Abstract
Clostridium difficile infections (CDI) are a growing concern in North America, because of their increasing incidence and severity. Using integrated approaches, we correlated pathogen genotypes and host clinical characteristics for 46 C. difficile infections in a tertiary care medical center during a 6-month interval from January to June 2010. Multilocus sequence typing (MLST) demonstrated 21 known and 2 novel sequence types (STs), suggesting that the institution's C. difficile strains are genetically diverse. ST-1 (which corresponds to pulsed-field gel electrophoresis strain type NAP1/ribotype 027) was the most prevalent (32.6%); 43.5% of the isolates were binary toxin gene positive, of which 75% were ST-1. All strains were ciprofloxacin resistant and metronidazole susceptible, and 8.3% and 13.0% of the isolates were resistant to clindamycin and tetracycline, respectively. The corresponding resistance loci, including potential novel mutations, were identified from the whole-genome sequencing (WGS) of the resistant strains. Core genome single nucleotide polymorphisms (SNPs) determining the phylogenetic relatedness of the 46 strains recapitulated MLST types and provided greater interstrain differentiation. The disease severity was greatest in patients infected with ST-1 and/or binary gene-positive strains, but genome-wide SNP analysis failed to provide additional associations with CDI severity within the same STs. We conclude that MLST and core genome SNP typing result in the same phylogenetic grouping of the 46 C. difficile strains collected in a single hospital. WGS also has the capacity to differentiate those strains within STs and allows the comparison of strains at the individual gene level and at the whole-genome level.
INTRODUCTION
Clostridium difficile infections (CDI) are the most common infectious antibiotic-associated gastrointestinal disorders. C. difficile colonization of the intestine results in a range of clinical states, ranging from asymptomatic carriage to self-limited diarrhea to life-threatening colitis. CDI was the leading cause of gastroenteritis- and gastrointestinal tract infection-associated deaths between 1999 and 2007 in the United States (1). Risk factors for CDI include antibiotic exposures (especially fluoroquinolones [FQ] and cephalosporins), advanced age, and the severity of the underlying illness (2, 3, 4).
The most common C. difficile strain that has emerged in the past decade in North America and some areas in Europe has been classified as 027 by ribotyping, NAP1 by pulsed-field gel electrophoresis (PFGE), BI by restriction endonuclease analysis, and ST-1 by multilocus sequence typing (MLST). ST-1 strains account for half of the sporadic hospital-associated CDI in some settings (5). Some studies have reported that ST-1 strains elaborate C. difficile toxins (TCDs) at high concentrations; its purported hypervirulence is plausibly related to this trait. This strain has single and 18-bp deletions of tcdC, a negative regulator of tcdA and tcdB. These mutations cause premature stops, and this truncation is believed to cause toxin overproduction (6, 7). However, this assumption was challenged by recent studies showing no significant difference in toxin production between hypervirulent and nonhypervirulent C. difficile strains, and no association of the tcdC genotype and toxin production (8, 9).
C. difficile strains containing cdtA and cdtB binary toxin genes are associated with greater mortality in their hosts than strains in which these genes are absent (10). However, it is not clear if the binary toxin genes increase the virulence of ST-1 or if they are simply epidemiologic markers of C. difficile strains with increased virulence (i.e., guilt by association). It is also notable that other ribotypes with binary toxin, such as 078 (ST-11), can also cause severe CDI, especially in young adults. These C. difficile ribotype 078 strains were highly related to animals and food-borne C. difficile strains (11). It is concerning that 078 strains have increased in prevalence from 3% (2008) to 13% (2011) (1). CDI caused by both 027/ST-1 and 078/ST-11 are associated with an increased risk of death (12).
Our understanding of the pathogenesis of C. difficile is based largely on studies in outbreak strains. While the epidemiology of CDI is changing, analysis of C. difficile, especially the strains causing severe CDI, in a nonoutbreak setting might shed light on the mechanism of the pathogenicity of sporadic C. difficile and possibly produce more generalizable data. The objective of this study, therefore, was to characterize the phenotypes and genotypes of 46 nonoutbreak C. difficile isolates from a large academic medical center using conventional microbiological analyses and whole-genome sequencing and to investigate the associations between strain phenotypes and genotypes and clinical outcomes.
MATERIALS AND METHODS
CDI severity, bacterial strains, and ribotyping and binary toxin characterization.
This study was approved by the Washington University Human Research Protection Office. All subjects were prospectively interviewed and examined as part of a C. difficile assay comparison evaluation (13). The presence of clinically significant diarrhea and the severity of CDI were determined. Patients without clinically significant diarrhea or those who were colonized with a nontoxigenic strain of C. difficile were not classified as having CDI. Severe CDI was defined according to the clinical practice guidelines for CDI in adults (14): subjects with a white blood cell count of ≥15,000 cells/mm3 and/or serum creatinine of ≥1.5 times the premorbid level at the time of CDI diagnosis. Specimens were collected, and C. difficile strains were isolated and characterized as part of a C. difficile laboratory method study (13). Ribotyping (15) and detection of the binary toxin genes from the isolates were performed by PCR as previously described (16, 17).
Antibiotic susceptibility testing.
