Abstract
Shiga toxin-producing Escherichia coli (STEC) is a heterogeneous group of bacteria causing disease ranging from asymptomatic carriage and mild infection to hemolytic uremic syndrome (HUS). Here, we describe patients with STEC infection and characterize the STEC strains detected in our laboratory by use of PCR for stx1, stx2, and eae from 1996 through 2011. Patient information was collected from referral forms and from the Norwegian Surveillance System for Communicable Diseases. STEC isolates were characterized with respect to serogroup or serotype, selected potential virulence genes, and multilocus variable-number tandem-repeat analysis (MLVA) genotype. STEC strains were isolated from 138 (1.09%) of 12,651 patients tested. STEC strains of serogroups O26, O103, O121, O145, and O157 were the most frequent. These serogroups, except non-sorbitol-fermenting O157, were also the most frequent among the 11 patients (all ≤5 years old) who developed HUS. Twenty-four STEC strains were classified as being HUS associated based on an epidemiological link to a HUS case, including an MLVA genotype identical to that of the STEC strain. The age of the patient (≤5 years) and the genes eae and stx2a were significantly associated with HUS-associated STEC (P < 0.05 for each parameter), while stx1 was associated with non-HUS-associated STEC (P < 0.05). All of the potential virulence genes analyzed, except ehxA, were significantly more frequent among HUS-associated than non-HUS-associated strains (P < 0.05 for each gene). However, these genes were also present in some non-HUS-associated STEC strains and could therefore not reliably differentiate between HUS-associated and non-HUS-associated STEC strains.
INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC) was recognized as a cause of bloody diarrhea and hemolytic uremic syndrome (HUS) for the first time in two independent studies in 1982 (1, 2). Later this pathogen was found to be the main cause of diarrhea-associated HUS with a high number of cases worldwide. Non-sorbitol-fermenting STEC (NSF) O157:H7 was the first STEC serotype that was isolated in association with HUS and has been the most frequently reported cause of diarrhea-associated HUS (3). However, STEC strains of other serogroups like O26, O103, O111, O121, and O145 have also been recognized to cause severe disease and outbreaks (4, 5).
Shiga toxins 1 and 2 (Stx1 and Stx2) are essential virulence factors of STEC. The term STEC is used to describe any E. coli-producing Shiga toxin, whereas the term enterohemorrhagic Escherichia coli (EHEC) is often used to describe the subset of STEC strains responsible for causing hemorrhagic colitis and HUS (3). Shiga toxins are encoded by the stx1 and stx2 genes located in lambdoid bacteriophages integrated into the bacterial host genome. The toxins exist as various subtypes, in which the Stx2 subtypes Stx2a, Stx2c, and Stx2d are more often associated with HUS than are the other Stx subtypes (6–9). In addition to the Shiga toxins, most STEC strains possess the locus of enterocyte effacement (LEE) pathogenicity island in which the virulence gene eae encoding the adherence factor intimin is located (3, 10).
In addition to stx1, stx2, and eae, the presence and absence of various other genes have been investigated as potential virulence markers for HUS and outbreaks. Karmali et al. reported that several genes located on the genomic O island OI-122 were present in 60 to 100% of STEC strains of serotypes highly associated with severe disease and outbreaks, while the same genes were detected in only 0 to 15% of strains of serotypes not associated with severe disease or outbreaks (11). In addition, the presence of non-LEE-encoded (nle) genes from the genomic O islands OI-71 and OI-57 has been associated with STEC virulence (12–14). Pathogenic STEC also harbors a large virulence plasmid (frequently termed pO157) encoding factors involved in STEC virulence (3, 15).
Since STEC was first recognized as a cause of diarrhea-associated HUS, the microbial detection of this pathogen has in most laboratories until recently relied on culturing on sorbitol-MacConkey agar (SMAC), which is a selective medium for NSF O157:H7 (16), with subsequent agglutination with anti-O157 antiserum. While a selective medium for NSF O157:H7 STEC is a sensitive method for detection of this specific STEC serotype, a major drawback is that other STEC serotypes and sorbitol-fermenting (SF) O157:H- not will be detected. Therefore, a more suitable strategy for the diagnosis of STEC infection is to combine culture and nonculture methods detecting both O157 and non-O157 STEC serotypes (17, 18). In recent years, many diagnostic laboratories have switched to PCR for detection of stx1 and stx2 and other putative virulence genes (e.g., eae and ehxA) in STEC strains. Although the use of PCR improves the detection of non-O157 STEC serotypes which may be the cause of HUS, it may also lead to detection of a range of STEC strains with a probable low potential for causing severe disease in humans (10, 17, 19). In many cases, we therefore still do not know how to differentiate between high- and low-risk STEC strains, and it is a challenge to make a reliable assessment of the clinical and public health risk related to the diagnosis of non-O157 STEC infections.
The aim of this study was to present the results of PCR-based diagnosis of STEC infection from patient stool specimens during the period 1996-2011 and to search for differences between HUS and HUS-associated compared to non-HUS-associated STEC isolates based on serotypes and analysis of selected potential virulence genes.
MATERIALS AND METHODS
Bacterial strains and clinical information.
All STEC isolates included in the present study were retrieved from patient stool specimens in the years 1996 through 2011 at the Department of Medical Microbiology, St. Olavs Hospital, Trondheim, Norway. From 1996 to 2011 the laboratory routine was to analyze stool specimens from children <2 years old for stx1 and stx2 (and eae from 2000) irrespective of clinical information by PCR and to analyze stool specimens from patients in age groups >2 years old if there was information on HUS or bloody diarrhea. In addition, specimens from persons epidemiologically associated with a HUS case or a STEC outbreak were analyzed for STEC. Based on data from the laboratory information system, stx1 and/or stx2 was detected in mixed stool cultures from 150 patients during the study period. Among these, 20 patients were excluded from the study because the laboratory did not succeed in obtaining STEC isolates in pure cultures, whereas for the remaining 130 patients, STEC isolates were identified in pure cultures. Another eight stx-negative (eae-positive) E. coli isolates were included in the study because they were isolated from patients with HUS or bloody diarrhea or were epidemiologically linked to a HUS case and were of the same MLVA genotype as the STEC isolate from that case (Table 1). STEC strains that have lost stx genes are often termed EHEC/STEC-lost Shiga toxin (LST) (20). In total, 138 strains were included in the study.
TABLE 1.
