Abstract
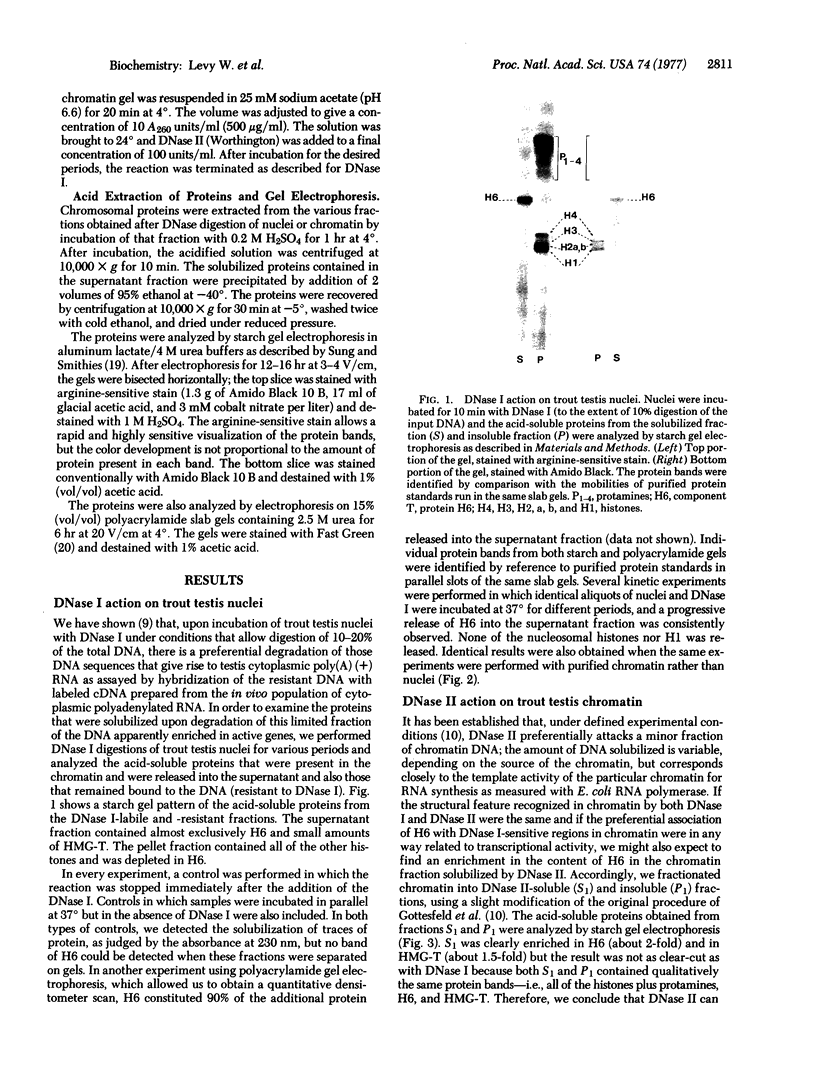
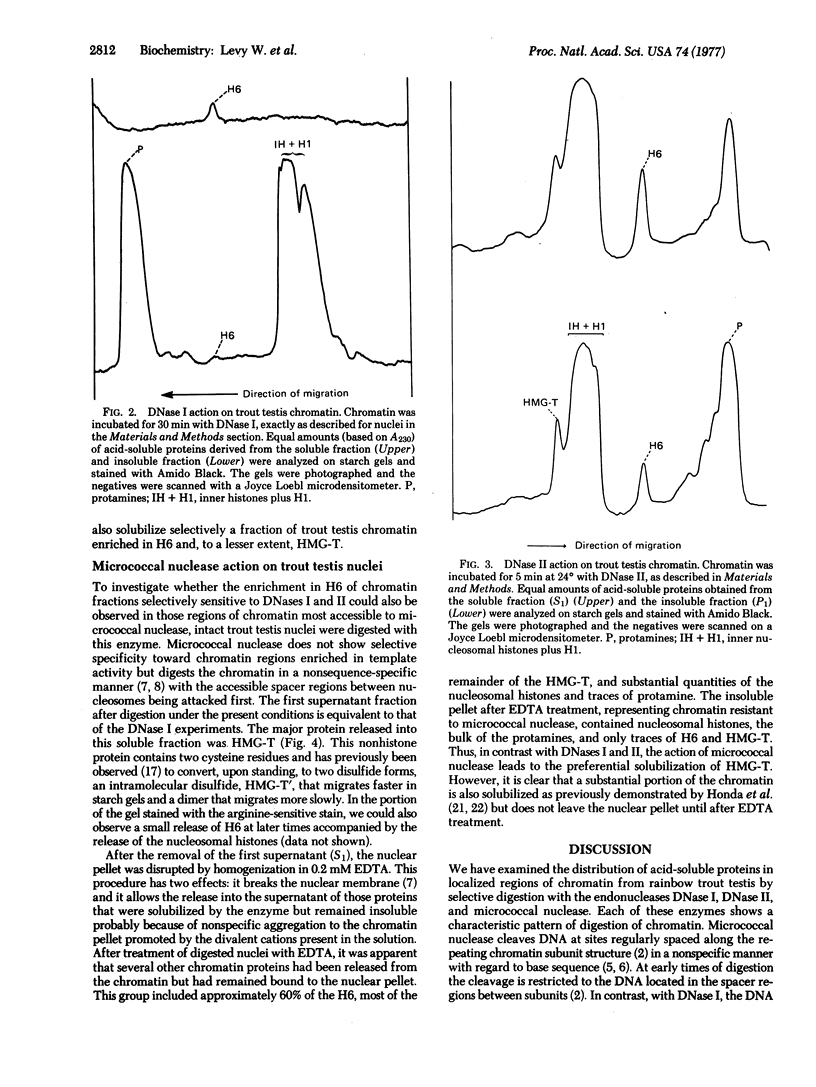
Nuclei and chromatin from trout testis cells were digested with three different nucleases (DNase I, DNase II, and micrococcal nuclease), and the acid-soluble proteins that were solubilized and those remaining bound to the nuclease-resistant DNA were compared electrophoretically. With the conditions described by H. Weintraub and M Groudine [(1976) science, 193, 848-856], which we previously found to be selective in digesting actively transcribed regions in trout testis chromatin, a single chromosomal protein, H6, was solubilized. The nucleosomal histones and H1 remained insoluble, bound to the resistant DNA. In contrast, digestion with micrococcal nuclease led to a preferential solubilization of a second protein, HMG-T, together with the release of some nucleosomal histones and H1 into the soluble fraction. DNase II also discriminated between "active" and "inactive" chromatins; when a DNase II-solubilized "active" chromatin fraction was prepared, it too was enriched in H6 and HMG-T. Thus, both H6 and HMG-T, the two major low-salt extractable chromosomal nonhistone the two major low-salt extractable chromosomal nonhistone proteins from trout testis, are associated with chromatin regions selectively sensitive to nucleases. The preferential solubilization of HMG-T by micrococcal nuclease action suggests that it might be located at the internucleosomal "spacer" region.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axel R., Cedar H., Felsenfeld G. Synthesis of globin ribonucleic acid from duck-reticulocyte chromatin in vitro. Proc Natl Acad Sci U S A. 1973 Jul;70(7):2029–2032. doi: 10.1073/pnas.70.7.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axel R., Cedar H., Felsenfield G. The structure of the globin genes in chromatin. Biochemistry. 1975 Jun 3;14(11):2489–2495. doi: 10.1021/bi00682a031. [DOI] [PubMed] [Google Scholar]
