Abstract
Mammalian CYP4B1 enzymes are cytochrome P450 monooxygenases that are responsible for the bioactivation of several exogenous pro-toxins including 4-ipomeanol (4-IPO). In contrast to the orthologous rabbit enzyme, we show here that native human CYP4B1 with a serine at position 427 is unable to bio-activate 4-IPO and does not cause cytotoxicity in HepG2 cells and primary human T-cells that overexpress these enzymes. We also demonstrate that a proline residue in the meander region at position 427 in human CYB4B1 and 422 in rabbit CYP4B1 is important for protein stability and rescues the 4-IPO bioactivation of the human enzyme, but is not essential for the catalytic activity of the rabbit CYP4B1 protein. Systematic substitution of native and p.S427P human CYP4B1 with peptide regions from the highly active rabbit enzyme reveals that 18 amino acids in the wild-type rabbit CYP4B1 protein are key for conferring high 4-IPO metabolizing activity. Introduction of 12 of the 18 amino acids that are also present at corresponding positions in other human CYP4 family members into the p.S427P human CYP4B1 protein results in a mutant human enzyme (P+12) that is as stable and as active as the rabbit wild-type CYP4B1 protein. These 12 mutations cluster in the predicted B–C loop through F-helix regions and reveal new amino acid regions important to P450 enzyme stability. Finally, by minimally re-engineering the human CYP4B1 enzyme for efficient activation of 4-IPO, we have developed a novel human suicide gene system that is a candidate for adoptive cellular therapies in humans.
Keywords: CYP4B1, 4-ipomeanol, Cytochrome P450, human, rabbit, suicide gene system
INTRODUCTION
Cytochrome P450 (CYP) proteins form a large superfamily of monooxygenases that can process a wide variety of endogenous and also exogenous substrates [1–6]. This superfamily of enzymes is systematically classified into subfamilies based on their protein homologies. In humans, eighteen families with 57 genes and 24 pseudogenes have been identified so far [4, 7]. While the different members of the CYP family 4 in mammals usually catalyze β-hydroxylase reactions towards fatty acids and alkyl hydrocarbons, the CYP family 4 subfamily B polypeptide 1 (CYP4B1) is the only CYP4 family member with significant activity towards structurally and chemically unrelated xenobiotics [8–10]. This unique substrate specificity in combination with the fact that the human CYP4B1 enzyme is a predominantly extrahepatic protein with 70% of transcripts expressed in lung tissue (and low levels in heart, skeletal musculature and kidney) [8, 9, 11–13] attracted the attention of oncologists [14] who intended to specifically target an alkylator chemotherapy for a specific solid cancer to the originating organ, the lung.
This chemotherapeutic approach was based on the discovery that the high toxicity associated with feeding mold-affected sweet potatoes (Ipomoea batatas) to animals [15] was mainly due to the presence of the phytoalexin, 4-ipomeanol (4-IPO). The predominantly lung-toxic furan is produced as a defense mechanism by sweet potatoes that were infected with the common mold Fusarium javanicum [16, 17]. Animal experiments subsequently revealed that numerous mammals, including cows, dogs, rats, rabbits and mice, die of acute pulmonary toxicity after administration of 4-IPO, while significant histological lesions or toxicities in other organ systems (except in the kidneys in male mice) were not detected [14, 18, 19]. The pulmonary toxicity was due to highly tissue-selective activation of 4-IPO by pulmonary microsomes of bronchiolar Clara cells and, to a lesser degree, in type II pneumocytes [14]. In 1993, Verschoyle et al. demonstrated that pulmonary toxicity of 4-IPO in rats was attenuated by co-administration of a chemical inhibitor of CYP4B1, which is the predominant cytochrome P450 enzyme in lung tissues in the different species [12]. Finally, in 2005 Baer et al. showed that purified recombinant rabbit CYP4B1 efficiently catalyzed the bioactivation of 4-IPO to generate a reactive intermediate that could be trapped by N-acetylcysteine and N-acetyllysine [20]. By transferring an oxygen atom to the furan ring, the CYP4B1 enzyme generates a highly toxic alkylating metabolite which causes DNA-protein cross-links and DNA strand breaks and thus triggers an apoptosis-mediated cell death [8].
Based on sensitivity data in human non-small cell lung cancer cell lines [21], 4-IPO was thought to be an ideal agent for lung-targeted chemotherapy regimens, as it would be activated predominantly in human lung tissue due to the tissue-specific CYP4B1 enzyme expression profile [14]. Remarkably, three phase I/II studies in humans with 4-IPO showed no significant activity/toxicity in the lungs and revealed an absence of any anti-tumor effects in lung or liver tumors [22–24]. Biochemical characterization of CYP4B1 enzymes from different species provided the scientific explanation for the failed human trials [11, 25], which originates in a large species difference in CYP4B1 activity between the human and experimental animal forms of the enzyme. Although the human protein has a high sequence similarity of 84% to the highly active rabbit CYP4B1 protein (Figure S1), human CYP4B1 is the only enzyme which has a serine at position 427 in the meander region. All other tested CYP4B1 enzymes – including those from rabbit, gorilla and chimpanzee – and also all other human P450 proteins have a proline at the corresponding position (Figure S2). This serine 427 is specific to human CYP4B1 and is thought to render the enzyme incapable of processing 4-IPO [26]. However, conflicting data showed that the human native CYP4B1 is active when expressed in liver cells in a transgenic mouse model [27].
Based on the idea that the endogenous human CYP4B1 enzyme is enzymatically inactive, Rainov et al. suggested exploitation of 4-IPO in combination with the very active rabbit CYP4B1 enzyme as a suicide gene system for the treatment of brain tumors [28]. To this end, the rabbit CYP4B1 cDNA was fused 3’ to EGFP and retrovirally expressed in human and rodent glioma cell lines. Incubation of the transgene expressing cells with 4-IPO resulted in highly efficient tumor cell killing in vitro. For in vivo studies, the authors established epidural tumors in Fisher 344 rats with syngenic rat 9L glioma cells and used a recombinant HSV amplicon with the CYP4B1-EGFP fusion protein under the control of a CMV promoter to successfully target these tumors [28]. However, the authors did not report the use of 4-IPO in these animals in vivo, presumably in order to avoid the acute lung toxicities of 4-IPO administration to rodents.
In the present study, in order to develop a novel suicide gene system based on 4-IPO with a human enzyme, we first systematically characterized the activity of human and rabbit wild-type and mutant CYP4B1 proteins, thus demonstrating that several distinct amino acids in both enzymes determine the activity towards 4-IPO. Based on this data, we then re-engineered the endogenous human CYP4B1 protein for maximal 4-IPO metabolizing activity, while introducing as few amino acid changes as possible in order to avoid immunogenicity. The final activity-optimized human CYP4B1 protein carries the proline at position 427 as well as 12 single amino acid substitutions that are also present in other human CYP family 4 members at corresponding positions. This mutant protein is an expression-optimized enzyme that, when introduced into human liver cells and in primary human T-cells, is as active as the native rabbit enzyme for inducing 4-IPO-dependent cell death.
EXPERIMENTAL
Plasmid construction
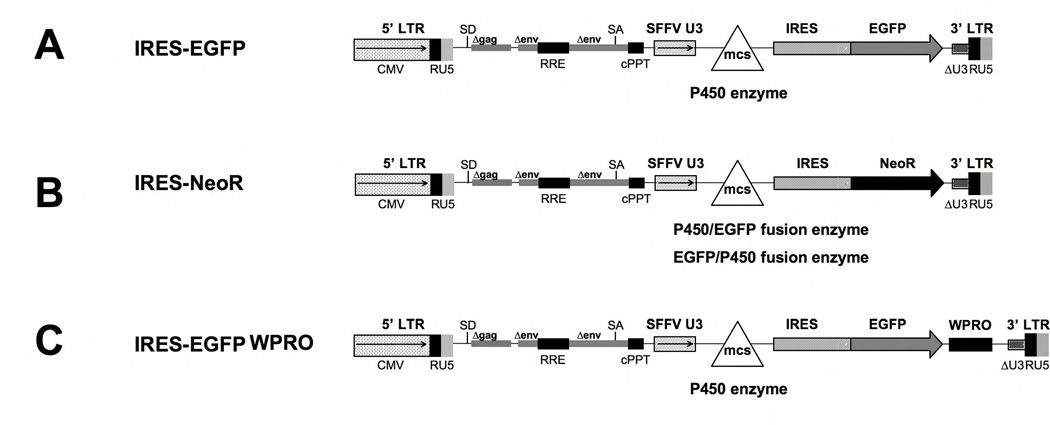
The lentiviral vectors (Figure 1) contain a viral spleen focus forming (SFFV) promoter driving both P450 isoforms and the marker EGFP or neomycin phosphotransferase npt II (NEO) genes separated by a ribosome entry site (IRES). For transient virus production the expression of the packaging RNA is driven by the CMV promoter. All vectors are derivatives of the puc2CL6 vector [29] and were produced by using standard cloning techniques. The codon-optimized cDNA from human CYP4B1 (native and P427 mutation) were created by GeneArt according to their unpublished algorithms (Regensburg, Germany). The P450/EGFP (3’-EGFP) and EGFP/P450 (5’-EGFP) fusion enzymes were constructed after deletion of the stop codons of the 5’ cDNAs.
Figure 1. Schematic outline of the lentiviral vectors used.
CMV: CMV promoter; SD: splice donor; LTR: long terminal repeat; SA: splice acceptor; RRE: Rev responsive element, cPPT: central polypurine binding tract; SFFV U3: U3 promoter of the spleen focus forming virus; mcs: multicloning site; IRES: internal ribosomal entry site; EGFP: enhanced green fluorescent protein; neoR: neomycin phosphotransferase (nptII) resistance cDNA; WPRO: woodchuck hepatitis virus post-transcriptional regulatory element optimized.
Cell culture
Human embryonic kidney cells (HEK293T, ATCC, Manassas, VA, U.S.A.) and human liver cancer cells (HepG2, ATCC) were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco, Karlsruhe, Germany) with 4.5 mM glucose and 2 mM GlutaMAX supplemented with 10% fetal bovine serum (FBS, PAA), 100 U/ml penicillin and 100 µg/ml streptomycin (GIBCO).