C. difficile strains were tested for antibiotic susceptibility using a gradient diffusion method according to the manufacturer's recommendations. Isolates of C. difficile were grown in an anaerobic environment on prereduced sheep blood agar (BBL; BD, Sparks, MD). A bacterial suspension was prepared in 0.9% saline to a 1 McFarland standard and then applied as a lawn of growth to Brucella agar with hemin and vitamin K (Hardy Diagnostics, Santa Maria, CA). Etest strips for metronidazole, clindamycin, moxifloxacin, ciprofloxacin, and tetracycline (bioMérieux) were applied to the agar and incubated with quality control strains according to the manufacturer's recommendations. The resulting MIC values were interpreted according to the Clinical and Laboratory Standards Institute guidelines (18).
Whole-genome sequencing and analysis.
Genomic DNA was extracted from the 46 isolates by a BiOstic bacteremia DNA isolation kit (MO BIO Laboratories). A genome paired-end library was constructed with average insert lengths from 150 to 200 bp, following the Illumina library construction protocol. The libraries were sequenced at an Illumina 2 × 100 bp platform. The genome assembly was performed by Velvet (version 1.1.04-64) (19). All assemblies were subjected to host contamination screening and met the criterion for draft genomes used in the Human Microbiome Project (20). Gene annotation employed the online RAST annotation pipeline with manual inspection (http://www.nmpdr.org/FIG/wiki/view.cgi/FIG/RapidAnnotationServer). The open reading frames (ORFs) were at least 300 bp long. Core gene sets were determined by pan-genome analysis pipeline (PGAP) with default parameters using all of the 46 C. difficile strains and reference strain 630 (21). The contigs from the draft genomes were aligned to the C. difficile MLST database (http://pubmlst.org/cdifficile/) to determining the sequence type (ST) using Mummer (22). The mutations from the novel ST types, regulatory genes in the pathogenicity locus (PaLoc), binary genes, and resistance genes were verified by manual inspection of the read alignment to reference alleles. Targeted PCR was performed to close the gaps in specific genes such as tcdE that were not fully covered in a subset of isolates.
Single nucleotide polymorphisms (SNPs) were identified with the SNP detection pipeline developed at the Washington University Genome Institute (TGI) by aligning the reads to the C. difficile reference strain 630 using BWA aligner (version 0.5.9) and SAMtools (version 0.1.12). SNP calling was performed as previously described but with increased stringency (23) as follows: (i) the coverage of a SNP is at least 10 reads and (ii) the number of reads supporting a SNP calling/the number of reads not supporting a SNP calling is ≥ 10 (3) ≥ 12 bp between two SNP sites. A phylogenetic tree based on the SNPs from the core gene sets was constructed using the neighbor-joining algorithms in Phylip (http://evolution.genetics.washington.edu/phylip.html).
Statistical analysis.
Fisher's exact test or the chi-square test was used to assess whether ST-1 and the presence of binary genes in C. difficile isolates are associated with CDI severity. P values <0.05 were considered statistically significant.
RESULTS
Isolate characterization.
The 46 C. difficile isolates represented 23 STs and 20 ribotypes. ST-1 (which corresponds to NAP1/027) accounted for 32.6% of the isolates. Other STs with at least two representatives in the collection were STs 2, 6, and 8 (Table 1). Three strains were not assignable to any STs in the current MLST database. One was a new allele profile (adk 3, atpA 7, dxr 14, glyA 8, recA 6, sodA 25, tpi 10), and the other two had an identical SNP at nucleotide position 198 in sodA. The correlation of ribotypes and STs was observed: ST-1 corresponded with ribotypes 027 and WU42, ST-2 with ribotypes 001/VPI/77/87 and 014/020, ST-6 with ribotype 005, and ST-8 with ribotypes WU22 and WU25. In addition, ribotypes 027 and WU42 were exclusively found in isolates belonging to ST-1, whereas ribotypes 001/VPI/77/87 were found among four STs (Table 1, Fig. 1). Thus, the ribotypes and ST types did not have a 1:1 correlation.
TABLE 1.