- Burgoyne L. A., Mobbs J. D., Marshall A. J. Chromatin structure: a property of the higher structures of chromatin and in the time course of its formation during chromatin replication. Nucleic Acids Res. 1976 Dec;3(12):3293–3304. doi: 10.1093/nar/3.12.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour R. S., Paul J. Tissue-specific transcription of the globin gene in isolated chromatin. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3440–3442. doi: 10.1073/pnas.70.12.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin G. H., Johns E. W. Isolation and characterisation of two calf-thymus chromatin non-histone proteins with high contents of acidic and basic amino acids. Eur J Biochem. 1973 Dec 3;40(1):215–219. doi: 10.1111/j.1432-1033.1973.tb03188.x. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M., Murphy R. F., Bonner J. Structure of transcriptionally active chromatin. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4404–4408. doi: 10.1073/pnas.72.11.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Honda B. M., Baillie D. L., Candido E. P. Properties of chromatin subunits from developing trout testis. J Biol Chem. 1975 Jun 25;250(12):4643–4647. [PubMed] [Google Scholar]
- Honda B. M., Baillie D. L., Candido E. P. The subunit structure of chromatin: characteristics of nucleohistone and nucleoprotamine from developing trout testis. FEBS Lett. 1974 Nov 1;48(1):156–159. doi: 10.1016/0014-5793(74)81086-0. [DOI] [PubMed] [Google Scholar]
- Huntley G. H., Dixon G. H. The primary structure of the NH 2 -terminal region of histone T. J Biol Chem. 1972 Aug 10;247(15):4916–4919. [PubMed] [Google Scholar]
- Lacy E., Axel R. Analysis of DNA of isolated chromatin subunits. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3978–3982. doi: 10.1073/pnas.72.10.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Wilson B., Gjerset R. A., McCarthy B. J. Acetylation and phosphorylation of Drosophila histones. Distribution of acetate and phosphate groups in fractionated chromatin. Biochim Biophys Acta. 1977 Mar 2;475(1):168–175. doi: 10.1016/0005-2787(77)90351-3. [DOI] [PubMed] [Google Scholar]
