The DNA polymerase II subunit B3-1 interacts with DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN2A and NUCLEAR FACTOR Y proteins to activate gene expression during heat stress responses.
Abstract
DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN2A (DREB2A) is a key transcription factor for drought and heat stress tolerance in Arabidopsis thaliana. DREB2A induces the expression of dehydration- and heat stress-inducible genes under the corresponding stress conditions. Target gene selectivity is assumed to require stress-specific posttranslational regulation, but the mechanisms of this process are not yet understood. Here, we identified DNA POLYMERASE II SUBUNIT B3-1 (DPB3-1), which was previously annotated as NUCLEAR FACTOR Y, SUBUNIT C10 (NF-YC10), as a DREB2A interactor, through a yeast two-hybrid screen. The overexpression of DPB3-1 in Arabidopsis enhanced the expression of a subset of heat stress-inducible DREB2A target genes but did not affect dehydration-inducible genes. Similarly, the depletion of DPB3-1 expression resulted in reduced expression of heat stress-inducible genes. Interaction and expression pattern analyses suggested the existence of a trimer comprising NF-YA2, NF-YB3, and DPB3-1 that could synergistically activate a promoter of the heat stress-inducible gene with DREB2A in protoplasts. These results suggest that DPB3-1 could form a transcriptional complex with NF-YA and NF-YB subunits and that the identified trimer enhances heat stress-inducible gene expression during heat stress responses in cooperation with DREB2A. We propose that the identified trimer contributes to the target gene selectivity of DREB2A under heat stress conditions.
INTRODUCTION
Environmental stresses, such as low or high temperatures, drought, or high salinity, have adverse effects on plant growth and productivity. Plants have developed many mechanisms to adapt to such environmental changes. The perception of a specific environmental stress triggers the transduction of stress signals, which induce stress-specific changes in gene expression. Although overlapping sets of genes are induced by different stress conditions, many genes show stress-specific expression patterns (Thomashow, 1999; Zhu, 2002; Chinnusamy et al., 2004; Bartels, 2005; Yamaguchi-Shinozaki and Shinozaki, 2006). Because the response mechanisms differ in response to various environmental conditions, the correct expression of genes is assumed to be necessary for plants to survive stress.
The dehydration-responsive element (DRE), a 9-bp cis-acting sequence motif (TACCGACAT), was identified from a subset of dehydration-inducible genes in Arabidopsis thaliana (Yamaguchi-Shinozaki and Shinozaki, 1994). DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN2A (DREB2A), an APETALA2/ethylene-responsive element binding factor-type (AP2/ERF-type) transcription factor, was isolated as a protein that recognizes DRE (Stockinger et al., 1997; Liu et al., 1998). Subsequent research has revealed that the activity of DREB2A is posttranslationally regulated, and a 30-amino acid region adjacent to the AP2/ERF DNA binding domain plays a critical role in the posttranslational regulation of this protein. The deletion of this region converts the DREB2A protein into a constitutively active form (DREB2A CA), and DREB2A CA-overexpressing Arabidopsis shows a dwarf phenotype and a significant increase in drought stress tolerance (Sakuma et al., 2006a). Previous studies revealed that wild-type DREB2A was unstable and degraded by the 26S proteasome pathway under normal conditions, while the conversion into DREB2A CA increased protein stability (Sakuma et al., 2006a; Qin et al., 2008). Additionally, DREB2A was stabilized under stress conditions, together demonstrating that regulation of stability is one of the posttranslational regulatory mechanisms affecting DREB2A (Qin et al., 2008; Morimoto et al., 2013). A microarray analysis revealed that the overexpression of DREB2A CA causes the expression of heat-inducible genes in addition to dehydration-inducible genes. Also, DREB2A expression increases in response to heat stress. The overexpression of DREB2A CA significantly increases heat stress tolerance, and DREB2A knockout mutants exhibit a significant decrease in heat stress tolerance (Sakuma et al., 2006b). Thus, the DREB2A transcription factor functions in both dehydration and heat stress responses by inducing the expression of specific gene sets under both conditions. However, the mechanism that determines the stress-specific expression pattern of DREB2A target genes, i.e., how the expression of a subset of DREB2A target genes is activated under a specific stress condition, remains unknown. We assume that the posttranslational regulation of DREB2A activity plays an important role in the correct expression of each stress-inducible gene via unknown mechanisms.
H2A/H2B-like histonefold domain (HFD)-containing proteins are highly conserved among eukaryotes, and the HFD is responsible for the dimerization of H2A- and H2B-like subunits and DNA binding (Gnesutta et al., 2013). Previous studies have revealed that these proteins are involved in various molecular biological processes, including transcription, replication, and remodeling. DNA POLYMERASE II SUBUNIT B3 (DPB3) and DPB4 subunits, which are major H2A/H2B-like proteins, have been identified as components of DNA polymerase II (Araki et al., 1991; Ohya et al., 2000); however, additional experiments have suggested that DPB3 and DPB4 are also associated with the histone acetylating complex and the chromatin remodeling complex (Hartlepp et al., 2005; Wang et al., 2008). Additionally, DPB3 and DPB4 are suggested to bind double-stranded DNA without sequence specificity (Tsubota et al., 2006). NF-YB and NF-YC subunits are members of other extensively analyzed families of H2A/H2B-like proteins. These subunits form a trimer with an NF-YA subunit, and the trimer binds to a cis-acting element, CCAAT, on gene promoters and regulates transcription in concert with other transcription factors (McNabb et al., 1995; Edwards et al., 1998; Mantovani, 1999). The negative cofactors 2α (NC2α) and NC2β are also H2A/H2B-like proteins and were originally characterized as corepressors of the TATA box binding protein (Mermelstein et al., 1996; Zhou et al., 2009). However, a subsequent study revealed that NC2 subunits are involved in activating promoters with downstream promoter elements (Willy et al., 2000). These protein families (DPB3/DPB4, NF-YB/NF-YC, and NC2α/NC2β) in Arabidopsis were correctly reclassified in a recent study, although all had previously been classified as NF-YB/NF-YC family proteins (Petroni et al., 2012). The number of genes encoding each subunit of H2A/H2B-like HFD-containing proteins in plants is much larger than those in animals (Petroni et al., 2012), which typically encode each subunit as a single gene. As described above, previous studies have revealed the function and characteristics of each family of H2A/H2B-like HFD-containing proteins. However, it is mostly unknown whether and how these protein families act in a coordinated manner to regulate molecular processes. In particular, very little research has been performed on H2A/H2B-like HFD-containing proteins other than NF-YB/NF-YC, such as DPB3/DPB4 and NC2α/NC2β in plants.
In this study, we identified an Arabidopsis DPB3 homolog, DPB3-1, which interacts with DREB2A. We confirmed the interaction between DREB2A and DPB3-1 and characterized functions of DPB3-1 in Arabidopsis. We identified that DPB3-1 could interact with NF-Y subunits, NF-YA2 and NF-YB3, and found that the trimer of these proteins enhanced the transcriptional activity of DREB2A in protoplasts. Our results suggest that the identified trimer comprising DPB3-1, NF-YA2, and NF-YB3 is involved as an enhancer in the heat stress-specific expression, but not the dehydration-specific expression, of DREB2A target genes. The function of the identified trimer may be important in the target selectivity of DREB2A in the adaptive responses of plants to heat stress.
RESULTS
Isolation of DPB3-1 as a DREB2A-Interacting Protein and Localization of the DPB3-1-Interacting Region
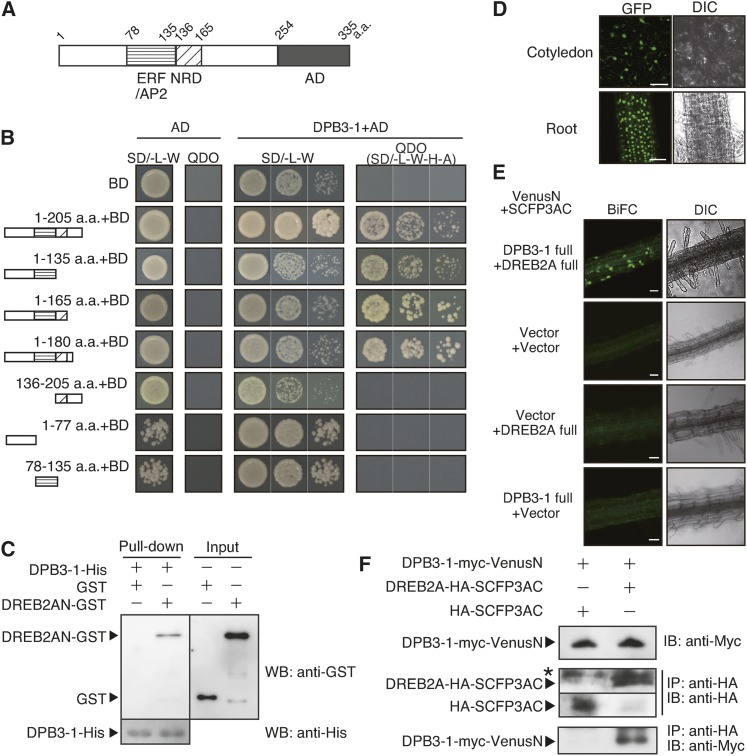
We performed a yeast two-hybrid screen to identify the proteins that interact with DREB2A. A fragment of DREB2A consisting of residues 1 to 205 (DREB2A 1–205; Figure 1A) was fused to the yeast GAL4 DNA binding domain and used as bait. Approximately 3.8 × 107 yeast transformants were screened, and after the verification of growth on a synthetic defined medium lacking Leu, Trp, His, and adenine (SD/-L-W-H-A), four clones containing the DPB3-1 cDNA, which was previously annotated as NF-YC10, were identified (Figure 1B). Considering its potential for forming a transcriptional complex with DREB2A to regulate target gene expression, we further examined this clone.
Figure 1.
DPB3-1 Interacts with DREB2A in Yeast Cells, in Vitro and in Vivo.
(A) Schematic diagram of DREB2A. AP2/ERF, DNA binding domain; NRD, negative regulatory domain; AD, activation domain. Numbers indicate the positions of amino acid residues starting from the N terminus.
(B) The growth of yeast cells harboring various truncated forms of DREB2A fused to the GAL4 DNA binding domain (BD). SD/-L-W is the nonselective medium, and SD-L-W-H-Ade (quadruple drop out [QDO]) is the selective medium. DPB3-1 was expressed as a fusion protein with the GAL4 activation domain (AD). a.a., amino acids.
(C) In vitro pull-down assays using GST or a DREB2AN (amino acids 1 to 165 of DREB2A)-GST fusion protein and the DPB3-1-His protein. The fractions pulled down by DPB3-1-His were analyzed with an anti-GST antibody and an anti-His antibody.
(D) Nuclear localization of sGFP-DPB3-1. The cotyledon and root tissues of 35S:sGFP-DPB3-1 plants were observed under a microscope. Confocal images of GFP fluorescence and differential interference contrast (DIC) images are shown. Bars = 100 μm.
(E) Verification of in vivo interaction between DREB2A and DPB3-1 by the BiFC system in transgenic Arabidopsis. The root tissues of the plants expressing DREB2A-SFCP3AC and DPB3-1-VenusN were observed after incubation at 37°C for 2 h. Confocal images of BiFC fluorescence (left) and DIC images (right) are shown. Bars = 50 μm.
(F) Coimmunoprecipitation of DPB3-1-myc-VenusN and DREB2A-HA-SCFP3AC in the transgenic Arabidopsis used in (C) after incubation at 37°C for 2 h. The total protein extracts (upper panel) or immunoprecipitated (IP) fractions using an anti-hemagglutinin (anti-HA) antibody (middle and lower panels) were analyzed by immunoblotting (IB) using anti-HA or anti-myc antibodies. The triangles and the asterisk indicate specific and nonspecific signals, respectively.
We conducted additional yeast two-hybrid assays to identify the region of DREB2A that interacts with DPB3-1. Several DREB2A fragments were designed based on the domain structure of DREB2A (Figure 1B). The yeast cells expressing the DREB2A fragment that included residues 1 to 135 could grow on the SD/-L-W-H-A medium (Figure 1B, 1–135 a.a.+BD), but the cells expressing further truncated fragments could not. Therefore, a region of DREB2A that includes both the AP2/ERF domain and the adjacent N-terminal sequence is sufficient for mediating the protein-protein interaction. We also tested the interactions between DREB2A and the other 12 H2A-like HFD-containing proteins in Arabidopsis, and we found that none of these proteins interacted with DREB2A (Supplemental Figure 1A).
DPB3-1 Interacts with DREB2A in Vitro and in Vivo
We further tested the direct interaction between DREB2A and DPB3-1 using in vitro pull-down assays. The N-terminal region (amino acids 1 to 165, containing the AP2/ERF domain and the NRD) of DREB2A and full-length DPB3-1 were expressed as fusion proteins of a glutathione S-transferase (GST) tag (DREB2AN-GST) or a 6× His tag (DPB3-1-His) and were purified. As a result of the pull-down assay, the DREB2AN-GST was detected in the pulled down fraction, whereas GST alone was not detected (Figure 1C). These data indicate that DPB3-1 interacts directly with the N-terminal region of DREB2A in vitro.
We then analyzed the in vivo interaction between DREB2A and DPB3-1 in Arabidopsis using the bimolecular fluorescence complementation (BiFC) system (Waadt et al., 2008). Prior to the analysis, we confirmed the subcellular localization of DPB3-1. A fusion protein of green fluorescent protein (GFP) and DPB3-1 was localized in the nuclei (Figure 1D). A similar localization pattern of DPB3-1 was detected in Arabidopsis mesophyll protoplasts (Supplemental Figure 1B). For the BiFC tests, we expressed one fusion protein consisting of DREB2A and the C-terminal half of SCFP3A with a hemagglutinin (HA) tag (DREB2A-HA-SCFP3AC) and one fusion protein consisting of DPB3-1 and the N-terminal half of Venus with a myc tag (DPB3-1-myc-VenusN) in transgenic Arabidopsis. No positive fluorescent signals were observed under the normal growth condition in any transgenic plants (Supplemental Figure 1C), but after heat shock (37°C, 2 h), fluorescence was detected in the nuclei of root cells in the transgenic plants coexpressing DREB2A-HA-SFCP3AC and DPB3-1-myc-VenusN (Figure 1E). Similar results were also obtained in BiFC experiments in Arabidopsis mesophyll protoplasts (Supplemental Figure 1D). We also performed coimmunoprecipitation assays using transgenic Arabidopsis plants expressing DREB2A-HA-SFCP3AC and/or DPB3-1-myc-VenusN after heat stress treatment. DPB3-1-myc-VenusN was detected after immunoprecipitation of DREB2A-HA-SCFP3AC using anti-HA antibody microbeads (Figure 1F). These results demonstrate that DREB2A and DPB3-1 colocalize and interact in the nuclei of Arabidopsis cells.
