Genetic and biochemical data reveal that the plastid RNA binding protein RHON1 terminates rbcL transcription, using an ATP-driven mechanism similar to that of Rho factor of bacteria.
Abstract
Although transcription termination is essential to generate functional RNAs, its underlying molecular mechanisms are still poorly understood in plastids of vascular plants. Here, we show that the RNA binding protein RHON1 participates in transcriptional termination of rbcL (encoding large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase) in Arabidopsis thaliana. Inactivation of RHON1 leads to enhanced rbcL read-through transcription and to aberrant accD (encoding β-subunit of the acetyl-CoA carboxylase) transcriptional initiation, which may result from inefficient transcription termination of rbcL. RHON1 can bind to the mRNA as well as to single-stranded DNA of rbcL, displays an RNA-dependent ATPase activity, and terminates transcription of rbcL in vitro. These results suggest that RHON1 terminates rbcL transcription using an ATP-driven mechanism similar to that of Rho of Escherichia coli. This RHON1-dependent transcription termination occurs in Arabidopsis but not in rice (Oryza sativa) and appears to reflect a fundamental difference between plastomes of dicotyledonous and monocotyledonous plants. Our results point to the importance and significance of plastid transcription termination and provide insights into its machinery in an evolutionary context.
INTRODUCTION
Transcription termination, which involves arrest of RNA biosynthesis, release of the transcript, and dissociation of the RNA polymerase from the DNA template, is essential for proper gene expression. It helps avoid interference with transcription of downstream genes. Moreover, it maintains a pool of RNA polymerases available for reinitiation or transcription, prevents formation of antisense RNAs that can interfere with normal pre-RNA production, and thereby avoids aberrant gene expression (Greger and Proudfoot, 1998; Peters et al., 2012). Termination mechanisms vary considerably between different organisms, ranging from relatively simple to exceptionally complex processes. There are at least two different ways to terminate transcription in Escherichia coli and other prokaryotes (Richardson, 2003; Peters et al., 2011). Intrinsic termination requires mainly elements located on the mRNA, whereas Rho-dependent termination relies on both mRNA elements and trans-acting factors (Richardson, 2003; Peters et al., 2011). In eukaryotes, two different pathways for transcriptional termination by RNA polymerase II have been proposed: One uses the Nrd1 complex, whereas the other uses 3′ cleavage and polyadenylation factors together with Rat1 exonuclease (Rondon et al., 2008; Richard and Manley, 2009). By contrast, less is known about the molecular mechanisms that mediate transcription termination in organelles.
Plastids of vascular plants originated from an ancestral cyanobacterial endosymbiont (Timmis et al., 2004). Regulation of plastid gene expression is distinct from that of eubacteria as well as from that in the eukaryotic cytosol and reflects a unique chimeric system assembled from multiple components whose origins can be traced to endocytobiosis (Stern et al., 2010; Barkan, 2011). Plastid transcription is performed by a nucleus-encoded phage-type RNA polymerase (NEP) in addition to the cyanobacterium-derived, plastid-encoded RNA polymerase (PEP) (Barkan, 2011; Liere et al., 2011). However, how transcription termination occurs in plastids remains obscure. Most plastid genes contain inverted repeat sequences at their 3′ ends that are able to form stem-loop structures (Sugita and Sugiura, 1984; Kirsch et al., 1986). Such structures were initially assumed to act as transcription terminators, in a similar way as Rho-independent terminators (Sugita and Sugiura, 1984; Kirsch et al., 1986). However, both in vitro and in vivo assays have shown that in plastids, 3′ inverted repeat sequences function as mRNA processing and stabilizing elements rather than as efficient terminators (Stern and Gruissem, 1987; Monde et al., 2000). Nevertheless, both PEP and NEP polymerases terminate in vitro at strong intrinsic bacterial terminators containing GC-rich RNA hairpins followed by several contiguous uridines (Chen and Orozco, 1988; Jeng et al., 1990; Lerbs-Mache, 1993; Kühn et al., 2007). Therefore, it seems likely that RNA polymerases of plastids recognize bacterial terminator sequences with specific features under in vivo conditions. However, no factors involved in plastid transcription termination have been identified and characterized in vascular plants.
Proper expression of plastid genes relies on coordinated interplay between cis-acting elements at the plastid DNA/RNA level and trans-acting factors, most of which are nuclear encoded and specifically bind to their cis-acting targets. Comparative analysis of the plastid genomic evolution of vascular plants has indicated that vascular plants underwent frequent plastome rearrangements, such as large inversions, gene and intron gains and losses, expansion, contraction, and loss of the inverted repeats that vary between plant species (Wicke et al., 2011). The intergenic regions of plastid genomes containing numerous cis-elements for plastid gene expression evolved especially rapidly (Wicke et al., 2011). In parallel to the intensive plastid genome variation, some novel species-specific mechanisms have been acquired for plastid gene expression. However, how these processes were established during evolution still remains elusive.
Here, we studied the mechanism of transcription termination of plastid genes mediated by the plastid RHON1 protein. RHON1 was originally identified in a screen for interaction partners of the endonuclease RNase E (RNE) and was proposed to be involved in RNE-mediated plastid RNA processing (Stoppel et al., 2012). We show that RHON1 is also involved in the transcription termination of rbcL, encoding the large subunit of ribulose bisphosphate carboxylase oxygenase. Like Rho factors in E. coli, the catalytic activity of RHON1is based on the use of an ATP-driven mechanism. In addition, we found that RHON1-dependent transcription termination occurs in Arabidopsis thaliana but not in rice (Oryza sativa), which appears to be related to a fundamental difference in the rbcL-accD intergenic region of eudicotyledonous and monocotyledonous plants.
RESULTS
Mutation of RHON1 Affects Transcriptional Termination of rbcL
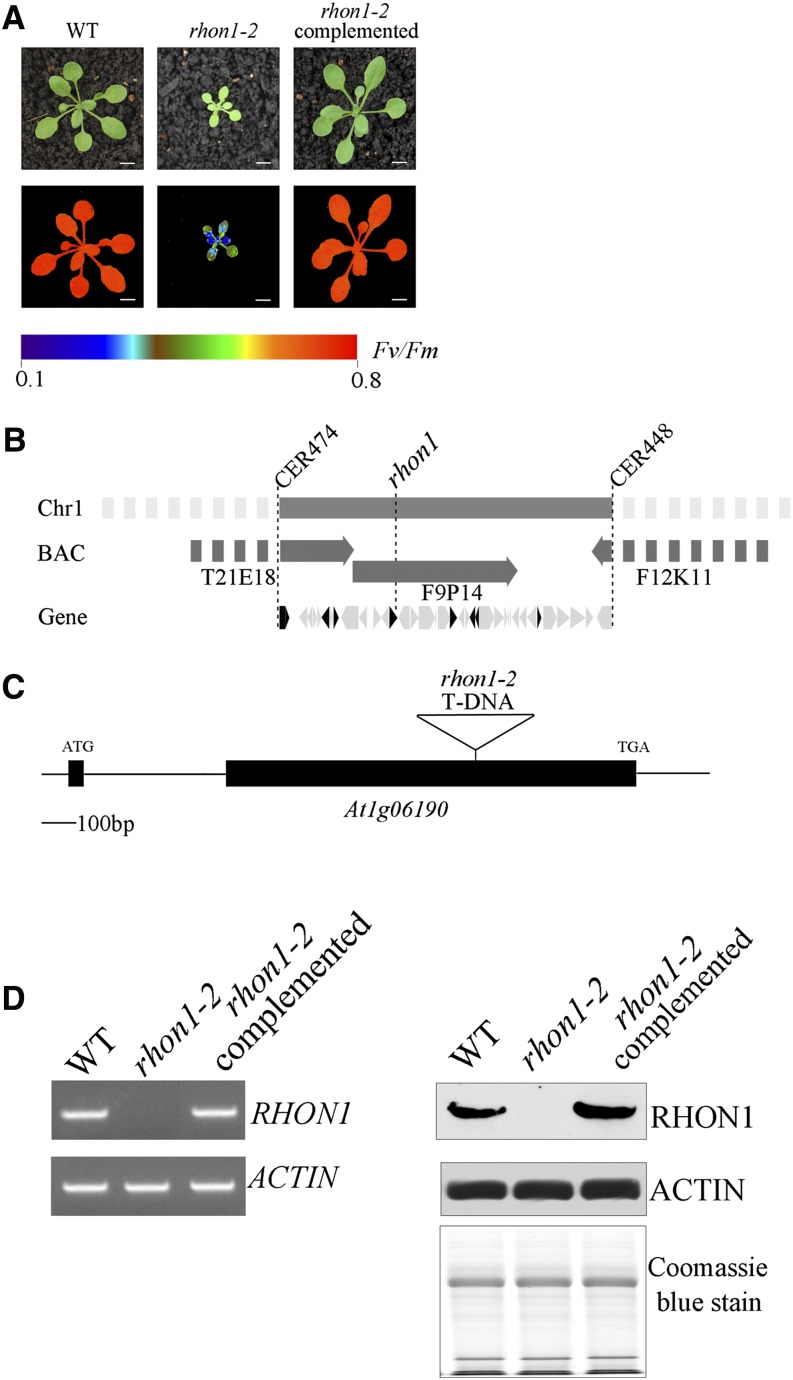
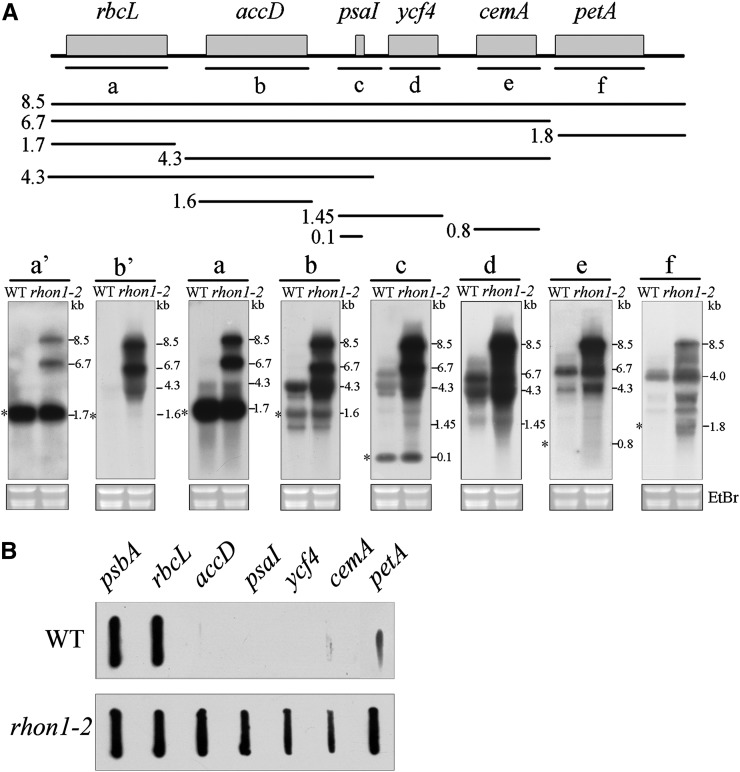
A mutant with a high chlorophyll fluorescence phenotype was isolated from the Scheible and Somerville T-DNA Arabidopsis lines (Figure 1A). The mutation was mapped to a 100-kb region between markers CER474 and CER448 on BAC F9P14 of chromosome 1 (Figure 1B). Sequencing of the 33 genes in the interval revealed a T-DNA insertion within the second exon of At1g06190 (Figure 1C). In a previous study, the AT1G06190 protein was given the name RHON1 because its C-terminal domain is similar to the N-terminal part of the bacterial transcription terminator Rho (Stoppel et al., 2012). Accordingly, we designated our high chlorophyll fluorescence mutant as rhon1-2. RT-PCR and immunoblot analysis showed expression of RHON1 in wild-type plants, but not in rhon1-2 (Figure 1D). Expression of full-length RHON1 cDNA under the control of the constitutive 35S promoter in rhon1-2 mutants fully restored the wild-type phenotype (Figure 1A). This functional complementation indicates that the RHON1 (At1g06190) gene is indeed responsible for the rhon1-2 phenotype. To explore the possible function of RHON1 in chloroplast gene expression, we examined the plastid gene transcription profile in the rhon1-2 mutant by microarray hybridization. This analysis revealed that the abundance of five plastid transcripts (accD, psaI, ycf4, cemA, and petA) was increased ∼20- to 40-fold in rhon1-2 (Supplemental Figure 1). Two NEP promoters (PaccD-172, 252) have been found to drive transcription of the Arabidopsis accD gene (promoters are specified by the gene name and the position of the initiation nucleotide with respect to the start of the coding sequence) (Swiatecka-Hagenbruch et al., 2007). However, a large polycistronic precursor transcript spanning the rbcL, accD, psaI, ycf4, cemA, and petA genes was also reported (Walter et al., 2010; Stoppel et al., 2012). This large polycistronic precursor is barely detectable in wild-type plants. This raised the question whether the dramatically increased levels of these five transcripts in rhon1-2 were associated with transcriptional and/or posttranscriptional processes related to the rbcL-accD-psaI-ycf4-cemA-petA operon. To analyze this operon in detail, RNA gel blot analysis using probes specific for the individual mature RNAs was performed. As shown in Figure 2A, the levels of unprocessed precursors (8.5-, 6.7-, 4.3-, and 1.45-kb fragments) were increased, as were those of the mature RNAs except for mature rbcL, which was not significantly changed in rhon1-2.
