Abstract
Hepatitis E virus (HEV) is a leading cause of waterborne acute hepatitis in developing countries. In Europe, HEV causes a zoonotic disease and is hyperendemic in southern France. Four HEV genotypes (1 to 4) have been defined, and the most used classification divides them into 24 subtypes. Autochthonous European HEV strains belong in majority to genotype 3. Subtypes 3c, 3f, and 3e are representative of the HEV diversity in France. HEV causes chronic hepatitis in solid-organ transplant recipients in Europe, and viral characteristics associated with chronicity are poorly documented. We sequenced 343-nucleotide-long HEV genomic fragments from the serum of eight chronically infected kidney transplant recipients and a near-full-length genome in one case. We identified in four patients (50%) HEV of subtype 3i, not described previously in France. If shorter genomic fragments were used in phylogenetic analyses, these HEV sequences were clustered with open reading frame 2 (ORF2) fragments labeled as subtype 3c. At least five of the eight HEV 3i sequences recovered from humans in our phylogenetic analyses were from chronically infected kidney transplant recipients. These data show that the description of the prevalence and geographical distribution of HEV subtypes may be partially inaccurate and that criteria for classification as 3i and 3c should be clarified. Extended molecular virology analyses are required to improve knowledge of HEV epidemiology and determinants of chronic HEV infection.
INTRODUCTION
Hepatitis E virus (HEV) is a leading cause of waterborne acute hepatitis in developing countries, whereas it causes a zoonotic disease in Europe and is hyperendemic in southern France (1, 2). All HEV strains belong to a single serotype (1), but four HEV genotypes (1 to 4) have been defined, and the most used classification divides these four genotypes into 24 subtypes (3). Since 2008, HEV genotype 3 has been known to cause acute and chronic hepatitis in solid-organ transplant recipients (1, 2). In the Marseille geographical area, southeastern France, autochthonous HEV infection is a clinical concern in kidney transplant recipients, as its incidence was estimated to be at least 1% and chronic progression occurred in 80% of the cases (2). Factors potentially leading to chronic HEV infection in solid-organ transplant recipients have been investigated (1). Chronic outcome was described in several studies as associated with some host immunological patterns and immunosuppressive drugs (1). In addition, this outcome was recently associated with a higher genetic heterogeneity at the time of acute infection in regions of the HEV genome encoding the polyproline region and the macrodomain in open reading frame 1 (ORF1) and the M and P capsid domains in ORF2 (1). Moreover, it has been noticed that all chronic HEV infections reported so far have involved genotype 3 HEV (1).
HEV genome fragments sequenced from kidney transplant recipients in our center have been described based on HEV subtype reference sequences as belonging to subtypes 3c, 3f, or 3e (2, 3). These subtypes are representative of the HEV diversity in France; however, they were more evenly distributed in our center (2). Here, we sequenced both HEV ORF2 fragments that were larger than those previously obtained in routine from the serum of eight chronically infected kidney transplant recipients and the near-full-length viral genome from one of these patients. We identified that HEV sequences from four patients were the most closely related to subtype 3i, which has not been previously described as circulating in France, and noticed that a majority of HEV sequences of subtype 3i detected through phylogenetic analyses and recovered from humans were from chronically infected kidney transplant recipients.
MATERIALS AND METHODS
Patients.
Virological investigations were conducted for eight HEV chronically infected kidney transplant recipients who received follow-up care in Marseille University hospitals. Their mean age (± standard deviation [SD]) was 53.7 ± 10.2 years, and most (88%) were men (see Table SA1 in the supplemental material). Hepatitis E was diagnosed based on increased levels of liver enzymes and HEV RNA detection in serum using an in-house real-time PCR assay and sequencing of a 192-nucleotide-long fragment of ORF2 (see Table SA1 in the supplemental material) (2). Chronic HEV infection was defined by HEV RNA persistence in serum for >6 months (2).
HEV sequencing and phylogenetic analysis.
