A helicase is required to preserve genome stability in plants, which is likely due to its role in destabilizing various types of DNA loop recombination intermediates.
Abstract
In humans, mutations in the DNA helicase Regulator of Telomere Elongation Helicase1 (RTEL1) lead to Hoyeraal-Hreidarsson syndrome, a severe, multisystem disorder. Here, we demonstrate that the RTEL1 homolog in Arabidopsis thaliana plays multiple roles in preserving genome stability. RTEL1 suppresses homologous recombination in a pathway parallel to that of the DNA translocase FANCM. Cytological analyses of root meristems indicate that RTEL1 is involved in processing DNA replication intermediates independently from FANCM and the nuclease MUS81. Moreover, RTEL1 is involved in interstrand and intrastrand DNA cross-link repair independently from FANCM and (in intrastrand cross-link repair) parallel to MUS81. RTEL1 contributes to telomere homeostasis; the concurrent loss of RTEL1 and the telomerase TERT leads to rapid, severe telomere shortening, which occurs much more rapidly than it does in the single-mutant line tert, resulting in developmental arrest after four generations. The double mutant rtel1-1 recq4A-4 exhibits massive growth defects, indicating that this RecQ family helicase, which is also involved in the suppression of homologous recombination and the repair of DNA lesions, can partially replace RTEL1 in the processing of DNA intermediates. The requirement for RTEL1 in multiple pathways to preserve genome stability in plants can be explained by its putative role in the destabilization of DNA loop structures, such as D-loops and T-loops.
INTRODUCTION
Efficient DNA repair is essential to prevent genome instability in all organisms, and several types of DNA repair mechanisms exist to preserve the genome. The basic mechanisms of DNA repair and recombination developed early during evolution and are highly conserved. However, detailed analysis of eukaryotes has revealed that significant variations exist among these mechanisms. For example, certain genes associated with the DNA repair processes are present in some organisms and absent in others. A classic example is the photolyase genes, which are found in plants but are absent in yeast and mammals (Schröpfer et al., 2014a). Furthermore, certain homologous proteins play different roles in either the same or alternative repair pathways in distantly related species.
DNA helicases and translocases are important for DNA replication, repair, and recombination. Mechanistically, these enzymes are involved in the processing of different types of DNA structures that arise during replication, repair, and recombination. Thus, the loss of helicase or translocase activity is typically correlated with a deficiency in the respective pathway. Interestingly, this is not always the case. Specifically, certain helicases and translocases limit the efficiency of homologous recombination (HR) in all organisms. Loss of these helicases results in hyperrecombination and adverse genome rearrangements. The importance of this type of helicase is documented by the fact that their loss of function causes severe genetic diseases and results in a predisposition to different forms of cancer in mammals.
Plants, mammals, and yeast differ considerably in the machinery involved in suppressing hyperrecombination. In yeast, three different helicases have been described that perform this function: the RecQ family homolog Small Growth Supressor1 (SGS1), the DNA helicase SRS2, and the DNA helicase Mutator Phenotype1 (MPH1) (Lawrence and Christensen, 1979; Aguilera and Klein, 1988; Gangloff et al., 1994; Scheller et al., 2000; Prakash et al., 2005). Two of these enzymes also have homologs in mammals: Bloom Syndrome Helicase (BLM; a RecQ helicase and homolog of SGS1) and Fanconi Anemia Group M (FANCM; a homolog of MPH1). Mutation of BLM or FANCM in humans results in severe genetic disorders, including Bloom syndrome and Fanconi anemia, respectively (Fanconi, 1927; German et al., 1965; Ellis et al., 1995; Meetei et al., 2005). Moreover, two additional helicases that have no obvious homolog in yeast have been characterized in mammals: F-Box Helicase1 (FBH1) and Regulator of Telomere Elongation Helicase1 (RTEL1) (Zhu et al., 1998; Ding et al., 2004; Morishita et al., 2005; Barber et al., 2008).
By contrast, plants contain a unique combination of homologs from yeast and mammals. Previously, we identified a homolog of SGS1 and BLM that suppresses HR in Arabidopsis thaliana, RECQ4A (Hartung et al., 2007; Schröpfer et al., 2014b), and a homolog of MPH1/FANCM, At-FANCM (Knoll et al., 2012). Moreover, plants lack an FBH1 homolog, but they contain homologs of SRS2 (found in yeast) and RTEL1 (found in mammals) (Blanck et al., 2009; Knoll and Puchta, 2011).
Interestingly, there are also major differences in the machinery used by yeast, mammals, and plants to repair DNA cross-links (CLs). In mammals, mutations in a number of genes lead to the hereditary disease Fanconi anemia, which is associated with a severe defect in CL repair. Most of these factors are not present in either the Arabidopsis or the yeast genome. Interestingly, FANCM is one of the few Fanconi anemia homologs found in the Arabidopsis genome. However, compared with other factors, it plays only a minor role in CL repair (Knoll et al., 2012; Dangel et al., 2014). We previously showed that the antirecombinase RECQ4A and the nuclease MUS81 are prominent factors involved in CL repair in plants, and they define different subpathways for DNA CL repair (Mannuss et al., 2010). In contrast with yeast, plants have an RTEL1 homolog; therefore, it was of special interest to define the biological function of this protein in Arabidopsis in relation to the suppression of hyperrecombination and CL repair.
It was previously demonstrated in animals that the DNA helicase RTEL1 plays important roles in both DNA repair and telomere stability (Ding et al., 2004; Barber et al., 2008). RTEL1 was first identified in mice by genomic mapping of regions of chromosomes that determine differences in telomere length between Mus musculus and Mus spretus (Zhu et al., 1998). In M. musculus, the loss of RTEL1 leads to embryonic lethality (Ding et al., 2004). The embryos show abnormal cell proliferation, particularly in rapidly proliferating tissues. Moreover, RTEL1 is important for telomere homeostasis. The mutants have shorter telomeres and show end-to-end fusions. Recently, it was shown that RTEL1 dismantles T-loops and counteracts telomeric G4-DNA, which can form G quadruplexes via noncanonical hydrogen bonds in G-rich DNA strands (Vannier et al., 2012). The absence of RTEL1 leads to an inadequate resolution of T-loops via the SLX4 nuclease complex, resulting in telomeric circles and telomere loss. Furthermore, RTEL1 acts in a pathway independent of the RecQ family helicase BLM to suppress G4-DNA structures, facilitate replication of the telomeres, and prevent telomere fragility.
The presence of an iron-sulfur domain places RTEL1 in a small subclass of iron-sulfur cluster-containing DNA helicases together with the helicases XPD, FANCJ, and CHL1 (Rudolf et al., 2006). Mutations in Xeroderma Pigmentosum Group D (XPD), Fanconi Anemia Group J (FANCJ) and Chromosome Loss1 (CHL1) cause the genetic disorders xeroderma pigmentosum, Fanconi anemia, and Warsaw breakage syndrome, respectively (Levitus et al., 2005; Rudolf et al., 2006; van der Lelij et al., 2010). Recently, it was shown that mutations in RTEL1 cause Hoyeraal-Hreidarsson syndrome, a severe variant of dyskeratosis congenita, which is associated with bone marrow failure and a predisposition to cancer (Ballew et al., 2013; Deng et al., 2013; Le Guen et al., 2013; Walne et al., 2013).
In this article, we report the characterization of an RTEL1 homolog in Arabidopsis and its relationship to RECQ4A, FANCM, and MUS81 in replicative and CL DNA repair and HR. Moreover, we also analyze its role in telomere homeostasis. Our data indicate that RTEL1 plays a central role in the preservation of genome stability in plants.
RESULTS
Identification of an RTEL1 Homolog in Arabidopsis
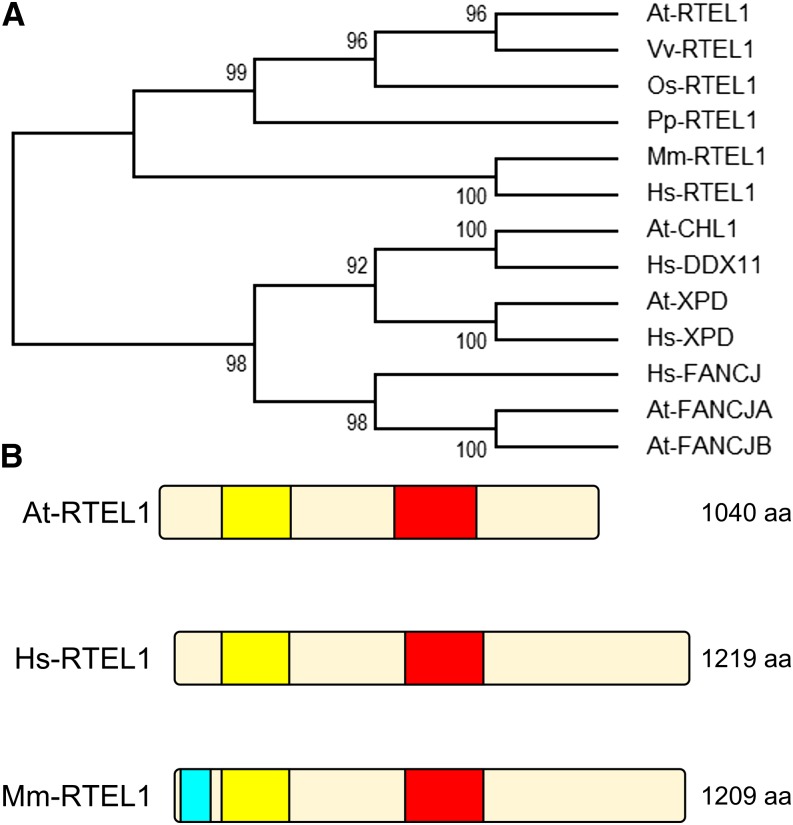
Based on our search for homologs of helicases and translocases involved in the suppression of hyperrecombination, we previously reported the identification of a homolog of RTEL1 in the Arabidopsis genome (Knoll and Puchta, 2011). We extended our analysis and found that this homolog is conserved throughout the plant kingdom and is represented in dicotyledonous Arabidopsis and grape (Vitis vinifera), in the monocotyledonous rice (Oryza sativa), and in the moss Physcomitrella patens (Figure 1A). RTEL1 has a length of 5958 bp and comprises 21 exons and 20 introns (Supplemental Figure 1A). The predicted protein NP_178113.3 has a length of 1040 amino acids, and it contains a bipartite helicase domain, consisting of a DEAD_2 domain (PF06733) near its N terminus and a Helicase_C_2 domain (PF13307) near its C terminus (Figure 1B). To examine the function of RTEL1 in Arabidopsis, we characterized the T-DNA insertion line rtel1-1 (SALK_113285) (Alonso et al., 2003). Based on the sequence of the insertion site, we determined that the T-DNA is inserted in the seventh exon, accompanied by deletions in both left borders (97 and 73 bp) and a deletion of 20 bp in the seventh exon (Supplemental Figure 1B). Furthermore, we identified two additional insertions that flank the T-DNA and have lengths of 24 and 18 bp.
Figure 1.
Identification of RTEL1 in Plants.