Phenotypic and genotypic characterization of 46 C. difficile strainsa
| Strain | Ribotype | ST | Presence (+) or absence (−) of: |
Strain response tob: |
Disease severityc | |||
|---|---|---|---|---|---|---|---|---|
| tcdA and tcdB | cdtA and cdtB | Clindamycin | Tetracycline | Moxifloxacin | ||||
| e01 | 5 | 6 | + | − | S | S | R | 0 |
| a02 | 5 | 6 | + | − | S | S | R | 0 |
| b09 | 5 | 6 | + | − | S | S | R | 1 |
| b01 | 27 | 1 | + | + | R | S | R | 1 |
| a01 | 27 | 1 | + | + | S | S | R | 0 |
| a08 | 27 | 1 | + | + | S | S | R | 0 |
| b10 | 27 | 1 | + | + | S | S | R | 1 |
| d09 | 27 | 1 | + | + | S | S | R | 1 |
| b07 | 27 | 1 | + | + | S | S | R | 1 |
| d02 | 53 | 103 | + | − | S | S | R | 0 |
| d01 | 77 | 3 | + | − | S | S | I | 0 |
| a12 | 78 | 11 | + | + | S | R | R | 1 |
| d10 | 2/75 | 55 | + | − | S | S | R | 0 |
| e09 | 001/VPI/77/87 | 46 | + | − | S | S | I | 0 |
| a11 | 001/VPI/77/87 | 48 | − | − | R | R | R | 0 |
| b04 | 001/VPI/77/87 | 7 | − | − | S | R | R | 0 |
| c04 | 001/VPI/77/87 | 2 | + | − | S | S | S | 0 |
| a09 | 106/174 | 36 | + | − | S | S | R | 0 |
| a03 | 106/174 | 42 | + | − | S | S | R | 0 |
| c12 | 014/020 | 2 | + | − | S | S | R | 0 |
| d04 | 014/020 | 2 | + | − | S | S | R | 0 |
| d03 | 014/020 | 2 | + | − | S | S | R | 1 |
| d08 | 014/020 | 14 | + | − | S | S | R | 0 |
| b08 | 014/020 | 110 | + | − | S | S | R | 0 |
| b05 | 15/46 | 10 | + | − | S | S | I | 1 |
| c11 | WU1 | 53 | + | − | S | S | I | 0 |
| d12 | WU22 | 35 | + | − | R | R | I | 0 |
| c03 | WU22 | 8 | + | − | S | S | R | 0 |
| a04 | WU22 | 8 | + | − | S | S | S | 0 |
| a05 | WU24 | NAd | + | + | S | S | R | 1 |
| e04 | WU25 | 8 | + | − | S | S | R | 0 |
| b11 | WU26 | 41 | + | + | S | S | I | 1 |
| b12 | WU26 | 10 | + | − | S | S | R | 0 |
| e07 | WU40 | 67 | + | + | S | S | I | 0 |
| d11 | WU42 | 1 | + | + | S | I | R | 0 |
| c05 | WU42 | 1 | + | + | S | S | R | 0 |
| c08 | WU42 | 1 | + | + | S | S | R | 0 |
| c10 | WU42 | 1 | + | + | S | S | R | 0 |
| e03 | WU42 | 1 | + | + | S | S | R | 0 |
| d05 | WU42 | 1 | + | + | S | S | R | 0 |
| c01 | WU42 | 1 | + | + | S | S | R | 1 |
| e08 | WU42 | 1 | + | + | S | S | R | 1 |
| c09 | WU42 | 1 | + | + | S | S | R | 1 |
| e02 | WU54 | 54 | + | − | S | R | I | 0 |
| d07 | WU60 | NAd | + | + | S | S | R | 1 |
| a07 | WU63 | NAe | − | − | S | S | R | 0 |
Organized by the ribotypes in alphabetical order.
S, susceptible; R, resistant; I, intermediate resistant.
0, not severe CDI; 1, severe CDI.
Mutation in sodA gene.
New allele profile.
FIG 1.
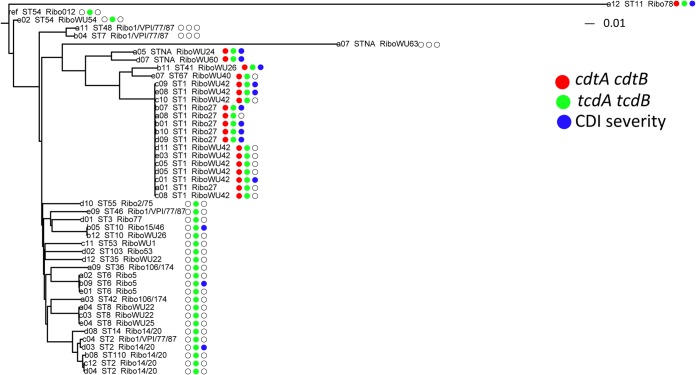
Phylogenetic tree of the 46 C. difficile strains based on the genome-wide SNPs. The neighbor-joining phylogenetic tree was constructed based on the SNPs from the core gene sets. C. difficile strain 630 is used as the reference strain for SNP calling. The STs and ribotypes are appended after the strain labels. The tree is annotated by the presence of the tcdA and tcdB genes (green), binary genes (red), and disease severity (blue) at the right side of the dendrogram. The white circles represent the absence of the toxin genes or nonsevere CDI.
We next studied the toxin-related genes, including tcdA, tcdB, and binary genes cdtA and ctdB. tcdA and tcdB genes were detected by conventional PCR and were further validated by sequencing. PCR results were perfectly correlated with the sequence data for detecting tcdA and tcdB. Binary toxin genes were also detected by PCR and successfully reconstructed from whole-genome sequencing. Isolates were grouped into three categories based on the presence of the toxin genes: (i) positive for tcdA, tcdB, cdtA, and cdtB, which comprised 43.5% (20 of 46) of the C. difficile isolates; (ii) positive for tcdA and tcdB + and negative for cdtA and cdtB, which comprised 50.0% of the strains (23 of 46); and (iii) negative for tcdA, tcdB, cdtA, and cdtB, which accounted for only three isolates (Fig. 1). Of the 20 tcdA-, tcdB-, cdtA-, and cdtB-positive strains, 15 belonged to ST-1. Other STs containing tcdA, tcdB and binary toxin genes were ST-11, ST-41, ST-67, and the two novel STs.
