Abstract
The lack of a sufficiently discriminatory molecular subtyping tool for Salmonella enterica serovar Enteritidis has hindered source attribution efforts and impeded regulatory actions required to disrupt its food-borne transmission. The underlying biological reason for the ineffectiveness of current molecular subtyping tools such as pulsed-field gel electrophoresis (PFGE) and phage typing appears to be related to the high degree of clonality of S. Enteritidis. By interrogating the organism's genome, we previously identified single nucleotide polymorphisms (SNP) distributed throughout the chromosome and have designed a highly discriminatory PCR-based SNP typing test based on 60 polymorphic loci. The application of the SNP-PCR method to DNA samples from S. Enteritidis strains (n = 55) obtained from a variety of sources has led to the differentiation and clustering of the S. Enteritidis isolates into 12 clades made up of 2 to 9 isolates per clade. Significantly, the SNP-PCR assay was able to further differentiate predominant PFGE types (e.g., XAI.0003) and phage types (e.g., phage type 8) into smaller subsets. The SNP-PCR subtyping test proved to be an accurate, precise, and quantitative tool for evaluating the relationships among the S. Enteritidis isolates tested in this study and should prove useful for clustering related S. Enteritidis isolates involved in outbreaks.
INTRODUCTION
Salmonella enterica serovar Enteritidis is an important cause of food-borne illness in humans all over the world (1, 2, 3). Although poultry products, including eggs, are very common sources of S. Enteritidis, the organism thrives in a variety of other food sources and in the environment, and this obligates regulatory agencies tasked with controlling the infection in humans and animals to effectively target the source incriminated in an outbreak. Regulatory interventions aimed at protecting consumers from an ongoing food-borne outbreak require that an isolate causing illnesses in people be matched with a contaminant in the food consumed by the patients, and this effort is achieved by a combination of epidemiological investigation and subtyping analysis in a laboratory. The subtyping of S. Enteritidis, which ideally should demonstrate a subserovar differentiation among unrelated strains and the clustering of related strains, is commonly done using one of two techniques, namely, pulsed-field gel electrophoresis (PFGE) and phage typing (4). PFGE subtyping relies on the sizes of DNA fragments resulting from the digestion of the organism's chromosomal and plasmid DNA by one or two restriction enzymes, namely, XbaI or BlnI, to produce a molecular fingerprint. Despite the presence of hundreds of different PFGE types among field S. Enteritidis isolates, the majority of isolates found in the PulseNet databases are of one (United States) to three (Canada) PFGE types. The three commonest Canadian S. Enteritidis PFGE types, namely, XAI.0003, XAI.0038, and XAI.0006, represent 32, 15, and 13% of Canadian S. Enteritidis isolates documented in the PulseNet database. The commonest S. Enteritidis PFGE type in Canada (XAI.0003) is identical to the one in the United States (JEGX01.0004) based on the electrophoretic mobility patterns of the restriction fragments (5; D. Ogunremi, unpublished observation). Poor discriminatory power of PFGE for S. Enteritidis (6) is responsible for the grouping of the majority of isolates into only a few molecular patterns. Phage typing is based on the pattern of susceptibility of different strains of the bacteria to a bacteriophage or a combination of bacteriophages, resulting in lysis of the organism (7, 8). Phage typing has sometimes provided better discrimination between strains than antimicrobial susceptibility testing, plasmid analysis, ribotyping, or pulsed-field gel electrophoresis (9). Historically, phage typing of S. Enteritidis has been the most widely used tool for establishing relationships between isolates from different sources (10, 11). However, the dependability of phage typing is limited by the potential for the conversion of isolates to different phage types through different mechanisms, including plasmid acquisition or loss, removal of the lipopolysaccharide layer, introduction of temperate phages, and mutation (12, 13). Occasional disagreement about the merits of different Salmonella phage typing schemes introduces additional uncertainties about using this approach as a universal method for subtyping and strain classification (14). Laboratories sometimes report different results from testing the same isolates because of variable test performance despite very rigorous quality control for phage typing reagents (15). There are documented cases where the same isolate has yielded disparate results on retesting in the same laboratory (16). In essence, the attributes of a bacterium that influence its phage type may not be stable from one generation to another. In this scenario, two isolates with the same phage type may in fact be unrelated, and conversely, two isolates that show distinct phage types may be closely related. Consequently, Van Belkum and colleagues (17) concluded that phage typing shows inadequate discriminatory power and displays partial typeability and poor reproducibility.
The documented limitations of both conventional S. Enteritidis subtyping approaches have prompted a concerted and rigorous effort to develop a new reliable subtyping method, but the outcomes have been so far unsuccessful or unimplementable. The multiple locus variable-number tandem repeat analysis (MLVA) was developed for S. Enteritidis (18), but the performance does not appear to be superior to that of PFGE, and the required discrimination among strains is yet to be attained. A newly published sequence typing scheme following PCR amplification of two loci is an example of the latest generation of methods for S. Enteritidis subtyping (16). A vast majority of the isolates tested (83%; n = 102), obtained from diverse geographical locations, hosts, types of environments, and time periods, grouped into only 3 sequence types out of a total of 16. The restricted distribution of putative subtypes using this newly described procedure mirrors the limitation already observed with PFGE typing.
Detailed genetic studies of Salmonella Enteritidis have consistently shown the underlying causes of the poor discriminatory abilities of existing subtyping tools: isolates of Salmonella Enteritidis are extremely similar (i.e., are highly clonal), and this poses a difficulty in finding a definitive, distinguishing trait that could be used to track lineages (5, 19). An ideal subtyping procedure should lead to a high level of discrimination among isolates, in contrast to existing methods, which tend to group apparently unrelated isolates together. To guarantee success, any strategy aimed at developing a tool capable of differentiating S. Enteritidis lineages will require interrogating a significant amount of the DNA information in each S. Enteritidis isolate. The use of massively parallel sequencing technology (20) to deduce the entire nucleotide sequence of S. Enteritidis may well be the only viable option to assess targets that could be used for developing an ideal subtyping test.
Unlike the other molecular methods, which investigate only very small portions of the entire bacterial genome, whole-genome sequencing determines and uses the entire genome as a basis for discrimination and can thus identify extremely small differences, such as single nucleotide polymorphisms (SNPs), which can be used for subtyping provided that such differences are consistently preserved in a particular bacterial lineage. The recent publication and release of multiple draft genomes of S. Enteritidis have added significantly to the resources available to pursue molecular typing of the organisms (21). Notably, Allard and colleagues (22) have carried out bioinformatics analysis of a total of 104 S. Enteritidis genomes belonging to the predominant PFGE pattern (JEGX01.0004) and some historical isolates. They described a total of 9 clades and found 366 genes that showed variation, i.e., presence or absence, in S. Enteritidis genomes. The above-mentioned report complemented and expanded on an earlier study by another laboratory which showed that two isolates of S. Enteritidis with the same phage type, PT 13a, were differentiated by a relatively large number of loci, i.e., 250, showing single nucleotide polymorphisms (SNPs) (5). Thus, genetic variation that could allow the development of a routine subtyping tool for tracking purposes is present within the S. Enteritidis genome.