Figure 1.
Phenotype and Gene Cloning of rhon1-2.
(A) Photographs and chlorophyll fluorescence images of rhon1-2 mutants and wild-type plants grown for 3 weeks in a growth chamber. Fluorescence is shown using the pseudocolor index indicated at the bottom.
(B) Map-based cloning places RHON1 on chromosome 1 between the markers CER474 and CER448. The interval contains 33 genes (illustrated as arrows), of which eight gene products are predicted to be localized in plastids (illustrated as solid arrows).
(C) Schematic illustration of the exon-intron structure of RHON1 with the solid boxes representing exons. The T-DNA insertion sites are indicated with triangles.
(D) RT-PCR (left) and immunoblot analysis (right) of RHON1 expression in wild-type, rhon1-2 mutants, and complemented plants.
Figure 2.
Plastid Gene Expression of the rhon1-2 Mutants.
(A) RNA gel blot analysis of rbcL polycistronic transcript. A diagram of the rbcL polycistronic operon and the locations of the probes (a to f) are shown at the top. Blots with reduced exposure time to x-ray film for probes a and b are also included (indicated as a’ and b’). The mature forms of RNA are indicated with asterisks to the left of each blot.
(B) Run-on transcription assay of plastid genes in rhon1-2 and wild-type plants. DNA probe (200 ng) was used for hybridization with labeled transcripts. Another run-on experiment with 100 ng DNA probe was also performed and similar results were observed (Supplemental Figure 2).
Levels of both the precursor and mature forms of accD, psaI, ycf4, cemA, and petA mRNAs were enhanced, suggesting increased transcription of these genes and/or increased stability of the mRNAs in rhon1-2. A run-on assay showed that the transcription rates of accD, psaI, ycf4, cemA, and petA were increased in rhon1-2 plants, whereas the control (psbA) did not show obvious changes (Figure 2B; Supplemental Figure 2). This increase observed for these five gene transcripts could theoretically be due to augmented transcription of the large polycistronic rbcL transcript in rhon1-2 However, our results showed that the transcription rate of rbcL itself was not increased in rhon1-2, indicating that the transcription of the rbcL-accD-psaI-ycf4-cemA-petA operon as whole is not enhanced. We therefore conclude that transcription terminates efficiently after rbcL in wild-type plants but not in rhon1-2, which leads to the increased transcription of downstream genes of rbcL. The unchanged abundance of monocistronic rbcL in rhon1 might be due to endonucleolytic cleavage of the extended polycistronic RNAs downstream of rbcL, followed by 3′ exonuclease trimming to the stabilizing 3′ hairpins. Nevertheless, the possibility of compensation by increased mRNA stability as suggested by Shiina et al. (1998) for the unaltered level of monocistronic rbcL cannot be completely excluded.
Since accD is transcribed by NEP, we reasoned that its increased levels in rhon1-2 might be a response to defects in PEP activity, as plastid transcripts generated by NEP are known to be upregulated in response to impaired PEP activity (Pfalz et al., 2006). However, RNA gel blot analysis showed that the levels of mRNAs transcribed by PEP in rhon1-2 were comparable to those in wild-type plants (Supplemental Figure 3A). In addition, PEP abundance of rhon1-2 was not changed as demonstrated by an immunoblotting assay with RpoB antiserum (Supplemental Figure 3B). This result indicates that PEP activity is not affected in rhon1-2.
Aberrant Transcription Initiation within the rbcL-accD Intergenic Region in rhon1-2
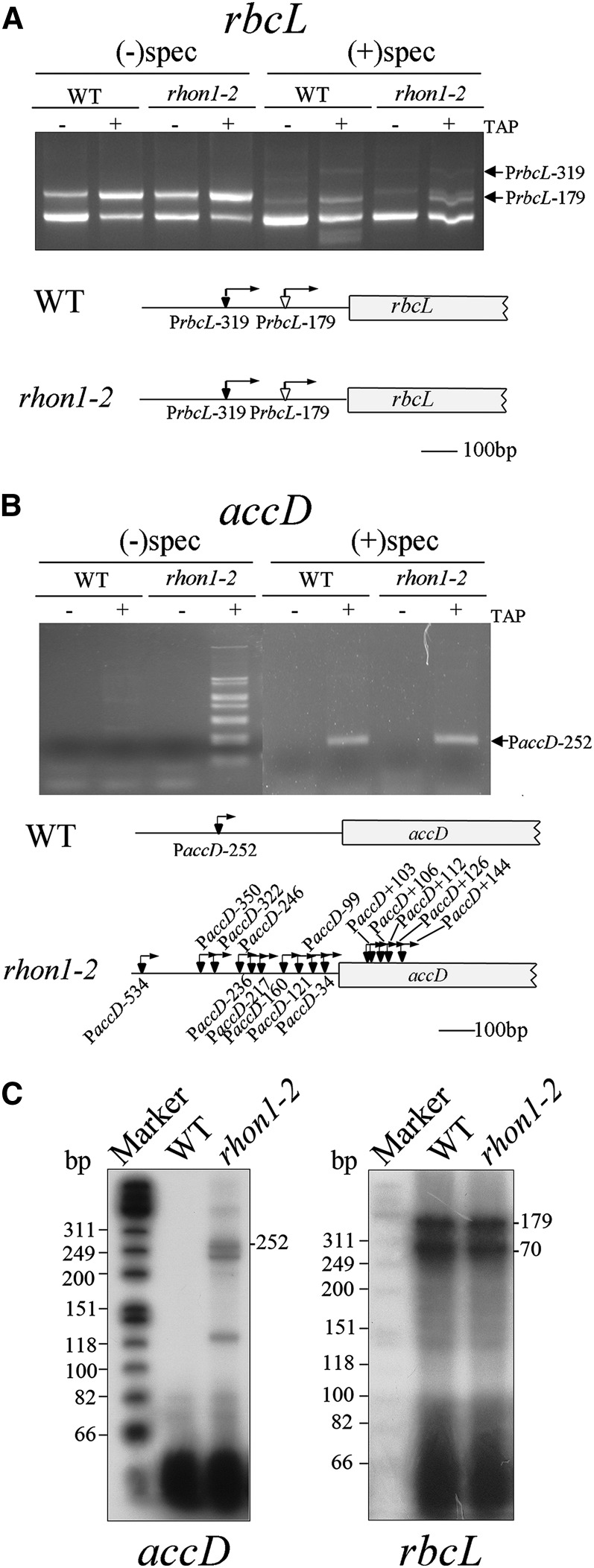
It was reported that the overaccumulation of some plastid polycistronic transcripts is associated with a change of promoter usage (Haley and Bogorad, 1990; Sexton et al., 1990; Schweer et al., 2006). Overaccumulation of polycistronic rbcL transcripts could be due to the activation of an unknown promoter(s) in rhon1-2. To address this possibility, we compared promoter usage in wild-type and rhon1-2 plants by mapping the transcription initiation site(s) using 5′-rapid amplification of cDNA ends (RACE) analysis combined with enzymatic treatment of RNA with tobacco acid pyrophosphatase (TAP). This approach not only can identify primary and secondary 5′-ends of plastid transcripts but also allows discrimination between NEP and PEP promoters by comparing the 5′-ends of transcripts from spectinomycin-treated seedlings with those of untreated seedlings (Swiatecka-Hagenbruch et al., 2007).