Characteristics of stx-negative strains, isolated at St. Olavs Hospital, Trondheim, Norway, during the 1996-2011 period, classified as STEC-lost Shiga toxin in this study
| Strain | stx2 subtype | eae | Serotype | Reason the strain was included in the study |
|---|---|---|---|---|
| St. Olav49 | STEC-LSTa | + | O103:H25 | HUSb |
| St. Olav59 | STEC-LST | + | O103:H25 | HUS |
| St. Olav75 | 2ac | + | SF O157:H- | Outbreak investigation |
| St. Olav77 | STEC-LST | + | SF O157:H- | HUS |
| St. Olav84 | STEC-LST | + | SF O157:H- | Outbreak investigation |
| St. Olav97 | STEC-LST | + | O145:H?d | Outbreak investigation |
| St. Olav131 | STEC-LST | + | O103:H25 | BDe |
| St. Olav154 | 2b | − | 0f | Previously positive for stx2b |
| St. Olav156 | STEC-LST | + | 0f | BD |
| St. Olav165 | STEC-LST | + | O145:H28 | BD |
eae-positive E. coli isolate classified as STEC that may have lost its stx genes (STEC-lost Shiga toxin [LST]).
HUS, hemolytic uremic syndrome.
Strain previously positive for stx2, which after storage was found to be stx2 negative.
H?, motile but unknown H-type.
BD, bloody diarrhea.
Serotype 0, the strains did not belong to any of the serogroups tested for in the study.
In Norway, STEC infection is notifiable to the Norwegian Surveillance System for Communicable Diseases (MSIS) at the Norwegian Institute of Public Health (NIPH), where clinical information on the patients and results from laboratory analyses of the bacterial strains are stored. We collected data from the MSIS on clinical symptoms (HUS, bloody diarrhea, diarrhea, or no symptoms), age, and sex and correlated these data with laboratory results.
The study was approved by the Regional Committee for Medical and Health Research Ethics, REC South-East (REC number 2011/2314).
Primary detection and identification of STEC.
stx1, stx2, and eae (from the year 2000) were detected by a two-step procedure where PCRs for the stx1, stx2, and eae genes first were done in mixed cultures from a stool specimen and thereafter repeated on subcultures of discrete colonies from positive specimens with the aim of identifying STEC strains in pure cultures. STEC isolate culturing was done by standard methods, including SMAC agar, and E. coli was identified by standard biochemical tests (API 10S/20E; bioMérieux, Marcy l'Etoile, France). During the period 1996-2004, screening for stx1 and stx2 was performed using primers and amplification conditions as described by Brian et al. (21). In 2004, conventional PCRs for stx1 and stx2 were replaced by multiplex real-time PCRs (for primers, see Table S1 in the supplemental material). DNA isolation methods, amplification conditions, PCR reagents, and PCR instruments varied in the years 2004-2011.
PCR for eae was done using the AE13 and AE14 primers, and amplification conditions were as described by Gannon et al. (22) from 2000 to 2004 and as described by Nielsen and Andersen (23) from 2004 to 2008. Thereafter detection of eae was done by real-time PCR with the primers described in Table S1 in the supplemental material. Confirmation of stx1, stx2, and eae was done at the National Reference Laboratory for Enteropathogenic Bacteria (NRL) at the NIPH (24, 25).
Serotyping.
Initial serogrouping was performed with O antisera using polyspecific anti-coli I, II, and III and monospecific O antisera for the O serogroups O26, O103, O111, O145, and O157, as described by the manufacturer (Sifin, Germany). Later, more extensive serotyping was done at the NRL, NIPH, using monospecific O:K and H antisera covering altogether 44 O serogroups, including O26, O103, O111, O121, O145, and O157 and 8 H antigens (in-house antisera and antisera from Sifin and SSI, Denmark).
stx2 subtyping and MLVA genotyping.
The stx2 subtype was determined at the NRL, NIPH, using modifications of previously published methods for PCR-restriction fragment length polymorphism (RFLP) and sequencing (8, 26, 27) and by PCR (9).
Two MLVA protocols, one for the O157 serogroup and one for all E. coli isolates, were used for MLVA typing (28–30) at the NRL, NIPH.
Verification of stx1, stx2, and eae and detection of potential virulence genes.
To verify the primary PCR results, we repeated PCRs for stx1, stx2, and eae for all strains included in the study. For PCR analyses, bacterial strains were grown overnight on MacConkey agar. One colony of bacterial cells was suspended in 100 μl lysis buffer (50 mM KCl, 10 mM Tris-HCl [pH 8.3], 2.5 mM MgCl2, 0.1 mg/ml gelatin, 0.45% NP-40, and 0.45% Tween 20) and 100 μl Tris-EDTA (TE) buffer solution (pH 7.4) and boiled for 15 min at 95°C. After centrifugation at 13,000 rpm for 1 min, the supernatant was used directly for PCR analysis.
Real-time PCRs for stx1, stx2, and eae were done using the primers described in Table S1 in the supplemental material and PerfeCTa Multiplex qPCR Supermix, UNG (Quanta Biosciences, Gaithersburg, MD, USA) as described by the manufacturer. Real-time PCR was performed in a CFX instrument (Bio-Rad, Hercules, CA, USA), in a 20-μl volume with cycling conditions for stx1 and stx2 as follows: 95°C for 3 min and then 40 cycles with denaturation at 95°C for 10 s and annealing at 58°C for 10 s before elongation at 72°C for 10 s. The PCR for eae was done using the following cycling conditions: 95°C for 3 min and then 40 cycles with denaturation at 95°C for 10 s and annealing at 50°C for 10 s before elongation at 72°C for 10 s.
Analyses for the ehxA, ent/espL2, nleB, nleE, nleF, nleH1-2, and nleA genes were done using the primers described by Bugarel et al. (31). In addition, PCR for the espK gene was done using the primers described by Bugarel et al. (12), and the efa-1/lifA gene was analyzed with the 88AT and 88TN primers from Nicholls et al. (32). A subset of strains was also tested for efa-1/lifA using an alternative efa-1 primer pair (see Table S1 in the supplemental material). The PCRs were done in singleplex in a 20-μl volume using the SsoFast EvaGreen Supermix (Bio-Rad), as described by the manufacturer. The cycling conditions for the nleB and nleE genes were 98°C for 2 min and then 40 cycles with 95°C for 5 s and 60°C for 5 s, and the cycling conditions for ehxA, ent/espL2, nleF, nleH1-2, nleA, espK, and efa-1/lifA were 95°C for 3 min and then 40 cycles with 95°C for 10 s and 57°C for 30 s.
HUS-associated strains.
A STEC strain was classified as HUS associated if it either was isolated from a patient with HUS or was epidemiologically linked to a HUS case and was of the same MLVA genotype as the STEC isolate from that case.
Statistical methods.
Fisher's exact test was used for statistical calculations. A P value of ≤0.05 was considered statistical significant.