(A) Phylogenetic tree of XPD helicase family members. Following the alignment of 13 protein sequences with Clustal Omega, the evolutionary history was inferred using the maximum parsimony method. The support values near the branches represent the percentages of replicate trees in which the associated taxa clustered together in the bootstrap test. At, Arabidopsis thaliana; Hs, Homo sapiens; Mm, M. musculus; Os, O. sativa; Pp, P. patens; Vv, V. vinifera.
(B) Domain composition of Arabidopsis, human, and M. musculus RTEL1 proteins. All three proteins contain a DEAD_2 (yellow) and a Helicase_C_2 (red) domain. In mouse RTEL1, a type III restriction enzyme, res subunit (light blue), can also be found at the N terminus. aa, amino acids.
Insertion of the T-DNA in the RTEL1 locus should disrupt gene expression. To test this, total RNA of 2-week-old wild-type and rtel1-1 plantlets was extracted and reverse transcribed. Relative expression was tested by quantitative PCR (qPCR) at three positions in the RTEL1 locus: 5′ of the insertion site, 3′ of the insertion site, and across the inserted T-DNA (Supplemental Figure 1C). Compared with the wild type, hardly any expression across the insertion could be measured for rtel1-1. 5′ of the T-DNA, RTEL1 expression in rtel1-1 was at 85% of the wild-type level, while expression 3′ of the insertion was at ∼9% in rtel1-1. The T-DNA insertion in rtel1-1, therefore, abolished the expression of a large part of the gene. In principle, it is possible that a fragment of RTEL1 corresponding to the first six exons is expressed at a level slightly lower than in the wild type in rtel1-1.
To demonstrate that the phenotype of the mutation is indeed due to the insertion within the RTEL1 open reading frame, we cloned a complementation construct with the RTEL1 full-length gene under the control of the natural promoter and terminator, including the 5′ and 3′ untranslated regions, respectively. We transformed the mutant line rtel1-1 with this RTEL1 wild-type construct. Three complemented lines (rtel1-1:RTEL1#1, rtel1-1:RTEL1#2, and rtel1-1:RTEL1#3) were used as controls for complementation in the experiments described below.
In these lines, the expression of RTEL1 was also analyzed by qPCR. The primers were placed 5′ and 3′ of the T-DNA insertion site in rtel1-1 so that amplification of a PCR product was possible from the wild-type RTEL1 locus as well as the transgenic RTEL1 genes in the complementation lines, but not from the mutant locus. We found that RTEL1 expression in all three complementation lines was at a similar level to that of the wild type (Supplemental Figure 2A).
It has been shown before in transcriptome studies that RTEL1 expression is not altered by the induction of genotoxic stress (Chen et al., 2003; Kilian et al., 2007; Winter et al., 2007). Nevertheless, to exclude the possibility that the mutation of other factors involved in genome stability might affect the expression of RTEL1 directly or indirectly, gene expression analysis of this locus was also performed by qPCR in the background of all other mutant lines used in this study: recq4A-4, mus81-1, fancm-1, and the telomerase-deficient mutant line tert. We did not find significant changes in the expression of RTEL1 when any of these genes were mutated (Supplemental Figure 2B).
RTEL1 Suppresses Spontaneous HR
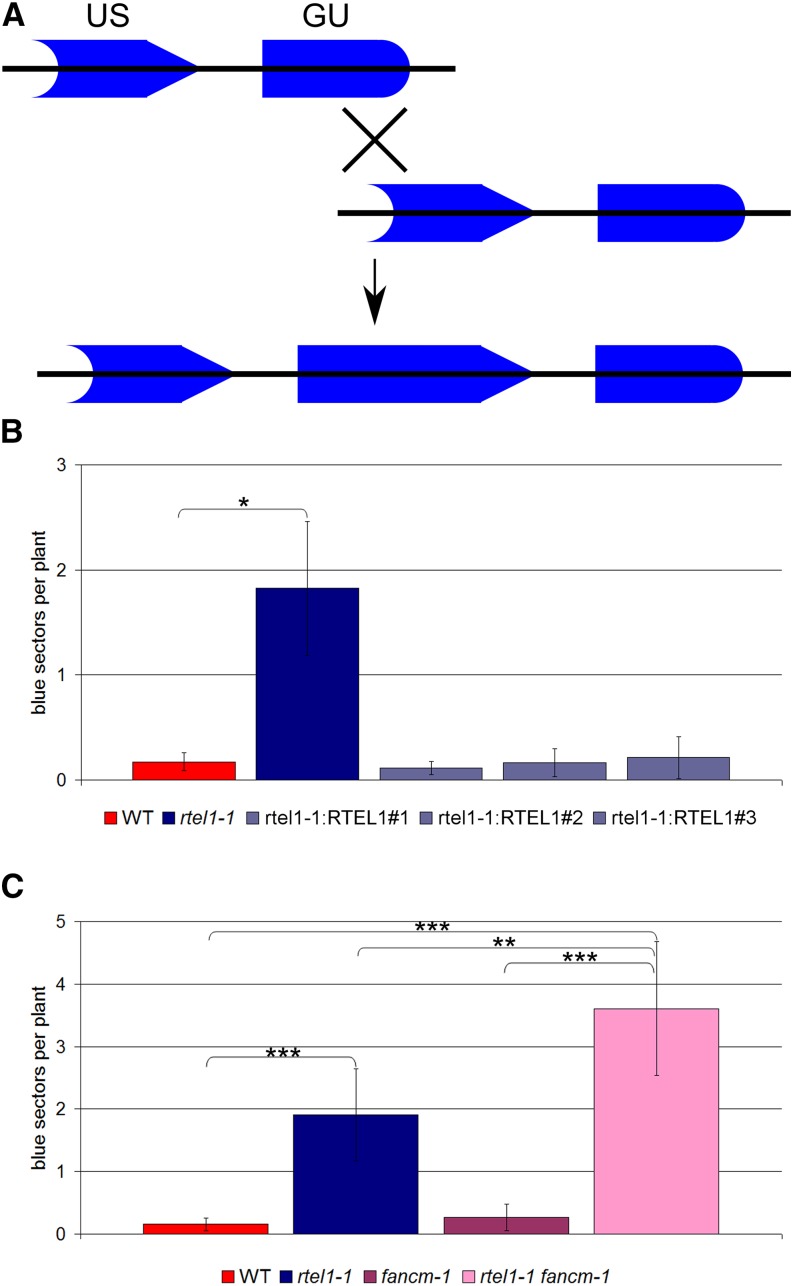
To examine the function of RTEL1 in spontaneous somatic HR events, we crossed the rtel1-1 line with the reporter line IC9 (Molinier et al., 2004). The reporter construct is composed of two nonfunctional parts of the GUS (for β‑glucuronidase) gene with overlapping homologous regions (Figure 2A). Only interchromosomal HR can restore the GUS gene. After histological staining, the recombination frequency was quantified by determining the number of blue sectors on the plantlets.
Figure 2.
Somatic HR Frequencies.
(A) Structure of the HR reporter construct of the IC9 line. Two fragments of the GUS gene (GU and US) with homology to each other are separated by a spacer sequence. Through intermolecular recombination events, a functional GUS gene can be restored. All recombined cells and their daughter cells can be quantified by histological staining.
(B) Mean recombination frequencies of the wild type, rtel1-1, and three complemented lines, rtel1-1:RTEL1#1, rtel1-1:RTEL1#2, and rtel1-1:RTEL1#3 (n = 3).
(C) Mean recombination frequencies of the wild type, rtel1-1, fancm-1, and the double mutant rtel1-1 fancm-1 (n = 8).
Columns in (B) and (C) correspond to mean values, and error bars represent sd. Statistical differences were calculated by the Mann-Whitney test: *P < 0.05, **P < 0.01, ***P < 0.001.
In comparison with the wild-type line, the recombination frequency of the mutant line rtel1-1 was elevated by 1 order of magnitude, whereas the rtel1-1:RTEL1 complemented lines exhibited HR frequencies comparable to that of the wild type (Figure 2B). Thus, loss of RTEL1 increased the recombination frequency in the mutant line, and we demonstrated that the observed phenotype is due to the loss of function of the RTEL1 gene and not to a secondary mutation.
RTEL1 and FANCM Act in Parallel Pathways to Suppress HR
We showed previously that the translocase FANCM and the helicase RECQ4A suppress somatic HR events in two different pathways in Arabidopsis (Hartung et al., 2007; Knoll et al., 2012). In this study, we were interested in defining the function of RTEL1 in relation to RECQ4A and FANCM in Arabidopsis. Therefore, we created the double mutants rtel1-1 fancm-1 and rtel1-1 recq4A-4 in the IC9 reporter background. The HR frequency in rtel1-1 fancm-1 was higher than that in either of the single mutants (Figure 2C). Thus, RTEL1 and FANCM counteract HR through two different pathways.
To examine whether RTEL1 and RECQ4A also act independently, we created the double mutant line rtel1-1 recq4A-4 by crossing the individual mutants. However, the double mutant exhibited severe developmental defects (see below) that made it impossible to examine the HR frequency in comparison with the wild type and both single mutants.
RTEL1 Is Involved in Intrastrand CL Repair
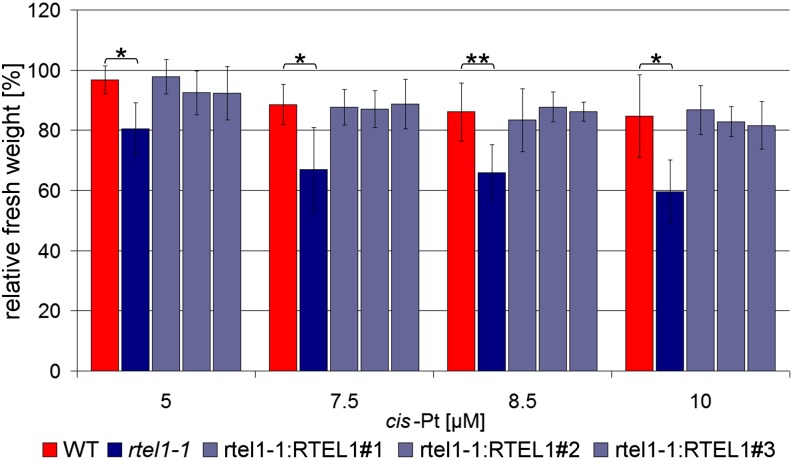
To investigate whether RTEL1 plays a role in CL repair, we tested rtel1-1 for hypersensitivity to the CL-inducing agents mitomycin C (MMC) and cis-platin. While MMC mostly induces interstrand CLs, cis-platin mainly causes intrastrand CLs (Rink et al., 1996; Boulikas and Vougiouka, 2003). To examine the sensitivity to the cross-linking agents, we performed a liquid medium assay and determined the relative fresh weight of 3-week-old plantlets, in which the fresh weight of the treated plantlets was normalized to the fresh weight of untreated plantlets of the same line. We did not observe a hypersensitivity of rtel1-1 against MMC. However, the mutant line rtel1-1 displayed a growth defect that was more severe than the wild type after cis-platin treatment. The complemented lines exhibited a relative fresh weight similar to that of the wild type (Figure 3). Thus, we demonstrated that the hypersensitive phenotype against intrastrand CLs is indeed caused by defective RTEL1.
