Significance
The fossil record shows that the wrist and digits have an aquatic origin, becoming recognizable in a group of (mostly extinct) fish that contained robust fins. Do the fins of living fishes have the equivalent of these structures? Because comparisons of fin and limb morphology have been inconclusive, we sought to investigate this question using developmental and molecular data. By utilizing a nonmodel fish (the spotted gar), we find that the regulatory networks that control “wrist and digit”-building genes (Hox) are deeply conserved between fish and tetrapods. The genomic architecture described here defines Hox gene activity in fins and limbs as equivalent, in turn suggesting equivalence between the distal bones of fish fins and the wrist and/or digits of tetrapods.
Keywords: evolution, development, autopod, gene regulation, Hox
Abstract
There is no obvious morphological counterpart of the autopod (wrist/ankle and digits) in living fishes. Comparative molecular data may provide insight into understanding both the homology of elements and the evolutionary developmental mechanisms behind the fin to limb transition. In mouse limbs the autopod is built by a “late” phase of Hoxd and Hoxa gene expression, orchestrated by a set of enhancers located at the 5′ end of each cluster. Despite a detailed mechanistic understanding of mouse limb development, interpretation of Hox expression patterns and their regulation in fish has spawned multiple hypotheses as to the origin and function of “autopod” enhancers throughout evolution. Using phylogenetic footprinting, epigenetic profiling, and transgenic reporters, we have identified and functionally characterized hoxD and hoxA enhancers in the genomes of zebrafish and the spotted gar, Lepisosteus oculatus, a fish lacking the whole genome duplication of teleosts. Gar and zebrafish “autopod” enhancers drive expression in the distal portion of developing zebrafish pectoral fins, and respond to the same functional cues as their murine orthologs. Moreover, gar enhancers drive reporter gene expression in both the wrist and digits of mouse embryos in patterns that are nearly indistinguishable from their murine counterparts. These functional genomic data support the hypothesis that the distal radials of bony fish are homologous to the wrist and/or digits of tetrapods.
The origin of novel features is a key question in evolutionary biology, and the autopod—wrists, fingers, ankles, and toes—is a hallmark example (1). Although paleontological data, such as that from the Devonian lobe fin Tiktaalik roseae, reveal a sequence of changes in the elaboration of the bony elements of fins into limbs (2), in living taxa there is a lack of obvious homology between the wrist and digits of tetrapod limbs and the pectoral fin skeleton of extant fish (3). Tetrapod forelimbs are generally composed of a series of long bones (upper arm and forearm), followed by small nodular bones (wrist), and ending in another group of long bones (digits). Ray-finned (Actinopterygian) pectoral fins are diverse but are usually composed of a series of long proximal radials, followed by a set of smaller distal radials. The open question remains: do extant fish have the equivalent of wrists or digits?
The molecular mechanisms governing the development of mammalian limbs have been approached in mouse models through multiple levels of analysis, from chromatin dynamics, to enhancer sequence, to gene expression patterns (4). Murine limbs display two successive phases of gene expression of the HoxD and HoxA gene clusters. The initial or “early” phase of expression begins with members at the 3′ end of the clusters being expressed broadly, and members at the 5′ end of the cluster being activated in an increasingly restricted number of cells (5). This “early” phase of Hox expression is associated with the development of the upper arm (stylopod) and forearm (zeugopod). The initial wave of Hox expression is followed by a temporally distinct second activation of Hox genes, this time beginning with members of the 5′ end of the cluster being expressed most broadly and in the presumptive digits. This second, “late” phase of expression is necessary for autopod formation, as evidenced by a loss of this domain resulting in deletion of the wrist and digits (5). The genomic regulatory elements and chromatin dynamics responsible for enacting these two phases have been studied in detail in the HoxD cluster, where the “early” and “late” phases are governed by enhancers that lie on opposite sides of the HoxD cluster—3′ and 5′, respectively—and activated in turn by shifting domains of open chromatin (6, 7). In addition, recent work has identified a series of enhancers that drive late phase expression of the HoxA cluster in the developing mouse autopod in a fashion similar to that of HoxD (8).
To what extent are the regulatory mechanisms that drive autopodial development present in fish fins and, if they are present, what is their developmental role? Previous work has shown that at least one of the “autopod” enhancers (CsB) is present and active in the common ancestor of gnathostomes (9). Additionally, recent work in zebrafish has shown that the early and late topological chromatin domains are indeed observed in bony fish (10). However, teleost fish enhancer domains were unable to drive reporter gene expression in the developing digits of transgenic mice, suggesting that although bony fish do contain a version of the autopod regulatory apparatus, these enhancers are not responsive to the regulatory program present in murine digits (10). Thus, the number, extent, and function of “limb” enhancers in fish remain to be fully explored, especially in fish species outside those of traditional model systems that might resemble ancestral characters more closely than teleosts.
