Abstract
Killifish survive and reproduce in the New Bedford Harbor (NBH) in Massachusetts (MA), USA, a site severely contaminated with polychlorinated biphenyls (PCBs) for decades. Levels of 22 different PCB congeners were analyzed in liver from killifish collected in 2008. Concentrations of dioxin-like PCBs in liver of NBH killifish were ~400 times higher, and the levels of non-dioxin-like PCBs ~3000 times higher than in killifish from a reference site, Scorton Creek (SC), MA. The NBH killifish are known to be resistant to the toxicity of dioxin-like compounds and to have a reduced aryl hydrocarbon receptor (AhR) signaling response. Little is known about the responses of these fish to non-dioxin-like PCBs, which are at extraordinarily high levels in NBH fish. In mammals, some non-dioxin-like PCB congeners act through nuclear receptor 1I2, the pregnane-X-receptor (PXR). To explore this pathway in killifish, a PXR cDNA was sequenced and its molecular phylogenetic relationship to other vertebrate PXRs was determined. Killifish were also collected in 2009 from NBH and SC, and after four months in the laboratory they were injected with a single dose of either the dioxin-like PCB 126 (an AhR agonist) or the non-dioxin-like PCB 153 (a mammalian PXR agonist). Gills and liver were sampled three days after injection and transcript levels of genes encoding PXR, cytochrome P450 3A (CYP3A), P-glycoprotein (Pgp), AhR2 and cytochrome P450 1A (CYP1A) were measured by quantitative PCR. As expected, there was little effect of PCB exposure on mRNA expression of AhR2 or CYP1A in liver and gills of NBH fish. In NBH fish, but not in SC fish, there was increased mRNA expression of hepatic PXR, CYP3A and Pgp upon exposure to either of the two PCB congeners. However, basal PXR and Pgp mRNA levels in liver of NBH fish were significantly lower than in SC fish. A different pattern was seen in gills, where there were no differences in basal mRNA expression of these genes between the two populations. In SC fish, but not in NBH fish, there was increased mRNA expression of branchial PXR and CYP3A upon exposure to PCB126 and of CYP3A upon exposure to PCB153. The results suggest a difference between the two populations in non-AhR transcription factor signaling in liver and gills, and that this could involve killifish PXR. It also implies possible cross-regulatory interactions between that factor (presumably PXR) and AhR2 in liver of these fish.
Keywords: PXR, NR1I2, CYP3A, P-glycoprotein, fish, PCB
1. Introduction
The non-migrating Atlantic killifish (Fundulus heteroclitus) can survive and successfully reproduce in severely polluted environments (Nacci et al., 2010). A key example is the stable population of killifish that inhabits a part of the New Bedford Harbor (NBH) in Massachusetts (MA), USA, which is heavily contaminated by polychlorinated biphenyls (PCBs) (Weaver, 1984). In the 1990s, levels of ortho-substituted PCBs (non-dioxin-like) and non-ortho-substituted PCBs (dioxin-like) were more than 1000 times higher in NBH fish than in fish from reference sites such as Scorton Creek (SC) on Cape Cod, MA (Lake et al., 1995; Black et al., 1998; Bello, 1999). The dioxin-like PCB congeners normally induce cytochrome P450 1A (CYP1A) gene expression, which is a sensitive biomarker commonly used to assess exposures to dioxin-like compounds and other planar aromatic hydrocarbons in the aquatic environment (Stegeman and Hahn, 1994). The NBH killifish have a heritable resistance to the toxicity of dioxin-like PCBs, which is accompanied by greatly reduced induction of CYP1A protein levels and/or catalytic activities (i.e. ethoxyresorufin-O-deethylase) (Nacci et al., 1999; Bello et al., 2001). The terms resistance and have both been used to describe the ability of NBH fish to withstand the toxicity of dioxin-like compounds. In this paper we use the term resistance.
The toxicity of dioxin-like compounds and induction of CYP1A genes are mediated by activation of the aryl hydrocarbon receptor (AhR) (Schmidt and Bradfield, 1996). It seems advantageous to have decreased activation of AhR for the fish that reside in heavily polluted areas such as NBH. The mechanisms of adaptation to dioxin-like compounds are not fully understood, and AhR and CYP1A could be involved. A protective effect of reduced CYP1A expression against 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) toxicity was seen in brain of CYP1A morpholino knock-down zebrafish (Teraoka et al., 2003). However, knock-down of AhR2, but not CYP1A, was protective against the broader toxicity of TCDD in zebrafish (Carney et al., 2004). Other CYP1 genes that are responsive to dioxin-like PCB exposure in zebrafish are also AhR targets (Jönsson et al., 2007), and therefore may be involved in AhR mediated toxicity. Previous studies in killifish suggest that the resistance to the toxicity of dioxin-like compounds is likely at the level of the AhR (Clark et al., 2010; Whitehead et al., 2010; 2012; Oleksiak et al., 2011). Differences in allelic compositions at the AhR and CYP1A loci have recently been reported among PCB resistant and PCB responsive killifish populations (Rietzel et al., 2014). Furthermore, AhRs and especially AhR2 have been proposed targets for selection in killifish for dioxin-like contaminants (Rietzel et al., 2014; Proestou et al., 2014). Regardless of the mechanism of resistance, there is evidence that the lack of induction of AhR target genes, such as CYP1A, in fish tolerant to dioxin-like-PCBs can reduce the capacity for elimination of chemicals that are substrates to the CYP1A enzyme. For example, reduced production of benzo[a]pyrene-DNA adducts accompanied with reduced CYP1A enzyme activities was shown in NBH killifish, when compared to that in reference fish from West Island, MA (Nacci et al., 2002). This illustrates the advantage of having a reduced capacity to generate CYP1A-mediated activation of pro-carcinogens.
Elimination of aromatic hydrocarbons in vertebrates also depends on other biotransformation enzymes, predominantly CYP2 and CYP3A isoforms, and on efflux pumps such as ATP-binding cassette transporters, including the P-glycoprotein (Pgp or multidrug resistance protein 1; MDR1). In mammals, induction of CYP3A and Pgp expression is largely controlled by the nuclear receptor 1I2 (NR1I2), also known as pregnane-X-receptor (PXR) or steroid-xenobiotic-receptor (SXR) (e.g. Geick et al., 2001; Wang et al., 2012). The non-dioxin-like and ortho-substituted PCB congener 153 is able to activate PXR and the constitutive androstane receptor (CAR, NR1I3), and induces expression of CYP2 and CYP3 genes in vitro in human hepatocytes and in vivo in mouse liver (Jacobs et al., 2005; Kopec et al., 2010; Al-Salman and Plant, 2012). However, non-dioxin-like PCBs, including PCB 153, could also be antagonists of human PXR (Tabb et al., 2004).
Teleost fish have a functional PXR but appear to lack a CAR homologue (Krasowski et al., 2011; Bainy et al., 2013). Exposure to prototypical PXR agonists resulted in up-regulation of CYP3A and Pgp genes in zebrafish liver and in rainbow trout hepatocytes (Bresolin et al., 2005; Wassmur et al., 2010). It has not been established how killifish from NBH respond to non-dioxin-like PCBs, and whether the PXR signaling pathway, including possibly CYP3A or Pgp expression, is affected in fish with impaired AhR signaling. We hypothesize that there could be cross-talk between AhR and PXR signaling pathways. The aim of the present study was therefore to identify the PXR in killifish, to examine PXR, CYP3A and Pgp mRNA levels in killifish from NBH and from the reference site SC, and to determine how these fish populations respond to exposure to additional PCB 126 (an AhR agonist) and PCB 153 (a mammalian PXR agonist) in the laboratory. The results suggest differential regulation of PXR, CYP3A and Pgp in liver and gills in the PCB-resistant NBH population compared to that in the SC reference population.
2. Materials and Methods
2.1 Chemicals
Dimethylsulphoxide (DMSO) was purchased from Sigma-Aldrich Chemical, St. Louis MO, USA. PCB 126 (3,3',4,4',5-pentachlorobiphenyl) and PCB 153 (2,2',4,4',5,5'-hexachlorobiphenyl) were purchased from Ultra Scientific, Kingston, RI, USA.