All 46 isolates were susceptible to metronidazole (MICs from 0.032 to 4 μg/ml) and resistant to ciprofloxacin (MICs of >32 μg/ml). The 46 strains had MICs between 2 and >32 μg/ml to moxifloxacin (overall MIC50 and MIC90 of 8 and >32 μg/ml, respectively), and 36, 8, and 2 isolates were resistant, intermediate, or susceptible to moxifloxacin, respectively. Of the isolates, 8.3% and 13.0% were resistant to clindamycin and tetracycline, respectively. Two (from ST35 and ST48) of the 46 isolates were resistant to all tested antibiotics except metronidazole; these two isolates were binary toxin gene negative and one was tcdA and tcdB negative. Five isolates were resistant to at least three antibiotics, including ciprofloxacin, tetracycline, and moxifloxacin.
Phylogenetic concordance between ST and WGS SNP typing.
WGS was performed on an Illumina HiSeq platform with 2 × 100 bp read lengths at 100× coverage on average. Read assemblies yielded 193 ± 53 contigs per genome. The contigs were annotated to provide the gene calling for each isolate. The gene content ranged from 3,612 to 4,054 ORFs per genome, indicating significant genetic variations across C. difficile strains. ST-1 isolates had between 3,700 and 3,768 genes, corresponding to 121 fewer genes on average than the other ST types in this study (see Table S1 in the supplemental material).
To determine the core genes used for phylogeny inference, we computed the shared genes from the 46 C. difficile strains and the C. difficile reference strain 630 (an ST-54 strain first isolated from a patient with pseudomembranous colitis). The C. difficile strain 630 was chosen as the reference because its genome was well annotated, and it has been widely used as the reference for SNP identification. We identified a total of 2,871 core genes across the isolates in this study, accounting for 64.0% of their gene content.
Between 1,096 and 44,935 SNPs were identified from the whole genomes of these isolates, of which 60.4% to 80.5% were distributed among the core genes. The median (interquartile range [IQR]) number of core genome SNPs was 6,926 per stain. No strains were identical at the SNP level, and 46 SNPs were identified between the two closest strains. The phylogenetic tree constructed using the SNPs from core genes from the 46 strains and the C. difficile reference strain 630 demonstrated a heterogeneous genetic nature of C. difficile strains in this collection (Fig. 1). Clades 1, 2, and 5 from a previous study (24) were identified. Clade 1 was composed of several STs. The ST-11 strain from clade 5 was most distant from the rest of lineages with 36,039 SNPs compared to the reference stain 630. The number of SNPs identified among the ST-11 strains was 3.3-fold higher than those of the other STs on average. Clade 2 was dominated by the ST-1/NAP1 strains. Two novel ST strains were genetically most similar to the ST-1 strains, and the third (from a nontoxigenic isolate) was distantly related to all other strains. The cluster of the SNPs from the core gene sets recapitulated the ST phylogeny, indicating the correspondence of the ST type with WGS (25).
WGS can offer an improved resolution compared to MLST characterization of isolates. For example, 99 to 656 SNP differences were detected within ST-1 strains, representing 1.3% to 4.4% of the differences in core gene sets. Within STs 6, 8, and 10, we identified 26 to 112 SNPs in the core genes, while 1,016 SNPs were detected for ST-2. The numbers of SNPs between ST types ranged from 1,568 to 44,204. However, the number of strains within a single ST type can change the degree of divergence within a ST type and may affect the pattern we observed here.
Genetic heterogeneity of toxin-related and antibiotic resistance genes.
Contigs were mapped to the six genes spanning across the PaLoc, two binary toxin genes, and resistance genes to determine the genetic variation of these regions.
Compared to the C. difficile reference strain 630, 3 strains lacked tcdA, tcdB, tcdC, tcdE, and tcdR genes and CD630_06620 (coding for a hypothetical protein). All PaLoc genes were present in the remaining 43 strains. A phylogenetic tree constructed from SNPs across the PaLoc revealed that diversity at this locus recapitulated ST typing, except for the PaLoc associated with ST-2, which appeared to be mixed with other STs (see Fig. S1 in the supplemental material). tcdA, tcdB, and tcdC genes had the greatest degrees of conservation within the same ST and high variation between STs as indicated in the circular plot (see Fig. S2 in the supplemental material). Other genes, such as tcdR and tcdE, were conserved even between STs, indicating different evolutionary changes in the PaLoc. The tcdC genes in these isolates were 92.7% to 100% identical to those in reference strain 630. Sequence alignment of the 43 tcdC genes demonstrated 12 tcdC variants. Based on the deletion pattern, we categorized the tcdC gene variants into 5 groups: (i) a single base pair deletion at nucleotide position 117 and an 18-bp deletion at nucleotide positions 330 to 347, which included all of the ST-1 strains; (ii) a single base pair deletion at nucleotide position 117 and no accompanying 18-bp deletion, which included only the ST-41 strain; (iii) a single base pair deletion at 117 bp, an insertion of T at 213 bp, and an 18-bp deletion, which included the two novel ST strains and has not been previously reported; (iv) a 39-bp deletion at nucleotide position 333, which included the ST-11 strain; and (v) single mutations without deletions, which occurred among heterogeneous STs.