Cluster analysis of virulence genes with construction of dendrograms was performed by BioNumerics version 6.6 (Applied Maths NV, Sint-Martens-Latem, Belgium) using the Dice correlation and the unweighted-pair group method using average linkages (UPGMA).
RESULTS
The present study included 138 patients among a total of 12,651 patients tested in whom STEC infection had been diagnosed by PCR, and STEC strains had successfully been isolated in pure cultures (positive rate, 1.09%) in the period 1996-2011 at the Department of Medical Microbiology, St. Olavs Hospital, Trondheim, Norway. Stool specimens from children <2 years old were most frequently tested for STEC, and this age group was also the most common among the 138 patients diagnosed with STEC infection (36.3%) (Table 2). Sixty-nine of the patients with STEC infections were female and 69 were male. Eleven patients had HUS, while bloody diarrhea was recorded for 9 patients, and nonbloody diarrhea was recorded for 68 patients. All HUS patients were ≤5 years old (P = 0.007) (Table 2). Six of the HUS patients were female and 5 were male (Table 3).
TABLE 2.
Age distribution and other characteristics of patients with STEC infections diagnosed at St. Olavs Hospital during the 1996-2011 period
| Age group (yr) | No. (%) of patients: |
Positive rate | Sex (no.) |
Clinical presentation in patients with STEC (no.): |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| With STEC | Tested | Female | Male | HUSa | BDb | Diarrhea | Asymptomaticc | NDd | ||
| <2 | 50 (36.3) | 6,860 (54.2) | 0.73 | 25 | 25 | 6 | 5 | 25 | 5 | 9 |
| 2–4 | 32 (23.2) | 1,301 (10.3) | 2.46 | 17 | 15 | 4 | 0 | 15 | 6 | 7 |
| 5–9 | 9 (6.5) | 344 (2.7) | 2.62 | 2 | 7 | 1 | 1 | 1 | 2 | 4 |
| ≥10 | 47 (34.0) | 4,146 (32.8) | 1.13 | 25 | 22 | 0 | 3 | 27 | 7 | 10 |
| Total | 138 (100.0) | 12,651 (100.0) | 1.09 | 69 | 69 | 11 | 9 | 68 | 20 | 30 |
HUS, hemolytic uremic syndrome.
BD, bloody diarrhea.
Some of the strains were from persons tested in outbreak investigations.
ND, no clinical information. Some of these strains were from outbreak investigations.
TABLE 3.
Characteristics of STEC strains from patients with hemolytic uremic syndrome and information from the patients with STEC infections diagnosed at St. Olavs Hospital during the 1996-2011 period
| Strain | yr | Sexa | Age (yr) | Serotype | stx2 subtype |
|---|---|---|---|---|---|
| St. Olav26 | 2002 | F | 4 | O26:H- | 2a |
| St. Olav49 | 2006 | F | 1 | O103:H25 | —b |
| St. Olav56 | 2006 | M | 2 | SF O157:H- | 2a |
| St. Olav59 | 2006 | M | 1 | O103:H25 | — |
| St. Olav77 | 2008 | F | 1 | SF O157:H- | — |
| St. Olav80 | 2009 | F | 3 | SF O157:H- | 2a |
| St. Olav81 | 2009 | M | 5 | SF O157:H- | 2a |
| St. Olav91 | 2009 | F | 1 | O121:H19 | 2a |
| St. Olav100 | 2009 | M | 1 | O145:H?c | 2a |
| St. Olav164 | 2011 | F | 2 | O145:H? | 2a |
| St. Olav166 | 2011 | M | 0 | SF O157:H- | 2a |
F, female; M, male.
Strains were negative for stx2a. The O103:H25 strains (St. Olav49 and St. Olav59) were part of a national outbreak in 2006 where stx2a was identified in some of the other strains included in the same MLVA genotype cluster (50). St. Olav77 was part of a small family outbreak with St. Olav75 (see Table 1).
H?; motile but unknown H-type.
The STEC strains included in the present study had the following distribution of serogroups: O157, n = 29 (21.0%); other common STEC serogroups, n = 69 (50.0%), including O145 (n = 28), O103 (n = 21), and O26 (n = 11); and less common STEC serogroups, n = 17 (12.3%). Twenty-three (16.7%) strains, one of which was Orough, did not belong to any of the serogroups tested (Table 4).
TABLE 4.
Serogroups of 138 HUS-associated and non-HUS-associated STEC strains isolated by PCR and culture at St. Olavs Hospital, Trondheim, Norway, during the 1996-2011 period
| Serogroup | No. (%) of STEC strains |
||
|---|---|---|---|
| HUS associated | Non-HUS associated | Total | |
| O145 | 7 (29.1) | 21 (18.4) | 28 (20.3) |
| O103 | 2 (8.4) | 19 (16.6) | 21 (15.3) |
| O157 | 0 (0) | 20 (17.5) | 20 (14.5) |
| O26 | 1 (4.2) | 10 (8.8) | 11 (7.9) |
| SF O157 | 9 (37.5) | 0 (0) | 9 (6.5) |
| O121 | 5 (20.8) | 4 (3.5) | 9 (6.5) |
| Othera | 0 (0) | 40 (35.1) | 40 (28.9) |
| Total | 24 (100) | 114 (100) | 138 (100) |
Other serogroups: O2 (n = 1), O76 (n = 1), O91 (n = 1), O104 (n = 3), O111 (n = 1), O113 (n = 3), O117 (n = 1), O118 (n = 1), O119 (n = 1), O128 (n = 3), O177 (n = 1), and unknown O serogroups (n = 23), of which one strain was Orough.
A total of 128 strains contained the stx1 and/or stx2 gene, and 108 strains contained eae (Table 5). A combination of stx1 and stx2 was found in 21 (15.2%) strains, while stx1 only was found in 57 (41.3%) strains and stx2 only in 50 (36.2%) strains. The stx2 subtypes most frequently detected were stx2a (n = 36) and stx2c (n = 18) (Table 5). Two strains which had previously been confirmed to be stx2 positive were negative for stx after frozen storage (Table 1).
TABLE 5.
