Significance
Fc gamma receptor I (FcγRI) contributes to protective immunity against bacterial infections, but exacerbates certain autoimmune diseases. It is the sole high-affinity receptor for IgG and plays a significant role in immunotherapy. To date, there is no structural information available on how the receptor recognizes its antibody ligands, however. Consequently, the mechanism of its high-affinity IgG binding remains unclear. We report the first structure of the high-affinity Fc receptor in complex with IgG-Fc. The structural work reveals a direct receptor recognition of Fc glycan as a major factor in receptor affinity. This is the first example of Fc receptor making direct glycan contact through protein residues. The results have implications for the use of glycan engineering in immunotherapy.
Keywords: CD64, FcgRI, IgG recognition, crystal structure, glycan recognition
Abstract
Fc gamma receptor I (FcγRI) contributes to protective immunity against bacterial infections, but exacerbates certain autoimmune diseases. The sole high-affinity IgG receptor, FcγRI plays a significant role in immunotherapy. To elucidate the molecular mechanism of its high-affinity IgG binding, we determined the crystal structure of the extracellular domains of human FcγRI in complex with the Fc domain of human IgG1. FcγRI binds to the Fc in a similar mode as the low-affinity FcγRII and FcγRIII receptors. In addition to many conserved contacts, FcγRI forms additional hydrogen bonds and salt bridges with the lower hinge region of Fc. Unique to the high-affinity receptor-Fc complex, however, is the conformation of the receptor D2 domain FG loop, which enables a charged KHR motif to interact with proximal carbohydrate units of the Fc glycans. Both the length and the charge of the FcγRI FG loop are well conserved among mammalian species. Ala and Glu mutations of the FG loop KHR residues showed significant contributions of His-174 and Arg-175 to antibody binding, and the loss of the FG loop–glycan interaction resulted in an ∼20- to 30-fold decrease in FcγRI affinity to all three subclasses of IgGs. Furthermore, deglycosylation of IgG1 resulted in a 40-fold loss in FcγRI binding, demonstrating involvement of the receptor FG loop in glycan recognition. These results highlight a unique glycan recognition in FcγRI function and open potential therapeutic avenues based on antibody glycan engineering or small molecular glycan mimics to target FcγRI for certain autoimmune diseases.
IgGs and pentraxins are circulating immune components that directly recognize pathogens. On formation of immune complexes or opsonization, they activate cellular response through Fc receptors (FcRs) (1, 2). The FcRs for IgGs include FcγRI (CD64); FcγRII (CD32) with A, B, and C isoforms; and FcγRIII (CD16) with two isoforms (3). Most of these are activating receptors either containing an intracellular immunoreceptor tyrosine-based activation motif or associated with an FcR common γ chain (4). FcγRIIB is an inhibitory receptor that contains an intracellular immunoreceptor tyrosine-based inhibitory motif. FcγRIIIB does not have a cytosolic domain and is anchored to the plasma membrane through glycosylphosphatidylinositol linkage. The binding affinity to IgG ranges from 10−8 M for FcγRI to 10−5–10−7 M for FcγRII and III (3).
FcγRI plays an important role in the protection against bacterial infections, but also exacerbates certain autoimmune diseases (5). Owing to its high-affinity antibody binding, FcγRI is important in antibody therapy as well (6, 7). To date, the structure of the ligand-bound high-affinity receptor has not been determined, however. Consequently, the mechanism of its high-affinity antibody recognition remains to be elucidated. The role of glycan in antibody function has been a subject of intense study. Differential glycosylation of Fc, notably fucosylated Fc, is known to affect Fc receptor binding (8, 9). Furthermore, sialylated IgGs have been shown to be anti-inflammatory components of intravenous immunoglobulin (10, 11), and glycosylation affects their binding to the low-affinity FcγRIIB and FcγRIII (11–13). Structural evidence suggests that the conserved glycosylation at Asn-297 of the constant region of IgG1 is important to maintain the conformation of Fc for receptor binding (12, 14). Whether Fc receptors make significant glycan contacts for their IgG affinity is not clear, however.
Structures of the low-affinity Fcγ receptors have been determined with bound IgG-Fc (15–18). Unlike the other two-domain FcγRs, FcγRI contains three extracellular Ig-like domains, designated D1, D2, and D3. Earlier mutational analysis suggests that D2 and D3 domains are important to confer high-affinity antibody binding (19). Recently, the structure of human FcγRI showed a close packing of the FcγRI D1 and D2 domains resembling that of FcεRI, and mutations in the FG loop of the FcγRI D2 domain reduced its IgG binding affinity (20). The mechanism of the high-affinity FcγRI ligand recognition remains unresolved. To provide further insight into the high-affinity antibody recognition by FcγRI, and to facilitate the development of FcγRI-mediated immunotherapy, we determined the structure of the extracellular domains of human FcγRIA in complex with the Fc domain of human IgG1. Our study identifies a structural mechanism for high-affinity IgG binding by the receptor.
Results and Discussions
Overall Structure of the FcγRI-Fc Complex.
The nonglycosylated extracellular region of human FcγRI (residues 16–289) was expressed in Escherichia coli as described previously, and then purified using sequential Ni-NTA and IgG1 affinity columns. Human IgG1-Fc (residues 216–446) was expressed in stable transfected CHO cells (7). The FcγRI and IgG1-Fc complex was crystallized in a P1 space group and diffracted to 3.5-Å resolution. The structure was solved by the molecular replacement method using Phaser and refined using Phenix to final R factors of 0.245 for Rcryst and 0.295 for Rfree (Table S1).