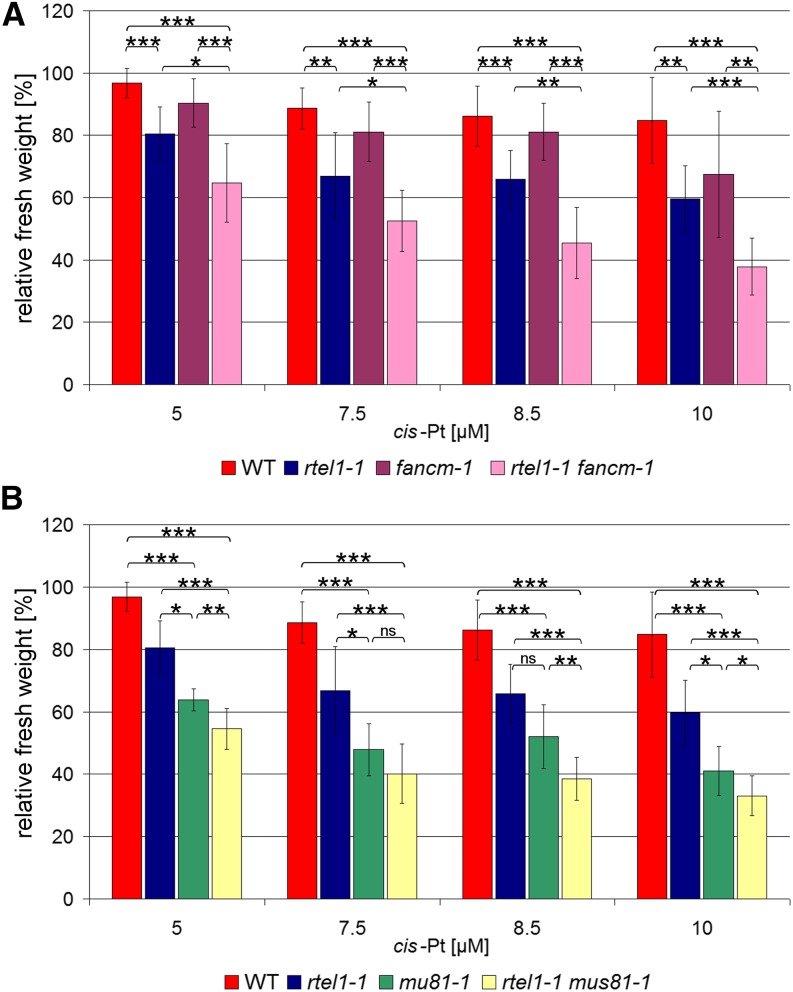
Figure 3.
Complementation of rtel1-1 Sensitivity against Treatment with cis-Platin.
The fresh weight relative to untreated controls after treatment with 5, 7.5, 8.5, and 10 μM cis-platin is shown. In the rtel1-1 single mutant, the fresh weight is significantly reduced compared with the wild type in all concentrations. The complementation lines rtel1-1:RTEL1#1, rtel1-1:RTEL1#2, and rtel1-1:RTEL1#3 have fresh weights similar to the wild type but different from rtel1-1. Columns correspond to mean values (n = 3), and error bars represent sd. Statistical differences were calculated by the Mann-Whitney test: *P < 0.05, **P < 0.01.
RTEL1 and MUS81 Act in the Same Pathway during Interstrand CL Repair, Parallel to FANCM
Previously, we showed that RECQ4A, MUS81, and FANCM are involved in the repair of interstrand CLs in Arabidopsis and that all of these proteins act in different pathways (Mannuss et al., 2010; Dangel et al., 2014). To examine the role of RTEL1, we examined the double mutants rtel1-1 mus81-1 and rtel1-1 fancm-1 after MMC treatment. To analyze the sensitivity of rtel1-1 fancm-1, we determined the relative fresh weight of the double mutant and compared it with the relative fresh weights of both single mutants and the wild type. After MMC treatment (5, 10, 15, and 20 μg/mL), we did not observe any statistically significant differences between rtel1-1, fancm-1, and the wild type. Nevertheless, we did detect hypersensitivity against MMC in the double mutant (Figure 4A). Therefore, although the phenotype was hidden in the single mutants, our results indicate that both RTEL1 and FANCM are involved in interstrand CL repair and that they act in parallel pathways. For Arabidopsis, it is already known that the mutant line mus81-1 is hypersensitive against treatment with MMC (Hartung et al., 2006). After MMC treatment, the relative fresh weight of the double mutant rtel1-1 mus81-1 was similar to that of the single mutant mus81-1 (Figure 4B). These results suggest that RTEL1 and MUS81 work in the same pathway to repair interstrand CLs in Arabidopsis.
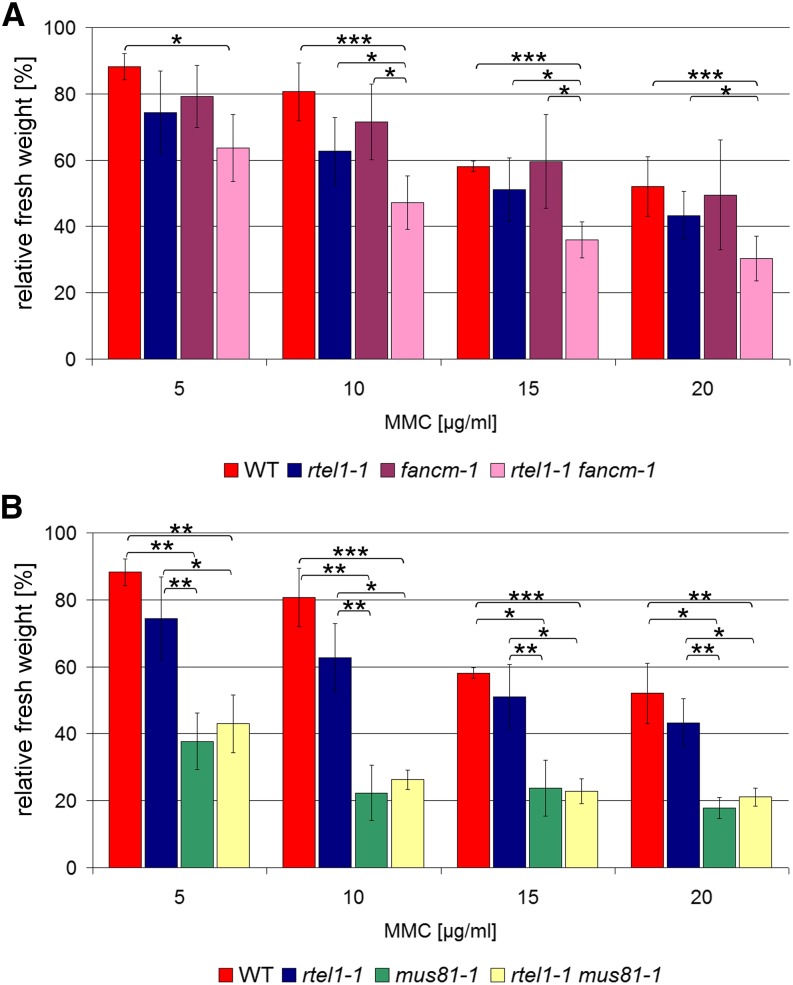
Figure 4.
Sensitivity against MMC-Induced DNA Interstrand CLs.
Fresh weights relative to untreated controls after treatment with 5, 10, 15, and 20 μg/mL MMC are shown.
(A) No statistically significant difference was detected between the wild type and rtel1-1 or fancm-1, but the rtel1-1 fancm-1 double mutant had a significantly lower fresh weight than did either single mutant or the wild type (n = 4).
(B) At all tested concentrations, the single mutant mus81-1 and the double mutant rtel1-1 mus81-1 had similar relative fresh weights that were significantly lower than those of the wild type and rtel1-1 single mutants (n = 3).
Columns correspond to mean values, and error bars represent sd. Statistical differences were calculated by the Mann-Whitney test: *P < 0.05, **P < 0.01, ***P < 0.001.
RTEL1, FANCM, and MUS81 Act in Different Pathways during Intrastrand CL Repair
To investigate the function of RTEL1 in intrastrand CL repair in relation to FANCM and MUS81, we examined the double mutants and the corresponding single mutants after cis-platin treatment (5, 7.5, 8.5, and 10 μM). The double mutant rtel1-1 fancm-1 exhibited an increased sensitivity compared with the hypersensitive phenotype of rtel1-1 (Figure 5A). Therefore, RTEL1 and FANCM appear to act in parallel pathways during intrastrand CL repair in Arabidopsis.
Figure 5.
Sensitivity against cis-Platin-Induced DNA Intrastrand CLs.
Fresh weights relative to untreated controls after treatment with 5, 7.5, 8.5, and 10 μM cis-platin are shown.
(A) While there was no difference in sensitivity against cis-platin in fancm-1 compared with the wild type, the single mutant rtel1-1 was more sensitive than the wild type at all tested concentrations. The rtel1-1 fancm-1 double mutant was significantly more sensitive against cis-platin treatment than either of the corresponding single mutants (n = 8).
(B) Both the rtel1-1 and mus81-1 single mutants were more sensitive against cis-platin at all tested concentrations. In contrast with treatment with MMC, after cis-platin treatment the rtel1-1 mus81-1 double mutant line was significantly more sensitive than either of the single mutants at three of the four tested concentrations (n = 8).
Columns correspond to mean values, and error bars represent sd. Statistical differences were calculated by the Mann-Whitney test: *P < 0.05, **P < 0.01, ***P < 0.001.
Previously, it was demonstrated that MUS81 is involved in the repair of interstrand as well as intrastrand CLs (Hartung et al., 2006; Mannuss et al., 2010). We determined the relative fresh weight of rtel1-1 mus81-1 after cis-platin treatment and compared it with that of the single mutants and that of the wild type. Contrary to the phenotype after MMC treatment, the double mutant showed an increased hypersensitivity compared with both single mutants (Figure 5B). Thus, RTEL1 and MUS81 act in different pathways during intrastrand CL repair.
RTEL1 Is Important for Proper Root Development and the Repair of Replication-Associated DNA Damage
We demonstrated that RTEL1 is involved in processing DNA repair intermediates that arise during artificially induced DNA CLs. It was tempting to speculate that under such conditions, stalled replication forks might arise and RTEL1 might also participate in the repair of replication-associated DNA damage in general. Therefore, we analyzed whether any specific features could be detected in dividing tissues from the mutant plant by a microscopic examination. The root meristem is composed of undifferentiated dividing cells. Therefore, this tissue is particularly suitable for investigating the effect of the loss of specific genes on cell division and cell differentiation. In Arabidopsis, root stem cells are located in a stem cell niche (SCN), adjacent to which is a region of transiently amplifying (TA) cells. The rate of root growth depends on the number of cycling cells (Beemster and Baskin, 1998). TA cells arise from cell divisions in the SCN. The different types of stem cells and the TA cells are determined based on their positions relative to the quiescent center (QC) in the SCN. The stem cells are located approximately five to six cell tiers above the root tip and remain undifferentiated due to contact with the QC. Cells of the QC rarely divide. Differentiated root cells arise from four different types of stem cells: epidermal/lateral root cap stem cells, cortical/endodermal stem cells, columella stem cells, and vascular stem cells (Scheres et al., 2002).
To analyze the involvement of RTEL1 in root development and replication, we stained the roots of 5-d-old plantlets with propidium iodide (PI). PI is excluded from live cells, dead cells are stained intensely red, and the intercellular space appears slightly red (Curtis and Hays, 2007). We examined root tissues from the previously described mutant lines and from wild-type plants.