Characteristics of DPB3-1
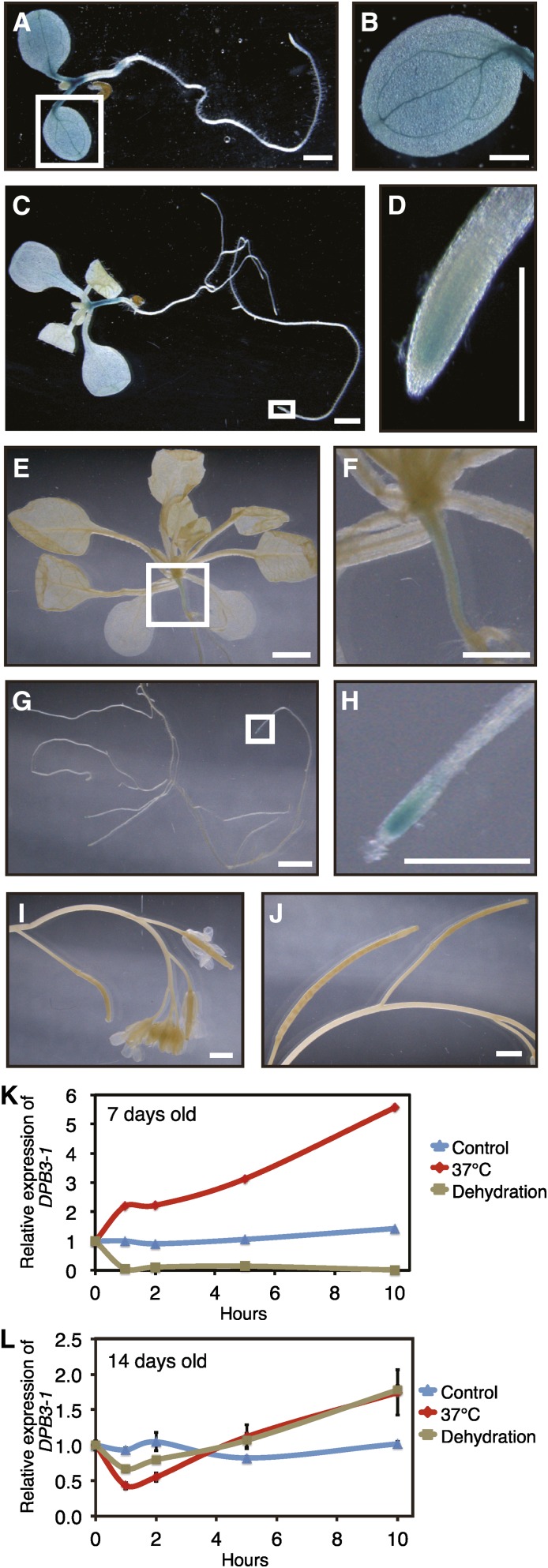
Next, we examined the tissue-specific expression pattern of DPB3-1. The 1-kb upstream sequence of DPB3-1 before the translational start site was inserted in front of the β-glucuronidase (GUS) coding sequence, and the tissue specificity of GUS activity was examined by histochemical staining. In 7-d-old seedlings, GUS activity was clearly detected in the petioles and leaf blades of the cotyledons as well as tops and bottoms of the hypocotyls (Figures 2A and 2B). In a previous study, the GUS staining pattern in the DPB3-1 (NF-YC10) 1-kb promoter:GUS transgenic plants at 6 d old was more vascular specific (Siefers et al., 2009). The difference in the expression patterns might result from the difference in the promoter region or in the experimental conditions used in those studies. In 10-d-old plants, GUS activity was detected mainly in whole hypocotyls and weakly in the cotyledons and root tips (Figures 2C and 2D). In 3-week-old plants, GUS activity was observed in the root tips and weakly in the hypocotyls (Figures 2E to 2H) but not in other tissues, including the reproductive tissues (Figures 2I and 2J). We also examined the changes in the expression levels of DPB3-1 during dehydration and heat stress treatments in 7- or 14-d-old plants. The expression of DPB3-1 was enhanced by heat stress and repressed by dehydration stress in 7-d-old plants (Figure 2K), whereas in 14-d-old plants, the expression level of DPB3-1 was almost stable under both stress conditions (Figure 2L). We also analyzed the tissue specificity of DPB3-1 expression under heat stress in the 7-d-old plants. DPB3-1 was expressed similarly under heat stress and the control condition, while GUS activity was detected more strongly under heat stress (Supplemental Figures 2A and 2B). These results suggest that DPB3-1 functions primarily in conjunction with DREB2A in the early seedling stage under heat stress.
Figure 2.
Tissue- and Stress-Specific Expression of DPB3-1.
(A) to (J) GUS staining of DPB3-1pro:GUS transgenic plants at different growth stages. Whole 7-d-old (A) and 10-d-old (C) seedlings, aerial (E) and root (G) tissues of 3-week-old seedlings, and flowers (I) and siliques (J) from 7-week-old plants are shown. Higher magnification images of (A), (C), (E), and (G) are shown in (B), (D), (F), and (H), respectively. Bars = 1 mm.
(K) and (L) Expression patterns of the DPB3-1 gene under control and stress conditions in 7-d-old (K) and 14-d-old (L) plants.
To analyze the phylogenetic relationships between DPB3-1 and other H2A-like HFD-containing proteins (NF-YC, NC2α, and DPB3 family proteins), we searched for proteins that contain the H2A-like HFD in other plant species for which whole genomic sequences are available, based on the literature (Petroni et al., 2012; Laloum et al., 2013). Based on the amino acid sequences of the conserved regions, we found 23 candidate proteins in soybean (Glycine max), 16 in rice (Oryza sativa), 12 in the moss (Physcomitrella patens), and three in each of the green algae Chlamydomonas reinhardtii and Volvox carteri (Supplemental Table 1). We also found that Arabidopsis At5g19490 encodes a protein that contains the conserved H2A-like HFD. A phylogenetic tree of the H2A-like HFD-containing proteins was constructed based on an alignment of the domains that are conserved around HFDs (Supplemental Data Set 1 and Supplemental Figure 2C). According to the phylogenetic tree, G. max has 14 NF-YC, five NC2α, and four DPB3 family proteins, O. sativa has six NF-YC, three NC2α, and three DPB3 family proteins, P. patens has six NF-YC, three NC2α, and three DPB3 family proteins, and C. reinhardtii and V. carteri each have one NF-YC, one NC2α, and one DPB3 family protein. We found that At5g19490 is a member of the NC2α subfamily and that Arabidopsis harbors two duplicated NC2α genes. We named this protein NC2α2. DREB2A and NC2α2 did not interact in yeast cells (Supplemental Figure 2D). The phylogenetic analysis also indicated that Arabidopsis DPB3-1 and DPB3-2 belong to a distinct monophyletic subgroup that is conserved among land plants (Supplemental Figure 2C). Therefore, it is possible that Arabidopsis DPB3-1 proteins have conserved and unique roles in land plants.
Overexpression of DPB3-1 Improves Heat Stress Tolerance
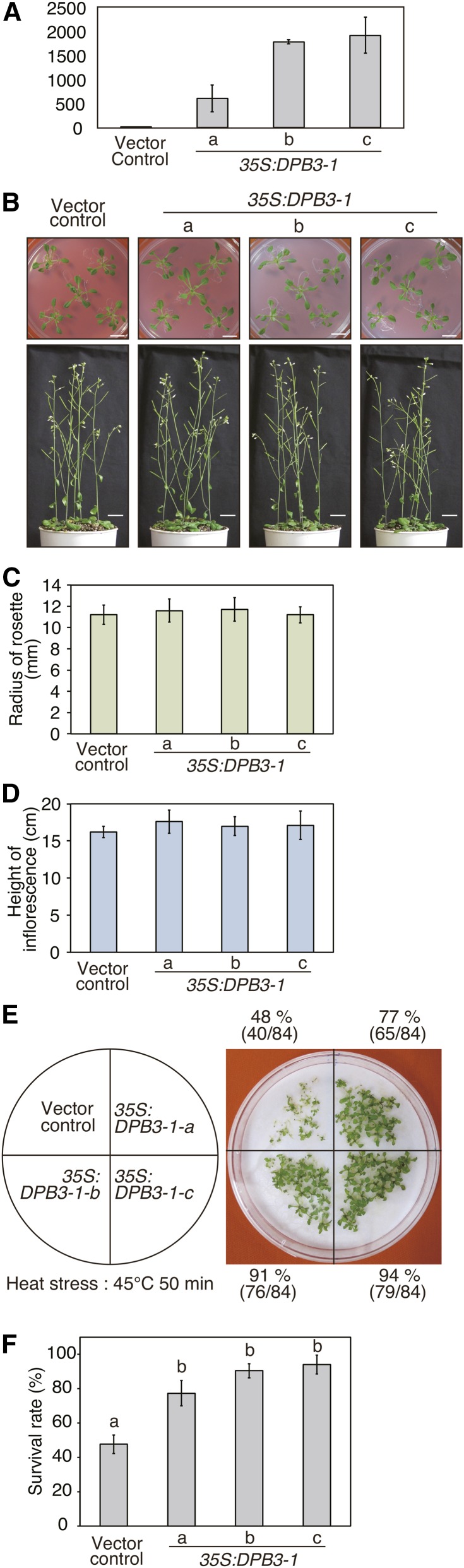
To examine the functions of DPB3-1, we generated transgenic Arabidopsis plants that overexpress DPB3-1 under the control of the enhanced cauliflower mosaic virus 35S promoter (Mitsuhara et al., 1996). Three transgenic lines were selected based on the expression levels of DPB3-1 (35S:DPB3-1-a, -b, and -c) (Figure 3A). These transgenic lines did not exhibit changes in growth or other visible phenotypes compared with the vector control plants under normal conditions (Figures 3B to 3D; Supplemental Figures 3C and 3D). Then, we evaluated whether the 35S:DPB3-1 plants have improved tolerance to heat or drought stress. The vector control and 35S:DPB3-1 plants were subjected to heat stress at 45°C for 50 min. As shown in Figures 3E and 3F, most of the 35S:DPB3-1 seedlings survived, while approximately half of the vector control plants survived after a subsequent 14-d recovery at 22°C. Therefore, the overexpression of DPB3-1 improved the heat stress tolerance of Arabidopsis as with the overexpression of DREB2A CA (Sakuma et al., 2006b). Conversely, tolerance to drought or salt was not significantly different between the vector control and 35S:DPB3-1 plants (Supplemental Figures 3A and 3B). These results raised the possibility that DPB3-1 might function in cooperation with DREB2A in the heat stress response but not in the drought or salt stress response. The enhanced stress tolerance without any growth retardation also indicates that the overexpression of DPB3-1 might be useful method for generating stress-tolerant crops.
Figure 3.
DPB3-1-Overexpressing Plants Showed Increased Heat Tolerance.
(A) Expression levels of DPB3-1 mRNA in the three transgenic lines overexpressing DPB3-1.
(B) Growth of DPB3-1-overexpressing plants under normal conditions. The seedlings were grown on agar medium for 21 d and then on soil for an additional 2 weeks. Photographs of 21- and 35-d-old plants are shown. Bars = 1 cm.
(C) and (D) Average radius of the rosette (C) and height of the inflorescence (D) calculated from the plants grown as in (B). The error bars indicate the sd (n = 25). The data were evaluated using one-way ANOVA, and no significant differences were detected (P > 0.05).
(E) Heat stress tolerance of the DPB3-1-overexpressing plants. Seven-day-old seedlings of the vector control and two transgenic lines were treated at 45°C for 50 min. The photograph was taken after a 7-d recovery period at 22°C.
(F) Percentages of surviving plants after the heat stress tolerance test. Each data point is the mean of three experiments, and each experiment included 28 plants. The letters above the bars indicate significant differences between the plants (P < 0.05 according to Tukey’s multiple range test).
[See online article for color version of this figure.]
Overexpression of DPB3-1 Enhances the Expression of DREB2A Target Genes and Other Genes during Heat Stress
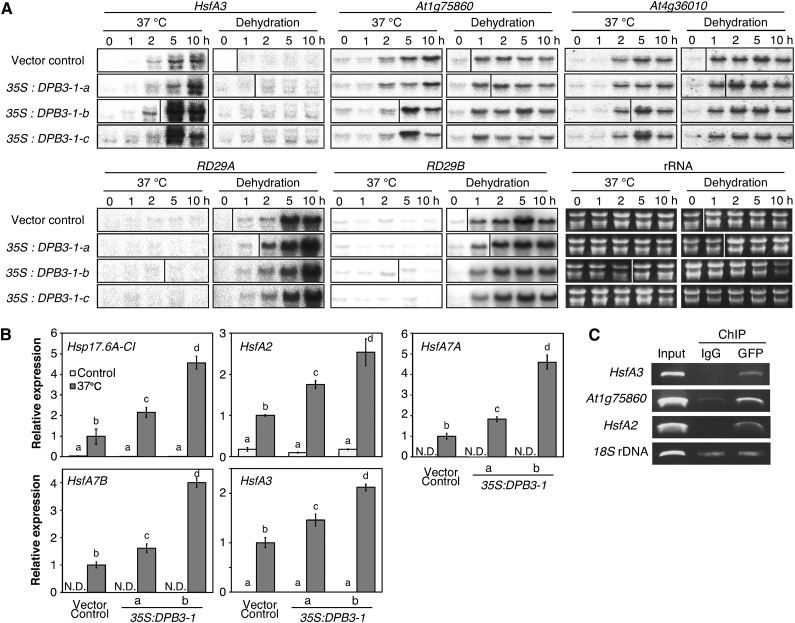
We analyzed the expression patterns of stress-inducible DREB2A target genes in the 35S:DPB3-1 and vector control plants under heat and dehydration stress conditions. Under the heat stress condition, the expression levels of the DREB2A target genes HEAT SHOCK TRANSCRIPTION FACTOR A3 (HsfA3), At1g75860, and At4g36010 were enhanced in the transgenic plants, especially after 5 h of heat stress (Figure 4A). The levels of the DREB2A mRNA and the DREB2A protein were comparable in the 35S:DPB3-1 and vector control plants (Supplemental Figures 4A and 4B). In contrast, under the dehydration stress condition, the expression levels of the target genes RESPONSIVE TO DESSICATION 29A (RD29A), RD29B, At1g75860, and At4g36010 in the 35S:DPB3-1 plants did not obviously differ from the levels in the vector control plants (Figure 4A). These results suggest that the overexpression of DPB3-1 positively affects the expression of heat stress-inducible DREB2A target genes under heat stress. We next overexpressed DPB3-1 in the dreb2a-1 knockout mutant (Supplemental Figures 4C and 4D) and examined the expression levels of the three heat-inducible target genes. The expression of these genes was not obviously enhanced by overexpression of DPB3-1 in the dreb2a-1 mutant background (Supplemental Figure 4E). This result confirmed that the increased gene expression of DREB2A target genes by the overexpression of DPB3-1 was dependent on DREB2A.
Figure 4.
Overexpression of DPB3-1 Enhanced the Expression Levels of Heat Stress-Inducible Genes in Arabidopsis.
(A) Expression levels of several genes downstream of DREB2A in the vector control plants and the three lines of DPB3-1-overexpressing plants during heat or dehydration stress. Ethidium bromide staining of the rRNA bands is shown to demonstrate equal loading.
(B) Quantitative RT-PCR analysis of the expression levels of several genes found to be upregulated by the microarray analysis (Supplemental Data Set 1) and the HsfA3 gene under heat stress (37°C, 5 h). The expression level of each gene in the vector control plant was defined as 1.0. The error bars indicate sd (n = 3). The letters above the bars indicate significant differences between treatments (P < 0.05 according to Tukey’s multiple range test).