We recently completed the whole-genome sequencing of 11 S. Enteritidis isolates obtained in Canada, and by comparing them with S. Enteritidis P125109 reference strain phage type 4, we identified 1,361 loci where the S. Enteritidis genome shows single nucleotide polymorphism (SNP). We have chosen a total of 60 SNPs spread throughout the genome and distributed among different gene types and in intergenic locations to develop a rapid, inexpensive fluorescence-based PCR assay. This report covers the development of a new molecular subtyping test, SNP-PCR, and its application to a group of 55 S. Enteritidis isolates obtained in Canada, which has now led to the recognition of 12 clades of S. Enteritidis.
MATERIALS AND METHODS
Strains of Salmonella serovar Enteritidis.
Isolates of Salmonella serovar Enteritidis (n = 55) used in the study were retrieved from the Canadian Food Inspection Agency (CFIA) inventory of bacterial glycerol stocks. The isolates came from a variety of sources: poultry environment, 23; food, 14; food processing equipment, 1; autopsied domestic animals, 3 (hogs and a chicken); wild bird, 1 (pigeon); animal feed, 1; reference strains from American Tissue Culture Collection (ATCC), 2; undetermined sources, 10 (see Data Set S1 in the supplemental material). Frozen stocks were inoculated into brain heart infusion broth and incubated at 37°C overnight with agitation at 200 rpm, and cultured bacteria were used for phage typing, pulsed-field gel electrophoresis (PFGE), or DNA extraction for PCR (see “Salmonella Enteritidis single nucleotide polymorphism-PCR subtyping procedure” below). Phage typing was carried out at the Office International des Epizooties (OIÉ) Reference Laboratory for Salmonellosis, Public Health Agency of Canada (PHAC), following a standardized protocol (23). The phage typing assay was inadvertently carried out twice in the same laboratory for half of the isolates (n = 27), and when the outcomes of testing the same isolate were different, both results are shown, although the last result was used when the different subtyping assays were compared. PFGE analysis was performed at the Ottawa Laboratory Fallowfield, Canadian Food Inspection Agency, by using the restriction enzymes XbaI and BlnI to create signature molecular patterns, which were electronically submitted to PulseNet Canada (National Microbiology Laboratory, Public Health Agency of Canada, Winnipeg, Canada) for PFGE subtype designation following a standardized protocol (http://www.pulsenetinternational.org/protocols/).
Single nucleotide polymorphism.
Single nucleotide changes among 11 S. Enteritidis genomes sequenced in an earlier study (24) relative to the reference S. Enteritidis strain P125109 were detected by means of the SNPsfinder software (25) (http://snpsfinder.lanl.gov/; Los Alamos National Laboratory, Los Alamos, NM). Using a table of annotated genomes (24), all the loci showing single nucleotide polymorphism were compiled (see Data Set S2 in the supplemental material), and 60 loci were identified for this study based on a number of criteria, which included good dispersal across the entire genome, a mix of genes to include a wide group of protein functions, and the presence of noncoding intergenic loci (Table 1; also, see Data Set S1 in the supplemental material).
TABLE 1.
Locations of 60 single nucleotide polymorphic sites used to develop the genotyping assay for Salmonella Enteritidis
| Reference position | Region or corresponding protein | Intergenic | Enzyme | Membrane | Regulatory | Transport | Phage | Secretory | Other | Unknown |
|---|---|---|---|---|---|---|---|---|---|---|
| 95192 | Intergenic space or other non-protein-coding region | • | ||||||||
| 204450 | Hypothetical ABC transporter ATP-binding protein | • | ||||||||
| 562921 | Putative allantoin permease | • | ||||||||
| 684875 | Putative hydrolase C terminus | • | ||||||||
| 738097 | Intergenic space or other non-protein-coding region | • | ||||||||
| 818905 | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase | • | ||||||||
| 833026 | Putative membrane protein | • | ||||||||
| 838275 | Intergenic space or other non-protein-coding region | • | ||||||||
| 928779 | NADH oxidoreductase Hcr | • | ||||||||
| 1035267 | Intergenic space or other non-protein-coding region | • | ||||||||
| 1218767 | Intergenic space or other non-protein-coding region | • | ||||||||
| 1249431 | Putative phage terminase; small subunit | • | ||||||||
| 1284118 | Putative membrane protein | • | ||||||||
| 1328395 | Hydrogenase-1 large chain (NifE hydrogenase) | • | ||||||||
| 1541966 | Intergenic space or other non-protein-coding region | • | ||||||||
| 1579797 | Putative integral membrane protein | • | ||||||||
| 1610591 | Membrane transport protein | • | ||||||||
| 1659052 | Putative ABC transporter membrane protein | • | ||||||||
| 1679797 | Intergenic space or other non-protein-coding region | • | ||||||||
| 1702656 | Putative proton/oligopeptide symporter | • | ||||||||
| 1723077 | Putative integral membrane transport protein | • | ||||||||
| 1730438 | Putative type III secretion protein | • | ||||||||
| 1738692 | Putative pathogenicity island lipoprotein | • | ||||||||
| 1934221 | ABC transporter ATP-binding subunit | • | ||||||||
| 1987706 | Glutaredoxin 2 | • | ||||||||
| 2008113 | Intergenic space or other non-protein-coding region | • | ||||||||
| 2008553 | csg operon transcriptional regulator protein | • | ||||||||
| 2081720 | Intergenic space or other non-protein-coding region | • | ||||||||
| 2084629 | Conserved hypothetical protein | • | ||||||||
| 2381848 | Glycerophosphoryl diester phosphodiesterase periplasmic precursor | • | ||||||||
| 2389182 | Conserved hypothetical protein | • | ||||||||
| 2526004 | Putative membrane protein | • | ||||||||
| 2557022 | Conserved hypothetical protein | • | ||||||||
| 2575052 | Putative cobalamin adenosyltransferase | • | ||||||||
| 2642024 | Intergenic space or other non-protein-coding region | • | ||||||||
| 2664170 | Dimethyl sulfoxide reductase | • | ||||||||
| 2664521 | Dimethyl sulfoxide reductase | • | ||||||||
| 2690699 | Putative inner membrane protein | • | ||||||||
| 2824211 | Putative GAB DTP gene cluster repressor | • | ||||||||
| 2835443 | Putative transcriptional regulator | • | ||||||||
| 2901286 | Pathogenicity 1 island effector protein | • | ||||||||
| 3135538 | Putative GntR-family regulatory protein | • | ||||||||
| 3153778 | l-Asparaginase | • | ||||||||
| 3446987 | Acriflavin resistance protein F | • | ||||||||
| 3455766 | Conserved hypothetical protein | • | ||||||||
| 3507751 | Nitrite reductase [NAD(P)H] small subunit | • | ||||||||
| 3793026 | Lipopolysaccharide core biosynthesis protein | • | ||||||||
| 3872863 | Intergenic space or other non-protein-coding region | • | ||||||||
| 3944202 | ATP synthase epsilon subunit | • | ||||||||
| 3995713 | Intergenic space or other non-protein-coding region | • | ||||||||
| 4014537 | Conserved hypothetical protein | • | ||||||||
| 4018775 | Adenylate cyclase | • | ||||||||
| 4042373 | Putative regulatory protein | • | ||||||||
| 4232942 | Elongation factor Tu (EF-Tu) (p-43) | • | ||||||||
| 4232945 | Elongation factor Tu (EF-Tu) (p-43) | • | ||||||||
| 4257290 | Uroporphyrinogen decarboxylase | • | ||||||||
| 4436213 | Transcriptional regulatory protein | • | ||||||||
| 4480295 | Intergenic space or other non-protein-coding region | • | ||||||||
| 4576384 | Putative GerE-family regulatory protein | • | ||||||||
| 4576396 | Putative GerE-family regulatory protein | • | ||||||||
| Total | 13 | 15 | 6 | 9 | 5 | 1 | 3 | 3 | 5 |
Salmonella Enteritidis single nucleotide polymorphism-PCR subtyping procedure.