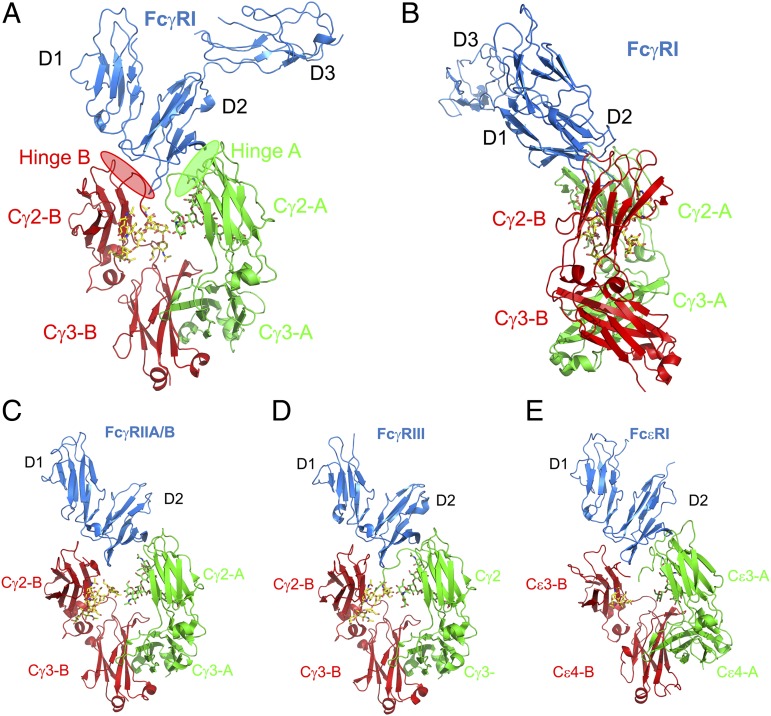
There are two complexes in each crystallographic unit cell. Electron densities are well defined for the two N-terminal domains of the receptor and for both chains of Fc (Fig. S1). The densities for the C-terminal D3 domain of FcγRI are less well defined, indicating flexibility in the receptor D3 domain orientation. The conformation of the Fc-bound FcγRI is essentially identical to that of the ligand-free receptor, resulting a root mean square difference (rmsd) of 0.7 Å among all Cα atoms. The human IgG1-Fc (residues 224–446) contains the lower hinge peptide (Cys-229–Pro-238), Cγ2 (Ser-239–Lys-338) and Cγ3 (Ala-339–Gly-446) domains. Both the Cγ2 and Cγ3 domains of Fc have well defined electron densities, including the glycosylation associated with Asn-297. The overall conformation of Fc resembles that of FcγRIII bound Fc with a small but asymmetric rotation induced by the bound receptor to the Cγ2 domains, resulting in a loss of the twofold symmetry observed in the receptor-free Fc structure.
FcγRI bound to the lower hinge region of the Fc with its D1 and D2 domains resembling the conformation of the two-domain FcγRII, FcγRIII, and FcεRI ligand complexes (15–17, 21) (Fig. 1). Except for a ∼10° rotation in the Cγ2 domains of each Fc to accommodate receptors, the entire complexes are superimposable, resulting in an rmsd of 1.0 Å between FcγRI-Fc and FcγRII and an rmsd of 1.3 Å between FcγRI-Fc and FcγRIII. The FcγRI-Fc complex buries a total solvent-accessible surface area of 2,161 Å2, significantly greater than the 1,800 Å2 buried in the low-affinity receptor-Fc complexes (15). Previous work has suggested the importance of FcγRI D3 domain in IgG binding (19). The D3 domain is not in direct contact with Fc in the current complex structure; however, the receptor D3 domain packs against the D2 domain, forming a hydrophobic hinge core, suggesting that the D3 domain is important for maintaining receptor conformation and stability.
Fig. 1.
(A and B) Ribbon representations of the overall structure of FcγRI (blue; A) and IgG1-Fc complex (B) in two orthogonal views. (C–E) Structures of FcγRIIA/B (PDB ID codes 3WJJ and 3RY6) (C), FcγRIII (PDB 1T83) (D), and FcεRI (PDB ID code 1F6A) (E) complexes with their Fc. The two chains of Fc are shown in green (chain A) and red (chain B). Carbohydrates associated with Asn-297 on both the A-chain and B-chain of Fc are shown as stick models. FcγRI contact regions with the hinge A and B of Fc are shaded in green and red, respectively.
Interface of FcγRI with the Lower Hinge Fc Region Is Conserved.
The interface between the receptor and the two Fc chains can be divided into three regions: between the FcγRI D2 domain C-strand (Tyr-133–Leu-136), C′-strand (Lys-142–His-148), and C′E loop (His-148–Trp-149) and the lower hinge region from the Fc A-chain; between the receptor D1-D2 interdomain hinge (Arg-102–Trp-104), D2 domain BC loop (Trp-127–Tyr-133), and lower hinge from the Fc B-chain; and between the FcγRI D2 domain FG loop (Met-171–Tyr-176) and the Fc glycans.