A 343-nucleotide-long fragment of the HEV ORF2 was recovered from the serum of the 8 patients by using previously described primers (2). HEV RNA was extracted from 200 μl of serum samples using EZ1 virus minikit v2.0 on the BioRobot EZ1 workstation (Qiagen, Courtaboeuf, France), by following the manufacturer's instructions. Prepared RNA was directly analyzed or stored at 80°C until processing. Reverse transcription (RT)-PCR amplification was performed in a single step or in two steps to produce large quantities of cDNA. In the latter case, contaminating DNA was removed from RNA preparations with DNase treatment (DNA-free kit; Applied Biosystems Ambion) before RT-PCR, by following the manufacturer's instructions. RT used 0.75 μl of reverse transcriptase (Roche, Meylan, France) and 3.1 μl of primer (20 μM). RT-PCR amplification in one step used the SuperScript One-Step RT-PCR system (Invitrogen Life Technologies, Carlsbad, CA) under the following conditions: 5 μl of RNA extract was added to 20 μl of reaction solution containing 0.5 μl (10 μM) of each outer sense and antisense primer, 12.5 μl of 2× reaction buffer, and 0.5 μl RT/Platinum Taq DNA polymerase (Life Technologies). Reaction conditions were as follows: 30 min at 45°C for RT, 2 min at 94°C for initial denaturation, and then 40 thermal cycles under conditions adapted to the primers' melting temperature (Tm), including denaturation for 30 s, annealing for 45 s, elongation for 2 min, and then a final elongation step of 5 min. Nested PCR was most often required and was performed by mixing 5 μl of first-round amplification product with 1 μl (10 μM) of each inner primer, 10 μl of 10× Master mix reaction buffer (Roche), and 0.25 μl of Taq DNA polymerase (Roche) in a final volume of 50 μl.
For one of the patients (case no. 2), overlapping nucleotide fragments covering the near-full-length HEV genome were obtained using 21 PCR primers selected from the literature (see Table SA2 in the supplemental material) (4–8) and then were aligned with the most closely related HEV genomes identified by BLAST (9) to design 11 additional PCR primers, which allowed us to close the HEV genome (see Table SA2). All PCR products were purified with Sephadex G-50 Superfine on MAHVN 45-50 plates (Millipore, Molsheim, France) and then sequenced by using the primers used in the first round (if a single round was performed for PCR amplification) or the second round with BigDye Terminator cycle sequencing kit version 1.1 on the ABI Prism 3130 genetic analyzer (Applied Biosystems, Branchburg, NJ). To obtain the near-full-length HEV genome (GenBank accession no. KJ701409), overlapping sequences were assembled manually and by the CLC Bio software by mapping on closely related genomes. For phylogeny reconstructions, HEV sequences obtained in the present study were aligned using the MUSCLE software with a comprehensive set of 245 full-length genome sequences that were downloaded from the Virus Pathogen Resource database (http://www.viprbrc.org/brc/home.do?decorator=hepe) (10). Then, phylogeny reconstruction was performed by the neighbor-joining method, or the maximum likelihood method, using the MEGA software version 5.0 (http://www.megasoftware.net/). HEV sequences that were analyzed included those obtained in the present study together with their 10 best BLAST hits in the NCBI GenBank nucleotide sequence database. Then, the HEV genotype and subtype were determined by phylogenetic analyses with the comprehensive set of 245 HEV genomes (10). HEV classification was guided by subtyping by Lu et al. (3) and retrieved from publications for the HEV sequences most closely related to those presented here.
RESULTS
Partial HEV ORF2 sequences.