For the purpose of addressing the need for a rapid, inexpensive, robust, and highly discriminatory molecular subtyping tool for S. Enteritidis, we developed a PCR test that could differentiate alternate nucleotide composition at specific loci in the genome of an S. Enteritidis isolate, i.e., S. Enteritidis SNP-PCR. Loci targeted included specific genes (i.e., based on essential role in the metabolism of S. Enteritidis) and intergenic locations (because of a reported high frequency of mutations compared to coding sequences) as well as sites that help achieve a relatively uniform spread across the entire genome. The developed S. Enteritidis SNP-PCR is an allele-specific, single-amplification, fluorescence-based PCR assay. Briefly, two similar primers were developed to bind to 18 to 20 nucleotides from a 5′ location up to the base option constituting the SNP at a specific locus (see Data Set S3 in the supplemental material). Thus, the two primers differed only at the terminal base complementary to the SNP, ensuring a competitive but specific binding. Each primer is designed with a specific tail that allows a complementary binding with another proprietary sequence (LGC Genomics, Beverly, MA), which is labeled with a fluorescent dye, 6-carboxyfluorescein (FAM) or hexaclorofluorescein (HEX) for allele 1 or 2, respectively. Thus, the first cycle of amplification ensures that the specific forward primer in the primer mix binds to the sequence containing the SNP and excludes the other primer. The reverse primer, also 18 to 20 nucleotides long, binds and elongates the fragment during amplification, ensuring that the tail sequence is present. The use of 5′ fluorescence-labeled oligonucleotides (FAM and HEX) which are complementary to the unique tail sequences, each containing a 3′ quencher, allowed an amplification-driven reporter system such that only the specific forward primer binds competitively, leading to the accumulation of amplicons detectable as the presence of one fluorescent dye but not the other and consequent designation of the locus as either allele 1 or allele 2. Bacterial DNA (3 to 10 ng in 5 μl of Tris-HCl, pH 7.5) was added to a PCR master mix which contained two labeled primers (2× master mix; 330 μl; LGC Genomics) and was combined with the forward and reverse primer mix (9.2 μl). The touchdown PCR procedure consisted of a denaturation step at 94°C for 15 min followed by an initial annealing step at 65°C for 60 s (ABI 7500Fast; Life Technologies, Burlington, Ontario, Canada). Nine additional cycles were carried out whereby the annealing temperature was dropped 0.9°C per cycle to attain a final annealing temperature of 57°C over a further 25 cycles by alternating with a denaturation step at 94°C for 10 s. After the final PCR cycle, the plate was read by the PCR machine at room temperature.
Statistical analysis.
Cluster analysis of SNP-PCR genotyping results was done by means of the unweighted pair group method using arithmetic averages (UPGMA) with the aid of Bionumerics software (version 7, Applied Math, Austin, TX), and the clade groupings were compared to PFGE and phage typing results. Clade designation was based on the hierarchical clustering of the isolates on the phylogenetic tree. The presence of a unique mutation shared by a number of isolates provided the threshold to assign them to a single clade. Clades lacking unique mutations were grouped based on a uniform pattern of mutations that distinguished them from other isolates and clades. The clade classification with the aid of the Bionumerics software was in agreement with the manual classification into groupings based on the total number or preponderance of shared SNPs. We assumed that the underlying genetic or phenotypic variation responsible for the subtypes of an organism is a continuous biological property and proceeded to test for the normality of the distribution of isolates into the different subgroups (i.e., clades, PFGE types or phage types). With the aid of a standard statistical program (MedCalc Software, Mariakerke, Belgium), we tested for normal distribution of the sample population using the Kolmogorov-Smirnov test and assessed significance at the level where P is <0.05. Simpson's reciprocal index (1/D) was estimated using the Biodiversity calculator developed by J. Danoff-Burg and X. Chen (www.columbia.edu/itc/cerc/danoff-burg/Biodiversity%20Calculator.xls) as a means of measuring and comparing the discriminatory ability of a subtyping test. A high 1/D value implies that the test is very discriminatory in differentiating subgroups present in the sampled population (26). To evaluate the concordance among the tests, comparison of the three subtyping tests was carried out by estimating the adjusted Rand index between pairs of tests and by a bidirectional estimation of the probability that two isolates grouped together by one test will also cluster with a second test using the adjusted Wallace coefficient (www.comparingpartitions.info) (27, 28).
RESULTS
Molecular subtyping of Salmonella Enteritidis by single nucleotide polymorphism using a PCR assay (S. Enteritidis SNP-PCR).
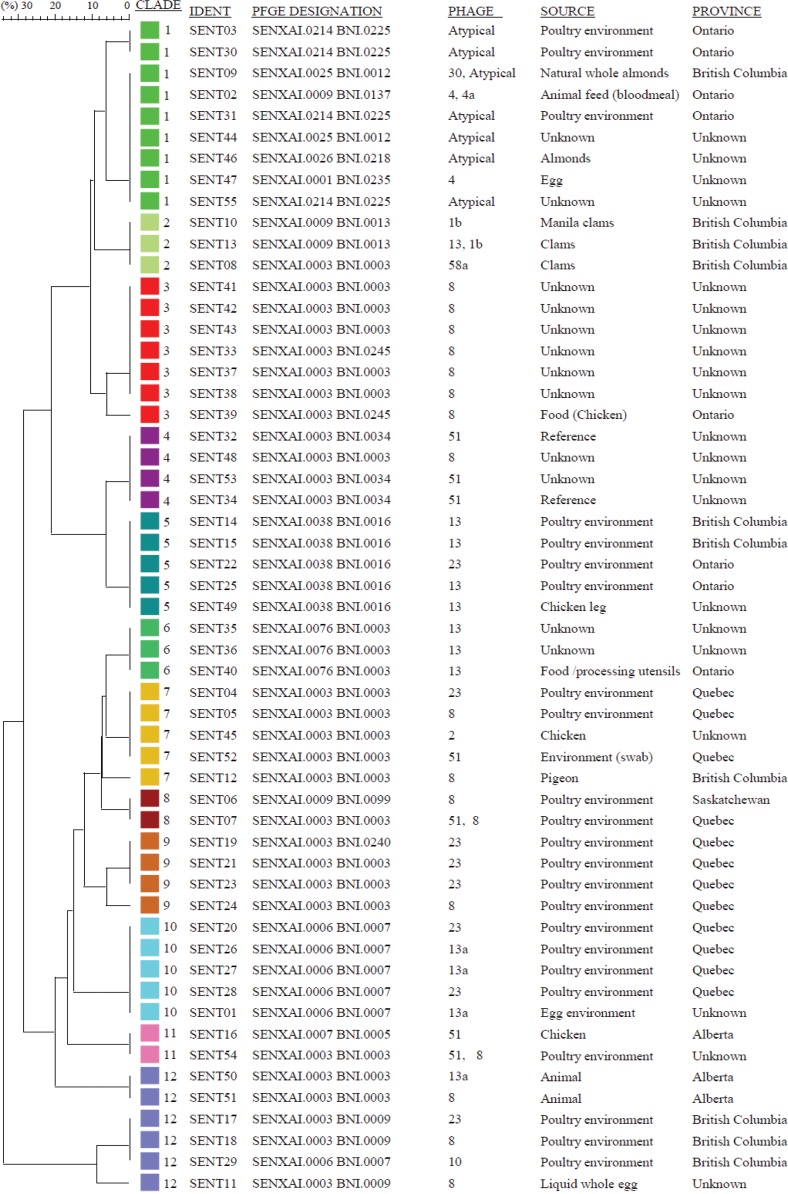