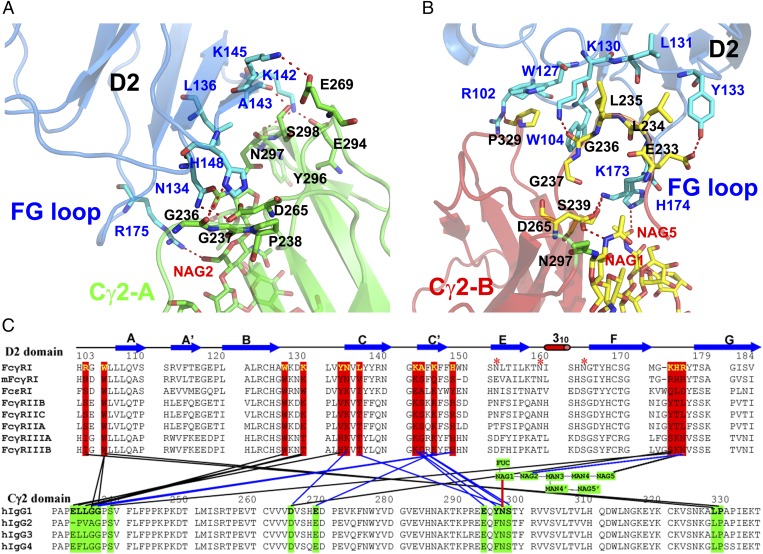
At the Fc A-chain lower hinge region, the receptor contacts are primarily salt bridges and hydrogen bonds. Specifically, Lys-142 and Lys-145 of FcγRI form two pairs of salt bridges with Fc residues, Glu-294 and Glu-269, respectively (Fig. 2A). Both lysine residues are also present in the low-affinity FcγRIIA, FcγRIIB, and FcγRIII (Fig. 2C). Two hydrogen bonds are observed, between Asn-134 of the receptor and Asp-265 of Fc and between His-148 of the receptor and the carbonyl group of Gly-236 of Fc. Asn-134 is replaced with a Lys in the low-affinity Fcγ receptors. His-148 is conserved in FcγRIIA and FcγRIII, but replaced with an Arg in FcγRIIB. In addition, Leu-136 and Phe-146 of FcγRI form van der Waals interactions with the lower hinge LLGG motif of Fc (15). The majority of FcγRI contacts with the A chain of Fc are preserved in the low-affinity Fcγ receptors. Likewise, Asp-265, Glu-269, and Glu-294 are conserved among subclasses of IgGs.
Fig. 2.
Detailed receptor-Fc contacts at the hinge A (A) and hinge B (B) interface regions. FcγRI residues are shown as stick models in blue, and residues from the A and B chains of Fc are shown in green and yellow, respectively. Glycans associated with the A and B chains of Fc are shown in green and yellow, respectively. (C) Sequences of Fc receptors and IgG-Fc, with contacts colored in blue and black for the receptor interactions with chain A and chain B Fc, respectively.
At the Fc B-chain interface, FcγRI forms primarily van der Waals interactions with the Fc (Fig. 2B). Trp-104 and Trp-127 from FcγRI form a hydrophobic cluster with a pair of conserved leucine and proline residues, Leu-328 and Pro-329, from the Cγ2 domain of the Fc. This cluster, also referred to as a Trp-Pro-Trp sandwich, is present in all low- affinity receptor complexes, including that of the IgE-Fc-FcεRI complex (15, 16, 21). Unique to FcγRI is a hydrogen bond between Arg-102 of the receptor and the carbonyl group of Pro-329 of Fc. A second hydrophobic cluster is observed between the side chains of Lys-130, Leu-131, and Tyr-133 of FcγRI and the lower hinge residues Leu-234 and Leu-235 of the Fc. This second hydrophobic cluster is smaller in the low affinity receptor-Fc complexes as both Leu-131 and Tyr-133 of FcγRI are replaced by smaller Pro, Ala, Val, and His residues in FcγRII and FcγRIII. Tyr-133 also forms a hydrogen bond with Glu-233 of Fc-B. Glu-233 is conserved in IgG1, IgG3, and IgG4 but is replaced by a Pro residue in IgG2, suggesting that the lack of this Glu-233–Tyr-133 hydrogen bond in IgG2 contributes to its lower receptor binding affinity. Interestingly, this hydrogen bond is absent in FcγRIIA because the receptor contains a Val-133 in place of Tyr, suggesting that FcγRIIA is less discriminative against IgG2 (22–24). As noted previously, other residues at the lower hinge of IgG2 also differ from those of IgG1 and may affect its receptor binding affinity (15).
The FcγRI D2 Domain FG Loop Forms Unique Contacts with Fc Glycan.