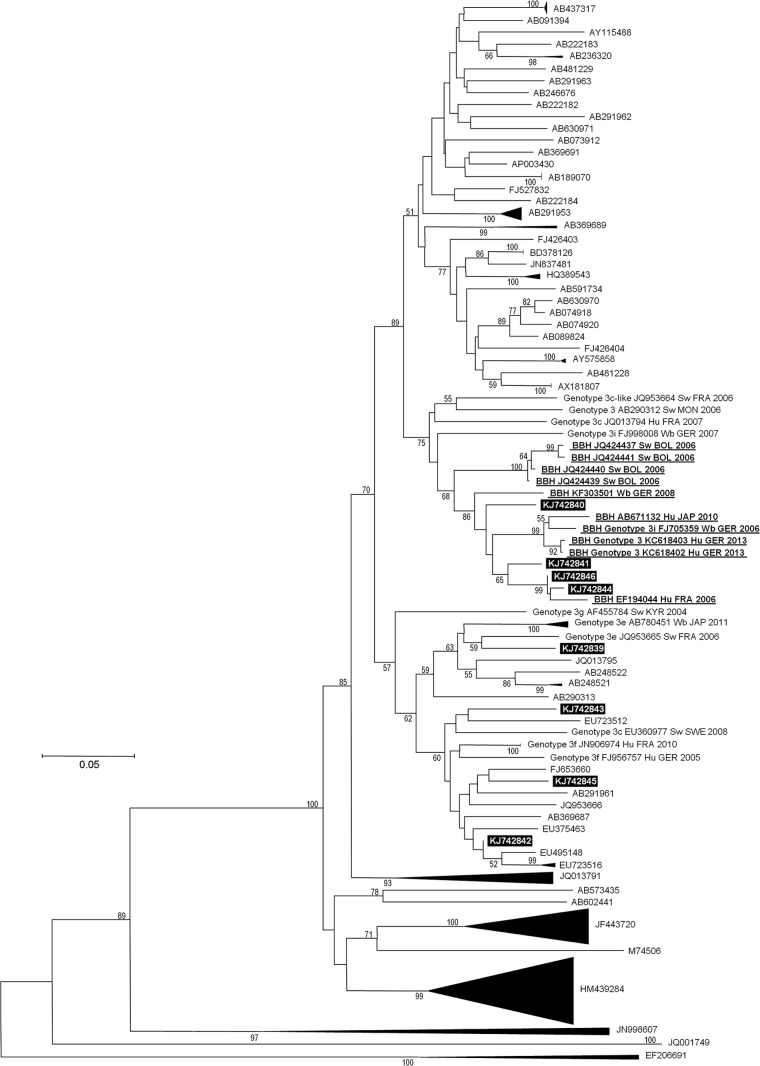
Phylogenetic reconstruction based on a 343-nucleotide-long HEV ORF2 region showed that the HEV sequences from four chronically infected kidney transplant recipients (case no. 2, 3, 6, and 8 from Table SA1 in the supplemental material) were clustered with HEV sequences previously classified as belonging to subtype 3i (Fig. 1; see also Fig. SA1 in the supplemental material), including partial sequences obtained in 2006 in Bolivia from swine (11), and HEV genomes FJ705359 and FJ998008, obtained in 2006 and 2007 in Germany from wild boars (12, 13), and KC618402 and KC618403, obtained in 2013 in Germany from a chronically infected kidney transplant recipient (14). The HEV sequences obtained here from the four chronically infected kidney transplant recipients were also clustered with HEV genomes of porcine origin, including JQ953664, a 3c-like sequence recovered in 2006 in France from a pig (15), AB290312, an unclassified genome recovered in 2006 in Mongolia from a pig (16), and KF303501, an unclassified genome recovered in 2008 in Germany from a wild boar (unpublished). Among HEV subtype 3i sequences from human origin from the present phylogenetic analysis, chronically infected kidney transplant recipients were known to be the source for at least 5/8 (62%). Nucleotide identities within the cluster encompassing HEV 3i genomes and the four sequences recovered here were 83.9 to 99.7% (mean ± SD, 90.5% ± 3.3%). HEV fragments from the four kidney transplant recipients of our center exhibited 90.9 to 96.2% identity (mean ± SD, 93.3% ± 1.9%) between each other and 85.7 to 94.4% identity (mean ± SD, 89.6% ± 3.2%) with the two first full-length HEV genomes labeled as belonging to subtype 3i. These levels of identity are greater than the lower limit at the level of subtypes defined by Lu et al. for ORF2 and HEV genotypes 3/4 (80.2 to 87.4%) (3). Noteworthy, if a shorter HEV ORF2 fragment (231 nucleotides) from the 343-nucleotide-long fragment was analyzed, ORF2 sequences classified as 3c by Lu et al. (3) were clustered with the HEV 3i genome FJ705359 (mean ± SD nucleotide identity, 91.6% ± 0.8%), while their counterparts in the HEV sequences from the four kidney transplant recipients exhibited 88.0 to 92.6% identity with HEV 3i genomes (see Fig. SA2 in the supplemental material).