2.2 Fish collection and PCB analyses
Killifish for analyses of PCB concentrations in liver were collected from New Bedford Harbor (NBH) and Scorton Creek (SC) in the summer of 2008 and the PCB concentrations were analyzed as previously described (Nacci et al., 2002). Immediately post-capture, fish were sacrificed by cervical section and livers were excised, pooled, and analyzed for concentrations of selected non-dioxin-like and dioxin-like PCBs using established methods (Gutjahr-Gobell, 1999; Jayaraman, 2001). Four non-ortho-substituted PCB and eighteen ortho-substituted PCB congeners were analyzed. The PCB concentrations were measured in pooled livers from three individuals for both sexes from the two populations, and reported as wet weight values. The protocols used here were approved by the Woods Hole Oceanographic Institution Animal Care and Use Committee (IACUC).
2.3 Fish collection and PCB exposure experiment
Killifish for the PCB-exposure experiment were collected from NBH and SC in summer 2009, using minnow traps as described before (Powell et al., 2000; Bello et al., 2001). The male NBH fish had an average body weight of 6.4 g, which was significantly larger than the average body weight for male SC fish that was 3.8 g. However, both groups were reproductively mature adults and there were no differences in body weights in the control compared to the treated groups within each site. The levels of PCBs in liver of killifish collected in 2009 are expected to approximate the hepatic levels in the killifish collected in 2008.
The fish were transported to the Woods Hole Oceanographic Institution and maintained in clean continuously flowing seawater at ambient temperature for a minimum of four months under natural light conditions. Fish were fed TetraMin® flakes (Tetra, Blacksburg, VA, USA). Next, male killifish with body weights ranging between 2.4 and 9.7 g were injected intraperitoneally with either PCB 126 (10 ng/g) or PCB 153 (6200 ng/g) dissolved in DMSO or DMSO alone (5 μl/g) as a vehicle control. These doses were chosen based on prior experience with effects in fish including killifish (Zanette et al., 2009). Three days post injection, fish were killed by cervical transection and tissues for mRNA analysis were snap frozen in liquid nitrogen and stored in −80°C prior to shipment to Sweden. Procedures used in the studies were approved by the Woods Hole Oceanographic IACUC.
2.4 Isolation of total RNA
Liver and gill tissues were shipped on dry-ice to Sweden, and stored upon arrival in −80°C until RNA isolation. Total RNA was isolated from liver and gills (10-30 mg) using the RNeasy plus mini kit (including DNA removal) from Qiagen, Solna, Sweden. The quality of RNA was analyzed in randomly selected samples using Experion system from BioRad (Sundbyberg, Sweden) and the quantity was determined using a NanoDrop 2000 (Fisher Scientific, Göteborg, Sweden).
2.5 Cloning of full-length PXR cDNA from killifish liver
Total RNA was isolated from a single liver or pooled guts of six adult female fish from SC, using the Stat60 protocol (Tel-Test), and the polyA+ RNA (mRNA) fraction was purified with the MicroPoly(A)Purist kit (Ambion). The Omniscript RT kit (Qiagen) was used to synthesize cDNA with random hexamers. Degenerate primers targeting partial PXR coding sequences were designed based on an alignment of all available vertebrate PXR amino acid sequences. The degree of degeneracy was reduced when possible with the use of inosine and the fish codon usage table as a guide. Gut cDNA was amplified with the degenerate primers PX-F4 (5'-GGITACCACTTYAAYGC-3') and PX-R5 (5'-ACICCIGGICGRTCIGG-3') using AmpliTaq Gold DNA polymerase (Promega, Madison, WI, USA). The PCR conditions were [95°C, 10 min], [95°C, 30 sec; 52°C, 15 sec; 72°C, 1 min] for 35 cycles, [72°C, 7 min]. The remaining coding sequence was obtained via 5'- and 3'-RACE following manufacturer's protocols using the gene-specific primers FhPX-5A, FhPX-5B, FhPX-3A, and FhPX-3B, along with adaptor primers AP1 and AP2 (Marathon cDNA amplification kit, Clontech, Mountainview, CA, USA). The RACE primer sequences were:
FhPX-5A: 5'-ATTACCGCTTCCTCTGACATGACC-3'
FhPX-3A: 5'-ATACTGCATACACGACGCTGTTCG-3'
FhPX-5B: 5'-ATGGACATCGGAGCTGTGTTGACC-3'
FhPX-3B: 5'-ACCACAATCTGCGCAAGCTGGACC-3'
Full-length PXR cDNA was amplified from killifish liver with Advantage DNA polymerase (Clontech, Mountainview, CA, USA) and the primer sequences were:
FhPXR-F: 5'-CACCATGAGTAGGAAGGCTGCTGG-3'
FhPXR-R: 5'-AAATAACAGCCAAGTCCTCCAAGC-3'
The PCR conditions were [94°C, 1 min], [94°C, 30 sec; 68°C, 2 min] for 35 cycles. All PCR amplified fragments were ligated into the pGEM-T Easy vector (Promega, Madison, WI, USA), and multiple clones were sequenced to ensure accuracy (University of Maine DNA Sequencing Facility, Orono, ME, USA).
2.6 Phylogenetic analyses of PXR
Phylogenetic relationships among the vertebrate PXRs were inferred based on their ligand-binding domains. The region that corresponds to amino acid residues 237-428 of the human PXR protein was aligned by ClustalX, followed by phylogenetic analysis with the Maximum Parsimony method using the Molecular Evolutionary Genetics Analysis (MEGA 6.06) software (Tamura et al., 2013). Gaps in the alignment were excluded, and bootstrap analyses were done with 1000 replications to determine the confidence in the topologies of the resulting trees.
2.7 Quantitative PCR (qPCR)
The cDNA was produced with qScript™ cDNA synthesis kit (Quanta BioSciences, VWR, Stockholm, Sweden), using 1 μg total RNA in a 20 μl reaction volume. Annealing temperatures and primer concentrations were optimized for AhR2, CYP1A, PXR, CYP3A and Pgp mRNA, as well as for the 18S rRNA, which was used as a reference gene for normalization. The primer sequences and PCR conditions are provided in Table 1. The primers used for CYP3A mRNA analyses recognize both CYP3A30 and CYP3A56 in killifish (Hegelund and Celander, 2003), and therefore the amplicon is referred to as CYP3A in the present study. The qPCR reactions were performed with 25 ng transcribed total RNA as template in a 20 μl reaction-volume, using IQSYBR green supermix and MyIQ machine with IQ5 software from BioRad. The qPCR efficiencies [Egene = (10(−1/slope) – 1) × 100] were determined by serial dilutions of pooled cDNA samples using cycle threshold (Ct) numbers for each gene analyzed. The EPXR was 80%, ECYP3A 90%, EPgp/gills 80%, EPgp/liver 90%, EAhR2 120%, ECYP1A 110% and E18S was 100%. Specificities of all PCR reactions were confirmed by melt curve analyses. The relative mRNA levels for each gene analyzed were presented as (E18SCt 18S)/(EgeneCt gene) ratios times 106.
Table 1.
The PCR primers used for mRNA analyses using quantitative PCR.
| Primer | Sequence 5’-3’ | Concentration (nM) |
Annealing Temp. (°C) |
Reference/ GenBank Accession Number |
|---|---|---|---|---|
| PXR Fw | TCACCGCAGTCGTTCTTA | 800 | 58 | EF626947 |
| PXR Rev | GCAGCCTTCCTACTCATC | 800 | ||
|
| ||||
| CYP3A30/56 Fw | GCCAGCCAGCAGAAGAGT | 600 | 58 | Hegelund and Celander, 2003 |
| CYP3A30/56 Rev | GGATTCGTAGCCAGATTGTAAGC | 600 | AF105068/AY143428 | |
|
| ||||
| Pgp Fw | GGCTTCACCTTCTCCTTCTC | 1000 | 58 | AF099732 |
| Pgp Rev | ATACTGCTTCCACATCCATCC | 1000 | ||
|
| ||||
| AhR2 Fw | GCAGTGATGTACAACCCTGAGC | 1000 | 62 | Aluru et al., 2011 |
| AhR2 Rev | CCCGTGGAACTTCAGTGCCAGG | 1000 | ||
|
| ||||
| CYP1A Fw | CTTTCACAATCCCACACTGCTC | 500 | 62 | Zanette et al., 2009 |
| CYP1A Rev | GGTCTTTCCAGAGCTCTGGG | 500 | ||
|
| ||||
| 18S Fw | TGGTTAATTCCGATAACGAACGA | 800 | 55 | Patel et al., 2006 |
| 18S Rev | CGCCACTTGTCCCTCTAAGAA | 800 | ||
2.8 Statistics
The statistical analyses were performed using the SPSS 18.0 software from SPSS, Sundbyberg, Sweden. The relative mRNA levels were compared between the different treatment groups within SC and NBH, respectively, using Kruskall Wallis followed by Mann-Whitney U-test. The basal levels of each transcript analyzed in vehicle-control fish were compared between SC and NBH using the Mann-Whitney U-test. The α-value (0.05) was not adjusted for multiple testing, although two Mann-Whitney U-tests were performed between the treatment groups within each sampling site. The sample sizes in the measurements were n=9 or 10, except for gills of NBH fish treated with PCB 153, for which n=5. Significance levels of p≥0.05 are indicated in the diagrams.