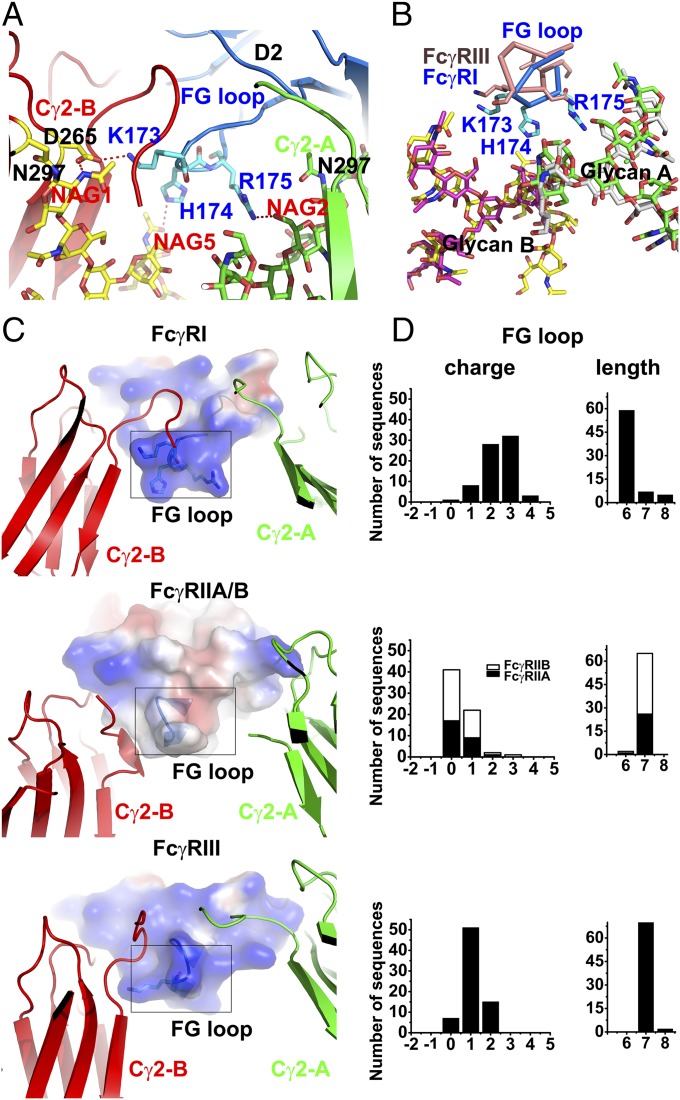
The third interface is from the receptor D2 domain FG loop, 171MGKHRY176, to Fc and its associated glycan. In particular, Lys-173 of the receptor forms a salt bridge with a conserved Asp-265 on the B-chain of Fc (Fig. 3A). A similar salt bridge is present between the Asp-265 on the B-chain of Fc and Lys-120 on the C-strand of FcγRIII in the low-affinity receptor complex structure (15); however, Lys-120 of FcγRIII is not conserved in FcγRI and is replaced by an asparagine. Instead, Lys-173 of FcγRI serves as a surrogate salt bridge partner.
Fig. 3.
(A) Detailed contacts between the FcγRI D2 domain FG loop and Fc with glycans associated with the Fc A and B chains in green and yellow, respectively. (B) Structural comparison of the FG loop conformation between FcγRI (blue) and FcγRIII (wheat). The structures are superimposed on their Fc A chain. The glycans (magenta and white) from FcγRIII-Fc complex (PDB ID code 1T83) and the glycans (yellow and green) from the current FcγRI-Fc complex are well superimposed. The FG loop of FcγRI is positioned closer to the carbohydrates. (C) Electrostatic potential [−50, 0, 50] of the D2 domain FG loop calculated for FcγRI (Top), FcγRIIA/B (Middle; PDB ID codes 3WJJ and 3RY6), and FcγRIII (Bottom; PDB ID code 1T83). (D) Distribution of the FG loop charge and length among homologous mammalian FcγRI (Top), FcγRIIA (Middle; solid bars), FcγRIIB (Middle, shaded bars), and FcγRIII (Bottom) receptors.
His-174 and Arg-175 of the receptor extend into the Fc opening toward the glycans, such that their closest contacts are the carbohydrates attached to Asn-297. The His-174 side chain is close to the C4 hydroxyl group on a branched mannose. Both Lys-173 and His-174 are close to the glycan attached to the B-chain of Fc. The side chain of Arg-175 is located within hydrogen-bonding distance to the O3-hydroxyl of the first N-acetyl glucosamine (GlcNAc) and to the O6-hydroxyl of the second GlcNAc attached to the A-chain of Asn-297 (Fig. 3A). Thus, the FG loop links glycans from both chains of Fc. Interestingly, all three FG loop residues contact the proximal rather than distal carbohydrate units of the Fc glycan, suggesting that the receptor interacts with the conserved and less flexible part of the glycan. This is also consistent with the finding that EndoS-treated IgG1, which leaves the first GlcNac and its associated branched core fucose, retains better FcγRI binding than IgG1 treated with peptide-N-glycosidase F, which removes all glycan attached to Asn-297 (23).
In comparison, the D2 domain FG loops of the low-affinity FcγRs are 8–10 Å away from the Fc glycans and make little contact with Fc, except for a hydrogen bond between the FcγRIII FG loop Lys Nζ and the backbone carbonyl of the lower hinge Gly-236 on Fc (Fig. 3B) (15, 17, 25). Whereas the FcγRI contacts to both hinge A and hinge B of Fc are largely conserved in the low-affinity Fcγ receptors, the contacts from the FG loop of FcγRI are unique to the high-affinity receptor, because the FG loop KHR motif is not conserved in other Fcγ receptors, except for the presence of a Lys in FcγRIII in the His-174 position of FcγRI (Fig. 2C).
Structural Mechanism for High-Affinity FcγRI Binding to Fc.
Although the overall receptor-ligand docking mode is similar in all Fcγ receptors, FcγRI is the sole high-affinity receptor for monomeric IgG. Two of the three FcγRI-Fc contact sites involve receptor residues interacting with the lower hinge regions of the Fc chains. Both the hinge A and hinge B contacts are quite conserved in the low-affinity FcγRII and FcγRIII ligand complexes, suggesting that they are the critical contacts to define the common FcγR receptor and IgG binding mode. Interestingly, FcγRI forms a more extensive hydrophobic contact at hinge B, thus favoring its binding to IgG. In contrast, the interface between the FG loop of FcγRI and IgG is unique to the high-affinity receptor. The equivalent receptor FG loops in the low-affinity receptor-ligand complexes do not form significant contacts to Fc.