To address these issues, we used a combination of epigenetic profiling in zebrafish and phylogenetic footprinting using the genome assembly of the recently sequenced spotted gar (Lepisosteus oculatus) (11) to investigate the enhancers that control hoxd and hoxa expression in bony fish. The phylogenetic position of gar is crucial to our investigation, in that gar represents a lineage that diverged from teleost fishes before the teleost genome duplication, an event that may cloud studies of regulatory evolution (Fig. 1A) (12–14). The data presented here reveal an unprecedented and previously undescribed level of deep conservation of the vertebrate autopod regulatory apparatus, suggesting homology between the distal radials of bony fish and the autopod of tetrapods.
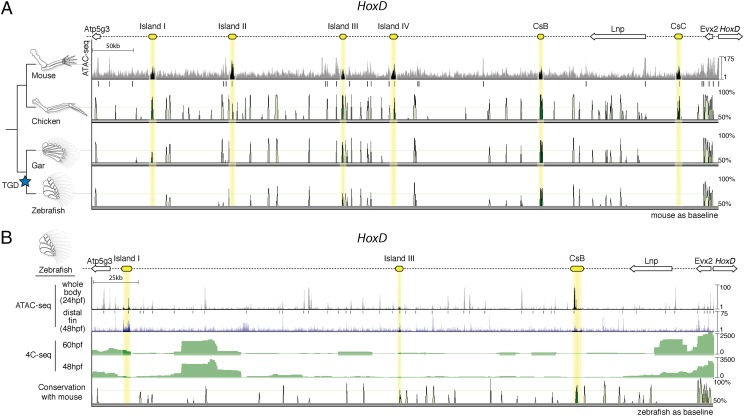
Fig. 1.
Chromatin state and sequence conservation of the HoxD autopod “regulatory archipelago” gene desert among select vertebrates. (A) (Top) Schematic representation of the HoxD centromeric gene desert, with cis-regulatory “islands” active in mouse denoted in yellow. ATAC-seq data are shown for mouse autopods at e12.5, providing a view of open chromatin. Statistically significant peaks are denoted by black bars. Sequence conservation is shown below for chicken, gar, and zebrafish. Note that sequence conservation for Island I is only found in gar, a nonteleost actinopterygian, and not in the teleost zebrafish. A blue star marks the teleost-specific genome duplication (TGD). (B) The zebrafish hoxD regulatory archipelago, with candidate “autopod” enhancers shown in yellow. ATAC-seq results for 24 hpf whole-body and 48 hpf distal fin are shown, identifying areas of open chromatin. 4C-seq results on whole-body 48 and 60 hpf embryos using hoxd13a as the target are shown in green. The putative teleost ortholog of Island I shows significant interaction with the hoxd13a promoter at 60 hpf. Vista plot with zebrafish as the baseline shows no sequence conservation with mouse for autopod enhancers other than Island III and CsB.
Results
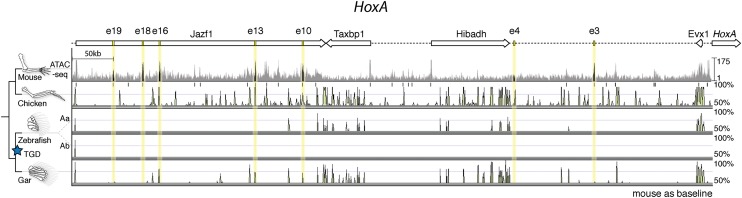
To identify orthologs of murine Hox limb enhancers in bony fish, we performed a multiple sequence alignment of the genomic region upstream of the HoxD and HoxA clusters from a number of tetrapod and teleost species (Figs. 1A and 2). Our initial survey revealed a dearth of sequence conservation for late phase enhancers in teleost species, aside from previously studied CsB, and an additional limb enhancer previously called Island III (6, 9). As we broadened our taxonomic input by including the genome of the spotted gar, we found peaks of conserved noncoding elements for the HoxD late phase digit enhancer Island I (Fig. 1A) and the HoxA late phase enhancers e16, e13, and e10 (Fig. 2) (6, 8). We reasoned that the unduplicated nature of the gar genome makes its Hox clusters a better representative of the common ancestor of bony fish (Osteichthyes), revealing sequences that have diverged beyond recognition in derived, duplicated teleost genomes (13).
Fig. 2.
Chromatin state and sequence conservation of HoxA autopod enhancers among select vertebrates. A schematic of the HoxA “autopod” enhancer region is shown at the top, highlighting autopod-specific enhancers, with ATAC-seq data from the mouse autopod directly below. Sequence conservation for chicken, zebrafish, and gar are provided. The gar genome uniquely reveals sequence conservation of three hoxA enhancers (e16, e13, e10) identified previously in mouse (8).
