Zinchenko N, et al. Fukutin-related protein is essential for mouse muscle, brain and eye development and mutation recapitulates the wide clinical spectrums of dystroglycanopathies. Hum Mol Genet. 2010;19:3995–4006. doi: 10.1093/hmg/ddq314. [DOI] [PubMed] [Google Scholar]
- 73.Bellion A, Baudoin JP, Alvarez C, Bornens M, Metin C. Nucleokinesis in tangentially migrating neurons comprises two alternating phases: forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J Neurosci. 2005;25:5691–5699. doi: 10.1523/JNEUROSCI.1030-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schaar BT, McConnell SK. Cytoskeletal coordination during neuronal migration. Proc Natl Acad Sci U S A. 2005;102:13652–13657. doi: 10.1073/pnas.0506008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Umeshima H, Hirano T, Kengaku M. Microtubule-based nuclear movement occurs independently of centrosome positioning in migrating neurons. Proc Natl Acad Sci U S A. 2007;104:16182–16187. doi: 10.1073/pnas.0708047104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina. II. Autoradiographic analysis of cell generation using tritiated thymidine. J Comp Neurol. 1979;188:263–272. doi: 10.1002/cne.901880205. [DOI] [PubMed] [Google Scholar]
- 77.Rich KA, Zhan Y, Blanks JC. Migration and synaptogenesis of cone photoreceptors in the developing mouse retina. J Comp Neurol. 1997;388:47–63. [PubMed] [Google Scholar]
- 78.Razafsky D, Blecher N, Markov A, Stewart-Hutchinson PJ, Hodzic D. LINC complexes mediate the positioning of cone photoreceptor nuclei in mouse retina. PLoS One. 2012;7:e47180. doi: 10.1371/journal.pone.0047180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsujikawa M, Omori Y, Biyanwila J, Malicki J. Mechanism of positioning the cell nucleus in vertebrate photoreceptors. Proc Natl Acad Sci U S A. 2007;104:14819–14824. doi: 10.1073/pnas.0700178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fan SS, Ready DF. Glued participates in distinct microtubule-based activities in Drosophila eye development. Development. 1997;124:1497–1507. doi: 10.1242/dev.124.8.1497. [DOI] [PubMed] [Google Scholar]
- 81.Kracklauer MP, Banks SM, Xie X, Wu Y, Fischer JA. Drosophila klaroid encodes a SUN domain protein required for Klarsicht localization to the nuclear envelope and nuclear migration in the eye. Fly (Austin) 2007;1:75–85. doi: 10.4161/fly.4254. [DOI] [PubMed] [Google Scholar]
- 82.Patterson K, Molofsky AB, Robinson C, Acosta S, Cater C, et al. The functions of Klarsicht and nuclear lamin in developmentally regulated nuclear migrations of photoreceptor cells in the Drosophila eye. Mol Biol Cell. 2004;15:600–610. doi: 10.1091/mbc.E03-06-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mosley-Bishop KL, Li Q, Patterson L, Fischer JA. Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the Drosophila eye. Curr Biol. 1999;9:1211–1220. doi: 10.1016/s0960-9822(99)80501-6. [DOI] [PubMed] [Google Scholar]
- 84.Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Coffinier C, Jung HJ, Nobumori C, Chang S, Tu Y, et al. Deficiencies in lamin B1 and lamin B2 cause neurodevelopmental defects and distinct nuclear shape abnormalities in neurons. Mol Biol Cell. 2011;22:4683–4693. doi: 10.1091/mbc.E11-06-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Grady RM, Starr DA, Ackerman GL, Sanes JR, Han M. Syne proteins anchor muscle nuclei at the neuromuscular junction. Proc Natl Acad Sci U S A. 2005;102:4359–4364. doi: 10.1073/pnas.0500711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang X, Xu R, Zhu B, Yang X, Ding X, et al. Syne-1 and Syne-2 play crucial roles in myonuclear anchorage and motor neuron innervation. Development. 2007;134:901–908. doi: 10.1242/dev.02783. [DOI] [PubMed] [Google Scholar]
- 88.Zhang J, Felder A, Liu Y, Guo LT, Lange S, et al. Nesprin 1 is critical for nuclear positioning and anchorage. Hum Mol Genet. 2009;19:329–341. doi: 10.1093/hmg/ddp499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Puckelwartz MJ, Kessler E, Zhang Y, Hodzic D, Randles KN, et al. Disruption of nesprin-1 produces an Emery Dreifuss muscular dystrophy-like phenotype in mice. Hum Mol Genet. 2009;18:607–620. doi: 10.1093/hmg/ddn386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lei K, Zhang X, Ding X, Guo X, Chen M, et al. SUN1 and SUN2 play critical but partially redundant roles in anchoring nuclei in skeletal muscle cells in mice. Proc Natl Acad Sci U S A. 2009;106:10207–10212. doi: 10.1073/pnas.0812037106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gros-Louis F, Dupre N, Dion P, Fox MA, Laurent S, et al. Mutations in SYNE1 lead to a newly discovered form of autosomal recessive cerebellar ataxia. Nat Genet. 2007;39:80–85. doi: 10.1038/ng1927. [DOI] [PubMed] [Google Scholar]