Virulence genes identified by PCR analysis and stx2 subtypes of 138 HUS-associated and non-HUS-associated STEC strains isolated at St. Olavs Hospital, Trondheim, Norway, during the 1996-2011 period
| Virulence gene | No. ([%]) of genes in: |
P | ||
|---|---|---|---|---|
| HUS-associated STEC (n = 24) | Non-HUS associated STEC (n = 114) | Total | ||
| stx1 | 0 (0) | 57 (50) | 57 (41.3) | <0.0001 |
| stx1 + stx2 | 0 (0) | 21 (18.4) | 21 (15.2) | 0.02 |
| stx2 | 18 (75)a | 32 (28)b | 50 (36.2) | <0.0001 |
| eae | 24 (100) | 84 (74) | 108 (78.3) | 0.002 |
| ehxA | 24 (100) | 99 (87) | 123 (89.1) | 0.073 |
| nleB | 24 (100) | 82 (72) | 106 (76.8) | 0.001 |
| nleE | 24 (100) | 81 (71) | 105 (76.1) | 0.001 |
| ent | 24 (100) | 82 (72) | 106 (76.8) | 0.001 |
| efa-1/lifA | 24 (100) | 73 (64) | 97 (70.3) | 0.0001 |
| nleA | 24 (100) | 63 (55) | 87 (63) | <0.0001 |
| nleF | 24 (100) | 84 (74) | 108 (78.3) | 0.002 |
| nleH1-2 | 24 (100) | 85 (75) | 109 (79) | 0.004 |
| espK | 23 (96) | 83 (73) | 106 (76.8) | 0.002 |
| Total | 24 (100) | 114 (100) | 138 (100) | |
| stx2 subtype | ||||
| stx2a | 18 (75)a | 18 (15.8) | 36 (50.7) | P < 0.0001 |
| stx2a + stx2c | 0 (0) | 4 (3.5) | 4 (5.6) | NDc |
| stx2b | 0 (0) | 9 (7.9)b | 9 (12.7) | ND |
| stx2c | 0 (0) | 18 (15.8) | 18 (25.4) | ND |
| stx2d | 0 (0) | 2 (1.8) | 2 (2.8) | ND |
| stx2g | 0 (0) | 1 (0.9) | 1 (1.4) | ND |
| ND | 0 (0) | 1 (0.9)d | 1 (1.4) | ND |
| Total | 18 (75) | 53 (47) | 71 100 | |
This number does not include one SF O157:H- strain (St. Olav75; see Table 1) previously found to be positive for stx2a.
This number does not include one strain of unknown serotype (St. Olav154; see Table 1) previously found to be positive for stx2b.
ND, not determined.
The stx2 phage was lost in the STEC strain at arrival at the Norwegian Institute of Public Health and therefore was not stx2 subtyped.
PCRs for ehxA, nleB, nleE, ent, efa-1/lifA, nleA, nleF, nleH1-2, and espK revealed that ehxA was the most frequent (n = 123, 89.1%), whereas nleA was the least frequent (n = 87, 63.0%) in the 138 STEC strains examined (Table 5). All of the potential virulence genes were found among strains of serogroups O145, O103, O157, O26, O121, SF O157, and O111 (see Table S2 in the supplemental material), whereas 11 strains with other serogroups contained none of the genes analyzed in the study.
The STEC strains isolated from patients with HUS belonged to the serotypes SF O157:H-, O145:H? (H unknown), O103:H25, O26:H-, and O121:H19 (Table 3). Another 13 STEC strains were epidemiologically linked to an HUS case and of the same MLVA genotype as the STEC strain isolated from that case. Altogether 24 strains were therefore classified as HUS associated. Among these strains SF O157:H- and O145:H? were the most common (Table 4). The age of the patient (≤5 years old) was significantly associated with HUS-associated STEC infection, as 20 of the 24 HUS-associated strains were isolated from children within this age group (P = 0.035). The eae gene was present in all of the HUS-associated strains compared with its presence in 84/114 (73.7%) non-HUS-associated strains (P = 0.002) (Table 5). stx2 was significantly more frequent among HUS-associated than non-HUS-associated strains (P = 0.013) (Table 5), especially the stx2a subtype, which was present in 18/24 HUS-associated strains compared with 22/114 non-HUS-associated strains (P < 0.0001) (Table 5). In contrast, stx1 was not detected in any of the HUS-associated strains, but was present in 78 of the non-HUS-associated strains (P < 0.0001) (Table 5). The genes ehxA, nleB, nleE, ent, efa-1/lifA, nleA, nleF, and nleH1-2 were present in all HUS-associated strains, whereas espK was present in all but one of these strains. All of the potential virulence genes analyzed, except ehxA, were significantly more frequent among HUS-associated than non-HUS-associated strains (P < 0.05 for each gene) (Table 5).
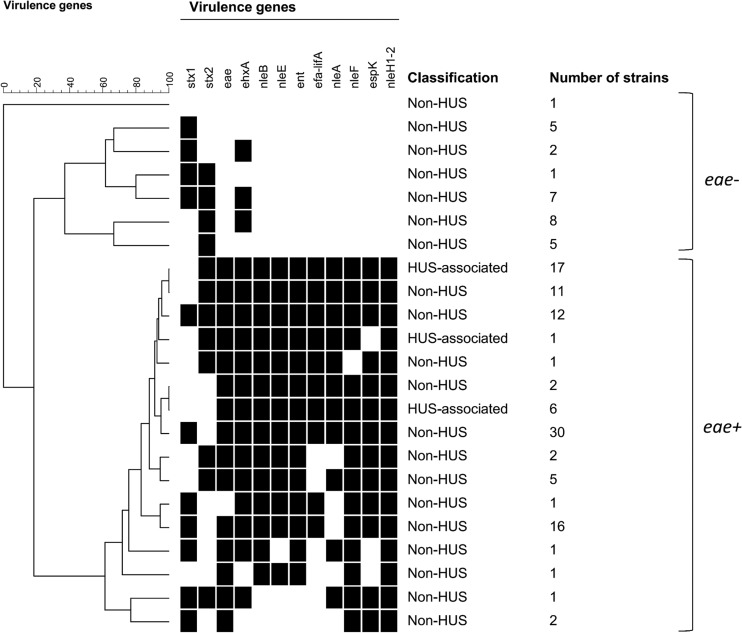
By cluster analysis of potential virulence genes, eae-positive and eae-negative STEC strains were separated into two main clusters (Fig. 1; see also Fig. S1 in the supplemental material). One exception was an eae-negative, stx1-positive strain (St. Olav12) that clustered with the group of eae-positive strains due to the presence of some of the potential virulence genes in this strain. In the cluster of 29 eae-negative STEC strains, ehxA was the only potential virulence gene present (17 strains) (Fig. 1; see also Fig. S1). All eae-positive strains harbored more than three of the potential virulence genes investigated (Fig. 1; see also Fig. S1). Although all the HUS-associated strains clustered among strains which were eae and stx2 positive but stx1 negative, non-HUS-associated strains were also found in the same cluster (Fig. 1; see also Fig. S1).
FIG 1.