To identify potential orthologs in the zebrafish genome that are not revealed by sequence conservation, we performed the “Assay for Transposable Accessible Chromatin” (ATAC-Seq) on 24 hours postfertilization (hpf) zebrafish embryos, as well as embryonic day 12.5 (e12.5) mouse autopods as a control for the assay (15, 16). ATAC-seq on mouse autopods significantly identified the majority of validated HoxD and HoxA enhancers with autopod activity (Figs. 1A and 2). Although not all previously defined Hox autopod enhancers were identified (e.g., Island I, CsB) in the mouse sample, we found substantial overlap of our ATAC-seq peaks with validated limb enhancers from other studies (Table S1), making us confident that the assay would be able to identify enhancers in zebrafish. We performed ATAC-seq on whole-body zebrafish embryos and noticed an area of open chromatin in the genome near the atp5g3a gene that roughly matched the genomic coordinates of the late phase enhancer Island I (Fig. 1B). To determine whether this region interacts with the zebrafish hoxd13a promoter, we performed “Circular Chromatin Conformation Capture” (4C-seq) to detect contact with enhancers up to ∼1 Mb away, on 48 and 60 hpf whole-body zebrafish embryos. We found that the area containing the putative ortholog of Island I in zebrafish significantly interacts with the hoxd13a locus at 60 hpf (Fig. 1B). Because the assay was performed on whole embryos, it is possible that these interactions may not be specific to the developing fin. We did not observe significant contact between Island I and hoxd13a before 60 hpf, possibly owing to limitations on the numbers of fin cells available when using whole-body embryos for 4C-seq. Although the ATAC and 4C-seq data revealed genomic areas in zebrafish that could be orthologous to HoxA enhancers (Fig. S1), these areas contained multiple candidates, and thus we sought to characterize only the gar orthologs of HoxA where sequence orthology was clear.
Having identified potential enhancers according to chromatin structure and sequence conservation, we sought to characterize their activity by cloning the sequences from gar and zebrafish into reporter vectors and injecting them into zebrafish embryos to assay for domains of expression (17). We performed transient injections of gar Island III, as well as the genomic “areas” from the gar genome that could potentially contain cryptic autopod enhancers (potential orthologs of mouse Islands II and IV not found by sequence alignment), and detected no fin signal for these regions (Table S2). In contrast, we were able to detect GFP in the pectoral fin of zebrafish embryos injected with constructs containing either gar Island I, CsB, or e16, which we raised to sexual maturity to obtain stable transgenic lines. The late phase enhancer gar Island I exhibited little activity at 31 hpf but increased and became distally restricted by 38 hpf (Fig. 3A and Fig. S2). At 48 hpf, gar Island I drove a pattern of expression that was restricted to the anterior portion of the distal pectoral fin (Fig. 3A and Fig. S2). Gar CsB drove a pattern of activity that was similar to that of Island I, with a peak of expression at 48 hpf but with a posterior–distal restriction to its expression. Of the three hoxA autopod enhancers that were identified in the gar genome by sequence conservation (Fig. 2), e16 from mouse has been shown to drive the most robust native pattern of expression throughout the autopod of transgenic mice (8). As a result, we chose to make stable transgenic zebrafish lines of the gar e16 regulatory element and found that it drove a distally restricted pattern of gene expression at 48 hpf, much like the late phase hoxD enhancers from gar (Fig. 3A and Fig. S2).
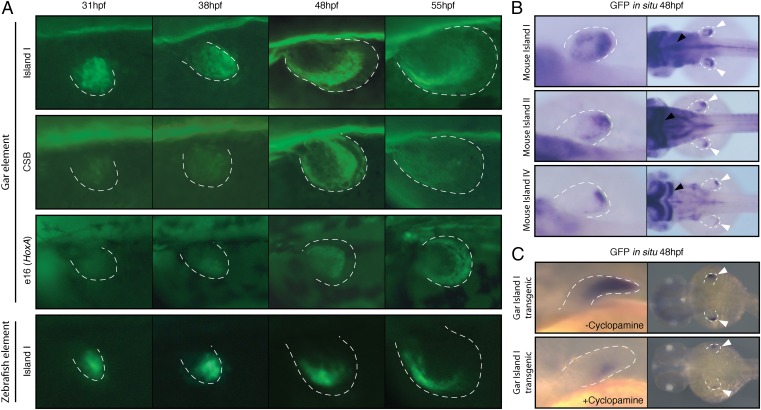
Fig. 3.
Transgenic zebrafish reveal the expression dynamics of multiple “autopod” regulatory elements present in fish genomes. (A) Left pectoral fins of stable zebrafish lines transgenic for putative late phase enhancers identified in Fig. 1 and Fig. S1. The gar Island I, CsB, and e16 enhancers all drove reporter expression at 48 hpf in a strip of expression at the distal edge of the fin. Island I from zebrafish, which was identified through ATAC-seq and 4C-seq, drove a pattern of expression nearly identical to its ortholog in gar. All views are dorsal, with anterior to the left, posterior to the right, except for e16 which shows a lateral view. (B) The expression patterns of mouse autopod enhancers Island I, Island II, and Island IV in transgenic zebrafish. (Left) Dorsal views of right pectoral fins; (Right) dorsal views of the embryo. Island I from mouse drove a pattern of expression that was strongest at 48 hpf, with most expression at the distal compartment of the pectoral fin. Similarly, mouse Islands II and IV were active distally in the pectoral fin. Expression in the brain (black arrows) is due to a strong hindbrain enhancer present in the vector that serves as a positive control for transgenesis. (C) Like their murine counterparts, fish late phase enhancers depend on Shh signaling. (Upper) Lateral view of an in situ hybridization for the GFP transgene at 48 hpf on an embryo transgenic for the gar Island I enhancer. (Lower) Reduced GFP expression in a transgenic embryo that was treated with the Shh inhibitor cyclopamine from 31 to 48 hpf. Distal edge of the fin is marked by a dotted white line in all pictures.
