Significance
Daily rhythms of gene expression ensure that biological processes occur at the optimal time of day. In plants, temporally regulated processes include traits of ecological and agricultural importance, and understanding how changes in daily rhythms of expression modify such traits has broad implications. We find that natural genetic variation can accurately modify temporal gene expression waveforms during the day by influencing light signaling pathways, rather than circadian rhythms. We further show that changes in transcriptional patterns induced by natural alleles are sufficient to affect downstream molecular outputs and cause phenotypic diversity. Such natural alleles could provide an advantage by adjusting the activity of temporally regulated processes while avoiding the pleiotropic effects associated with severe disruptions of the circadian system.
Keywords: diurnal, circadian, rhythms, Arabidopsis, GIGANTEA
Abstract
Daily rhythms of gene expression provide a benefit to most organisms by ensuring that biological processes are activated at the optimal time of day. Although temporal patterns of expression control plant traits of agricultural importance, how natural genetic variation modifies these patterns during the day and how precisely these patterns influence phenotypes is poorly understood. The circadian clock regulates the timing of gene expression, and natural variation in circadian rhythms has been described, but circadian rhythms are measured in artificial continuous conditions that do not reflect the complexity of biologically relevant day/night cycles. By studying transcriptional rhythms of the evening-expressed gene GIGANTEA (GI) at high temporal resolution and during day/night cycles, we show that natural variation in the timing of GI expression occurs mostly under long days in 77 Arabidopsis accessions. This variation is explained by natural alleles that alter light sensitivity of GI, specifically in the evening, and that act at least partly independent of circadian rhythms. Natural alleles induce precise changes in the temporal waveform of GI expression, and these changes have detectable effects on PHYTOCHROME INTERACTING FACTOR 4 expression and growth. Our findings provide a paradigm for how natural alleles act within day/night cycles to precisely modify temporal gene expression waveforms and cause phenotypic diversity. Such alleles could confer an advantage by adjusting the activity of temporally regulated processes without severely disrupting the circadian system.
In plants, many aspects of physiology and development, including metabolism, growth, flowering, and plant defense, are controlled by genes whose expression pattern oscillates on a daily basis (1, 2). These genes usually show peaks of expression around the time at which their function is required to regulate downstream processes. The timing of expression of most temporally regulated genes is at least partly determined by the circadian clock, an endogenous time-keeping mechanism that generates internal rhythms of ∼24 h (3). When synchronized to the external day/night cycle, circadian clocks confer an advantage to plants and other organisms by improving fitness (4, 5). Importantly, circadian rhythms are generally studied under conditions of continuous light (LL) or continuous dark (DD), in which they are not influenced by environmental transitions. These constant conditions, however, do not reflect the complexity of biologically relevant day/night cycles that organisms experience in nature. During the day, fluctuations in external cues such as light and temperature also contribute to defining the timing and amplitude of biologic processes. These cues influence rhythms of gene expression either indirectly, by synchronizing endogenous circadian rhythms to the external day/night cycle (6–8), and/or directly, by activating signaling pathways that regulate transcription (9–11). Thus, the precise timing and amplitude of daily gene expression patterns are defined by a combination of endogenous and external signals.
Temporal rhythms of expression control plant traits of ecological and agricultural importance (12–16), and understanding how precisely these rhythms vary and how this variation influences phenotypes has broad implications for plant biology. Natural diversity in daily transcriptional rhythms has mostly been analyzed by comparing gene expression between limited numbers of selected genotypes and by using temporal resolutions of relatively low precision (14, 17). To date, there has been no extensive survey describing how rhythms of expression vary at the intraspecies level, at more informative temporal resolutions, and during biologically relevant day/night cycles. The latter point is of particular relevance because natural variation in rhythms has mainly been studied in artificial continuous conditions that are used to determine certain circadian parameters. Natural variation of period length, defined as the length of the circadian cycle, was quantified in constant environmental conditions by measuring rhythms of leaf movements or oscillations of gene expression (4, 18–21). Phase, or the time at which an event occurs within a cycle, also varies extensively when determined in constant conditions (4, 22). Although changes in period length would be expected to influence phase, the relationship between both parameters is still unclear in natural genotypes (4). In summary, it is not known how much daily rhythms of expression vary in natural genotypes, what mechanisms generate this variation, and to what extent this variation influences phenotypic outputs.
These questions were addressed by using GIGANTEA (GI) as a model temporally regulated gene. GI is conserved within the plant kingdom and regulates a variety of phenotypes such as growth of the hypocotyl, flowering time, cold resistance, and starch accumulation (23–29). The peak of GI expression occurs in the evening in various plant species and is regulated by the circadian clock (12, 25, 26, 30–32). To monitor GI expression at high temporal resolution and in a large number of genotypes, we fused a 2.5-Kb fragment of the GI promoter to the luciferase (LUC) marker gene. Similar GI::LUC fusions had already been shown to faithfully track the rhythmic expression pattern of the endogenous transcript (33–35). With the luciferase system, we could accurately determine the timing of GI expression during day/night cycles and detect genetic loci that cause precise changes in the GI expression waveform. This genetic information was then exploited to create lines that precisely differ in their GI expression patterns and to test whether changes in these patterns affect downstream phenotypes.
Results and Discussion
Natural Genetic Variation Regulates the Timing of GI Expression Within Long-Day Cycles.