FIG 1.
Phylogenetic tree based on a 343-nucleotide partial sequence corresponding to nucleotides 5999 to 6342 of open reading frame 2 (ORF2) of the HEV genome (GenBank accession no. AF082843). The HEV sequences obtained in our laboratory are indicated by a black frame. The 10 sequences with the highest BLAST scores (excluding those from our laboratory or with a query coverage of <98%) recovered from the NCBI GenBank nucleotide sequence database (indicated in boldface, underlined, labeled with BBH [for “best BLAST hit”], GenBank accession number, host, country, and year of sample collection or sequence submission) with the ORF2 fragment from the near-full-length genome obtained in this study have been incorporated into the phylogeny reconstruction in addition to a comprehensive set of 245 full-length genome sequences that were downloaded from the Virus Pathogen Resource database (10) (labeled, if they are clustered with sequences from the present study, with genotype and subtype, GenBank accession number, host, country, and year of sample collection or sequence submission). To allow for the better legibility of the tree, branches were collapsed when they did not directly link to sequences from the present study and when the bootstrap value at the node was >90%. Nucleotide alignments were performed using the MUSCLE software (http://www.ebi.ac.uk/Tools/msa/muscle/). The tree was constructed using the MEGA 5 software (http://www.megasoftware.net/) and the neighbor-joining method. Branches with bootstrap values were obtained from 1,000 resamplings of the data, and values greater than 50% are labeled on the tree. The EF206691 avian HEV sequence was used as an outgroup. A single sequence name is indicated for collapsed branches. A single sequence name is indicated for collapsed branches. The scale bar indicates the number of nucleotide substitutions per site. BBH, best BLAST hit; BOL, Bolivia; GER, Germany; FRA, France; Hu, Human; JAP, Japan; KYR, Kyrgyzstan; MON, Mongolia; NLD, the Netherlands; Sw, Swine; SWE, Sweden; Wb, wild boar. See also Figure SA1 in the supplemental material.
Full-length HEV sequences.
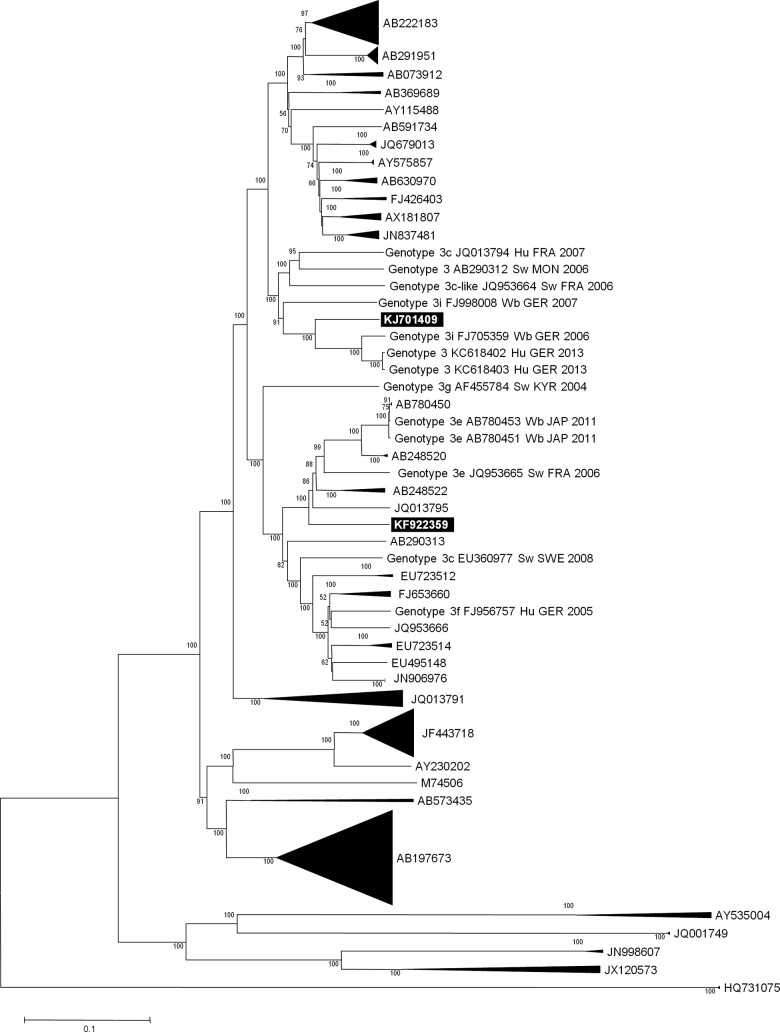
A phylogenetic reconstruction based on the HEV full-length genomes clearly classified the near-full-length genome obtained here (GenBank accession no. KJ701409) as belonging to subtype 3i, with a bootstrap value of 91%, and the cluster encompassing genomes FJ705359, KC618402, and KC618403 had a bootstrap value of 100% (Fig. 2). Among HEV 3i full-length genomes retrieved in the present phylogenetic analysis, sequences from human origin represented only 3/8, but at least 2/3 were known to originate from chronically infected kidney transplant recipients. The KJ701409 genome obtained here shared 84.2 to 88.1% identity with HEV 3i genomes FJ705359 and FJ998008, which is greater than the lower limit of identity at the level of subtype defined by Lu et al. for HEV 3/4 genomes (82.0 to 87.9%) (3), and 82.8% identity with genome JQ013794, labeled as 3c; identity with the EU360977 HEV genome, also labeled as 3c, was 78.8%. Finally, we analyzed 285-nucleotide-long fragments from ORF2 (corresponding to nucleotides 6000 to 6284 of genome FJ705359) that were described in Uruguay as belonging to subtype 3i (17) and observed that these sequences were clustered with the HEV 3i genomes (mean ± SD identity, 88.8% ± 0.3%) and with HEV sequences recovered from our four kidney transplant recipients (86.5% ± 1.3%) (data not shown).