A PCR assay for each of 60 polymorphic loci in the S. Enteritidis genome led to the successful detection of single nucleotide variants in our culture collection of 55 S. Enteritidis isolates. Based on the distribution of SNPs, the isolates clustered into a total of 12 S. Enteritidis clades, consisting of 2 to 9 isolates per clade (Fig. 1; also, see Data Set S1 in the supplemental material). Twenty-one of the 60 SNPs were observed among members of only one clade and were designated “signature SNPs” because they exclusively assigned an isolate to a specific clade (Table 2). Clades 2, 5, 9, 10, 11, and 12 have two, one, three, four, four, and seven signature SNPs, respectively (Table 2). The presence of signature SNPs not only provided a very tight grouping for clades whose members possessed one but also allowed what could have been large clusters to be broken down into smaller clades and thereby improved the discriminatory power of the SNP-PCR (see Data Set S1 in the supplemental material). Clade 10 has four signature SNPs, and at the same time, all the remaining SNPs among the five members were in total agreement, creating an overall pattern not reproduced in any of the remaining 11 clades of Canadian origin or in the reference strain P125109 (Table 2; also, see Data Set S1 in the supplemental material). Similarly, members of clade 12 (n = 6) also had signature SNPs—seven in all, which is the highest number of signature SNPs observed among any of the clades. The members of clade 12 all showed complete agreement at an additional 40 loci; however, there was diversity at the remaining 13 loci. Six clades identified in this study, namely, 1, 3, 4, 6, 7, and 8, did not possess any signature SNP; nevertheless, the pattern of shared SNPs present among the 60 loci, including the lack of mutations at specific loci of interest (i.e., relative to the reference genome P125109), sufficiently and unambiguously defined membership in each clade.
FIG 1.
Phylogenetic analysis of Salmonella Enteritidis isolates of Canadian origin based on single nucleotide polymorphism (PCR) subtyping. Isolates of Salmonella Enteritidis obtained from food, animals, and poultry environments (n = 55) were tested by SNP-PCR at 60 loci (see Materials and Methods). The phylogenetic tree was constructed by means of cluster analysis of alleles at each locus using the Bionumerics software program.
TABLE 2.
Unique single nucleotide polymorphisms present among all members of the same clade (clade-specific signature)
| Reference genome location | Nucleotide | Mutation | Clade | Locusa | Locus description |
|---|---|---|---|---|---|
| 204450 | G | A | 12 | SEN0177 | Hypothetical ABC transporter ATP-binding protein |
| 928779 | G | A | 10 | SEN0844 | NADH oxidoreductase Hcr |
| 1035267 | A | G | 12 | IGS | Intergenic space or other non-protein-coding region |
| 1702656 | G | A | 11 | SEN1595 | Putative proton/oligopeptide symporter |
| 1730438 | T | A | 2 | SEN1626 | Putative type III secretion protein |
| 2008553 | T | C | 12 | SEN1906 | csg operon transcriptional regulator protein |
| 2081720 | G | A | 9 | IGS | Intergenic space or other non-protein-coding region |
| 2381848 | C | T | 11 | SEN2264 | Glycerophosphoryl diester phosphodiesterase periplasmic precursor |
| 2526004 | A | G | 12 | SEN2396 | Putative membrane protein |
| 2575052 | T | C | 2 | SEN2447 | Putative cobalamin adenosyltransferase |
| 2642024 | G | A | 12 | IGS | Intergenic space or other non-protein-coding region |
| 2901286 | G | A | 11 | SEN2716 | Pathogenicity 1 island effector protein |
| 3446987 | G | A | 12 | SEN3225 | Acriflavin resistance protein F |
| 3507751 | G | A | 12 | SEN3302 | Nitrite reductase [NAD(P)H] small subunit |
| 3944202 | C | T | 10 | SEN3678 | ATP synthase epsilon subunit |
| 4014537 | C | T | 10 | SEN3737 | Conserved hypothetical protein |
| 4042373 | C | T | 11 | SEN3761 | Putative regulatory protein |
| 4232945 | C | T | 10 | SEN3930 | Elongation factor Tu (EF-Tu) (p-43) |
| 4257290 | T | C | 9 | SEN3953 | Uroporphyrinogen decarboxylase |
| 4436213 | A | C | 5 | SEN4107 | Transcriptional regulatory protein |
| 4480295 | A | C | 9 | IGS | Intergenic space or other non-protein-coding region |
IGS, intergenic sequence.
Genes of Salmonella Enteritidis showing single nucleotide polymorphisms.
The choice of 60 polymorphic loci for developing the SNP-PCR (see Materials and Methods) was made from a list of 1,360 loci (see Data Set S2 in the supplemental material) identified from the analysis of the genomes of 11 Canadian S. Enteritidis isolates (28). Thirteen of the loci were located in noncoding, intergenic sites. The remaining loci were distributed according to gene function or group as follows: enzymes (15/60), regulatory (9/60), membrane or membrane associated (6/60), transport or transport associated (5/60), secretory (3/60), pathogenicity island associated (2/60), lipopolysaccharide core synthesis (1/60), phage protein (1/60), and hypothetical proteins of unknown function (5/60) (Table 1).
Comparison of three subtyping methods: phage typing, pulsed-field gel electrophoresis, and SNP genotyping.
Phage typing of the isolates revealed a total of 12 phage types in our collection (Table 3; also, see Data Set S1 in the supplemental material). Phage type 8 was the most common (17 of 55 isolates, or 31%), followed by phage type 23 (8 of 51, or 15%). Phage types 13a and 13 were also fairly common. Together, the two commonest phage types represented approximately half (45%) of the isolates in our collection. PFGE analysis revealed a more striking dominance of a subgroup by any of the three subtyping methods. Using the primary restriction enzyme XbaI as the basis for classification, one particular PFGE type, XAI.0003, was seen in a majority of our isolates, i.e., 28 out of 55, or 51% (Tables 1 and 3). The remaining nine primary PFGE types each had 1 to 5 isolates (Table 3). In contrast to both PFGE and phage typing, there was no dominance among the clade groupings by SNP analysis (range = 2 to 9 isolates per clade). The distribution of the isolates followed a normal distribution when classified into clades by SNP typing (KS test, P = 0.2561) but not when classified according to phage typing (P = 0.0008) or PFGE (P < 0.0001) (see Fig. S1 in the supplemental material).
TABLE 3.