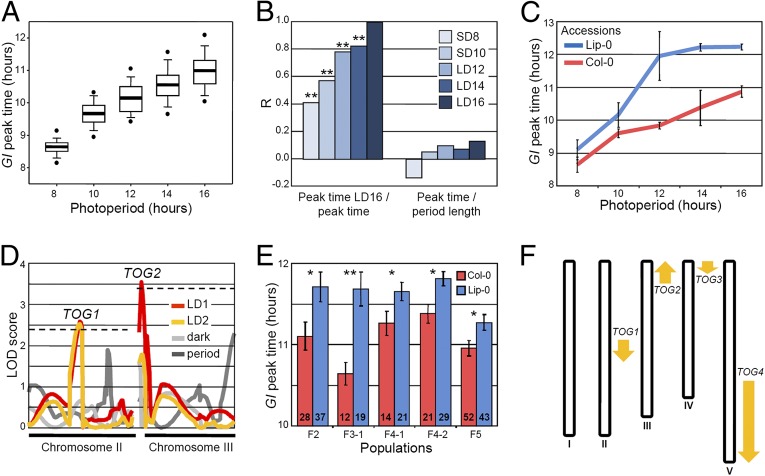
Natural variation in the waveform of GI transcription was tested for by introducing GI::LUC into 77 Arabidopsis accessions. Temporal patterns of luciferase activity were recorded under five day lengths and used to determine the peak time of GI::LUC expression (GI peak time, or sidereal phase) in each accession and condition. GI peak time varied in all day lengths, but the range of peak times was broader in long photoperiods, and the genetic contribution to peak time variation was more significant in long days (LDs) compared with in short days (SDs) (Fig. 1A and SI Appendix, Fig. S1 and Tables S1 and S2). GI peak times measured in LDs of 16 h strongly correlated with peak times measured in LDs of 14 h and LDs of 12 h, but not as strongly with peak times measured in SDs (Fig. 1B). These data suggest the existence of mechanisms that cause variability in the timing of GI expression specifically in LDs.
Fig. 1.
Natural variation and genetic basis of the timing of GI expression during day/night cycles. (A) Box plots representing the variation and average (horizontal bars) of GI peak time of expression in 77 accessions. (B) Correlations between GI peak time measured in LDs of 16 h, with GI peak time measured in other photoperiods, and between GI peak time and period length (DD) measured after entrainment in the same photoperiod. The Pearson correlation coefficient (R) indicates the strength of the correlations, with 1 and −1 indicating perfect positive and negative correlations, respectively. **P ≤ 0.01. (C) GI peak time in Col-0 and Lip-0 accessions (mean ± SD of two biological replicates). (D) QTL mapping in Col-0 × Lip-0 GI::LUC F2 progenies. QTLs were detected for GI peak time in two consecutive LDs of 16 h (LD1 and LD2), for GI peak time in the first day in darkness after the shift to constant conditions (dark), and for period length in constant darkness (DD). Dashed lines represent LOD thresholds. (E) Allelic effect of the TOG1 QTL in F2, F3, F4, and F5 progenies that were Col-0 or Lip-0 homozygous at TOG1. Seedlings were grown in LDs of 16 h. The populations are described in SI Appendix, Fig. S4A (mean ± SEM; n is indicated inside the bars; *P ≤ 0.05, **P ≤ 0.01 with a two-tailed Student t test). (F) Location of the TOG QTLs. Upward and downward arrows indicate that the Lip-0 allele advances or delays GI peak time, respectively.
Daily patterns of gene expression are controlled by endogenous and environmental inputs. Pathways that convey information from these internal or external signals could therefore contribute to the day length-dependent variation of GI peak time observed in the accessions. External light signals, on the one hand, directly influence the timing of GI expression in the evening because an extension of the light period after dusk in SDs is sufficient to cause an immediate delay in GI peak time (SI Appendix, Fig. S2A). The earlier onset of darkness in SDs might explain why GI expression and other rhythms are advanced in SDs compared with LDs (Fig. 1A) (9, 26, 36) and might also explain why natural variation of GI peak time is limited under SDs (Fig. 1A and SI Appendix, SI Discussion). Endogenous circadian rhythms, on the other hand, did not seem to be related to GI peak time variation in any of the photoperiods (SI Appendix, Fig. S2 B and C and SI Discussion). Circadian rhythms measured in LL do not correlate with phase in Arabidopsis accessions (4), and we report a similar trend for GI peak time and period length measured in DD, where circadian rhythms are not influenced by light (Fig. 1B). Although these results do not exclude that period length influences phase in particular accessions (SI Appendix, Fig. S2D), they do suggest that natural variation of GI peak time in LDs might generally be determined by natural alleles that regulate light signaling, rather than endogenous rhythms. Searching for such alleles was the goal of this study.
A cluster analysis identified Lipowiec (Lip-0) as belonging to a group of accessions that showed a late peak of GI::LUC expression under LDs (Fig. 1C and SI Appendix, Tables S3 and S4). Lip-0 GI::LUC was crossed to Columbia (Col-0), and extensive phenotyping of the Col-0 X Lip-0 F2 population in different photoperiods confirmed that maximum variability of GI peak time was observed in LDs of 16 h (SI Appendix, Fig. S3A). The F2 population (135 individuals) and subsequently selected F3, F4, and F5 families were used to detect and confirm four TIMING OF GI (TOG) quantitative trait loci (QTL) of moderate effect that precisely regulated the timing of GI expression during the LD 16-h cycle but had no significant effect on period length or on GI peak time in darkness (Fig. 1 D–F and SI Appendix, Figs. S3 and S4 and Table S5). The size and direction of the TOG effects were consistent with the phenotype of the Lip-0 parent. Allelic variation at the TOGs modified the timing of GI expression by ∼30 min, and the Lip-0 alleles of all TOGs except one (TOG2) delayed GI peak time (Fig. 1F and SI Appendix, Fig. S4). Together with the confirmation of the TOGs in near isogenic lines (NILs), these data collectively demonstrate the existence of natural alleles of moderate effect that precisely regulate the timing of GI expression within LD day/night cycles (SI Appendix, Fig. S5). Previous studies had reported natural variation of daily transcriptional rhythms in the range of hours (14, 17), but our experiments reveal that natural alleles can cause significant variation of a higher level of precision.