Using the data from ATAC-seq, 4C-seq, and gar conservation as a guide, we cloned the putative region of Island I from zebrafish and tested its potential activity by cloning it into a reporter vector and producing stable zebrafish transgenic lines. We found that zebrafish Island I drives strong expression of GFP in the distal pectoral fin at 48 hpf, in a pattern similar to that of its gar ortholog (Fig. 3A and Fig. S2). To confirm that the activity of zebrafish Island I was present in the distal fin (and not simply active elsewhere in the body), we grew multiple F1 fish from one founder and collected a pool of distal fin cells from embryos at 48 hpf using FACS. We subjected these distal fin cells to ATAC-seq and found that open chromatin signal for Island I was enriched in this sample (Fig. 1B). Combined with ATAC-seq and 4C-seq data, our transgenic zebrafish analysis indicates that zebrafish Island I functions endogenously as a distal fin enhancer acting upon the hoxd13a gene.
Because Island I from gar seemed to mark a distal compartment of the zebrafish pectoral fin, we sought to investigate whether Island I from mouse would elicit a similar pattern in zebrafish. To test this hypothesis, we cloned Island I from mouse into a zebrafish enhancer detection vector and again created independent stable lines of transgenic zebrafish. Mouse Island I drove a pattern of robust expression in the distal portion of the endochondral disk at 48 hpf, marking a distal segment of the fin much like its gar ortholog (Fig. 3B). To further assess the activity of the other murine HoxD “autopod” enhancers when injected into zebrafish, we cloned mouse Islands II and IV and created stable zebrafish lines (at least three independent lines/construct). Both of these mouse enhancers drove GFP in the distal compartment of the zebrafish pectoral fin (Fig. 3B). These experiments demonstrate that the mouse autopod enhancer Island I, when transgenic in zebrafish, drives expression in the same pattern and area (the distal endochondral compartment of the fin) as the fish ortholog of this enhancer. Furthermore, mouse autopod enhancers that may not be present in fish genomes (Islands II and IV) are active in this same distal portion of the zebrafish pectoral fin.
The late phase of Hox expression in the mouse autopod and zebrafish distal pectoral fin depend on sonic hedgehog (Shh) (18–20). To test whether the expression driven by the gar late phase enhancers also depends on Shh signaling, we inhibited Shh function by adding its antagonist cyclopamine (20) to transgenic zebrafish embryos from 31 to 48 hpf. Embryos transgenic for the gar late phase enhancer Island I that were treated with cyclopamine had markedly decreased expression of the reporter at 48 hpf, whereas the overall fin morphology remained normal (Fig. 3C). We repeated these experiments on five independent transgenic lines of gar Island I and found that nearly all embryos for all lines exhibited a loss of late phase GFP expression when treated with cyclopamine (n = 21 of 25, 5 embryos per line). These findings show that both mouse and gar late phase enhancers depend on Shh signaling and suggest that Shh control of distal Hoxd expression is an ancestral characteristic of bony fish.
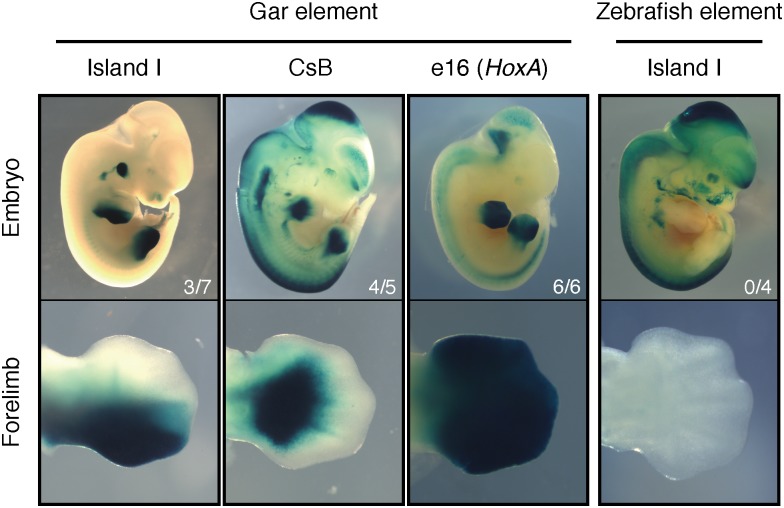
Finally, to test whether the fish enhancers could respond to the trans-acting environment of developing tetrapod digits, we cloned Island I from zebrafish, and Island I, Island III, CsB, and e16 from gar, each into an Hsp68-LacZ reporter vector for mouse transgenesis. In line with previous findings using teleost enhancers (9, 10), zebrafish Island I did not elicit activity in the developing mouse digits at e12.5 (Fig. 4 and Fig. S3). The gar Island III construct, which failed to produce fin reporter expression in zebrafish fins, also failed to elicit reporter expression in transgenic mouse limbs (Fig. S3). However, the gar late phase enhancer Island I drove a pattern of expression that was restricted to the posterior half of the limb, extending throughout the autopod and into the digits (Fig. 4). In addition, the late phase element CsB from gar drove robust expression in the developing autopod and in the neural tube. Furthermore, we found that the e16 (hoxA) enhancer from gar drove strong expression in the entire autopod of transgenic mice at e12.5, with a sharp boundary at the zeugopod (Fig. 4). In sum, gar Island I, CsB, and e16 evoked late phase patterns of activity in developing mouse digits that were nearly indistinguishable from those driven by the orthologous elements from the mouse genome (Fig. 4 and Fig. 5) (6, 8, 21).