(C) ChIP-PCR assays in heat-stressed GFP-DPB3-1-overexpressing plants. The enrichment of specific promoter regions was determined by RT-PCR. The results show the recovery of immunoprecipitated material using an anti-GFP antibody (GFP) or IgG (negative control) relative to the input DNA.
To analyze the impact of DPB3-1 overexpression on the transcriptome under heat stress, we performed a microarray analysis using an Agilent Arabidopsis 3 Oligo Microarray at the 5-h time point. In the 35S:DPB3-1 plants, 110 genes were upregulated more than 2-fold in comparison to vector control plants. Among the 110 genes, 24 genes were also upregulated in 35S:DREB2A CA plants (Sakuma et al., 2006b; Mizoi et al., 2013), and 14 genes were induced after 5 h of heat stress at 37°C (Sakuma et al., 2006b) (Supplemental Data Set 2). HsfA1s are master regulators of the heat shock response in Arabidopsis, and most heat stress-inducible genes, including DREB2A, are drastically impaired in quadruple hsfa1 mutants (hsfa1a hsfa1b hsfa1d hsfa1e) (Liu et al., 2011; Yoshida et al., 2011). We found that 25 out of the 110 genes were downregulated in the hsfa1 quadruple mutant under heat stress (Yoshida et al., 2011). The proportions of the candidate DREB2A downstream genes, heat stress-inducible genes, and candidate HsfA1 downstream genes among the 110 upregulated genes were significantly higher than their proportions in the entire Arabidopsis genome (P < 0.0001) (Supplemental Data Set 2). We also analyzed the 110 upregulated genes for enrichment using the MapMan ontology (Berardini et al., 2004; Usadel et al., 2005) and found that the terms heat stress and heat shock transcription factor were significantly enriched compared with the entire Arabidopsis genome (P < 0.0001) (Supplemental Table 2). The enhanced expression of several genes in Supplemental Data Set 2 and HsfA3 under the heat stress condition was confirmed by quantitative RT-PCR (Figure 4B). It is noteworthy that the overexpression of DPB3-1 enhanced the expression of HsfA2, which is an important HSF in the heat stress response. The expression of HsfA2 is regulated by HsfA1s in a similar manner to DREB2A (Nishizawa-Yokoi et al., 2011; Yoshida et al., 2011). These findings suggest that DPB3-1 can enhance the expression of heat stress-inducible genes not only through the DREB2A-regulated pathway but also through other pathways. Because DPB3-1 was initially annotated as an NF-YC subunit, we calculated the percentage of genes with the CCAAT element, which is the binding sequence for NF-Y (Mantovani, 1999), within a 1-kb region of their promoters. However, this percentage was not different between all genes in Arabidopsis and the upregulated genes in the DPB3-1 (NF-YC10)-overexpressing plants under heat stress (Supplemental Data Set 2). No enrichment of the CCAAT motif was also observed in the promoters of differentially expressed genes in NF-YA2-overexpressing Arabidopsis in a previous study (Leyva-González et al., 2012).
To verify the ability of DPB3-1 to bind the promoters of the upregulated genes, we performed chromatin immunoprecipitation (ChIP) assays in transgenic Arabidopsis that expressed sGFP-fused DPB3-1 under the control of the 35S promoter (Figure 1D) using an anti-GFP antibody or mouse IgG (negative control). The promoter fragments of the heat-inducible DREB2A target genes HsfA3 and At1g75860 were enriched with the anti-GFP antibody relative to the negative control (Figure 4C). The promoter fragments of the HsfA2 gene was also coprecipitated with the sGFP-DPB3-1 protein in agreement with the microarray result. These results confirmed that DPB3-1 binds directly to the promoters of heat stress-inducible genes. We performed additional ChIP assays with an anti-DREB2A antibody using the DPB3-1-overexpressing and vector control plants to analyze the effect of DPB3-1 overexpression on the DNA binding ability of DREB2A, but the levels of enrichment were not different between the vector control and the DPB3-1-overexpressing plants (Supplemental Figure 4F).
Knockdown of DPB3-1 Results in Reduced Expression of Heat-Inducible Genes and Enhanced Sensitivity to Heat Stress
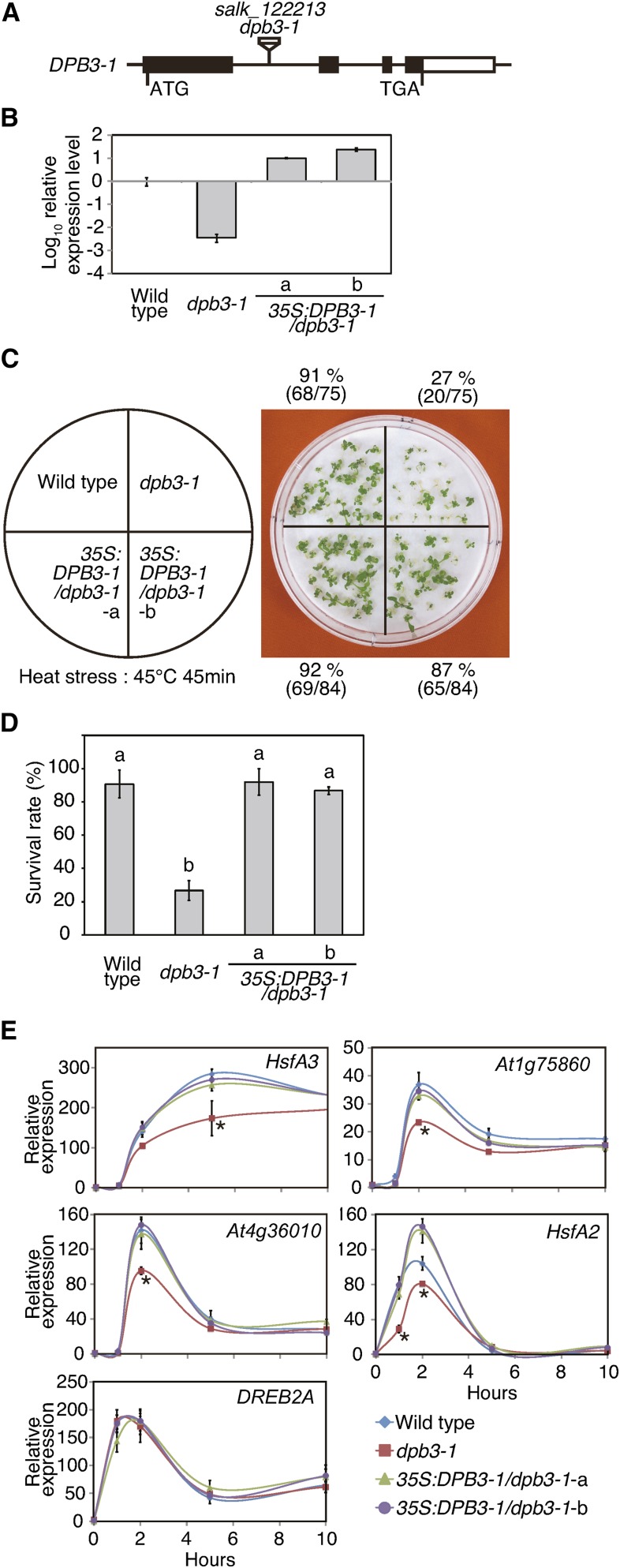
To further understand the function of DPB3-1, we obtained a T-DNA insertion mutant of DPB3-1 (dpb3-1; Figure 5A). The expression level of DPB3-1 was markedly reduced in the dpb3-1 mutant (Figures 5B), although a low amount of the mature DPB3-1 mRNA by splicing was detected (Supplemental Figure 5A). These results indicate that the dpb3-1 was a DPB3-1 knockdown mutant. We also transformed the dpb3-1 mutant with the 35S:DPB3-1 construct and selected two transgenic lines that exhibited relatively low levels of DPB3-1 expression (35S:DPB3-1/dpb3-1-a and -b) (Figure 5B). The dpb3-1 knockdown mutant did not differ from the wild type in the growth under nonstress conditions (Supplemental Figures 5B and 5C) or tolerance to drought or salt stress (Supplemental Figures 5D and 5E). However, the survival rate after heat stress was significantly decreased in the dpb3-1 mutant compared with the survival rate of the wild type, and the heat stress sensitivity was rescued by the expression of DPB3-1 (Figures 5C and 5D). The increased sensitivity to heat stress in the dpb3-1 mutant indicates a critical role of DPB3-1 in the heat stress response in Arabidopsis.
Figure 5.
The dpb3-1 Mutant Is Sensitive to Heat Stress and Exhibited Impaired Expression of Heat-Inducible Genes.
(A) A gene model of DPB3-1 and the T-DNA insertion in the dpb3-1 mutant. The coding region, untranslated regions, and introns are indicated as a closed box, open boxes, and lines, respectively.
(B) Relative DPB3-1 gene expression in wild-type, dpb3-1, and 35S:DPB3-1/dpb3-1 plants as determined by quantitative RT-PCR. The error bars indicate sd. The values are shown in the log scale.
(C) Heat stress tolerance of the dpb3-1 mutant. The experiments were performed as described in Figure 3E, except that the plants were treated at 45°C for 45 min.
(D) Percentages of surviving plants after the heat stress tolerance test. Each data point is the mean of three experiments, and each experiment comprised 28 plants. The letters above the bars indicate significant differences between the plants (P < 0.05 according to Tukey’s multiple range test). The mutant plants showed significantly higher sensitivity to heat than the wild-type plants.
(E) Quantitative RT-PCR analysis of the expression levels of several heat stress-inducible genes during heat stress (37°C). The expression level of each gene in the wild-type plant before the stress treatment was defined as 1.0. The error bars indicate sd (n = 3). The asterisk indicates the time point at which the expression level of each gene in the dpb3-1 mutant was significantly decreased compared with the wild-type and 35S:DPB3-1/dpb3-1 plants (P < 0.05 according to Tukey’s multiple range test).
Next, we analyzed the expression patterns of heat stress-inducible genes in 7-d-old dpb3-1 mutant and wild-type plants during heat stress. The expression levels of the three heat stress-inducible DREB2A target genes, HsfA3, At1g75860, and At4g36010, were reduced in the dpb3-1 mutant compared with the wild type, and the decreases in their expression levels were restored by the expression of DPB3-1 (Figure 5E). A similar result was obtained for HsfA2, which is not a DREB2A target (Figure 5E). The levels of the DREB2A transcript and the DREB2A protein during heat stress were not significantly different between the dpb3-1 mutant and wild-type plants (Figure 5E; Supplemental Figure 5F). Moreover, the expression levels of the dehydration-inducible DREB2A target genes RD29A and RD29B did not significantly differ between the dpb3-1 mutant and wild-type plants (Supplemental Figure 5G). These results suggest that DPB3-1 is necessary for the full induction of DREB2A target genes specifically under heat stress. Also, they support the idea that DPB3-1 contributes to gene expression under heat stress through multiple pathways, including the DREB2A-dependent pathway. We also analyzed the expression patterns of DREB2A target genes under heat and dehydration stress conditions using 14-d-old plants in which DPB3-1 was more weakly expressed (Figures 2A to 2H) and found that the expression levels of these genes were not obviously different between the mutant and wild-type plants under either stress condition (Supplemental Figure 5H).
NF-YA2 and NF-YB3 Are Candidate Components of a Transcriptional Complex That Regulates the Activity of DREB2A under Heat Stress Conditions
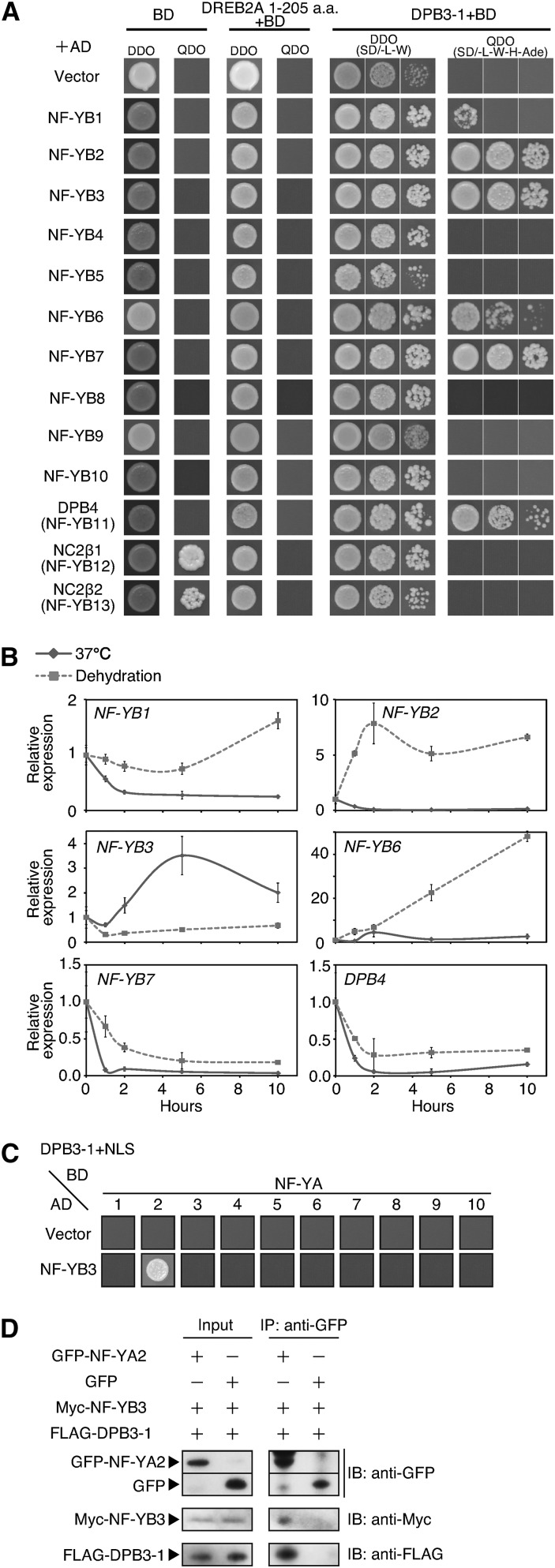
We tested whether DPB3-1 has a positive effect on the transactivation of target genes by DREB2A in protoplasts prepared from Arabidopsis mesophyll cells. However, DPB3-1 expression did not affect the transactivation of an HsfA3 promoter-GUS reporter gene by DREB2A (data not shown). We hypothesized that DPB4 or other H2B-like HFD-containing proteins in Arabidopsis may be necessary to enhance the transactivation of DREB2A in protoplasts. We tested the interactions of DREB2A and DPB3-1 with DPB4 and the other 12 Arabidopsis H2B-like HFD-containing proteins (Siefers et al., 2009; Petroni et al., 2012) using yeast two-hybrid analysis. No proteins interacted with DREB2A; however, six proteins (NF-YB1, NF-YB2, NF-YB3, NF-YB6, NF-YB7, and DPB4) interacted with DPB3-1 in yeast cells (Figure 6A). A previous study suggested that DPB3-1 (NF-YC10) does not interact with any H2B-like proteins, including DPB4 (NF-YB11), in yeast cells (Hackenberg et al., 2012). The discrepancy between these studies might be caused by differences in the vector constructs and yeast strains used or in other experimental conditions. However, our result clearly indicated that DPB3-1 and DPB4 interact, as reported previously for yeast, human, and other eukaryotic cells (Kukimoto et al., 2004; Hartlepp et al., 2005; Asturias et al., 2006). We also confirmed that interactions occur between DPB3-1 and these six H2B-like proteins using the BiFC system analysis in Arabidopsis mesophyll protoplasts and concluded that these proteins could interact with DPB3-1 (Supplemental Figure 6A). Yeast DPB3/DPB4 and human CHRAC-15/CHRAC-17 reportedly interact with other factors to form protein complexes (Kukimoto et al., 2004; Asturias et al., 2006), and some studies have suggested that an NF-Y dimer comprising NF-YB and NF-YC might form a trimer with a transcription factor in Arabidopsis (Yamamoto et al., 2009; Liu and Howell, 2010; Li et al., 2011). Therefore, we tested whether DPB4 or other H2B-like HFD-containing proteins could form a trimer with DREB2A using yeast three-hybrid analysis. However, no H2B-like proteins formed a trimer with DREB2A and DPB3-1 (Supplemental Figure 6B). Moreover, the coexpression of each DPB3-1-interacting H2B-like protein with DREB2A and DPB3-1 in Arabidopsis mesophyll protoplasts did not enhance the transactivation of the HsfA3 promoter-GUS reporter gene (Supplemental Figures 6C and 6D). Therefore, we assumed that additional factors might be necessary to enhance reporter gene transactivation.