Distribution of isolates of Salmonella Enteritidis based on comparative analysis of single nucleotide polymorphism (SNP)-PCR, phage typing, and pulsed-field gel electrophoresis (PFGE) subtyping procedures
| Phage type | Frequency | Rank | No. of isolates in SNP type group (clade) |
Total | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||||
| 1b | 2 | 2 | |||||||||||||
| 2 | 1 | 1 | |||||||||||||
| 4 | 1 | 1 | |||||||||||||
| 4a | 1 | 1 | |||||||||||||
| 8 | 17 | 1st | 7 | 1 | 2 | 2 | 1 | 1 | 3 | ||||||
| 10 | 1 | 1 | |||||||||||||
| 13 | 7 | 3rd | 4 | 3 | |||||||||||
| 13a | 4 | 3 | 1 | ||||||||||||
| 23 | 8 | 2nd | 1 | 1 | 3 | 2 | 1 | ||||||||
| 51 | 5 | 5th | 3 | 1 | 1 | ||||||||||
| 58a | 1 | 1 | |||||||||||||
| Atypical | 7 | 3rd | 7 | ||||||||||||
| Total | 9 | 3 | 7 | 4 | 5 | 3 | 5 | 2 | 4 | 5 | 2 | 6 | 55 | ||
| PFGE | |||||||||||||||
| SENXAI.0001 | 1 | 1 | |||||||||||||
| SENXAI.0003 | 28 | 1st | 1 | 7 | 4 | 5 | 1 | 4 | 1 | 5 | |||||
| SENXAI.0006 | 6 | 2nd | 5 | 1 | |||||||||||
| SENXAI.0007 | 1 | 1 | |||||||||||||
| SENXAI.0009 | 4 | 4th | 1 | 2 | 1 | ||||||||||
| SENXAI.0025 | 2 | 2 | |||||||||||||
| SENXAI.0026 | 1 | 1 | |||||||||||||
| SENXAI.0038 | 5 | 3rd | 5 | ||||||||||||
| SENXAI.0076 | 3 | 3 | |||||||||||||
| SENXAI.0214 | 4 | 4th | 4 | ||||||||||||
| Total | 9 | 3 | 7 | 4 | 5 | 3 | 5 | 2 | 4 | 5 | 2 | 6 | 55 | ||
The SNP typing procedure showed a higher discriminatory ability in that the predominant PFGE type XAI.0003 and phage type 8 could be subdivided into different clades. XAI.0003 isolates were distributed among 8 clades (Table 3). Similarly, the most common phage type, PT 8, was distributed among 7 clades (Table 3).
We noticed a high degree of uniformity in the isolates grouping as clade 3 by the SNP genotyping analysis (n = 7 isolates): they were all PT 8 and PFGE XAI.0003, although two of them had a different secondary BlnI pattern (specifically, 5 were BNI.0003 and 2 were BNI.0245). In contrast, all 5 members of clade 10 belonged to the same PFGE type—XAI.0006 BNI.0007—but two different phage types, namely, 13a and 23. Two of the isolates in the clade were obtained from the same poultry environment on the same day, supporting (but not necessarily confirming) the SNP and PFGE typing results, yet the phage type results were different.
All five members of clade 7 belonged to only one PFGE type (XAI.0003 BNI.0003) but four different phage types (i.e., only 2 of the 5 members share the same phage type). Three of the isolates were collected in the same province in central Canada, yet all had different phage types. One of them shared the same phage type (PT 8) with an isolate from a pigeon from a western Canadian province. Members of clade 5 were quite homogenous in their SNP profile, with the only exception being a mutation in the gene coding for a putative transcriptional regulator (gene SEN2647) at reference position 2835443, where the pigeon isolate had a T, in contrast to a G being present in all the remaining isolates from a chicken (1 isolate) and from poultry environments (3 isolates).
Although the majority of isolates found to have the PFGE pattern XAI.0003 also carried phage type 8 (16 isolates), five other phage types were also represented within the predominant PFGE type, namely, PT 2 (1 isolate), PT 13a (1 isolate), 23 (5 isolates), 51 (4 isolates), and 58a (1 isolate), indicating a substantial discordance between PFGE and phage typing results (Table 3).
The congruence between any two tests as measured by the adjusted Rand index was low, with a range between 0.210 (SNP-PCR and PFGE) and 0.365 (PFGE and phage type) (Table 4). Similarly, the adjusted Wallace coefficient (W) was low, with average values ranging from 0.129 for WPFGE→SNP-PCR to 0.588 for WPFGE→phage type. Although the WSNP-PCR→PFGE value was higher at 0.566 than WSNP-PCR→phage type at 0.47, there was an overlap in the confidence intervals of the two values. The highest coefficient was obtained for Wphage type→PFGE but was not significantly different from those for higher values of WSNP-PCR→phage type or WSNP-PCR→PFGE.
TABLE 4.
Measures of congruency and diversity among subtyping tests for Salmonella Enteritidis
| Subtyping method | Adjusted Rand index |
Adjusted Wallace coefficient (95% confidence interval)a |
Simpson's reciprocal index (1/D) | ||||
|---|---|---|---|---|---|---|---|
| SNP-PCR | PFGE | Phage type | SNP-PCR | PFGE | Phage type | ||
| SNP-PCR | 0.566 (0.440–0.692) | 0.470 (0.307–0.632) | 12.17 | ||||
| PFGE | 0.210 | 0.129 (0.067–0.190) | 0.264 (0.081–0.447) | 3.54 | |||
| Phage type | 0.316 | 0.365 | 0.238 (0.123–0.353) | 0.588 (0.396–0.780) | 6.55 | ||
Wallace coefficient is the probability that two isolates grouped together when tested by A (e.g., SNP-PCR) will remain clustered when tested by B (e.g., PFGE).
Clade 9 consisted of 4 isolates, which provided a good illustration of the clustering ability of the three subtyping tests. All isolates were from the same poultry establishment; three were obtained in the same year (2009) and the fourth during the subsequent year. All isolates had the same primary PFGE type (XAI.0003); however, one of the isolates (from 2009) had a different secondary enzyme PFGE result, namely, BNI.0024 (compared to BNI.0003 for the other three isolates). The phage type results were the same for the first three isolates (PT 23), but the last isolate had a different phage type (PT 8). The four isolates had identical SNPs at all loci tested, including the three signature SNPs, except for the last isolate (2010), which had a unique mutation at position 2824211 (SEN 2634; putative γ-aminobutyrate di/tripeptide permease gene cluster repressor). This mutation was very unusual, as it distinguished this isolate from all the other 54 isolates and the reference P125109 strain. SNP testing appropriately clustered the isolates together while portraying a unique mutation acquired by the strain before the last sampling event.
In general, the comparison of the three typing procedures revealed a number of trends which are best illustrated by examining the data from poultry establishments, which collectively were the largest contributor of isolates tested (23 out of 55 isolates). Multiple isolates from each of three poultry establishments show clustering, as detected by SNP PCR and to some extent PFGE, even when the phage type results were dissimilar. Three members of clade 12, namely, SENT 17, 18, and 29, illustrated this point very well. All three isolates were obtained from the same poultry establishment on the same day and grouped together in the same clade. Two of the isolates had the same PFGE type, namely, XAI.0003 BNI.0009, but the third isolate tested differently on both the primary and secondary enzymes (XAI.0006 BNI.0009). The phage typing results were even more disparate: all three isolates had different phage types (PT 8, 10, and 23).