3. Results
3.1 Hepatic PCB concentrations in killifish
The levels of eighteen non-dioxin-like and four dioxin-like PCBs were measured in pooled livers of killifish collected in 2008 (Table 2). The levels of total non-dioxin-like (ortho-substituted) PCB concentrations in liver pooled from fish from NBH were 91202 ng/g wet weight in males and 95469 ng/g wet weight in females. In comparison, the SC males had 45 ng/g wet weight and females had 27 ng/g wet weight of non-dioxin-like PCBs. Thus, levels of total non-dioxin-like PCBs in liver of NBH fish were 2005 times higher in males and 3554 times higher in females, compared to those in SC. Consistent with prior studies (Lake et al., 1995; Black et al., 1998; Bello, 1999), concentrations of dioxin-like (non-ortho) PCBs were much lower than those of ortho-substituted PCBs, but also showed a dramatic difference between sites. For the dioxin-like PCBs, the differences between the sites were 272 and 487 times [NBH>SC] for males and females, respectively (Table 2).
Table 2.
PCB concentrations in liver of killifish of both sexes collected in 2008.
| Collection site | ||||
|---|---|---|---|---|
|
| ||||
| Scorton Creek | New Bedford Harbor | |||
|
| ||||
| PCB Congeners | concentration (ng/g wet weight)a | |||
| Dioxin-like | Male | Female | Male | Female |
| PCB 77 | 1.54 | 0.887 | 618 | 672 |
| PCB 81 | 0.456 | 0.094 | 32.5 | 36.5 |
| PCB 126 | 0.587 | 0.589 | 50.3 | 55.4 |
| PCB 169 | (< 0.124)b | (< 0.124) | 1.05 | 1.15 |
| Σ non-ortho-PCB | 2.58 | 1.57 | 701.85 | 765.05 |
| Non-Dioxin-like | Male | Female | Male | Female |
| PCB 8 | 0.39 | (< 0.032) | 3746 | 4455 |
| PCB 18 | (< 0.057)c | (< 0.025) | 9417 | 10006 |
| PCB 28 | 4.05 | 2.88 | 12958 | 13874 |
| PCB 44 | (< 0.055) | 0.12 | 5992 | 6643 |
| PCB 52 | 2.28 | 2.02 | 19000 | 19713 |
| PCB 66 | 0.63 | 0.45 | 6170 | 6516 |
| PCB 101 | 5.09 | 3.17 | 9160 | 8456 |
| PCB 105 | 0.73 | 0.61 | 1691 | 2389 |
| PCB 118 | 6.92 | 3.94 | 7873 | 7619 |
| PCB 128 | (<0.045) | (<0.020) | 1003 | 1295 |
| PCB 138 | 9.15 | 4.99 | 4508 | 4898 |
| PCB 153 | 12.83 | 7.18 | 7002 | 6560 |
| PCB 170 | (< 0.077) | (< 0.034) | 459 | 631 |
| PCB 180 | 0.78 | 0.63 | 923 | 1037 |
| PCB 187 | 2.20 | 0.67 | 1205 | 1213 |
| PCB 195 | (< 0.061) | (< 0.027) | 43.5 | 51.4 |
| PCB 206 | (< 0.065) | (< 0.028) | 42.3 | 44.8 |
| PCB 209 | (< 0.073) | (< 0.032) | 7.83 | 6.93 |
|
| ||||
| Σ ortho-PCBs | 45.5 | 26.9 | 91202 | 95469 |
Concentrations are averages for liver pooled from three fish per collection site and sex.
The value in parentheses for congener PCB 169 is the instrument detection limit.
Values in parentheses give ½ the method detection limits for those congeners as measured, and which were below the limits of detection.
3.2 Sequencing of PXR from killifish
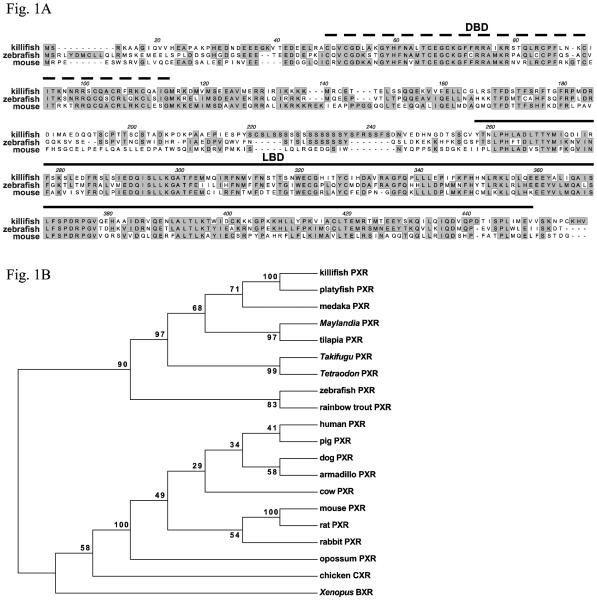
A partial killifish PXR cDNA fragment of 941 base pairs was amplified from gut cDNA using degenerate primers. The deduced protein sequence of this fragment shared 44.5% sequence identity with mouse PXR amino acid residues 47-353. The remaining coding sequence for the killifish PXR was obtained via 5'- and 3'-RACE. The 379 base pair long 5'-RACE fragment included 153 base pairs of the upstream untranslated sequence. The 3'-RACE yielded a 872 base pair long fragment, which contained 537 base pairs of the 3'-untranslated sequence. The complete PXR cDNA was amplified from liver. The full-length killifish PXR protein has 442 amino acid residues, and a calculated molecular weight of 50.2 kDa (Fig. 1A).
Fig. 1.
A) Complete coding sequence of killifish PXR aligned with that of zebrafish and mouse. The DNA binding domain (DBD) and the ligand binding domain (LBD) are highlighted with dashed and solid lines, respectively. B) Phylogenetic analyses of the PXR LBD in killifish and other vertebrates. The LBD (amino acid residues corresponding to 237-428 of the human PXR) were aligned and the phylogenetic relationships were determined using the Maximum Parsimony method.
The DNA-binding and ligand-binding domains of the killifish PXR have sequence similarity to that of PXR proteins in other fish and mammals. Overall, the complete coding sequence shares 40.6% and 50.2% predicted amino acid identity with mouse and zebrafish PXR proteins, respectively. Since these cloning efforts, the killifish genome has been sequenced and a draft assembly became available. This allowed us to examine the genomic organization of the PXR gene, which is arranged in 8 exons, spanning 13.6 kilobases. The number and locations of the exon junctions are highly similar to that of the mouse PXR gene, including the presence of an intron in the 5'-untranslated region.
Phylogenetic analysis was performed to compare the killifish PXR with other vertebrate PXR amino acid sequences (Fig. 1B). Given the diverse ligand-binding specificities among PXRs from different species, we explored the phylogenetic relationship of the killifish PXR ligand-binding domain with that of mammals and other fish. To this end, the region corresponding to residues 237-428 of the human PXR protein was aligned for this analysis. We found that the topology of the resulting tree with the ligand binding domains was essentially identical to the one constructed using full-length PXR sequences (data not shown). Among the fish species, killifish PXR is most closely related to the platyfish PXR, and least similar to the zebrafish and rainbow trout PXR sequences.