Figure 6.
NF-YA2 and NF-YB3 Were Identified as Candidate Components of the Trimer That Interacts with DREB2A under Heat Stress Conditions.
(A) The growth of yeast cells harboring the N-terminal region of DREB2A or full-length DPB3-1 fused to the GAL4 DNA binding domain (BD). SD/-L-W was used as the nonselective medium, and SD-L-W-H-Ade (quadruple drop out [QDO]) was the selective medium. H2B-like proteins were expressed as fusion proteins with the GAL4 activation domain (AD).
(B) Expression patterns of the NF-YB1, 2, 3, 6, and 7 and DPB4 genes under heat and dehydration stress conditions in 14-d-old plants.
(C) The growth of yeast cells harboring NF-YA family proteins fused to the GAL4 DNA binding domain (BD), NF-YB3 protein fused to the GAL4 activation domain (AD), and DPB3-1 fused to a nuclear localization signal (NLS) on selective medium (SD/-L-W-H-M-Ade).
(D) Coimmunoprecipitation of GFP-NF-YA2, Myc-NF-YB3, and FLAG-DPB3-1 in N. benthamiana leaves. Total protein extracts (Input) or immunoprecipitated (IP) fractions using an anti-GFP antibody were analyzed by immunoblotting (IB) using anti-GFP, anti-myc, or anti-FLAG antibodies. The triangles indicate specific signals for each expressed protein.
We examined the expression levels of the six genes encoding H2B-like HFD-containing proteins that interacted with DPB3-1 during dehydration and heat stress treatments in 14-d-old plants. DPB4 was found to be repressed by both heat and dehydration stress treatment (Figure 6B). A previous study revealed that the relative expression levels of human CHRAC-15 were correlated with those of CHRAC-17 in several tissues (Poot et al., 2000). The different and opposite expression patterns of Arabidopsis DPB3-1 and DPB4 suggested that DPB4 might not act in concert with DREB2A and DPB3-1 during heat stress. Expression analysis revealed that only NF-YB3 exhibited heat stress-specific gene induction (Figure 6B). The expression patterns of these six genes were similar in 7-d-old plants (Supplemental Figure 7A). These results suggest that NF-YB3 formed a complex with DREB2A and DPB3-1 under heat stress conditions, and we hypothesized that an additional NF-YA subunit was necessary to enhance reporter gene transactivation in protoplasts. We next performed yeast three-hybrid assays to identify NF-YA proteins that form a trimer with NF-YB3 and DPB3-1; only NF-YA2 was identified among the ten Arabidopsis NF-YA proteins (Figure 6C). Additional interaction assays in yeast cells confirmed that NF-YA2 did not directly interact with NF-YB3 (Supplemental Figure 7B). These results suggest that dimerization of DPB3-1 and NF-YB3 is required for the interaction with NF-YA2. Moreover, myc-NF-YB3 and FLAG-DPB3-1 were coimmunoprecipitated with GFP-NF-YA2 using anti-GFP microbeads, when each protein was transiently expressed in tobacco (Nicotiana benthamiana) leaves (Figure 6D), and BiFC analysis in Arabidopsis mesophyll cells also confirmed that NF-YA2, NF-YB3, and DPB3-1 interact in vivo (Supplemental Figure 7C).
Analysis of the crystal structure of human NF-Y identified several amino acid residues that are essential for NF-Y trimerization (Nardini et al., 2013). Several essential amino acid residues (Asp-175, Gln-176, Phe-180, and Asp-183) in Arabidopsis DPB3-1 were conserved with those in human NF-YC, whereas Glu-179 and Ser-184 in Arabidopsis DPB3-1 correspond to Asp-113 and Ile-117 in human NF-YC, respectively (Supplemental Figure 7D). The partial conservation of amino acid residues between human NF-YC and Arabidopsis DPB3-1 also suggests that NF-YA2, NF-YB3, and DPB3-1 might trimerize. NF-YA2 gene expression was relatively stable under heat stress conditions but decreased under drought stress conditions (Supplemental Figure 7E). Based on these results, we concluded that NF-YA2 and NF-YB3 might form a heat stress-specific transcription factor complex with DPB3-1 and DREB2A.
To reveal the phylogenetic relationships of NF-YA2 and NF-YB3 with each family protein, we searched for NF-YA and H2B-like HFD-containing proteins in other plant species using the same method as that used for the H2A-like HFD-containing proteins, based on the literature (Petroni et al., 2012; Laloum et al., 2013). Based on the amino acid sequences of the conserved domain of NF-YA and H2B-like proteins, we found 11 candidate NF-YA family proteins in O. sativa, two in P. patens, and one and two in the green algae Coccomyxa subellipsoidea C-169 and Micromonas pusilla CCMP1545, respectively (Supplemental Table 3). We also found 13 candidate H2B-like proteins in O. sativa, nine in P. patens, and three each in the green algae C. reinhardtii and V. carteri (Supplemental Table 4). Phylogenetic trees of the NF-YA and H2B-like family proteins were constructed based on alignments of the conserved domains (Supplemental Data Sets 3 and 4 and Supplemental Figures 8A and 8B). According to the phylogenetic tree, O. sativa has 11 NF-YB, one NC2β, and one DPB4 family proteins; P. patens has six NF-YB, two NC2β, and one DPB4 family proteins; and C. reinhardtii and V. carteri each have one NF-YB, one NC2β, and one DPB4 family protein. Arabidopsis NF-YA2 and NF-YB3 are conserved in eukaryotes. Thus, we assume that Arabidopsis NF-YA2 and NF-YB3 have functions that are conserved across eukaryotes.
Subcellular Localization and Tissue-Specific Expression Patterns of NF-YA2 and NF-YB3
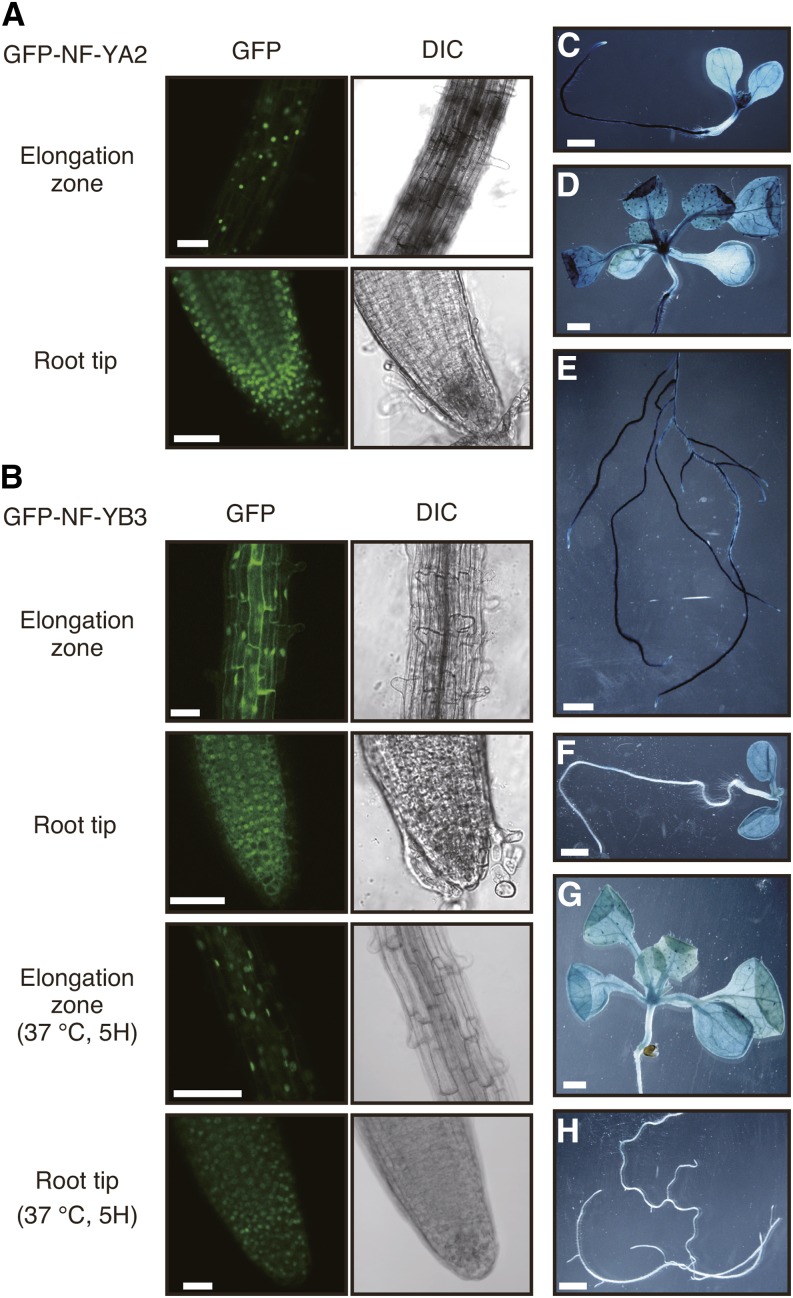
To reveal the subcellular localization of the NF-YA2 and NF-YB3 proteins, we generated transgenic plants carrying a 35S:sGFP-NF-YA2 or 35S:sGFP-NF-YB3 construct. Microscopy revealed that the sGFP-NF-YA2 protein was located in the nucleus under nonstress conditions, as suggested in a previous study (Hackenberg et al., 2012) (Figure 7A), and that the localization did not change under heat stress conditions (Supplemental Figure 9A). The fluorescence of the sGFP-NF-YB3 protein was detected in both the cytosol and the nucleus under nonstress conditions (Figure 7B). It was previously reported that NF-YB3 was located mainly in the cytosol in DMSO-treated (negative control) seedlings (Liu and Howell, 2010). This difference in localization from this study might result from the difference between the experimental conditions or protein accumulation levels. Heat stress-induced nuclear translocation of NF-YB3 in root cells as was also seen in tunicamycin-treated seedlings in the previous study (Liu and Howell, 2010) (Figure 7B). This translocation suggests that NF-YB3 has functions in the nucleus under heat stress conditions.
Figure 7.
Subcellular Localization of NF-YA2 and NF-YB3 Subunits and Tissue-Specific Expression Patterns of the NF-YA2 and NF-YB3 Genes.
(A) Nuclear localization of sGFP-NF-YA2. The root tissues of 35S:sGFP-NF-YA2 plants were observed under a microscope. Confocal images of GFP fluorescence and differential interference contrast (DIC) images are shown. Bars = 100 μm.
(B) Nuclear translocation of sGFP-NF-YB3 under heat stress conditions. The root tissues of 35S:sGFP-NF-YA2 plants were observed under a microscope. Confocal images of GFP fluorescence and differential interference contrast (DIC) images are shown. Bars =100 μm.
(C) to (H) GUS staining of NF-YA2pro:GUS ([C] to [E]) or NF-YB3pro:GUS ([F] to [H]) transgenic plants at different growth stages. Whole 7-d-old ([C] and [F]) and aerial ([D] and [G]) and root ([E] and [H]) tissues of 3-week-old seedlings. Bars = 1 mm.
We also examined the tissue-specific expression pattern of NF-YA2 and NF-YB3 to identify the tissues in which these proteins were coexpressed with DPB3-1. The 3- and 1-kb upstream sequences before the translational start sites of NF-YA2 and NF-YB3, respectively, were inserted upstream of the GUS gene, and then transgenic plants carrying each construct were generated. Histochemical staining of 7-d-old plants showed that the NF-YA2 and NF-YB3 genes are expressed in the petioles and leaf blades of the cotyledons and the tops of hypocotyls, similarly to DPB3-1 (Figures 7C and 7F). These expression patterns suggested that the trimer comprising NF-YA2, NF-YB3, and DPB3-1 mainly functions in these tissues. Additional observation of GUS activity at later developmental stages revealed that NF-YA2 and NF-YB3 were also expressed in true leaves and that NF-YA2 was strongly expressed in whole root tissues excluding the root tip (Figures 7D, 7E, 7G, and 7H; Supplemental Figures 9B and 9E). Moreover, NF-YA2 and NF-YB3 were coexpressed in several reproductive organ tissues (Supplemental Figures 9C, 9D, 9F, and 9G). NF-YA2 and NF-YB3 may have different functions in these tissues in which no GUS activity was detected in the DPB3-1 promoter-GUS plants.
A Trimer Comprising NF-YA2, NF-YB3, and DPB3-1 Enhanced the Transactivation of the HsfA3 Promoter by DREB2A in Protoplasts
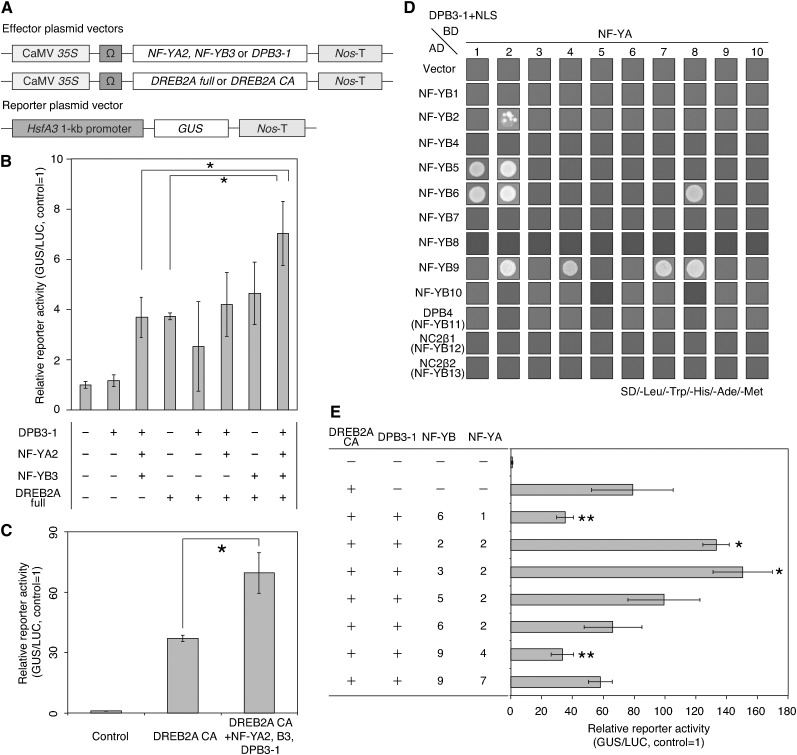
We performed transactivation assays using an HsfA3 promoter-GUS reporter gene with various combinations of the NF-Y subunits DPB3-1 and DREB2A in Arabidopsis mesophyll protoplasts (Figure 8A). The HsfA3 promoter fragment is located 1 kb from the translational start site and contains two DREs, one abscisic acid-responsive element-like (ABRE-like) motif and four CCAAT boxes as putative cis-acting elements (Supplemental Figure 10A). Full-length DREB2A alone weakly activated the reporter gene; however, when the trimer components were coexpressed together with full-length DREB2A, the reporter activity was significantly enhanced by almost 2-fold (Figure 8B). In contrast, coexpression of one or two subunits with DREB2A did not affect the transactivation of the reporter gene. This result suggests that the identified trimer activates the HsfA3 promoter in cooperation with DREB2A in protoplasts. We found that coexpression of the trimer components without DREB2A also induced activation of the HsfA3 promoter (Figure 8B). It is assumed that the trimer itself or its interacting factor is responsible for the transcriptional activity of the HsfA3 promoter or, alternatively, that the trimer functions with endogenous DREB2A in protoplasts. To test whether enhancement of the reporter transactivation by the trimer was additive or synergistic, we examined the effect of the trimer on the transactivation of the same HsfA3 promoter-GUS reporter by DREB2A CA. The result indicated that coexpression of the trimer with DREB2A CA also enhanced the transactivation of the reporter gene by almost 2-fold (Figure 8C). This result implies that DREB2A and the trimer of NF-YA2, NF-YB3, and DPB3-1 synergistically activate the HsfA3 promoter.