For statistical analysis, we determined the frequency of roots showing at least one dead vascular stem cell, TA cell, or nonvascular stem cell. Furthermore, we also checked for roots with aberrant SCNs.
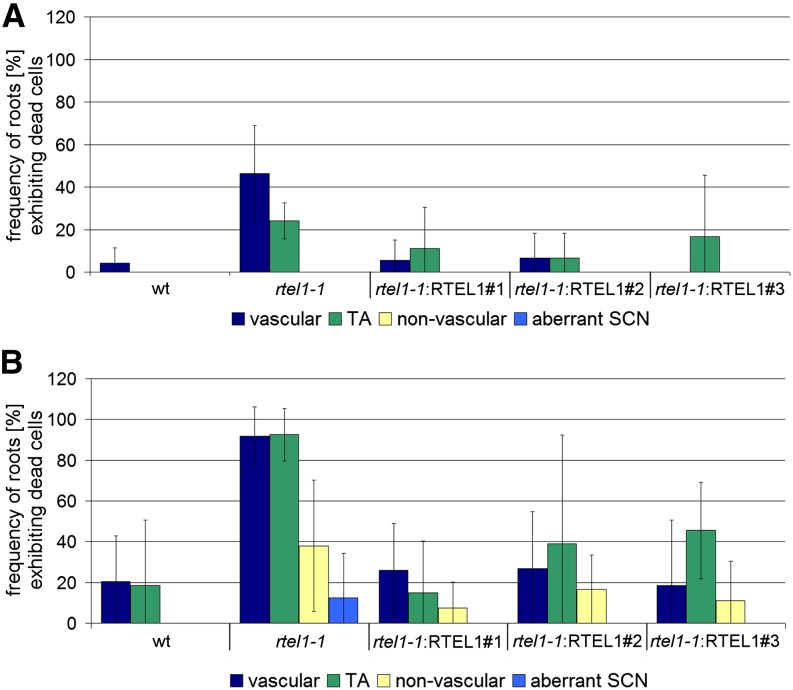
Very few of the roots from the wild-type plants presented dead vascular stem cells (∼4%), and no dead TA cells were observed. By contrast, an increased frequency of roots with at least one dead vascular stem cell was detected in rtel1-1 (∼46%). Furthermore, we observed an increased frequency of roots with dead TA cells (∼24%; Figure 6A). To test whether we could reproduce the defects in the repair of intrastrand CLs using the fresh weight of whole seedlings, we also analyzed the roots of plants treated with cis-platin. As expected, after cis-platin exposure, the wild-type and rtel1-1 plants showed a higher frequency of roots with dead stem cells compared with the corresponding untreated lines. Additionally, we observed roots with dead nonvascular stem cells in the rtel1-1 line. Interestingly, in rtel1-1 roots but not in wild-type roots treated with cis-platin, the regular layering of the SCN, including the QC, was lost. Again, it was possible to complement these phenotypes of rtel1-1: after transformation with the wild-type RTEL1 construct, the lines behaved like wild-type plants and exhibited a similar frequency of roots with dead stem cells (Figure 6B).
Figure 6.
Complementation of Stem Cell Death in the rtel1-1 Root Tip.
After staining root tips with PI, the frequency of roots containing at least one dead stem cell belonging to the vascular, TA, or nonvascular stem cell pools was determined. In addition, roots with aberrant layering of the SCN were quantified. Experiments were performed either without (A) or with 35 μM cis-platin (B). The complementation lines rtel1-1:RTEL1#1, rtel1-1:RTEL1#2, and rtel1-1:RTEL1#3 showed a lower frequency of stem cell death, comparable to wild-type roots. In rtel1-1, the numbers of dead vascular cells and TA cells were higher than in the wild type. After cis-platin treatment, dead nonvascular stem cells and aberrant layering of the SCN were detected in rtel1-1 but not in the wild type. Columns correspond to mean values (n = 3), and error bars represent sd.
RTEL1 Is Involved in a Different Pathway from FANCM in the Repair of DNA Replication Defects
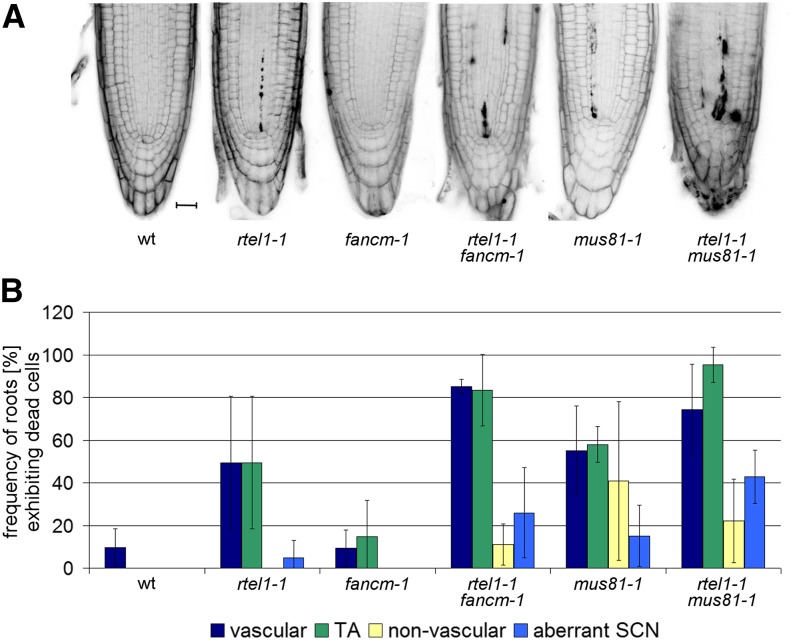
We conducted a cytological analysis of the root meristem in double mutants of rtel1-1 with fancm-1 or mus81-1. The mutant line fancm-1 behaved similarly to the wild type, and we did not observe an increased frequency of roots with dead cells in this mutant (wild type, ∼10%; fancm-1, ∼9%). The rtel1-1 fancm-1 double mutant had a higher proportion of roots with dead vascular stem cells (∼85%) and dead TA cells (∼83%) than the rtel1-1 single mutant (∼50% vascular and ∼50% TA cells). Furthermore, in the rtel1-1 single mutant, we observed more roots with dead nonvascular stem cells (∼5%), and we identified more roots with aberrant SCNs (∼26%). Thus, RTEL1 and FANCM are likely to be involved in root development and to contribute to the repair of aberrant replication structures through two different pathways (Figures 7A and 7B).
Figure 7.
Epistasis Analysis of Root Tip Stem Cell Death.
To elucidate the interplay between RTEL1, FANCM, and MUS81 in replication-associated DNA repair, root tip stem cell death was analyzed in single and double mutants of the respective genes.
(A) Representative micrographs of PI-stained root tips from the wild type, rtel1-1, fancm-1, and mus81-1 single mutants, and rtel1-1 fancm-1 and rtel1-1 mus81-1 double mutants under uninduced conditions. Bar = 20 μm.
(B) Quantification of roots containing at least one dead stem cell belonging to the vascular, TA, or nonvascular stem cell pool under uninduced conditions. Columns correspond to mean values (n = 3), and error bars represent sd.
We also tested whether we could reproduce the findings reported above that both enzymes act independently in the repair of intrastrand CLs. As expected, this was indeed the case. After cis-platin treatment, the frequency of roots with vascular stem cells in the fancm-1 line (∼30%) was comparable to that of the wild-type line (∼24%). However, we observed an increased proportion of roots with dead TA cells (∼59%) and roots with dead nonvascular cells (∼29%). In the rtel1-1 fancm1-1 double mutant, every root examined showed at least one dead vascular stem cell and one dead TA cell after treatment with cis-platin. The frequency of roots with dead nonvascular stem cells was also increased (∼54%; Supplemental Figure 3).
RTEL1 Is Involved in a Different Pathway from MUS81 in the Repair of DNA Replication Defects
The rtel1-1 and mus81-1 single mutants showed a comparable frequency of roots with dead vascular stem cells (∼50 and ∼55%, respectively) and dead TA cells (∼50 and ∼58%, respectively). In addition, the mus81-1 mutant line exhibited an increased frequency of roots with dead nonvascular stem cells (∼41%). Interestingly, we observed more roots with aberrant SCNs than in the rtel1-1 line. In the double mutant, ∼40% of the roots exhibited aberrant SCNs. The frequencies of roots with at least one dead vascular stem cell and one dead TA cell were ∼75 and ∼95%, respectively (Figures 7A and 7B).
We also tested the interaction between RTEL1 and MUS81 in intrastrand CL repair using cis-platin. Indeed, in the rtel1-1 mus81-1 double mutant, every root examined had at least one dead vascular stem cell and one dead TA cell. The frequencies of roots with dead nonvascular stem cells and roots with aberrant SCNs were higher than in either single mutant. This result confirms that MUS81 and RTEL1 have nonredundant functions in CL repair (Supplemental Figure 3).
Loss of RTEL1 and RECQ4A Causes Severe Developmental Defects and Very Early Stem Cell Death
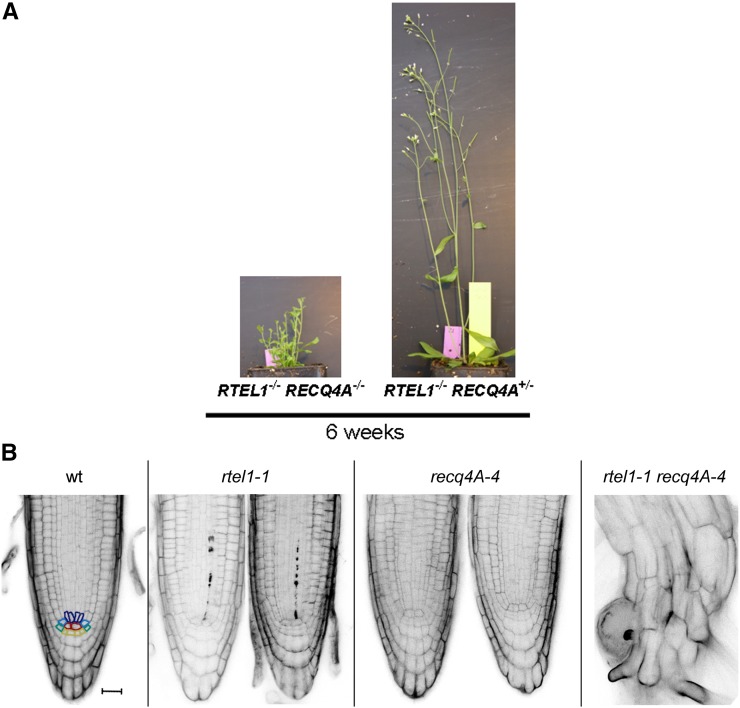
Surprisingly, the rtel1-1 recq4A-4 double mutant presented severe developmental defects. During vegetative growth, the rosette leaves were visibly smaller than those of both single mutants and the wild type. After 6 weeks of cultivation in soil, the wild-type line, both single mutant lines, and the homozygous double mutant line had developed shoots. However, the rtel1-1 recq4A-4 double mutant plants developed much shorter shoots without any flowers or siliques. Because the growth of the rtel1-1−/− recq4A-4+/− plants was not different from that of the wild-type plants, only the loss of both helicases leads to such severe defects (Figure 8A). Therefore, it was not possible to determine the HR frequencies or the relative fresh weights after genotoxin treatment of these plants. Nevertheless, it was possible to characterize the double mutant by studying the root meristem. Therefore, we stained the roots with PI and compared them with those of the corresponding single mutants and those of the wild type (Figure 8B). Contrary to the rtel1-1 mutant line, we did not observe an increased frequency of dead stem cells in the recq4A-4 mutant. The single mutant line recq4A-4 had an increased frequency of dead cells in the roots compared with the wild-type line only after cis-platin treatment (Supplemental Figure 4). The rtel1-1 recq4A-4 double mutant line exhibited extremely short roots. The root morphology in these plants was totally compromised, and a comparison with the other plant lines was not possible. Loss of both RTEL1 and RECQ4A apparently caused very early stem cell death, and there were very few cells remaining that could be stained. Differentiation between live and dead cells was not possible. Due to this severe phenotype, we could not quantify the frequency of roots with at least one dead stem cell. The massive degeneration of the roots indicates that RECQ4A and RTEL1 have no overlapping essential functions during DNA replication.