3.3 Comparisons of basal mRNA levels between the two killifish populations
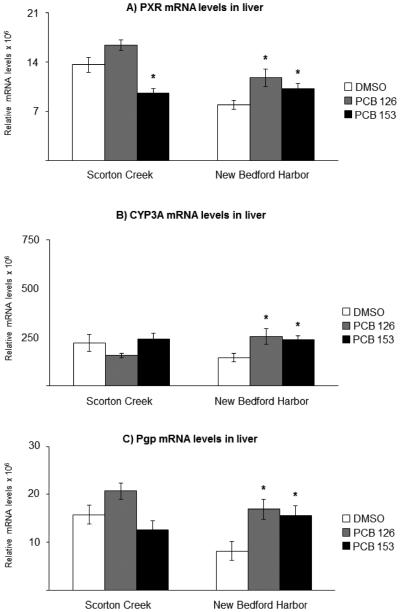
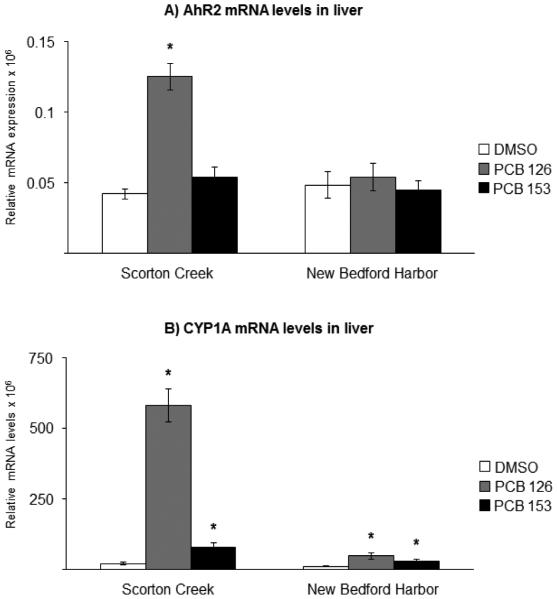
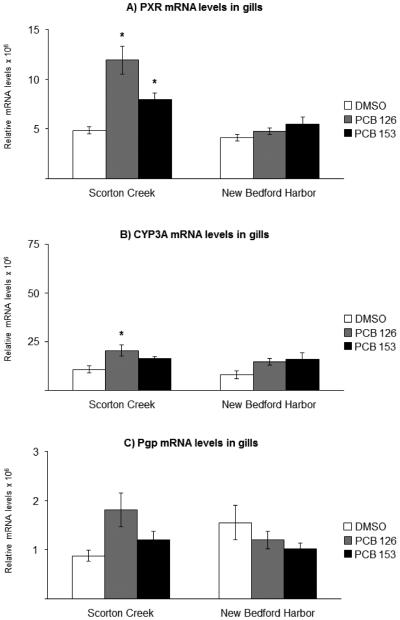
The mRNA levels of target genes in vehicle-exposed fish, here referred to as basal levels, were compared between NBH fish and SC fish and are summarized in Table 3. In liver from NBH fish, the PXR mRNA levels were 40% lower compared to basal levels in liver from SC fish (Table 3, Fig. 2A). There was no difference in hepatic CYP3A basal mRNA levels between the two populations (Table 3, Fig. 2B). The Pgp mRNA basal levels were 50% lower in liver of NBH fish compared to that in SC fish (Table 3, Fig. 2C). For the AhR2 and CYP1A genes, there were no significant differences in basal hepatic levels between the two killifish populations (Table 3, Fig. 3). In gills, there were no differences between the two populations in basal levels of expression, for any of the five genes analyzed (Table 3, Fig. 4, Fig. 5).
Table 3.
Basal PXR, CYP3A, Pgp, AhR2 and CYP1A mRNA levels in liver and gills from vehicle-control treated fish from Scorton Creek (SC) and New Bedford Harbor (NBH).
| Liver | Gills | |||
|---|---|---|---|---|
|
|
||||
| Target gene | SC | NBH | SC | NBH |
| PXR | 14 ± 1 | 8 ± 1* | 5 ± 0.4 | 4 ± 1 |
| CYP3A | 220 ± 44 | 144 ± 22 | 11 ± 2 | 8 ± 7 |
| Pgp | 16 ± 2 | 8 ± 2* | 1 ± 0.1 | 2 ± 1 |
| AhR2 | 0.04 ± 0.004 | 0.05 ± 0.009 | 0.2 ± 0.02 | 0.2 ± 0.02 |
| CYP1A | 22 ± 4 | 12 ± 2 | 0.6 ± 0.03 | 0.6 ± 0.1 |
The mRNA levels are presented as relative mRNA levels times 106 and each value represents the mean of 5-10 individual killifish ± standard error. The difference between NBH and SC on CYP1A mRNA levels has a p-value of 0.06.
Fig. 2.
Effect of exposure to the vehicle DMSO (white bars), 10 ng/g PCB 126 (grey bars) or 6 μg/g PCB 153 (black bars) on A) PXR; B) CYP3A; and C) Pgp mRNA levels in liver from killifish from Scorton Creek and New Bedford Harbor. Each bar represents the mean of 9-10 individual fish ± standard error. * Indicates statistically different (p≥0.05) compared to vehicle-carrier (DMSO) treated fish from each site.
Fig. 3.
Effect of exposure to the vehicle DMSO (white bars), 10 ng/g PCB 126 (grey bars) or 6 μg/g PCB 153 (black bars) on A) AhR2; and B) CYP1A mRNA levels in liver from killifish from Scorton Creek and New Bedford Harbor. Each bar represents the mean of 9-10 individual fish ± standard error. * Indicates statistically different (p≥0.05) compared to vehicle-carrier (DMSO) treated fish from each site.
Fig. 4.
Effect of exposure to the vehicle DMSO (white bars), 10 ng/g PCB 126 (grey bars) or 6 μg/g PCB 153 (black bars) on A) PXR; B) CYP3A; and C) Pgp mRNA levels in gills from killifish populations from Scorton Creek and New Bedford Harbor. Each bar represents the mean of 9-10 individual fish ± standard error, except for fish from New Bedford Harbor treated with PCB 153 (n=5). * Indicates statistically different (p≥0.05) compared to vehicle-carrier (DMSO) treated fish from each site.
Fig. 5.
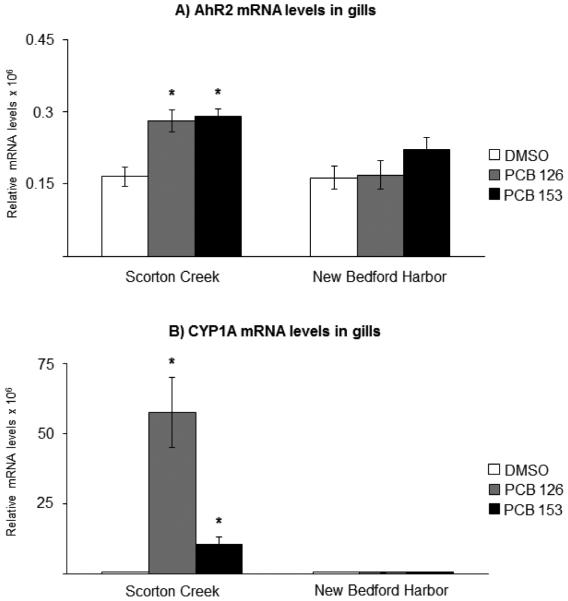
Effect of exposure to the vehicle DMSO (white bars), 10 ng/g PCB 126 (grey bars) or 6 μg/g PCB 153 (black bars) on A) AhR2; and B) CYP1A mRNA levels in gills from killifish populations from Scorton Creek and New Bedford Harbor. Each bar represents the mean of 9-10 individual fish ± standard error, except for fish from Scorton Creek treated with PCB 126 (n=8) and PCB 153 (n=7). * Indicates statistically different (p≥0.05) compared to vehicle-carrier (DMSO) treated fish from each site.
3.4 Effects of PCB exposures on hepatic mRNA levels
In SC fish, there was no significant induction of hepatic PXR, CYP3A or Pgp mRNA levels upon exposure to either of the two PCB congeners tested, compared to vehicle-control fish (Fig. 2A-C). In NBH fish, in contrast, the hepatic PXR, CYP3A and Pgp mRNA levels were induced by exposure to both PCB congeners. Hence, exposure to PCB 126 and PCB 153 resulted in 1.5 and 1.3-fold induction of PXR mRNA, respectively, compared to the vehicle-control treated fish (Fig. 2A). In addition, the corresponding CYP3A mRNA levels were increased 2-fold (Fig. 2B). The Pgp mRNA levels were also increased 2-fold in NBH fish by both PCB congeners (Fig. 2C).