FIG 2.
Phylogenetic tree of HEV full-length or near-full-length genomes. The legend of this figure is the same as that for Fig. 1, except the abbreviation “BBH” is not used in this figure and the avian HEV sequence AY535004 was used as an outgroup.
DISCUSSION
Our analyses revealed discrepant HEV 3 subtype classifications according to the HEV fragments and reference sequences used for phylogeny. Thus, HEV sequences from four chronically infected kidney transplant recipients were most closely related to HEV 3i genomes when the analysis was based on a 343-nucleotide-long ORF2 fragment and a comprehensive set of HEV genomes, whereas they were classified as 3c based on shorter ORF2 fragments and comparison to partial HEV sequences. Using short HEV ORF2 fragments may therefore generate misclassification. HEV 3i has been identified in five countries (Austria, Argentina, Bolivia, Germany, and Uruguay) and from various hosts, including humans (3, 14, 17), wild boars (12, 13), and swine (11, 17). HEV sequences from strains wbGER27 (FJ705359) and BB02 (FJ998008), which were recovered from wild boars in Germany, were fully sequenced and considered the first HEV 3i genomes (12, 13). HEV 3c was first described based on partial sequences of the HEV genome obtained from pigs in the Netherlands (3) and then was reported in humans and pigs in France and Germany (18, 19). In 2008, the first HEV genome classified as 3c was described from a Swedish swine (EU360977) (20). A second genome, obtained from a human in France in 2011 (JQ013794), was labeled as 3c (1); it shows 85 to 86% nucleotide identity with the HEV 3i genomes. In addition, an HEV 3c-like genome (JQ953664), which displays 85 to 86% identity with HEV 3i genomes, was described in a pig in France and was initially classified as 3c based on a 5′ ORF2 fragment (EF494703) (15), whereas this fragment displays 86 to 89% identity with HEV 3i genomes. In addition, some HEV ORF1 fragments (including FR846455 and FN995001) were classified as 3c, although they show ∼94 to 98% identity with the FJ705359 HEV 3i genome (19). The present findings suggest that the criteria for classification as 3i and 3c should be clarified. In France, only HEV 3 subtypes f, c, and e have been described (2), and HEV 3c was described in 2011 as the second most prevalent after HEV 3f, encompassing 15% and 9% of the HEV sequences obtained from humans and pigs, respectively (2). Previous data indicate that more extensive HEV molecular analysis and appropriate target genome fragments are required to accurately identify strains that circulate in a given geographical area and worldwide and to improve our knowledge of HEV epidemiology.