Figure 8.
Trimer Comprising NF-YA2, NF-YB3, and DPB3-1 Enhanced Reporter Activity in Cooperation with DREB2A.
(A) Schematic diagram of the effector and reporter constructs for the transactivation experiment. The HsfA3 1-kb promoter:GUS fusion gene was used as the reporter. The plasmids containing the CaMV 35S promoter and the tobacco mosaic virus Ω sequence fused to NF-YA2, NF-YB3, DPB3-1, full-length DREB2A, or DREB2A CA coding sequence were cotransfected into protoplasts. Nos-T indicates the terminator sequence of the gene for nopaline synthetase.
(B) Transactivation of the HsfA3 1-kb promoter:GUS reporter gene by various combinations of full-length DREB2A, NF-YA2, NF-YB3, and DPB3-1. The effector plasmids cotransfected are indicated as plus signs (+) in the table, and minus signs (-) indicate the cotransfection of empty effector plasmids. Bars indicate the mean values from assays performed in triplicate and the sd, and asterisks indicate significant differences between the reporter activities (P < 0.05 according to Bonferroni-corrected Student’s t test). The 35S:luciferase (LUC) plasmid was also cotransfected in each experiment as an internal control.
(C) Transactivation of the HsfA3 1-kb promoter:GUS reporter gene by DREB2A CA and the trimer of NF-YA2, NF-YB3, and DPB3-1. Bars indicate the mean values from assays performed in triplicate and the sd, and asterisks indicate significant differences between the reporter activities (P < 0.05 according to Student’s t test). The 35S:LUC plasmid was also cotransfected in each experiment as an internal control.
(D) Yeast three-hybrid assay to identify candidate trimers containing DPB3-1. The growth of yeast cells harboring NF-YB family proteins fused to the GAL4 DNA binding domain (BD), NF-YA family proteins fused to the GAL4 activation domain (AD), and DPB3-1 fused to a nuclear localization signal (NLS) on selective medium (SD/-L-W-H-M-Ade) are shown.
(E) Transactivation of the HsfA3 1-kb promoter:GUS reporter gene by various combinations of trimers and DREB2A CA. The effector plasmids cotransfected are indicated with the plus signs (+) or numbers in the table; for example, the number “2” in the “NF-YA” column of the table indicates that NF-YA2 was cotransfected. The minus signs (−) indicate the cotransfection of empty effector plasmids. Values represent means from assays performed in triplicate, and the bars indicate the sd. The asterisks indicate significant high (*) or low (**) differences between the reporter activities (P < 0.05 according to Bonferroni-corrected Student’s t test). The 35S:LUC plasmid was also cotransfected in each experiment as an internal control.
To examine the effect of other trimers comprising DPB3-1, H2B-like, and NF-YA proteins on the transcriptional activity of DREB2A, we performed comprehensive yeast three-hybrid assays. Consequently, we identified several candidate combinations of the trimer including DPB3-1 (Figure 8D; Supplemental Figure 10B); however, the transactivation assays with some of the identified trimer combinations showed that only the combination of NF-YA2, NF-YB2, and DPB3-1, as well as the combination of NF-YA2, NF-YB3, and DPB3-1, activated the HsfA3 promoter (Figure 8E). Because the NF-YB2 protein is phylogenetically located near NF-YB3 (Supplemental Figure 8B), these two proteins might function similarly in protoplasts. However, the NF-YB2 gene was repressed by heat stress and induced by dehydration stress (Figure 6B; Supplemental Figure 7A); thus, we speculate that NF-YB2 does not function under heat stress conditions in seedlings. On the other hand, two combinations of the trimers (NF-YA1, NF-YB6, and DPB3-1, and NF-YA4, NF-YB9, and DPB3-1) repressed reporter activity (Figure 8E). We also examined the effect of the trimer combinations on the transactivation of the RD29A promoter. RD29A is a dehydration-inducible DREB2A target gene, and the expression of this gene was not enhanced by DPB3-1 overexpression (Figure 4A). The RD29A promoter contains four DREs, one ABRE-like motif, and two CCAAT boxes (Supplemental Figure 10C). The DPB3-1 and NF-Y trimer did not affect or even repressed the transactivation of the RD29A promoter-GUS reporter gene by DREB2A CA (Supplemental Figures 10D and 10E). This result is consistent with the heat stress-specific function of the NF-Y trimer of NF-YA2, NF-YB3, and DPB3-1.
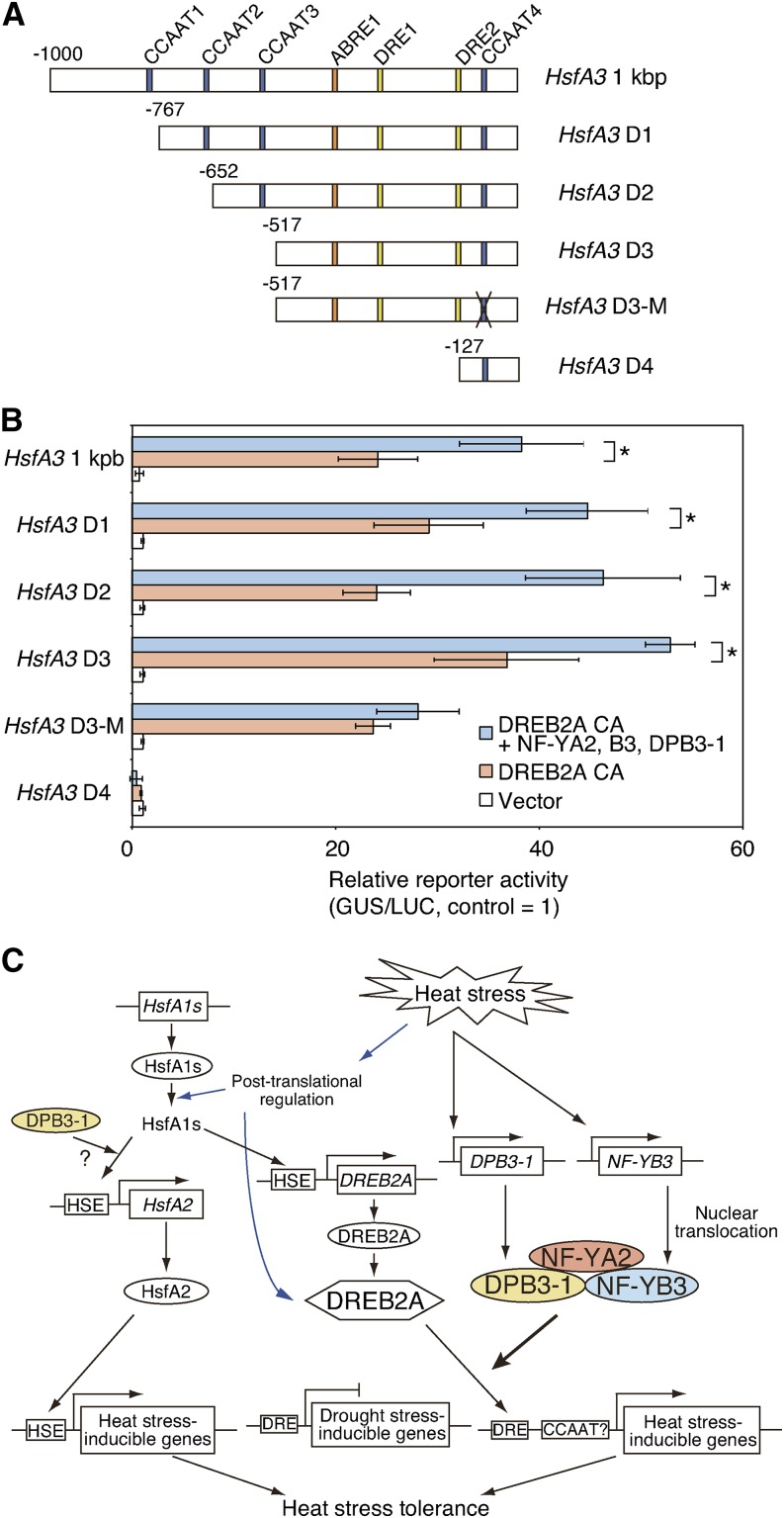
Synergistic Activation of the HsfA3 Promoter by DREB2A and the Identified Trimer Depends on the CCAAT Box
Because the identified trimer comprising NF-YA2, NF-YB3, and DPB3-1 (NF-YC10) functioned as a coactivator of DREB2A, we tested whether this trimer behaves like an NF-Y trimer in Arabidopsis mesophyll protoplasts. Many studies have reported that an NF-Y trimer binds to the CCAAT box and coactivates the target gene with a transcription factor (Yamamoto et al., 2009; Liu and Howell, 2010). Therefore, we constructed a series of HsfA3 promoter-GUS reporter genes containing deletions or mutations and tested the occurrence of the synergistic transactivation by DREB2A and the trimer of NF-YA2, NF-YB3, and DPB3-1 (Figure 9A). The deletion of three CCAAT boxes (CCAAT1, 2, and 3) in the promoters did not affect the enhancement of the reporter activity by the trimer (HsfA3 D1, D2, and D3); however, when base-change mutations were introduced into CCAAT4 (HsfA3 D3-M), the coexpression of NF-YA2, NF-YB3, and DPB3-1 together with DREB2A CA did not further enhance reporter activity (Figure 9B). These results suggested that the synergistic effect of the identified trimer in protoplasts was dependent on the CCAAT box on the HsfA3 promoter. To examine the functional redundancy among DPB3 and other H2A-like HFD-containing proteins, we performed additional transactivation assays employing H2A-like proteins instead of DPB3-1 using the HsfA3 1-kb promoter-GUS reporter gene. The trimer comprising NC2α1 (NF-YC11), NF-YC12, and DPB3-2 (NF-YC13) also enhanced the reporter activity with DREB2A CA, as was the case with DPB3-1 (NF-YC10) (Supplemental Figure 11A). These results suggested that functional redundancy might exist among the Arabidopsis H2A-like HFD-containing proteins, at least in protoplasts. We therefore tested whether these three H2A-like proteins interacted with DREB2A in protoplasts using BiFC analysis, and relatively weak fluorescence signals were detected compared with DPB3-1 (NF-YC10) (Supplemental Figure 11B). These data suggest that the three H2A-like proteins might interact with DREB2A in protoplasts, although such interactions were not detected in yeast cells (Supplemental Figure 1A).
Figure 9.
The Importance of an Upstream CCAAT Motif for the Enhancement of DREB2A-Dependent Reporter Activation by the Trimer of NF-YA2, NF-YB3, and DPB3-1 and a Model of the Function of the Identified Trimer under Heat Stress Conditions.
(A) Schematic diagram of the promoter fragments used in the reporter constructs. Each HsfA3 promoter fragment was fused to the GUS reporter gene. The positions of the DREs, CCAAT boxes, and an ABRE-like motif on the promoter are indicated. Crosses (×) indicate the mutation at the CCAAT motif. The effector plasmids used in this analysis are described in Figure 8A.
(B) Transactivation of each HsfA3 promoter:GUS reporter gene indicated in (A) by DREB2A CA and the trimer of NF-YA2, NF-YB3, and DPB3-1. Bars indicate mean values obtained from assays performed in triplicate and sd, and the asterisks indicate significant differences between the reporter activities (P < 0.05 according to Student’s t test). The 35S:LUC plasmid was also cotransfected in each experiment as an internal control.
(C) A model of the function of NF-YA2, NF-YB3, and DPB3-1 under heat stress conditions. The DREB2A, DPB3-1, and NF-YB3 genes are induced by heat stress. The DREB2A protein is stabilized under heat stress, and the NF-YB3 protein is translocated into the nucleus. NF-YA2, NF-YB3, and DPB3-1 form a trimer in the nucleus; thereafter, the trimer forms a transcriptional complex with DREB2A at the promoters of heat stress-inducible DREB2A target genes. Finally, the trimer enhances the efficiency of transcription by DREB2A. The effects of DPB3-1 levels on HsfA2 expression and the direct binding of DPB3-1 to the HsfA2 promoter under heat stress imply that DPB3-1 is involved in the transcription of HsfA2 by HsfA1s.
DISCUSSION
Arabidopsis DREB2A is an important transcription factor that specifically activates the expression of dehydration- and heat stress-responsive genes via the DRE sequence in promoters (Yamaguchi-Shinozaki and Shinozaki, 1994; Liu et al., 1998; Sakuma et al., 2002). The mechanisms that regulate the selective activation of stress-specific target genes by DREB2A remain to be elucidated. In this study, we identified DPB3-1 (NF-YC10), an H2A-like HFD-containing protein, as a potential coactivator of DREB2A that specifically functions under heat stress (Figure 1B). Arabidopsis has 14 H2A-like HFD-containing proteins (Petroni et al., 2012), and our phylogenetic analysis revealed that the Arabidopsis DPB3-1 belongs to a monophyletic subgroup conserved among land plants (Supplemental Figure 2C). This result suggests that DPB3-1 has a unique function among Arabidopsis H2A-like proteins. In accordance with this hypothesis, yeast two-hybrid assays revealed that, of the 14 H2A-like proteins, only DPB3-1 showed the clear interaction with DREB2A (Supplemental Figures 1A and 2D), although the BiFC experiments suggested the weak interactions between DREB2A and other H2A-like proteins (Supplemental Figure 11B). Orthologs of Arabidopsis DPB3-1 in other land plant species might interact with DREB2A homologs and have similar conserved roles.