Because binary toxin genes are not present in the C. difficile reference strain 630, the gene variation was determined by aligning the contigs to the binary genes in C. difficile strain CD196, which is an epidemic ST-1 strain harboring both type of toxin genes (26). In 15 of 21 strains, the sequences of cdtA and cdtB were identical to those of the CD196 strains. A transition from T to C at nucleotide position 813 of the cdtA gene was discovered in the remaining six strains containing binary toxin genes. Two ST-11 strains exhibited significant polymorphisms (21 SNPs) in comparison to those for CD196 in the 1,391-bp cdtA gene. Similarly, the relation of the cdtB gene in ST-11 strains was distant compared to those of other STs as indicated by 49 SNPs in this 2,630-bp region (see Table S2 in the supplemental material).
Fluoroquinolone (FQ) resistance is typically attributed to mutations in gyrA and gyrB, encoding DNA gyrase subunits. Among the 46 ciprofloxacin-resistant strains, 17 (36.9%) had a mutation at nucleotide position 82 (substitution T→I), as in a prototype FQ-resistant strain R20291 (ST-1). This mutation was common to all study ST-1 strains and to one ST-54 and one ST-55 strain. This substitution at position 82 in gyrA is the cause of FQ resistance in most European strains (27). We also identified eight other nonsynonymous mutations in gyrA from other ST strains, and these mutations tended to be conserved in the same clades. In some cases, these mutations are also ST specific. gyrB was intact in most (64.6%) strains. The precise contributions to ciprofloxacin resistance at additional mutations (see Table S2 in the supplemental material) are not yet known, but the gyrA and gyrB genes from two moxifloxacin-susceptible strains were identical to these loci in some of the moxifloxacin- and ciprofloxacin-resistant strains.
tetM and ermB are the genetic determinants of resistance to tetracycline and clindamycin, respectively. Multiple polymorphisms were observed in tetM genes compared to those in the reference strain 630. tetM genes displayed different degrees of heterogeneity between STs but were conserved within STs. Compared to the rest of the STs, in the ST-11 strains tetM contained an additional amino acid substitution at position 490 (M→T). We did not detect any other tet genes in the tetracycline-resistant strains. Three strains were resistant to clindamycin. The ermB gene of one strain (ST48) was 100% identical to the reference strain 630. The other two strains were from ST-1 and ST-35, sharing four nonsynonymous mutations at 454 (K→Q), 511 (A→V), 649 (Y→H), and 664 (D→N).
No link between SNPs and disease severity within ST-1.
Severe CDI accounted for 30.4% (14 of 46) of the CDI cases. The strains associated with severe CDI were from 7 different STs with ST-1 being predominant (46.7%). ST-6, ST-41, ST-11, ST-2, a novel ST with a mutation in the sodA gene, and ST-10 accounted for the remaining severe CDI cases (Fig. 1). ST-1 strains were not significantly different in their association with severe CDI compared to that of non-ST-1 strains (χ2 = 1.2, df = 1, P = 0.28). Among ST-1 strains, regulatory PaLoc genes (i.e., tcdC, tcdR, and tcdE) were identical in the seven and eight isolates from patients with severe and nonsevere CDI, respectively. Because of the highly repeated sequences in tcdA and tcdB, full ORFs could not be constructed with accuracy. Thus, we compared the SNPs identified in those two genes between ST-1 strains causing severe CDI and nonsevere CDI. The majority of the SNPs (88.5%) were shared for all ST-1 strains independent of disease severity in the patients from whom they were isolated. The rest of the SNPs were shared in either all ST-1 strains causing severe CDI/nonsevere CDI or a proportion of the ST-1 strains causing nonsevere CDI/severe CDI. Thus, genetic heterogeneity of the PaLoc did not distinguish disease severity within ST-1 strains (see Fig. S1 in the supplemental material). Similarly, SNPs within the PaLoc between non-ST-1 strains (ST-6 and ST-10) failed to identify associations with CDI severity, indicating no strong role for the genetic composition of pathogenic loci and disease phenotype.
Interestingly, 11 of 14 (78.6%) strains causing severe CDI were binary gene positive. Eleven of 20 (55%) binary toxin gene-positive strains were recovered from patients with severe CDI. The chi-square test showed that binary toxin gene-positive strains were significantly associated with severe CDI compared to binary toxin gene-negative strains (Fisher's exact test, P = 0.003). Thus, the presence of binary genes might be a marker for strains that cause severe CDI, but the current genetic data do not support the role of binary genes in causing severe disease, as within the STs, binary gene sequences were identical in strains causing severe CDI and nonsevere CDI.
Whole-genome-wide SNP analysis showed that 92.3% and 92.7% of the SNPs were conserved within all of the ST-1 strains causing severe CDI and nonsevere CDI, respectively. However, no single SNP distinguished the ST-1 strains causing severe CDI from those causing nonsevere CDI.
DISCUSSION
Using integrated approaches, we delineated the phenotypes and genotypes of C. difficile strains causing CDI from a single institution over a 6-month interval in 2010. Our data most notably indicate that ST-1 strains remain predominant in nonoutbreak settings, comparable to a statewide strain collection (548 strains) conducted from 2006 to 2009 (5). The vast majority of the strains were tcdA- and tcdB positive, and no tcdB-positive and tcdA-negative strains were detected in our data. This is likely in line with the low prevalence (∼2%) of this type of stain in the United States, despite the high prevalence rates in Japan, Israel, and Argentina (28).