When we retested 27 isolates in our culture collection using phage typing, 22 (81%) of them agreed with original results, while the remaining 5 showed inconsistent results (see Data Set S1 in the supplemental material). The PFGE testing system does not lend itself easily to retesting of samples because of the centralized system that will inevitably lead to double entry in the PulseNet database; however, in our experience, PFGE fingerprinting patterns are quite reproducible, although band assignment could be subjective. We carried out a preliminary assay reproducibility analysis for the SNP-PCR by testing 52 isolates at 29 loci in two laboratories. There was complete agreement on 1,493 of the 1,508 individual test results, or 99%.
The Simpson reciprocal indices (1/D) for the three S. Enteritidis subtyping tests were 3.54 for PFGE, 6.55 for phage typing, and 12.17 for SNP typing (Table 4). Thus, the S. Enteritidis SNP-PCR subtyping test showed a very high degree of discrimination and reproducibility. Unlike the conventional subtyping procedures (PFGE and phage typing), the new SNP typing procedure grouped the isolates into smaller subgroups with no obvious, predominant clade among our isolates. The highly discriminatory nature of the SNP-PCR still permitted putatively related strains to be clustered.
DISCUSSION
We have developed a highly discriminatory PCR test for identifying the nucleotide variation at specific loci in the genome of S. Enteritidis as a robust laboratory tool for the genotyping of isolates of this food-borne pathogen. The test reliably subtyped a collection of 55 isolates and clustered them into 12 clades, with each clade being made up of 2 to 9 isolates. Although the isolates tested are relatively few (n = 55), they appear to be sufficiently diverse and mirror, to some degree, the diversity of the S. Enteritidis isolates in Canada as seen in the Canadian PulseNet database. In addition, a total of 10 PFGE types were present in our collection, based on the primary restriction enzyme XbaI, representing a considerably diverse set of samples. Guard et al. (5) reported a high number of SNPs (38 out 250) located in the noncoding areas of the genome, i.e., intergenic, while comparing two S. Enteritidis isolates. In this study, we report a total of 285 intergenic SNPs in S. Enteritidis (see Data Set S2 in the supplemental material), thereby improving on the previous report (5), and have incorporated 13 of the intergenic SNPs (Table 1), dispersed throughout the S. Enteritidis genome, into our SNP-PCR design. The remaining 47 SNPs used in the SNP-PCR design were located among coding sequences and were selected to (i) represent a broad range of protein families and (ii) provide good discrimination among a group of 11 sequenced genomes consisting of different PFGE and phage types (24).
The 12 clades covered isolates obtained from Canadian sources but included four isolates from imported food, three of which clustered together (clade 2) (Fig. 1; also, see Data Set S1 in the supplemental material).
Clade 1 isolates were indistinguishable from the reference S. Enteritidis strain P125109, which was isolated about 3 decades ago in the United Kingdom (36). In contrast, clade 12 was the most genetically distant from the reference strain among our collection, as shown by the high number of mutations. The presence of 7 unique mutations among clade 12 isolates (n = 5) justified their inclusion in a single clade. As many more isolates are identified, the possibility that the members could form distinct subclades may become evident. It is also possible that continuing selection pressure could lead to further mutations in the clade and eventual emergence of more distinct clades from this group, each of which will show less intraclade diversity. To that end, we expect to detect more clades as we test more isolates of Canadian origin in the future. However, we do not expect that the number of clades will grow indefinitely, given that Salmonella Enteritidis is a very clonal organism; nevertheless, it will be important to test isolates from other areas of the world to ascertain geographically related differences. In this study, we observed that that the classification of isolates into clades followed a normal distribution (see Fig. S1 in the supplemental material), suggesting that the test population may be a good and unbiased sampling of the natural S. Enteritidis population. Notably, we have completed the testing of an additional 250 S. Enteritidis isolates of human, animal, and environmental origins from 7 Canadian provinces; no new clade was identified, and all isolates clustered into one clade or another from the list of 12 reported in this study. Thus, the SNP-PCR shows excellent typeability and is expected to become a very useful tool for source attribution.
An accurate prediction of mutation rates in Salmonella Enteritidis could shed light on how long it might take an S. Enteritidis isolate to accumulate enough mutations that may alter its clade designation.
If the rate is similar to that reported for Salmonella Typhimurium at 3 to 5 SNPs per year per chromosome (29), it would appear that mutations are infrequent in Salmonella and clades may be quite stable. Two isolates obtained from the same premises 1 year apart, namely, SENT 27 and 28, which were found to cluster in this study, i.e., clade 10, showed very little difference based on whole-genome analysis and annotation (28). There was a total of 82 single nucleotide changes (ΔSNP) between the two isolates, implying a very close relationship (28). The gene compositions of both isolates were nearly identical (4,703 genes versus 4,712 genes). However, we observed that a total of 15 genes present in both isolates showed mutations only in the isolate obtained 12 months later. Thus, it appears that over a period of many months, a small number of mutations accumulated in a significant number of genes in this strain (data not shown). Two of the affected genes, rfbU and rfaL code for enzymes involve in the synthesis and processing of the O antigen of lipopolysaccharide, one of the most important virulence factors for S. Enteritidis and all the other Gram-negative bacterial pathogens (30). Many phages use the O group antigen as a receptor to enter Salmonella (31), and mutational changes in the lipopolysaccharide could alter the phage infection phenotype. We noticed a similar change in another pair of genomes studied (SENT 17 and 18) (Fig. 1) in which a mutation was found in another gene in the rfb operon, rfbP, which codes for galactose phosphotransferase (data not shown), which is also involved in O antigen biosynthesis (32). This last pair of genomes had a ΔSNP of 29, suggesting an even closer relationship, which was not surprising, because the isolates were obtained from the same poultry hatchery operation on the same day. The rfbP mutation was one of 3 genes that showed differences between the two isolates. In both pairs of genomes (SENT 17/18 pair and SENT 27/28 pair), there was a conversion of phage type results from PT 13a (SENT 27) or PT 8 (SENT 18) to PT 23 (both SENT 28 and SENT 17, respectively) (see Data Set S1 in the supplemental material), suggesting that a mutation in an enzyme that acts on the lipopolysaccharide could lead to an alteration in S. Enteritidis phage type results and may explain the mechanism whereby closely related bacteria may produce different phage type results, as proposed by Van Belkum et al. (17).
Many authors have concluded that S. Enteritidis isolates show a limited genetic diversity (19, 33). Observations about the identification of predominant PFGE types of S. Enteritidis have reinforced this point of view. Our observations also agree with previous studies that reported a remarkably high degree of homogeneity in the genomes of Salmonella Enteritidis (5, 22); however, we also found considerable variability at the level of single nucleotides. In addition, we found many indels among our study of 11 S. Enteritidis genomes which, together with SNPs, could lead to pseudogene formation (D. Ogunremi, S. Belanger, E. Lerat, K. Omidi, and I. Galvan Marquez, unpublished observations). Another source of variation in S. Enteritidis genomes is the frequent incorporation of bacteriophages (24). Future work is needed to explain how the observed genetic diversity in S. Enteritidis may mechanistically explain the phenotypic and behavioral differences seen when different isolates are used in studies such as animal infection trials, in vitro cell invasion assays, and studies of the ability of various isolates to contaminate internal contents of eggs and survive (5, 34).