Primary human T-lymphocytes were collected from peripheral blood of healthy adult volunteers and separated by Ficoll-Paque (GE Healthcare, Freiburg, Germany) centrifugation. All studies were approved by the local ethics committee. The T-cells were stimulated with CD3 (OKT3, Ortho Biotech, Neuss, Germany) and CD28 (BD Bioscienses Pharmingen, San Diego, CA, USA) in combination with IL-2 (Chiron, Marburg, Germany) and were grown in Isocove´s Modified Dulbecco´s Medium (IMDM; Sigma-Aldrich, Taufkirchen, Germany) with the same additives as DMEM.
Production of lentiviral vectors
HEK 293T cells were transfected using polyethyleneimine transfection reagent (Sigma-Aldrich, Deisenhofen, Germany) with 6 µg of an HIV1 helper plasmid expression construct for HIV1 gag/pol/rev (pCD/NL-BH), kindly obtained from Jakob Reiser, New Orleans, U.S.A., 6 µg of the envelope vector (pczVSV-G) kindly obtained from Dirk Lindemann, Dresden, Germany, and 6 µg of the vector plasmids, as described [29]. Viral supernatants were harvested 48 hours after transfection, filtered through a 0.45-µm filter (Sartorius AG, Germany) and then used to transduce the HepG2 and primary T cells. After 24 hours the viral supernatant was replaced with fresh medium. Human T lymphocytes were transduced on the fibronectin fragment CH296 (Takara Bio Inc., Otsu, Japan) with 100 U/ml IL-2 similarly as described [30–32].
Cell proliferation assays
In order to investigate the cytotoxic effects of 4-IPO, transduced HepG2 (primary T) cells were plated in 12- (24-) well plates at a density of 2 × 105 (1 × 105) cells/well for day 1, 1 × 105 (0.5 × 105) cells/well for day 2 and 0.5 × 105 (0.25 × 105) cells/well for day 3. The cells were treated with varying concentrations of 4-IPO (range: 0.5–5 µg/ml; provided by the National Cancer Institute, Drug Synthesis and Chemistry Branch, Bethesda, MD). After 24, 48 and 72 hours, the percent of surviving cells was determined with a FACScan (Becton Dickinson, Heidelberg). To this end, the cells were pelleted and stained with propidium iodide (PI; 50 µg/ml; Sigma). Flow cytometric data on EGFP and PI fluorescence were analyzed in parallel to discriminate between living, dead, transduced and non-transduced cells. The number of live cells were counted and calculated relative to the untreated controls (=100%), respectively. Data are expressed as the mean ± SEM (standard error of the mean). Statistical analysis was performed using the R version 3.1.1 software by the R Foundation for Statistical Computing (Vienna, Austria) [33]. We limited the statistical analysis of the cell survival data (Figures 2, 3A, 4A, 5A–C, 6B, 8 and S3) to the 90 µM 4-IPO concentration, as this was the plasma concentration of 4-IPO that had been achieved without major toxicity in vivo in humans in the phase I/II dose escalation studies [22–24]. A one-way ANOVA was calculated for survival rates followed by a posthoc pairwise comparison using Tukey's Honest Significant Difference (Tukey HSD) method (Figures S4–23). In the ANOVA blots, statistical differences between two survival curves exist, if the confidence interval is located to the left or right of the vertical line through 0. For the protein half-life experiments, the predicted values for the decrease of the MFIs (Figure 3C) were calculated by linear regression (stats package). Confidence intervals around predicted values were obtained via the 'geom_smooth' function (ggplot2 package) of the R software [34] which then calculates the values by using the 'predict' function in the stats package.
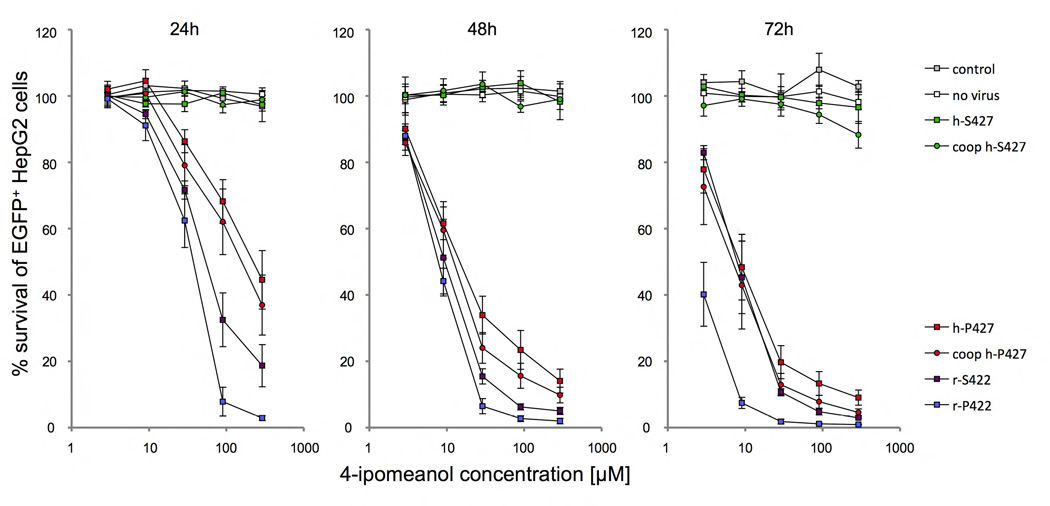
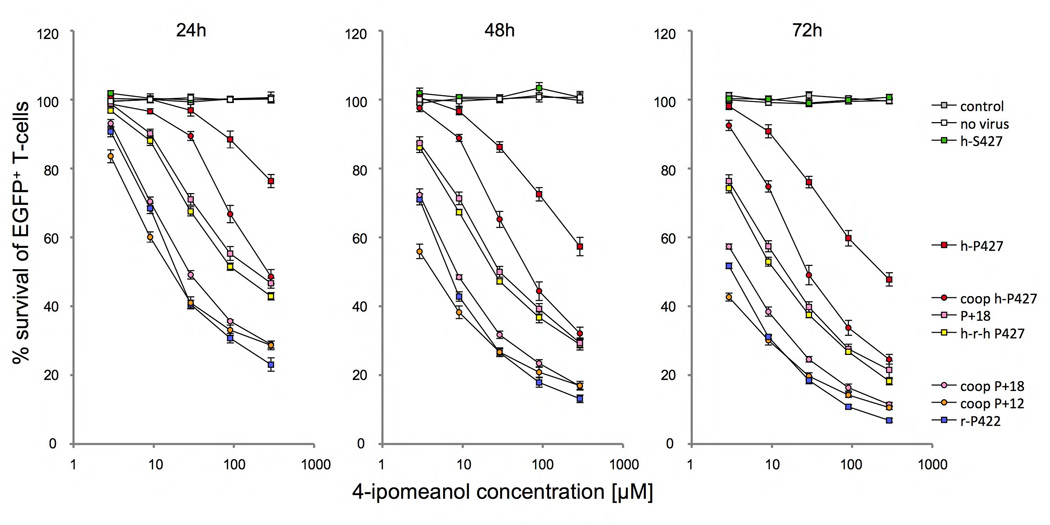
Figure 2. Toxicity of HepG2 cells after 4-IPO exposure.
Survival of HepG2 cells after 24, 48 and 72h of incubation with increasing doses of 2.9, 9, 29, 90 and 290 µM 4-IPO, transduced with S → P (h-P427) amino acid substitution in the human wild-type (h-S427) and P → S (r-S422) in the wild-type rabbit (r-P422) CYP4B1 enzymes. For each construct, the mean ± SEM is shown from at least three experiments. The transduction efficiency in HepG2 (data not shown) was ≥90%, as determined by EGFP expression in flow cytometry analysis. Shown is also the impact of mRNA codon optimization (coop) on enzyme activities of h-S427 and h-P427 in HepG2 cells.
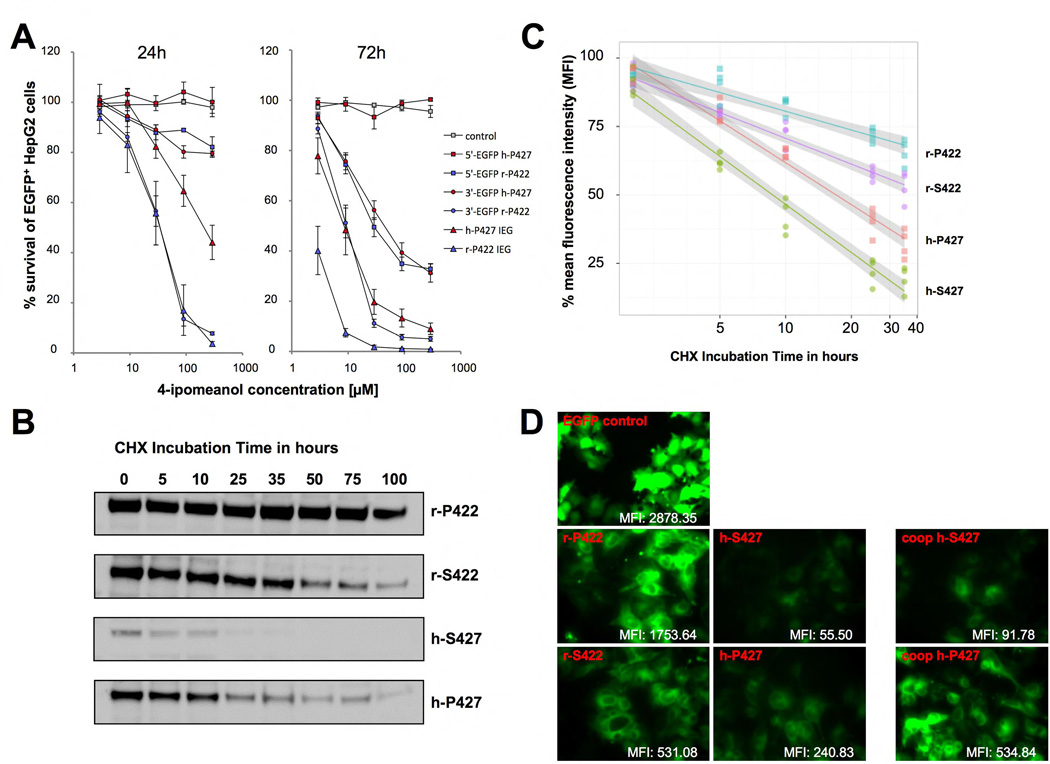
Figure 3. Determining the protein stability of the human and the rabbit CYP4B1 enzymes.