Our data suggest that MLST and ribotyping are robust approaches for identifying phylogenic relationships in C. difficile strains (25, 29), but the most precise resolution requires WGS. WGS allows single nucleotide-level resolution for strain comparisons, thus serving as a powerful tool for outbreak investigations and clarifying institutional versus community acquisition. As demonstrated by outbreaks of Escherichia coli O104:H4 in Europe, the resolution of single nucleotide differences using WGS data led to the distinction of lineages from German and French isolates, which standard tests failed to distinguish (30). Along the same line, whole-genome sequencing of methicillin-resistant Staphylococcus aureus (MRSA) in a special care nursery unit successfully tracked the transmission within the unit (31). Recent studies have proved the feasibility of using WGS to track C. difficile transmission (32). Importantly, more efforts are needed to conduct prospective epidemiologic studies since the current studies are retrospective in response to a perceived outbreak, and, thus, early shifts in local epidemiology may not be detected (5). With the dropping of sequencing cost and establishment of streamlined analysis pipeline, WGS is becoming an advantageous approach in real-time pathogen surveillance and outbreak detection (33). Close surveillance and the prospective epidemiology of the C. difficile strains, especially those associated with severe CDI such as ST-1, ST-11, and ST-6, might prevent future C. difficile outbreaks.
In addition, the antibiotic resistance genes identified from WGS data matched well with the antibiotic susceptibility testing, providing another application for WGS data in clinical settings. However, correlation of antibiotic resistance by WGS relies on a database of well-curated resistance genes. With the potential antimicrobial resistance suggested by novel mutations identified from WGS, phenotypic testing and genetic engineering are still indispensable for validating the potential resistance from the novel mutations identified from WGS. The novel mutations in gyrA and gyrB from non-ST-1 strains revealed diverse genetic compositions in conferring potential resistance. The mutations require further verification to determine their role in fluoroquinolone resistance. The virulence genes reconstructed from WGS reads were consistent with the toxin gene testing in the clinic, demonstrating the versatile potentials of WGS in the clinic.
Interestingly, the phylogenetic topology of C. difficile is reflected in the PaLoc. Specifically, tcdC exhibited identical mutations across all of the ST-1 strains, as indicated by 1- and 18-bp deletions. Along the same line, two binary toxin genes, fluoroquinolone resistance genes (especially gyrA), also had identical sequences in all of the ST-1 strains studied and differed from those found in other STs. These findings suggest coevolution of the MLST genes with toxin genes and gyrA.
Although the clonal nature of the C. difficile strains might provide genetic insights into pathogenicity and antibiotic resistance, it did not shed light on the likelihood of causing severe disease. Previous reports suggested a poor correlation between the presence of tcdC or binary genes with clinical outcomes in nonepidemic settings (34, 35). These studies were based on comparisons of NAP1 strains in different severity groups without a genetic comparison, or the genetic comparison was limited to tcdC and binary toxin genes. In our study, we found that the presence of binary genes in C. difficile significantly increased the risk of severe CDI, but we could not attribute such increased risk to binary gene sequence differences in the strains that caused severe CDI and nonsevere CDI. Moreover, among the 40,000+ SNPs throughout these strains, no single nucleotide variation correlated with disease severity. However, larger sample sizes and future multicenter studies are needed to validate these findings. Disease presentation and outcome are determined by multiple aspects of the host-pathogen interaction, including toxin production, intestinal microbial ecology, host immune response, and timing and selection of treatment. The lack of association between C. difficile genetics and disease severity suggests that the bacterial genome itself only partly contributes to disease severity. Future RNA sequence analysis might provide more insight into the C. difficile virulence at a bacterial gene expression level. Also, investigation of the role of gut microbiota in CID severity will add an ecological perspective for understanding the disease, especially in the antibiotic-disturbed ecological niche that predominates in patients with CDI.
Although this study demonstrated the potential for application of WGS in clinical diagnosis and linked the genotype, phenotype, and clinical outcome of a collection of C. difficile strains, the major limitations of the study were small sample size and a single-center cohort. Future work with a larger sample size will allow a more complete picture of C. difficile epidemiology and confirm our findings of genotypic and clinical data. Additional evaluations of a regional or national C. difficile network will provide collective resources and both facilitate epidemiological surveillance and inform genetic determinants linked to pathogenesis and specific-disease phenotypes in C. difficile.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Centers for Disease Control and Prevention (grant 5U01C1000333) and the National Institute of Allergy and Infectious Diseases (grant K23AI065806). P. I. Tarr was supported by the National Institute of Diabetes and Digestive and Kidney Disease (NIDDK) Digestive Diseases Research Core Center (grant P30DK052574). Y. Zhou and G. L. Weinstock were supported by grant U54HG004968 from the NIH Common Fund.
Footnotes
Published ahead of print 1 October 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02115-14.