The Waveform of GI Expression Is Regulated by Light Signaling During LD Cycles.
The timing of GI expression is influenced by light signaling (SI Appendix, Fig. S2A), and the related mechanism might explain part of the GI peak time variation observed between accessions in LDs. Consistent with this idea, the gene encoding the red light photoreceptor PHYTOCHROME B (PHYB) was present in the TOG1 region and was a candidate for this QTL. The Lip-0 allele of PHYB contains a deletion in the N-terminal part of the protein that is associated with longer hypocotyls and reduced PHYB activity (37) (SI Appendix, Fig. S6). Light signaling was previously shown to regulate GI (38, 39), but how changes in PHYB activity could modify the timing of GI expression in LD day/night cycles was not known (35, 40).
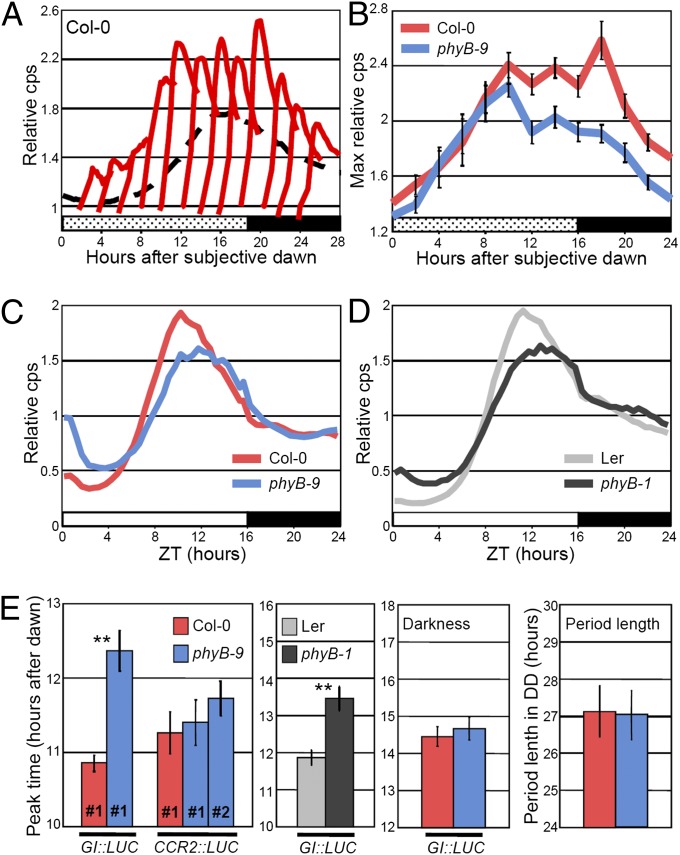
Detailed analysis of GI expression in phyB mutants revealed that PHYB activity shapes the GI waveform by mediating light signals that activate GI transcription in the evening of a LD (Fig. 2 and SI Appendix, Figs. S7–S9 and SI Discussion). On the basis of the results of a mathematical modeling study in which rapid responses to light were predicted to modulate the phase of circadian outputs (36), we first tested how GI expression responded to 30 min white or red light pulses applied in darkness after entrainment in LDs. The light pulses triggered an immediate response of GI::LUC that was maximal in the evening of the subjective day and that was significantly reduced in the phyB-9 mutant (Fig. 2 A and B and SI Appendix, Fig. S8 A and B). During LD cycles, substitution of white light by darkness in the evening suppressed the evening peak of GI::LUC, whereas substitution by red light was sufficient for full activation of GI (SI Appendix, Fig. S8C). Importantly, reduced activation of GI expression in phyB mutants was accompanied by a rightward shift of the GI waveform (negative skewness) and by a delay of GI peak time that was consistent with TOG1 Lip-0 delaying GI::LUC expression (Fig. 2 C–E and SI Appendix, Figs. S7 A, D, F and S8 D and E). This effect was specific to LDs of 16 h (SI Appendix, Figs. S7 B and C and S10) and had been reported for rhythms of cytosolic Ca2+ (36), but was not detected with a circadian marker that was not regulated by light (Fig. 2E). These results provide a mechanistic understanding of how light signaling shapes the waveform of GI expression in LDs and support a role for rapid responses to light in determining the phase of circadian outputs (36). The circadian clock is implicated in this mechanism not by modifying endogenous rhythms in DD or in LL but by gating (constraining) light activation of GI transcription in the evening (Fig. 2E and SI Appendix, Figs. S6 C and D and S7E and SI Discussion).
Fig. 2.
Light signaling defines the temporal waveform of GI expression during LDs. In all experiments, plants were entrained during 9 d in LDs of 16 h and GI::LUC expression was monitored on day 10 unless otherwise stated. (A and B) Col-0 and phyB-9 plants were entrained in LDs of 16 h, transferred to DD, and exposed to 30-min red light pulses of 60 µmol⋅m−2⋅s−1 applied every 2 h during the first subjective day. The response of GI::LUC expression to each pulse (A) is expressed relative to the expression level (cps, counts per second) measured before the pulse. The dashed line represents the nontreated control in DD (not to scale). Maximum relative luminescence after each pulse was plotted in (B). (C and D) Waveform of GI::LUC expression in (C) Col-0 and phyB-9 and in (D) Ler and phyB-1 in LDs of 16 h. (E) Peak time of GI::LUC expression in LDs of 16 h in Col-0 and phyB-9, of CCR2::LUC expression in Col-0, and in two independent phyB-9 transgenic lines, of GI::LUC expression in the Ler and phyB-1 background, of GI::LUC expression measured in the first day in DD and of period length measured in DD. Confirmation of these results with more transgenic lines is provided in SI Appendix, Fig. S7. Mean ± SEM; n = 12–24; *P ≤ 0.05, **P ≤ 0.01 with a two-tailed Student t test.