In a previous study, we found no association between HEV subtype and chronic outcome of HEV infection in kidney transplant recipients (2). Nevertheless, present analyses revealed that approximately two-thirds of HEV 3i sequences from humans were from known chronically infected kidney transplant recipients. In addition, Johne et al. recently obtained a persistently infected cell line after inoculation with the serum sample of a kidney transplant recipient chronically infected with HEV 3i (genome KC618402) (14). In this study, two additional HEV-positive serum samples from patients acutely infected with a virus of unknown genotype and a wild boar liver sample infected with HEV 3i genome FJ705359 were also inoculated onto A549 cells, and a persistent infection was obtained only with the serum sample of the chronically infected patient. Interestingly, a complex insertion within the hypervariable region of the genome was identified, as in two other cell culture-adapted HEV 3a strains that also originated from chronically infected patients (14). These three insertions differed by their sequences but had similar lengths and positions in the HEV genome, and such insertions were absent from the genome described here. Further studies, including more patients and accurate HEV subtyping, are needed to determine if HEV genotypic patterns may be associated with progression to chronic HEV infection in immunocompromised patients, particularly solid-organ transplant recipients.
Supplementary Material
ACKNOWLEDGMENTS
We have no potential conflicts of interest.
We did not receive financial support for this work.
Footnotes
Published ahead of print 3 September 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02028-14.
REFERENCES
- 1.Kamar N, Dalton HR, Abravanel F, Izopet J. 2014. Hepatitis E virus infection. Clin. Microbiol. Rev. 27:116–138. 10.1128/CMR.00057-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moal V, Legris T, Burtey S, Morange S, Purgus R, Dussol B, Garcia S, Motte A, Gerolami R, Berland Y, Colson P. 2013. Infection with hepatitis E virus in kidney transplant recipients in southeastern France. J. Med. Virol. 85:462–471. 10.1002/jmv.23469. [DOI] [PubMed] [Google Scholar]
- 3.Lu L, Li C, Hagedorn CH. 2006. Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Rev. Med. Virol. 16:5–36. 10.1002/rmv.482. [DOI] [PubMed] [Google Scholar]
- 4.Zhai L, Dai X, Meng J. 2006. Hepatitis E virus genotyping based on full-length genome and partial genomic regions. Virus Res. 120:57–69. 10.1016/j.virusres.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 5.Erker JC, Desai SM, Mushahwar IK. 1999. Rapid detection of hepatitis E virus RNA by reverse transcription-polymerase chain reaction using universal oligonucleotide primers. J. Virol. Methods 81:109–113. 10.1016/S0166-0934(99)00052-X. [DOI] [PubMed] [Google Scholar]
- 6.Peralta B, Mateu E, Casas M, de Deus N, Martin N, Pina S. 2009. Genetic characterization of the complete coding regions of genotype 3 hepatitis E virus isolated from Spanish swine herds. Virus Res. 139:111–116. 10.1016/j.virusres.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Legrand-Abravanel F, Mansuy JM, Dubois M, Kamar N, Peron JM, Rostaing L, Izopet J. 2009. Hepatitis E virus genotype 3 diversity, France. Emerg. Infect. Dis. 15:110–114. 10.3201/eid1501.080296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaba M, Davoust B, Marie JL, Barthet M, Henry M, Tamalet C, Raoult D, Colson P. 2009. Frequent transmission of hepatitis E virus among piglets in farms in Southern France. J. Med. Virol. 81:1750–1759. 10.1002/jmv.21553. [DOI] [PubMed] [Google Scholar]