REFERENCES
- 1. Hall AJ, Curns AT, McDonald LC, Parashar UD, Lopman BA. 2012. The roles of Clostridium difficile and norovirus among gastroenteritis-associated deaths in the United States, 1999-2007. Clin. Infect. Dis. 55:216–223. 10.1093/cid/cis386. [DOI] [PubMed] [Google Scholar]
- 2. Dubberke ER, Reske KA, Yan Y, Olsen MA, McDonald LC, Fraser VJ. 2007. Clostridium difficile-associated disease in a setting of endemicity: identification of novel risk factors. Clin. Infect. Dis. 45:1543–1549. 10.1086/523582. [DOI] [PubMed] [Google Scholar]
- 3. Brown KA, Khanafer N, Daneman N, Fisman DN. 2013. Meta-analysis of antibiotics and the risk of community-associated Clostridium difficile infection. Antimicrob. Agents Chemother. 57:2326–2332. 10.1128/AAC.02176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Deshpande A, Pasupuleti V, Thota P, Pant C, Rolston DD, Sferra TJ, Hernandez AV, Donskey CJ. 2013. Community-associated Clostridium difficile infection and antibiotics: a meta-analysis. J. Antimicrob. Chemother. 68:1951–1961. 10.1093/jac/dkt129. [DOI] [PubMed] [Google Scholar]
- 5. Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, Kuijper EJ, Wilcox MH. 2010. The changing epidemiology of Clostridium difficile infections. Clin. Microbiol. Rev. 23:529–549. 10.1128/CMR.00082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dupuy B, Govind R, Antunes A, Matamouros S. 2008. Clostridium difficile toxin synthesis is negatively regulated by TcdC. J. Med. Microbiol. 57:685–689. 10.1099/jmm.0.47775-0. [DOI] [PubMed] [Google Scholar]
- 7. MacCannell DR, Louie TJ, Gregson DB, Laverdiere M, Labbe AC, Laing F, Henwick S. 2006. Molecular analysis of Clostridium difficile PCR ribotype 027 isolates from Eastern and Western Canada. J. Clin. Microbiol. 44:2147–2152. 10.1128/JCM.02563-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cartman ST, Kelly ML, Heeg D, Heap JT, Minton NP. 2012. Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcdC genotype and toxin production. Appl. Environ. Microbiol. 78:4683–4690. 10.1128/AEM.00249-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Merrigan M, Venugopal A, Mallozzi M, Roxas B, Viswanathan VK, Johnson S, Gerding DN, Vedantam G. 2010. Human hypervirulent Clostridium difficile strains exhibit increased sporulation as well as robust toxin production. J. Bacteriol. 192:4904–4911. 10.1128/JB.00445-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bacci S, Molbak K, Kjeldsen MK, Olsen KE. 2011. Binary toxin and death after Clostridium difficile infection. Emerg. Infect. Dis. 17:976–982. 10.3201/eid/1706.101483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jhung MA, Thompson AD, Killgore GE, Zukowski WE, Songer G, Warny M, Johnson S, Gerding DN, McDonald LC, Limbago BM. 2008. Toxinotype V Clostridium difficile in humans and food animals. Emerg. Infect. Dis. 14:1039–1045. 10.3201/eid1407.071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Walker AS, Eyre DW, Wyllie DH, Dingle KE, Griffiths D, Shine B, Oakley S, O'Connor L, Finney J, Vaughan A, Crook DW, Wilcox MH, Peto TE. 2013. Relationship between bacterial strain type, host biomarkers, and mortality in Clostridium difficile infection. Clin. Infect. Dis. 56:1589–1600. 10.1093/cid/cit127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dubberke ER, Han Z, Bobo L, Hink T, Lawrence B, Copper S, Hoppe-Bauer J, Burnham CA, Dunne WM., Jr 2011. Impact of clinical symptoms on interpretation of diagnostic assays for Clostridium difficile infections. J. Clin. Microbiol. 49:2887–2893. 10.1128/JCM.00891-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox MH. 2010. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect. Control Hosp. Epidemiol. 31:431–455. 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 15. Westblade LF, Chamberland RR, MacCannell D, Collins R, Dubberke ER, Dunne WM, Jr, Burnham CA. 2013. Development and evaluation of a novel, semiautomated Clostridium difficile typing platform. J. Clin. Microbiol. 51:621–624. 10.1128/JCM.02627-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alasmari F, Seiler SM, Hink T, Burnham CA, Dubberke ER. 2014. Prevalence and risk factors for asymptomatic Clostridium difficile carriage. Clin. Infect. Dis. 59:216–222. 10.1093/cid/ciu258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Antikainen J, Pasanen T, Mero S, Tarkka E, Kirveskari J, Kotila S, Mentula S, Kononen E, Virolainen-Julkunen AR, Vaara M, Tissari P. 2009. Detection of virulence genes of Clostridium difficile by multiplex PCR. APMIS 117:607–613. 10.1111/j.1600-0463.2009.02509.x. [DOI] [PubMed] [Google Scholar]
- 18.Clinical and Laboratory Standards Institute. 2007. Methods for antimicrobial susceptibility testing of anaerobic bacteria: approved standard–7th ed. Approved standard M11-A7. Clinical and Laboratory Standards Institute; Wayne, PA. [Google Scholar]