We then asked whether natural TOG alleles regulate GI through the same mechanism. Similar to the phyB-9 mutation, the less active TOG1 Lip-0 allele reduced GI::LUC evening expression levels in F2 progenies (SI Appendix, Fig. S11 A–C). TOG1 Lip-0 also reduced GI::LUC expression in segregating populations generated by crossing phyB-9 with two NILs that carried the TOG1 Col-0 or Lip-0 alleles (SI Appendix, Fig. S11C). The effects of the different allelic combinations obtained in these populations were consistent with PHYB being the gene underlying TOG1. We next combined Lip-0 alleles of TOG1-4 in the Col-0 background and created a population of 12 NILs that we used to more generally address how the TOGs were regulating GI (SI Appendix, Fig. S12A). Again similar to the analysis of phyB mutants, GI peak time significantly and negatively correlated with maximum GI expression levels in the NIL population grown in LDs of 16 h, and changes in peak time occurred at least partly independent of circadian rhythms in DD or in LL (SI Appendix, Fig. S12 B and C, Table S7, and SI Discussion). Thus, the detailed description of GI expression patterns in various populations supports a role for the TOGs in mediating a direct effect of light on the GI promoter through a mechanism that involves PHYB activity.
Precise Changes in the Waveform of GI Expression Are Sufficient to Alter a Downstream Phenotype.
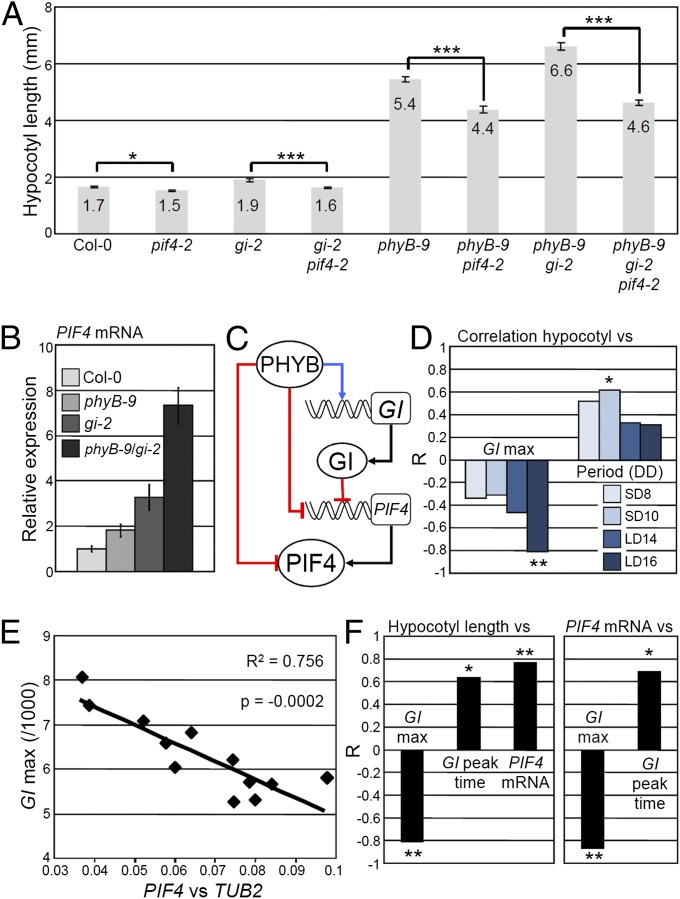
The TOGs cause precise changes in the daily pattern of GI expression, but it remained to be determined whether changes of such magnitude were biologically relevant and could affect overt phenotypes. We used hypocotyl growth as a phenotypic output of GI activity and tested whether alterations of this trait could be a result of precise changes in the GI expression waveform. A major advantage of using growth as a trait was that it can be precisely quantified in conditions directly comparable to the ones used for the GI::LUC activity assays. GI represses growth of the hypocotyl (27), but how GI function contributes to the molecular network that regulates growth in day/night conditions, and particularly in LDs, was not known.
GI acts in the hypocotyl growth repression pathway activated by PHYB (27), which, according to our results, could at least partially be explained by PHYB-mediated activation of GI expression. GI is also known to reduce mRNA levels of the transcription factor PHYTOCHROME INTERACTING FACTOR 4 (PIF4) during the night when PIF4 contributes to the promotion of growth (41–43). Loss of GI function in the Col-0 background also enhanced PIF4 expression in our conditions, and the long hypocotyl phenotype of gi-2 required PIF4 activity (Fig. 3 A and B). We further found that GI and PHYB act synergistically to inhibit growth and repress PIF4 during the night. Enhanced hypocotyl growth of phyB-9 gi-2 compared with phyB-9 required functionally active PIF4 and was associated with increased PIF4 expression levels (Fig. 3 A and B). As PHYB also promotes degradation of PIF4 at dawn (41), the synergy between GI and PHYB probably acts at both the transcriptional and posttranscriptional levels (Fig. 3C).
Fig. 3.