For comparison, the AhR2 and CYP1A mRNA levels were also analyzed in liver. Consistent with earlier reports (Nacci et al., 1999; Bello et al., 2001; Whitehead et al., 2012; Oleksiak et al., 2011), in the fish from NBH, hepatic AhR2-CYP1A signaling was considerably less responsive to PCB exposure, compared to that in SC fish. Thus, in NBH fish, exposure to either PCB 126 or PCB 153 had no effect on AhR2 mRNA levels, compared to vehicle-control. In SC fish, on the other hand, the AhR2 mRNA levels were induced 3-fold by PCB 126, whereas PCB 153 had no effect on AhR2 mRNA expression (Fig. 3A). Exposure to PCB 126 and PCB 153 induced CYP1A mRNA levels 4 and 2-fold, respectively, in NBH fish compared to vehicle-control. In SC fish, the CYP1A mRNA levels were induced 26-fold by PCB 126 and 4-fold in liver by PCB 153 (Fig. 3B).
3.5 Effects of PCB exposures on branchial mRNA levels
In contrast to effects in liver, there were no significant effects of treatment with either PCB 126 or PCB 153 on branchial PXR, CYP3A and Pgp mRNA levels in NBH fish (Fig. 4A-C). In SC fish, PXR mRNA levels were induced 2.5-fold upon exposure to PCB 126 and 1.7-fold upon exposure to PCB 153 (Fig. 4A). The CYP3A mRNA levels were induced 2-fold by PCB 126, while exposure to PCB 153 had no effect on CYP3A mRNA levels. The Pgp mRNA levels in gills were not affected by exposure to either of the two PCB congeners, compared to vehicle-control treated fish, in fish from either of the two populations (Fig. 4C).
The AhR2 and CYP1A mRNA levels were also analyzed in the gills. In gills of NBH fish, neither AhR2 nor CYP1A mRNA levels responded to treatment with either of the two PCB congeners (Fig. 5A-B). This is in sharp contrast to SC fish, in which exposure to PCB 126 resulted in a 2-fold induction of AhR2 and a 95-fold induction of CYP1A mRNA levels in gills, compared to vehicle-control treated fish. Moreover, exposure to PCB 153 also resulted in 2-fold induction of AhR2 and a 17-fold induction of CYP1A mRNA levels in gills in SC fish, but not in NBH fish (Fig. 5).
4. Discussion
There are reports of wild fish populations of several species that reside in heavily PCB contaminated areas and that have impaired CYP1A induction responses (Förlin and Celander, 1995; Elskus et al., 1999; Wirgin and Waldman, 2004). The impaired AhR-CYP1A response phenomenon has been studied extensively in the killifish population from NBH, defining a heritable resistance to halogenated AhR agonists (Nacci et al., 1999; Powell et al., 2000; Bello et al., 2001; Oleksiak et al., 2011; Whitehead et al., 2012). The ortho-substituted, non-dioxin-like PCB congeners are also present at extraordinarily high levels in NBH killifish (Table 2; Lake et al., 1995; Bello, 1999). These high levels of ortho-PCBs in the NBH fish argue for examining possible adaptation to non-dioxin-like congeners. Some non-dioxin-like congeners are known agonists for mammalian PXR and CAR (Jacobs et al., 2005; Kopec et al., 2010; Al-Salman and Plant 2012). Compared to AhR and the AhR-CYP1A signaling pathway, there is a lack of knowledge on function and regulation of PXR and PXR target genes in PCB-resistant fish.
4.1 PXR Phylogeny
Teleost fish do not have a distinct CAR, but most possess a single PXR/CAR-like receptor (Krasowski et al., 2011). To pursue the questions of PXR responses in killifish, we first established the identity of a sequence cloned from killifish as a PXR, and examined molecular phylogenetic relationships. The functional domains of killifish PXR showed close relationships to other PXR proteins, consistent with what has been found for rainbow trout and zebrafish PXR sequences (Wassmur et al., 2010; Bainy et al., 2013). Thus, the DNA binding domain shows a greater similarity to the DNA binding domain of PXR from other vertebrates, while the ligand-binding domain show less similarity to those of other species’ PXR genes. There is evidence for PXR alleles in zebrafish, with variation occurring in the coding region (Bainy et al., 2013). For AhR2, there is extensive single nucleotide polymorphism variation with frequencies differing between the SC and NBH killifish populations (Reitzel et al., 2014; Proestou et al., 2014). We anticipate that there will be allelic variation in PXR of killifish as well. Whether PXR alleles in fish are functional is not known. It has been shown that PXR alleles in mammals often occur in non-coding regions, and do affect PXR function (e.g. Lamba et al., 2008).
4.2 Responses to PCB treatment in liver and gills and effects on PXR signaling
In mammals, CYP3A and Pgp expression is induced in response to PXR activation. However, the role of PXR in Pgp and CYP3A regulation in killifish is not yet demonstrated. In zebrafish, induced expression of CYP3A and of PXR itself appears to be dependent, at least in part, on the PXR (Kubota and Stegeman, unpublished data). In the present study, NBH killifish responded to both PCB 126 and PCB 153 exposure with elevation in hepatic PXR, CYP3A and Pgp mRNA levels. This is in contrast to SC fish, in which exposure to these two PCB congeners had no effect on any of these genes in the liver. This is an intriguing, but not yet understood, difference between the two populations. Interestingly, basal hepatic levels of PXR and Pgp expression (i.e. mRNA levels in vehicle-control fish) differed between the populations, with basal expression of these genes being significantly lower in liver in NBH fish compared to that in SC fish. In a previous study using primary cultures of rainbow trout hepatocytes, there was an inverted relationship between basal mRNA levels of PXR-CYP3A/Pgp and the responsiveness to prototypical PXR agonists (Wassmur et al., 2010). Higher CYP3A inducibility was reported in zebrafish larvae that had the lowest basal CYP3A levels (Tseng et al., 2005). Hence, it is possible that the observed induction-response pattern of hepatic PXR signaling between killifish from NBH and SC is a consequence of differences in basal expression between the two populations. The mechanisms underlying these differences in basal expression, and whether they involve genetic, epigenetic, or exposure-related differences between populations, remain to be determined.
In the gills, the basal mRNA levels of these genes did not differ between the two populations, although there was a different response to PCB in gills compared to the response in liver. In NBH fish, exposure to either PCB 126 or PCB 153 had no effect on PXR, CYP3A and Pgp mRNA levels in gills. However, in SC fish there was a 2-fold induction of branchial PXR mRNA levels by both PCB congeners and a 2-fold induction of CYP3A mRNA levels by PCB 126. In the present study, the vehicle-control fish were exposed to 5 μl DMSO/g body weight. In a human colon cell-line, exposure to 2% (v/v) DMSO in the media induced Pgp levels (Mickley et al., 1989). The possibility that the vehicle carrier affects regulation of PXR, CYP3A and Pgp genes in the present study cannot yet be ruled out. In addition to plausible different responsiveness between different cell types, PCB kinetics (e.g. uptake, tissue-distribution and metabolism) can result in different PCB tissue concentrations. Conceivably, this could influence PXR expression and signaling differently in distinct tissues. In gills, CYP3A proteins are found in filament cells, and in liver the CYP3A proteins are found in hepatocytes in killifish (Hegelund and Celander, 2003). The reasons for the observed tissue and population differences in responses to PCB treatment on PXR-CYP3A/Pgp mRNA levels are not clear and require further studies before it can be fully understood.
The doses of PCB 126 and PCB 153 used in the present study are much less than the body burden of these congeners (or of total dioxin-like or total non-dioxin-like congeners) in NBH killifish collected in 2008, and much greater than the amounts in SC fish. This inevitably raises questions about what might be expected from an additional dose, especially for the NBH fish. While the fish used experimentally were depurated for four months, this actually would not be expected to greatly reduce the content of PCBs in the NBH fish. In this context, the differences between the NBH and SC fish are intriguing, and again argue strongly for examination of responses to PCB 153 in F2 and F3 fish from both sites. Such fish would not be expected to differ in PCB residue levels, or to have elevated levels of PCBs.