Previous work has identified a PEP promoter for rbcL in Arabidopsis, PrbcL-179, which is responsible for the generation of monocistronic rbcL (Hanaoka et al., 2003). Our results revealed two transcription initiation sites for the rbcL gene. Initiation at PrbcL-179 was detected at high levels in green seedlings but at low levels in seedlings treated with spectinomycin (Figure 3A). This suggests that PrbcL-179 is PEP dependent, consistent with the earlier report (Hanaoka et al., 2003). Meanwhile, another transcript initiation site, PrbcL-319, was detected only in spectinomycin-treated seedlings and not in green seedlings (Figure 3A), suggesting that this transcription initiation site is NEP dependent. Recently, a rbcL NEP promoter was identified in barley (Hordeum vulgare) as well (Zhelyazkova et al., 2012). The usage of the PrbcL-179 and PrbcL-319 promoters did not differ between the wild type and rhon1-2, suggesting that the accumulation of rbcL polycistronic transcript in rhon1-2 is not related to rbcL promoter usage. Sequencing of 5′-3′-end ligation products of the 8.5-kb rbcL polycistronic transcript obtained by circularized RNA (CR) RT-PCR indicated that its 5′ end corresponds to the rbcL PEP transcription initiation site (Supplemental Figure 4), which is compatible with the assumption that the rbcL polycistronic mRNA could be the by-product of inefficient transcriptional termination of rbcL. In addition, no difference in 3′- or 5′-ends of rbcL monocistronic transcript between wild-type and mutant plants was observed (Supplemental Figure 4).
Figure 3.
The Transcription Initiation Sites of Plastid Genes in Wild-Type and rhon1-2 Plants.
(A) Identification of the transcription initiation site of rbcL by 5′-RACE. Total RNA from green (without spectinomycin treatment) and white (with spectinomycin treatment) leaf tissue was ligated to an RNA linker after TAP treatment and, along with untreated RNA as a control, was subjected to RT-PCR with specific primers. The transcription initiation sites identified by sequencing of cloned RT-PCR products are underlined and marked by arrows in the diagram of the 5′ region of rbcL. The expression of the smallest PCR product in TAP-treated leaves was lower than that in leaves without TAP treatment. Therefore this product most likely corresponds to a processing product rather than to a primary transcript (Hanaoka et al., 2003; also refer to the −70 band of [C]).
(B) Identification of the transcription initiation sites of accD. The procedure was similar to that used for rbcL.
(C) Primer extension analysis of rbcL and accD from wild-type and rhon1-2 plants. The positions of bands corresponding to transcripts with distinct 5′-ends are indicated at the right.
Analysis of accD transcripts in wild-type plants revealed one PCR fragment that was amplified only from TAP-treated RNA isolated from spectinomycin-treated seedlings (Figure 3B). Sequence analysis showed that it corresponds to one of the two reported NEP-dependent transcript initiation sites, PaccD-252 (Swiatecka-Hagenbruch et al., 2007). The other transcript initiation site, PaccD-172, was not detected in wild-type plants under our conditions and was probably not active at this developmental stage, as described by Courtois et al. (2007). However, the amplified products from TAP-treated RNA isolated from rhon1-2 green seedlings displayed a ladder-like pattern, which was markedly different from that of wild-type plants (Figure 3B). This ladder-like pattern of accD transcript initiation sites in rhon1-2 was further confirmed by primer extension assays (Figure 3C). Extensive sequencing of those PCR products indicated that there were multiple transcript initiation sites of accD scattered all over the rbcL-accD intergenic region. In addition, some transcript initiation sites were even mapped within the 5′-end of the accD coding region (Figure 3B). To our surprise, these transcript initiation sites disappeared when rhon1-2 seedlings were treated with spectinomycin: As shown in Figure 3B, the rhon1-2 and wild-type plant RNAs showed similar banding patterns in spectinomycin-treated plants. Our results clearly show that absence of RHON1 not only increases transcriptional read-through of the rbcL terminator but also leads to aberrant transcription initiation in the rbcL-accD intergenic region.
In agreement with aberrant transcription initiation, the mature accD transcripts of rhon1-2 plants showed differences in their 5′-ends but had similar 3′ ends when detailed end-mapping of accD transcripts by (CR)RT-PCR was performed (Supplemental Figure 4).
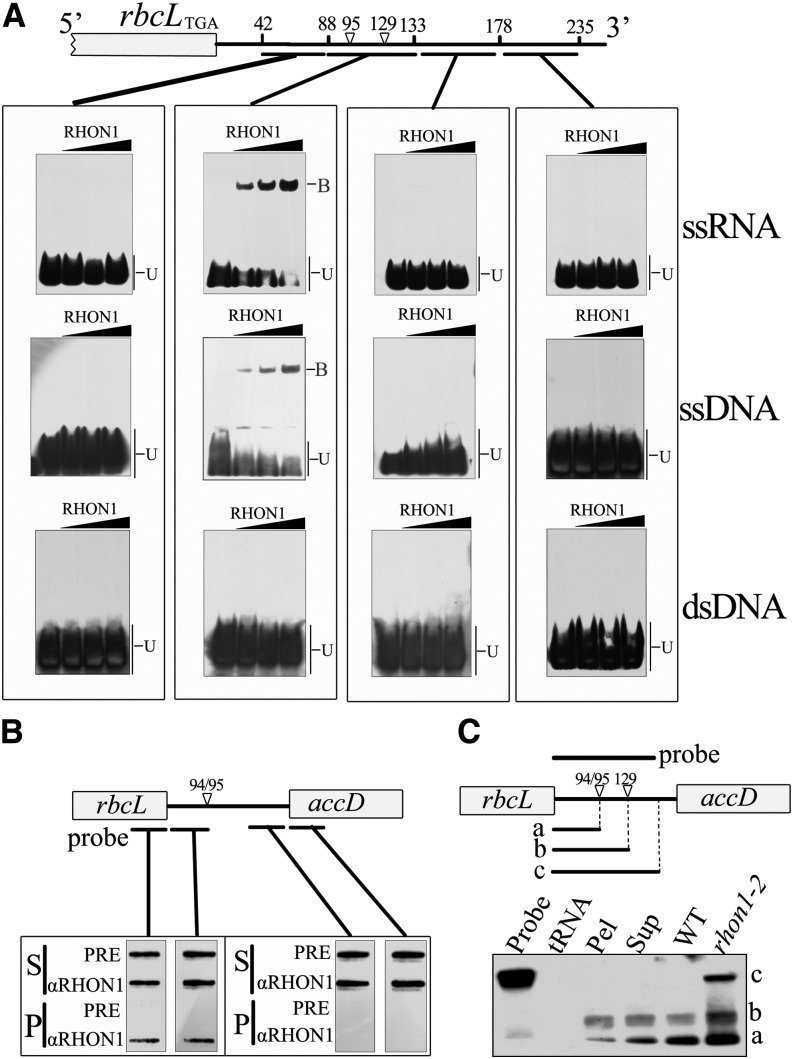
RHON1 Binds to the 3′-Untranslated Region of rbcL
The overaccumulation of rbcL-accD polycistronic transcript in rhon1-2 suggested that RHON1 could be involved in the regulation of rbcL transcription termination, acting as a trans-factor. If that were the case, it seemed likely that RHON1 would interact with the mRNA of rbcL. The binding activity of the RHON1 protein to rbcL mRNA was investigated by electrophoretic mobility shift assays (EMSAs). The C-terminal fragment of 100 amino acids (301 to 401 amino acids) comprising the potential Rho-N domain was expressed for EMSA analysis. Indeed, this fragment was able to bind to the 3′-untranslated region (UTR) of rbcL (Figure 4A). Our results place the RHON1 binding site between 88 and 133 nucleotides downstream of the rbcL termination codon (Figure 4A). This RHON1 binding site covers the region between the two 3′ ends of monocistronic rbcL assayed by (CR)RT-PCR (−94/95 and −129 with respect to the termination codon of rbcL). RHON1 protein was also found to bind to the corresponding single-stranded DNA of its RNA target (Figure 4A), which is similar to the nucleotide binding activity of Rho factors (Galluppi and Richardson, 1980; Skordalakes and Berger, 2006).
Figure 4.
Binding of RHON1 to the rbcL 3′-UTR.
(A) EMSA assay. At the top is a diagram showing the positions of the oligonucleotide probes (underlined). Numbers represent the positions of the nucleotide with respect to the termination codon of rbcL. The substrates, single-stranded RNA (ssRNA), single-stranded DNA (ssDNA), and double-stranded DNA (dsDNA) were mixed with increasing concentrations of RHON1 (125, 250, and 500 nM). The positions of protein-bound DNA/RNA (B) and unbound DNA/RNA (U) are indicated.
(B) Coimmunoprecipitation of RHON1 and rbcL 3′-UTR. The RNA from each immunoprecipitation (P) and supernatant (S) was applied to a nylon membrane with a slot-blot manifold and hybridized with probes as indicated. Immunoprecipitation with preimmune serum (PRE) was used as a negative control.
(C) RNase protection analysis of the 3′-end of the rbcL in the immunoprecipitation pellet (Pel) and supernatant (Sup) samples. The probe is indicated above the diagram of the rbcL-accD intergenic region. The positions of the protected bands, corresponding to rbcL transcripts with distinct 3′-ends, are indicated.
To further test the RHON1-RNA interaction in vivo, RNAs that coimmunoprecipitated with the anti-RHON1 antibody were analyzed by slot-blot hybridization using probes corresponding to the different regions of the rbcL-accD intergenic spaces (Figure 4B). The coding region and 3′-UTR of rbcL mRNA were detected in substantial amounts in the precipitate, whereas the coding region and 5′-UTR of accD were not detected (Figure 4B). This analysis suggests that RHON1 may bind to the monocistronic rbcL mRNA but not to the rbcL-accD polycistronic precursor or the monocistronic accD mRNA. Subsequently, a ribonuclease-protection assay was used to map the 3′-ends of the RNAs that coimmunoprecipitated with RHON1. Our results showed that the RHON1-coimmunoprecipitated RNA was enriched in monocistronic rbcL rather than polycistronic rbcL precursor (Figure 4C). In addition, the ratio of rbcL mRNA ending at position 129 compared with that ending at 94/95 was increased in the coimmunoprecipitated RNAs.