Cluster analysis of potential virulence genes in STEC strains. eae-negative and eae-positive STEC strains were mainly separated into two clusters. One exception was one eae-negative strain that clustered among the eae-positive strains due to the presence of some of the potential virulence genes. All HUS-associated strains clustered among the eae-positive strains and harbored all of the potential virulence genes investigated in the study. For further details (serotype, stx2 subtype, etc.), see Fig. S1 in the supplemental material.
The 109 non-O157 STEC strains were distributed in 48 distinct MLVA genotypes (see Fig. S2 in the supplemental material). Thirty-one of these MLVA genotypes were represented by only one strain each, including one O26:H- strain from an HUS patient. The other 17 MLVA genotypes, with 78 non-O157 strains, were found in 2 to 15 strains, respectively. Some of these MLVA genotypes included STEC strains from local or national outbreaks. Among the 29 O157 and SF O157 STEC strains, 17 distinct MLVA genotypes were observed (see Fig. S3 in the supplemental material). Twelve of these MLVA genotypes were present in single strains, including two SF O157:H- strains from HUS patients, while five genotypes included two to five strains. For the two MLVA methods, some of the HUS-associated strains showed the same MLVA genotype as non-HUS-associated strains.
DISCUSSION
In this study, we present the results of STEC infection diagnosis in our laboratory based on PCRs for the stx1, stx2, and eae genes in cultures from stool samples in the years 1996-2011. During this period stx genes were detected in stool samples from 150 patients, and STEC or STEC-LST strains were isolated in pure cultures from 138 patients (Table 2). Similar to what has been reported elsewhere, the highest number of STEC infections was diagnosed in children <5 years old (4, 33). In our study, this may partly be explained by the routine of the laboratory to analyze all stool specimens from children <2 years old for STEC, while samples from older age groups were analyzed only if there was a clinical suspicion of HUS or bloody diarrhea or the samples were part of a STEC outbreak investigation. However, the fact that a high number of STEC infections were also detected in children 2 to 4 years old (Table 2) in whom tests for STEC were done only because of a specific suspicion, supports the notion that STEC infection is most common in young age groups.
As shown in Table 4, the STEC serogroups isolated most often in this study, including strains associated with HUS, belonged to the STEC serogroups frequently implicated in severe disease and outbreaks described elsewhere (5, 34). However, only 20 (14.5%) of the 138 STEC strains belonged to serotype NSF O157:H7, the only STEC serotype that is selected for by SMAC agar. Seventy-eight (56.5%) STEC strains belonged to other common STEC serogroups, including SF O157, that would easily have been missed on SMAC agar since they could not be differentiated from the majority of commensal E. coli, and 40 strains belonged to serogroups not common for STEC or unknown serogroups. Consequently, use of PCRs resulted in detection of a high number of non-O157 STEC strains, both of STEC serogroups that based on current knowledge may be viewed as high-risk strains (35), and of non-O157 STEC serogroups that most likely do not represent a high risk for HUS development.
More than half (55%) of the patients infected with SF O157:H- STEC developed HUS, and this serotype was the most common serotype isolated from HUS patients in this study (Table 3). These results are in line with previous reports suggesting that there is a high risk for development of HUS with SF O157 infection (36, 37). Six (54.5%) of the 11 HUS cases were caused by non-O157 STEC serotypes, while NSF O157, which is the most common STEC serotype causing HUS worldwide (3–5), was not the cause of any of the HUS cases in this study. During the same time period, this serotype was isolated from four cases of HUS in other parts of Norway (MSIS).
In this study, HUS-associated STEC strains contained the following characteristics: (i) all of them, except STEC-LST strains, carried stx2 (stx2a), (ii) all harbored eae, (iii) all but one contained the other nine potential virulence genes tested, and (iv) all belonged to STEC serogroups frequently associated with severe disease, many of them non-O157. Stx2a and intimin are important virulence factors in STEC strains that have been associated with severe disease (3, 38). Analysis of the presence or absence of virulence genes in this study revealed that eae and stx2a were significantly more frequent in HUS-associated than non-HUS-associated strains, whereas STEC strains containing stx1 were exclusively associated with non-HUS infection (Table 5). While the ehxA gene has been regarded as an important virulence marker in STEC infection and has been reported to be a marker of “typical EHEC” (14), it was the only potential virulence gene analyzed that was not significantly more frequent in HUS-associated than non-HUS-associated strains in this study. ehxA was also the sole potential virulence gene present among the subset of eae-negative STEC strains (Fig. 1; see also Fig. S1 in the supplemental material). However, although most of the potential virulence genes were significantly more present among HUS-associated than non-HUS-associated strains, several of the non-HUS-associated STEC strains contained a virulence gene profile similar to that for the group of HUS-associated strains. In particular, non-HUS-associated STEC strains of the serogroups O145, O103, O157 (NSF and SF), O26, O121, and O111 contained a high number of the potential virulence genes, which is in line with previous reports (39, 40). This made reliable differentiation between HUS-associated and non-HUS-associated STEC strains based on serotype and potential virulence genes impossible in this strain collection.
Two of the STEC strains were negative for stx after frozen storage. Although in general only stx-positive strains are regarded as STEC, it is well known that STEC may lose its stx encoding prophage, either in the course of an infection or upon handling in the laboratory (20, 41, 42).
Although eae- and stx1-positive, stx2-negative STEC strains have been isolated from patients with HUS (43, 44), there are to our knowledge no reports of outbreaks of severe disease with such bacteria. Furthermore, in line with reports from other regions in Norway (L. T. Brandal, A. L. Wester, H. Lange, I. Løbersli, B.-A. Lindstedt, G. Kapperud, and L. Vold, unpublished data) and other countries (5, 45–47), our results support the notion that infections with STEC strains that do not belong to common STEC serogroups and lack eae and/or stx2a even if stx1 is present represent a low risk for HUS development. Therefore, based on the results from the present study, it seems safe to suggest a classification of stx1-positive, stx2-negative STEC strains with a low risk for HUS development.
In the current study, it was difficult to assess the risk profile of STEC strains with a serotype and virulence profile similar to that of HUS-associated strains. It may be that such strains are actually virulent and should be interpreted as STEC with a high risk for HUS development. Alternatively, there may be other bacterial characteristics not analyzed in these strains that may be of importance for the virulence potential of STEC. In addition to bacterial virulence, young age (48) of the infected person is a risk factor for HUS development. This is also evident in this study where all HUS cases were in children ≤5 years old. Most likely, development of HUS may be influenced by other host factors as well.