(A) Activity of CYP4B1/EGFP (3’-EGFP) and EGFP/CYP4B1 (5’-EGFP) fusion enzymes for 4-IPO. The IRES-EGFP constructs were used as reference for the activity of the non-fused enzymes. Shown is the % survival of EGFP+ HepG2 cells after incubation for 24 and 72 hours with increasing doses of 2.9, 9, 29, 90 and 290 µM 4-IPO. For each construct, the mean ± SEM is shown from three independent experiments. (B,C) CYP4B1 stability assessed by Western blotting and by flow cytometry. The studies on protein expression and stability were performed with the rabbit and the human CYP4B1/EGFP (3’-EGFP) fusion construct in the LV vector with the additional IRES-neomycin cassette. Protein half-life analysis was performed by adding of 50 µg/ml cycloheximide to the transduced and G418-selected HepG2 cells. Samples were collected at the indicated time points. (B) Western Blot analysis. Samples were lysed and equal numbers of cells analyzed by Western blot. The blots were taken with identical exposure time. The detection of the different CYP4B1 constructs was performed with anti-EGFP antibody. A β-actin antibody was used as loading control (not shown). (C) Flow cytometry analysis. The mean fluorescence intensity (MFI) of cells was assessed at various time points by FACS. Linear regression of the decrease in the MFI over time is blotted and the confidence intervals for each construct are shown in gray. (D) Microscope images. Each microscope image was performed with the same setting and exposure time. The MFI levels were determined by FACS.
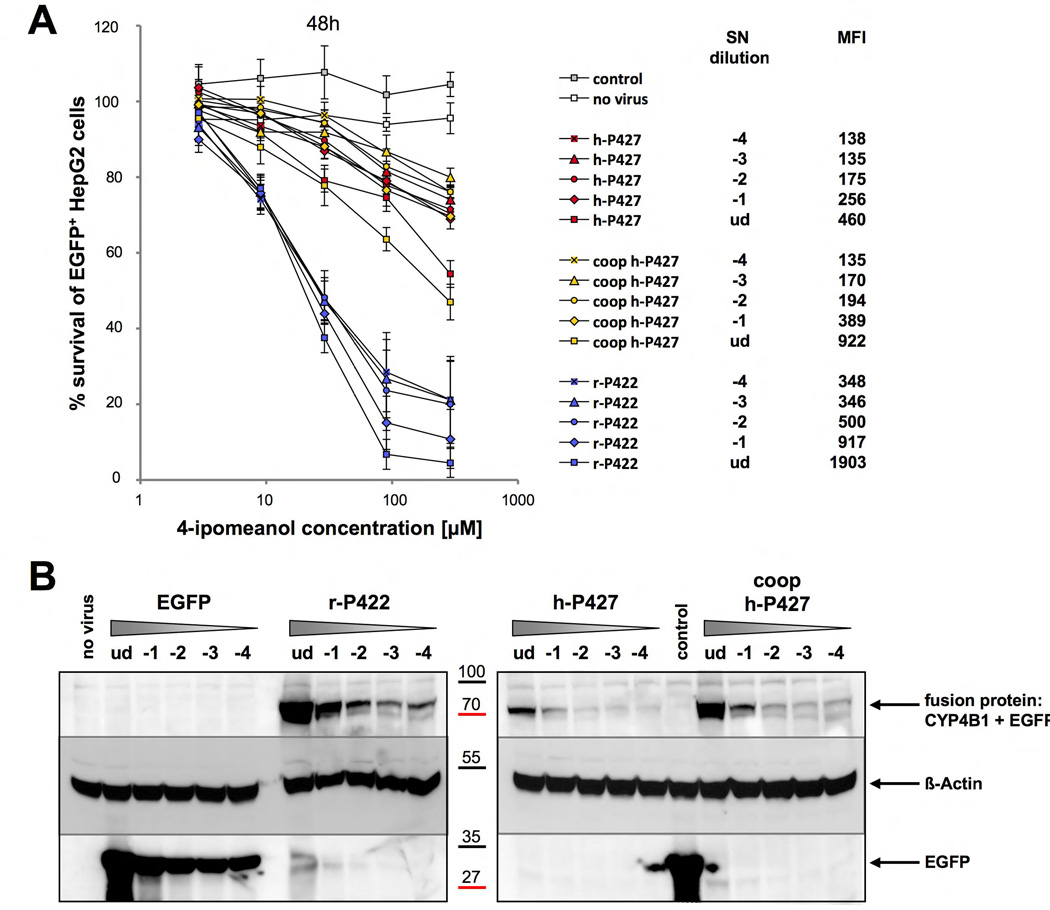
Figure 4. Impact of the number of viral integration on the toxification potential.
The CYP450 constructs are created as 3'-EGFP fusions with additional IRES-neoR cassette. (A) Shown is the survival of EGFP+ HepG2 cells in percent after two days of incubation with 2.9, 9, 29, 90 and 290 µM 4-IPO. For each construct the mean ± SEM is shown from three experiments. (B) Western blot analysis of transduced HepG2 cells. Detection of the fusion proteins was performed with anti-EGFP antibody. ud: undiluted, −1: virus dilution 10−1; −2 virus dilution 10−2; −3: 10−3 virus dilution; −4: virus dilution 10−4; coop: codon-optimized.
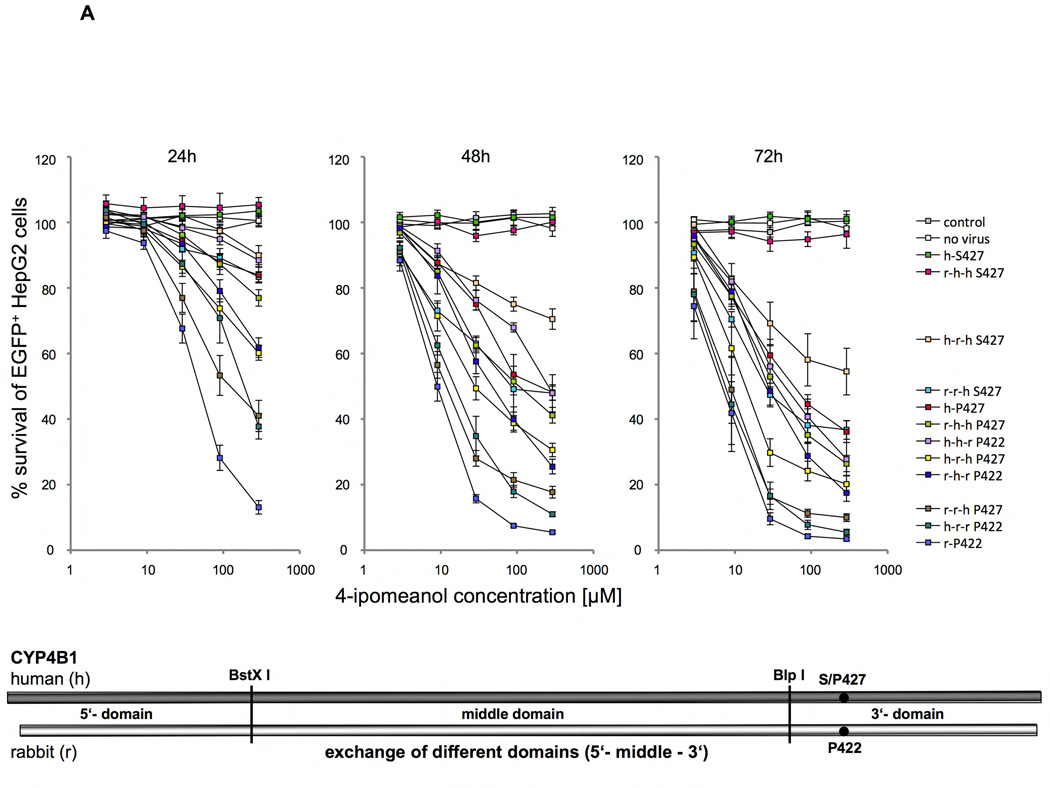
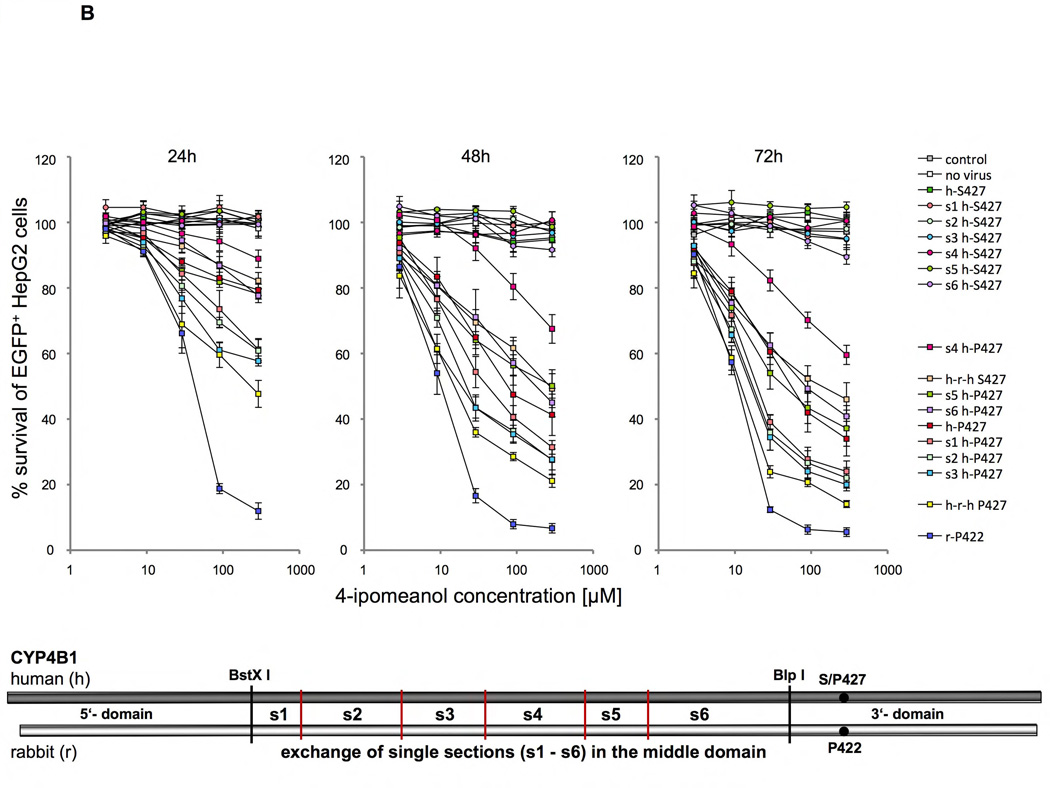
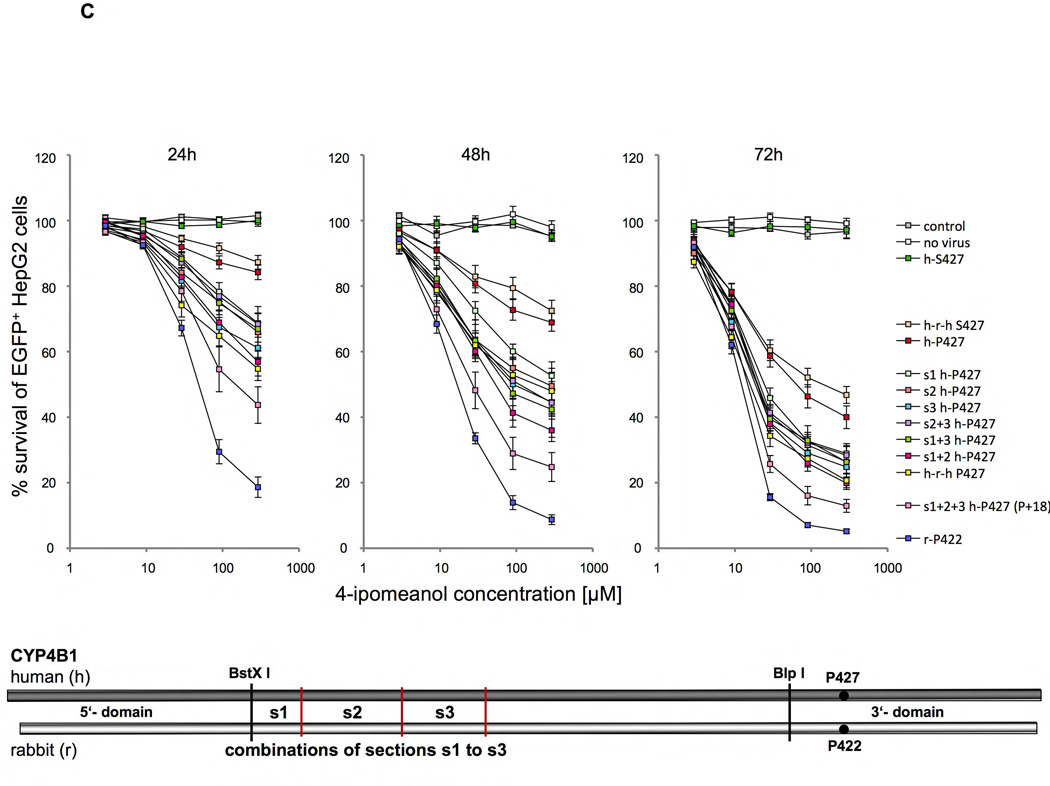
Figure 5. Identifying amino acid determinants in the human and rabbit CYP4B1 enzymes critical for the activity to process 4-IPO.