RHON1 Has Transcription Termination Catalytic Activity
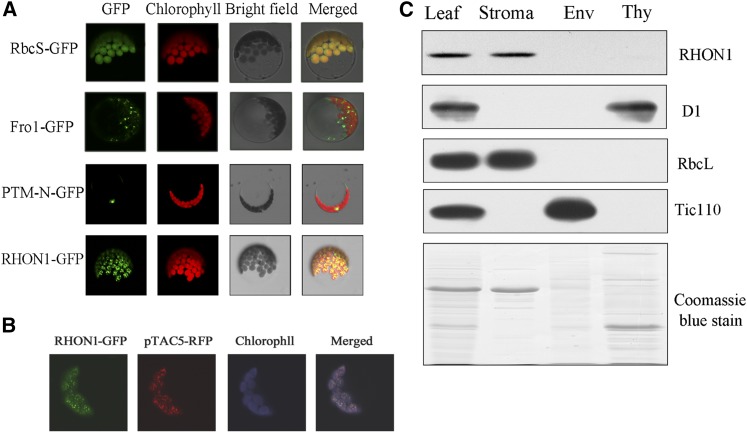
Transient expression of RHON1-green fluorescent protein (GFP) fusions in protoplasts of Arabidopsis gave rise to fluorescence signal exclusively in plastids, suggesting that RHON1 is targeted to the plastids (Figure 5A). Nevertheless, the fluorescence signal of RHON1 inside chloroplasts showed punctuate patterns resembling the distribution of nucleoids visualized by GFP constructs fused with nucleoid proteins on the red background of the chlorophyll autofluorescence (Terasawa and Sato, 2005; Zhong et al., 2013). When RHON1-GFP fusions were transiently coexpressed with red fluorescent protein fusions of pTAC5 protein, a plastid nucleoid-localized protein in protoplasts of Arabidopsis (Zhong et al., 2013), we found that green and red fluorescence signals within the chloroplasts were merged (Figure 5B), indicating that RHON1 as well as pTAC5 are localized in chloroplast nucleoids. Immunoblot analysis of chloroplast subfractions showed that RHON1 is mainly localized in the stromal fractions but is absent or present only at low levels in thylakoid and envelope fractions (Figure 5C). This result agrees with previous reports indicating that some stromal proteins accumulate in nucleoids based on GFP assays or mass spectrometry analysis of nucleoids (Majeran et al., 2012; Pfalz and Pfannschmidt, 2013).
Figure 5.
Protein Subcellular Localization of RHON1.
(A) Subcellular localization of RHON1-GFP fusion protein. The fluorescence signals were visualized using confocal laser scanning microscopy. Green indicates GFP fluorescence, red shows chloroplast autofluorescence, and orange/yellow indicates regions where the two types of fluorescence merged. GFP signals from the RHON1-GFP fusion protein (bottom row) as well as three controls, one with the transit peptide of the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (RbcS-GFP), one with FROSTBITE1 (FRO1) of mitochondria (FRO1-GFP; Lee et al., 2002), and one with the nuclear localization signal of PTM-N (Sun et al., 2011), are shown.
(B) Colocalization of the RHON1-GFP with pTAC5-dsRED protein in chloroplast nucleoids. For better contrast, red fluorescence of chlorophyll was converted into blue.
(C) Immunoblot analysis of RHON1. Total and chloroplast subcellular fractions were separated by SDS-PAGE. After gel electrophoresis, the resolved proteins were probed with antibodies to the proteins indicated on the right. D1, RbcL, and Tic110 (translocon at inner envelop membrane of chloroplasts 110) were used as markers for thylakoid (Thy), stroma, and envelope (Env), respectively.
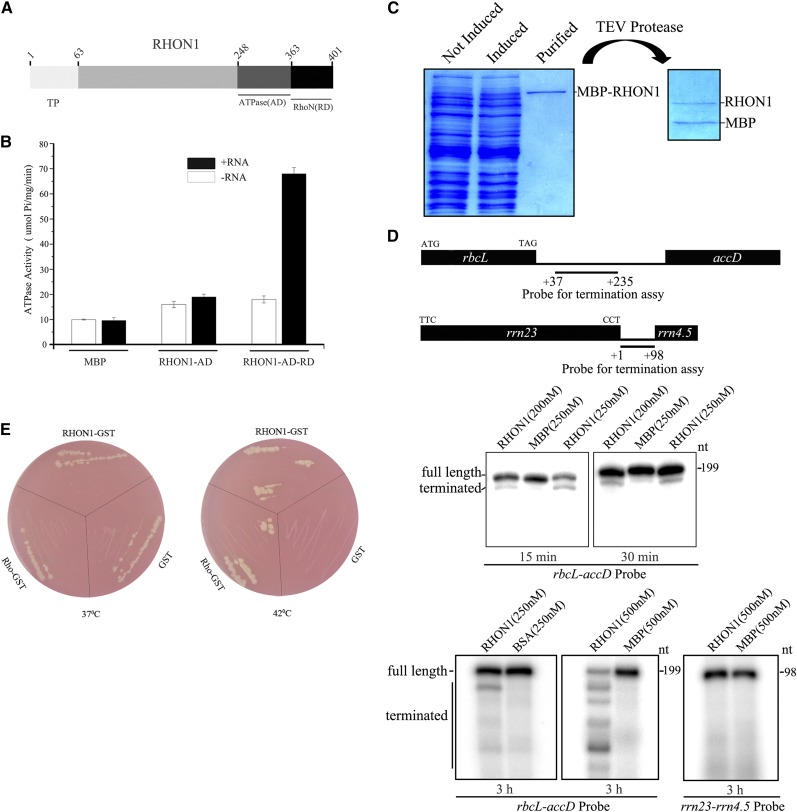
Rho is a homohexameric protein with RNA-dependent ATPase and RNA/DNA helicase activities and is distributed throughout the bacterial world with very few exceptions (Allison et al., 1998; Aravind and Koonin, 2000). The first 125 residues of E. coli Rho comprise a separate domain that by itself can bind to oligonucleotides, whereas the remaining 290 residues are closely related to the sequences of the corresponding parts of the α- and β-subunits of the F1ATPase (Skordalakes and Berger, 2006). The C terminus of RHON1 displays high similarity to the RNA binding region of Rho, whereas no domain related to F1ATPase was found in this protein. However, a domain of ∼100 residues close to the Rho-like domain showed low similarity to the P-type ATPase (Figure 6A).
Figure 6.
RHON1 Has Transcription Termination Activity.
(A) Schematic diagram of ATPase domain and RNA binding domain of RHON1.
(B) The RNA-dependent ATPase activity of RHON1 protein. The C-terminal region of RHON1 containing ATPase domain (RHON1-AD) and the ATPase domain followed with RNA binding domain (RHON1-AD-RD) were each fused with a MBP tag and expressed in E. coli BL21 (DE3). The purified recombinant proteins with or without poly (rC) were used for the ATPase activity assay by measuring phosphate release. The MBP protein was used as a control. Error bars represent the se of the mean (n = 6).
(C) Preparation of recombinant RHON1 proteins used for the in vitro transcription termination assay. The MBP-RHON1 recombinant protein was expressed in BL21 (DE3) pLysS cells. Following purification, the MBP tag of the recombinant protein was removed with TEV protease.
(D) In vitro transcription termination assay of RHON1. The transcription reaction was performed by the T7 RNA polymerase for 15 min, 30 min, and 3 h. The localization of the probe on the plastid genome is shown on the top lane, and the numbers indicate the positions of the nucleotides with respect to the termination codon of rbcL and the 3′ end of rrn 23. MBP and BSA proteins were used as controls.
(E) Complementation of the rho mutant of E. coli with RHON1. The rho mutant of E. coli (DH152) is temperature sensitive and dies at 42°C due to the loss of Rho protein but survives at 37°C (Inoko et al., 1977). Rho-GST, RHON1-GST, and GST were introduced into the DH152 strain and growth at 37 and 42°C was measured.
[See online article for color version of this figure.]
The Rho factor of E. coli has RNA-dependent ATPase activity, which provides free energy to wrest the transcript from RNA polymerase and the DNA template. The presence of the potential P-type ATPase domain in RHON1 prompted us to investigate whether RHON1 possesses a similar ATPase activity as Rho factors. We first expressed the ATPase domain (AD) of RHON1 alone and examined its ATPase catalytic activity of hydrolysis of ATP to ADP and inorganic phosphate. No obvious ATPase activity of the AD domain was found, whether or not RNA substrates were added. However, when the AD domain of RHON1 was expressed together with the RNA binding domain, a high ATPase activity could be detected in the presence of RNA substrates (Figure 6B). In addition, when the conserved residues for RNA binding were mutated, the ATPase activity of RHON1 was completely eliminated (Supplemental Figure 5). These results clearly indicate that RHON1 indeed has RNA-dependent ATPase activity resembling that of the Rho factor.
We next assayed the transcription termination activity of RHON1 using an in vitro transcription termination assay. In this experiment, full-length RHON1 in which the maltose binding protein (MBP) tag was removed was incubated with DNA templates in an in vitro transcription system with the T7 RNA polymerase (Figures 6C and 6D). When the rbcL-accD intergenic region was used as template, the terminated transcript could be clearly observed among the transcription products in the presence of RHON1, whereas no terminated transcript was detected with the MBP proteins (Figure 6D). When the rrn 23-rrn 4.5 intergenic region was used as template, no terminated transcript was observed even when the reaction time was extended to 3 h (Figure 6D). Taken together, these results suggest that RHON1 can specifically terminate rbcL transcription in vitro.
The similarity between the activities of RHON1 and Rho raised the possibility that RHON1 could be a functional analog of the E. coli Rho factor. To test this hypothesis, we introduced RHON1 into a conditional-lethal rho mutant of E. coli termed HD152. The HD152 strain is temperature sensitive; it dies at the restrictive temperature of 42°C and survives at the permissive temperature of 37°C (Das et al., 1976; Inoko et al., 1977). As shown in Figure 6E, the HD152 strain that was transformed with RHON1 as well as with Rho could grow at both 42 and 37°C. As a control, the HD152 strain transformed with GST (glutathione S-transferase) protein could only grow at 37°C. This result clearly demonstrates that RHON1 can functionally replace the E. coli Rho protein in vivo.