- 92.Horn HF, Brownstein Z, Lenz DR, Shivatzki S, Dror AA, et al. The LINC complex is essential for hearing. J Clin Invest. 2013 doi: 10.1172/JCI66911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hale CM, Chen WC, Khatau SB, Daniels BR, Lee JS, et al. SMRT analysis of MTOC and nuclear positioning reveals the role of EB1 and LIC1 in single-cell polarization. J Cell Sci. 2011;124:4267–4285. doi: 10.1242/jcs.091231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, et al. A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci U S A. 2009;106:19017–19022. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim DH, Khatau SB, Feng Y, Walcott S, Sun SX, et al. Actin cap associated focal adhesions and their distinct role in cellular mechanosensing. Sci Rep. 2012;2:555. doi: 10.1038/srep00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Khatau SB, Bloom RJ, Bajpai S, Razafsky D, Zang S, et al. The distinct roles of the nucleus and nucleus-cytoskeleton connections in three-dimensional cell migration. Sci Rep. 2012;2:488. doi: 10.1038/srep00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fraley SI, Feng Y, Giri A, Longmore GD, Wirtz D. Dimensional and temporal controls of three-dimensional cell migration by zyxin and binding partners. Nat Commun. 2012;3:719. doi: 10.1038/ncomms1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hale CM, Shrestha AL, Khatau SB, Stewart-Hutchinson PJ, Hernandez L, et al. Dysfunctional connections between the nucleus and the actin and microtubule networks in laminopathic models. Biophys J. 2008;95:5462–5475. doi: 10.1529/biophysj.108.139428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee JS, Hale CM, Panorchan P, Khatau SB, George JP, et al. Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization, and migration. Biophys J. 2007;93:2542–2552. doi: 10.1529/biophysj.106.102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Khatau SB, Kim DH, Hale CM, Bloom RJ, Wirtz D. The perinuclear actin cap in health and disease. Nucleus. 2010;1:337–342. doi: 10.4161/nucl.1.4.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee JS, Chang MI, Tseng Y, Wirtz D. Cdc42 mediates nucleus movement and MTOC polarization in Swiss 3T3 fibroblasts under mechanical shear stress. Mol Biol Cell. 2005;16:871–880. doi: 10.1091/mbc.E03-12-0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fraley SI, Feng Y, Krishnamurthy R, Kim DH, Celedon A, et al. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat Cell Biol. 2010;12:598–604. doi: 10.1038/ncb2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Khatau SB, Kusuma S, Hanjaya-Putra D, Mali P, Cheng L, et al. The differential formation of the LINC-mediated perinuclear actin cap in pluripotent and somatic cells. PLoS One. 2012;7:e36689. doi: 10.1371/journal.pone.0036689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wirtz D, Konstantopoulos K, Searson PC. The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nat Rev Cancer. 2011;11:512–522. doi: 10.1038/nrc3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Konstantopoulos K, Wu PH, Wirtz D. Dimensional control of cancer cell migration. Biophys J. 2013;104:279–280. doi: 10.1016/j.bpj.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fraley S, Feng Y, Wirtz D, Longmore G. Reply: reducing background fluorescence reveals adhesions in 3D matrices. Nature Cell Biology. 2011;13:5–U254. doi: 10.1038/ncb0111-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Balzer EM, Tong Z, Paul CD, Hung WC, Stroka KM, et al. Physical confinement alters tumor cell adhesion and migration phenotypes. FASEB J. 2012;26:4045–4056. doi: 10.1096/fj.12-211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Karra D, Dahm R. Transfection techniques for neuronal cells. J Neurosci. 2010;30:6171–6177. doi: 10.1523/JNEUROSCI.0183-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Taverna E, Haffner C, Pepperkok R, Huttner WB. A new approach to manipulate the fate of single neural stem cells in tissue. Nat Neurosci. 2012;15:329–337. doi: 10.1038/nn.3008. [DOI] [PubMed] [Google Scholar]