Precise changes in GI expression modify PIF4 expression and growth. (A) Hypocotyl length and (B) PIF4 mRNA levels at zeitgeber time (ZT) 20 h, quantified by quantitative RT-PCR (qRT-PCR) in LDs of 16 h in the indicated mutant backgrounds. (C) Working model for how PHYB and GI interact to regulate growth. Red lines indicate repression, blue lines activation, and black lines translation. Rectangles and circles represent genes and proteins, respectively. (D) Correlation of hypocotyl length of the NILs grown in LDs of 16 h with GI::LUC expression level at peak time (GI max) and period length in DD after entrainment in four photoperiods. (E) Correlation between GI::LUC expression level at peak time (GI max) and PIF4 mRNA levels quantified by qRT-PCR in the NILs entrained in LDs of 16 h and sampled at ZT 20 h. (F) Correlation among growth, GI::LUC expression level at peak time (GI max), GI peak time, and PIF4 mRNA levels at ZT 20 h. The Pearson correlation coefficient (R) indicates the strength of the correlations, with 1 and −1 indicating perfect positive and negative correlations, respectively. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001, with (A) a two-tailed Student t test or (D and F) the Pearson test. The correlations were also tested with the Spearman test and yielded similar results.
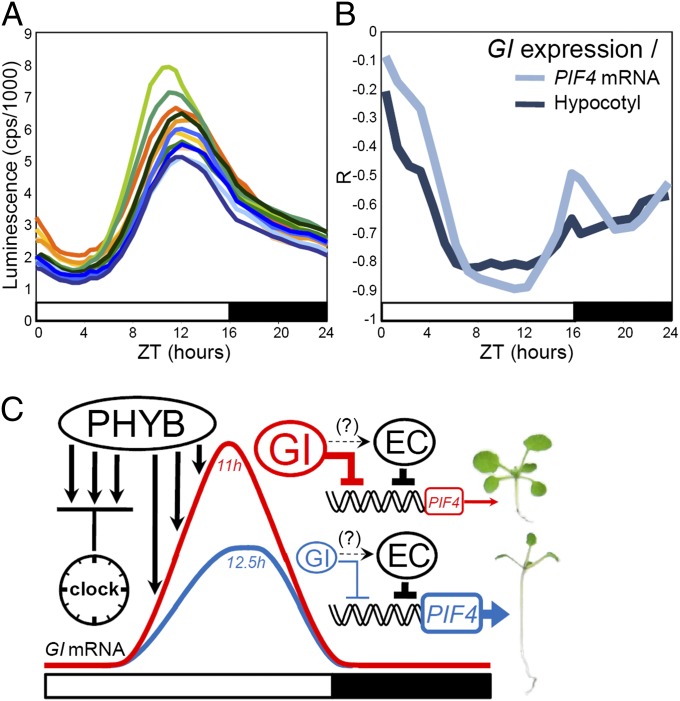
In the NILs, growth was affected through the same mechanism. GI::LUC expression levels and peak time, but not period length, strongly correlated with hypocotyl length and PIF4 mRNA levels measured in LDs of 16 h (Fig. 3 D–F). The correlations between GI expression levels and hypocotyl length or PIF4 mRNA were negative, which was consistent with GI being a repressor of growth. The data also confirmed the model for the regulation of GI transcription by light via the TOGs. If the TOGs regulate GI expression in the evening, phenotypic changes downstream of GI in the NILs should be induced by variations of GI expression at this time. As anticipated, the correlations between GI::LUC expression levels and PIF4 mRNA or growth were strongest during the second part of the day, which was also the time when the differences in GI::LUC activity between NILs were more pronounced (Fig. 4 A and B). Thus, natural TOG alleles regulate PIF4 expression and growth in LDs, at least partly by modifying the waveform of GI transcription in the evening and in a way that would be enhanced by changes in PHYB activity (Fig. 4C and SI Appendix, SI Discussion).
Fig. 4.
GI regulates growth by acting predominantly in the evening. (A) Waveforms of GI::LUC expression in the NILs in LDs of 16 h. (B) Pearson coefficient (R) of correlations between GI::LUC expression at different times of the day with hypocotyl length and PIF4 mRNA levels at ZT 20 h in the NILs. R indicates the strength of the correlations, with 1 and −1 indicating perfect positive and negative correlations, respectively. (C) A model for the regulation of GI expression by natural alleles during the day, and how this affects PIF4 expression and growth. Light signaling mediated by PHYB is repressed by the circadian clock in the morning of a long day. Clock repression is released later during the day, so that light activates GI expression until it reaches its peak in the evening. GI then contributes to the repression of PIF4 early in the night, so that growth is less efficiently promoted. Weak PHYB alleles cause less GI accumulation (blue line), more PIF4 transcription, and more growth. GI could hypothetically regulate PIF4 transcription by interacting with the EC, as represented by the dashed lines. Blue and red lines represent the GI waveform when influenced by weak or strong PHYB alleles, respectively. Numbers in italics indicate representative GI peak times.
These data additionally provide novel insights on the function of GI and on the growth regulation model. First, they show that circadian-gated expression of GI in the evening contributes to the temporal regulation of hypocotyl length. Second, they reveal how light signaling regulates PIF4 expression during day/night cycles. The underlying mechanism might involve coexpression and functional interactions of GI with components of the EVENING COMPLEX (EC), a protein complex that directly represses PIF4 (42–44) (Fig. 4C). Interestingly, we detected no significant relationship between GI expression and flowering in the NILs, despite an important function of GI being the promotion of flowering through the regulation of CONSTANS (CO) (45, 46). GI regulates diverse traits through distinct molecular pathways (47–49), and it is possible that these pathways are not all equally sensitive to precise changes in GI expression. GI-mediated promotion of flowering might be more robust than growth to small perturbations of GI expression and function, an idea supported by a previous study in which an induced mutation of GI altered growth but not GI-dependent promotion of CO (48). A similar scenario could explain why precise changes in GI expression do not alter flowering time in the NILs but do affect growth through the regulation of PIF4 transcription.