- Levy B., Dixon G. H. Renaturation kinetics of cDNA complementary to cytoplamic polyadenylated RNA from rainbow trout testis. Accessibility of transcribed genes to pancreatic DNase. Nucleic Acids Res. 1977 Apr;4(4):883–898. doi: 10.1093/nar/4.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Corden J., Tatchell K., Kovacic R. T., Van Holde K. E. Comparative subunit structure of HeLa, yeast, and chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1977 Jan;74(1):79–83. doi: 10.1073/pnas.74.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marushige K., Dixon G. H. Transformation of trout testis chromatin. J Biol Chem. 1971 Sep 25;246(18):5799–5805. [PubMed] [Google Scholar]
- McMaster-Kaye R., Kaye J. S. Staining of histones on polyacrylamide gels with amido black and fast green. Anal Biochem. 1974 Sep;61(1):120–132. doi: 10.1016/0003-2697(74)90339-x. [DOI] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974 Sep 20;251(5472):249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Felsenfeld G. A comparison of the digestion of nuclei and chromatin by staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2915–2920. doi: 10.1021/bi00684a019. [DOI] [PubMed] [Google Scholar]
- Sung M., Smithies O. Differential elution of histones from gel-trapped nuclei. Biopolymers. 1969;7(1):39–58. doi: 10.1002/bip.1969.360070105. [DOI] [PubMed] [Google Scholar]
- Tien Kuo M., Sahasrabuddhe C. G., Saunders G. F. Presence of messenger specifying sequences in the DNA of chromatin subunits. Proc Natl Acad Sci U S A. 1976 May;73(5):1572–1575. doi: 10.1073/pnas.73.5.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. M., Goodwin G. H., Johns E. W. The similarity between the primary structures of two non-histone chromosomal proteins. Eur J Biochem. 1976 Mar 1;62(3):461–469. doi: 10.1111/j.1432-1033.1976.tb10179.x. [DOI] [PubMed] [Google Scholar]
- Watson D. C., Peters E. H., Dixon G. H. The purification, characterization and partial sequence determination of a trout testis non-histone protein, HMG-T. Eur J Biochem. 1977 Mar 15;74(1):53–60. doi: 10.1111/j.1432-1033.1977.tb11365.x. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Wigle D. T., Dixon G. H. A new histone from trout testis. J Biol Chem. 1971 Sep 25;246(18):5636–5644. [PubMed] [Google Scholar]