Fig. 4.
Late phase enhancers from the nonteleost gar drive expression throughout the autopod in transgenic mice. Island I from gar drove expression in a posterior strip throughout the autopod and to the distal tip of the developing digits. Gar CsB drove strong expression in the autopod and in the presumptive digits. The hoxA late phase enhancer e16 from gar drove robust expression in the entire autopod, with a sharp boundary at the zeugopod. In accordance with previous reports, Island I from the teleost zebrafish had no activity in the autopod of transgenic mice.
Fig. 5.
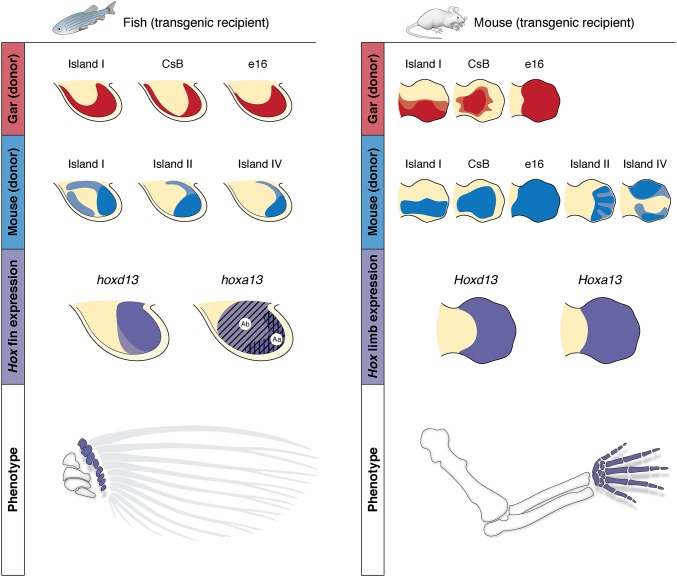
Summary and comparison of transgenic animals and their implications for the origin and evolution of the autopod. Transgenics and expression dynamics for fish (Left) and mouse (Right). Patterns driven by gar enhancers are drawn in red; mouse enhancer activity is drawn in blue. Late phase enhancers from gar drive distal fin expression in transgenic zebrafish, whereas late phase enhancers from mouse drive various patterns of autopod expression in mouse (row 1). When introduced into zebrafish, autopod enhancers from mouse (row 2, Left) drive reporter expression in the same distal compartment of the pectoral fin as the endogenous fish enhancers (compare rows 1 and 2, Left). Transgenic mice carrying late phase enhancers from gar (row 1, Right) show patterns of reporter expression throughout the autopod that are nearly identical to the endogenous activity from mouse (compare rows 1 and 2, Right). The comparative regulatory data shown here define the late phase Hox expression from fish and mouse (row 3) as homologous. Row 4 depicts a hypothesis of homology between the area of future distal radials in fish fins and the autopod of tetrapods (shown in purple), supported by the expression and chromatin conformation data reported here.
Discussion
Using a combination of model and nonmodel systems, functional genomics, and transgenesis, we have identified “autopod” building enhancers of the hoxD and hoxA cluster in the genomes of bony fish. We find that these enhancers drive distal expression in fish fins and respond to the same functional cue as their murine counterparts. There are likely to be three classes of “autopod” enhancers that emerge in comparisons between fish and tetrapods: those that are either fish or tetrapod specific, and those that are shared between the two groups. Here we have defined a potential minimum complement of enhancers that are shared between fish and tetrapods and were present in their common ancestor. Our investigations suggest that a set of enhancers are tetrapod-specific (6, 8) and may have been assembled during the fin to limb transition to elaborate the autopod (22). However, a complete investigation of the total number of late phase Hox enhancers in fish is necessary to determine the pattern of regulatory elements gained or lost over evolutionary time. Overall, our results provide regulatory support for an ancient origin of the “late” phase of Hox expression that is responsible for building the autopod.