Conclusions and Perspectives
Collectively, our findings provide a paradigm for how natural alleles cause phenotypic diversity by precisely altering daily waveforms of gene expression. We also show that natural variation in temporal rhythms of expression during the day can be determined by changes in sensitivity to input signals, and not only by changes in circadian rhythms. The LD-specific mechanism of GI regulation we describe is part of a more general external coincidence model for the global control of phase in day/night conditions (36). The model predicts that the evening phase of processes dual-regulated by light and by the circadian clock adjusts to seasonal changes by responding predominantly to rapid light inputs. Natural alleles implicated in the perception of input signals that influence rhythms might explain why period length and phase generally do not correlate in Arabidopsis accessions. Such natural alleles could confer an advantage by optimizing the activity of temporally regulated processes while avoiding the pleiotropic effects associated with severe disruptions of the circadian system (5).
Rhythms of gene expression were analyzed within day/night cycles, at high temporal resolution, and in a population of natural accessions large enough to estimate the range of variation that exists at the intraspecies level. We then revealed the precision with which natural alleles modify daily expression patterns and demonstrated that these modifications have detectable effects on growth, a complex quantitative trait known to be under the control of many small effect loci (50, 51). Theoretical models predict that loci of small effect are a major source of phenotypic variation (52), but understanding how these loci modify phenotypes has been limited by the technical difficulties of their detection and validation. The exploitation of marker gene technology such as luciferase to identify alleles with small phenotypic effects may represent a general approach to uncovering such variation.
Methods
Plant Material and Growth Conditions.
The 77 accessions used in this work were a donation from Thomas Altmann, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), Gatersleben, Germany, and a subset of these accessions was previously described elsewhere (53). The phyB-9 and gi-2 mutant alleles also were described previously (54, 55). To obtain the GI::LUC transgene, a 2,513-bp fragment of the GI promoter was amplified, using primer 5′-attB1-accagcatatctctaatcag-3′ and primer 5′-attB2-accgaaactaaaccccaac-3′, and recombined with the pGWLuc vector (GeneBank: AM295157). The GI::LUC transgene was inserted into Arabidopsis by Agrobacterium-mediated transformation (56). Col-0 and phyB-9 GI::LUC lines 2 and 3 were obtained after transformation with a vector containing 2,755 bp of the GI promoter and were described elsewhere (35). Transgenic lines were made homozygous before use. The first 39 accessions were scored using at least two transgenic lines per accession. A significant contribution of the genotype (accessions) to variations of GI::LUC expression was detected in this data set (SI Appendix, Table S2). Only one line per accession was subsequently used to reduce the workload. Seedlings were grown in different photoperiods at 22 °C under 100 μmol⋅m−2⋅s−1 fluorescent white light (Philips TL741), or in a Percival growth chamber equipped with E-30LED for monochromatic light experiments (Percival).
Bioluminescence Imaging.
Plants were entrained for 9 d in different photoperiods and light conditions, and measurements were started on day 10. For experiments performed in white light, seedlings were transferred to 96-well black optiplates (Perkin-Elmer Life Sciences) containing Murashige and Skoog medium with 2% (mass/vol) sucrose and 20 μL d-luciferin (5 mM). Luminescence of individual seedlings was monitored in a TopCount Microplate Scintillation Counter (Perkin-Elmer) by manual feeding, which allowed the study of GI expression in white light conditions that contained the whole spectrum of wavelengths. In constant darkness (DD) conditions, feeding of the plates to the TopCount was automatic. LUC activity in monochromatic red light was monitored with a CCD camera in 24-well plates containing approximately 10 seedlings of the same genotype and supplied with 150 μL of 10 mM Luciferin. The resolution of the assays was 30 min. The images generated by the CCD camera were analyzed with Metamorph (Molecular Devices). Rhythms of GI::LUC expression during day/night cycles or in constant conditions were analyzed with BRASS (www.amillar.org).
QTL Detection and Statistical Analyses.
The QTLs were detected in 135 F2 progenies scored for GI::LUC expression and genotyped by Sequenom Inc. Linkage maps were created using JoinMap 4 (Kyazma B.V.), and QTL analysis was performed with MapQTL 5 (Kyazma B.V.), using the multiple QTL mapping (MQM) procedure. A thousand permutations were used to determine chromosome-specific logarithm of odds (LOD) thresholds. Markers used as cofactors were chosen by backward selection. More detailed information on the phenotyping and genotyping procedures and on the QTL validation in segregating populations and in the NILs is provided in SI Appendix, SI Methods. Hierarchical clustering was performed using Cluster version 3. The raw data were mean centered and normalized across the different day lengths. Clustering was performed with an uncentered correlation matrix and average linkage clustering. Self-organizing maps were generated before hierarchical clustering to determine the best orientation of the tree nodes. The resulting trees were displayed using Treeview version 3. All other statistical analyses were performed with SigmaStat version 3 or R.
Hypocotyl Measurements.
Hypocotyl length of the seedlings grown under different photoperiods and light conditions was measured after 9 days so the data would be directly comparable to the luciferase data. A high-resolution photograph of the seedlings was taken with a digital camera, and hypocotyl length was measured with Image J (National Institutes of Health). In all experiments, ∼20–30 seedlings per genotype were analyzed. Hypocotyl data for the NILs were obtained from five independent biological replicates performed in each of the four photoperiods tested (4,632 seedlings total).