Survival of transduced EGFP+ HepG2 cells expressing different P450/EGFP (3’-EGFP) fusion enzymes in percent after 24, 48 and 72 hours of incubation with 2.9, 9, 29, 90 and 290 µM 4-IPO. For each construct the mean ± SEM is shown from at least three experiments. Schematics drawing in each figure A–C visualize the substitutions made in the human wild-type (h-S427), the human mutant (h-P427) and the wild-type rabbit (r-P422) CYP4B1 enzymes. (A) Domain exchanges (5’, middle, 3’) between the human wild-type (h-S427) and mutant (h-P427) CYP4B1 enzymes and the wild-type rabbit (r-P422) CYP4B1 protein and the influences on enzyme activity in transduced HepG2 cells. The human domains are depicted by h and the rabbit domains by r. (B) Exchange of six sections (s1 to s6) in the middle domain between the human wild-type (h-S427) and mutant (h-P427) and the wild-type rabbit (r-P422) CYP4B1 proteins and the influence on enzyme activity in transduced HepG2 cells. (C) Effects of the combinations of sections s1+2, s1+3, s2+3 and s1+2+3 on the activity of the human P427 proteins towards 4-IPO when stably expressed in HepG2 cells.
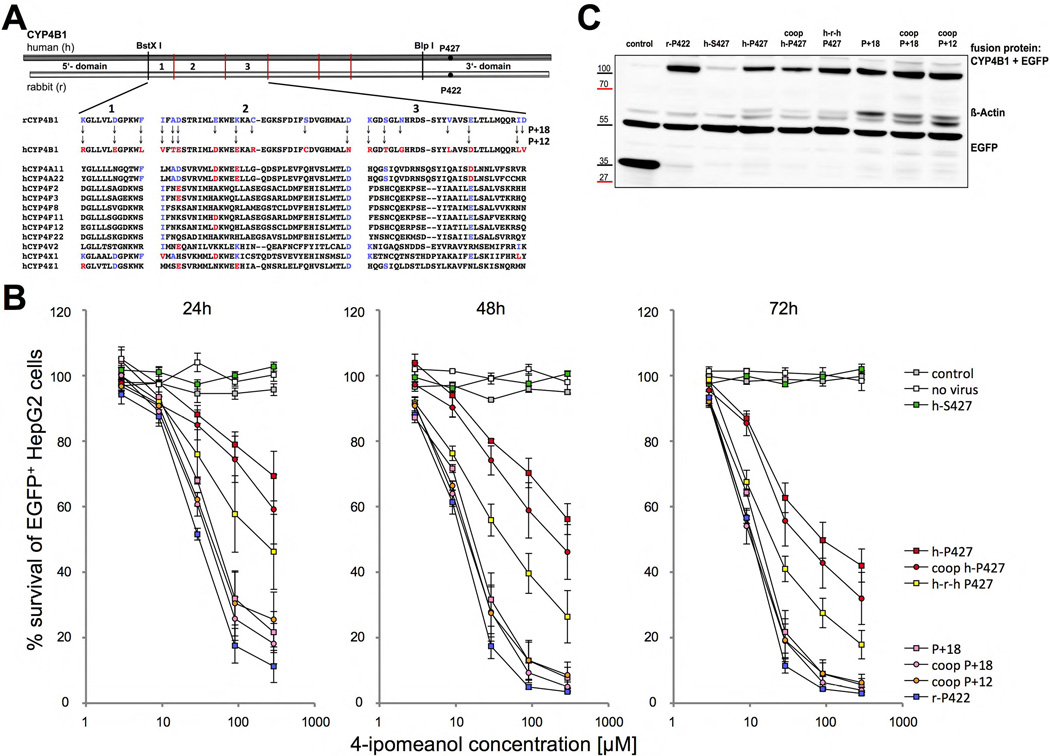
Figure 6. Activity of the human CYP4B1 enzymes with 13 (P+12) and 19 (P+18) amino acid exchanges in HepG2 cells towards 4-IPO exposure.
(A) Multiple sequence alignments of corresponding sections s1–s3 of other human CYP4 enzymes with the wild-type human and rabbit CYP4B1 proteins. In the P+18 CYP4B1 protein, 18 amino acids present in the wild-type rabbit enzyme in sections s1–s3 were introduced into the h-P427 CYP4B1. The P+12 h-P427 CYP4B1 carries 12 amino acid substitutions present in sections s1–s3 in the rabbit enzyme but is devoid of the six amino acids that are uniquely present in the rabbit enzyme and not in other human CYP4 family members at corresponding positions. (B) Influence of different P450/EGFP fusion enzymes on the survival of EGFP+ HepG2 cells (in %) after 24, 48 and 72 hours of incubation with increasing doses of 4-IPO. For each construct the mean ± SEM is shown from at least three experiments. (C) Representative western blot on HepG2 cells expressing the different CYP4B1/EGFP fusion proteins. The fusion proteins were detected with an anti-EGFP antibody. Actin detected by an anti-β-actin antibody was utilized to ensure equal protein loading.
Figure 8. Survival of primary human T-cells expressing the re-engineered P+12 and P+18 CYP4B1 proteins in 4-IPO.
Survival of EGFP+ primary T cells (in %) after 24, 48 and 72 hours of incubation with 2.9, 9, 29, 90 and 290 µM 4-IPO assessed by flow cytometry. For the experiments shown here, co-expression of the CYP4B1 enzymes and EGFP was achieved with the lentiviral vector depicted in Figure 1C. For each construct, the mean ± SEM is shown from at least six experiments. coop: codon-optimized.
Imaging
HepG2 cells that had been transduced with vectors expressing P450/3'-EGFP fusions enzymes with an additional IRES-NEO cassette were selected with G418 and stable cells examined microscopically with an inverted microscope (Axiovert 200M, Zeiss, Objective 40×). The fluorescent images of the different fusion constructs were taken with a digital camera (AxioCam MRm, Zeiss) using the same setting and exposure time for all probes. The determination of the mean fluorescence intensity (MFI) was performed by FACS.
Determination of Protein Stability
After transduction with the lentiviral P450/EGFP (3’-EGFP) fusion constructs, HepG2 cells were exposed to 50 µg/ml cycloheximide (CHX, Sigma-Aldrich) in growth medium. After different incubation times, cells were trypsinized and either fixed with 4% paraformaldehyde solution and directly measured by FACS or lysed directly for subsequent Western blot analysis with anti-EGFP. Microsoft Excel was used for a semi-logarithmic blot to determine the protein half-lives.
Western blot analysis
We performed immunoblots with samples of whole cell lysates on 4–12% NuPage Bis-Tris polyacrylamide gels (Invitrogen). Membranes were probed with mouse monoclonal anti-EGFP (1:20000, 632375 Clontech) or mouse monoclonal anti–β-actin (1:5000; A2228 Sigma-Aldrich). Secondary horseradish peroxidase–linked sheep anti-mouse IgG (RPN4201 GE Healthcare) at a dilution of 1:10000 was used to detect the primary antibodies by chemiluminescence using the ECL system (Pierce, Thermo Fisher Scientific). The chemiluminescence signal was digitally recorded using a LAS-3000 imager (Fuji Film).
Modeling of human CYP4B1
Structural modeling of human CYP4B1 was performed on the Robetta server [35, 36] using the 1.95 Å resolution crystal structure of human microsomal P450 1A2 (pdb id: 2HI4) [37] as a template. The sequence identity between human CYP4B1 and human CYP1A2 is ~21%. 5,000 models were generated followed by model clustering. The best CYP4B1 models were chosen as centers of the largest clusters, defined as having the lowest standard mean deviation value (between corresponding positions of C-α atoms of all residues) in comparison to all other models in a cluster.