4.3 Putative receptor cross-talk
In mammals, cross-talk between PXR, CAR and AhR have been reported (Köhle and Bock, 2009). For example, AhR agonists induce CAR expression and function in both mouse and humans (Patel et al., 2007). Exposure to polyaromatic hydrocarbons induces CYP3A4 transcription in vitro through activation of human PXR (Kumagai et al., 2012). In human hepatocytes, activation of PXR leads to induction of AhR-CYP1A1/CYP1A2 signaling (Maglich et al., 2002). Cross-talk between AhR and PXR was recently described in zebrafish (Kubota et al., 2014). The NBH killifish have an AhR-CYP1A signaling with reduced sensitivity to PCB exposure, but these fish responded to PCB treatment by induction of PXR, CYP3A and Pgp mRNA levels in liver. This suggests that the contribution of AhR is low or insignificant in the regulation of PXR, CYP3A and Pgp genes in fish from NBH, or that there could be an inhibiting cross-talk between PXR and AhR in NBH fish, because PXR, CYP3A and Pgp mRNA levels are higher and CYP1A mRNA levels are lower in NBH fish exposed to PCBs in the laboratory. The fact that both a dioxin-like PCB (AhR agonist) and a non-dioxin-like PCB (putative PXR agonist) up-regulated mRNA levels of hepatic CYP3A/Pgp and at the same time these fish display reduced CYP1A inducibility in liver from NBH fish suggests that both receptors are activated, which implies the possibility of cross-regulatory interactions between PXR and AhR signaling pathways in the NBH killifish. In SC fish, the opposite was seen with no effect on PXR-CYP3A/Pgp signaling by either of these congeners and a powerful induction of AhR-CYP1A signaling upon treatment with PCB 126, and weaker induction of CYP1A with PCB 153. There are clear differences in responsiveness to PCB treatment between the two killifish populations and further studies are warranted to elucidate the extent and mechanisms of AhR-PXR cross-talk in killifish, in comparison to the cross-talk reported in zebrafish, and to address the observed tissue-differences in response to PCB exposures.
4.4 Pgp and CYP3A and PCB toxicity
The basal mRNA levels of hepatic Pgp were significantly lower (i.e. 50%) in killifish from NBH compared to fish from SC. This is in accordance with earlier findings, showing lower Pgp protein levels in liver of killifish from NBH compared to the SC population (Bard et al., 2002). Elevated Pgp protein levels have been reported in another killifish population from the highly creosote contaminated Elizabeth River, VA, USA. Those fish had a higher tolerance to exposure to creosote-contaminated sediments compared to killifish from a reference site. Interestingly, the creosote-resistant killifish population displayed increased frequencies of liver tumors and the Pgp protein levels were higher in more advanced stage liver tumors in these fish (Cooper et al., 1999). Those studies and our results suggest that Pgp could participate in the resistance to toxicity in PCB- and creosote-exposed killifish populations. In porcine kidney cells, that over-express human or mouse Pgp genes, the dioxin-like PCB 77 was shown to bind to Pgp. However, no transport of either PCB 77 or PCB 153 was seen in this cell system (Tampal et al., 2003). In addition, PCB 77 and PCB 126 acted as strong inhibitors of the Pgp-mediated vinblastine transport in these cells. It was further proposed that, in addition to interaction with AhR signaling, toxicity of dioxin-like PCBs could be mediated by interaction with efflux function in this cell system (Sasawatari et al., 2004). Taken together, these findings suggest that Pgp is affected by exposure to dioxin-like PCBs. In mammals, Pgp expression is regulated via PXR activation (Synold et at., 2001), and there is evidence that PXR regulates both Pgp and CYP3A genes in fish (Wassmur et al., 2010; Bainy et al., 2013). For example, in cultured rainbow trout hepatocytes PXR mRNA levels correlated with CYP3A and Pgp mRNA and these genes were all up-regulated in cells exposed to prototypical PXR agonists (Wassmur et al., 2010).
A genetic survey of the killifish population from NBH and a reference population at Block Island, RI revealed differences at the CYP3A30 locus, in addition to the differences at the AhR2 and CYP1A loci (Proestou et al., 2014). This implies that CYP3A could be involved in the adaptive response to non-dioxin-like as well as dioxin-like PCBs. In the present study, the CYP3A gene responded differently to PCB exposure between the NBH and SC populations and low basal levels of PXR mRNA coincided with low basal CYP3A and Pgp mRNA levels in killifish from NBH, distinctive from SC fish. Hence, it is possible that the low basal expression of hepatic CYP3A and Pgp genes is a result of low basal expression of PXR in killifish from NBH. The apparent down-regulation of hepatic PXR signaling may be beneficial for the fish in addition to the non-responsiveness of AhR signaling pathways in highly PCB contaminated areas. Experiments to test these ideas are underway.
Conclusions and future perspectives
There were clear differences between PXR and putative target genes and AhR2-CYP1A signaling between gill and liver, as well as between the different killifish populations. There are both population and tissue-specific differences in responses of PXR, CYP3A and Pgp genes. There also was an inverted relationship between inducibility and basal levels of PXR and Pgp mRNA in liver of fish from NBH. Exposures to both dioxin-like and non-dioxin-like PCB congeners induced hepatic PXR, CYP3A and Pgp genes in this PCB resistant population. The possibility that PXR, CYP3A and Pgp proteins participate in PCB resistance cannot be ruled out. In gills in SC fish, expression of targets of both PXR and AhR were elevated by dioxin-like and non-dioxin-like PCB congeners. Assuming that killifish PXR is activated by PCBs, this suggests that there could be cross-talk between PXR and AhR signaling, but it may function differently in the NBH and SC fish populations. The non-dioxin-like PCB 153 (a mammalian PXR agonist) showed some site differences in effects on expression of possible target genes for PXR (i.e. PXR, CYP3A and Pgp). This was similar to the response to the dioxin-like PCB 126, which suggests that there also may be adaptation to the non-dioxin-like congeners in the NBH population. Recently a functional difference in ryanodine receptors (i.e. Ca2+-channels in sarcoplasmic and endoplasmic reticulum), which is also responsive to non-dioxin-like PCBs, was observed between NBH and SC killifish (Fritsch et al., unpublished data). The differences between the NBH and SC populations could reflect an adaptation of NBH fish to the ortho-substituted, non-dioxin-like PCBs that are hugely abundant in that site. Identifying associated phenotypic differences in fish in these populations would indicate the importance of such biochemical differences. Determining whether gene expression or phenotypic differences occur in F1 or F2 generation fish, which should have no residual PCB content, will be necessary to determine any heritability of the changes described here.
Highlights.