Quantification of mRNA Expression.
RNA was isolated from 10-day-old seedlings, using the RNeasy Plant Mini kit (Qiagen), following the recommendations of the manufacturer. Genomic DNA was removed with the DNA-free kit (Ambion). For cDNA synthesis, 1 μg total RNA was primed using the oligodT primer and reverse transcribed with the SuperScript II kit (Invitrogen). The PCR mix was composed of 3 μL diluted cDNA and 7 μL iQ SYBR Green Supermix (Biorad). The thermocycles used for amplification were 3 min at 95 °C, followed by 40 cycles of 10 s at 95 °C, 10 s at 60 °C, and 10 s at 72 °C. TUB2 (At5g62690) and IPP2 (At3g02780) were used as housekeeping genes to normalize the expression data and yielded similar results. Primer sequences are provided in SI Appendix, Table S8.
Supplementary Material
Acknowledgments
We thank M. Koornneef, S. Davis, J. Jimenez Gomez, and F. Fornara for critical reading of the manuscript. We also thank E. de Ansorena for excellent technical assistance. This work was supported by a Marie Curie fellowship from the European Commission (A.d.M.) and by a core funding of the Max Planck Society (G.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1422242112/-/DCSupplemental.
References
- 1.de Montaigu A, Tóth R, Coupland G. Plant development goes like clockwork. Trends Genet. 2010;26(7):296–306. doi: 10.1016/j.tig.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Wang W, et al. Timing of plant immune responses by a central circadian regulator. Nature. 2011;470(7332):110–114. doi: 10.1038/nature09766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60:357–377. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 4.Michael TP, et al. Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science. 2003;302(5647):1049–1053. doi: 10.1126/science.1082971. [DOI] [PubMed] [Google Scholar]
- 5.Dodd AN, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309(5734):630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 6.Somers DE, Devlin PF, Kay SA. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. 1998;282(5393):1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- 7.Michael TP, Salome PA, McClung CR. Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proc Natl Acad Sci USA. 2003;100(11):6878–6883. doi: 10.1073/pnas.1131995100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salomé PA, To JP, Kieber JJ, McClung CR. Arabidopsis response regulators ARR3 and ARR4 play cytokinin-independent roles in the control of circadian period. Plant Cell. 2006;18(1):55–69. doi: 10.1105/tpc.105.037994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Millar AJ, Kay SA. Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc Natl Acad Sci USA. 1996;93(26):15491–15496. doi: 10.1073/pnas.93.26.15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fowler SG, Cook D, Thomashow MF. Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 2005;137(3):961–968. doi: 10.1104/pp.104.058354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li G, et al. Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat Cell Biol. 2011;13(5):616–622. doi: 10.1038/ncb2219. [DOI] [PubMed] [Google Scholar]
- 12.Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature. 2003;422(6933):719–722. doi: 10.1038/nature01549. [DOI] [PubMed] [Google Scholar]
- 13.Turner A, Beales J, Faure S, Dunford RP, Laurie DA. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005;310(5750):1031–1034. doi: 10.1126/science.1117619. [DOI] [PubMed] [Google Scholar]
- 14.Böhlenius H, et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312(5776):1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- 15.Izawa T, et al. Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell. 2011;23(5):1741–1755. doi: 10.1105/tpc.111.083238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kloosterman B, et al. Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature. 2013;495(7440):246–250. doi: 10.1038/nature11912. [DOI] [PubMed] [Google Scholar]
- 17.Slotte T, Holm K, McIntyre LM, Lagercrantz U, Lascoux M. Differential expression of genes important for adaptation in Capsella bursa-pastoris (Brassicaceae) Plant Physiol. 2007;145(1):160–173. doi: 10.1104/pp.107.102632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swarup K, et al. Natural allelic variation identifies new genes in the Arabidopsis circadian system. Plant J. 1999;20(1):67–77. doi: 10.1046/j.1365-313x.1999.00577.x. [DOI] [PubMed] [Google Scholar]
- 19.Edwards KD, Lynn JR, Gyula P, Nagy F, Millar AJ. Natural allelic variation in the temperature-compensation mechanisms of the Arabidopsis thaliana circadian clock. Genetics. 2005;170(1):387–400. doi: 10.1534/genetics.104.035238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boikoglou E, et al. Environmental memory from a circadian oscillator: The Arabidopsis thaliana clock differentially integrates perception of photic vs. thermal entrainment. Genetics. 2011;189(2):655–664. doi: 10.1534/genetics.111.131417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anwer MU, et al. Natural variation reveals that intracellular distribution of ELF3 protein is associated with function in the circadian clock. eLife. 2014;3:3. doi: 10.7554/eLife.02206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darrah C, et al. Analysis of phase of LUCIFERASE expression reveals novel circadian quantitative trait loci in Arabidopsis. Plant Physiol. 2006;140(4):1464–1474. doi: 10.1104/pp.105.074518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eimert K, Wang SM, Lue WI, Chen J. Monogenic Recessive Mutations Causing Both Late Floral Initiation and Excess Starch Accumulation in Arabidopsis. Plant Cell. 1995;7(10):1703–1712. doi: 10.1105/tpc.7.10.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurepa J, Smalle J, Van Montagu M, Inzé D. Oxidative stress tolerance and longevity in Arabidopsis: The late-flowering mutant gigantea is tolerant to paraquat. Plant J. 1998;14(6):759–764. doi: 10.1046/j.1365-313x.1998.00168.x. [DOI] [PubMed] [Google Scholar]