The DPB3-1 gene was specifically induced by heat stress in 7-d-old plants (Figure 2K). This result suggests that DPB3-1 has a heat stress-specific function. The enhanced expression of the heat stress-inducible DREB2A target genes HsfA3, At1g75860, and At4g36010 in DPB3-1-overexpressing Arabidopsis (Figure 4A) and the direct binding of the DPB3-1 protein to the corresponding promoters under heat stress (Figure 4C) support this hypothesis. Moreover, the knockdown of DPB3-1 resulted in decreased expression of the heat stress-responsive genes of interest in the dpb3-1 mutant (Figure 5E). Conversely, the expression levels of the dehydration-inducible DREB2A target genes were not different under dehydration in 35S:DPB3-1 plants or in the dpb3-1 mutant (Figures 4A; Supplemental Figure 5G). These data indicate that DPB3-1 has a positive role in the heat stress response. Together with the interaction between DREB2A and DREB2A-dependent enhancement of the target gene expression (Supplemental Figures 4C to 4E), DPB3-1 most likely positively regulates its transcriptional activity specifically under heat stress. Previous studies have suggested that DPB3 family proteins that are associated with DNA polymerase represent part of the histone acetylating complex or the chromatin remodeling complex (Hartlepp et al., 2005; Asturias et al., 2006; Wang et al., 2008); however, it remains largely unknown whether and how DPB3 is involved in the regulation of gene expression. Here, we found that a DPB3 protein can regulate the activity of a transcription factor in Arabidopsis.
Because the coexpression of the DPB3-1 subunit alone or dimers of the DPB3-1 and H2B-like proteins with DREB2A did not affect the transactivation of the HsfA3 promoter-GUS reporter gene (Figure 8B; Supplemental Figure 6D), we identified the NF-YA2 subunit as a candidate protein that forms a trimer with DPB3-1 and NF-YB3 (Figure 6C) and NF-YB3 is encoded by a heat stress-inducible gene (Figure 6B). The identified trimer comprising NF-YA2, NF-YB3, and DPB3-1 enhanced HsfA3 promoter transactivation by DREB2A (Figure 8B), although this combination of trimer components did not further increase the activity of the RD29A promoter in combination with DREB2A (Supplemental Figure 10E). These results are consistent with the hypothesis that this trimer is involved in the heat stress-specific positive regulation of DREB2A. Many studies have shown that a trimer or a subunit of NF-Y is involved in the promoter binding or transcriptional efficiency of specific transcription factors (Wright et al., 1994; Li et al., 1998; Yamamoto et al., 2009; Liu and Howell, 2010; Schiavoni et al., 2010; Li et al., 2011). Our current results indicated that DPB3 family proteins could also affect gene expression, together with NF-YA and NF-YB subunits. Our data suggest a complex mechanism of H2A/H2B-like HFD-containing proteins in eukaryotic cells involving the possible combination of subunits comprising the protein complex to regulate gene expression levels in cooperation with another transcription factor. The transactivation assay revealed that a CCAAT box was necessary for HsfA3 promoter activation in protoplasts by the trimer of NF-YA2, NF-YB3, and DPB3-1 in protoplasts (Figure 9B). Therefore, it is assumed that this trimer of NF-YA2, NF-YB3, and DPB3-1 binds to the CCAAT box as the NF-Y trimer (Figure 9B). Analysis of the crystal structure of animal NF-Y trimer on the CCAAT box revealed that the CCAAT motif was mostly recognized by NF-YA and partially by NF-YB (Nardini et al., 2013). Therefore, the DPB3-1 subunit might not critically contribute to the binding of the trimer to the CCAAT motif. Further studies are required to understand the detailed mechanism by which the identified trimer enhances target gene expression.
Interestingly, some of the trimers (Figure 8D) containing DPB3-1 did not significantly enhance HsfA3 promoter-GUS reporter gene activation by DREB2A (Figure 8E). These analyses provide some suggestive evidence for the existence of a novel and complex relationship among H2A/H2B-like proteins and NF-YA subunits in Arabidopsis. The yeast three-hybrid analysis indicated that a H2A/H2B dimer is selective for NF-YA subunits, although the NF-YA subunits are highly similar to each other in Arabidopsis (Figure 8D). A previous study suggested that the NF-YB6/NF-YC3 dimer did not form a trimer with NF-YA2 (Calvenzani et al., 2012), although a recent study indicated that the NF-YB3/NF-YC9 dimer formed a trimer with NF-YA2 (Hou et al., 2014). Additionally, among the H2B-like proteins, DPB4, NC2β1, and NC2β2 did not form trimers with DPB3-1 and NF-YA (Figure 8D), suggesting that NF-YB proteins, among the H2B-like proteins, are necessary components in the formation of a trimer containing DPB3-1 and NF-YA. These facts imply that NF-YA subunits exhibit a preference to form trimers. Additionally, our results suggest functional diversity among the differently combined H2A/H2B and NF-YA subunits in land plants. The two types of trimers observed (NF-YA1, NF-YB6, and DPB3-1, and NF-YA4, NF-YB9, and DPB3-1) repressed HsfA3 promoter-GUS reporter activity in protoplasts (Figure 8E). Some reports have suggested that NF-Y demonstrates both activation and repression of gene expression (Willy et al., 2000; Dalvai et al., 2011; Zhu et al., 2012). Therefore, these two trimers, which repressed the HsfA3 promoter-GUS reporter, might function in Arabidopsis under non-heat-stressed conditions or during the attenuation of gene expression to regulate the heat stress-specific expression pattern. Intriguing data have been presented indicating that NF-YA2 overexpression in Arabidopsis enhanced heat stress tolerance; however, a significant number of heat stress-related genes in the MapMan ontology were repressed under the nonstress condition (Leyva-González et al., 2012). Some transcriptional complexes, including NF-YA2, might also repress the expression of heat stress-inducible genes under non-stress conditions. Upregulated genes in NF-YA2-overexpressing plants under nonstress conditions did not overlap with those in DPB3-1-overexpressing plants under heat stress conditions: This difference might result from the presence or absence of the heat stress. Moreover, our three-hybrid analysis revealed that NF-YB9 and NF-YB6 formed different trimers with DPB3-1 and NF-YA subunits (Figure 8D). The NF-YB9 and NF-YB6 genes (alias LEAFY COTYLEDON1 and LEAFY COTYLEDON1-LIKE, respectively) are reportedly required for development during the early and late phases of embryogenesis (Lotan et al., 1998; Kwong et al., 2003). According to the plant microarray database (Genevestigator; https://www.genevestigator.com/gv/), DPB3-1 is also expressed strongly in embryos. These results suggest that DPB3-1 might act in concert with NF-YB9 and NF-YB6 during embryogenesis.
Our microarray analysis of the 35S:DPB3-1 plants confirmed that some DREB2A target genes are upregulated upon heat stress (Supplemental Data Set 2), and the enhanced expression of those genes was confirmed using quantitative RT-PCR (Figure 4B). However, several heat stress-inducible genes that were not DREB2A targets were also identified as potential downstream of DPB3-1 (Supplemental Data Set 2). Interestingly, the expression of HsfA2 was enhanced in the 35S:DPB3-1 plants, while it was reduced in the dpb3-1 mutant (Figures 4B and 5E). Moreover, ChIP assays confirmed the direct binding of DPB3-1 to the HsfA2 promoter under heat stress (Figure 4C). Previous studies have revealed that HsfA2 is a major transcription factor that regulates numerous target genes in response to heat stress and that HsfA2 is not a DREB2A target gene (Sakuma et al., 2006b). Instead, it is induced by HsfA1 master transcription factors in response to heat shock (Yoshida et al., 2011). Therefore, instead of the effects on DREB2A, DPB3-1 (NF-YC10) may positively regulate the expression of HsfA2 together with HsfA1s. Other studies in human and other animal cells suggested that an NF-Y trimer induces the transcription of heat stress-inducible genes together with heat shock factors (Li et al., 1998; Imbriano et al., 2001; Sasi et al., 2014). Further analysis is necessary to confirm whether Arabidopsis DPB3-1 and other NF-Y subunits form a transcriptional complex with HsfA1s to regulate HsfA2 expression.
Consistent with the increased expression levels of the various heat stress-inducible genes in the DPB3-1-overexpressing Arabidopsis, these transgenic plants showed significantly enhanced heat stress tolerance (Figure 3E). Moreover, DPB3-1-overexpressing plants did not exhibit any growth retardation compared with the wild-type plants under normal conditions (Figures 3B to 3D; Supplemental Figures 3C and 3D). The overexpression of transcription factors that have constitutive activity, such as DREB1A, DREB2A CA, and HsfA2, often causes severe growth retardation under normal conditions (Liu et al., 1998; Sakuma et al., 2006a; Ogawa et al., 2007). Therefore, the application of these strong transcription factors to the molecular breeding of crops requires an optimized stress-inducible promoter for the control of gene expression to minimize negative effects on plant growth (Kim et al., 2003; Sakuma et al., 2006a; Nakashima et al., 2007; Pino et al., 2007). However, the optimization of stress-inducible promoters for each gene and plant species is time-consuming. Due to its simplicity, the overexpression of DPB3-1 or its orthologs could be a useful strategy for generating stress-tolerant crops. According to the plant microarray database (Genevestigator; https://www.genevestigator.com/gv/), a DPB3-1 ortholog in O. sativa, LOC_Os03g63530 (OsDPB3-2), is also induced upon heat stress (>3-fold change; P < 0.001).
Although our study strongly suggests that DPB3-1 enhances the transactivation of target promoters by DREB2A and raises the possibility that the trimer of NF-YA2, NF-YB3, and DPB3-1 forms a transcriptional complex with DREB2A specifically under heat stress, further analysis is necessary to elucidate the detailed mechanisms. First, the mechanisms underlying the dehydration stress-specific target gene expression of DREB2A remain unknown. It is possible that another combination of a trimer or enhancers other than H2A/H2B-like proteins may regulate dehydration stress-specific target gene expression. Recent studies revealed the cell type-specific expression of enhancers and cell type-specific chromatin connectivity and suggested the existence of cell type-specific transcriptional complexes in human and mouse cells (Jin et al., 2013; Zhang et al., 2013; Andersson et al., 2014). Future research should focus on the identification of various stress-specific transcriptional complexes in eukaryotes, including Arabidopsis. In addition, the detailed mechanisms by which the identified trimer enhanced the transcriptional activity of DREB2A were not elucidated. The overexpression of DPB3-1 did not affect the amount of protein accumulation or DNA binding activity of DREB2A (Supplemental Figures 4 and 4F). Therefore, the trimer may enhance the transcriptional efficiency of DREB2A via other mechanisms. One possibility is that the identified trimer regulates histone modifications for gene activation. DPB3/DPB4 family proteins are reportedly components of the histone-acetylating complex (Wang et al., 2008) and exhibit nucleosome remodeling activity (Hartlepp et al., 2005). Additionally, NF-Y trimers have been suggested to act in concert with a histone acetyltransferase (Li et al., 1998; Caretti et al., 2003; Peng et al., 2007), affecting the acetylation of histone H2A.Z on promoters (Li et al., 1998; Caretti et al., 2003; Peng et al., 2007; Gatta and Mantovani, 2011). Moreover, a recent study revealed that a NF-Y trimer is also involved in histone methylation to regulate the target gene expression of a transcription factor (Hou et al., 2014). Therefore, further study is required to reveal the regulation of histone modifications during drought or heat stress responses.
Figure 9C shows our current model. When Arabidopsis cells receive a heat stress signal, the expression levels of the DREB2A, DPB3-1, and NF-YB3 genes are increased. In addition, the stress signal also induces the stabilization of DREB2A and the nuclear translocation of NF-YB3. NF-YB3 and DPB3-1 form a trimer with NF-YA2 in the nucleus, and the trimer comprises a transcriptional complex with stabilized DREB2A on the promoters of target genes. We also hypothesize that DPB3-1 might be involved in the induction of the HsfA2 gene, which is mainly controlled by the HsfA1 transcription factors, and that the CCAAT box on the target promoter has an important role for gene induction. Thus, DPB3-1 enhances the induction of heat-inducible genes and contributes to the acquisition of heat stress tolerance. In conclusion, we found that DPB3-1 is a DREB2A-interacting factor and identified the candidate trimer that regulates heat stress-specific target gene expression of DREB2A. Our findings provide insight into the elaborate mechanisms that regulate stress-specific gene expression through the formation of a specific transcriptional complex.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia plants were grown on germination medium (GM) agar plates at 22°C under a 16-h photoperiod at a photon flux density of 40 μmol m−2 s−1 as described previously (Sakuma et al., 2006a, 2006b). At 16 d of age they were transferred to perlite-containing soil (Dio Chemicals) in a plastic pot with a diameter of 6 cm and continued to be grown under the conditions described above. For the analysis of root growth, the seedlings were grown on agar medium for 3 d and then in square Petri dishes containing 35 mL of 1.2% solid medium (0.5× Murashige and Skoog medium). The T-DNA insertion line for DPB3-1 was obtained from the ABRC (SALK_122213). The T-DNA insertion was confirmed by amplifying the left border-flanking region of the genomic DNA using the primer set given in Supplemental Table 5. The T-DNA insertion line for DREB2A was described previously (Sakuma et al., 2006b). Transgenic plants were generated by the floral dip method (Clough and Bent, 1998) using Agrobacterium tumefaciens GV3101 (pMP90) cells. More than two independent transgenic lines were analyzed for each construct.
Yeast Two-Hybrid Screening and Interaction Assays in Yeast
An Arabidopsis cDNA library was prepared in the pGADT7 vector in the MatchMaker GAL4 two-hybrid system 3 (Clontech) using RNA isolated from 2-week-old seedlings grown on solid GM agar plates according to the manufacturer’s instructions. Yeast two-hybrid screening and two-hybrid or three-hybrid interaction assays were performed as described in the user manual supplied with the Matchmaker Gold Two-Hybrid System (Clontech). The bait vector harboring the DREB2A fragment for the screening was used previously (Qin et al., 2008).
Stress Treatment
The dehydration treatment was performed as described previously (Liu et al., 1998). For the heat treatment, 8- or 16-d-old plants on GM agar plates with or without antibiotics were transferred to 37°C in a hybridization oven (HB-80; TAITEC). The plant heat or drought stress tolerance test was performed as previously described (Sakuma et al., 2006a, 2006b). For the salt stress tolerance test, the seedlings were grown on solid GM for 9 d and then transferred onto 0.8% agar plates (0.5× Murashige and Skoog medium) supplemented with or without 175 mM NaCl. They were grown for ∼9 to 10 d after the transfer at 22°C.
Pull-Down Assay
The DPB3-1-His protein was expressed in 2× YT broth and purified using His-Bind Kits (Novagen) as described in the supplier’s instructions. The GST and DREB2A N-GST proteins were expressed in Overnight Express Instant LB Medium (Novagen) and purified using glutathione Sepharose 4B according to the handbook for the GST gene fusion system (Amersham Biosciences). The purified DPB3-1-His protein (1 μg) was bound to 100 μL of the His-Bind Resin and washed as described in the supplier’s instructions. The resin was then mixed with 1 μg of the purified GST or DREB2AN-GST proteins in 1 mL of the PBS buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4) and rotated at 4°C for 1 h. The resin was washed with the PBS buffer and incubated in 100 μL of the elution buffer (1 M imidazole, 0.5 M NaCl, and 20 mM Tris-HCl, pH 7.9). The DPB3-1-His proteins and GST or DREB2AN-GST proteins were detected using the Anti-His-tag and Anti-GST-tag HRP-DirecT antibody (MBL). SuperSignal West Dura Extended Duration Substrate (Thermo Scientific) and an Image Quant LAS 4000 system (GE Healthcare) were used to visualize the signals.