The RHON1 Homolog of Rice Cannot Replace RHON1 Function in rbcL Transcription Termination
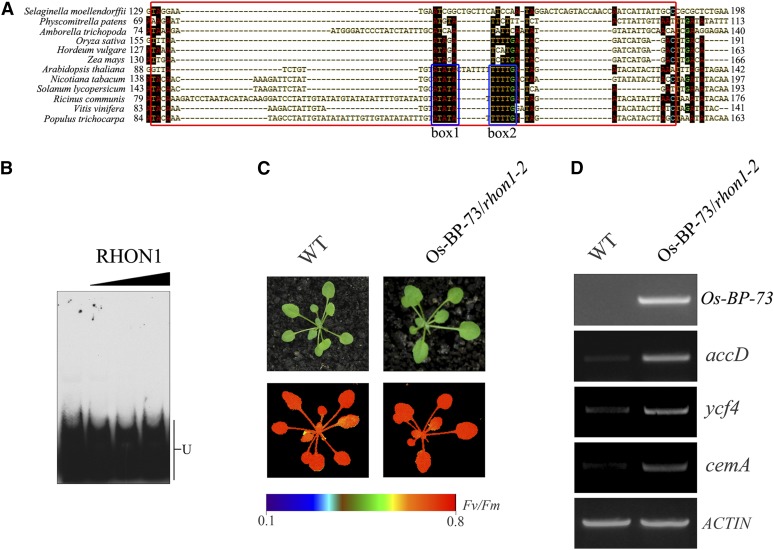
There is a major difference related to the operon organization associated with the rbcL and accD loci between eudicotyledonous and monocotyledonous plants, as the accD gene has been transferred to the nuclear genome in monocots (Bryant et al., 2011; Supplemental Figure 6A). This raises the question of whether this transcriptional interplay between rbcL and its downstream gene occurs in other plant species besides Arabidopsis. To answer this question, we first compared the 3′-UTRs in a range of plants for which the plastid genome sequence is available. This analysis revealed that two conserved boxes of RHON1 binding nucleotides exist in the examined dicotyledonous but not in monocotyledonous plants (Figure 7A). In addition, the EMSA analysis showed that RHON1 does not bind to the 3′-UTR of rice rbcL (Figure 7B). These results suggest that the cis-elements of rbcL involved in the binding of RHON1 were lost in monocotyledonous plants.
Figure 7.
The RHON1 Homolog of Rice Cannot Replace RHON1 Function in rbcL Transcription Termination.
(A) Comparison of RHON1 binding site sequences in the rbcL 3′-UTR between selected monocots and eudicots. Two conserved regions are boxed.
(B) EMSA assay of the interaction of RHON1 with the rbcL 3′-UTR of rice.
(C) Photographs and chlorophyll fluorescence images of Os-BP-73/rhon1-2 and wild-type plants grown for 3 weeks in a growth chamber. Fluorescence is shown using the pseudocolor index indicated at the bottom.
(D) Accumulation of accD, ycf4, cemA, and Os-BP-73 in Os-BP-73/rhon1-2 and wild-type plants.
BLAST analysis revealed that RHON1 homologs exist ubiquitously in vascular plants, but no corresponding RHON1 homolog was found in the unicellular alga Chlamydomonas reinhardtii. This suggests that RHON1 is a vascular plant-specific protein. In correspondence with the evolution of rbcL 3′-UTRs, phylogenetic analysis of the aligned putative homologs of RHON1 revealed two well-supported clades which correspond to eudicotyledons and monocotyledons (Supplemental Figure 6B and Supplemental Data Set 1). This phylogenetic analysis points to a substantial biochemical difference in RHON1 homologs from eudicotyledons and monocotyledons. Indeed, the detailed analysis of the protein sequence of the rice RHON1 homolog Os-BP-73 revealed that the ATPase domain was lost and only the RNA binding domain was retained in Os-BP-73. Accordingly, no ATPase activity of Os-BP-73 was detected under the same conditions used for the At-RHON1 assay (Supplemental Figure 7A). In addition, Os-BP-73 was unable to complement the temperature-sensitive phenotype of the rho mutant strain of E. coli (Supplemental Figure 7B), which suggests that it cannot substitute for the Rho factor of E. coli in vivo.
The distinct biochemical features of Os-BP-73 suggest that it acts in a different manner in vivo compared with RHON1. To address this possibility, we expressed Os-BP-73 in the rhon1 background and investigated whether it was able to complement the molecular defects of rhon1 plants completely. As shown in Figure 7C, expression of full-length Os-BP-73 cDNA under the control of the constitutive 35S promoter in rhon1-2 mutants was able to rescue the high chlorophyll fluorescence phenotype of rhon1-2, suggesting that Os-BP-73 was functionally expressed in rhon1-2. However, the abundance of transcripts from genes downstream of rbcL in the Os-BP-73/rhon1-2 plant was still maintained at a high level, rather than the wild-type level (Figure 7C), suggesting that inefficient transcriptional termination still occurred in Os-BP-73/rhon1-2 plants. In contrast to Os-BP-73/rhon1-2 plants, the accumulation of those transcripts was restored to wild-type levels in plants complemented with RHON1 (Supplemental Figure 8). Taken together with the in vitro functional analysis of Os-BP-73, we propose that unlike At-RHON1, Os-BP-73 has lost its ability to terminate transcription of rbcL but retained the function related to RNA processing during evolution.
DISCUSSION
RHON1 Is Involved in Transcription Termination of rbcL in Addition to RNA Processing
This study and that of Stoppel et al. (2012) show that a large rbcL polycistronic transcript normally undetectable in wild-type plants is present in two rhon1 mutants from different genetic backgrounds (Figure 2A). The accumulation of the rbcL polycistronic precursor might result from processing defects in the rbcL-accD intercistronic region, as suggested by Stoppel et al. (2012). Based on the data presented here, we propose that inefficient transcription termination may account for the overaccumulation of rbcL polycistronic transcripts in rhon1-2. At present, neither of these possibilities can be excluded, suggesting that RHON1 might have a function in processing as well as in transcription termination. In fact, RNA polymerase II transcription termination is coupled to 3′-end processing of the pre-mRNA in eukaryotes, and 3′ cleavage and polyadenylation factors are also critical for efficient transcriptional termination of RNA polymerase II (Rondon et al., 2008; Richard and Manley, 2009). Although the coupling of plastid RNA polymerase transcription termination and plastid RNA processing has not been addressed, it is possible that RHON1 acts as a factor coupling these two processes in plastids.
It has been proposed that RHON1 forms a distinct 800-kD complex with RNE and supports RNE function, by conferring sequence specificity to the endonuclease (Stoppel et al., 2012). However, the rhon1 rne double mutant displayed a more severe pleiotropic phenotype than that of rne, suggesting that RHON1 has additional functions compared with RNE (Stoppel et al., 2012). Indeed, investigation of accD promoter usage in rne mutant plants showed that the aberrant transcription initiation observed in rhon1-2 did not occur in rne (Supplemental Figure 9). This difference suggests that RHON1 also acts on the rbcL-accD intercistronic region independently of RNE. In addition to the 3′-UTR of rbcL, RHON1 was able to bind multiple RNA species, including rRNAs and petB, in vitro (Stoppel et al., 2012). These multiple functions of RHON1 might be related to its ability to bind multiple protein/RNA partners. RHON1 may form different complexes by interacting with distinct protein/RNA partners, as revealed by the multiple RHON1 complexes fractionated by native acrylamide gradient gel electrophoresis (Stoppel et al., 2012). Those complexes may act independently or cooperate with each other to function in two steps of plastid gene expression.
Possible Mechanisms of Transcription Termination in Plastids
There are conflicting observations concerning transcription termination in plastids of vascular plants. The spinach (Spinacia oleracea) RNA polymerase has been shown to recognize prokaryotic Rho-independent transcription terminators in vitro, suggesting that equivalent sequences might function as terminators of plastid transcription in vivo (Chen and Orozco, 1988). However, transcription termination assays using an in vitro transcription system from spinach showed that plastid 3′ inverted repeats of psbA, rbcL, and rpoB do not function as transcription terminators (Stern and Gruissem, 1987). Instead, two tRNAs, which are not followed by 3′ inverted repeats, do effectively terminate transcription under the same conditions (Stern and Gruissem, 1987). These discordant results raise the possibility that site-specific components are required for proper termination processes in vivo (Blowers et al., 1993). Alternatively, plastid transcription termination could also be mediated in as yet unknown ways that are not associated with 3′ inverted repeats.
The termination of rbcL transcription was originally inferred from the transformation of the plastid genome of tobacco (Nicotiana tabacum) with the rbcL gene (Staub and Maliga, 1994). The tobacco rbcL mRNA is always monocistronic when transcribed in its native context (Sinagawa-García et al., 2009). However, when chimeric genes were introduced into the intergenic region between rbcL and accD of tobacco, rbcL read-through transcripts were readily detectable (Staub and Maliga, 1994; Monde et al., 2000; Sinagawa-García et al., 2009). In addition, when the insertion site of the chimeric genes was moved further downstream, read-through transcription did not occur (Sinagawa-García et al., 2009). These results suggest the presence of a termination or processing site upstream of accD.
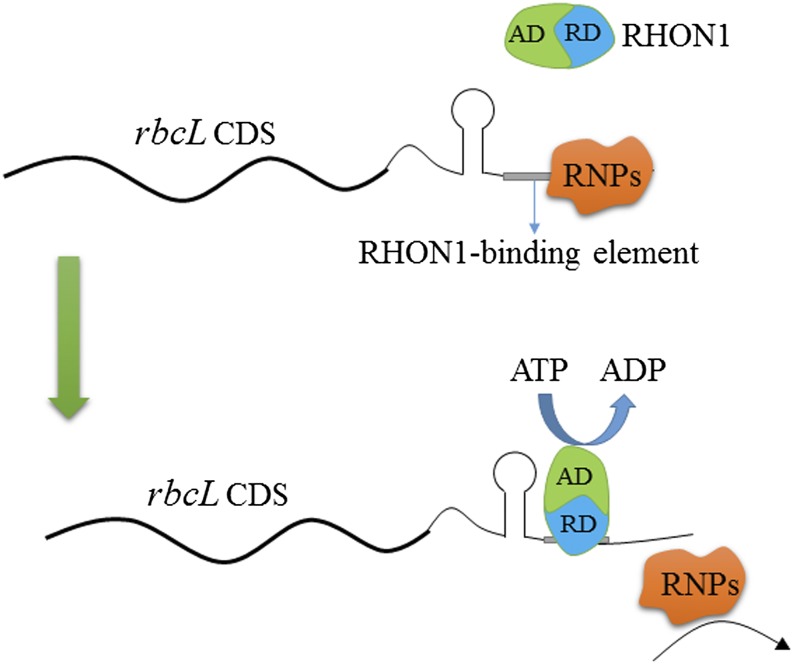
Considering the transcriptional independence of rbcL and accD in wild-type Arabidopsis plants, together with the data presented here, we suggest that there is a termination site in the intergenic region between rbcL and accD, although the presence of a processing site cannot be completely excluded. Our results show that efficient transcription termination of rbcL requires the RNA binding protein RHON1, related to bacterial Rho factors. Nucleotide binding assays and ATPase activity analysis suggest that RHON1 acts like Rho factors and is involved in transcription termination of plastid genes. Rho factors usually bind to a wide range of mRNA sequences but have a strong preference for mRNA sequences enriched in rC residues (Graham, 2004). However, the RHON1 binding mRNA region contains few rC residues, which is clearly different from the RNA binding sites of Rho factors. In addition, the ATPase domain of RHON1 does not belong to F-type ATPase and the protein structure is distinct from that of Rho. Therefore, it appears that the mechanism of action of RHON1 is not totally identical to that of Rho factors. Although the Rho-dependent mechanism has been established only in eubacteria, a similar ATP-driven termination machine was also discovered in eukaryotic cells. For instance, a Drosophila melanogaster ATPase, called factor2, and the Sen1p helicase in budding yeast both employ this ATP-driven mechanism to terminate RNA Polymerase II transcription (Xie and Price, 1997; Porrua and Libri, 2013). Therefore it is quite possible that RHON1 uses an unknown ATP-driven device to mediate the dissociation of nascent rbcL transcripts from PEP. A postulated mechanism for RHON1 action is schematically depicted in Figure 8.