The importance of the FG loop to high-affinity receptor-IgG binding was demonstrated in previous mutational work (26). Placement of the FG loop of FcγRI into FcγRIII resulted in a 15-fold increase in IgG binding affinity, and the mutant FcγRIII bound to IgG only fivefold weaker than FcγRI. As noted previously, the FG loop of FcγRI is one residue shorter than the FG loops of other Fcγ receptors (Fig. 2); however, this affinity increase is lost when a Val is inserted into the FG loop to match the length of those in the low-affinity Fcγ receptors. Structurally, this shorter FG loop of FcγRI is positioned ∼5 Å closer to the glycan in the center of the horseshoe opening than that of FcγRIII (Fig. 3B). Interestingly, the D2 domain FG loop of FcγRI is the most positively charged FG loop among all three Fcγ receptors, owing to the presence of the KHR motif (Fig. 3C).
To further address the significance of a shorter but positively charged FG loop to the high-affinity Fcγ receptor, we compared the length and the number of charged residues present in the FG loop sequences among homologous Fcγ receptors in mammalian species with Lys, Arg, and His each as +1 and Asp and Glu each as −1. The sequences of majority homologous FcγRI receptors have six amino acid FG loops and carry +2 or +3 charges (Fig. 3D). In contrast, most of the D2 domain FG loops in all homologous low-affinity receptors are seven residues long. The FG loop sequences of FcγRIIA and FcγRIIB carry +1 or less charge, whereas those of FcγRIII carry 0 to +2 charges, with majority +1. Furthermore, the D1 domain FG loop of FcγRI is also longer and carries a −1 net charge, similar to the corresponding loops in the low-affinity receptors. Thus, the shorter and positively charged FG loop is a unique feature of the high-affinity receptor. Structurally, the shorter FG loop enables it to move closer toward the Fc glycans and to adopt a glycan binding conformation.
Mutations in the FG Loop of FcγRI Reduce Its IgG Binding Affinity.
To further investigate the contribution of the FG loop KHR motif to high-affinity receptor-IgG binding, we generated both single and triple alanine mutations of the KHR motif and measured the mutant receptor binding affinities to all three subclasses of IgGs. K173A and H174A mutant FcγRI exhibited only minor reductions in their binding affinities (twofold to fivefold) to IgG1, IgG3, and IgG4 (Table 1). In contrast, R175A mutation, the most disruptive mutant, resulted in ∼20 fold reductions to IgG1, IgG3, and IgG4 binding. The triple AAA mutant (K173A/H174A/R175A) bound to all three IgGs with 0.18–0.3 µM affinity, similar to that of R175A.
Table 1.
Dissociation constants for FcγRI and IgG binding
| FcγRI | Affinity, KD, nM | ||
| IgG1 | IgG3 | IgG4 | |
| WT, CHO | 8.8 ± 7 | 3.3 ± 3 | 26.2 ± 2 |
| WT, E. coli | 7.0 ± 1 | 8.1 ± 7 | 16.7 ± 7 |
| K173A | 25.9 ± 4 | 40.3 ± 10 | 49.4 ± 10 |
| H174A | 16.8 ± 8 | 29.4 ± 3 | 32.0 ± 10 |
| R175A | 166.0 ± 35 | 157.0 ± 37 | 314.0 ± 74 |
| AAA | 180.0 ± 42 | 189.0 ± 67 | 320.0 ± 83 |
| K173E | 44.0 ± 6 | 45.6 ± 22 | 200.0 ± 15 |
| H174E | 250.6 ± 115 | 285 ± 89 | 303.0 ± 152 |
| R175E | 26.0 ± 2 | 27.0 ± 8 | 51.3 ± 7 |
To evaluate the effect of charge contribution to FcγRI and IgG binding, we replaced the positively charged KHR each with a negatively charged Glu in the FG loop. K173E and R175E resulted in mild reductions (threefold to 10-fold) in IgG affinity. In contrast to the Ala mutants, R175E is the least disruptive among the three mutations, and H174E is the most disruptive, with 15- to 30-fold reductions in IgG binding affinities. The differential mutational effects on IgG binding, in which Glu is the most disruptive at residue 174 and Ala is the least favored at position 175, suggest that charge interaction is more important to H174, whereas hydrogen bond interaction is more important to R175. Both Ala and Glu mutations showed that glycan contact by the FG loop contributes 20- to 30-fold in receptor-IgG binding affinity, consistent with the earlier results from the FG loop swapping (26).
To further investigate the interaction of glycan with the FG loop, we measured FcγRI binding to IgG1 treated with peptide-N-glycosidase F. Removal of the Fc glycan reduced receptor binding to 0.35 µM, a 40-fold decrease in affinity. The greater loss in IgG1 binding affinity by FcγRI deglycosylation compared with the R175A, H174E, and AAA mutations is likely related to an additional conformational destabilization in Fc as the result of glycan removal (12, 14). Thus, both the receptor mutations and glycan removal resulted in a loss of binding affinity, demonstrating the specific glycan binding by the FG loop KHR motif of FcγRI. Given that deglycosylation reduced the IgG1 binding affinity of FcγRI to a level comparable to that of the lower-affinity Fcγ receptors, it is conceivable that the receptor can still recognize aglycosylated IgG in the context of multivalent immune complexes (23).
Potential Contribution of Fucosylated and Sialylated Fc Glycan to FcγRI Binding.