GFP and BiFC Fluorescence Observation and Histochemical GUS Staining
Sixteen-day-old seedlings that expressed the fluorescent proteins were observed using a confocal laser scanning microscope (LSM5 PASCAL; Zeiss). To accumulate the DREB2A protein, the transfected protoplasts were treated with 25 μM MG132, a 26S proteasome inhibitor that promotes the accumulation of the DREB2A protein (Morimoto et al., 2013), for 2 h in the dark. Histochemical GUS staining was performed by soaking the plants in a solution containing 0.26 mg/mL 5-bromo-4-chloro-3-indolyl β-d-glucuronide, 50 mM phosphate buffer, pH 7.0, and 20% (v/v) methanol at 28°C for 12 to 24 h.
Coimmunoprecipitation Experiments in Arabidopsis Plants
One gram of the transgenic plants was ground in liquid nitrogen and homogenized in 3 mL of an extraction buffer containing 100 mM Tris-HCl, pH 8.0, 400 mM NaCl, 1% (v/v) Triton X-100, and a protease inhibitor mixture (cOmplete, EDTA-free; Roche Applied Science). The homogenate was sonicated on ice and treated with 25 units of Benzonase Nuclease (Merck) for 2 h on ice. The solution was filtered through Miracloth (Merck) and centrifuged at 500g for 2 min; then, the supernatant was further centrifuged at 10,000g for 10 min. The resulting supernatant was mixed with 6 mL of a binding buffer containing 100 mM Tris-HCl, pH 8.0, 1% (v/v) Triton X-100, and the protease inhibitor mixture. The SCFP3AC-HA-DREB2A protein was immunoprecipitated using anti-HA agarose beads (Sigma-Aldrich). The resin was washed with wash buffer (100 mM Tris-HCl, pH 8.0, 100 mM NaCl, and 1% [v/v] Triton X-100) and eluted with 100 μL of the 2× SDS sample buffer (100 mM Tris-HCl, pH 6.8, 4 mM EDTA, 2% [w/v] SDS, 20 mM DTT, and 2% [v/v] glycerol). The resulting immunoprecipitates were subjected to immunoblot analysis using an anti-HA antibody (Sigma-Aldrich). The coimmunoprecipitated VenusN-myc-DPB3-1 protein was analyzed by immunoblotting using an anti-myc antibody (Millipore). The signals were visualized as described above.
Analysis for Stabilization of the DREB2A Protein
Detection of the DREB2A protein was performed as described previously (Morimoto et al., 2013) with minor modifications. For the signal detection, SuperSignal West Dura Extended Duration Substrate (Thermo Scientific) and an Image Quant LAS 4000 system (GE Healthcare) were used. The Rubisco large subunit (rbcL) was visualized with Coomassie Brilliant Blue staining solution (0.25% [w/v] Coomassie Brilliant Blue R 250, 40% [v/v] methanol, and 10% [v/v] acetic acid) to demonstrate equal loading.
RNA Preparation, Quantitative RT-PCR, and RNA Gel Blot Analysis
The total RNA was isolated with RNAiso plus (TaKaRa) from 7- or 14-d-old plants according to the supplier’s instructions. RNA gel blot analyses were performed as described previously (Yamaguchi-Shinozaki and Shinozaki, 1994). For quantitative RT-PCR, cDNA was synthesized from the total RNA using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) as described in the supplier’s instructions. Quantitative RT-PCR analyses were performed using an Applied Biosystems 7500 real-time PCR system. The Power SYBR Green PCR Master Mix (Applied Biosystems) was used for the reactions. The primers used for quantitative RT-PCR are listed in Supplemental Table 4. Triplicate measurements were made for each cDNA sample, and the obtained values were normalized according to the amounts of 18S rRNA.
Sequence Alignment and Phylogenic Analysis
The peptide sequences of the each family protein were obtained from Phytozome (Phytozome v9.1, http://www.phytozome.net/). Alignment of the family proteins was performed using eBioX software (http://macdownload.informer.com/ebiox/download/) and the ClustalW program. Neighbor-joining phylogenetic trees were constructed using MEGA6 (http://www.megasoftware.net/) with the Poisson model, 1000 bootstrap replicates, and pairwise deletion for gap/missing data treatment.
Microarray Analysis
The total RNA was isolated from the vector control and two lines of transgenic plants overexpressing DPB3-1 (35S:DPB3-1-b, -c) and used to prepare Cy5- and Cy3-labeled cRNA probes. All microarray experiments, including the data analysis, were performed by the two-color method according to Agilent’s instructions, as described previously (Qin et al., 2008). All microarray data have been deposited to ArrayExpress (http://www.ebi.ac.uk/arrayexpress/; E-MTAB-2144).
ChIP
ChIP assays were performed using an EpiQuik Plant ChIP kit (Epigentek) according to the user guide. Samples of 1 g were collected from 35S:sGFP-DPB3-1 transgenic plants grown on GM agar plates for 2 weeks. To immunoprecipitate the DNA-protein complexes containing sGFP-DPB3-1, an anti-GFP antibody (Sigma-Aldrich) was used. The primer sets employed to amplify the promoter regions and 18S rDNA are shown in Supplemental Table 5.
Transient Expression Assays with Arabidopsis Mesophyll Protoplasts
Transient transformation of Arabidopsis mesophyll protoplasts was performed as described (Yoo et al., 2007) with minor modifications. Approximately 2 × 104 cells were added to a plasmid mixture (4 μg of internal control and 3 μg of other effector or reporter plasmid vectors) for transfection. After incubation, the transfected protoplasts were centrifuged at 150g for 2 min and resuspended in 150 μL of lysis buffer (19.5 mM NaH2PO4, 30.5 mM Na2HPO4, 1 mM EDTA, 0.1% [w/w] Triton X-100, and 0.07% [w/w] 2-mercaptoethanol), and the protoplasts were disrupted with a homogenizer (AS ONE) on ice. The solution was centrifuged at 20,000g for 5 min. Luciferase activity was measured with a Picagene BrillianStar-LT Luminescence Kit (Toyo B-Net) according to the manufacturer's instructions. For GUS assays, 10 μL of the protoplast lysate was added to 100 μL of methylumbelliferyl glucuronide substrate mix (19.5 mM NaH2PO4, 30.5 mM Na2HPO4, 1 mM EDTA, 0.1% [w/w] Triton X-100, 0.07% [w/w] 2-mercaptoethanol, 0.04% [w/v] 4-methylumbelliferyl glucuronide) and incubated for 60 min at 37°C. To stop the reaction, 800 μL of 0.2 M Na2CO3 was added. The LUC luminescence and MU fluorescence were measured by an ARVO MX plate reader (Perkin-Elmer).
Transient Expression and Coimmunoprecipitation Experiments in Nicotiana benthamiana
Agrobacterium infiltration experiments were performed as reported previously (Romeis et al., 2001). Briefly, Agrobacterium GV3101 (pMP90) cells harboring plasmids for protein expression were cultured and then harvested by centrifugation. The cells were resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES, pH 5.6, and 150 μM acetosyringone) to an OD600 of 0.5. After incubation for 2 h at room temperature, the resuspended cells were infiltrated into leaves using a syringe. Three days after infiltration, the leaves were harvested and immediately frozen in liquid nitrogen. Coimmunoprecipitation experiments were performed as described in the user manual supplied with the μMACS Epitope Tag Protein Isolation Kit (Miltenyi Biotec) using nuclear extracts obtained from the infiltrated leaves.
Construct Generation
To generate the 35S:DPB3-1 constructs, the DPB3-1 coding sequence was inserted into the XbaI and XhoI sites of the pGKX vector (Qin et al., 2008). This construct was used for transient expression assays and the generation of DPB3-1-overexpressing Arabidopsis.
The full-length DPB3-1 coding sequence was cloned into the ClaI and XhoI sites of the prey vector pGADT7 or into the SalI and PstI sites of the bait vector pGBKT7. The plasmids containing the various DREB2A fragments were generated by inserting cDNA fragments amplified by PCR into the pGBKT7 vector between the EcoRI and PstI sites. The NF-YB1 and NF-YB10 coding sequences were cloned into the BamHI and XhoI sites, NC2β1 was inserted between the EcoRI and ClaI sites, and other H2B-like genes were cloned into the ClaI and XhoI sites of the pGADT7 vector. The NF-YC1, NF-YC4, and NC2α2 coding sequences were cloned into the EcoRI and ClaI, ClaI and XhoI, SmaI and XhoI sites, respectively, and other H2A-like genes were inserted into the EcoRI and XhoI sites of the pGADT7 vector. For three-hybrid assays, the NF-YA1, 2, 3, 9, and 10 coding sequences were cloned into the SmaI and BamHI sites, and other NF-YA family genes were cloned into BamHI and PstI sites of the pBridge vector. The DREB2A fragment was cloned into the EcoRI and PstI sites of multi-cloning site I (MCSI) and the DPB3-1 coding sequence was cloned into the NotI site of MCSII of the pBridge vector. These constructs were used for yeast two-hybrid or three-hybrid assays.
The coding sequence of DPB3-1 was amplified by PCR and inserted into the pCold I vector (TaKaRa) between the BamHI and PstI sites, and the DREB2AN-GST expression construct was described previously (Liu et al., 1998). Each construct for recombinant protein production was transformed into Rosetta (DE3) pLysS cells (Novagen).
For GFP fluorescence observation, the coding sequences of NF-YA2, NF-YB3, and DPB3-1 were inserted into pGKX-NsGFP, and the coding sequence of DPB3-1 was also inserted into the pGKX-CsGFP vector between the XbaI and XhoI sites (Qin et al., 2008).
For the BiFC assays, the DREB2A and DPB3-1 coding sequences were inserted into the XbaI and XhoI sites of the pSCYCE vector and the SpeI and ClaI sites of the pVYNE vector (Waadt et al., 2008), respectively. In addition, the DPB3-1 and NF-YA2 coding sequences were inserted into the XbaI and XhoI sites and other H2A-like genes were inserted into the SalI and XhoI sites of the pUCSPYNE vector (Qin et al., 2008). DREB2A coding sequences were inserted into the SpeI and ClaI sites, and H2B-like genes were inserted into the XbaI and XhoI sites of the pUCSPYCE vector (Qin et al., 2008), respectively, for the BiFC assay in protoplasts.
For histochemical GUS staining, 3-kb NF-YA2 and 1-kb NF-YB3 or DPB3-1 promoter fragments upstream of the translational start site were amplified from Columbia plants. These fragments were positioned upstream of GUS in the pGK-GUS vector (Qin et al., 2008). The NF-YA2 and NF-YB3 promoters were inserted into the PstI and XhoI sites, and the DPB3-1 promoter was inserted into the SacI and PstI sites.
To generate the 35S:NF-YA, 35S:H2A-like, and 35S:H2B-like gene constructs, the coding sequences were inserted into the pGKX vector (Qin et al., 2008) between the XbaI and XhoI sites. NC2α1 and NF-YC12 were inserted into the SmaI and XhoI sites and DPB3-2 were inserted into the BamHI and XhoI sites of the pGKX vector. Various fragments of the HsfA3 promoter upstream of the translational start site were positioned upstream of GUS in the pGK-GUS vector between the SacI and PstI sites, and 1-kb HsfA3 and RD29A promoter fragments were inserted into the SpeI and PstI sites and XbaI and PstI sites, respectively. To create base change mutations in CCAAT boxes on the HsfA3 promoter, the megaprimer PCR method was used. The 35S:LUC reporter construct used as an internal control, and the 35S:DREB2A and 35S:DREB2A CA constructs were described previously (Liu et al., 1998; Sakuma et al., 2006b). These constructs were used for the transient expression assays.
For coimmunoprecipitation analysis in N. benthamiana leaves, the NF-YB3 and DPB3-1 coding sequences were inserted into the XbaI and XhoI sites of the pGKX-NMyc vector and pGHX-NSF vector (described below), respectively.
Construction of Vectors
To construct pGKX-Nmyc, a DNA fragment containing the Myc tag coding sequence was generated by dimerization and extension of the primer pair Myc-N/SpeI and Myc-C/XbaI. The ends of the DNA fragment were cleaved using SpeI and XbaI and inserted into the XbaI site of the pGKX vector (Qin et al., 2008).
For pGHX-NSF, first, the pGHX-NStrepII vector was generated from the pGHX vector (a hygromycin version of pGKX; Fujita et al., 2012) in the same way as pGKX-NMyc using the primer pair StrepII-N/SpeI and StrepII-C/XbaI. To construct the tandem tag vector pGHX-NSF, a DNA fragment containing the 3×FLAG tag was amplified using PCR with the primer pair 3×Flag-N/SpeI and 3×Flag-C/XbaI; p3XFLAG-CMV-8 (Sigma-Aldrich) was used as a template. The ends of the DNA fragment were cleaved using SpeI and XbaI and inserted into the XbaI site of the pGHX-NStrepII vector.
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource (http://www.arabidopsis.org/), GenBank, EMBL, or Phytozome database under the accession numbers listed in Supplemental Tables 1, 3, and 4.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Analysis of the Interactions between DREB2A and H2A-Like Proteins and Verification of the DREB2A-DPB3-1 Interaction by the BiFC System in Transgenic Arabidopsis under the Control Condition or in Mesophyll Protoplasts.
Supplemental Figure 2. Expression Patterns of DPB3-1 and a Phylogenetic Tree of the H2A-Like HFD-Containing Proteins.
Supplemental Figure 3. Stress Tolerance and Root Growth of DPB3-1-Overexpressing Arabidopsis.
Supplemental Figure 4. Expression Analysis of the Heat Stress-Inducible Genes in 35S:DPB3-1/dreb2a-1 Plants and Effect of the DPB3-1 Subunit on the Accumulation Level or DNA Binding Activity of DREB2A.
Supplemental Figure 5. Additional Analysis of the dpb3-1 Mutant.
Supplemental Figure 6. Confirmation of the Interaction between DPB3-1 and H2B-Like Proteins, Analysis of the Interaction between DREB2A and H2A/H2B Dimers Comprising DPB3-1 and H2B-Like Proteins, and the Effect of the H2A/H2B Dimers on the Transcriptional Activity of DREB2A.
Supplemental Figure 7. Expression Analysis of the NF-YA2 and H2B-Like Genes under Stress Conditions in 7-d-Old Plants, Interaction among NF-YA2 and NF-YB3 and DPB3-1, and Conservation of the Amino Acids Residues for the Trimerization of a NF-Y Trimer.
Supplemental Figure 8. Phylogenetic Trees of the NF-YA and H2B-Like HFD-Containing Proteins.
Supplemental Figure 9. Subcellular Localization of NF-YA2 under Heat Stress Conditions and Additional Analysis of the Tissue-Specific Expression Patterns of NF-YA2 and NF-YB3.
Supplemental Figure 10. Schematic Diagram of the HsfA3 1-kb and RD29A 1-kb Promoters, Promoter-Specific Effect of the Trimer Comprising NF-YA2, NF-YB3, and DPB3-1, and Yeast Three-Hybrid Assay on Nonselective Medium.
Supplemental Figure 11. Effect of Other H2A-Like Proteins on the Reporter Activity with DREB2A and Interaction between the H2A-Like Proteins and DREB2A in Protoplasts.