- 25.Park DH, et al. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science. 1999;285(5433):1579–1582. doi: 10.1126/science.285.5433.1579. [DOI] [PubMed] [Google Scholar]
- 26.Fowler S, et al. GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999;18(17):4679–4688. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huq E, Tepperman JM, Quail PH. GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc Natl Acad Sci USA. 2000;97(17):9789–9794. doi: 10.1073/pnas.170283997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dalchau N, et al. The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc Natl Acad Sci USA. 2011;108(12):5104–5109. doi: 10.1073/pnas.1015452108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim WY, et al. Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat Commun. 2013;4:1352. doi: 10.1038/ncomms2357. [DOI] [PubMed] [Google Scholar]
- 30.Dunford RP, Griffiths S, Christodoulou V, Laurie DA. Characterisation of a barley (Hordeum vulgare L.) homologue of the Arabidopsis flowering time regulator GIGANTEA. Theor Appl Genet. 2005;110(5):925–931. doi: 10.1007/s00122-004-1912-5. [DOI] [PubMed] [Google Scholar]
- 31.Zhao XY, Liu MS, Li JR, Guan CM, Zhang XS. The wheat TaGI1, involved in photoperiodic flowering, encodes an Arabidopsis GI ortholog. Plant Mol Biol. 2005;58(1):53–64. doi: 10.1007/s11103-005-4162-2. [DOI] [PubMed] [Google Scholar]
- 32.Hecht V, et al. Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol. 2007;144(2):648–661. doi: 10.1104/pp.107.096818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Onai K, et al. Large-scale screening of Arabidopsis circadian clock mutants by a high-throughput real-time bioluminescence monitoring system. Plant J. 2004;40(1):1–11. doi: 10.1111/j.1365-313X.2004.02191.x. [DOI] [PubMed] [Google Scholar]
- 34.Ding Z, Millar AJ, Davis AM, Davis SJ. TIME FOR COFFEE encodes a nuclear regulator in the Arabidopsis thaliana circadian clock. Plant Cell. 2007;19(5):1522–1536. doi: 10.1105/tpc.106.047241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palágyi A, et al. Functional analysis of amino-terminal domains of the photoreceptor phytochrome B. Plant Physiol. 2010;153(4):1834–1845. doi: 10.1104/pp.110.153031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dalchau N, et al. Correct biological timing in Arabidopsis requires multiple light-signaling pathways. Proc Natl Acad Sci USA. 2010;107(29):13171–13176. doi: 10.1073/pnas.1001429107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Filiault DL, et al. Amino acid polymorphisms in Arabidopsis phytochrome B cause differential responses to light. Proc Natl Acad Sci USA. 2008;105(8):3157–3162. doi: 10.1073/pnas.0712174105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Locke JC, et al. Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Syst Biol. 2005;1:2005.0013. doi: 10.1038/msb4100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paltiel J, Amin R, Gover A, Ori N, Samach A. Novel roles for GIGANTEA revealed under environmental conditions that modify its expression in Arabidopsis and Medicago truncatula. Planta. 2006;224(6):1255–1268. doi: 10.1007/s00425-006-0305-1. [DOI] [PubMed] [Google Scholar]
- 40.Salomé PA, et al. The out of phase 1 mutant defines a role for PHYB in circadian phase control in Arabidopsis. Plant Physiol. 2002;129(4):1674–1685. doi: 10.1104/pp.003418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nozue K, et al. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448(7151):358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- 42.Nusinow DA, et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475(7356):398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim Y, et al. GIGANTEA and EARLY FLOWERING 4 in Arabidopsis exhibit differential phase-specific genetic influences over a diurnal cycle. Mol Plant. 2012;5(3):678–687. doi: 10.1093/mp/sss005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim Y, et al. ELF4 regulates GIGANTEA chromatin access through subnuclear sequestration. Cell Reports. 2013;3(3):671–677. doi: 10.1016/j.celrep.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sawa M, Nusinow DA, Kay SA, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318(5848):261–265. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fornara F, et al. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell. 2009;17(1):75–86. doi: 10.1016/j.devcel.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 47.Mizoguchi T, et al. Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell. 2005;17(8):2255–2270. doi: 10.1105/tpc.105.033464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin-Tryon EL, Kreps JA, Harmer SL. GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol. 2007;143(1):473–486. doi: 10.1104/pp.106.088757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oliverio KA, et al. GIGANTEA regulates phytochrome A-mediated photomorphogenesis independently of its role in the circadian clock. Plant Physiol. 2007;144(1):495–502. doi: 10.1104/pp.107.097048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kroymann J, Mitchell-Olds T. Epistasis and balanced polymorphism influencing complex trait variation. Nature. 2005;435(7038):95–98. doi: 10.1038/nature03480. [DOI] [PubMed] [Google Scholar]
- 51.Joseph B, et al. Hierarchical nuclear and cytoplasmic genetic architectures for plant growth and defense within Arabidopsis. Plant Cell. 2013;25(6):1929–1945. doi: 10.1105/tpc.113.112615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rockman MV. The QTN program and the alleles that matter for evolution: All that’s gold does not glitter. Evolution. 2012;66(1):1–17. doi: 10.1111/j.1558-5646.2011.01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Narang RA, Bruene A, Altmann T. Analysis of phosphate acquisition efficiency in different Arabidopsis accessions. Plant Physiol. 2000;124(4):1786–1799. doi: 10.1104/pp.124.4.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rédei GP. Supervital Mutants of Arabidopsis. Genetics. 1962;47(4):443–460. doi: 10.1093/genetics/47.4.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell. 1993;5(2):147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.