The use of a fairly large number of SNPs (n = 60) provides extremely useful and robust data about genetic distances and relatedness among and between clades, which we anticipate using to construct an evolutionary map of S. Enteritidis after testing a large number of isolates. With the benefit of metadata such as time, location, and host from which the isolates were obtained, we hope to use the genetic distances to construct an evolutionary map of S. Enteritidis in Canada, which among other things could delineate the strings of mutation and the order in which they occurred from one isolate to a related but chronologically younger isolate. However, it is also important to appreciate that while testing for these large number of SNPs could be ideal for interpreting S. Enteritidis evolution, a much smaller number of SNPs is all that is required to demonstrate relatedness among isolates. The identification of signature SNPs among half the clades will suggest that testing at 1 to 5 loci may be sufficient to rule in or out a membership in those clades. For the remaining half of the clades, it is probable that testing of 10 to 12 loci will suffice for clade assignment.
Another important attribute of the S. Enteritidis SNP-PCR is the low cost of running the procedure. For equipment, any real-time PCR machine will suffice, and one that can handle a 384-well plate will be an advantage when high throughput is required. The cost of reagents can be as low as $0.25 per SNP per isolate. Testing an isolate at 60 SNP loci, many more than will ever be required for a subtyping procedure, would still make the test comparable to or cheaper than existing subtyping procedures ($26 for phage typing and $36 for two-enzyme PFGE analysis in reagent costs). The labor costs of running the S. Enteritidis SNP-PCR test (2 h PCR time) and analyzing the results are at least an order of magnitude lower than those of the other two subtyping tests, each of which takes days to complete and requires a high level of technical expertise for both setup and analysis of results. Testing of a few SNPs will certainly reduce the cost of reagents and labor and the time it takes to get results. The starting material for S. Enteritidis SNP-PCR could be a very crude DNA extract, which is in stark contrast to PFGE, which utilizes a very involved procedure of digesting intact or high-molecular-weight DNA in a gel bed. The entire S. Enteritidis SNP-PCR test can be completed in a single working day if a bacterial culture is available in the morning and the number of loci targeted is not extensive. In our laboratory, an analyst with 2 PCR machines at her disposal comfortably completed the SNP-PCR on 88 bacterial isolates at 8 loci within 8 h. The S. Enteritidis SNP-PCR test shows very good reproducibility (99%) in tests conducted in two laboratories. An ideal subtyping test is expected to meet seven criteria, including cost effectiveness, rapid performance, robust results, typeability, high discrimination, reproducibility, and epidemiological concordance (17, 35). We have described in various degrees above how the SNP-PCR test meets six of the seven characteristics of an ideal subtyping test. The last feature of an ideal typing procedure, epidemiological concordance, was not demonstrated in this report, since the data presented did not include isolates from humans. To that end, we have recently completed the testing of 121 human isolates, and the data provide strong evidence of the epidemiological usefulness of the test. First, all of the isolates, without exception, were assigned to a clade based on the SNP pattern observed for each isolate. Second, the clustering of isolates into the respective clades is in agreement with the exposure pattern of the sick individuals, suggesting that humans with S. Enteritidis isolates belonging to the same clade may have been exposed to the same contaminant (S. Bekal, personal communication; unpublished data).
This report shows that the newly developed SNP subtyping procedure is highly discriminatory and is a marked improvement over PFGE and phage typing as a means of showing quantifiable differences among unrelated isolates. The discrete distribution of SNPs leading to identification of signature SNPs among some of the isolates is a very intriguing observation that may shed more light on S. Enteritidis genetics and evolution. Our recent observations may be the first to shed light on the mechanism of why related isolates may have different phage types. Finally, the previously intractable problem of differentiating among unrelated S. Enteritidis isolates appears to have been resolved with the development of a new subtyping test for S. Enteritidis, paving the way for a more effective strategy for controlling the most prevalent Salmonella serovars causing food-borne diseases in humans.
Supplementary Material
ACKNOWLEDGMENTS
Financial support was provided by the Canadian Food Inspection Agency.
We acknowledge the contributions of Susan Nadin-Davis, Victoria Arling, Katie Eloranta, Neil Vary, Sean Quinlan, Sam Mohajer, Ray Allain, Mohamed Elmufti, Olga Andrievskaia, Katayoun Omidi, and Imelda Galvan Marquez of the Canadian Food Inspection Agency, Sadjia Bekal of Pathogènes entériques Laboratoire de santé publique du Québec, and Roger Johnson, Linda Cole, Ann Perets, Ketna Mistry, and Betty Wilkie of the OIÉ Reference Laboratory for Salmonellosis, Public Health Agency of Canada, Guelph, Canada. We are grateful to Ray Allain of the OLF PFGE unit for sharing the Canadian PulseNet data.
Footnotes
Published ahead of print 8 October 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01410-14.
REFERENCES
- 1. Vieira AR. 2009. WHO Global Foodborne Infections Network Country Databank–a resource to link human and non-human sources of Salmonella. ISVEE Conference, Durban, South Africa: http://www.who.int/gfn/activities/CDB_poster_Sept09.pdf Accessed 23 October 2013. [Google Scholar]
- 2. Nesbitt A, Ravel A, Murray R, McCormick R, Savelli C, Finley R, Parmley J, Agunos A, Majowicz S. Enteritidis, Gilmour M. 2012. Integrated surveillance and potential sources of Salmonella Enteritidis in human cases in Canada from 2003 to 2009. Epidemiol Infect 140:1757–1772. 10.1017/S0950268811002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rodrigue DC, Tauxe RV, Rowe B. 1990. International increase in Salmonella enteritidis: a new pandemic? Epidemiol. Infect. 105:21–27. 10.1017/S0950268800047609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sabat AJ, Budimir A, Nashev D, Sá-Leão R, van Dijl JM, Laurent F, Grundmann H, Friedrich AW, ESCMID Study Group of Epidemiological Markers (ESGGEM) 2013. Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Eurosurveillance 18:20380 http://www.eurosurveillance.org/ViewArticle.aspx?Articleid=20380. [DOI] [PubMed] [Google Scholar]
- 5. Guard J, Morales CA, Fedorka-Cray P, Gast RK. 2011. Single nucleotide polymorphisms that differentiate two populations of Salmonella enteritidis within phage type. BMC Res. Notes 4:369. 10.1186/1756-0500-4-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zheng J, Keys CE, Zhao S, Meng J, Brown EW. 2007. Enhanced subtyping scheme for Salmonella Enteritidis. Emerg. Infect. Dis. 13:1932–1935. 10.3201/eid1312.070185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rabsch W. 2007. Salmonella Typhimurium phage typing for pathogens. Methods Mol Biol 394:177–211. 10.1007/978-1-59745-512-1_10. [DOI] [PubMed] [Google Scholar]