RESULTS
Expression of the CYP4B1 proteins in human cells
In order to initially characterize the activities of the native human and rabbit CYP4B1 proteins for 4-IPO-induced cytotoxicity, both cDNAs were cloned into our lentiviral (LV) HIV1-based expression vector [29] with an IRES-EGFP expression cassette (Figure 1A). We also mutated the serine at position 427 in the human protein to proline (h-P427) that is present at this position in all CYP4B1 proteins of different species (Figure S2). The proline to serine exchange of the human CYP4B1 was introduced into the native rabbit CYP4B1 enzyme (r-P422) at the corresponding position 422 (r-S422) (Figure S1) because Zheng et al. had reported that the P422S exchange in the rabbit CYP4B1 enzyme expressed in insect cells results in loss of the P450 chromophore and the rabbit S422 mutant thereby is unable to metabolize lauric acid, a hallmark of CYP4B1 and other CYP4 proteins [26]. Subsequently, LV particles expressing the two rabbit and the two human cDNAs were produced and used to transduce the human liver HepG2 cell line.
Stably expressing cells were challenged with increasing concentrations of 4-IPO and the survival of EGFP+ cells was analyzed after 24, 48 and 72 hours by flow cytometry using propidium iodide for live/dead cell discrimination. The results demonstrated (Figure 2 plus statistical analysis in Figure S4) that the native human CYP4B1 S427 protein did not cause cytotoxicity, even at the highest concentration of 4-IPO used (290 µM), thus explaining the failed human clinical trials with 4-IPO. Although the S427P exchange in the human protein rendered this enzyme capable of processing 4-IPO, the activity of the native rabbit CYP4B1 protein (with P422) at 90 µM still was significantly superior to the human mutant protein after 24h and 48h. While the introduction of serine at position 422 into the rabbit CYP4B1 caused a decrease in sensitivity of cells to 4-IPO compared to the native rabbit protein, interestingly, the mutant rabbit enzyme still induced cell death in the HepG2 cells at 90 µM far more effectively than the mutant human P427 protein (Figures 2 plus S4) already after 24h. These results suggested that, while the S427P exchange is the main reason why the human enzyme cannot activate 4-IPO, other amino acid regions in the rabbit protein may also be important for the strong functional activity of the wild-type enzyme.
To exclude the possibility that the different activities of the human and rabbit enzymes are due to differences in mRNA stability, the human cDNA was optimized for human-codon usage by de novo synthesis with silent mutagenesis by GeneArt (Regensburg, Germany), which removes internal TATA-boxes, instability motifs, RNA secondary structures and (cryptic) splice donor/acceptor sites and ensures an average GC content of around 60%. As shown in Figures 2 plus S4, the codon-optimization did not lead to an improvement in the activity of the native human protein and only slightly increased the activity of the P427-containing enzyme. As the activity of the native rabbit enzyme was still much higher, these findings confirmed that mRNA stability is not the decisive factor that explains the differences in activity of the CYP4B1 proteins from the two species.
Protein stability of the different CYP4B1s
We then hypothesized that the different activities of the human and rabbit proteins in processing of 4-IPO may, at least partially, be due to differences in the protein half-life of the enzymes. As no antibody was available that could robustly detect both rabbit and human CYP4B1 proteins, EGFP fusion constructs were generated that allowed visualization and comparison of the expression levels of EGFP-tagged proteins in cells. An additional advantage of EGFP fusions is that flow cytometry can readily be utilized to assess the expression levels of the fusion proteins in non-fixed cells. 5'- and 3'-fusions of EGFP with the native rabbit P422 and the human P427 CYP4B1 proteins were cloned in LV vectors with an IRES-NEO (Figure 1B) and stably expressed in HepG2 cells. For comparison, cells expressing the CYP4B1s from the corresponding IRES-EGFP constructs (Figure 1A) were also included.
To confirm that the fusion proteins were still capable of processing 4-IPO, transduced HepG2 cells were incubated for 24, 48 and 72 hours with increasing concentrations of 4-IPO. Subsequently, the cultures were harvested and analyzed for survival of transduced EGFP+ cells by flow cytometry. The results demonstrated (Figures 3A plus S5) that the 5'-EGFP fusion strongly inhibits the activity of CYP4B1 proteins, while the 3'-EGFP fusion only slightly diminished the ability of the enzymes to metabolize 4-IPO compared to the IRES-EGFP constructs.
As the 5’ fusions with EGFP most likely disturbed the leader peptide and therefore interfered with the activity of CYP450 enzymes [38], we used 3’-EGFP fusions of both human and rabbit proteins for the subsequent half-life experiments. HepG2 cells were transduced at the same multiplicities-of-infections (MOIs) for all constructs and then selected for 100% transduced cells with G418 (Geneticin®, Gibco) as described [29]. After inhibition of de novo protein synthesis by cycloheximide (CHX), the half-life of the CYP4B1 proteins in G418-resistant HepG2 cells was assessed by determining the protein expression levels over time either by Western blot (Figure 3B) or by flow cytometry (Figure 3C). These analyses demonstrated profound differences in the half-life of the various proteins in our test systems. Wild-type rabbit CYP4B1 enzyme (r-P422) has the highest protein stability (t1/2 >75h) and the introduction of the proline-to-serine exchange decreases the half-life of the mutated rabbit enzyme (r-S422) to ~45 hours. The human native protein (h-S427) has the shortest half-life of only ~9 hours, however, the serine-to-proline exchange (h-P427) increases protein half-life of the mutated human enzyme to ~17 hours, which is still inferior to the half-life of the r-S422 rabbit CYP4B1 protein. Using the semi-quantitative assessment of EGFP by flow cytometry as readout (Figure 3D), HepG2 cells expressing the rabbit P422 protein have the highest fluorescence intensity of EGFP-tagged protein (mean fluorescent intensity (MFI): 1753). Compared to the human CYP4B1 forms, the rabbit P422 shows a 30-fold higher MFI value in relation to the wild-type human CYP4B1 (MFI: 56), and a 7-fold higher MFI with regard to the human P427 mutant (MFI: 240). Interestingly, codon optimization of the human CYP4B1 proteins h-P427 and h-S427 in both cases revealed only a doubling of the mean fluorescence intensity (MFIs: 535 and 92, respectively). The raw fluorescence microscope images clearly reflect these results (Figure 3D).
Finally, HepG2 cells were infected with limiting dilutions of vector particles (the 10−4 dilution corresponded to an MOI of approximately 0.01) and then selected with G418. Using this strategy, we obtained polyclonal dilutions of transduced cells at lower MOIs with only one proviral integration per cell, thereby allowing us to assess the intrinsic functional differences between the different CYP4B1 proteins more accurately. The 4-IPO survival curves of these transduced HepG2 cells (Figures 4A plus S6) demonstrated that the MOIs and the associated CYP4B1 expression levels only weakly influenced cytotoxicity. Finally, although the r-P422 (−2) and h-P427 (ud=undiluted) overexpressing cells have similar CYP4B1 protein expression levels by western blot (Figure 4B) or by MFI values (data not shown), the toxicity assay after 48h revealed a significantly better activation of 90 µM 4-IPO by the rabbit protein when compared with the human h-P427 enzyme (Figures 4A plus S6).
Re-engineering of the human CYP4B1 protein for high activity towards 4-IPO
Although the proline-to-serine exchange in the meander region of the native human protein is a prerequisite for high activity of the human enzyme towards 4-IPO, our results demonstrating robust activity for the rabbit Ser422 enzyme led us to hypothesize that other amino acid differences between the two proteins determine the huge differences in their bioactivation of 4-IPO.
In order to identify smaller segments between the human and the rabbit proteins that influence the processing of 4-IPO, we took advantage of two unique restriction enzyme recognition sites, BstX I and Blp I, that are present in both cDNAs in frame and therefore could be used to exchange whole regions between the two proteins. As shown in Figure 5, these two restriction sites facilitate division of the human and the rabbit CYP4B1 cDNAs into three domains, which were exchanged by cloning and the chimeric proteins then expressed with lentiviral vectors in HepG2 cells. These domain exchanges between the human and rabbit enzymes revealed that each rabbit domain (5'-, middle and 3'-) is associated with an increase in the enzyme activity of the human P427 and S427 CYP4B1 proteins and that introduction of each human domain decreased the activity of processing 4-IPO by the native rabbit P422 isoform (Figures 5A plus S7–9). The exchanges of the C-terminus or the N-terminus leader region of the proteins between the two species resulted in the most minimal changes in CYP4B1 enzyme activity, while the largest differences in the toxicity induced by 4-IPO were observed by exchanging the middle domains (Figures 5A plus S7–9). The difference in cell toxicity between the native human S427 (h-S427) enzyme and the human-rabbit-human-S427 (h-r-h S427) fusion protein was significant at 90 µM after 48h and 72h (Figures 5A plus S8–9).
We therefore decided to concentrate our efforts on modifying the middle domain of the human protein to achieve higher enzyme activity. To this end, the middle domain of the rabbit CYP4B1 enzyme was divided into six small sections (s1 to s6) and each section cloned individually into the cDNAs for the human native S427 and the mutant P427 CYP4B1 proteins. As shown in Figures 5B plus S10–12, only the first three sections of the middle domain of the rabbit protein produced an improvement in enzyme activity after 24, 48, and 72h, and only when using the P427 form of human CYP4B1. In the human native S427 CYP4B1 protein, none of the six sections from the rabbit protein restored any functional activity of the human protein towards 4-IPO. In addition, introducing all 18 different amino acid between the rabbit and the human proteins singly into the human P427 CYP4B1 protein did not result in an increase in enzyme activity at 90 µM 4-IPO (Figures S3 plus S18–23). We therefore additionally cloned the four possible combinations of sections 1 to 3 (s1+2, s2+3, s1+3, s1+2+3) of the rabbit protein into the human 4B1 P427 enzyme and compared the activity of the new constructs with the constructs that carried section 1, 2, or 3 alone and also with those expressing the native rabbit and human CYP4B1 proteins as well as the human P427 mutant protein (Figures 5C plus S13–15). Importantly, this approach revealed that the enzyme activity of the human P427 CYP4B1 protein with sections 1+2+3 of the middle domain of the native rabbit protein for 4-IPO was almost as high as the activity of native rabbit protein. This mutant protein was even more active than the human P427 protein with the complete middle rabbit domain (h-r-h P427) with sections 1–6 (Figures 5C plus S13–15).
