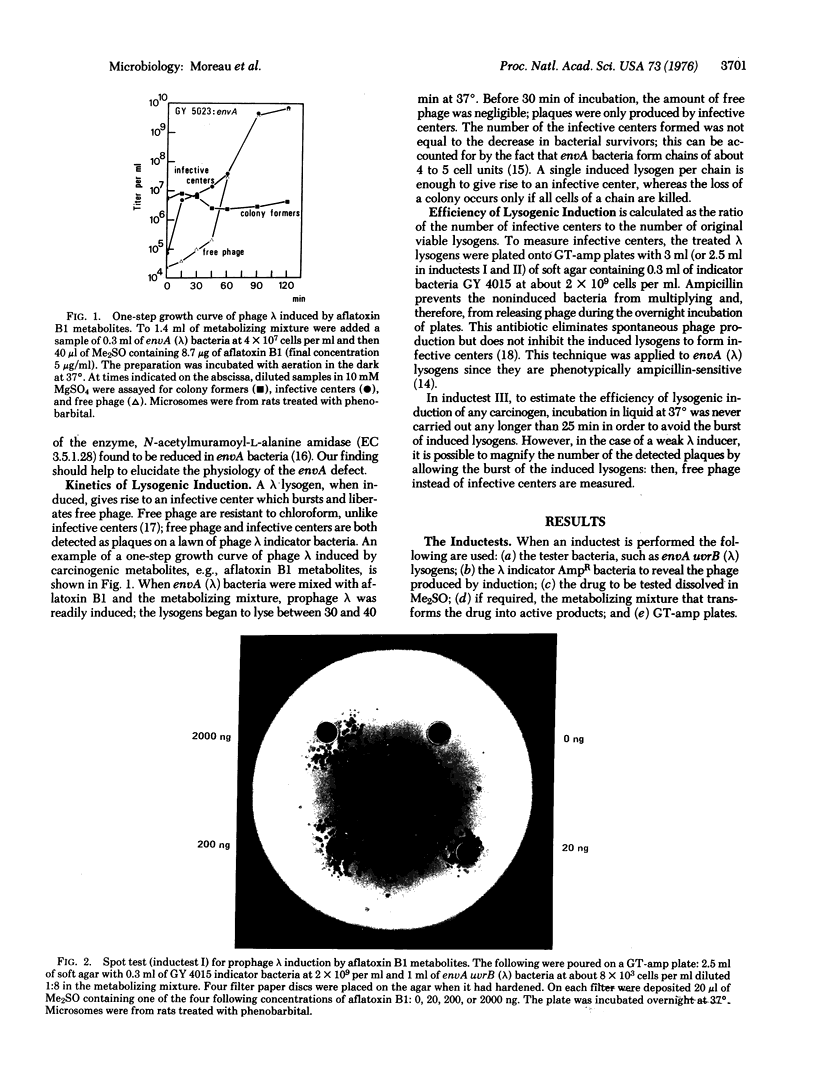
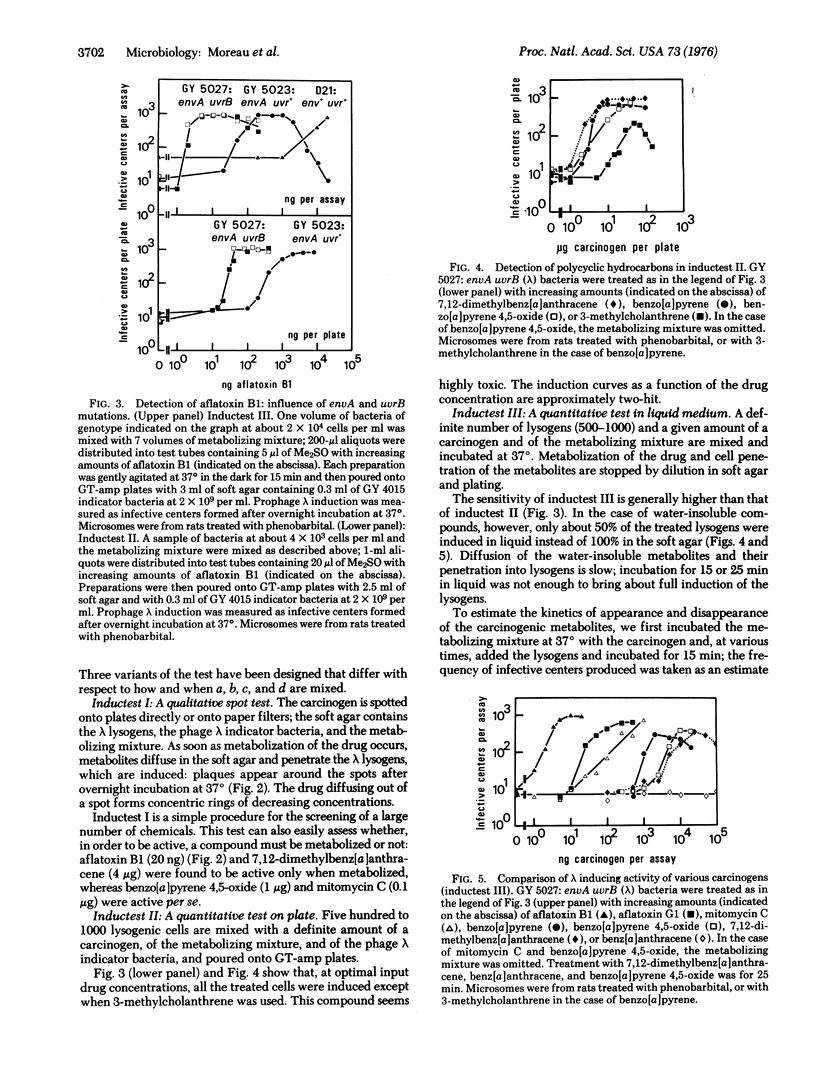
Abstract
A simple, inexpensive, and sensitive test for potential carcinogens based upon the property of carcinogens to induce prophage lambda is described. By using chemicals activated with microsomal enzymes and E. coli K12 permeable (envA) tester bacteria also deficient in DNA repair (uvrB), the range of carcinogens detected in a lysogenic induction test (inductest) has been extended. We have provided the evidence that, after activation, carcinogenic polycyclic hydrocarbons such as benzo[a5pyrene and 7,12-dimethylbenz[a]anthracene induce prophage lambda. Three variants of the test have been developed (inductests I, II, and III), which are as sensitive as the mutagenicity test of Ames et al. [Ames, B. N., McCann, J. and Yamasaki, E. (1975) Mutat. Res. 31, 347-364]. Inductests II and III provide a quantitative estimation of the inducing activity of a carcinogen. With the latter test, one can determine: (i) the cellular toxic effect of a carcinogen and (ii) the kinetics of appearance and disappearance of active metabolites. For two series of chemicals, aflatoxins and benz[a]anthracenes, there is a good correlation between their carcinogenic activity in rodents and their prophage inducing activity in bacteria. The fact that the majority of the cell population is induced makes it possible to test the inducing activity of carcinogens at the biochemical level, e.g., by measuring lambda repressor inactivation.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ames B. N., Mccann J., Yamasaki E. Methods for detecting carcinogens and mutagens with the Salmonella/mammalian-microsome mutagenicity test. Mutat Res. 1975 Dec;31(6):347–364. doi: 10.1016/0165-1161(75)90046-1. [DOI] [PubMed] [Google Scholar]