Basal levels of PXR and Pgp mRNA are lower in liver of fish from NBH than from SC
Hepatic PXR, CYP3A and Pgp mRNA levels are induced by PCB in fish from NBH
Both non-dioxin-like and dioxin-like PCBs induce PXR, CYP3A and Pgp in NBH fish
Branchial PXR and CYP3A mRNA levels are induced by PCB 126 in fish from SC
There is possible cross-talk between AhR and PXR signaling in killifish
Acknowledgements
This study was supported by grants from FORMAS (216-2007-468) and University of Gothenburg to MCC, and by the Superfund Research Program at Boston University, NIH grant P42ES007381 to JJS, MEH, and SIK. Data interpretation was aided by reference to a preliminary draft of the Fundulus heteroclitus genome sequence, which was supported by funding from the National Science Foundation (collaborative research grants DEB-1120512, DEB-1265282, DEB-1120013, DEB-1120263, DEB-1120333, DEB-1120398). This study was also supported by NOAA Grant No. NA16RG2273 (WHOI Sea Grant Project No. R/P-70 to SIK and MEH) and by funding from Adlerbertska Forskningsstiftelsen, Helge Ax:son Johnsons Stiftelse and Wilhelm och Martina Lundgrens Vetenskapsfond to BW and JG. We thank Dr. Joanna Y. Wilson, Department of Biology, McMaster University, Hamilton, Ontario, Canada for valuable discussions. We also thank Dr. Kerstin Wiklander, Department of Mathematical Sciences, Chalmers University of Technology, Göteborg, Sweden for statistical guidance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
- AhR
- arylhydrocarbon receptor
- CAR
- constitutive androstane receptor
- CYP
- cytochrome P450
- DMSO
- dimethylsulphoxide
- DBD
- DNA binding domain
- LBD
- ligand binding domain
- NBH
- New Bedford Harbor
- PCR
- polymerase chain reaction
- PXR
- pregnane-X-receptor
- Pgp
- P-glycoprotein
- PCB
- polychlorinated biphenyls
- RACE
- rapid amplification of cDNA ends
- SC
- Scorton Creek
- TCDD
- 2,3,7,8-tetrachlorodibenzo-p-dioxin
References
- Al-Salman F, Plant N. Non-coplanar polychlorinated biphenyls (PCBs) are direct agonists for the human pregnane-X receptor and constitutive androstane receptor, and activate target gene expression in a tissue-specific manner. Toxicol. Appl. Pharmacol. 2012;263:7–13. doi: 10.1016/j.taap.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Aluru N, Karchner SI, Hahn ME. Role of DNA methylation of AHR1 and AHR2 promoters in differential sensitivity to PCBs in Atlantic Killifish, Fundulus heteroclitus. Aquat. Toxicol. 2011;101:288–294. doi: 10.1016/j.aquatox.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainy ACD, Kubota A, Goldstone JV, Lille-Langøy R, Karchner SI, Celander MC, Hahn ME, Goksøyr A, Stegeman JJ. Functional characterization of a full length pregnane × receptor, expression in vivo, and identification of PXR alleles, in Zebrafish (Danio rerio) Aquat. Toxicol. 142. 2013;143:447–457. doi: 10.1016/j.aquatox.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard SM, Bello SM, Hahn ME, Stegeman JJ. Expression of P-glycoprotein in killifish (Fundulus heteroclitus) exposed to environmental xenobiotics. Aquat. Toxicol. 2002;59:237–251. doi: 10.1016/s0166-445x(01)00256-9. [DOI] [PubMed] [Google Scholar]
- Bello SM. Characterization of resistance to halogenated aromatic hydrocarbons in a population of Fundulus heteroclitus from a marine superfund site. 1999:211. Ph.D. Thesis, Woods Hole Oceanographic Institution/Massachusetts Institute of Technology Joint Program in Oceanography. [Google Scholar]
- Bello SM, Franks DG, Stegeman JJ, Hahn ME. Acquired resistance to Ah receptor agonists in a population of Atlantic killifish (Fundulus heteroclitus) inhabiting a marine superfund site: in vivo and in vitro studies on the inducibility of xenobiotic metabolizing enzymes. Toxicol. Sci. 2001;60:77–91. doi: 10.1093/toxsci/60.1.77. [DOI] [PubMed] [Google Scholar]
- Black DE, Gutjahr-Gobell R, Pruell RJ, Bergen B, Mills L, McElroy AE. Reproduction and polychlorinated biphenyls in Fundulus heteroclitus (Linnaeus) from New Bedford Harbor, Massachusetts, USA. Environ. Toxicol. Chem. 1998;17:1405–1414. [Google Scholar]
- Bresolin T, de Freitas Rebelo M, Bainy ACD. Expression of PXR, CYP3A and MDR1 genes in liver of zebrafish. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2005;140:403–407. doi: 10.1016/j.cca.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Carney SA, Peterson RE, Heideman W. 2,3,7,8-Tetrachlorodibenzo-p-dioxin activation of the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator pathway causes developmental toxicity through a CYP1A-independent mechanism in zebrafish. Mol. Pharmacol. 2004;66:512–521. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- Cooper PS, Vogelbein WK, Van Veld PA. Altered expression of the xenobiotic transporter P-glycoprotein in liver and liver tumours of mummichog (Fundulus heteroclitus) from a creosote-contaminated environment. Biomarkers. 1999;4:48–58. doi: 10.1080/135475099230994. [DOI] [PubMed] [Google Scholar]
- Clark BW, Matson CW, Jung D, Di Giulio RT. AHR2 mediates cardiac teratogenesis of polycyclic aromatic hydrocarbons and PCB-126 in Atlantic killifish (Fundulus heteroclitus) Aquat. Toxicol. 2010;99:232–240. doi: 10.1016/j.aquatox.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elskus AA, Monosson E, McElroy AE, Stegeman JJ, Woltering DS. Altered CYP1A expression in Fundulus heteroclitus adults and larvae: a sign of pollutant resistance? Aquat. Toxicol. 1999;45:99–113. [Google Scholar]
- Förlin L, Celander M. Studies of the inducibility of P450 1A in perch from the PCB contaminated Lake Järnsjön in Sweden. Mar. Environ. Res. 1995;39:85–88. [Google Scholar]
- Gutjahr-Gobell RE, Black DE, Mills LJ, Pruell RJ, Taplin BK, Jayaraman S. Feeding the mummichog (Fundulus heteroclitus) a diet spiked with non-ortho- and mono-ortho-substituted Polychlorinated biphenyls: Accumulation and effects. Environ. Toxicol. Chem. 1999;18:699–707. [Google Scholar]
- Geick A, Eichelbaum M, Burk O. Nuclear receptor response elements mediate induction of intestinal MDR1 by rifampin. J. Biol. Chem. 2001;276:14581–14587. doi: 10.1074/jbc.M010173200. [DOI] [PubMed] [Google Scholar]
- Hegelund T, Celander MC. Hepatic versus extrahepatic expression of CYP3A30 and CYP3A56 in adult killifish (Fundulus heteroclitus) Aquat. Toxicol. 2003;64:277–291. doi: 10.1016/s0166-445x(03)00057-2. [DOI] [PubMed] [Google Scholar]
- Jacobs MN, Nolan GT, Hood SR. Lignans, bacteriocides and organochlorine compounds activate the human pregnane × receptor (PXR) Toxicol. Appl. Pharmacol. 2005;209:123–133. doi: 10.1016/j.taap.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Jayaraman S, Pruell RJ, McKinney R. Extraction of organic contaminants from marine sediments and tissues using microwave energy. Chemosphere. 2001;44:181–191. doi: 10.1016/s0045-6535(00)00201-0. [DOI] [PubMed] [Google Scholar]
- Jönsson ME, Orrego R, Woodin BR, Goldstone JV, Stegeman JJ. Basal and 3,3',4,4',5-pentachlorobiphenyl-induced expression of cytochrome P450 1A, 1B and 1C genes in zebrafish. Toxicol. Appl. Pharmacol. 2007;221:29–41. doi: 10.1016/j.taap.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhle C, Bock KW. Coordinate regulation of human drug-metabolizing enzymes, and conjugate transporters by the Ah receptor, pregnane × receptor and constitutive androstane receptor. Biochem. Pharmacol. 2009;77:689–699. doi: 10.1016/j.bcp.2008.05.020. [DOI] [PubMed] [Google Scholar]
- Kopec AK, Burgoon LD, Ibrahim-Aibo D, Mets BD, Tashiro C, Potter D, Sharratt B, Harkema JR, Zacharewski TR. PCB153-elicited hepatic responses in the immature, ovariectomized C57BL/6 mice: comparative toxicogenomic effects of dioxin and non-dioxin-like ligands. Toxicol. Appl. Pharmacol. 2010;243:359–371. doi: 10.1016/j.taap.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasowski MD, Ni A, Hagey LR, Ekins S. Evolution of promiscuous nuclear hormone receptors: LXR, FXR, VDR, PXR, and CAR. Mol. Cell Endocrin. 2011;334:39–48. doi: 10.1016/j.mce.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota A, Goldstone JV, Lemaire B, Takata M, Woodin BR, Stegeman JJ. Pregnane × receptor and aryl hydrocarbon receptor both are involved in transcriptional regulation of pxr, CYP2 and CYP3 genes in developing zebrafish. Toxicol. Sci. 2014 doi: 10.1093/toxsci/kfu240. In press. pii: kfu240. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai T, Suzuki H, Sasaki T, Sakaguchi S, Miyairi S, Yamazoe Y, Nagata K. Polycyclic aromatic hydrocarbons activate CYP3A4 gene transcription through human pregnane × receptor. Drug Metab. Pharmacokinet. 2012;27:200–206. doi: 10.2133/dmpk.dmpk-11-rg-094. [DOI] [PubMed] [Google Scholar]