Figure 8.
Severe Growth Defect Due to the Concurrent Loss of RTEL1 and RECQ4A.
(A) After 6 weeks of cultivation in soil, the rtel1-1 recq4A-4 double mutant was much smaller than the rtel1-1−/− recq4A-4+/− line. Loss of both helicases led to smaller rosette leaves, shorter shoots, and the absence of true flowers or siliques.
(B) PI-stained root tips presented no stem cell death in the wild type or the recq4A-4 mutant, but a few dead stem cells were present in the rtel1-1 mutant. The double mutant had severely short roots with a complete loss of root structure. Bar = 20 μm.
[See online article for color version of this figure.]
RTEL1 Is Involved in Telomere Homeostasis
Previously, it was shown in humans and in mice that RTEL1 is important for telomere maintenance. In mice, RTEL1 dismantles T-loops and counteracts G4-DNA. Loss of RTEL1 leads to rapid telomere shortening (Ding et al., 2004; Vannier et al., 2012). In humans, mutations in RTEL1 cause Hoyeraal-Hreidarsson syndrome, which leads to telomere instability (Ballew et al., 2013; Deng et al., 2013; Le Guen et al., 2013; Walne et al., 2013).
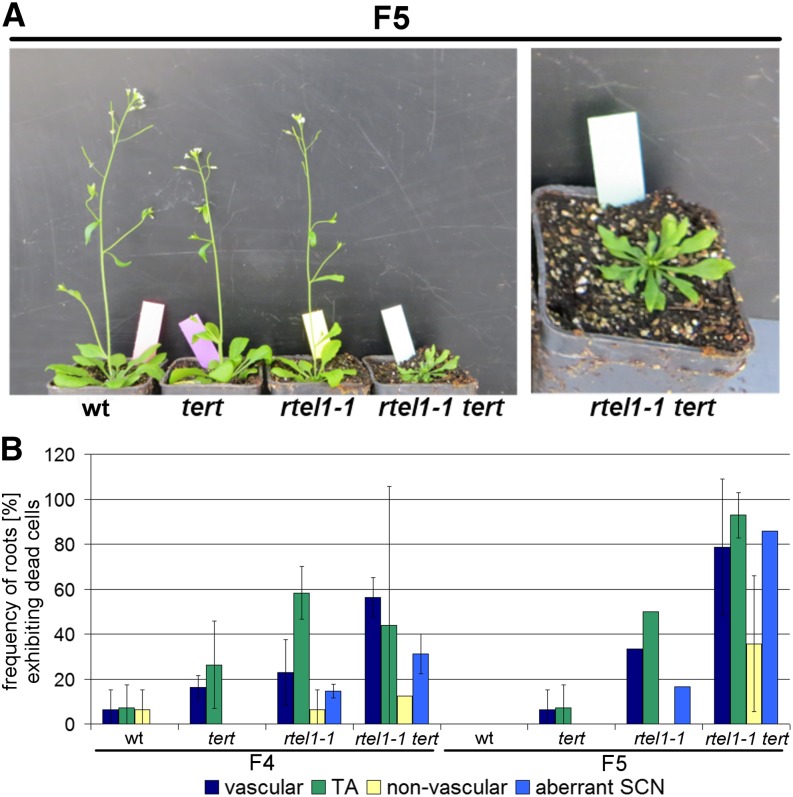
To define the role of RTEL1 in telomere maintenance in Arabidopsis, we crossed rtel1-1 with the telomerase-deficient mutant line tert (Fitzgerald et al., 1999). In Arabidopsis, the loss of TERT leads to gradual telomere shortening. After five generations, the plants present defects in shoot and leaf morphology. These developmental defects worsen gradually in successive generations and cause a total developmental arrest after 10 to 11 generations (Riha et al., 2001). Here, we studied the development of the double mutant line rtel1-1 tert in successive generations and compared it with the single mutant lines and the wild type of the corresponding generations. The F2 generation was the first generation evaluated in the experiment, in which all possible genotypes segregating from the F1 generation were selected. Thus, the first generation when seedlings with homozygous T-DNA insertions in RTEL1 and TERT could be obtained was the F2 generation. In generations F3 and F4, we did not observe any phenotypical differences between the double mutant, the single mutants, and the wild-type plants (Supplemental Figure 5). In the F5 generation, the single mutants and the wild type still developed similarly (Figure 9A). However, we observed severe developmental defects in the rtel1-1 tert double mutant line. These plants exhibited an aberrant shoot and leaf morphology and were considerably reduced in size. Thus, in contrast to the tert single mutant line, the development of the double mutant line was arrested already after the fourth homozygous generation (F5).
Figure 9.
Developmental Defect after Four Homozygous Generations in rtel1-1 tert.
(A) In the F5 generation, the fourth generation of plants homozygous for both the rtel1-1 and tert mutations, the plants never developed past the rosette stage, while both single mutants and the wild type grew normally in soil.
(B) Quantification of the roots of the wild-type and the tert, rtel1-1, and rtel1-1 tert mutants in F4 and F5 containing at least one dead vascular, TA, or nonvascular stem cell under uninduced conditions. Columns correspond to mean values (n = 3), and error bars represent sd.
Loss of RTEL1 and TERT Leads to Aberrant Development of the SCN
To further define the mutant phenotype, we analyzed cell death in the root meristem of the rtel1-1, tert, and rtel1-1 tert mutant lines and of the wild type (Figure 9B). Both the F4 and the F5 generations showed no differences in root development between wild-type and tert lines. In the F4 generation, the rtel1-1 tert double mutant exhibited a similar frequency of roots with dead stem cells to the rtel1-1 single mutant. However, the F5 generation of the rtel1-1 tert line, which displayed general and developmental growth defects, presented a higher frequency of roots with at least one dead stem cell than the rtel1-1 line. Moreover, we observed more roots with aberrant SCNs in both generations of the double mutant than in the corresponding single mutant line rtel1-1. The cis-platin treatment increased the proportion of roots with dead stem cells in all lines (Supplemental Figure 6).
Loss of RTEL1 and TERT Causes Rapid Telomere Shortening
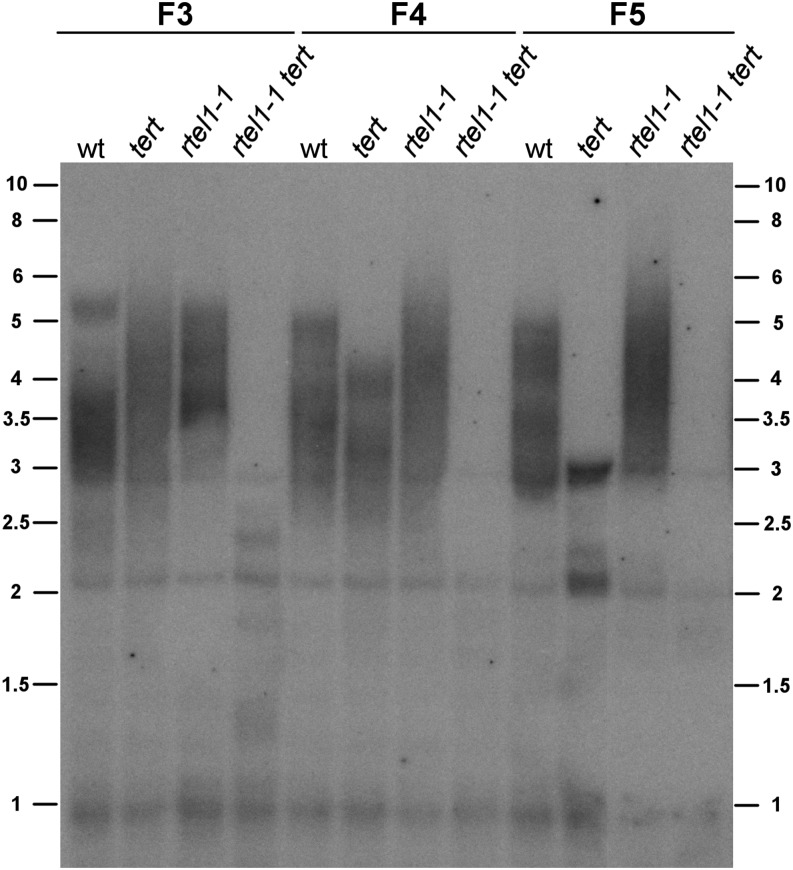
To investigate the role of RTEL1 in telomere stability in more detail, we performed terminal restriction fragment (TRF) analyses with the rtel1-1 tert double mutant line, the corresponding single mutant lines, and the wild-type line in the F3, F4, and F5 generations. Previously, TRF analysis demonstrated that the loss of TERT leads to gradual telomere shortening (Riha et al., 2001). We detected telomere lengths of ∼5 kb in the F3 generation and ∼4.5 kb in the F4 generation in the tert line. In the F5 generation of the tert line, we detected a telomere length of ∼3 kb. The wild-type plants exhibited a telomere length of ∼5 kb in all generations. In the rtel1-1 mutant line, the telomeres were even longer than in the wild type (∼6.5 kb), but they maintained their length in all generations. Due to a rapid loss of telomeres in the double mutant line, only very short TRF signals could be detected in the F3 generation (∼3 kb). Finally, in the F5 generation, we detected only weak TRF signals of ∼2 kb (Figure 10).
Figure 10.
Telomere Length Analysis by TRF.
Telomere length is shown for the wild type and the tert, rtel1-1, and rtel1-1 tert genomic DNA from generations F3, F4, and F5. The telomere length did not change in the wild type or in rtel1-1. The telomeres gradually shortened over the generations in the tert mutant, as reported previously. In the rtel1-1 tert double mutant, the telomere length shortened rapidly, even after one homozygous generation (F3). The bands at 2 and 3 kb are derived from interstitial telomeric sequences.