The final P+12 and P+18 human CYP4B1 proteins
The 1+2+3 construct differs at 19 positions between human and rabbit CYP4B1 (P+18, Figure 6A), 18 of which are amino acids located in sections 1–3 and the serine-to-proline substitution at position 427. In order to introduce as few as possible potentially immunological amino acids from the rabbit enzyme into the human CYP4B1 protein, we aligned the corresponding sections of all members of the human CYP450 family 4. As shown in Figure 6A,12 of the 18 aa positions in the P+18 protein are also present at corresponding positions in other human CYP4 family members and thus less likely to trigger immunogenicity. We therefore created a second modified human CYP4B1 protein (P+12, Figure 6A), which contained these 12 substitutions but is devoid of the six amino acids that are uniquely present in the native rabbit enzyme. When stably expressed in HepG2 cells, the CYP4B1 P+12 variant achieved a comparable killing curve to the P+18 variant (1+2+3, both with P427) (Figures 6B plus S16). We achieved a further improvement in 4-IPO processing by optimization of the mRNAs for human codon usage (coop), as is obvious in the direct comparison of h-P427 to coop h-P427 or P+18 to coop P+18 (Figure 6B). Finally, Western blotting of the HepG2 cells revealed that the codon-optimized P+12 and P+18 human CYP4B1 enzymes were expressed at similar protein levels as the rabbit enzyme (Figure 6C), thus confirming the functional analysis shown in Figure 6B.
Structural model of human CYP4B1
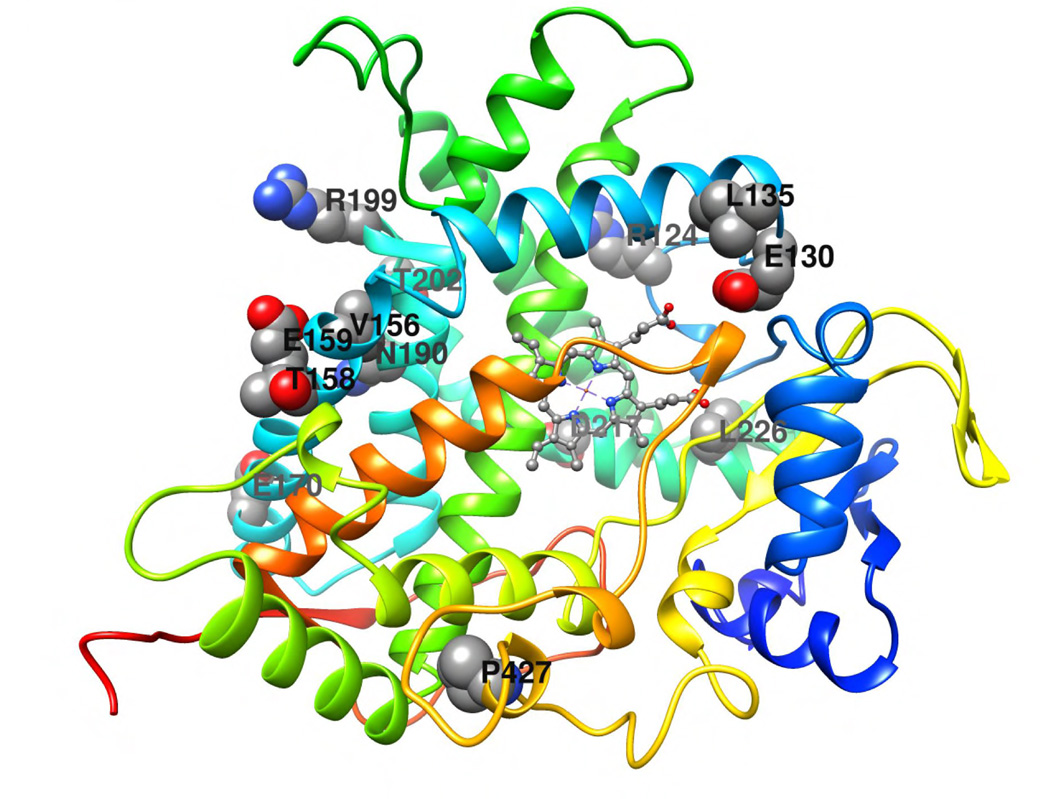
The exhaustive site-directed mutagenesis employed to resurrect the human CYP4B1 enzyme affords us a unique opportunity to assess the location of amino acid residues that contribute to P450 enzyme stability. However, since no crystal structure is yet available for CYP4B1, or any CYP4 protein, we used the Rosetta-based Robetta server to generate a homology model of the wild-type human CYP4B1, as described in Experimental Procedures. The model showed that the majority of the CYP4B1 residue side chains at the P+12 variant positions are located in the distal region of the enzyme, above the plane of the heme and in a contiguous area between the predicted B–C loop and the F-helix. All twelve residues other than the previously studied S427P are at least partially exposed to the soluble environment (Figure 7). A number of the P+12 mutations have conserved charge or polarity, including R124K, E130D, E159D, R199K, T202S, and D217E. In particular, R124K may be involved in salt bridge and hydrogen bond interactions with D258. E130D is buried within the protein core and may form a hydrogen bond with R451. T202S may also form a hydrogen bond with D208. It is possible that D217E forms a salt bridge and hydrogen bond interactions with R245. There are also several P+12 mutations that have conserved hydrophobicity, including L135F, V156I, and L226I. L135F is fully exposed to the outside of the protein and has no proximal residues within CYP4B1. V156I projects towards the protein core and is in proximity with T191, K194, and C195. L226I projects towards the protein core where it is in proximity with F117 and L119. The other P+12 mutations alter the charge or the polarity of the amino acids. The three alterations T158A, E170K, N190D are all exposed on the outside of the protein. N190 in the native human CYP4B1 is a particularly interesting residue, because the other human CYP4 family members and also the native rabbit CYP4B1 have aspartate at this position. However, functional testing of N190D in combination with S427P in the human CYP4B1 did not have any effect on the 4-IPO processing activity (Figures S3 plus S18–23).
Figure 7. Structural model of human CYP4B1.
The cartoon representation of the human CYP4B1 model was generated as described in Experimental Procedures. The model was colored using the rainbow scheme – from N-terminus (blue) to C-terminus (red). The CYP4B1 wild-type residues that are sites of the P+12 mutations are shown in space filling representation and labeled. The heme is shown in ball and stick representation. The figure was generated using UCSF Chimera [51].
A new human suicide gene system for human T-cells
In order to demonstrate that primary human T-cells can be efficiently eliminated with our novel suicide gene system, T-cells from healthy volunteers were stimulated by incubation on immobilized CD3 and CD28 antibodies for 3 days and then transduced once on the recombinant fibronectin fragment CH-296 in the presence of IL-2 with lentiviral vectors, similar to our previously described protocol [31]. For these experiments, we used an improved lentiviral vector (Figure 1C), where the CYP4B1 cDNAs were expressed together with EGFP off the internal SFFV promoter by an IRES-EGFP cassette. To increase the transcript levels in primary T-cells, we also included an optimized safety-modified version of the woodchuck hepatitis virus post-transcriptional regulatory element (WPRO) [39] in the vector. Five days after transduction, we challenged the primary T-cells with increasing doses of 4-IPO and subsequently analyzed the survival of the transduced T-cells after 24, 48 and 72h by flow cytometry. The results demonstrated that 4-IPO exposure can kill ≥60% of primary T-cells that expressed either the rabbit or the human codon-optimized (coop) P+12 and P+18 CYP4B1 P427 proteins within 24 hours (Figure 8). Within 3 days, ≥90% of the T-cells expressing the native rabbit or the human P+12/P+18 CYP4B1 proteins are killed by exposure to 29 or 90 µM 4-IPO. Interestingly, in T-cells, codon-optimization of the mRNA further increased the activities of CYP4B1 enzymes, as shown for h-P427 and P+18 at all time points and all 4-IPO concentrations (Figures 8 plus S17).
DISCUSSION
Twenty five years after the identification of the human CYP4B1 cDNA from lung tissue, the function(s) of the native human CYP4B1 protein is(are) still not known and thus the human CYP4B1 is classified as an orphan P450 enzyme [8]. Czerwinski et al. first demonstrated that the human CYP4B1 enzyme is unable to metabolize 4-IPO [11], a hallmark substrate for the activity of animal CYP4B1 orthologs [8], and we have generated healthy Cyp4b1−/− knock-out mice that are completely resistant to doses of 4-IPO known to induce acute respiratory distress and to cause high mortality in wild-type mice [40]. Therefore, while CYP4B1 is unambiguously a critical enzyme for 4-IPO bioactivation, a clear species difference in bioactivation capacity exists that is relevant to earlier failed efforts to use the enzyme as a suicide gene for cancer treatment in humans.
In our systematic analysis of the human CYP4B1 enzyme, we found that the simple serine to proline exchange at position 427 stabilizes the human protein and partially restores the enzymatic activity for 4-IPO. The proline 427 in the meander region is highly conserved in all CYP4B1 proteins from different species (Figure S2) and also in all members of the human xenobiotic-metabolizing CYP1–CYP4 P450 families [25]. Prior to the results reported here, the proline residue was thought to be part of an essential Pro-X-Arg/His motif in the meander region [41, 42] that is destroyed by the serine at position 427 in human CYP4B1 enzyme. This Ser-X-Arg/His motif could therefore influence the structural arrangement of the protein in relation to the glutamine-arginine-arginine (ERR) motif (Figure S1), which is conserved in all cytochrome P450 enzymes [43]. As this ERR triad might also play an important role in stabilizing and anchoring the heme in the enzyme’s active center, it was suggested to be essential for maintaining the activity of any cytochrome P450 enzyme [42, 44]. However, by substituting sections of human CYP4B1 with corresponding sections of the rabbit enzyme, we demonstrated here that the human S427 protein with the middle domain of the rabbit is still a functional enzyme. Therefore, while S427 in both human CYP4B1 and S422 in rabbit CYP4B1 are clearly important to enzyme stability, it does not alone control the functional activity of the enzyme. Earlier studies had suggested that the meander region proline was essential for heme binding to CYP4B1 [42, 44]. However, this is clearly not the case from the current data. Differences between the two studies may reflect the use of different expression systems, insect cells versus HepG2 cells, with enzyme stability enhanced in the latter perhaps due to the presence of mammalian chaperones.