With a largely conserved regulatory system, the question remains as to how (or whether) changes to HoxD and HoxA enhancers have contributed to appendage evolution. It has been suggested that although late phase Hox expression is present in fish pectoral fins, a type of genetic “rewiring” of the regulatory landscape may have occurred in the transition from fins into limbs to produce digits (10). Our data using zebrafish enhancers, together with previous reports, support the hypothesis that late phase enhancers are present in teleosts and are active during fin development but are unable to drive expression in the developing digits of transgenic mice (Fig. 4) (9, 10). Our results show that in contrast to teleosts, gar late phase enhancers Island I, CsB, and e16 are able to drive expression throughout the mouse autopod (including digits) in a pattern that is markedly similar to those of the mouse enhancers (Figs. 4 and 5) (6, 8). Taken together, these data suggest that at least a subset of regulatory elements that drive late phase expression of Hox genes was present in the last common ancestor of bony vertebrates (Osteichthyes) and have remained functionally conserved throughout evolution. Teleosts, perhaps owing to genome duplication followed by subfunctionalization (23), represent a derived state that has lost the ability in some taxa to respond to a distal program in the developing limb.
Regulatory data presented here define the late phases of Hoxd and Hoxa expression in the pectoral appendages of ray-finned fish and tetrapods as homologous domains. Thus, we suggest that the distal compartment marked by the late phase in fish, which will contribute to the distal radials, is equivalent to the autopod of tetrapods (Fig. 5). This relationship raises the intriguing possibility that digits and/or mesopodials are an ancient feature of fish fins represented by distal radials, and makes a specific prediction testable by future genomic manipulations. Our data point to a generally conserved regulatory network governing Hox genes in fins, emphasizing the need to study subtle modifications to Hox regulation as well as additional genomic regions that may influence fin morphology and that emerged during the fin to limb transition.
Materials and Methods
VISTA Alignments.
Genomic segments of interest were downloaded from the Ensembl and University of California, Santa Cruz genome databases and aligned using the mVista LAGAN program. The following parameters were used: calc window: 100 bps; Min Cons Width: 100 bps; Cons Identity: 70%.
Enhancer Cloning.
Zebrafish and gar.
Putative enhancers were amplified by PCR using primers found in Table S3, purified genomic DNA, and Platinum Taq DNA polymerase High Fidelity (Invitrogen). PCR fragments (∼3 kb in length to accommodate flanking sequence) were gel purified using a NucleoSpin Gel and PCR Clean-up Kit (Macherey-Nagel) and subcloned into PCR8GW/GW/TOPO vector (Invitrogen) according to the manufacturer’s protocols. Fragments were then moved via the Gateway system (Invitrogen) into either the pXIG-cFos-eGFP vector (17) for zebrafish transgenesis or to a Gateway-Hsp68-LacZ vector (kind gift of Marcelo Nobrega, The University of Chicago, Chicago) for mouse transgenesis. Final destination vectors were confirmed by restriction digest and sequencing.
Mouse.
Mouse islands were cloned in the PCR8GW/GW/TOPO vector as stated above. These enhancers were shuttled via Gateway into an enhancer detection vector composed of a gata2 minimal promoter, an enhanced GFP reporter gene, and a strong midbrain enhancer (z48) that works as an internal control for transgenesis. All these elements were previously reported (24) and were assembled with a tol2 transposon (25).
Zebrafish transgenesis.
All zebrafish and mouse work was performed according to standard protocols approved by The University of Chicago. Zebrafish embryos (strain *AB) were collected from natural spawning. Transposase RNA was synthesized from the pCS2-zT2TP vector using the mMessage mMachine SP6 kit (Ambion) (26). Injections solutions were made following Fisher et al. (17), and ∼2 nL were injected into the cytoplasm of embryos at the one- or two-cell stage. Injected embryos were raised to sexual maturity according to standard protocols (17, 26) and outcrossed to *AB fish to identify transgenic founders by visualization of GFP. Transgenic embryos from founders were visualized using a Leica M205FA microscope.
Mouse transgenesis.
Mouse transgenesis was performed by Cyagen Biosciences. Briefly, enhancer-Hsp68-LacZ vectors were linearized with SalI, gel purified, microinjected into fertilized mouse oocytes, and transferred to pseudopregnant females. Embryos were collected at e12.5, stained for β-galactosidase activity, and genotyped with LacZ primers using DNA extracted from yolk sacs. A summary of primers used for mouse constructs as well as injection results appears in Fig. S2 and Table S2.
Cyclopamine treatment and RNA in situ hybridization.
Cyclopamine was resuspended in 100% ethanol to make a stock of 10 mM and stored protected from light. Zebrafish embryos were dechorionated manually and placed into embryo medium containing 100 μM cyclopamine (LC Laboratories) in the dark from 31 hpf until fixation at 48 hpf. RNA in situ hybridization experiments targeting the GFP transcript were performed after 2 d of fixation in 4% PFA, according to standard protocols (27).
ATAC-Seq.
Mouse.
ATAC-seq experiments were performed as previously described (15). Mouse autopods at e12.5 were isolated by cutting the limb at the zeugopod/autopod boundary. Three autopods were placed in PBS supplemented with 0.125% collagenase (Sigma) and left shaking at 300 rpms for 20 min. The solution was then pipetted to make a homogeneous solution and filtered through a 0.45-µm cell strainer. A total of 50,000 cells were then subsequently used for transposition (15).
Zebrafish.