As expected MLVA genotyping revealed that STEC isolates of some MLVA genotypes clustered with isolates from other parts of the country, in relation to local or national outbreaks. The largest local outbreak occurred in a kindergarten in 2009 where 15 isolates of STEC serotype O145:H28 were of the same MLVA genotype. Although the index child presented with bloody diarrhea, none of the children affected in that outbreak developed HUS (49).
Our laboratory was one of the first in Norway to introduce PCR for detection of human STEC infection. During the period from 1996 through 2011, a higher number of STEC infections were detected in our laboratory than in other regions of Norway, as more than one-fourth of the cases of STEC infections on the national level were detected here (MSIS), although less than one-tenth of the population live in this region. The high detection rate of STEC infections in our laboratory compared to rates in other parts of Norway most likely was, at least to some extent, due to early introduction of PCR in our laboratory. However, the fact that during this period, a higher proportion than expected of HUS cases (11 of 53 [20.8%], data from MSIS) also were from central Norway where our hospital is located may indicate that there may be epidemiological differences in the risks of STEC infection and disease between different regions in Norway.
Starting with PCR for STEC detection in 1996, the routine in our laboratory was to analyze all stool specimens from children <2 years for STEC irrespective of the clinical diagnosis and only those from older children and adults on clinical suspicion. One might expect that this practice could have led to identification of less virulent STEC strains not related to clinical disease in the younger age group. However, a comparison of the virulence profile of STEC strains isolated from children <2 years of age compared with that of STEC strains isolated from older children and adults does not support this idea. Strains from the younger age group contained at least as many virulence genes as those isolated from older children and adults (see Table S3 in the supplemental material).
A limitation of this study was that the laboratory routine to use different indications for testing of samples for STEC dependent on the age of the patient was not always followed. This was reflected in that the 39 patients diagnosed with STEC infections in the present study who were >2 years of age had diagnoses other than HUS and bloody diarrhea and were not part of an outbreak investigation. Furthermore, information regarding bloody diarrhea may have been incomplete since bloody diarrhea was recorded for only 9 of the patients, while there was no information on bloody stools in 68 other patients with diarrhea. For these cases, the information given by the referring physician might have been incomplete, or such data were not always recorded or updated in the patient information database. Another weakness was that in the first part of the study period, only the stx1 and stx2 genes were analyzed, whereas eae was included from year 2000 onward. In addition, different primers, reagents, and equipment were used for PCR analysis through the study period, including a switch from conventional to real-time PCR. Although different primers were used for detection of stx1 and stx2, the primers used were designed to detect all variants of the stx genes, except stx2f. The use of different PCR methods for STEC detection might potentially have had an impact on which STEC strains were detected. However, despite the variations in the PCR protocols used throughout the study period, the strain collection represents an unselected group of STEC infections diagnosed by PCR analysis of the stx and eae genes in a hospital laboratory throughout a period of 16 years. In this study, we tested the STEC strains for a limited number of virulence genes. Although through this virulence characterization of the STEC strains, we did not disclose new knowledge, we were able to confirm results from previous studies regarding STEC virulence in a unselected collection of STEC isolates from hospital routine diagnosis based on PCR (12–14, 31).
In summary, STEC infection was diagnosed by PCR and STEC strains were isolated from stool samples from 138 (1.09%) of 12,651 patients tested at St. Olavs Hospital, Norway, during the period 1996-2011. More than half of the patients diagnosed with STEC infections were <5 years old. Eleven patients (all ≤5 years old) had HUS, but no one died. All HUS patients were infected with STEC strains of serogroups frequently involved in severe disease and outbreaks elsewhere. Six of the 11 HUS patients were infected with non-O157 serogroups. Twenty-four STEC strains were classified as HUS associated. Young age (≤5 years old) and STEC strains containing eae and stx2a were significantly associated with HUS (P < 0.05 for each parameter), while STEC strains containing stx1 were associated with non-HUS-associated STEC infections (P < 0.05). Also, the other potential virulence genes analyzed, except for ehxA, were significantly associated with HUS (P < 0.05 for each gene). However, as they were also present in some of the non-HUS-associated STEC strains, these genes could not reliably differentiate between HUS-associated and non-HUS-associated STEC strains.
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff of the Department of Medical Microbiology at St. Olavs Hospital and the Department of Foodborne Infections at the Norwegian Institute of Public Health for skillful technical assistance. We also thank Frode Width Gran and Hilde Fossum of the Department of Medical Microbiology at St. Olavs Hospital for help with information on STEC strains and patients included in the study, Anne Nor for design of the primers SLTI Rnew, SLTI TaqMan, SLTIIFnew, SLTIIRnew, and SLTII TaqMan, and Tonje Haukeberg for design of the primers eae-Fny, eae-Rny, and eae-P.
The funding for this project was part of Ph.D. grant 81723800 to K.H. from the Norwegian University of Science and Technology.
Footnotes
Published ahead of print 11 June 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00453-14.