We also demonstrated that combinations of several amino acids at key positions in the rabbit CYP4B1 protein are responsible for the high activity of the rabbit enzyme. Through extensive rounds of mutagenesis, we were finally able to create highly active enzymes, CYP4B1 P+12 and P+18, which differ in only 13 and 19 amino acids, respectively, from the native human CYP4B1 protein (98 and 96% identity). Upon optimization of the cDNAs for human codon usage, the P+12 and P+18 CYP4B1 proteins showed improved stability by western blotting (Figure 6C) and increased activities for 4-IPO dependent cytotoxicity, comparable to the rabbit enzyme, which led to efficient cell death induction in transduced human liver cells and human T-cells (Figure 6B and 8). Interestingly, based on the homology model shown in Figure 7, the 12 amino acid substitutions in the mutant P+12 protein are clustered in an area that maps to the B–C loop through the F-helix regions that are conserved in the membrane-bound eukaryotic P450s and are known to be important for enzyme activity [6]. These localizations, however, must be viewed as tentative because no CYP4 protein has yet been crystallized. Nevertheless, a large component of the increased enzyme activity of the P+12 enzyme is due to higher levels of expression of the enzyme, and so the B–C loop through F-helix regions are important for conferring enzyme stability to CYP4B1. However, whereas a great deal is known about amino acid determinants of P450 substrate specificity, much less has been established regarding P450 enzyme stability. One potential mechanism for enhanced enzyme stability is improved binding to accessory protein partners such as P450 reductase and/or cytochrome b5, which have been predicted to bind to basic amino acids in this general region of CYP2B4 [45, 46]. In support of this, Imaoka et al. [47] reported expression of functional human CYP4B1 as a fusion protein with P450 reductase in yeast. However, the same fusion did not rescue activity of the native human enzyme when stably expressed in HepG2 cells (Wiek, Hanenberg, unpublished data). Recently, Correia and coworkers have shown that phosphorylation of CYP2E1 and CYP3A4 occurs by protein kinases A and C, which can then accelerate ubiquitination [48]. Possibly, improvements in the activity/stability of the P+12 enzyme could reflect altered phosphorylation and subsequent ubiquitin-mediated degradation, but additional studies are required to test this scenario. It is also possible that these mutations contribute to an increase in functional activity of the P+12 enzyme due to altered membrane anchoring/substrate access and/or improvements in the intrinsic activity of the enzyme due to topographical changes in the enzyme’s binding cavity or substrate access/exit channels that affect substrate binding or product release, but additional studies are required to test these scenarios.
Based on the foregoing structure-function/activity associations, it is tempting to speculate that the human CYP4B1 enzyme was subject to rigorous negative selection in our ancestors as an evolutionary consequence of the introduction of sweet potatoes or other roots in our food chain, especially as neither cooking nor baking effectively destroys 4-IPO [49]. However, in contrast to our findings, two previous studies hint at a functional role for human CYP4B1. Imaoka et al. demonstrated that the CYP4B1 protein and also mRNA levels were significantly up-regulated in resection samples from patients with bladder cancer compared to patients without, suggesting that the native human enzyme might have a role in the mutagenic activation of pro-carcinogens in the bladder [47]. In a transgenic mouse model where the human CYP4B1 S427/S207 splice variant was expressed in the liver, the same group reported specific catalytic activities of the human protein in liver microsomes for 2-aminofluorene and for ω-hydroxylation of lauric acid [27]. Therefore, further studies e.g. by knocking-in the human native CYP4B1 enzyme and also our genetically re-engineered P+12 and P+18 variants into the Cyp4b1−/− locus of the knock-out mice might be necessary to ultimately decipher the physiological function(s) of the native human CYP4B1 enzyme and also to determine the range of substrate(s) that can be processed by both the human native and re-engineered CYP4B1s.
In conclusion, here we have further developed the idea of Rainov et al. [28] to use 4-IPO as pro-drug in a suicide gene system for clinical applications, but have substituted the rabbit wild-type CYP4B1 protein for a highly active human mutant CYP4B1 enzyme. The 13 amino acid alterations in this protein are all found in corresponding positions in other human CYP family 4 proteins (Figure 6A) and therefore unlikely to be recognized by the immune system. 4-IPO as a pro-drug for a highly toxic DNA alkylating intermediate has been administered to poor prognosis lung and liver cancer patients in three clinical phase I/II studies with dose escalation regimens [22–24]. Paramount for the intended use as pro-drug, however, is that the 5-day infusion regimens used in these trials readily achieved 4-IPO serum levels of up to 90 µM [22, 23], a concentration at which effective killing of human transduced HepG2 and primary T-cells expressing the P+12 mutant protein occurs already at 24 h and by >90% after 72 h (Figure 6 and 8). As there is no bystander effect of CYP4B1-activated 4-IPO for neighboring cells in vitro and 4-IPO mediates a proliferation-independent elimination of cells [50], we will next determine the elimination kinetics of circulating and also organ-infiltrating murine and human T-cells expressing the P+12 CYP4B1 in acute graft-versus-host disease (GvHD) models in vivo in mice. A key focus of these preclinical studies will be to determine how quickly the acute GvHD will clinically resolve after 4-IPO administration.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jakob Reiser and Dirk Lindemann for kindly providing reagents for the packaging of the lentiviral vectors and Michael Gombert for critical discussions.
FUNDING
This work was initially supported by the Deutsche José-Carreras-Leukämie-Stiftung e. V. (Munich, Germany), the Deutsche Krebshilfe e. V. (to CMK and HH) and the NIH grant, R01 GM49054 (AER). Later support was from the Forschungskommission of the Medical Faculty and the Strategische Forschungskommission of the Heinrich Heine University, Düsseldorf, Germany, (to CW), the UW School of Pharmacy Brady Fund for Natural Products Research (to AER and EJK) and the NIH R01s CA138237-01 and CA155294-01 (to HH). Helmut Hanenberg is supported by the Lilly Foundation Physician/Scientist initiative.
The abbreviations used are
- CYP450
cytochrome P450
- 4-IPO
4-ipomeanol
- CYP4B1
CYP family 4 subfamily B polypeptide 1
- VSV-G
glycoprotein G of the vesiculo stomatitis virus
- DLI
donor lymphocyte infusion
- GvHD
graft-versus-host disease
- IRES
internal ribosomal entry site
- EGFP
enhanced green fluorescent protein
- LV
lentiviral vector
- WPRO
woodchuck hepatitis virus post-transcriptional regulatory element optimized
Footnotes
AUTHOR CONTRIBUTIONS
The study was designed by CW, CMK, AER and HH. All experiments and/or analyses were performed by CW, EMS, KR, MF, MN, EJK, VYY, WK and HH. The manuscript was written by CW, AER and HH with essential help from the other authors.
CONFLICT OF INTEREST
HH may receive royalties based on a license agreement between Indiana University, Indianapolis, USA, and Takara Shuzo Inc., Kyoto, Japan, on the sales and usage of Retronectin® CH-296 in transduction protocols. The other authors declare no competing financial interests.
REFERENCES
- 1.Ortiz de Montellano PR, Nelson SD. Rearrangement reactions catalyzed by cytochrome P450s. Arch Biochem Biophys. 2011;507:95–110. doi: 10.1016/j.abb.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gotoh O. Evolution of cytochrome p450 genes from the viewpoint of genome informatics. Biol Pharm Bull. 2012;35:812–817. doi: 10.1248/bpb.35.812. [DOI] [PubMed] [Google Scholar]
- 3.Lamb DC, Waterman MR. Unusual properties of the cytochrome P450 superfamily. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120434. doi: 10.1098/rstb.2012.0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nebert DW, Wikvall K, Miller WL. Human cytochromes P450 in health and disease. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120431. doi: 10.1098/rstb.2012.0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munro AW, Girvan HM, Mason AE, Dunford AJ, McLean KJ. What makes a P450 tick? Trends Biochem Sci. 2013;38:140–150. doi: 10.1016/j.tibs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Johnson EF, Stout CD. Structural diversity of eukaryotic membrane cytochrome p450s. J Biol Chem. 2013;288:17082–17090. doi: 10.1074/jbc.R113.452805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW, Gunsalus IC, Nebert DW. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- 8.Baer BR, Rettie AE. CYP4B1: an enigmatic P450 at the interface between xenobiotic and endobiotic metabolism. Drug Metab Rev. 2006;38:451–476. doi: 10.1080/03602530600688503. [DOI] [PubMed] [Google Scholar]
- 9.Zhang JY, Wang Y, Prakash C. Xenobiotic-metabolizing enzymes in human lung. Curr Drug Metab. 2006;7:939–948. doi: 10.2174/138920006779010575. [DOI] [PubMed] [Google Scholar]
- 10.Edson KZ, Rettie AE. CYP4 Enzymes as potential drug targets: focus on enzyme multiplicity, inducers and inhibitors, and therapeutic modulation of 20-hydroxyeicosatetraenoic acid (20-HETE) synthase and fatty acid omega-hydroxylase activities. Curr Top Med Chem. 2013;13:1429–1440. doi: 10.2174/15680266113139990110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czerwinski M, McLemore TL, Philpot RM, Nhamburo PT, Korzekwa K, Gelboin HV, Gonzalez FJ. Metabolic activation of 4-ipomeanol by complementary DNA-expressed human cytochromes P-450: evidence for species-specific metabolism. Cancer Res. 1991;51:4636–4638. [PubMed] [Google Scholar]