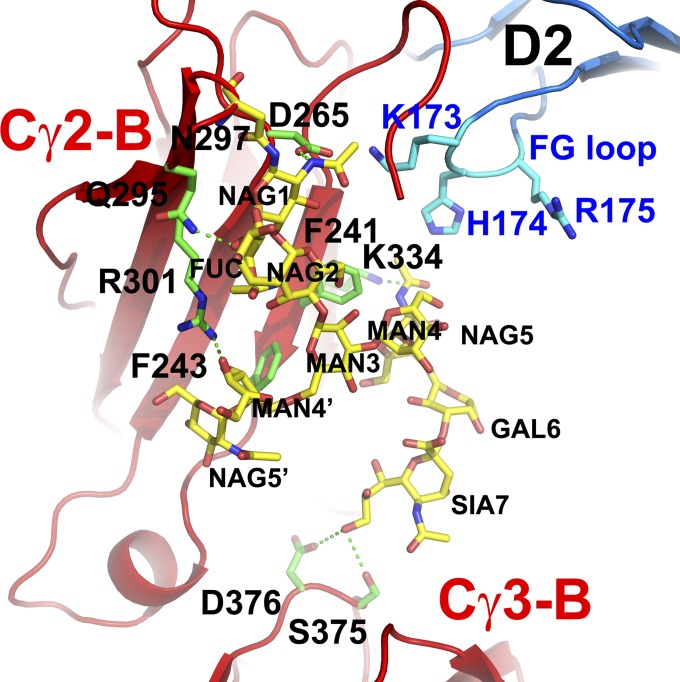
Even though the electron densities at the current resolution of the complex contain significant ambiguities, there are sufficient densities for much of the Fc glycan, although the proximal carbohydrates are better defined than the distal carbohydrate units. The N-linked glycans associated with Asn-297 are fucosylated biantennaed complex glycans. Receptor glycosylation on Asn-162 of FcγRIIIA was observed to interact with nonfucosylated glycan to stabilize the receptor complex, suggesting that a fucosylated glycan may inhibit such interaction (9, 18). Because Asn-162 is not present in FcγRI, the potential role of the branched fucose associated with the Asn-297 of Fc must be different for the high-affinity receptor. On both Fc chains, the branched fucose units form van der Waals contacts with Tyr-296 of Fc. On both Fc chains, the glycans extend along the Fc peptide, making multiple hydrogen bonds and van der Waals contacts with the Cγ2 domain residues, including Phe-241, Phe-243, Arg-301, and Lys-334 (Fig. 4). These interactions help reduce the conformational flexibility of the glycan. The direction of the glycan points from Asn-297 toward the Cγ3 domain of its Fc. The terminal sialic acid is visible only on one chain (Fig. 4), and is located close to hydrogen bonding distance to the side chain of Ser-375 and Asp-376 from the Cγ3 domain of Fc, suggesting that sialylated glycans may reduce glycan conformational flexibility by contacting the Cγ3 domain of Fc.
Fig. 4.
Detailed interaction between carbohydrates associated with Asn-297 and Fc.
In summary, the structure of the high-affinity human FcγRI bound to IgG1-Fc illustrates both a conserved receptor recognition to the lower hinge region of Fc that defines the common Fcγ receptor-IgG binding mode and a unique FcγRI FG loop interaction with Fc and its glycans that contributes to receptor high-affinity IgG binding. The contribution of Fc glycan to FcγRI high-affinity ligand binding opens potential new avenues in immunotherapy to apply glycan engineering in targeting FcγRI to achieve desired antibody efficacy. It also raises the possibility of using small-molecular glycan-based compounds or mimics to target FcγRI as effective treatment for certain autoimmune diseases.
Materials and Methods
Protein Expression, Purification, and Crystallization.
The ectodomain of FcγRI (residues 16–289) was engineered with 19 mutations as described previously, including T20P, T25K, T38S, L46P, T63I, S69T, R71H, V77E, N78D, I100V, F114L, T160M, N163S, N195T, N206T, L207P, N240D, L283H, and L285Q. This engineered FcγRI ectodomain was synthesized and cloned into a pET26b vector by gene synthesis (Genscript), solubly expressed in E. coli cells, and purified via Ni-NTA affinity chromatography followed by IgG affinity chromatography. The purified FcγRI from E. coli bound to human IgG1, IgG3, and IgG4 with nM affinities similar to those from recombinant mammalian expressed FcγRI (Table 1). Human IgG1-Fc protein (216–444) was expressed in CHO DXB-11 cells. The human tissue plasminogen activator prepro secretion signal sequence was used to direct secretion of the IgG1-Fc.
The recombinant Fc was purified by a protein A affinity column, followed by a Superdex 200 size exclusion column (GE Healthcare) in 50 mM sodium phosphate and 109 mM sodium chloride pH 7.3. The final product was filtered through a 0.22-μm syringe filter, aliquotted into polypropylene vials, and stored at below −60 °C. Purified IgG1-Fc was verified by N-terminal sequencing, was at least 95% pure, and contained <0.27 endotoxin units/mg. Before crystallization, FcγRI was mixed with Fc dimer at a 1.2:1 molar ratio, and the FcγRI-Fc complex was further purified by gel filtration chromatography in 10 mM Hepes (pH7.4) and 0.15 M NaCl. The crystals used for data collection were grown in hanging drops at 22 °C using 1 μL of protein with a final OD280nm of 6.7 and 1 μL of reservoir solution (10% PEG 8000, 10 mM Hepes pH 7.5, and 50 mM Li2SO4). Human plasma IgG1, IgG3, and IgG4 were purchased from Athens Research & Technology.