REFERENCES
- 1.Karmali MA, Steele BT, Petric M, Lim C. 1983. Sporadic cases of haemolytic-uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet i:619–620. [DOI] [PubMed] [Google Scholar]
- 2.Riley LW, Remis RS, Helgerson SD, McGee HB, Wells JG, Davis BR, Hebert RJ, Olcott ES, Johnson LM, Hargrett NT, Blake PA, Cohen ML. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681–685. 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 3.Gyles CL. 2007. Shiga toxin-producing Escherichia coli: an overview. J. Anim. Sci. 85(13 Suppl):E45−E62. 10.2527/jas.2006-508. [DOI] [PubMed] [Google Scholar]
- 4.European Centre for Disease Prevention and Control. 2013. Annual Epidemiological Report 2012. Reporting on 2010 surveillance data and 2011 epidemic intelligence data. European Centre for Disease Prevention and Control, Stockholm, Sweden. [Google Scholar]
- 5.Gould LH, Mody RK, Ong KL, Clogher P, Cronquist AB, Garman KN, Lathrop S, Medus C, Spina NL, Webb TH, White PL, Wymore K, Gierke RE, Mahon BE, Griffin RM, Emerging Infections Program Foodnet Working Group 2013. Increased recognition of non-O157 Shiga toxin-producing Escherichia coli infections in the United States during 2000-2010: epidemiologic features and comparison with E. coli O157 infections. Foodborne Pathog. Dis. 10:453–460. 10.1089/fpd.2012.1401. [DOI] [PubMed] [Google Scholar]
- 6.Bielaszewska M, Friedrich AW, Aldick T, Schurk-Bulgrin R, Karch H. 2006. Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clin. Infect. Dis. 43:1160–1167. 10.1086/508195. [DOI] [PubMed] [Google Scholar]
- 7.Friedrich AW, Bielaszewska M, Zhang WL, Pulz M, Kuczius T, Ammon A, Karch H. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74–84. 10.1086/338115. [DOI] [PubMed] [Google Scholar]
- 8.Persson S, Olsen KE, Ethelberg S, Scheutz F. 2007. Subtyping method for Escherichia coli Shiga toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J. Clin. Microbiol. 45:2020–2024. 10.1128/JCM.02591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheutz F, Teel LD, Beutin L, Pierard D, Buvens G, Karch H, Mellmann A, Caprioli A, Tozzoli R, Morabito S, Strockbine NA, Melton-Celsa AR, Sanchez M, Persson S, O'Brien AD. 2012. Multicenter evaluation of a sequence-based protocol for subtyping shiga toxins and standardizing stx nomenclature. J. Clin. Microbiol. 50:2951–2963. 10.1128/JCM.00860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karmali MA, Mascarenhas M, Shen S, Ziebell K, Johnson S, Reid-Smith R, Isaac-Renton J, Clark C, Rahn K, Kaper JB. 2003. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J. Clin. Microbiol. 41:4930–4940. 10.1128/JCM.41.11.4930-4940.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bugarel M, Martin A, Fach P, Beutin L. 2011. Virulence gene profiling of enterohemorrhagic (EHEC) and enteropathogenic (EPEC) Escherichia coli strains: a basis for molecular risk assessment of typical and atypical EPEC strains. BMC Microbiol. 11:142. 10.1186/1471-2180-11-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coombes BK, Wickham ME, Mascarenhas M, Gruenheid S, Finlay BB, Karmali MA. 2008. Molecular analysis as an aid to assess the public health risk of non-O157 Shiga toxin-producing Escherichia coli strains. Appl. Environ. Microbiol. 74:2153–2160. 10.1128/AEM.02566-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bugarel M, Beutin L, Martin A, Gill A, Fach P. 2010. Micro-array for the identification of Shiga toxin-producing Escherichia coli (STEC) seropathotypes associated with hemorrhagic colitis and hemolytic uremic syndrome in humans. Int. J. Food Microbiol. 142:318–329. 10.1016/j.ijfoodmicro.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 15.Johnson TJ, Nolan LK. 2009. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 73:750–774. 10.1128/MMBR.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.March SB, Ratnam S. 1986. Sorbitol-MacConkey medium for detection of Escherichia coli O157:H7 associated with hemorrhagic colitis. J. Clin. Microbiol. 23:869–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gould LH, Bopp C, Strockbine N, Atkinson R, Baselski V, Body B, Carey R, Crandall C, Hurd S, Kaplan R, Neill M, Shea S, Somsel P, Tobin-D'Angelo M, Griffin PM, Gerner-Smidt P. 2009. Recommendations for diagnosis of Shiga toxin-producing Escherichia coli infections by clinical laboratories. MMWR Recomm. Rep. 58(RR-12):1–14. [PubMed] [Google Scholar]
- 18.Gould LH. 2012. Update: recommendations for diagnosis of Shiga toxin-producing Escherichia coli infections by clinical laboratories. Clin. Microbiol. Newsl. 34:75–83. 10.1016/j.clinmicnews.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Karch H, Bielaszewska M, Bitzan M, Schmidt H. 1999. Epidemiology and diagnosis of Shiga toxin-producing Escherichia coli infections. Diagn. Microbiol. Infect. Dis. 34:229–243. 10.1016/S0732-8893(99)00031-0. [DOI] [PubMed] [Google Scholar]
- 20.Bielaszewska M, Kock R, Friedrich AW, von Eiff C, Zimmerhackl LB, Karch H, Mellmann A. 2007. Shiga toxin-mediated hemolytic uremic syndrome: time to change the diagnostic paradigm? PLoS One 2:e1024. 10.1371/journal.pone.0001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brian MJ, Frosolono M, Murray BE, Miranda A, Lopez EL, Gomez HF, Cleary TG. 1992. Polymerase chain reaction for diagnosis of enterohemorrhagic Escherichia coli infection and hemolytic-uremic syndrome. J. Clin. Microbiol. 30:1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gannon VP, Rashed M, King RK, Thomas EJ. 1993. Detection and characterization of the eae gene of Shiga-like toxin-producing Escherichia coli using polymerase chain reaction. J. Clin. Microbiol. 31:1268–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen EM, Andersen MT. 2003. Detection and characterization of verocytotoxin-producing Escherichia coli by automated 5′ nuclease PCR assay. J. Clin. Microbiol. 41:2884–2893. 10.1128/JCM.41.7.2884-2893.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brandal LT, Lindstedt BA, Aas L, Stavnes TL, Lassen J, Kapperud G. 2007. Octaplex PCR and fluorescence-based capillary electrophoresis for identification of human diarrheagenic Escherichia coli and Shigella spp. J. Microbiol. Methods 68:331–341. 10.1016/j.mimet.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 25.Brandal LT, Sekse C, Lindstedt BA, Sunde M, Lobersli I, Urdahl AM, Kapperud G. 2012. Norwegian sheep are an important reservoir for human-pathogenic Escherichia coli O26:H11. Appl. Environ. Microbiol. 78:4083–4091. 10.1128/AEM.00186-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jelacic JK, Damrow T, Chen GS, Jelacic S, Bielaszewska M, Ciol M, Carvalho HM, Melton-Celsa AR, O'Brien AD, Tarr PI. 2003. Shiga toxin-producing Escherichia coli in Montana: bacterial genotypes and clinical profiles. J. Infect. Dis. 188:719–729. 10.1086/376999. [DOI] [PubMed] [Google Scholar]