DISCUSSION
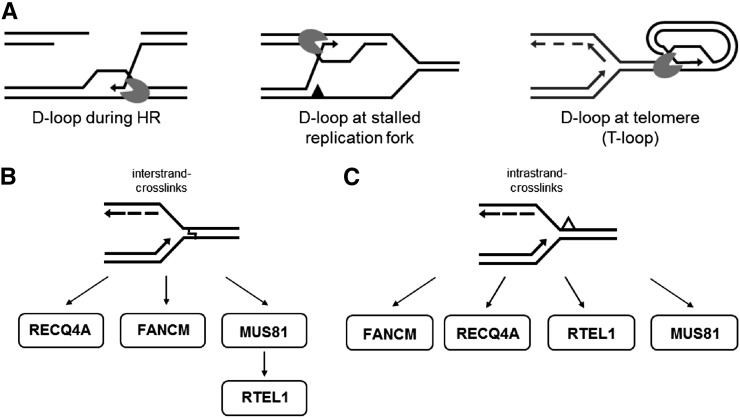
RTEL1 plays a crucial role in DNA repair and telomere homeostasis in humans and mice. Moreover, investigation of the RTEL1 homolog in Caenorhabditis elegans suggests that RTEL1 plays an important role in the suppression of hyperrecombination and is essential for the processing of DNA repair intermediates (Barber et al., 2008). Here, we demonstrate that the RTEL1 homolog in Arabidopsis is involved in preserving genome stability in plants through multiple pathways. In principle, all of its diverse roles can be explained by the ability of the helicase to disassemble D-loop-like recombination intermediates (Figure 11A).
Figure 11.
Model of RTEL1 Functions in Arabidopsis.
(A) At D-loop-like structures that can be found during HR, at stalled replication forks, and at the telomeres, antirecombinases such as RTEL1 disrupt the D-loop by rejecting the invaded single strand. This opens the end of the chromosome for replication in the case of telomeres, or it enables annealing to the second end of a double strand break for repair via the synthesis-dependent strand-annealing pathway in the case of HR.
(B) During interstrand CL repair, RTEL1 acts downstream of MUS81, in a parallel pathway to FANCM and RECQ4A.
(C) During intrastrand CL repair, RTEL1 and MUS81 function in parallel pathways. Other pathways involve FANCM and RECQ4A.
RTEL1 Suppresses HR Events in a Parallel Pathway to That of FANCM
The increased recombination frequency in the mutant line rtel1-1 compared with the wild type suggests that RTEL1 plays an important role in suppressing somatic HR. At-RTEL1 may act similarly to the RTEL1 homolog in C. elegans (Barber et al., 2008; Youds et al., 2010) and disassemble D-loop recombination intermediates to promote the synthesis-dependent strand-annealing pathway.
Previously, the helicase/translocase FANCM and the RecQ helicase RECQ4A were shown to be important HR antagonists, and the loss of both FANCM and RECQ4A results in hyperrecombination in Arabidopsis (Hartung et al., 2007; Knoll et al., 2012). Furthermore, the concurrent loss of FANCM and RECQ4A leads to a higher recombination frequency compared with both single mutants (Knoll et al., 2012). Our analyses of the double mutant rtel1-1 fancm-1 revealed a strongly increased HR frequency compared with both single mutants. According to these results, RTEL1 and FANCM appear to act in different pathways to suppress somatic HR. The severe developmental defects in the rtel1-1 recq4A-4 line precluded phenotypic analysis but might directly suggest that RTEL1 and RECQ4A also act in different pathways to suppress HR.
This analysis demonstrates that Arabidopsis has at least three unrelated DNA helicases with antirecombinase functions: RECQ4A, FANCM, and RTEL1. Indeed, we were also able to identify an SRS2 homolog in plants (Blanck et al., 2009). Biochemical analysis revealed that SRS2 and RTEL1 are able to resolve similar recombination intermediates, and they were postulated to be functional analogs of each other (Barber et al., 2008). We expressed At-SRS2 in Escherichia coli and reported that the protein is a very efficient 3′ to 5′ helicase (Blanck et al., 2009). However, we did not detect any defect in DNA repair or DNA recombination in many of the available SRS2 mutants. It is possible that the T-DNA insertion mutants used in these preliminary studies do not represent a null phenotype, and it may only be possible to produce true knockouts by applying the recently established CRISPR/Cas system (Fauser et al., 2014). It is also possible that the helicase SRS2 of Arabidopsis has developed new unrelated functions during evolution in comparison with RTEL1 that were not detected in our analysis.
RTEL1 Is Involved in Interstrand CL Repair Independently of FANCM and Shares One Pathway with MUS81
We did not find any differences between rtel1-1 and the wild type after MMC treatment. Nevertheless, we did find that RTEL1 participates in interstrand CL repair, because the double mutant rtel1-1 fancm-1 was hypersensitive against MMC treatment. These results indicate the existence of several pathways involved in the repair of interstrand CLs. The loss of one pathway is compensated for by an alternative pathway, so the single mutants show no difference from the wild-type line. Only the loss of at least two proteins leads to a notable phenotype. After MMC treatment, we determined a relative change in the fresh weight of between 60 and 30% in the rtel1-1 fancm-1 double mutant line. These results suggest that other pathways are still able to repair interstrand CLs. Interestingly, prior examinations showed that RECQ4A and MUS81 play an important role in CL repair (Mannuss et al., 2010). Furthermore, recent studies suggested that FANCM acts in a RECQ4A-independent pathway during interstrand CL repair (Dangel et al., 2014). The rtel1-1 mus81 double mutant did not show higher sensitivity than either of the single mutants, and the fresh weight was comparable to that of the mus81-1 mutant. Taking these results together, we propose that, during interstrand CL repair, RTEL1 acts in the same pathway as MUS81, which is parallel to the FANCM-dependent and RECQ4A-dependent pathways. A model of these pathways is depicted in Figure 11B.
During Intrastrand CL Repair, RTEL1 Acts in a Parallel Pathway to That of FANCM and MUS81
The hypersensitive phenotype of rtel1-1 after cis-platin treatment revealed that RTEL1 plays a direct role in intrastrand CL repair. The double mutant rtel1-1 fancm-1 exhibited a higher sensitivity than the single mutant rtel1-1. These findings suggest that RTEL1 and FANCM function in parallel pathways during intrastrand CL repair. Furthermore, we observed a higher sensitivity in the rtel1-1 mus81-1 double mutant than in the corresponding single mutants after cis-platin treatment. Thus, RTEL1 and MUS81 act in different pathways during intrastrand CL repair as well. This result may seem surprising at first, as our prior findings show a common pathway for RTEL1 and MUS81 during interstrand CL repair. However, interstrand and intrastrand CLs are different types of lesions in which either one or both DNA strands are severely damaged. Therefore, these structures should also require different types of DNA processing to resolve the damage, at least to a certain extent. Alternatively, one could also speculate that, during interstrand CL repair, the damage must be cleaved by MUS81 first to provide a possible substrate for RTEL1 or another helicase. This would also explain the phenotype of rtel1-1 mus81-1 after MMC treatment: due to the loss of MUS81, another pathway is chosen, and the loss of RTEL1 is not detectable.
RTEL1 Is Involved in the Repair of Aberrant DNA Replication Intermediates in Root Meristems
The root meristem consists of undifferentiated dividing cells. Hence, it is a suitable system to examine the tolerance to DNA damage during cell division and cell differentiation. In Arabidopsis, the stem cells, which are located in an SCN, divide to form TA cells. The rate of root growth depends on the number of cycling cells (Beemster and Baskin, 1998). If replication fork progression is blocked by DNA lesions, DNA repair can lead to cell cycle delay. If the damage persists, cell division could proceed, resulting in genetically unstable cells and cell death. Moreover, a cell cycle delay could cause the loss of synchronicity during cell division, which would lead to an aberrant root meristem architecture. Here, we found that RTEL1 is important for proper cell division. Without RTEL1, the cells were unable to correctly repair replication-associated lesions, and an increased number of dead stem cells was observed in the rtel1-1 mutant. The additional treatment with cis-platin increased the frequency of roots with dead stem cells in the rtel1-1 single mutant line. Furthermore, we observed more roots with aberrant SCNs. These rearrangements of the root meristem in rtel1-1 suggest that the stem cells attempt to replace the dead stem cells in the SCN (Heyman et al., 2014). Moreover, the loss of synchronized cell division could be a reason for a disruption of the root meristem architecture.
RTEL1 Acts Independently of FANCM and MUS81 to Repair Aberrant Replication Intermediates in Root Meristems
Our analysis of the root meristem in double mutants of RTEL1 with FANCM and MUS81 indicated that, in both cases, the meristems were more damaged in the double mutants than in the single mutants. Thus, there are multiple pathways for removing aberrant replication intermediates. Interestingly, the same type of genetic interaction was observed in plants that were treated with cis-platin in both the fresh weight and root assay, indicating that lesions arising during replicative repair are similar to CLs induced by the genotoxin.
The Role of RTEL1 in Telomere Homeostasis
Because eukaryotic chromosomes are linear, the cell must distinguish between chromosome ends and double strand breaks. Furthermore, the replication machinery is unable to fully replicate linear chromosomes to their ends (end replication problem). To address this issue, chromosome ends are protected by telomeres. In mice and humans, RTEL1 has been found to participate in telomere maintenance, and our data also reveal a role of RTEL1 in telomere homeostasis, but only in combination with a loss of TERT activity. TERT counteracts the end replication problem and extends the telomeres by reverse transcription. In Arabidopsis, the loss of TERT leads to gradual telomere shortening. After 10 generations, chromosome fusions in the tert mutant line finally lead to genome instability, which causes developmental arrest (Riha et al., 2001). The further loss of RTEL1 in the tert mutant causes these severe developmental defects to appear in the fourth homozygous generation (F5). These findings indicate that RTEL1 plays a role in telomere stability. Moreover, we observed a higher frequency of roots with dead stem cells in the double mutant compared with either single mutant. We suggest that the telomeres of the double mutant shorten much more quickly than those of the tert single mutant. Thus, the unprotected chromosome ends lead to chromosome fusions and chromosome breaks, which finally result in cell death and aberrant SCNs. Furthermore, TRF analysis of the rtel1-1 mutant revealed that the telomeres might even be longer than in the wild type. This result was unexpected, as in humans and mice, the loss of RTEL1 leads to shorter telomeres (Ding et al., 2004; Barber et al., 2008; Vannier et al., 2012; Ballew et al., 2013; Deng et al., 2013; Le Guen et al., 2013). Although the increase in telomere length might just be due to natural variation within a species (Shakirov and Shippen, 2004), it is obvious that there is no telomere shortening over the generations tested in our study in Arabidopsis.
The loss of both RTEL1 and TERT leads to a much more rapid telomere shortening than in the tert single mutant line. Besides the role of RTEL1 in telomerase regulation, RTEL1 also might participate in the resolution of T-loops. It is possible that RTEL1 suppresses HR and dismantles T-loops to facilitate replication of the telomeres, similar to the function of RTEL1 in mice (Vannier et al., 2012). Furthermore, the loss of RTEL1 leads to inadequate HR events at T-loops, which causes recombination-dependent formation of T-circles and rapid telomere shortening. If both RTEL1 and TERT are defective, TERT could not counteract the rapid telomere shortening, leading to an extremely rapid loss of telomeres.