Figure 8.
Model of RHON1 Action in rbcL Transcription Termination.
RHON1 binds to the elements adjacent to the stem-loop structure of the 3′-UTR of rbcL mRNA via its RNA binding domain (RD). After binding to mRNA, RHON1 catalyzes the dissociation of rbcL mRNA from genomic DNA and the RNA polymerases (RNPs) via its AD.
In rhon1-2 plants, we observed aberrant transcription initiation in the rbcL-accD intergenic region. It is likely that this is a consequence of inefficient transcription termination of rbcL because it did not occur when rhon1-2 plants were grown on spectinomycin, which leads to the inhibition of rbcL transcription. This suggests that these processes are coupled. Currently, the molecular basis for the aberrant accD transcription initiation in rhon1-2 remains unclear. The elongating RNA polymerase complex across the termination site of rbcL might interfere with the initiation transcription complex of the accD transcriptional unit and thus reduce the transcription accuracy especially since plastid transcription initiation sites in vascular plants are not very stringent (Holec et al., 2006; Swiatecka-Hagenbruch et al., 2007). Alternatively, because after termination of rbcL transcription, PEP cannot be released from the template due to the lack of RHON1, it might slide on the template and finally reinitiate transcription nonspecifically at different positions in the rbcL-accD intergenic region and even within the accD gene. Thus, one of the functions of RHON1 may be to avoid aberrant transcription initiation in the rbcL-accD intergenic region due to inefficient rbcL transcription termination.
Evolution of RHON1-Dependent Transcription Termination
It is believed that the evolution of land plant organelle genomes is inextricably linked to the evolution of nuclear-encoded trans-acting factors, and vice versa. Plastid RNA editing has been one of the most enigmatic processes in this respect (Takenaka et al., 2013). In this study, both in vitro and in vivo evidence showed that Os-BP-73, the rice ortholog of RHON1, appears to have lost the ability to terminate transcription due to the lack of the ATPase domain. The RNA processing activity of Os-BP-73 is mediated through its conserved RNA binding domain could support RNE functions presumably by conferring sequence specificity to the endonuclease. In agreement with the difference between At-RHON1 and Os-BP-73, the putative RNA binding sites of RHON1 within in rbcL-accD intergenic region of the Arabidopsis plastid genome are absent from the rice plastid genome (Figures 7A and 7B), which suggests coevolution between At-RHON1/Os-BP-73 and its target elements. Our results indicate that the loss of a cognate cis-element might not result in the complete loss of its corresponding trans-acting factor but lead to the elimination of its related domain, which might represent a more economical strategy for evolution.
One fundamental difference between plastid genomes in dicotyledons and monocotyledons is that three genes, ycf1, ycf2 (functions unknown), and accD, are present in dicotyledons but absent from monocotyledon plastids. The failure to translate one or more of these three genes is likely to be the reason for embryo lethality in Arabidopsis but not in maize (Zea mays), which suggests that the control of the expression of these genes is critical for plant development in dicotyledons (Asakura and Barkan, 2006; Schmitz-Linneweber and Small, 2008; Bryant et al., 2011). Therefore, it is not surprising that some specific factors have been recruited to serve the expression of these genes in dicotyledons, for instance, the species-specific editing factors for accD (Robbins et al., 2009; Yu et al., 2009). In rhon1 plants, the accumulation of rbcL transcripts was normal, whereas the transcription initiation of accD was affected. Our results thus point to an alternative strategy for regulating accD expression in dicotyledons, which relies on the efficient transcription termination of its upstream gene, rbcL, mediated by RHON1.
METHODS
Plant Materials and Growth Conditions
The rhon1-2 mutant was isolated from a collection of pSKI015 T-DNA-mutagenized Arabidopsis thaliana (ecotype Columbia) lines based on its high chlorophyll fluorescence phenotype. The seeds of wild-type and mutant plants were surface-sterilized, sown on Murashige and Skoog medium containing 2% sucrose, and grown under short-day conditions (10-h-light/14-h-dark cycles) with a photon flux density of 120 µmol m−2 s−1 at 22°C. To ensure synchronized germination, the seeds were incubated in darkness for 48 h at 4°C before sowing.
Complementation of the rhon1 Mutant
The coding sequences of RHON1 and Os-BP-73 were cloned into BamHI and KpnI sites of the pSN1301 vector, in which they were constitutively expressed under the control of the cauliflower mosaic virus 35S promoter. These two resultant constructs were transferred into Agrobacterium tumefaciens strain C58 and then introduced into rhon1-2 plants by the floral dip method (Clough and Bent, 1998), respectively. The transformants were selected on Murashige and Skoog medium containing 50 μg mL−1 hygromycin B.
Chlorophyll Fluorescence Measurements
Chlorophyll fluorescence measurements were performed using a CF Imager (Technologica) as described by the manufacturer. Before each measurement, leaves were dark adapted for 10 min. The minimum fluorescence yield (Fo) and the maximum fluorescence yield (Fm) were measured. The maximal photochemical efficiency of photosystem II was calculated from the ratio of Fv to Fm [Fv/Fm = (Fm − Fo)/Fm]. Image data acquired in each experiment were normalized to a false color scale, with arbitrarily assigned extreme values of 0.1 (lowest) and 0.8 (highest). This resulted in the highest and lowest Fv/Fm values being represented by the red and blue extremes of the color scale, respectively.
RT-PCR, Quantitative PCR, and RNA Gel Blotting
Total leaf RNA was extracted from Arabidopsis seedlings with the Qiagen RNeasy Plant Mini kit (Qiagen) and used to generate first-strand cDNA in a 50-μL reaction with the Superscript III cDNA synthesis system (Invitrogen) following the manufacturer’s instructions. Aliquots of the resulting cDNA samples were used as template for PCR analysis. Quantitative PCR was performed with SYBR PrimeScript Ready Mix (Takara) in an ABI 7900 sequence detection system (Applied Biosystems) according to the manufacturer's protocol. Total RNA (5 to 10 µg) from wild-type and rhon1-2 seedlings was separated on 1.2 to 2.0% (w/v) agarose-formaldehyde gels, blotted to positively charged nylon membrane by capillary blotting, fixed by cross-linking, and subsequently subjected to hybridization. Hybridization probes were prepared by PCR amplification of fragments of the plastid genes from Arabidopsis. The sequences of all primers used in this study are listed in Supplemental Table 1.
Run-on Transcription Assay
The chloroplast run-on assays were performed using DIG-11-UTP as the labeling nucleotide as described (Chi et al., 2010). Briefly, the leaves were homogenized in buffer containing 50 mM HEPES-KOH, pH 6.8, 0.33 M sorbitol, 2 mM EDTA, 1 mM MgCl2, 1 mM MnCl2, and 9 mM DTT. The homogenate was filtered through four layers cheesecloth and spun at 3000g for 5 min at 0°C. The pellet was fractionated in 20/50/80% Percoll gradients by centrifugation at 6500g for 15 min. Intact chloroplasts were collected at the interface between 50 and 80% Percoll, the integrity of chloroplasts was examined by CP-ISO kits supplied by Sigma-Aldrich, and the number of chloroplasts in the samples was determined by counting the organelles using light microscopy. Transcription was performed in 100-μL reactions containing 5.0 × 107 lysed plastids for 10 min, followed by RNA extraction with phenol/chloroform. Labeled transcripts were hybridized to Hybond N+ filters carrying 100 or 200 ng DNA probes of plastid genes. A slot-blotting apparatus (Bio-Dot SF; Bio-Rad) was used for filter preparation. Signals were analyzed using ALPHAVIEW 3.0 (Alpha Innotech).
GFP Assays
The coding sequences of RHON1 were amplified by RT-PCR. The PCR products were then cloned into XhoI/BamHI sites of the transient expression vector pSAT6-EGFP-N1 (Citovsky et al., 2006), which contained the mGFP5 coding sequence under the control of the cauliflower mosaic virus 35S promoter, to generate a RHON1 fusion protein with GFP in the C terminus. Similarly, two controls were then constructed: one in which the nuclear localization signal of the PTM-N (Sun et al., 2011) and the other in which the transit peptide of chloroplast inner envelop translocation component (TIC21) of Arabidopsis were also fused to the vector. The resulting constructs were transfected into Arabidopsis mesophyll protoplasts according to the method as described by Kovtun et al. (2000). Fluorescence analysis was performed on an LSM 510 META confocal laser scanning system (Zeiss).
Expression of Recombinant Proteins and Antibody Production
To produce recombinant MBP-RHON1-Strep proteins, the coding sequence of RHON1 lacking the transit peptide sequence was PCR amplified, digested with SalI and BamHI, and inserted into the SalI/BamHI sites of pMAL-TEV (kindly provided by Alice Barkan). BL21(DE3) pLysS cells (Novagen) harboring the resulting vector were grown at 37°C until OD600 of 0.6 was reached and transferred to 22°C for 30 min. Expression was then induced by 1 mM isopropyl β-d-1-thiogalactopyranoside, and cells were grown for 3 h at 22°C with shaking. All subsequent steps were performed at 4°C. Sedimented cells were lysed in lysis buffer (20 mM Tris-HCl, pH 7.4, 350 mM NaCl, and Complete Protease Inhibitor Cocktail Tablets [Roche]) by sonication and cleared by centrifugation (36,000g/20 min). Affinity chromatography was performed using amylose resin (NEB). After three washes with lysis buffer, proteins were eluted in Strep-washing buffer (IBA) supplemented with 150 mM maltose and subjected to a second affinity purification using Strep-Tactin-Macroprep (IBA). The column was washed three times with Strep-washing buffer, and proteins were eluted with Strep-elution buffer (IBA). RHON1-Strep was subsequently cleaved from the MBP moiety using AcTEV protease (Life Technologies). The DNA sequences coding for three RHON1 fragments (63 to 200, 201 to 300, and 301 to 401 amino acids) were subcloned into the EcoRI/XhoI sites of the expression vector pMAL-c5x (NEB) to produce MBP fusions. Induction, cell lysis, and purification of the fusion proteins using amylose affinity chromatography were performed according to the manufacturer’s protocol (NEB). A RHON1 fragment (64 to 200 amino acids) with a C-terminal 6 × histidine tag was expressed from a pET28a vector (Novagen) and purified with nickel-nitrilotriacetic acid agarose. The purified protein was used to generate polyclonal antibodies against RHON1 in rabbits.