Supplemental Table 1. Gene Names and Accession Numbers of the H2A-Like HFD-Containing Proteins.
Supplemental Table 2. GO Analysis of the Genes Upregulated in DPB3-1-Overexpressing Plants under Heat Stress.
Supplemental Table 3. Gene Names and Accession Numbers of the NF-YA Family Proteins.
Supplemental Table 4. Gene Names and Accession Numbers of the H2B-Like HFD-Containing Family Proteins.
Supplemental Table 5. Sequences of Primers Used in This Study.
Supplemental Data Set 1. Alignment of the H2A-Like HFD-Containing Proteins.
Supplemental Data Set 2. Genes Upregulated in the DPB3-1-Overexpressing Plants under the Heat Stress Condition.
Supplemental Data Set 3. Alignment of the NF-YA Family Proteins.
Supplemental Data Set 4. Alignment of the H2B-Like HFD-Containing Proteins.
Supplementary Material
Acknowledgments
We thank Y. Tanaka and S. Murasaki for technical assistance, K. Yoshiwara for microarray analysis, and E. Toma for skillful editorial assistance. This work was supported by a Grant-in-Aid for JSPS Fellows (25-4185 to H.S.) and Scientific Research on Innovative Areas (22119004 to K.Y.-S.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, the Program for the Promotion of Basic Research Activities for Innovative Biosciences (BRAIN) of Japan, and the Science and Technology Research Partnership for Sustainable Development (SATREPS) of the Japan Science and Technology Agency/Japan International Cooperation Agency (to K.Y.-S.).
AUTHOR CONTRIBUTIONS
H.S. designed the research, performed the experiments, analyzed data, and wrote the article. J.M. designed the research and edited the article. H.T. and K.S. designed the research. K. Maruyama performed the microarray analysis and analyzed the data. F.Q., T.O., K.K., K. Morimoto, M.K., and Y.O. contributed materials and methods. K.Y.S. designed the research and edited the article.
Glossary
- DRE
dehydration-responsive element
- BiFC
bimolecular fluorescence complementation
- ChIP
chromatin immunoprecipitation
- GM
germination medium
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Andersson R., et al. ; FANTOM Consortium (2014). An atlas of active enhancers across human cell types and tissues. Nature 507: 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H., Hamatake R.K., Morrison A., Johnson A.L., Johnston L.H., Sugino A. (1991). Cloning DPB3, the gene encoding the third subunit of DNA polymerase II of Saccharomyces cerevisiae. Nucleic Acids Res. 19: 4867–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asturias F.J., Cheung I.K., Sabouri N., Chilkova O., Wepplo D., Johansson E. (2006). Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo-electron microscopy. Nat. Struct. Mol. Biol. 13: 35–43. [DOI] [PubMed] [Google Scholar]
- Bartels D. (2005). Desiccation tolerance studied in the resurrection plant Craterostigma plantagineum. Integr. Comp. Biol. 45: 696–701. [DOI] [PubMed] [Google Scholar]
- Berardini T.Z., et al. (2004). Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 135: 745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvenzani V., Testoni B., Gusmaroli G., Lorenzo M., Gnesutta N., Petroni K., Mantovani R., Tonelli C. (2012). Interactions and CCAAT-binding of Arabidopsis thaliana NF-Y subunits. PLoS ONE 7: e42902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caretti G., Salsi V., Vecchi C., Imbriano C., Mantovani R. (2003). Dynamic recruitment of NF-Y and histone acetyltransferases on cell-cycle promoters. J. Biol. Chem. 278: 30435–30440. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Schumaker K., Zhu J.K. (2004). Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J. Exp. Bot. 55: 225–236. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Dalvai M., Mondesert O., Bourdon J.C., Ducommun B., Dozier C. (2011). Cdc25B is negatively regulated by p53 through Sp1 and NF-Y transcription factors. Oncogene 30: 2282–2288. [DOI] [PubMed] [Google Scholar]
- Edwards D., Murray J.A., Smith A.G. (1998). Multiple genes encoding the conserved CCAAT-box transcription factor complex are expressed in Arabidopsis. Plant Physiol. 117: 1015–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Fujita Y., Iuchi S., Yamada K., Kobayashi Y., Urano K., Kobayashi M., Yamaguchi-Shinozaki K., Shinozaki K. (2012). Natural variation in a polyamine transporter determines paraquat tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 6343–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatta R., Mantovani R. (2011). NF-Y affects histone acetylation and H2A.Z deposition in cell cycle promoters. Epigenetics 6: 526–534. [DOI] [PubMed] [Google Scholar]
- Gnesutta N., Nardini M., Mantovani R. (2013). The H2A/H2B-like histone-fold domain proteins at the crossroad between chromatin and different DNA metabolisms. Transcription 4: 114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg D., Wu Y., Voigt A., Adams R., Schramm P., Grimm B. (2012). Studies on differential nuclear translocation mechanism and assembly of the three subunits of the Arabidopsis thaliana transcription factor NF-Y. Mol. Plant 5: 876–888. [DOI] [PubMed] [Google Scholar]
- Hartlepp K.F., Fernández-Tornero C., Eberharter A., Grüne T., Müller C.W., Becker P.B. (2005). The histone fold subunits of Drosophila CHRAC facilitate nucleosome sliding through dynamic DNA interactions. Mol. Cell. Biol. 25: 9886–9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Zhou J., Liu C., Liu L., Shen L., Yu H. (2014). Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat. Commun. 5: 4601. [DOI] [PubMed] [Google Scholar]
- Imbriano C., Bolognese F., Gurtner A., Piaggio G., Mantovani R. (2001). HSP-CBF is an NF-Y-dependent coactivator of the heat shock promoters CCAAT boxes. J. Biol. Chem. 276: 26332–26339. [DOI] [PubMed] [Google Scholar]
- Jin F., Li Y., Dixon J.R., Selvaraj S., Ye Z., Lee A.Y., Yen C.A., Schmitt A.D., Espinoza C.A., Ren B. (2013). A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503: 290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.Y., Kwon S.Y., Lee H.S., Hur Y., Bang J.W., Kwak S.S. (2003). A novel oxidative stress-inducible peroxidase promoter from sweetpotato: molecular cloning and characterization in transgenic tobacco plants and cultured cells. Plant Mol. Biol. 51: 831–838. [DOI] [PubMed] [Google Scholar]
- Kukimoto I., Elderkin S., Grimaldi M., Oelgeschläger T., Varga-Weisz P.D. (2004). The histone-fold protein complex CHRAC-15/17 enhances nucleosome sliding and assembly mediated by ACF. Mol. Cell 13: 265–277. [DOI] [PubMed] [Google Scholar]
- Kwong R.W., Bui A.Q., Lee H., Kwong L.W., Fischer R.L., Goldberg R.B., Harada J.J. (2003). LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15: 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloum T., De Mita S., Gamas P., Baudin M., Niebel A. (2013). CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 18: 157–166. [DOI] [PubMed] [Google Scholar]
- Leyva-González M.A., Ibarra-Laclette E., Cruz-Ramírez A., Herrera-Estrella L. (2012). Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS ONE 7: e48138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Distelfeld A., Comis A., Dubcovsky J. (2011). Wheat flowering repressor VRN2 and promoter CO2 compete for interactions with NUCLEAR FACTOR-Y complexes. Plant J. 67: 763–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Herrler M., Landsberger N., Kaludov N., Ogryzko V.V., Nakatani Y., Wolffe A.P. (1998). Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the Xenopus hsp70 promoter in vivo. EMBO J. 17: 6300–6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.C., Liao H.T., Charng Y.Y. (2011). The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 34: 738–751. [DOI] [PubMed] [Google Scholar]
- Liu J.X., Howell S.H. (2010). bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22: 782–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T., Ohto M., Yee K.M., West M.A., Lo R., Kwong R.W., Yamagishi K., Fischer R.L., Goldberg R.B., Harada J.J. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205. [DOI] [PubMed] [Google Scholar]
- Mantovani R. (1999). The molecular biology of the CCAAT-binding factor NF-Y. Gene 239: 15–27. [DOI] [PubMed] [Google Scholar]
- McNabb D.S., Xing Y., Guarente L. (1995). Cloning of yeast HAP5: a novel subunit of a heterotrimeric complex required for CCAAT binding. Genes Dev. 9: 47–58. [DOI] [PubMed] [Google Scholar]
- Mermelstein F., Yeung K., Cao J., Inostroza J.A., Erdjument-Bromage H., Eagelson K., Landsman D., Levitt P., Tempst P., Reinberg D. (1996). Requirement of a corepressor for Dr1-mediated repression of transcription. Genes Dev. 10: 1033–1048. [DOI] [PubMed] [Google Scholar]
- Mitsuhara I., et al. (1996). Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol. 37: 49–59. [DOI] [PubMed] [Google Scholar]
- Mizoi J., Ohori T., Moriwaki T., Kidokoro S., Todaka D., Maruyama K., Kusakabe K., Osakabe Y., Shinozaki K., Yamaguchi-Shinozaki K. (2013). GmDREB2A;2, a canonical DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN2-type transcription factor in soybean, is posttranslationally regulated and mediates dehydration-responsive element-dependent gene expression. Plant Physiol. 161: 346–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto K., Mizoi J., Qin F., Kim J.S., Sato H., Osakabe Y., Shinozaki K., Yamaguchi-Shinozaki K. (2013). Stabilization of Arabidopsis DREB2A is required but not sufficient for the induction of target genes under conditions of stress. PLoS ONE 8: e80457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Tran L.S., Van Nguyen D., Fujita M., Maruyama K., Todaka D., Ito Y., Hayashi N., Shinozaki K., Yamaguchi-Shinozaki K. (2007). Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 51: 617–630. [DOI] [PubMed] [Google Scholar]
- Nardini M., Gnesutta N., Donati G., Gatta R., Forni C., Fossati A., Vonrhein C., Moras D., Romier C., Bolognesi M., Mantovani R. (2013). Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell 152: 132–143. [DOI] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A., Nosaka R., Hayashi H., Tainaka H., Maruta T., Tamoi M., Ikeda M., Ohme-Takagi M., Yoshimura K., Yabuta Y., Shigeoka S. (2011). HsfA1d and HsfA1e involved in the transcriptional regulation of HsfA2 function as key regulators for the Hsf signaling network in response to environmental stress. Plant Cell Physiol. 52: 933–945. [DOI] [PubMed] [Google Scholar]
- Ogawa D., Yamaguchi K., Nishiuchi T. (2007). High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J. Exp. Bot. 58: 3373–3383. [DOI] [PubMed] [Google Scholar]
- Ohya T., Maki S., Kawasaki Y., Sugino A. (2000). Structure and function of the fourth subunit (Dpb4p) of DNA polymerase epsilon in Saccharomyces cerevisiae. Nucleic Acids Res. 28: 3846–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Stewart D., Li W., Hawkins M., Kulak S., Ballermann B., Jahroudi N. (2007). Irradiation modulates association of NF-Y with histone-modifying cofactors PCAF and HDAC. Oncogene 26: 7576–7583. [DOI] [PubMed] [Google Scholar]
- Petroni K., Kumimoto R.W., Gnesutta N., Calvenzani V., Fornari M., Tonelli C., Holt B.F. III, Mantovani R. (2012). The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 24: 4777–4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino M.T., Skinner J.S., Park E.J., Jeknić Z., Hayes P.M., Thomashow M.F., Chen T.H. (2007). Use of a stress inducible promoter to drive ectopic AtCBF expression improves potato freezing tolerance while minimizing negative effects on tuber yield. Plant Biotechnol. J. 5: 591–604. [DOI] [PubMed] [Google Scholar]
- Poot R.A., Dellaire G., Hülsmann B.B., Grimaldi M.A., Corona D.F., Becker P.B., Bickmore W.A., Varga-Weisz P.D. (2000). HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J. 19: 3377–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20: 1693–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T., Ludwig A.A., Martin R., Jones J.D. (2001). Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J. 20: 5556–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y., Liu Q., Dubouzet J.G., Abe H., Shinozaki K., Yamaguchi-Shinozaki K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290: 998–1009. [DOI] [PubMed] [Google Scholar]
- Sakuma Y., Maruyama K., Osakabe Y., Qin F., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2006a). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18: 1292–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y., Maruyama K., Qin F., Osakabe Y., Shinozaki K., Yamaguchi-Shinozaki K. (2006b). Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. USA 103: 18822–18827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasi B.K., Sonawane P.J., Gupta V., Sahu B.S., Mahapatra N.R. (2014). Coordinated transcriptional regulation of Hspa1a gene by multiple transcription factors: crucial roles for HSF-1, NF-Y, NF-κB, and CREB. J. Mol. Biol. 426: 116–135. [DOI] [PubMed] [Google Scholar]
- Schiavoni G., Bennati A.M., Castelli M., Fazia M.A., Beccari T., Servillo G., Roberti R. (2010). Activation of TM7SF2 promoter by SREBP-2 depends on a new sterol regulatory element, a GC-box, and an inverted CCAAT-box. Biochim. Biophys. Acta 1801: 587–592. [DOI] [PubMed] [Google Scholar]
- Siefers N., Dang K.K., Kumimoto R.W., Bynum W.E. IV, Tayrose G., Holt B.F. III (2009). Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 149: 625–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger E.J., Gilmour S.J., Thomashow M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94: 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow M.F. (1999). PLANT COLD ACCLIMATION: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 571–599. [DOI] [PubMed] [Google Scholar]
- Tsubota T., Tajima R., Ode K., Kubota H., Fukuhara N., Kawabata T., Maki S., Maki H. (2006). Double-stranded DNA binding, an unusual property of DNA polymerase epsilon, promotes epigenetic silencing in Saccharomyces cerevisiae. J. Biol. Chem. 281: 32898–32908. [DOI] [PubMed] [Google Scholar]
- Usadel B., et al. (2005). Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol. 138: 1195–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R., Schmidt L.K., Lohse M., Hashimoto K., Bock R., Kudla J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56: 505–516. [DOI] [PubMed] [Google Scholar]
- Wang Y.L., Faiola F., Xu M., Pan S., Martinez E. (2008). Human ATAC Is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J. Biol. Chem. 283: 33808–33815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willy P.J., Kobayashi R., Kadonaga J.T. (2000). A basal transcription factor that activates or represses transcription. Science 290: 982–985. [DOI] [PubMed] [Google Scholar]
- Wright K.L., Vilen B.J., Itoh-Lindstrom Y., Moore T.L., Li G., Criscitiello M., Cogswell P., Clarke J.B., Ting J.P. (1994). CCAAT box binding protein NF-Y facilitates in vivo recruitment of upstream DNA binding transcription factors. EMBO J. 13: 4042–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57: 781–803. [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Kagaya Y., Toyoshima R., Kagaya M., Takeda S., Hattori T. (2009). Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J. 58: 843–856. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yoshida T., et al. (2011). Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol. Genet. Genomics 286: 321–332. [DOI] [PubMed] [Google Scholar]
- Zhang Y., et al. (2013). Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature 504: 306–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Lin I.J., Darst R.P., Bungert J. (2009). Maneuver at the transcription start site: Mot1p and NC2 navigate TFIID/TBP to specific core promoter elements. Epigenetics 4: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53: 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Wang Y., Pi W., Liu H., Wickrema A., Tuan D. (2012). NF-Y recruits both transcription activator and repressor to modulate tissue- and developmental stage-specific expression of human γ-globin gene. PLoS ONE 7: e47175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.