Diffraction Data Collection and Structure Determination.
The crystals were immersed in cryoprotectant (20% PEG 8000, 10 mM Hepes pH 7.5, 50 mM Li2SO4, and 15% glycerol) before being flash-cooled in liquid nitrogen. Five X-ray datasets were collected to 3.5-Å resolution at SER-CAT beamlines, processed, and merged with HKL2000 (27) (Table S1). FcγRI-Fc complex crystals belong to space group P1 with a pseudomerohedral twofold twinning. The structure of the FcγRI-Fc complex was solved by a molecular replacement method with Phaser (28) in the CCP4 package (29) using FcγRI (PDB ID code 3RJD) and Fc (PDB ID code 3AY4), respectively, as the search model. Model building and refinement were carried out using Coot (30) and Phenix (31) with noncrystallography symmetry (NCS) restraints and the twin law of (−h, −k, l). The overall electron density of the complex is of decent quality (Fig. S1).
Carbohydrate molecules were added manually using (2Fo-Fc) electron density maps contoured at 1.0σ (SD of the map) and refined. The residues are numbered consistent with FCGR1_HUMAN in the Swiss-Prot entry. The final model includes two FcγRI-Fc complexes (PDB ID code 4X4M). The Ramachandran statistics were generated and verified by Procheck of CCP4. Hinge angles were calculated using HINGE (32), and the buried surface area was calculated using the CCP4 package. The electrostatic charges were converted from atom coordinates using the PDB2PQR server with the CHARMM force field and calculated by APBS and displayed between –50 and 50 kT/e (33). All structure figures were generated with Pymol (34).
Site-Directed Mutagenesis of FcγRI.
FcγRI D2 domain FG loop (171MGKHRY176) mutations were generated by site-directed mutagenesis using a QuikChange II Site-Directed Mutagenesis Kit (Agilent) according to the manufacturer’s instructions. A total of eight mutants were generated, including three single Ala mutations (K173A, H174A, and R175A), a triple AAA mutant (K173A/H174A/R175A), three single Glu mutations (K173E, H174E, and R175E), and a triple EEE mutant (K173E/H174E/R175E). All mutations were confirmed by DNA sequencing (ACGT Inc.). The mutated FcγRI proteins were expressed as WT as described above and purified by Ni-NTA chromatography.
Surface Plasmon Resonance Solution Binding Experiments.
Surface plasmon resonance measurements were performed using a Biacore 3000 instrument and analyzed with BIAevaluation 4.1 software (GE Healthcare). Different human IgG subclasses were obtained from Athens Research & Technology. For measuring the affinity to WT or mutant FcγRI proteins, human IgG1, IgG3, and IgG4 were immobilized on Biacore carboxylated dextran CM5 chips (GE Healthcare) to 200–1,000 response units using primary amine coupling in 10 mM sodium acetate (pH 5.0). The analytes consisted of serial dilutions of WT or mutant FcγRI proteins between 2 µM and 63 nM in a buffer containing 10 mM Hepes (pH 7.4) and 0.15 M NaCl. The dissociation constants were obtained by kinetic curve fitting for the binding of FcγRI to IgG, and steady-state fitting for the binding of FcγRI mutants to IgG, using BIAevaluation 4.1 software. IgG1 (100 µg) was treated with 2,500 U of peptide-N-glycosidase F (New England Biolabs) in sodium phosphate buffer (pH 7.5) at 37 °C for 3 h to remove the glycan on Fc. Representative binding sensorgrams are shown in Fig. S2.
Supplementary Material
Acknowledgments
This work was supported by the intramural research funding from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4X4M).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1418812112/-/DCSupplemental.
References
- 1.Hulett MD, Hogarth PM. Molecular basis of Fc receptor function. Adv Immunol. 1994;57:1–127. doi: 10.1016/s0065-2776(08)60671-9. [DOI] [PubMed] [Google Scholar]
- 2.Lu J, et al. Structural recognition and functional activation of FcgammaR by innate pentraxins. Nature. 2008;456(7224):989–992. doi: 10.1038/nature07468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daëron M. Fc receptor biology. Annu Rev Immunol. 1997;15:203–234. doi: 10.1146/annurev.immunol.15.1.203. [DOI] [PubMed] [Google Scholar]
- 4.Ravetch JV, Kinet JP. Fc receptors. Annu Rev Immunol. 1991;9:457–492. doi: 10.1146/annurev.iy.09.040191.002325. [DOI] [PubMed] [Google Scholar]
- 5.Ioan-Facsinay A, et al. FcgammaRI (CD64) contributes substantially to severity of arthritis, hypersensitivity responses, and protection from bacterial infection. Immunity. 2002;16(3):391–402. doi: 10.1016/s1074-7613(02)00294-7. [DOI] [PubMed] [Google Scholar]
- 6.Bevaart L, et al. The high-affinity IgG receptor, FcgammaRI, plays a central role in antibody therapy of experimental melanoma. Cancer Res. 2006;66(3):1261–1264. doi: 10.1158/0008-5472.CAN-05-2856. [DOI] [PubMed] [Google Scholar]
- 7.Ellsworth JL, et al. Targeting immune complex-mediated hypersensitivity with recombinant soluble human FcgammaRIA (CD64A) J Immunol. 2008;180(1):580–589. doi: 10.4049/jimmunol.180.1.580. [DOI] [PubMed] [Google Scholar]