- 27.Rüssmann H, Schmidt H, Heesemann J, Caprioli A, Karch H. 1994. Variants of Shiga-like toxin II constitute a major toxin component in Escherichia coli O157 strains from patients with haemolytic uraemic syndrome. J. Med. Microbiol. 40:338–343. 10.1099/00222615-40-5-338. [DOI] [PubMed] [Google Scholar]
- 28.Lindstedt BA, Vardund T, Kapperud G. 2004. Multiple-Locus Variable-Number Tandem-Repeats Analysis of Escherichia coli O157 using PCR multiplexing and multi-colored capillary electrophoresis. J. Microbiol. Methods 58:213–222. 10.1016/j.mimet.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 29.Lindstedt BA, Brandal LT, Aas L, Vardund T, Kapperud G. 2007. Study of polymorphic variable-number of tandem repeats loci in the ECOR collection and in a set of pathogenic Escherichia coli and Shigella isolates for use in a genotyping assay. J. Microbiol. Methods 69:197–205. 10.1016/j.mimet.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Lindstedt BA. 2011. Genotyping of selected bacterial enteropathogens in Norway. Int. J. Med. Microbiol. 301:648–653. 10.1016/j.ijmm.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 31.Bugarel M, Beutin L, Fach P. 2010. Low-density macroarray targeting non-locus of enterocyte effacement effectors (nle genes) and major virulence factors of Shiga toxin-producing Escherichia coli (STEC): a new approach for molecular risk assessment of STEC isolates. Appl. Environ. Microbiol. 76:203–211. 10.1128/AEM.01921-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicholls L, Grant TH, Robins-Browne RM. 2000. Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol. Microbiol. 35:275–288. 10.1046/j.1365-2958.2000.01690.x. [DOI] [PubMed] [Google Scholar]
- 33.Centers for Disease Control and Prevention. 2012. National Shiga toxin-producing escherichia coli (STEC) surveillance annual summary, 2009. CDC, Atlanta, GA. [Google Scholar]
- 34.European Centre for Disease Prevention and Control, European Food Safety Authority. 2011. Shiga toxin/verotoxin-producing Escherichia coli in humans, food and animals in the EU/EEA, with special reference to the German outbreak strain STEC O104. European Centre for Disease Prevention and Control, Stockholm, Sweden. [Google Scholar]
- 35.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. 2013. Recent advances in understanding enteric pathogenic escherichia coli. Clin. Microbiol. Rev. 26:822–880. 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alpers K, Werber D, Frank C, Koch J, Friedrich AW, Karch H, An Der Heiden M, Prager R, Fruth A, Bielaszewska M, Morlock G, Heissenhuber A, Diedler A, Gerber A, Ammon A. 2009. Sorbitol-fermenting enterohaemorrhagic Escherichia coli O157:H− causes another outbreak of haemolytic uraemic syndrome in children. Epidemiol. Infect. 137:389–395. 10.1017/S0950268808001465. [DOI] [PubMed] [Google Scholar]
- 37.Rosser T, Dransfield T, Allison L, Hanson M, Holden N, Evans J, Naylor S, La Ragione R, Low JC, Gally DL. 2008. Pathogenic potential of emergent sorbitol-fermenting Escherichia coli O157:NM. Infect. Immun. 76:5598–5607. 10.1128/IAI.01180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melton-Celsa A, Mohawk K, Teel L, O'Brien A. 2012. Pathogenesis of Shiga-toxin producing Escherichia coli. Curr. Top. Microbiol. Immunol. 357:67–103. 10.1007/82_2011_176. [DOI] [PubMed] [Google Scholar]
- 39.Delannoy S, Beutin L, Fach P. 2013. Towards a molecular definition of enterohemorrhagic Escherichia coli (EHEC): detection of genes located on O island 57 as markers to distinguish EHEC from closely related enteropathogenic E. coli strains. J. Clin. Microbiol. 51:1083–1088. 10.1128/JCM.02864-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson KE, Thorpe CM, Sears CL. 2006. The emerging clinical importance of non-O157 Shiga toxin-producing Escherichia coli. Clin. Infect. Dis. 43:1587–1595. 10.1086/509573. [DOI] [PubMed] [Google Scholar]
- 41.Friedrich AW, Zhang W, Bielaszewska M, Mellmann A, Kock R, Fruth A, Tschape H, Karch H. 2007. Prevalence, virulence profiles, and clinical significance of Shiga toxin-negative variants of enterohemorrhagic Escherichia coli O157 infection in humans. Clin. Infect. Dis. 45:39–45. 10.1086/518573. [DOI] [PubMed] [Google Scholar]
- 42.Mellmann A, Lu S, Karch H, Xu JG, Harmsen D, Schmidt MA, Bielaszewska M. 2008. Recycling of Shiga toxin 2 genes in sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM. Appl. Environ. Microbiol. 74:67–72. 10.1128/AEM.01906-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mellmann A, Bielaszewska M, Kock R, Friedrich AW, Fruth A, Middendorf B, Harmsen D, Schmidt MA, Karch H. 2008. Analysis of collection of hemolytic uremic syndrome-associated enterohemorrhagic Escherichia coli. Emerg. Infect. Dis. 14:1287–1290. 10.3201/eid1408.071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karch H, Friedrich AW, Gerber A, Zimmerhackl LB, Schmidt MA, Bielaszewska M. 2006. New aspects in the pathogenesis of enteropathic hemolytic uremic syndrome. Semin. Thromb. Hemost. 32:105–112. 10.1055/s-2006-939766. [DOI] [PubMed] [Google Scholar]
- 45.Buvens G, Pierard D. 2012. Virulence profiling and disease association of verocytotoxin-producing Escherichia coli O157 and non-O157 isolates in Belgium. Foodborne Pathog. Dis. 9:530–535. 10.1089/fpd.2011.1073. [DOI] [PubMed] [Google Scholar]
- 46.Kawano K, Ono H, Iwashita O, Kurogi M, Haga T, Maeda K, Goto Y. 2012. stx genotype and molecular epidemiological analyses of Shiga toxin-producing Escherichia coli O157:H7/H− in human and cattle isolates. Eur. J. Clin. Microbiol. Infect. Dis. 31:119–127. 10.1007/s10096-011-1283-1. [DOI] [PubMed] [Google Scholar]
- 47.Pradel N, Bertin Y, Martin C, Livrelli V. 2008. Molecular analysis of Shiga toxin-producing Escherichia coli strains isolated from hemolytic-uremic syndrome patients and dairy samples in France. Appl. Environ. Microbiol. 74:2118–2128. 10.1128/AEM.02688-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khan F, Proulx F, Lingwood CA. 2009. Detergent-resistant globotriaosyl ceramide may define verotoxin/glomeruli-restricted hemolytic uremic syndrome pathology. Kidney Int. 75:1209–1216. 10.1038/ki.2009.7. [DOI] [PubMed] [Google Scholar]
- 49.Wahl E, Vold L, Lindstedt BA, Bruheim T, Afset JE. 2011. Investigation of an Escherichia coli O145 outbreak in a child day-care centre—extensive sampling and characterization of eae- and stx1-positive E. coli yields epidemiological and socioeconomic insight. BMC Infect. Dis. 11:238. 10.1186/1471-2334-11-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schimmer B, Nygard K, Eriksen HM, Lassen J, Lindstedt BA, Brandal LT, Kapperud G, Aavitsland P. 2008. Outbreak of haemolytic uraemic syndrome in Norway caused by stx2-positive Escherichia coli O103:H25 traced to cured mutton sausages. BMC Infect. Dis. 8:41. 10.1186/1471-2334-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.