- 19. Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, Wortman JR, Rusch DB, Mitreva M, Sodergren E, Chinwalla AT, Feldgarden M, Gevers D, Haas BJ, Madupu R, Ward DV, Birren BW, Gibbs RA, Methe B, Petrosino JF, Strausberg RL, Sutton GG, White OR, Wilson RK, Durkin S, Giglio MG, Gujja S, Howarth C, Kodira CD, Kyrpides N, Mehta T, Muzny DM, Pearson M, Pepin K, Pati A, Qin X, Yandava C, Zeng Q, Zhang L, Berlin AM, Chen L, Hepburn TA, Johnson J, McCorrison J, Miller J, Minx P, Nusbaum C, Russ C, Sykes SM, Tomlinson CM, Young S, et al. 2010. A catalog of reference genomes from the human microbiome. Science 328:994–999. 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhao Y, Jia X, Yang J, Ling Y, Zhang Z, Yu J, Wu J, Xiao J. 2014. PanGP: a tool for quickly analyzing bacterial pan-genome profile. Bioinformatics 30:1297–1299. 10.1093/bioinformatics/btu017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Turabelidze G, Lawrence SJ, Gao H, Sodergren E, Weinstock GM, Abubucker S, Wylie T, Mitreva M, Shaikh N, Gautom R, Tarr PI. 2013. Precise dissection of an Escherichia coli O157:H7 outbreak by single nucleotide polymorphism analysis. J. Clin. Microbiol. 51:3950–3954. 10.1128/JCM.01930-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Griffiths D, Fawley W, Kachrimanidou M, Bowden R, Crook DW, Fung R, Golubchik T, Harding RM, Jeffery KJ, Jolley KA, Kirton R, Peto TE, Rees G, Stoesser N, Vaughan A, Walker AS, Young BC, Wilcox M, Dingle KE. 2010. Multilocus sequence typing of Clostridium difficile. J. Clin. Microbiol. 48:770–778. 10.1128/JCM.01796-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kurka H, Ehrenreich A, Ludwig W, Monot M, Rupnik M, Barbut F, Indra A, Dupuy B, Liebl W. 2014. Sequence similarity of Clostridium difficile strains by analysis of conserved genes and genome content is reflected by their ribotype affiliation. PLoS One 9:e86535. 10.1371/journal.pone.0086535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley TD, Sebaihia M, Quail MA, Rose G, Gerding DN, Gibert M, Popoff MR, Parkhill J, Dougan G, Wren BW. 2009. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 10:R102. 10.1186/gb-2009-10-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Spigaglia P, Barbanti F, Mastrantonio P. 2011. Multidrug resistance in European Clostridium difficile clinical isolates. J. Antimicrob. Chemother. 66:2227–2234. 10.1093/jac/dkr292. [DOI] [PubMed] [Google Scholar]
- 28. Drudy D, Fanning S, Kyne L. 2007. Toxin A-negative, toxin B-positive Clostridium difficile. Int. J. Infect. Dis. 11:5–10. 10.1016/j.ijid.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 29. He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, Connor TR, Harris SR, Fairley D, Bamford KB, D'Arc S, Brazier J, Brown D, Coia JE, Douce G, Gerding D, Kim HJ, Koh TH, Kato H, Senoh M, Louie T, Michell S, Butt E, Peacock SJ, Brown NM, Riley T, Songer G, Wilcox M, Pirmohamed M, Kuijper E, Hawkey P, Wren BW, Dougan G, Parkhill J, Lawley TD. 2013. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat. Genet. 45:109–113. 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Grad YH, Lipsitch M, Feldgarden M, Arachchi HM, Cerqueira GC, Fitzgerald M, Godfrey P, Haas BJ, Murphy CI, Russ C, Sykes S, Walker BJ, Wortman JR, Young S, Zeng Q, Abouelleil A, Bochicchio J, Chauvin S, Desmet T, Gujja S, McCowan C, Montmayeur A, Steelman S, Frimodt-Moller J, Petersen AM, Struve C, Krogfelt KA, Bingen E, Weill FX, Lander ES, Nusbaum C, Birren BW, Hung DT, Hanage WP. 2012. Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in Europe, 2011. Proc. Natl. Acad. Sci. U. S. A. 109:3065–3070. 10.1073/pnas.1121491109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Köser CU, Holden MT, Ellington MJ, Cartwright EJ, Brown NM, Ogilvy-Stuart AL, Hsu LY, Chewapreecha C, Croucher NJ, Harris SR, Sanders M, Enright MC, Dougan G, Bentley SD, Parkhill J, Fraser LJ, Betley JR, Schulz-Trieglaff OB, Smith GP, Peacock SJ. 2012. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N. Engl. J. Med. 366:2267–2275. 10.1056/NEJMoa1109910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Eyre DW, Cule ML, Wilson DJ, Griffiths D, Vaughan A, O'Connor L, Ip CL, Golubchik T, Batty EM, Finney JM, Wyllie DH, Didelot X, Piazza P, Bowden R, Dingle KE, Harding RM, Crook DW, Wilcox MH, Peto TE, Walker AS. 2013. Diverse sources of C. difficile infection identified on whole-genome sequencing. N. Engl. J. Med. 369:1195–1205. 10.1056/NEJMoa1216064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM. 2014. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52:1501–1510. 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Goldenberg SD, French GL. 2011. Lack of association of tcdC type and binary toxin status with disease severity and outcome in toxigenic Clostridium difficile. J. Infect. 62:355–362. 10.1016/j.jinf.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 35. Sirard S, Valiquette L, Fortier LC. 2011. Lack of association between clinical outcome of Clostridium difficile infections, strain type, and virulence-associated phenotypes. J. Clin. Microbiol. 49:4040–4046. 10.1128/JCM.05053-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.