- Garner R. C., Miller E. C., Miller J. A. Liver microsomal metabolism of aflatoxin B 1 to a reactive derivative toxic to Salmonella typhimurium TA 1530. Cancer Res. 1972 Oct;32(10):2058–2066. [PubMed] [Google Scholar]
- Geissler E. Lysogenie und virale Kanzerogenese. Arch Geschwulstforsch. 1967;29(4):355–372. [PubMed] [Google Scholar]
- George J., Devoret R., Radman M. Indirect ultraviolet-reactivation of phage lambda. Proc Natl Acad Sci U S A. 1974 Jan;71(1):144–147. doi: 10.1073/pnas.71.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goze A., Sarsin A., Moulé Y., Devoret R. Induction and mutagenesis of prophage lambda in Escherichia coli K12 by metabolites of aflatoxin B1. Mutat Res. 1975 Apr;28(1):1–7. [PubMed] [Google Scholar]
- Gustafsson P., Nordström K., Normark S. Outer penetration barrier of Escherichia coli K-12: kinetics of the uptake of gentian violet by wild type and envelope mutants. J Bacteriol. 1973 Nov;116(2):893–900. doi: 10.1128/jb.116.2.893-900.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. Y., Kuntzman R., West S., Conney A. H. Reconstituted liver microsomal enzyme system that hydroxylates drugs, other foreign compounds and endogenous substrates. I. Determination of substrate specificity by the cytochrome P-450 and P-448 fractions. Biochem Biophys Res Commun. 1971 Mar 19;42(6):1200–1206. doi: 10.1016/0006-291x(71)90033-7. [DOI] [PubMed] [Google Scholar]
- Magee P. N. Activation and inactivation of chemical carcinogens and mutagens in the mammal. Essays Biochem. 1974;10:105–136. [PubMed] [Google Scholar]
- McCann J., Choi E., Yamasaki E., Ames B. N. Detection of carcinogens as mutagens in the Salmonella/microsome test: assay of 300 chemicals. Proc Natl Acad Sci U S A. 1975 Dec;72(12):5135–5139. doi: 10.1073/pnas.72.12.5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normark S., Boman H. G., Bloom G. D. Cell division in a chain-forming envA mutant of Escherichia coli K12. Fine structure of division sites and effects of EDTA, lysozyme and ampicillin. Acta Pathol Microbiol Scand B Microbiol Immunol. 