Protein Isolation and Immunoblot Analysis
Total protein of Arabidopsis leaves was isolated as previously described (Chi et al., 2010). Stroma, thylakoid, and envelope fractions were purified from lysed chloroplasts by sucrose gradient centrifugation as previously described (Williams and Barkan, 2003). The protein levels were quantified using the Dc protein assay kit (Bio-Rad). The proteins resolved by SDS-PAGE were blotted onto nitrocellulose membranes and incubated with specific antibodies, and the signals were detected using the enhanced chemiluminescence method.
Mapping of Transcription Initiation Sites
The transcription initiation sites of plastid genes were determined using a 5′-RACE Full-RACE kit (Takara) based on the methods of Swiatecka-Hagenbruch et al. (2007) with minor modifications. Briefly, the total RNA was treated with calf intestine alkaline phosphatase to remove free 5′ phosphates. After extraction, 2 µg purified RNA was then treated with 0.5 units of TAP at 37°C for 1 h in the presence of 40 units of RNase inhibitor. Control reactions were set up without TAP. The RNA was subsequently purified and ligated to a 5′ RNA adapter according to the supplier’s protocol. PCR products were analyzed on agarose gels after two successive PCR amplifications, using two outer primers and two inner primers, respectively. The inner and outer adapter-specific forward primers were included in the Full-RACE kit.
RNA Coimmunoprecipitation Analysis
The stroma was prepared by hypotonic lysis of purified chloroplasts and used for RNA coimmunoprecipitation analysis as described by Barkan (2009). RNA was recovered from the precipitated fractions in the presence of 1% SDS, 0.05 µg µL−1 yeast tRNA, and 5 mM EDTA. The RNA was isolated by phenol-chloroform isoamyl alcohol extraction and precipitated with 3 volumes of ethanol. RNAs from the supernatant fractions were extracted as the controls. Equal portions of each RNA sample were applied to a nylon membrane through a slot-blot manifold (Bio-Dot SF; Bio-Rad) and hybridized with radiolabeled DNA probes specific for the indicated rbcL and accD sequences. Probes were generated by PCR amplification of the relevant chloroplast sequences and labeled by the random priming method. The ribonuclease-protection assay of the RNA samples was performed as described by Chi et al. (2012).
Primer Extension
Primer extension was performed using the Primer Extension System of Promega. Sequence-specific oligonucleotides were end-labeled with [γ-32P]ATP and then were annealed with 10 µg total RNA for 20 min at the primer annealing temperature. Primer extension was performed for 30 min at 42°C with AMV reverse transcriptase. Extension products were heated at 90°C for 10 min and were separated on a 6 × 9-cm denaturing polyacrylamide gel containing 6% acrylamide (19:1 acrylamide:bis), 7 M urea, and 1× TBE buffer. The signals were visualized by autoradiography.
EMSA
DNA/RNA and protein interactions were detected using the LightShift Chemiluminescent EMSA kit (Pierce). Briefly, the target DNA/RNA was 5′end-labeled with biotin and terminal deoxynucleotidyl transferase and then incubated with the RHON1 fragments (63 to 200, 201 to 300, and 301 to 401 amino acids) in the DNA/RNA binding reaction mixture supplied by the kit. After incubation for 20 min at 25°C, the samples were resolved on a 6% Tris-borate gel in 0.5× TBE buffer and transferred to a nylon membrane. The biotin end-labeled DNA was detected using a streptavidin-horseradish peroxidase conjugate and chemiluminescent substrate.
ATPase Activity Assay
The ATPase activity was determined by measuring phosphate release in a mixture containing 50 mM Tris-HCl, pH 8.0, 50 mM NaCl, 4 mM ATP, and 2 mM MgCl2 incubated with 0.2 mL reconstitution mixture for 5 min at 37°C. After the reaction was stopped by adding 40 μL trichloroacetic acid, the supernatant was collected and the released Pi was determined by a microcolorimetric method (Taussky and Shorr, 1953).
Complementation of the rho Mutant of Escherichia coli
The rho mutant (HD152) was obtained from the E. coli Genetic Stock Center at Yale University. The coding sequence of RHON1 and Os-Bp-73 lacking the transit peptide sequences were inserted into the EcoRI and XhoI sites of pGEX-4T-2 (GE Healthcare) to produce GST fusions. RHO of E. coli was inserted into the XmaI and XhoI sites of pGEX-4T-2 and used as a control. These constructs then were transformed into HD152 cells by the CaCl2 method. The transformed cells were grown in the presence of 0.4 mM β-d-1-thiogalactopyranoside. The temperature sensitivity of E. coli cells was assayed as described by Inoko et al. (1977).
In Vitro Transcription Termination Assay
Rbcl-accD (nucleotides 56434 to 56632; GenBank accession number AP000423.1) and rrn23-rrn4.5 (nucleotides 107501 to 107598; GenBank accession number AP000423.1) intergenic regions were PCR amplified. Amplification products were gel purified. In vitro transcription was performed in 20-μL reactions containing 300 pM each PCR product, 0.5 mM each ATP, CTP, and GTP, 10 µM UTP, 10 µCi [α-32P]UTP, 20 units T7 RNA Polymerase (Fermentas), 4 μL 5× transcription buffer (200 mM Tris-HCl, pH 7.9, 30 mM MgCl2, 50 mM DTT, 50 mM NaCl, and 10 mM spermidine; Fermentas), 20 units RiboLoc RNase Inhibitor (Fermentas), and protein concentrations as indicated in the figure. Reaction mixtures were incubated for 15 min, 30 min and 3 h at 30°C, denatured by heating in the presence of 80% formaldehyde, and analyzed on 5% denaturing polyacrylamide gel containing 8 M urea and 1× TBE. Gels were exposed to a phosphor imager screen and analyzed with the Typhoon Variable Mode Imager (GE Healthcare).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: RHON1, AT1G06190; accD, ATCG00500; rbcL, ATCG00490; and OsBP-73, Os03g0183100.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Microarray Analysis of Plastid Gene Transcripts in rhon1-2 Compared with the Wild Type.
Supplemental Figure 2. Run-on Transcription Assay of Plastid Genes in rhon1-2 and Wild-Type Plants.
Supplemental Figure 3. Analysis of PEP-Dependent Transcription in rhon1-2.
Supplemental Figure 4. Mapping of 3′- and 5′-Ends of Plastid Transcripts in Wild-Type and rhon1-2 Plants.
Supplemental Figure 5. The ATPase Activities of Mutated RHON1 Proteins.
Supplemental Figure 6. Phylogenetic Analysis of RHON1 Homologs in Vascular Plants.
Supplemental Figure 7. Characterization of a RHON1 Homolog of Rice, Os-BP-73.
Supplemental Figure 8. The Accumulation of rbcL, accD, psaI, ycf4, cemA, and petA Transcripts in RHON1- Complemented Plants Assayed by RT-PCR.
Supplemental Figure 9. The Transcription Initiation Sites of accD in the rne Mutant.
Supplemental Table 1. List of Primers Used in This Study.
Supplemental Methods. Microarray, Circularized RNA RT-PCR, and Phylogenetic Analysis.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 6.
Supplementary Material
Acknowledgments
We thank Alice Barkan for kindly sending us pMAL-TEV. We also thank the reviewers for important insights and for suggesting key experiments that changed and improved our conclusions and insights regarding RHON1 function. This work was supported by the National Natural Science Foundation of China (Grant 31370273), the Major State Basic Research Development Program (Grant 2015CB150100), and the Solar Energy Initiative and Cooperative Team Project of the Chinese Academy of Sciences.
AUTHOR CONTRIBUTIONS
W.C. and L.Z. designed the research. W.C., B.H., N.M., J. Mao, and D.J. performed the research. W.C., B.H., N.M., J. Meurer, C.L., J.D.R., and L.Z. analyzed the data. W.C., J.D.R., and L.Z. wrote the article.