RTEL1 and RECQ4A: Two DNA Helicases in Arabidopsis That Might Process Similar D-Loop-Like Intermediates in Different Ways
Prior studies have shown that the RecQ helicase RECQ4A plays a role in intrastrand CL repair and that it acts in a parallel pathway to that of FANCM or MUS81 (Hartung et al., 2007; Mannuss et al., 2010; Knoll et al., 2012). The double mutant rtel1-1 recq4A-4 exhibited severe developmental defects. These plants showed aberrant leaf morphology, fasciated shoots, and a reduced size. Furthermore, the double mutant was infertile. Because of the dramatically compromised plant morphology, we could not analyze the double mutant using recombination assays and sensitivity assays. Nevertheless, it was possible to examine the root meristem and to compare it with the corresponding single mutants and the wild type. The double mutant exhibited severe developmental defects in the root meristem. We observed a completely disrupted root morphology. The roots were degenerated, and we could not identify the different types of stem cells. The loss of both RTEL1 and RECQ4A led to an extremely early stem cell death during development, explaining why few cells remained that could be identified as dead cells. Due to these severe defects in rtel1-1 recq4A-4, it was not possible to analyze the SCN in more detail.
Previously, it was shown that the concurrent loss of RTEL1 and the RecQ helicase HIM6 in C. elegans, as well as the concurrent loss of the RTEL1 analog SRS2 and the homologous RecQ helicase SGS1 in Saccharomyces cerevisiae, leads to lethality in both organisms (Lee et al., 1999; Barber et al., 2008). Due to the loss of both helicases, the cell is unable to resolve recombination intermediates either through Sc-SRS2/Ce-RTEL1 or through Sc-SGS1/Ce-HIM6. These defects result in an accumulation of toxic recombination intermediates, which finally causes lethality. Our results suggest that the loss of At-RTEL1 and At-RECQ4A leads to a similar effect. Thus, both helicases might process similar types of repair and replication intermediates, but in different ways. Indeed, RECQ helicases have been found in various organisms to be involved in telomere homeostasis. It was demonstrated before that At-RECQ4A is involved in removing interchromosomal telomeric connections that arise during meiotic recombination (Higgins et al., 2011).
The most promising speculation is that At-RTEL1 is an enzyme that can efficiently resolve different kinds of D-loop structures (Figure 11). There is no other known helicase in plants with the same substrate specificity as At-RTEL1. Nevertheless, depending on the nature of the D-loop-like structure, other enzymes, such as At-RECQ4A, might be able to substitute for At-RTEL1, at least to a certain extent.
METHODS
Plant Growth
To elucidate the role of RTEL1 in Arabidopsis thaliana, the mutant line rtel1-1 (SALK_ 113285) was used. For the generation of double mutants, the mutant lines fancm-1 (SALK_069784), mus81-1 (GABI_113F11), and recq4A-4 (GABI_203C07) were used, which have been described previously (Hartung et al., 2006, 2007; Knoll et al., 2012). For the cross with the tert mutant line (Fitzgerald et al., 1999), heterozygous seeds were used. For breeding of the double mutants, the plants were grown in a greenhouse in soil (1:1 mixture of Floraton 3 [Floragard] and Vermiculit [1 to 3 mm; Deutsche Vermiculite Dämmstoff]) at 22°C (16 h of light/8 h of dark). To examine the sensitivity against genotoxic agents and the HR frequency, the plants were grown under axenic conditions. After surface sterilization with 4% NaOCl and stratification at 4°C overnight, the seeds were sown on agar plates containing germination medium (GM; 4.9 g/L Murashige and Skoog medium including vitamins and MES [Duchefa Biochemie]), 10 g/L sucrose, and 7.6 g/L agar (adjusted to pH 5.7 with KOH) and were incubated in a CU-36L4 plant culture chamber (Percival Scientific) under stable conditions with 16 h of light at 22°C and 8 h of dark at 20°C.
Primers Used for PCR-Based Genotyping of the T-DNA Insertion Lines
To determine the genotypes of the T-DNA insertion lines, two primer pairs for each mutant line were used. One gene-specific primer pair targeted sequences located upstream and downstream of the T-DNA. The second primer pair comprised a gene-specific primer and one primer located on the T-DNA. The gene-specific primers for rtel1-1 were RTEL1-Fw1 (5′-GGGTTACCAAACGATTATAC-3′) and RTEL1-Rev1 (5′-CGACACAGAATATAAAGAACA-3′). For the T-DNA PCR, RTEL1-Fw1 and the T-DNA-specific primer LBd1 (5′-TCGGAACCACCATCAAAC-3′) were used. PCR-based genotyping of the mutant lines fancm-1, mus81-1, recq4A-4, and tert was performed as described previously (Fitzgerald et al., 1999; Hartung et al., 2006, 2007; Knoll et al., 2012).
Cloning and Transformation of RTEL1 Complementation Constructs
Using Arabidopsis Columbia-0 wild-type genomic DNA as a template, a 7505-bp-long fragment containing the natural promoter, open reading frame, and terminator of the RTEL1 gene was amplified by PCR using primers RTEL1-Prom-fw (5′-CTCAGGTAGCGGGTTTATC-3′) and RTEL1-Term-rev (5′-TAAGATACACGGATGAGCC-3′). The PCR product was transferred to the binary vector pSBO-2 (Bonnet et al., 2013) at the PacI site of the MCS with the In-Fusion HD Cloning Kit (Clontech). Transformation of wild-type and rtel1-1 plants was done by floral dip (Clough and Bent, 1998) with Agrobacterium tumefaciens GV3103:pMP90 (Koncz and Schell, 1986) carrying the RTEL1 plasmid. Using a PPT resistance cassette on the T-DNA, selection of transformants was possible. In the T2 generation, lines carrying the T-DNA at a single locus were identified by a 3:1 Mendelian segregation of the resistance cassette and were propagated. In the T3 generation, only lines homozygous for the resistance cassette, and therefore the RTEL1 complementation construct, were used in further experiments.
Gene Expression Analysis by qPCR
For four separate biological replicates, total RNA was extracted from 2-week-old plantlets using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. Five micrograms of RNA was used together with an oligo(dT)18 primer and the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) for the reverse transcription step. After a 1:5 dilution in TE buffer, 5 μL of this cDNA were used as a template in qPCR combined with 4.7 μL of water, 0.15 μL of each primer, and 10 μL of 2× LightCycler 480 SYBR Green I Master Mix (Roche Diagnostics). qPCR was run in a LightCycler 480 instrument with a 96-well block (Roche Diagnostics). All cDNAs were analyzed in triplicate for the expression of the reference genes ACT2 (At3g18780) and At4g34270 and amplified with primer pairs ACT2-fw (5′-ATTCAGATGCCCAGAAGTCTTGTTC-3′)/ACT2-rev (5′-GCAAGTGCTGTGATTTCTTTGCTCA-3′) and At4g34270-F1 (5′-AGATGAACTGGCTGACAATG-3′)/At4g34270-R1 (5′-TGTTGCTTCTCTCCAACAGT-3′), respectively. In addition, the expression of RTEL1 was tested at three positions in the gene: 5′, 3′, and across the T-DNA insertion present in line rtel1-1 using primer pairs RTEL1-RT-fw-A (5′-GAGGAGGAGGAGGAGGA-3′)/RTEL1-RT-rev-A (5′-TGACAGGCATTAGTGAGTG-3′), RTEL1-RT-fw-B (5′-AGTGCCAATTCCCAAAAGG-3′)/RTEL1-RT-rev-B (5′-GATTTCTAGTTTGCTTCCAG-3′), and RTEL1-RT-fw-C (5′-TGGAAAAATGGGTGTTACAG-3′)/RTEL1-RT-rev-C (5′-GACCTTACCGCGACAAAC-3′). For expression analysis in complementation lines and other mutant lines, the primer pair RTEL1-RT-fw-B/RTEL1-RT-rev-B was used. After normalization with the mean of both reference genes, the relative expression of RTEL1 was calculated compared with a Columbia-0 wild-type cDNA.
DNA Isolation
Isolation of genomic DNA was performed as described previously (Salomon and Puchta, 1998). The plants were ground in liquid nitrogen, and the powder was mixed with isolation buffer. After incubation at 65°C for 1 h, the DNA was extracted with chloroform, digested with RNase A, and precipitated with isopropanol. The dried DNA pellet was dissolved in 225 μL of TE buffer.
TRF Analysis
One microgram of genomic DNA was digested with AluI and precipitated with sodium acetate (3 M, pH 5.4) and ethanol (99.8%). The digested DNA was separated on a 1% agarose gel (2 V/cm for 16 h), transferred onto a neutral nylon membrane (Hybond-NX; GE Healthcare Europe), and hybridized to a 5′-labeled (T3AG3)4 probe. The probe was labeled with T4 polynucleotide kinase and [γ-32P]ATP (6000 Ci/mmol). After hybridization at 55°C for 18 h, the signals were detected on an imaging plate (Fujifilm) with a CR 35 Bio High Speed Image Plate Scanner (DÜRR NDT).
Sensitivity Assays
Sensitivity assays were performed as described previously (Hartung et al., 2007). Ten 7-d-old plantlets were transferred to six-well plates with 5 mL of liquid GM (untreated control) or 4 mL of liquid GM for treatment with genotoxic agents. The next day, 1 mL of liquid GM with different concentrations of the genotoxic agents was added. After 13 d of further incubation in the plant culture chamber, the relative fresh weight was quantified. The fresh weight of the treated plantlets was normalized to the fresh weight of the untreated plantlets of the same line.
HR Assays
To determine the frequency of the intermolecular recombination events with the IC9 reporter construct, HR assays were performed as described previously (Hartung et al., 2007). Forty 7-d-old plantlets were transferred to halved Petri plates with 10 mL of liquid GM. After 7 d of incubation in the plant culture chamber, the plantlets were histologically stained with a staining solution containing 5 mL/L X-GlcA (2.5% in DMF) and 2 mL/L NaN3 (1% in water) diluted in 100 mM phosphate buffer, for 2 d at 37°C, followed by extraction of plant pigments in 70% ethanol overnight at 60°C. The blue sectors on each plantlet were quantified using a binocular microscope (SZB300; VWR).
Root Growth Assays and PI Staining
To investigate the root development and cell death in the roots, the seeds were sown as described above. After 4 d, the plantlets were transferred to six-well plates with 5 mL of liquid GM supplemented with cis-platin (35 μM) and incubated at room temperature for 18 h. The next day, the plantlets were washed four times with liquid GM. Afterward, the roots were transferred to microscope slides (already wetted with 5 μg/mL PI solution) and covered with cover slips. The roots were analyzed using a confocal microscope (LSM 700 laser scanning microscope; Carl Zeiss Microscopy) and LSM Software ZEN 2011.
Phylogenetic Analysis
Protein sequences were aligned with Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) using default settings (Sievers et al., 2011). The resulting alignment file (Supplemental Data Set 1) was imported into MEGA 6.06 (Tamura et al., 2013) to calculate a maximum parsimony phylogenetic tree with the Subtree-Pruning-Regrafting algorithm with search level 1. All positions containing gaps and missing data were eliminated. Statistical support of the nodes was calculated with the bootstrap method with 1000 replicates.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Arabidopsis CHL1 (AT1G79890; NP_178107.1), Arabidopsis FANCJA (AT1G20750; NP_173498.3), Arabidopsis FANCJB (AT1G20720; NP_173495.5), Arabidopsis RTEL1 (AT1G79950; NM_106644.3; NP_178113.3), Arabidopsis XPD (AT1G03190; NP_849584.1), Homo sapiens DDX11 isoform 3 (NP_689651.1), H. sapiens FANCJ (NP_114432.2), H. sapiens RTEL1 isoform 1 (NP_057518.1), H. sapiens XPD (NP_000391.1), M. musculus RTEL1 isoform 1 (NP_001001882.3), rice RTEL1 (B9EXU8_ORYSJ), P. patens RTEL1 (A9RE00_PHYPA), and grape RTEL1 (F6HYG0_VITVI).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Characterization of the T-DNA Insertion Site of rtel1-1.