- 9.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 10.Pickett BE, Sadat EL, Zhang Y, Noronha JM, Squires RB, Hunt V, Liu M, Kumar S, Zaremba S, Gu Z, Zhou L, Larson CN, Dietrich J, Klem EB, Scheuermann RH. 2012. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 40:D593–D598. 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purdy MA, Dell'Amico MC, Gonzales JL, Segundo H, Tolari F, Mazzei M, Bartoloni A, Khudyakov YE. 2012. Human and porcine hepatitis E viruses, southeastern Bolivia. Emerg. Infect. Dis. 18:339–340. 10.3201/eid1802.111453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schielke A, Sachs K, Lierz M, Appel B, Jansen A, Johne R. 2009. Detection of hepatitis E virus in wild boars of rural and urban regions in Germany and whole genome characterization of an endemic strain. Virol. J. 6:58. 10.1186/1743-422X-6-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adlhoch C, Wolf A, Meisel H, Kaiser M, Ellerbrok H, Pauli G. 2009. High HEV presence in four different wild boar populations in East and West Germany. Vet. Microbiol. 139:270–278. 10.1016/j.vetmic.2009.06.032. [DOI] [PubMed] [Google Scholar]
- 14.Johne R, Reetz J, Ulrich RG, Machnowska P, Sachsenroder J, Nickel P, Hofmann J. 2014. An ORF1-rearranged hepatitis E virus derived from a chronically infected patient efficiently replicates in cell culture. J. Viral Hepat. 21:447–456. 10.1111/jvh.12157. [DOI] [PubMed] [Google Scholar]
- 15.Bouquet J, Cherel P, Pavio N. 2012. Genetic characterization and codon usage bias of full-length hepatitis E virus sequences shed new lights on genotypic distribution, host restriction and genome evolution. Infect. Genet. Evol. 12:1842–1853. 10.1016/j.meegid.2012.07.021. [DOI] [PubMed] [Google Scholar]
- 16.Lorenzo FR, Tsatsralt-Od B, Ganbat S, Takahashi M, Okamoto H. 2007. Analysis of the full-length genome of hepatitis E virus isolates obtained from farm pigs in Mongolia. J. Med. Virol. 79:1128–1137. 10.1002/jmv.20905. [DOI] [PubMed] [Google Scholar]
- 17.Mirazo S, Ramos N, Russi JC, Arbiza J. 2013. Genetic heterogeneity and subtyping of human hepatitis E virus isolates from Uruguay. Virus Res. 173:364–370. 10.1016/j.virusres.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Colson P, Borentain P, Motte A, Lagrange X, Kaba M, Henry M, Tamalet C, Gerolami R. 2007. First human cases of hepatitis E infection with genotype 3c strains. J. Clin. Virol. 40:318–320. 10.1016/j.jcv.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Wenzel JJ, Preiss J, Schemmerer M, Huber B, Plentz A, Jilg W. 2011. Detection of hepatitis E virus (HEV) from porcine livers in southeastern Germany and high sequence homology to human HEV isolates. J. Clin. Virol. 52:50–54. 10.1016/j.jcv.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Xia H, Liu L, Linde AM, Belak S, Norder H, Widen F. 2008. Molecular characterization and phylogenetic analysis of the complete genome of a hepatitis E virus from European swine. Virus Genes 37:39–48. 10.1007/s11262-008-0246-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.