Briefly, 10 zebrafish embryos were manually dechorionated, anesthetized with tricaine (Sigma), and disrupted in 500 µL of Ginzburg Fish Ringers (55 mM NaCl, 1.8 mM KCl, and 1.25 mM NaHCO3). The cells in the resulting solution were counted using a neubauer chamber, then pelleted at 500 × g in a tabletop centrifuge and washed with cold PBS. A total of 75,000 cells were lysed [lysis buffer: 10 µM Tris (pH 7.4), 10 µM NaCl, 3 µM MgCl2, and 0.1% IGEPAL (Sigma CA-630)] and incubated for 30 min at 37 °C with the TDE1 enzyme. The sample was then purified with a Qiagen Minelute kit, and PCR was performed with 13 cycles using Ad1F and Ad2.1R primers (15) with KAPA HiFi hotstart enzyme (Kapa Biosystems). The resulting library was sequenced in a HisEq 2000 pair end lane producing 180 M of 49-bp reads per end. Reads were aligned using zebrafish July 2010 assembly (danRer7) as the reference genome. Duplicated pairs or those ones separated by more than 2 kb were removed. The enzyme cleavage site was determined as the position −4 (minus strand) or +5 (plus strand) from each read start, and this position was extended 5 bp in both directions. ATAC-seq on FACS-sorted cells were performed and analyzed in the same manner with the following modifications: ∼400 embryos from the gar Island I transgenic line were grown to 48 hpf, anesthetized with tricaine (Sigma), and placed in PBS supplemented with 0.125% collagenase (Sigma). Embryos were incubated at 37 °C for 1 h shaking at 300 rpm, with gentle pipetting every 20 min to disrupt embryos. The solution was filtered twice with a 0.45-µm cell strainer to obtain a single cell suspension, and FACS sorted for GFP using an Avalon 2-4 machine at the University of Chicago. The resulting solution contained the 40,000 brightest GFP+ cells and was processed immediately for ATAC-seq as described above.
Statistically significant peaks were determined by extending ATAC-seq reads to 100 bp and using MACS software (28) with default parameters.
Circular chromosome conformation capture.
Circular chromosome conformation capture (4C-seq) assays were performed as previously reported (29–32). Five hundred zebrafish embryos at 48 or 60 hpf were dechorionated using pronase and disrupted in 1 mL of Ginzburg Fish Ringers (55 mM NaCl, 1.8 mM KCl, and 1.25 mM NaHCO3). Isolated cells were treated with lysis buffer [lysis buffer: 10 mM Tris⋅HCl (pH 8), 10 mM NaCl, 0.3% IGEPAL CA-630 (Sigma), and 1× protease inhibitor mixture (cOmplete, Roche)] and the DNA digested with DpnII (NEB) and Csp6I (Fermentas, Thermo Scientific) as primary and secondary enzymes, respectively. T4 DNA ligase (Promega) was used for both ligation steps. Specific primers were designed at gene promoters, and Illumina adaptors were included in primer sequence. Eight PCRs were performed with the Expand Long Template PCR System (Roche) and pooled together. This library was purified with a High Pure PCR Product Purification Kit (Roche) and its concentration measured using the Quanti-iTTM PicoGreen dsDNA Assay Kit (Invitrogen, P11496) in a Qubit machine and sent for deep sequencing in an Illumina HisEq 2000 machine multiplexing 10 samples per lane. 4C-seq data were analyzed as previously described (31). Briefly, raw sequencing data were demultiplexed using the primer sequences as barcodes and aligned using zebrafish July 2010 assembly (danRer7) as the reference genome. Reads located in fragments flanked by two restriction sites of the same enzyme, or in fragments smaller than 40 bp, were filtered out. Mapped reads were then converted to reads-per-first-enzyme-fragment-end units and smoothed using a 30-fragment mean running window algorithm.
Supplementary Material
Acknowledgments
We thank John Westlund for excellent illustration and assistance assembling the figures; Marcelo Nobrega, Mike Coates, Ilya Ruvinsky, and Tom Stewart for critical reading of the manuscript; Gokhan Dalgin, Noritaka Adachi, Joyce Pieretti, Darcy Ross, and Justin Lemberg for helpful discussions; and the Prince and Ho laboratories for reagents and help with zebrafish. This study was supported by The Brinson Foundation (N.H.S.), National Science Foundation Doctoral Dissertation Improvement Grant 1311436 (to A.R.G.), Brazilian National Council for Scientific and Technological Development Grants 402754/2012-3 and 477658/2012-1 (to I.S.), National Institutes of Health Grant R01RR020833 (R01OD011116) (to J.H.P), “Initiative Evolutionary Biology” from the Volkswagen Foundation, Germany and Alexander von Humboldt-Foundation (I.B.), and Spanish and Andalusian Governments Grants BFU2010-14839, BFU2013-41322-P, and Proyecto de Excelencia BIO-396 (to J.L.G.-S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE61063 and GSE61065).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1420208112/-/DCSupplemental.