Glossary
- PEP
plastid-encoded RNA polymerase
- NEP
nucleus-encoded phage-type RNA polymerase
- RNE
RNase E
- RACE
5′-rapid amplification of cDNA ends
- TAP
tobacco acid pyrophosphatase
- CR
circularized RNA
- EMSA
electrophoretic mobility shift assay
- UTR
untranslated region
- AD
ATPase domain
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Allison T.J., Wood T.C., Briercheck D.M., Rastinejad F., Richardson J.P., Rule G.S. (1998). Crystal structure of the RNA-binding domain from transcription termination factor rho. Nat. Struct. Biol. 5: 352–356. [DOI] [PubMed] [Google Scholar]
- Aravind L., Koonin E.V. (2000). SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25: 112–114. [DOI] [PubMed] [Google Scholar]
- Asakura Y., Barkan A. (2006). Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol. 142: 1656–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (2009). Genome-wide analysis of RNA-protein interactions in plants. Methods Mol. Biol. 553: 13–37. [DOI] [PubMed] [Google Scholar]
- Barkan A. (2011). Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 155: 1520–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blowers A.D., Klein U., Ellmore G.S., Bogorad L. (1993). Functional in vivo analyses of the 3′ flanking sequences of the Chlamydomonas chloroplast rbcL and psaB genes. Mol. Gen. Genet. 238: 339–349. [DOI] [PubMed] [Google Scholar]
- Bryant N., Lloyd J., Sweeney C., Myouga F., Meinke D. (2011). Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis. Plant Physiol. 155: 1678–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.J., Orozco E.M. Jr. (1988). Recognition of prokaryotic transcription terminators by spinach chloroplast RNA polymerase. Nucleic Acids Res. 16: 8411–8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W., Mao J., Li Q., Ji D., Zou M., Lu C., Zhang L. (2010). Interaction of the pentatricopeptide-repeat protein DELAYED GREENING 1 with sigma factor SIG6 in the regulation of chloroplast gene expression in Arabidopsis cotyledons. Plant J. 64: 14–25. [DOI] [PubMed] [Google Scholar]
- Chi W., He B., Mao J., Li Q., Ma J., Ji D., Zou M., Zhang L. (2012). The function of RH22, a DEAD RNA helicase, in the biogenesis of the 50S ribosomal subunits of Arabidopsis chloroplasts. Plant Physiol. 158: 693–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., Lee L.Y., Vyas S., Glick E., Chen M.H., Vainstein A., Gafni Y., Gelvin S.B., Tzfira T. (2006). Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J. Mol. Biol. 362: 1120–1131. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Courtois F., Merendino L., Demarsy E., Mache R., Lerbs-Mache S. (2007). Phage-type RNA polymerase RPOTmp transcribes the rrn operon from the PC promoter at early developmental stages in Arabidopsis. Plant Physiol. 145: 712–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Court D., Adhya S. (1976). Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc. Natl. Acad. Sci. USA 73: 1959–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluppi G.R., Richardson J.P. (1980). ATP-induced changes in the binding of RNA synthesis termination protein Rho to RNA. J. Mol. Biol. 138: 513–539. [DOI] [PubMed] [Google Scholar]
- Graham J.E. (2004). Sequence-specific Rho-RNA interactions in transcription termination. Nucleic Acids Res. 32: 3093–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger I.H., Proudfoot N.J. (1998). Poly(A) signals control both transcriptional termination and initiation between the tandem GAL10 and GAL7 genes of Saccharomyces cerevisiae. EMBO J. 17: 4771–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley J., Bogorad L. (1990). Alternative promoters are used for genes within maize chloroplast polycistronic transcription units. Plant Cell 2: 323–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka M., Kanamaru K., Takahashi H., Tanaka K. (2003). Molecular genetic analysis of chloroplast gene promoters dependent on SIG2, a nucleus-encoded sigma factor for the plastid-encoded RNA polymerase, in Arabidopsis thaliana. Nucleic Acids Res. 31: 7090–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holec S., Lange H., Kühn K., Alioua M., Börner T., Gagliardi D. (2006). Relaxed transcription in Arabidopsis mitochondria is counterbalanced by RNA stability control mediated by polyadenylation and polynucleotide phosphorylase. Mol. Cell. Biol. 26: 2869–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoko H., Shigesada K., Imai M. (1977). Isolation and characterization of conditional-lethal rho mutants of Escherichia coli. Proc. Natl. Acad. Sci. USA 74: 1162–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeng S.T., Gardner J.F., Gumport R.I. (1990). Transcription termination by bacteriophage T7 RNA polymerase at rho-independent terminators. J. Biol. Chem. 265: 3823–3830. [PubMed] [Google Scholar]
- Kirsch W., Seyer P., Herrmann R.G. (1986). Nucleotide sequence of the clustered genes for two P700 chlorophyll a apoproteins of the photosystem I reaction center and the ribosomal protein S14 of the spinach plastid chromosome. Curr. Genet. 10: 843–855. [Google Scholar]
- Kühn K., Bohne A.-V., Liere K., Weihe A., Börner T. (2007). Arabidopsis phage-type RNA polymerases: accurate in vitro transcription of organellar genes. Plant Cell 19: 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun Y., Chiu W.L., Tena G., Sheen J. (2000). Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 97: 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.H., Lee H., Xiong L., Zhu J.K. (2002). A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell 14: 1235–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerbs-Mache S. (1993). The 110-kDa polypeptide of spinach plastid DNA-dependent RNA polymerase: single-subunit enzyme or catalytic core of multimeric enzyme complexes? Proc. Natl. Acad. Sci. USA 90: 5509–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liere K., Weihe A., Börner T. (2011). The transcription machineries of plant mitochondria and chloroplasts: Composition, function, and regulation. J. Plant Physiol. 168: 1345–1360. [DOI] [PubMed] [Google Scholar]
- Majeran W., Friso G., Asakura Y., Qu X., Huang M., Ponnala L., Watkins K.P., Barkan A., van Wijk K.J. (2012). Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol. 158: 156–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monde R.A., Greene J.C., Stern D.B. (2000). The sequence and secondary structure of the 3′-UTR affect 3′-end maturation, RNA accumulation, and translation in tobacco chloroplasts. Plant Mol. Biol. 44: 529–542. [DOI] [PubMed] [Google Scholar]
- Pfalz J., Liere K., Kandlbinder A., Dietz K.J., Oelmüller R. (2006). pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18: 176–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J., Pfannschmidt T. (2013). Essential nucleoid proteins in early chloroplast development. Trends Plant Sci. 18: 186–194. [DOI] [PubMed] [Google Scholar]
- Peters J.M., Vangeloff A.D., Landick R. (2011). Bacterial transcription terminators: the RNA 3′-end chronicles. J. Mol. Biol. 412: 793–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.M., Mooney R.A., Grass J.A., Jessen E.D., Tran F., Landick R. (2012). Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev. 26: 2621–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrua O., Libri D. (2013). A bacterial-like mechanism for transcription termination by the Sen1p helicase in budding yeast. Nat. Struct. Mol. Biol. 20: 884–891. [DOI] [PubMed] [Google Scholar]
- Richard P., Manley J.L. (2009). Transcription termination by nuclear RNA polymerases. Genes Dev. 23: 1247–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J.P. (2003). Loading Rho to terminate transcription. Cell 114: 157–159. [DOI] [PubMed] [Google Scholar]
- Robbins J.C., Heller W.P., Hanson M.R. (2009). A comparative genomics approach identifies a PPR-DYW protein that is essential for C-to-U editing of the Arabidopsis chloroplast accD transcript. RNA 15: 1142–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon A.G., Mischo H.E., Proudfoot N.J. (2008). Terminating transcription in yeast: whether to be a ‘nerd’ or a ‘rat’. Nat. Struct. Mol. Biol. 15: 775–776. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Small I. (2008). Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 13: 663–670. [DOI] [PubMed] [Google Scholar]
- Schweer J., Loschelder H., Link G. (2006). A promoter switch that can rescue a plant sigma factor mutant. FEBS Lett. 580: 6617–6622. [DOI] [PubMed] [Google Scholar]
- Sexton T.B., Christopher D.A., Mullet J.E. (1990). Light-induced switch in barley psbD-psbC promoter utilization: a novel mechanism regulating chloroplast gene expression. EMBO J. 9: 4485–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina T., Allison L., Maliga P. (1998). rbcL Transcript levels in tobacco plastids are independent of light: reduced dark transcription rate is compensated by increased mRNA stability. Plant Cell 10: 1713–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinagawa-García S.R., Tungsuchat-Huang T., Paredes-López O., Maliga P. (2009). Next generation synthetic vectors for transformation of the plastid genome of higher plants. Plant Mol. Biol. 70: 487–498. [DOI] [PubMed] [Google Scholar]
- Skordalakes E., Berger J.M. (2006). Structural insights into RNA-dependent ring closure and ATPase activation by the Rho termination factor. Cell 127: 553–564. [DOI] [PubMed] [Google Scholar]
- Staub J.M., Maliga P. (1994). Translation of psbA mRNA is regulated by light via the 5′-untranslated region in tobacco plastids. Plant J. 6: 547–553. [DOI] [PubMed] [Google Scholar]
- Stern D.B., Goldschmidt-Clermont M., Hanson M.R. (2010). Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 61: 125–155. [DOI] [PubMed] [Google Scholar]
- Stern D.B., Gruissem W. (1987). Control of plastid gene expression: 3′ inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell 51: 1145–1157. [DOI] [PubMed] [Google Scholar]
- Stoppel R., Manavski N., Schein A., Schuster G., Teubner M., Schmitz-Linneweber C., Meurer J. (2012). RHON1 is a novel ribonucleic acid-binding protein that supports RNase E function in the Arabidopsis chloroplast. Nucleic Acids Res. 40: 8593–8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita M., Sugiura M. (1984). Nucleotide sequence and transcription of the gene for the 32,000 dalton thylakoid membrane protein from Nicotiana tabacum. Mol. Gen. Genet. 195: 308–313. [Google Scholar]
- Sun X., Feng P., Xu X., Guo H., Ma J., Chi W., Lin R., Lu C., Zhang L. (2011). A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat. Commun. 2: 477. [DOI] [PubMed] [Google Scholar]
- Swiatecka-Hagenbruch M., Liere K., Börner T. (2007). High diversity of plastidial promoters in Arabidopsis thaliana. Mol. Genet. Genomics 277: 725–734. [DOI] [PubMed] [Google Scholar]
- Takenaka M., Zehrmann A., Verbitskiy D., Härtel B., Brennicke A. (2013). RNA editing in plants and its evolution. Annu. Rev. Genet. 47: 335–352. [DOI] [PubMed] [Google Scholar]
- Taussky H.H., Shorr E. (1953). A microcolorimetric method for the determination of inorganic phosphorus. J. Biol. Chem. 202: 675–685. [PubMed] [Google Scholar]
- Terasawa K., Sato N. (2005). Visualization of plastid nucleoids in situ using the PEND-GFP fusion protein. Plant Cell Physiol. 46: 649–660. [DOI] [PubMed] [Google Scholar]
- Timmis J.N., Ayliffe M.A., Huang C.Y., Martin W. (2004). Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5: 123–135. [DOI] [PubMed] [Google Scholar]
- Walter M., Piepenburg K., Schöttler M.A., Petersen K., Kahlau S., Tiller N., Drechsel O., Weingartner M., Kudla J., Bock R. (2010). Knockout of the plastid RNase E leads to defective RNA processing and chloroplast ribosome deficiency. Plant J. 64: 851–863. [DOI] [PubMed] [Google Scholar]
- Wicke S., Schneeweiss G.M., dePamphilis C.W., Müller K.F., Quandt D. (2011). The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76: 273–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P., Barkan A. (2003). A chloroplast-localized PPR protein required for plastid ribosome accumulation. Plant J. 36: 675–686. [DOI] [PubMed] [Google Scholar]
- Xie, Z., and Price, D. (1997). Drosophila factor 2, an RNA polymerase II transcript release factor, has DNA-dependent ATPase activity. J. Biol. Chem. 272: 31902–31907. [DOI] [PubMed] [Google Scholar]
- Yu Q.B., Jiang Y., Chong K., Yang Z.N. (2009). AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana. Plant J. 59: 1011–1023. [DOI] [PubMed] [Google Scholar]
- Zhelyazkova P., Sharma C.M., Förstner K.U., Liere K., Vogel J., Börner T. (2012). The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell 24: 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L., Zhou W., Wang H., Ding S., Lu Q., Wen X., Peng L., Zhang L., Lu C. (2013). Chloroplast small heat shock protein HSP21 interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development in Arabidopsis under heat stress. Plant Cell 25: 2925–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.