- 12.Verschoyle RD, Philpot RM, Wolf CR, Dinsdale D. CYP4B1 activates 4-ipomeanol in rat lung. Toxicol Appl Pharmacol. 1993;123:193–198. doi: 10.1006/taap.1993.1237. [DOI] [PubMed] [Google Scholar]
- 13.Rettie AE, Sheffels PR, Korzekwa KR, Gonzalez FJ, Philpot RM, Baillie TA. CYP4 isozyme specificity and the relationship between omega-hydroxylation and terminal desaturation of valproic acid. Biochemistry. 1995;34:7889–7895. doi: 10.1021/bi00024a013. [DOI] [PubMed] [Google Scholar]
- 14.Christian MC, Wittes RE, Leyland-Jones B, McLemore TL, Smith AC, Grieshaber CK, Chabner BA, Boyd MR. 4-Ipomeanol: a novel investigational new drug for lung cancer. J Natl Cancer Inst. 1989;81:1133–1145. doi: 10.1093/jnci/81.15.1133. [DOI] [PubMed] [Google Scholar]
- 15.Hansen AA. Two Fatal Cases of Potato Poisoning. Science. 1925;61:340–341. doi: 10.1126/science.61.1578.340. [DOI] [PubMed] [Google Scholar]
- 16.Wilson BJ, Yang DT, Boyd MR. Toxicity of mould-damaged sweet potatoes (Ipomoea batatas) Nature. 1970;227:521–522. doi: 10.1038/227521a0. [DOI] [PubMed] [Google Scholar]
- 17.Wilson BJ, Boyd MR, Harris TM, Yang DT. A lung oedema factor from mouldy sweet potatoes (Ipomoea batatas) Nature. 1971;231:52–53. doi: 10.1038/231052a0. [DOI] [PubMed] [Google Scholar]
- 18.Boyd MR. Role of metabolic activation in the pathogenesis of chemically induced pulmonary disease: mechanism of action of the lung-toxic furan, 4-ipomeanol. Environ Health Perspect. 1976;16:127–138. doi: 10.1289/ehp.7616127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doster AR, Mitchell FE, Farrell RL, Wilson BJ. Effects of 4-ipomeanol, a product from mold-damaged sweet potatoes, on the bovine lung. Vet Pathol. 1978;15:367–375. doi: 10.1177/030098587801500312. [DOI] [PubMed] [Google Scholar]
- 20.Baer BR, Rettie AE, Henne KR. Bioactivation of 4-ipomeanol by CYP4B1: adduct characterization and evidence for an enedial intermediate. Chem Res Toxicol. 2005;18:855–864. doi: 10.1021/tx0496993. [DOI] [PubMed] [Google Scholar]
- 21.Falzon M, McMahon JB, Schuller HM, Boyd MR. Metabolic activation and cytotoxicity of 4-ipomeanol in human non-small cell lung cancer lines. Cancer Res. 1986;46:3484–3489. [PubMed] [Google Scholar]
- 22.Rowinsky EK, Noe DA, Ettinger DS, Christian MC, Lubejko BG, Fishman EK, Sartorius SE, Boyd MR, Donehower RC. Phase I and pharmacological study of the pulmonary cytotoxin 4-ipomeanol on a single dose schedule in lung cancer patients: hepatotoxicity is dose limiting in humans. Cancer Res. 1993;53:1794–1801. [PubMed] [Google Scholar]
- 23.Kasturi VK, Dearing MP, Piscitelli SC, Russell EK, Sladek GG, O'Neil K, Turner GA, Morton TL, Christian MC, Johnson BE, Kelley MJ. Phase I study of a five-day dose schedule of 4-Ipomeanol in patients with non-small cell lung cancer. Clin Cancer Res. 1998;4:2095–2102. [PubMed] [Google Scholar]
- 24.Lakhanpal S, Donehower RC, Rowinsky EK. Phase II study of 4-ipomeanol, a naturally occurring alkylating furan, in patients with advanced hepatocellular carcinoma. Invest New Drugs. 2001;19:69–76. doi: 10.1023/a:1006408803734. [DOI] [PubMed] [Google Scholar]
- 25.Zheng YM, Henne KR, Charmley P, Kim RB, McCarver DG, Cabacungan ET, Hines RN, Rettie AE. Genotyping and site-directed mutagenesis of a cytochrome P450 meander Pro-X-Arg motif critical to CYP4B1 catalysis. Toxicol Appl Pharmacol. 2003;186:119–126. doi: 10.1016/s0041-008x(02)00028-5. [DOI] [PubMed] [Google Scholar]
- 26.Zheng YM, Fisher MB, Yokotani N, Fujii-Kuriyama Y, Rettie AE. Identification of a meander region proline residue critical for heme binding to cytochrome P450: implications for the catalytic function of human CYP4B1. Biochemistry. 1998;37:12847–12851. doi: 10.1021/bi981280m. [DOI] [PubMed] [Google Scholar]
- 27.Imaoka S, Hayashi K, Hiroi T, Yabusaki Y, Kamataki T, Funae Y. A transgenic mouse expressing human CYP4B1 in the liver. Biochem Biophys Res Commun. 2001;284:757–762. doi: 10.1006/bbrc.2001.5055. [DOI] [PubMed] [Google Scholar]
- 28.Rainov NG, Sena-Esteves M, Fraefel C, Dobberstein KU, Chiocca EA, Breakefield XO. A chimeric fusion protein of cytochrome CYP4B1 and green fluorescent protein for detection of pro-drug activating gene delivery and for gene therapy in malignant glioma. Adv Exp Med Biol. 1998;451:393–403. doi: 10.1007/978-1-4615-5357-1_61. [DOI] [PubMed] [Google Scholar]
- 29.Nakano M, Kelly EJ, Wiek C, Hanenberg H, Rettie AE. CYP4V2 in Bietti’s crystalline dystrophy: ocular localization, metabolism of ω-3 polyunsaturated fatty acids and functional deficit of the p.H331P variant. Mol Pharmacol. 2012;82:679–686. doi: 10.1124/mol.112.080085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanenberg H, Xiao XL, Dilloo D, Hashino K, Kato I, Williams DA. Colocalization of retrovirus and target cells on specific fibronectin fragments increases genetic transduction of mammalian cells. Nat Med. 1996;2:876–882. doi: 10.1038/nm0896-876. [DOI] [PubMed] [Google Scholar]
- 31.Pollok KE, Hanenberg H, Noblitt TW, Schroeder WL, Kato I, Emanuel D, Williams DA. High-efficiency gene transfer into normal and adenosine deaminase-deficient T lymphocytes is mediated by transduction on recombinant fibronectin fragments. J Virol. 1998;72:4882–4892. doi: 10.1128/jvi.72.6.4882-4892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanenberg H, Hashino K, Konishi H, Hock RA, Kato I, Williams DA. Optimization of fibronectin-assisted retroviral gene transfer into human CD34+ hematopoietic cells. Hum Gene Ther. 1997;8:2193–2206. doi: 10.1089/hum.1997.8.18-2193. [DOI] [PubMed] [Google Scholar]
- 33.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. http://docs.ggplot2.org/current/ [Google Scholar]
- 34.Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer; 2009. http://had.co.nz/ggplot2/book. [Google Scholar]
- 35.Chivian D, Baker D. Homology modeling using parametric alignment ensemble generation with consensus and energy-based model selection. Nucleic acids research. 2006;34:e112. doi: 10.1093/nar/gkl480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic acids research. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sansen S, Yano JK, Reynald RL, Schoch GA, Griffin KJ, Stout CD, Johnson EF. Adaptations for the oxidation of polycyclic aromatic hydrocarbons exhibited by the structure of human P450 1A2. The Journal of biological chemistry. 2007;282:14348–14355. doi: 10.1074/jbc.M611692200. [DOI] [PubMed] [Google Scholar]
- 38.Lengler J, Holzmuller H, Salmons B, Gunzburg WH, Renner M. FMDV-2A sequence and protein arrangement contribute to functionality of CYP2B1-reporter fusion protein. Anal Biochem. 2005;343:116–124. doi: 10.1016/j.ab.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 39.Schambach A, Bohne J, Baum C, Hermann FG, Egerer L, von Laer D, Giroglou T. Woodchuck hepatitis virus post-transcriptional regulatory element deleted from X protein and promoter sequences enhances retroviral vector titer and expression. Gene Ther. 2006;13:641–645. doi: 10.1038/sj.gt.3302698. [DOI] [PubMed] [Google Scholar]
- 40.Parkinson OT, Liggitt HD, Rettie AE, Kelly EJ. Generation and characterization of a Cyp4b1 null mouse and the role of CYP4B1 in the activation and toxicity of ipomeanol. Toxicol Sci. 2013;134:243–250. doi: 10.1093/toxsci/kft123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hasemann CA, Kurumbail RG, Boddupalli SS, Peterson JA, Deisenhofer J. Structure and function of cytochromes P450: a comparative analysis of three crystal structures. Structure. 1995;3:41–62. doi: 10.1016/s0969-2126(01)00134-4. [DOI] [PubMed] [Google Scholar]
- 42.Peterson JA, Graham SE. A close family resemblance: the importance of structure in understanding cytochromes P450. Structure. 1998;6:1079–1085. doi: 10.1016/s0969-2126(98)00109-9. [DOI] [PubMed] [Google Scholar]
- 43.Krone N, Riepe FG, Grotzinger J, Partsch CJ, Bramswig J, Sippell WG. The residue E351 is essential for the activity of human 21-hydroxylase: evidence from a naturally occurring novel point mutation compared with artificial mutants generated by single amino acid substitutions. J Mol Med (Berl) 2005;83:561–568. doi: 10.1007/s00109-005-0655-3. [DOI] [PubMed] [Google Scholar]
- 44.Werck-Reichhart D, Feyereisen R. Cytochromes P450: a success story. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-6-reviews3003. REVIEWS3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bridges A, Gruenke L, Chang YT, Vakser IA, Loew G, Waskell L. Identification of the binding site on cytochrome P450 2B4 for cytochrome b5 and cytochrome P450 reductase. J Biol Chem. 1998;273:17036–17049. doi: 10.1074/jbc.273.27.17036. [DOI] [PubMed] [Google Scholar]
- 46.Lehnerer M, Schulze J, Achterhold K, Lewis DF, Hlavica P. Identification of key residues in rabbit liver microsomal cytochrome P450 2B4: importance in interactions with NADPH-cytochrome P450 reductase. J Biochem. 2000;127:163–169. doi: 10.1093/oxfordjournals.jbchem.a022578. [DOI] [PubMed] [Google Scholar]
- 47.Imaoka S, Yoneda Y, Sugimoto T, Hiroi T, Yamamoto K, Nakatani T, Funae Y. CYP4B1 is a possible risk factor for bladder cancer in humans. Biochem Biophys Res Commun. 2000;277:776–780. doi: 10.1006/bbrc.2000.3740. [DOI] [PubMed] [Google Scholar]
- 48.Correia MA, Wang Y, Kim SM, Guan S. Hepatic cytochrome P450 ubiquitination: conformational phosphodegrons for E2/E3 recognition? IUBMB Life. 2014;66:78–88. doi: 10.1002/iub.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson BJ, Yang DT, Boyd MR. Toxicity of mould-damaged sweet potatoes (Ipomoea batatas) Nature. 1970;227:521–522. doi: 10.1038/227521a0. [DOI] [PubMed] [Google Scholar]
- 50.Frank S, Steffens S, Fischer U, Tlolko A, Rainov NG, Kramm CM. Differential cytotoxicity and bystander effect of the rabbit cytochrome P450 4B1 enzyme gene by two different prodrugs: implications for pharmacogene therapy. Cancer Gene Ther. 2002;9:178–188. doi: 10.1038/sj.cgt.7700422. [DOI] [PubMed] [Google Scholar]
- 51.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.