1971;79(5):651–664. doi: 10.1111/j.1699-0463.1971.tb00093.x. [DOI] [PubMed] [Google Scholar]
- Normark S., Boman H. G., Matsson E. Mutant of Escherichia coli with anomalous cell division and ability to decrease episomally and chromosomally mediated resistance to ampicillin and several other antibiotics. J Bacteriol. 1969 Mar;97(3):1334–1342. doi: 10.1128/jb.97.3.1334-1342.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Tomizawa J. Absortive lysogenization of bacteriophage lambda b2 and residual immunity of non-lysogenic segregants. J Mol Biol. 1967 Jan 28;23(2):225–245. doi: 10.1016/s0022-2836(67)80030-5. [DOI] [PubMed] [Google Scholar]
- SECHAUD J., KELLENBERGER E. Lyse précoce, provoquée par le chloroforme, chez les bactéries infectées par du bactériophage. Ann Inst Pasteur (Paris) 1956 Jan;90(1):102–106. [PubMed] [Google Scholar]
- Witkin E. M. Thermal enhancement of ultraviolet mutability in a tif-1 uvrA derivative of Escherichia coli B-r: evidence that ultraviolet mutagenesis depends upon an inducible function. Proc Natl Acad Sci U S A. 1974 May;71(5):1930–1934. doi: 10.1073/pnas.71.5.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wogan G. N. Mycotoxins. Annu Rev Pharmacol. 1975;15:437–451. doi: 10.1146/annurev.pa.15.040175.002253. [DOI] [PubMed] [Google Scholar]
- Yang S. K., Selkirk J. K., Plotkin E. V., Gelboin H. V. Kinetic analysis of the metabolism of benzo(a)pyrene to phenols, dihydrodiols, and quinones by high-pressure chromatography compared to analysis by aryl hydrocarbon hydroxylase assay, and the effect of enzyme induction. Cancer Res. 1975 Dec;35(12):3642–3650. [PubMed] [Google Scholar]