- 8. Rabsch W, Truepschuch S, Windhorst D, Gerlach RG. 2011. Typing phages and prophages of Salmonella, p 25–48 In Porwollik S. (ed), Salmonella—from genome to function. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 9. Valdezate S, Echeita A, Díez R, Usera MA. 2000. Evaluation of phenotypic and genotypic markers for characterisation of the emerging gastroenteritis pathogen Salmonella hadar. Eur. J. Clin. Microbiol. Infect. Dis. 19:275–281. 10.1007/s100960050475. [DOI] [PubMed] [Google Scholar]
- 10. Henzler DJ, Ebel JE, Sanders J, Kradel D, Mason J. 1994. Salmonella enteritidis in eggs from commercial chicken layer flocks implicated in human outbreaks. Avian Dis. 38:37–43. 10.2307/1591834. [DOI] [PubMed] [Google Scholar]
- 11. Hogue A, White P, Guard-Petter J, Schlosser W, Gast R, Ebel E, Farrar J, Gomez T, Madden J, Madison M, McNamara AM, Morales R, Parham D, Sparling P, Sutherlin W, Swerdlow D. 1997. Epidemiology and control of egg-associated Salmonella Enteritidis in the United States of America. Rev. Sci. Tech. 16:542–553. [DOI] [PubMed] [Google Scholar]
- 12. Chen C-L, Wang C-Y, Chu C, Su L-H, Chiu C-H. 2009. Functional and molecular characterization of pSE34 encoding a type IV secretion system in Salmonella enterica serotype Enteritidis. FEMS Immunol. Med. Microbiol. 57:274–283. 10.1111/j.1574-695X.2009.00612.x. [DOI] [PubMed] [Google Scholar]
- 13. Íçgen B, Gürakan GC, Özcengiz G. 2001. Effects of plasmid curing on antibiotic susceptibility, phage type, lipopolysaccharide and outer membrane profiles in local Salmonella isolates. Food Microbiol. 18:631–635. 10.1006/fmic.2001.0437. [DOI] [Google Scholar]
- 14. Glosnicka R, Dera-Tomaszewska B. 1999. Comparison of two Salmonella enteritidis phage typing schemes. Eur J. Epidemiol. 15:395–401. [PubMed] [Google Scholar]
- 15. Baggesen DL, Sørensen G, Nielsen EM, Wegener HC. 2010. Phage typing of Salmonella Typhimurium—is it still a useful tool for surveillance and outbreak investigation? Eurosurveillance 15:19471 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19471. [PubMed] [Google Scholar]
- 16. Tankouo-Sandjong B, Kinde H, Wallace I. 2012. Development of a sequence typing scheme for differentiation of Salmonella Enteritidis strain. FEMS Microbiol. Lett. 331:165–175. 10.1111/j.1574-6968.2012.02568.x. [DOI] [PubMed] [Google Scholar]
- 17. Van Belkum A, Tassios PT, Dijkshoorn L, Haeggman S, Cookson B, Fry NK, Fussing V, Green J, Feil E, Gerner-Smidt P, Brisse S, Struelens M. 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13(Suppl 3):1–46. 10.1111/j.1469-0691.2007.01786.x. [DOI] [PubMed] [Google Scholar]
- 18. Boxrud D, Pederson-Gulrud K, Wotton J, Medus C, Lyszkowicz E, Besser J, Bartkus JM. 2007. Comparison of multiple-locus variable-number tandem repeat analysis, pulsed-field gel electrophoresis, and phage typing for subtype analysis of Salmonella enterica serotype Enteritidis. J. Clin. Microbiol. 45:536–543. 10.1128/JCM.01595-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Olson AB, Andrysiak AK, Tracz DM, Guard-Bouldin J, Demczuk W, Ng LK, Maki A, Jamieson F, Gilmour MW. 2007. Limited genetic diversity in Salmonella enterica serovar Enteritidis PT13. BMC Microbiol. 7:87. 10.1186/1471-2180-7-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kircher M, Kelso J. 2010. High-throughput DNA sequencing—concepts and limitations. Bioessays 32:524–536. 10.1002/bies.200900181. [DOI] [PubMed] [Google Scholar]
- 21. Timme RE, Allard MW, Luo Y, Strain E, Pettengill J, Wang C, Keys CE, Zheng J, Stones R, Wilson MR, Musser SM, Brown EW. 2012. Draft genome sequences of 21 Salmonella enterica serovar Enteritidis strains. J. Bacteriol. 194:5994–5995. 10.1128/JB.01289-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Allard MW, Luo Y, Strain E, Pettengill J, Timme R, Wang C, Li C, Keys CE, Zheng J, Stones R, Wilson MR, Musser SM, Brown EW. 2013. On the evolutionary history, population genetics and diversity among isolates of Salmonella Enteritidis PFGE pattern JEGX011.0004. PLoS One 8:e55254. 10.1371/journal.pone.0055254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ward LR, de Sa JDH, Rowe B. 1987. A phage-typing scheme for Salmonella enteritidis. Epidemiol. Infect. 99:291–294. 10.1017/S0950268800067765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ogunremi D, Devenish J, Amoako K, Kelly H, Dupras A, Belanger S, Wang L. 2014. Whole genome sequencing and characterization of eleven isolates of Salmonella Enteritidis obtained in Canada. BMC Genomics 15:713. 10.1186/1471-2164-15-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Song J, Xu Y, White S, Miller KWP, Wolinsky M. 2005. SNPsFinder—a web-based application for genome-wide discovery of single nucleotide polymorphisms in microbial genomes. Bioinformatics 21:2083–2084. 10.1093/bioinformatics/bti176. [DOI] [PubMed] [Google Scholar]
- 26. Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carriço JA, Silva-Costa C, Melo-Cristino J, Pinto FR, de Lencastre H, Almeida JS, Ramirez M. 2006. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J. Clin. Microbiol. 44:2524–2532. 10.1128/JCM.02536-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Severiano A, Pinto FR, Ramirez M, Carriço JA. 2011. Adjusted Wallace coefficient as a measure of congruence between typing methods. J. Clin. Microbiol. 49:3997–4000. 10.1128/JCM.00624-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hawkey J, Edward DJ, Dimovski K, Hiley L, Billman-Jacobe H, Hogg G, Holt KE. 2013. Evidence of microevolution of Salmonella Typhimurium during a series of egg-associated outbreaks linked to a single chicken farm. BMC Genomics 14:800. 10.1186/1471-2164-14-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Munford RS. 2008. Sensing gram-negative bacterial lipopolysaccharide: a human disease determinant. Infect. Immun. 76:454–465. 10.1128/IAI.00939-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim M, Ryu S. 2012. Spontaneous and transient defence against bacteriophage by phase-variable glucosylation of O antigen in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 86:411–425. 10.1111/j.1365-2958.2012.08202.x. [DOI] [PubMed] [Google Scholar]
- 32. Wang L, Reeves PR. 1994. Involvement of the galactosyl-1-phosphate transferase encoded by the Salmonella enterica rfbP gene in O-antigen subunit processing. J. Bacteriol. 176:4348–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Botteldoorn N, Van Coillie E, Goris J, Werbourck H, Piessens V, Godard C, Scheldeman P, Herman L, Heyndricks M. 2010. Limited genetic diversity and gene expression differences between egg- and non-egg-related Salmonella enteritidis strains. Zoonoses Public Health 57:345–357. 10.1111/j.1863-2378.2008.01216.x. [DOI] [PubMed] [Google Scholar]
- 34. Guard J, Shah D, Morales CA, Call D. 2011. Evolutionary trend associated with niche specialization as modelled by whole genome analysis of egg-contaminating Salmonella enterica serovar Enteritidis, p 91–106 In Porwollik S. (ed), Salmonella—from genome to function. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 35. Kruczkiewicz P. 2013. A comparative genomic framework for in silico design and assessment of molecular typing methods using whole-genome sequence data with application to Listeria monocytogenes. M.Sc. thesis University of Lethbridge, Lethbridge, Canada. [Google Scholar]
- 36. Hooper SA, Mawer S. 1988. Salmonella enteritidis in a commercial layer flock. Vet. Rec. 123:351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.