Supplemental Figure 2. Gene Expression Analysis of RTEL1.
Supplemental Figure 3. Epistasis Analysis of Root Tip Stem Cell Death after cis-Platin Induction.
Supplemental Figure 4. Root Tip Stem Cell Death after cis-Platin Induction in recq4A-4.
Supplemental Figure 5. Normal Development of rtel1-1 tert in F3 and F4.
Supplemental Figure 6. Analysis of Root Tip Stem Cell Death after cis-Platin Induction in rtel1-1 tert.
Supplemental Data Set 1. Clustal Omega Alignment File of Protein Sequences from XPD Family Helicases.
Supplementary Material
Acknowledgments
This work was funded by the European Research Council Advanced Grant COMREC and the Deutsche Forschungsgemeinschaft (Grant Pu 137-11) to H.P.
AUTHOR CONTRIBUTIONS
A.K. and H.P. designed the research. J.R. and A.K. performed the research. J.R., A.K., and H.P. analyzed data. J.R., A.K., and H.P. wrote the article.
Glossary
- HR
homologous recombination
- CL
cross-link
- qPCR
quantitative PCR
- MMC
mitomycin C
- SCN
stem cell niche
- TA
transiently amplifying
- QC
quiescent center
- PI
propidium iodide
- TRF
terminal restriction fragment
- GM
germination medium
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Aguilera A., Klein H.L. (1988). Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics 119: 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Ballew B.J., Yeager M., Jacobs K., Giri N., Boland J., Burdett L., Alter B.P., Savage S.A. (2013). Germline mutations of regulator of telomere elongation helicase 1, RTEL1, in Dyskeratosis congenita. Hum. Genet. 132: 473–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber L.J., et al. (2008). RTEL1 maintains genomic stability by suppressing homologous recombination. Cell 135: 261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster G.T., Baskin T.I. (1998). Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol. 116: 1515–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanck S., Kobbe D., Hartung F., Fengler K., Focke M., Puchta H. (2009). A SRS2 homolog from Arabidopsis thaliana disrupts recombinogenic DNA intermediates and facilitates single strand annealing. Nucleic Acids Res. 37: 7163–7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet S., Knoll A., Hartung F., Puchta H. (2013). Different functions for the domains of the Arabidopsis thaliana RMI1 protein in DNA cross-link repair, somatic and meiotic recombination. Nucleic Acids Res. 41: 9349–9360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulikas T., Vougiouka M. (2003). Cisplatin and platinum drugs at the molecular level. Review.Oncol. Rep. 10: 1663–1682. [PubMed] [Google Scholar]
- Chen I.-P., Haehnel U., Altschmied L., Schubert I., Puchta H. (2003). The transcriptional response of Arabidopsis to genotoxic stress: A high-density colony array study (HDCA). Plant J. 35: 771–786. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Curtis M.J., Hays J.B. (2007). Tolerance of dividing cells to replication stress in UVB-irradiated Arabidopsis roots: Requirements for DNA translesion polymerases eta and zeta. DNA Repair (Amst.) 6: 1341–1358. [DOI] [PubMed] [Google Scholar]
- Dangel N.J., Knoll A., Puchta H. (2014). MHF1 plays Fanconi anaemia complementation group M protein (FANCM)-dependent and FANCM-independent roles in DNA repair and homologous recombination in plants. Plant J. 78: 822–833. [DOI] [PubMed] [Google Scholar]
- Deng Z., et al. (2013). Inherited mutations in the helicase RTEL1 cause telomere dysfunction and Hoyeraal-Hreidarsson syndrome. Proc. Natl. Acad. Sci. USA 110: E3408–E3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H., et al. (2004). Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell 117: 873–886. [DOI] [PubMed] [Google Scholar]
- Ellis N.A., Groden J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., German J. (1995). The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell 83: 655–666. [DOI] [PubMed] [Google Scholar]
- Fanconi G. (1927). Familiäre infantile perniziosartige Anämie (perniziöses Blutbild und Konstitution). In Jahrbuch für Kinderheilkunde. (Berlin: Karger; ), pp. 257–280. [Google Scholar]
- Fauser F., Schiml S., Puchta H. (2014). Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 79: 348–359. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M.S., Riha K., Gao F., Ren S., McKnight T.D., Shippen D.E. (1999). Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl. Acad. Sci. USA 96: 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S., McDonald J.P., Bendixen C., Arthur L., Rothstein R. (1994). The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: A potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14: 8391–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German J., Archibald R., Bloom D. (1965). Chromosomal breakage in a rare and probably genetically determined syndrome of man. Science 148: 506–507. [DOI] [PubMed] [Google Scholar]
- Hartung F., Suer S., Bergmann T., Puchta H. (2006). The role of AtMUS81 in DNA repair and its genetic interaction with the helicase AtRecQ4A. Nucleic Acids Res. 34: 4438–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Suer S., Puchta H. (2007). Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 18836–18841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyman J., Kumpf R.P., De Veylder L. (2014). A quiescent path to plant longevity. Trends Cell Biol. 24: 443–448. [DOI] [PubMed] [Google Scholar]
- Higgins J.D., Ferdous M., Osman K., Franklin F.C. (2011). The RecQ helicase AtRECQ4A is required to remove inter-chromosomal telomeric connections that arise during meiotic recombination in Arabidopsis. Plant J. 65: 492–502. [DOI] [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D'Angelo C., Bornberg-Bauer E., Kudla J., Harter K. (2007). The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50: 347–363. [DOI] [PubMed] [Google Scholar]
- Knoll A., Puchta H. (2011). The role of DNA helicases and their interaction partners in genome stability and meiotic recombination in plants. J. Exp. Bot. 62: 1565–1579. [DOI] [PubMed] [Google Scholar]
- Knoll A., Higgins J.D., Seeliger K., Reha S.J., Dangel N.J., Bauknecht M., Schröpfer S., Franklin F.C., Puchta H. (2012). The Fanconi anemia ortholog FANCM ensures ordered homologous recombination in both somatic and meiotic cells in Arabidopsis. Plant Cell 24: 1448–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Schell J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204: 383–396. [Google Scholar]
- Lawrence C.W., Christensen R.B. (1979). Metabolic suppressors of trimethoprim and ultraviolet light sensitivities of Saccharomyces cerevisiae rad6 mutants. J. Bacteriol. 139: 866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.K., Johnson R.E., Yu S.L., Prakash L., Prakash S. (1999). Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science 286: 2339–2342. [DOI] [PubMed] [Google Scholar]
- Le Guen T., et al. (2013). Human RTEL1 deficiency causes Hoyeraal-Hreidarsson syndrome with short telomeres and genome instability. Hum. Mol. Genet. 22: 3239–3249. [DOI] [PubMed] [Google Scholar]
- Levitus M., et al. (2005). The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat. Genet. 37: 934–935. [DOI] [PubMed] [Google Scholar]
- Mannuss A., Dukowic-Schulze S., Suer S., Hartung F., Pacher M., Puchta H. (2010). RAD5A, RECQ4A, and MUS81 have specific functions in homologous recombination and define different pathways of DNA repair in Arabidopsis thaliana. Plant Cell 22: 3318–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meetei A.R., et al. (2005). A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 37: 958–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J., Ries G., Bonhoeffer S., Hohn B. (2004). Interchromatid and interhomolog recombination in Arabidopsis thaliana. Plant Cell 16: 342–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita T., Furukawa F., Sakaguchi C., Toda T., Carr A.M., Iwasaki H., Shinagawa H. (2005). Role of the Schizosaccharomyces pombe F-box DNA helicase in processing recombination intermediates. Mol. Cell. Biol. 25: 8074–8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R., Krejci L., Van Komen S., Anke Schürer K., Kramer W., Sung P. (2005). Saccharomyces cerevisiae MPH1 gene, required for homologous recombination-mediated mutation avoidance, encodes a 3′ to 5′ DNA helicase. J. Biol. Chem. 280: 7854–7860. [DOI] [PubMed] [Google Scholar]
- Riha K., McKnight T.D., Griffing L.R., Shippen D.E. (2001). Living with genome instability: Plant responses to telomere dysfunction. Science 291: 1797–1800. [DOI] [PubMed] [Google Scholar]
- Rink S.M., Lipman R., Alley S.C., Hopkins P.B., Tomasz M. (1996). Bending of DNA by the mitomycin C-induced, GpG intrastrand cross-link. Chem. Res. Toxicol. 9: 382–389. [DOI] [PubMed] [Google Scholar]
- Rudolf J., Makrantoni V., Ingledew W.J., Stark M.J., White M.F. (2006). The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol. Cell 23: 801–808. [DOI] [PubMed] [Google Scholar]
- Salomon S., Puchta H. (1998). Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J. 17: 6086–6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller J., Schürer A., Rudolph C., Hettwer S., Kramer W. (2000). MPH1, a yeast gene encoding a DEAH protein, plays a role in protection of the genome from spontaneous and chemically induced damage. Genetics 155: 1069–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B., Benfey P., Dolan L. (2002). Root development. The Arabidopsis Book 1: e0101, doi/10.1199/tab.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröpfer S., Knoll A., Trapp O., Puchta H. (2014a). Nucleus and genome: DNA recombination and repair. In Molecular Biology, S.H. Howell, ed (New York: Springer; ), pp. 1–37. [Google Scholar]
- Schröpfer S., Kobbe D., Hartung F., Knoll A., Puchta H. (2014b). Defining the roles of the N-terminal region and the helicase activity of RECQ4A in DNA repair and homologous recombination in Arabidopsis. Nucleic Acids Res. 42: 1684–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov E.V., Shippen D.E. (2004). Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell 16: 1959–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J.D., Higgins D.G. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lelij P., Chrzanowska K.H., Godthelp B.C., Rooimans M.A., Oostra A.B., Stumm M., Zdzienicka M.Z., Joenje H., de Winter J.P. (2010). Warsaw breakage syndrome, a cohesinopathy associated with mutations in the XPD helicase family member DDX11/ChlR1. Am. J. Hum. Genet. 86: 262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannier J.-B., Pavicic-Kaltenbrunner V., Petalcorin M.I., Ding H., Boulton S.J. (2012). RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 149: 795–806. [DOI] [PubMed] [Google Scholar]
- Walne A.J., Vulliamy T., Kirwan M., Plagnol V., Dokal I. (2013). Constitutional mutations in RTEL1 cause severe dyskeratosis congenita. Am. J. Hum. Genet. 92: 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youds J.L., Mets D.G., McIlwraith M.J., Martin J.S., Ward J.D., ONeil N.J., Rose A.M., West S.C., Meyer B.J., Boulton S.J. (2010). RTEL-1 enforces meiotic crossover interference and homeostasis. Science 327: 1254–1258.PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Hathcock K.S., Hande P., Lansdorp P.M., Seldin M.F., Hodes R.J. (1998). Telomere length regulation in mice is linked to a novel chromosome locus. Proc. Natl. Acad. Sci. USA 95: 8648–8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.