- 8.Niwa R, et al. IgG subclass-independent improvement of antibody-dependent cellular cytotoxicity by fucose removal from Asn297-linked oligosaccharides. J Immunol Methods. 2005;306(1-2):151–160. doi: 10.1016/j.jim.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 9.Ferrara C, et al. Unique carbohydrate–carbohydrate interactions are required for high-affinity binding between FcgammaRIII and antibodies lacking core fucose. Proc Natl Acad Sci USA. 2011;108(31):12669–12674. doi: 10.1073/pnas.1108455108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaneko Y, Nimmerjahn F, Ravetch JV. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science. 2006;313(5787):670–673. doi: 10.1126/science.1129594. [DOI] [PubMed] [Google Scholar]
- 11.Nimmerjahn F, Ravetch JV. Divergent immunoglobulin g subclass activity through selective Fc receptor binding. Science. 2005;310(5753):1510–1512. doi: 10.1126/science.1118948. [DOI] [PubMed] [Google Scholar]
- 12.Radaev S, Sun PD. Recognition of IgG by Fcgamma receptor: The role of Fc glycosylation and the binding of peptide inhibitors. J Biol Chem. 2001;276(19):16478–16483. doi: 10.1074/jbc.M100351200. [DOI] [PubMed] [Google Scholar]
- 13.Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu Rev Immunol. 2007;25:21–50. doi: 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]
- 14.Feige MJ, et al. Structure of the murine unglycosylated IgG1 Fc fragment. J Mol Biol. 2009;391(3):599–608. doi: 10.1016/j.jmb.2009.06.048. [DOI] [PubMed] [Google Scholar]
- 15.Radaev S, Motyka S, Fridman WH, Sautes-Fridman C, Sun PD. The structure of a human type III Fcgamma receptor in complex with Fc. J Biol Chem. 2001;276(19):16469–16477. doi: 10.1074/jbc.M100350200. [DOI] [PubMed] [Google Scholar]
- 16.Sondermann P, Huber R, Oosthuizen V, Jacob U. The 3.2-A crystal structure of the human IgG1 Fc fragment-Fc gammaRIII complex. Nature. 2000;406(6793):267–273. doi: 10.1038/35018508. [DOI] [PubMed] [Google Scholar]
- 17.Ramsland PA, et al. Structural basis for Fc gammaRIIa recognition of human IgG and formation of inflammatory signaling complexes. J Immunol. 2011;187(6):3208–3217. doi: 10.4049/jimmunol.1101467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizushima T, et al. Structural basis for improved efficacy of therapeutic antibodies on defucosylation of their Fc glycans. Genes Cells. 2011;16(11):1071–1080. doi: 10.1111/j.1365-2443.2011.01552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hulett MD, Hogarth PM. The second and third extracellular domains of FcgammaRI (CD64) confer the unique high-affinity binding of IgG2a. Mol Immunol. 1998;35(14-15):989–996. doi: 10.1016/s0161-5890(98)00069-8. [DOI] [PubMed] [Google Scholar]
- 20.Anthony RM, Wermeling F, Ravetch JV. Novel roles for the IgG Fc glycan. Ann N Y Acad Sci. 2012;1253:170–180. doi: 10.1111/j.1749-6632.2011.06305.x. [DOI] [PubMed] [Google Scholar]
- 21.Garman SC, Wurzburg BA, Tarchevskaya SS, Kinet JP, Jardetzky TS. Structure of the Fc fragment of human IgE bound to its high-affinity receptor Fc epsilonRI alpha. Nature. 2000;406(6793):259–266. doi: 10.1038/35018500. [DOI] [PubMed] [Google Scholar]
- 22.Bruhns P, et al. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood. 2009;113(16):3716–3725. doi: 10.1182/blood-2008-09-179754. [DOI] [PubMed] [Google Scholar]
- 23.Lux A, Yu X, Scanlan CN, Nimmerjahn F. Impact of immune complex size and glycosylation on IgG binding to human FcγRs. J Immunol. 2013;190(8):4315–4323. doi: 10.4049/jimmunol.1200501. [DOI] [PubMed] [Google Scholar]
- 24.Ravetch JV. In: in Fundamental Immunology. 5th Ed. Paul W, editor. Lippincott Raven; Philadelphia: 2008. pp. 685–700. [Google Scholar]
- 25.Mimoto F, et al. Engineered antibody Fc variant with selectively enhanced FcγRIIb binding over both FcγRIIa(R131) and FcγRIIa(H131) Protein Eng Des Sel. 2013;26(10):589–598. doi: 10.1093/protein/gzt022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu J, Ellsworth JL, Hamacher N, Oak SW, Sun PD. Crystal structure of Fc{gamma}RI and its implication in high-affinity {gamma}-immunoglobulin G binding. J Biol Chem. 2011;286(47):40608–40613. doi: 10.1074/jbc.M111.257550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Afonine PV, Grosse-Kunstleve RW, Adams PD. The Phenix refinement framework. CCP4 Newsletter. 2005;42(8) [Google Scholar]
- 32.Snyder GA, Brooks AG, Sun PD. Crystal structure of the HLA-Cw3 allotype-specific killer cell inhibitory receptor KIR2DL2. Proc Natl Acad Sci USA. 1999;96(7):3864–3869. doi: 10.1073/pnas.96.7.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dolinsky TJ, et al. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35(Web Server issue):W522–525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeLano W. The PyMOL Molecular Graphics System, Version 1.3. Schrödinger; New York: 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.