- Lake JL, McKinney R, Lake CA, Osterman FA, Heltshe J. Comparisons of patterns of polychlorinated biphenyl congeners in water, sediment, and indigenous organisms from New Bedford harbor, Massachusetts. Arch. Environ. Contam. Toxicol. 1995;29:207–220. [Google Scholar]
- Lamba J, Lamba V, Strom S, Venkataramanan R, Schuetz E. Novel single nucleotide polymorphisms in the promoter and intron 1 of human pregnane × receptor/NR1I2 and their association with CYP3A4 expression. Drug Metab. Dispos. 2008;36:169–181. doi: 10.1124/dmd.107.016600. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, Kliewer SA. Nuclear pregnane × receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol. Pharmacol. 2002;62:638–646. doi: 10.1124/mol.62.3.638. [DOI] [PubMed] [Google Scholar]
- Mickley LA, Bates SE, Richert ND, Currier S, Tanaka S, Foss F, Rosen N, Fojo A. Modulation of the expression of a multidrug resistance gene (mdr-1/P-glycoprotein) by differentiating agents. J. Biol. Chem. 1989;264:18031–18040. [PubMed] [Google Scholar]
- Nacci DE, Champlin D, Jayaraman S. Adaptation of the estuarine fish Fundulus heteroclitus (Atlantic killifish) to polychlorinated biphenyls (PCBs) Estuaries and Coasts. 2010;33:853–864. [Google Scholar]
- Nacci D, Coiro L, Champlin D, Jayaraman S, McKinney R, Gleason TR, Munns WR, Specker JL, Cooper KR. Adaptations of wild populations of the estuarine fish Fundulus heteroclitus to persistent environmental contaminants. Mar. Biol. 1999;134:9–17. [Google Scholar]
- Nacci DE, Kohan M, Pelletier M, George E. Effects of benzo[a]pyrene exposure on a fish population resistant to the toxic effects of dioxin-like compounds. Aquat. Toxicol. 2002;57:203–215. doi: 10.1016/s0166-445x(01)00196-5. [DOI] [PubMed] [Google Scholar]
- Oleksiak MF, Karchner SI, Jenny MJ, Franks DG, Welch DB, Hahn ME. Transcriptomic assessment of resistance to effects of an aryl hydrocarbon receptor (AHR) agonist in embryos of Atlantic killifish (Fundulus heteroclitus) from a marine Superfund site. BMC Genomics. 2011;12:263. doi: 10.1186/1471-2164-12-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RD, Hollingshead BD, Omiecinski CJ, Perdew GH. Aryl-hydrocarbon receptor activation regulates constitutive androstane receptor levels in murine and human liver. Hepatol. 2007;46:209–218. doi: 10.1002/hep.21671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MR, Scheffler BE, Wang L, Willett KL. Effects of benzo(a)pyrene exposure on killifish (Fundulus heteroclitus) aromatase activities and mRNA. Aquat. Toxicol. 2006;77:267–78. doi: 10.1016/j.aquatox.2005.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell WH, Bright R, Bello SM, Hahn ME. Developmental and tissue-specific expression of AHR1, AHR2, and ARNT2 in dioxin-sensitive and -resistant populations of the marine fish, Fundulus heteroclitus. Toxicol. Sci. 2000;57:229–239. doi: 10.1093/toxsci/57.2.229. [DOI] [PubMed] [Google Scholar]
- Proestou DA, Flight P, Champlin D, Nacci D. Targeted approach to identify genetic loci associated with evolved dioxin tolerance in Atlantic killifish (Fundulus heteroclitus) BMC Evol. Biol. 2014;14:7. doi: 10.1186/1471-2148-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzel AM, Karchner SI, Franks DG, Evans BR, Nacci DE, Champlin D, Vieira VM, Hahn ME. Genetic variation at aryl hydrocarbon receptor (AHR) loci in populations of Atlantic killifish (Fundulus heteroclitus) inhabiting polluted and reference habitats. BMC Evol. Biol. 2014;14:6. doi: 10.1186/1471-2148-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasawatari S, Toki M, Horie T, Nakano Y, Ikeda T, Ueda K, Fujise H. Effect of PCB-126 on intracellular accumulation and transepithelial transport of vinblastine in LLC-PK1 and its transformant cells expressing human P-glycoprotein. J. Vet. Med. Sci. 2004;66:1079–1085. doi: 10.1292/jvms.66.1079. [DOI] [PubMed] [Google Scholar]
- Schmidt JV, Bradfield CA. Ah receptor signaling pathways. Annu. Rev. Cell Dev. Biol. 1996;12:55–89. doi: 10.1146/annurev.cellbio.12.1.55. [DOI] [PubMed] [Google Scholar]
- Stegeman JJ, Hahn ME. Aquatic toxicology: molecular, biochemical and cellular perspectives. D. C. Malins, Ostrander, G.K., Lewis Publishers; 1994. Biochemistry and molecular biology of monooxygenases: Current perspectives on forms, functions, and regulation of cytochrome P450 in aquatic species; pp. 87–206. [Google Scholar]
- Synold TW, Dussault I, Forman BM. The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat. Med. 2001;7:584–590. doi: 10.1038/87912. [DOI] [PubMed] [Google Scholar]
- Tabb MM, Kholodovych V, Grün F, Zhou C, Welsh WJ, Blumberg B. Highly chlorinated PCBs inhibit the human xenobiotic response mediated by the steroid and xenobiotic receptor (SXR) Environ. Health Perspect. 2004;112:163–169. doi: 10.1289/ehp.6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tampal NM, Robertson LW, Srinivasan C, Ludewig G. Polychlorinated biphenyls are not substrates for the multidrug resistance transporter-1. Toxicol. Appl. Pharmacol. 2003;187:168–177. doi: 10.1016/s0041-008x(02)00069-8. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teraoka H, Dong W, Tsujimoto Y, Iwasa H, Endoh D, Ueno N, Stegeman JJ, Peterson RE, Hiraga T. Induction of cytochrome P450 1A is required for circulation failure and edema by 2,3,7,8-tetrachlorodibenzo-p-dioxin in zebrafish. Biochem. Biophys. Res. Commun. 2003;304:223–228. doi: 10.1016/s0006-291x(03)00576-x. [DOI] [PubMed] [Google Scholar]
- Tseng HP, Hseu TH, Buhler DR, Wang WD, Hu CH. Constitutive and xenobiotics-induced expression of a novel CYP3A gene from zebrafish larva. Toxicol. Appl. Pharmacol. 2005;205:247–258. doi: 10.1016/j.taap.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Wang YM, Ong SS, Chai SC, Chen T. Role of CAR and PXR in xenobiotic sensing and metabolism. Expert Opin. Drug Metab. Toxicol. 2012;8:803–817. doi: 10.1517/17425255.2012.685237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmur B, Gräns J, Kling P, Celander MC. Interactions of pharmaceuticals and other xenobiotics on hepatic pregnane × receptor and cytochrome P450 3A signaling pathway in rainbow trout (Oncorhynchus mykiss) Aquat. Toxicol. 2010;100:91–100. doi: 10.1016/j.aquatox.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Weaver G. PCB contamination in and around New Bedford, Mass. Environ. Sci. Technol. 1984;18:22A–27A. doi: 10.1021/es00119a721. [DOI] [PubMed] [Google Scholar]
- Whitehead A, Pilcher W, Champlin D, Nacci D. Common mechanism underlies repeated evolution of extreme pollution tolerance. Proc. R. Soc. B. 2012;279:427–433. doi: 10.1098/rspb.2011.0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A, Triant DA, Champlin D, Nacci D. Comparative transcriptomics implicates mechanisms of evolved pollution tolerance in a killifish population. Molec. Ecol. 2010;19:5186–5203. doi: 10.1111/j.1365-294X.2010.04829.x. [DOI] [PubMed] [Google Scholar]
- Wirgin I, Waldman JR. Resistance to contaminants in North American fish populations. Mut. Res. 2004;552:73–100. doi: 10.1016/j.mrfmmm.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Zanette J, Jenny MJ, Goldstone JV, Woodin BR, Watka LA, Bainy ACD, Stegeman JJ. New cytochrome P450 1B1, 1C2 and 1D1 genes in the killifish Fundulus heteroclitus: Basal expression and response of five killifish CYP1s to the AHR agonist PCB126. Aquat. Toxicol. 2009;93:234–243. doi: 10.1016/j.aquatox.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]