References
- 1.Wagner GP . Homology, Genes, and Evolutionary Innovation. Princeton Univ Press; Princeton: 2014. pp. 327–384. [Google Scholar]
- 2.Shubin NH, Daeschler EB, Jenkins FA., Jr The pectoral fin of Tiktaalik roseae and the origin of the tetrapod limb. Nature. 2006;440(7085):764–771. doi: 10.1038/nature04637. [DOI] [PubMed] [Google Scholar]
- 3.Schneider I, Shubin NH. The origin of the tetrapod limb: From expeditions to enhancers. Trends Genet. 2013;29(7):419–426. doi: 10.1016/j.tig.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 4.Montavon T, Duboule D. Chromatin organization and global regulation of Hox gene clusters. Philos Trans R Soc Lond B Biol Sci. 2013;368(1620):20120367. doi: 10.1098/rstb.2012.0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zakany J, Duboule D. The role of Hox genes during vertebrate limb development. Curr Opin Genet Dev. 2007;17(4):359–366. doi: 10.1016/j.gde.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 6.Montavon T, et al. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147(5):1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 7.Andrey G, et al. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science. 2013;340(6137):1234167. doi: 10.1126/science.1234167. [DOI] [PubMed] [Google Scholar]
- 8.Berlivet S, et al. Clustering of tissue-specific sub-TADs accompanies the regulation of HoxA genes in developing limbs. PLoS Genet. 2013;9(12):e1004018. doi: 10.1371/journal.pgen.1004018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneider I, et al. Appendage expression driven by the Hoxd Global Control Region is an ancient gnathostome feature. Proc Natl Acad Sci USA. 2011;108(31):12782–12786. doi: 10.1073/pnas.1109993108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woltering JM, Noordermeer D, Leleu M, Duboule D. Conservation and divergence of regulatory strategies at Hox Loci and the origin of tetrapod digits. PLoS Biol. 2014;12(1):e1001773. doi: 10.1371/journal.pbio.1001773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braasch I, et al. A new model army: Emerging fish models to study the genomics of vertebrate Evo-Devo. J Exp Zoolog B Mol Dev Evol. 2014 doi: 10.1002/jez.b.22589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 2003;13(3):382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amores A, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282(5394):1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 14.Amores A, Catchen J, Ferrara A, Fontenot Q, Postlethwait JH. Genome evolution and meiotic maps by massively parallel DNA sequencing: Spotted gar, an outgroup for the teleost genome duplication. Genetics. 2011;188(4):799–808. doi: 10.1534/genetics.111.127324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10(12):1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lara-Astiaso D, et al. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345(6199):943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher S, et al. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc. 2006;1(3):1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- 18.Chiang C, et al. Manifestation of the limb prepattern: Limb development in the absence of sonic hedgehog function. Dev Biol. 2001;236(2):421–435. doi: 10.1006/dbio.2001.0346. [DOI] [PubMed] [Google Scholar]
- 19.Litingtung Y, Dahn RD, Li Y, Fallon JF, Chiang C. Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature. 2002;418(6901):979–983. doi: 10.1038/nature01033. [DOI] [PubMed] [Google Scholar]
- 20.Ahn D, Ho RK. Tri-phasic expression of posterior Hox genes during development of pectoral fins in zebrafish: Implications for the evolution of vertebrate paired appendages. Dev Biol. 2008;322(1):220–233. doi: 10.1016/j.ydbio.2008.06.032. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez F, Duboule D, Spitz F. Transgenic analysis of Hoxd gene regulation during digit development. Dev Biol. 2007;306(2):847–859. doi: 10.1016/j.ydbio.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 22.Freitas R, Gómez-Marín C, Wilson JM, Casares F, Gómez-Skarmeta JL. Hoxd13 contribution to the evolution of vertebrate appendages. Dev Cell. 2012;23(6):1219–1229. doi: 10.1016/j.devcel.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 23.Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151(4):1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bessa J, et al. Zebrafish enhancer detection (ZED) vector: A new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Dev Dyn. 2009;238(9):2409–2417. doi: 10.1002/dvdy.22051. [DOI] [PubMed] [Google Scholar]
- 25.Kawakami K, Shima A, Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc Natl Acad Sci USA. 2000;97(21):11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suster ML, Abe G, Schouw A, Kawakami K. Transposon-mediated BAC transgenesis in zebrafish. Nat Protoc. 2011;6(12):1998–2021. doi: 10.1038/nprot.2011.416. [DOI] [PubMed] [Google Scholar]
- 27.Thisse C, Thisse B, Postlethwait JH. Expression of snail2, a second member of the zebrafish snail family, in cephalic mesendoderm and presumptive neural crest of wild-type and spadetail mutant embryos. Dev Biol. 1995;172(1):86–99. doi: 10.1006/dbio.1995.0007. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295(5558):1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 30.Hagège H, et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat Protoc. 2007;2(7):1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 31.Noordermeer D, et al. The dynamic architecture of Hox gene clusters. Science. 2011;334(6053):222–225. doi: 10.1126/science.1207194. [DOI] [PubMed] [Google Scholar]
- 32.Splinter E, de Wit E, van de Werken HJ, Klous P, de Laat W. Determining long-range chromatin interactions for selected genomic sites using 4C-seq technology: From fixation to computation. Methods. 2012;58(3):221–230. doi: 10.1016/j.ymeth.2012.04.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.