Significance
Both Hippo and c-Jun N-terminal kinase (JNK) signaling have well-established roles in promoting tissue growth, while it remains elusive whether and how the two pathways interact or coordinate in growth control. We report in the present study that the Hpo–Yki pathway modulates Rho1–JNK signaling-mediated growth in Drosophila wing development. We show that the impaired Hpo pathway activates JNK through transcriptional up-regulation of Rho1. These findings expands our understanding about the cross-talk between two important signaling pathways, namely Hippo–Yki and Rho1–JNK, in regulating growth.
Keywords: Hippo, JNK, growth, Rho1, Drosophila
Abstract
The Hippo and c-Jun N-terminal kinase (JNK) pathway both regulate growth and contribute to tumorigenesis when dysregulated. Whereas the Hippo pathway acts via the transcription coactivator Yki/YAP to regulate target gene expression, JNK signaling, triggered by various modulators including Rho GTPases, activates the transcription factors Jun and Fos. Here, we show that impaired Hippo signaling induces JNK activation through Rho1. Blocking Rho1–JNK signaling suppresses Yki-induced overgrowth in the wing disk, whereas ectopic Rho1 expression promotes tissue growth when apoptosis is prohibited. Furthermore, Yki directly regulates Rho1 transcription via the transcription factor Sd. Thus, our results have identified a novel molecular link between the Hippo and JNK pathways and implicated the essential role of the JNK pathway in Hippo signaling-related tumorigenesis.
Tumor progression often involves deregulated signaling pathways that lead to unchecked proliferation and evasion of apoptosis, which are considered two important hallmarks of cancer development (1). Recently, the Hippo pathway was shown to play an evolutionarily conserved role in regulating cell growth, proliferation, and survival in tumorigenesis (2, 3). In Drosophila, the core Hippo pathway acts through a kinase cascade consisting of Hippo (Hpo) and Warts (Wts) to inactivate the transcriptional coactivator Yorkie (Yki; YAP/TAZ in mammals) (4, 5). Yki/YAP interacts with different DNA-binding transcription factors to activate transcription of growth-regulating genes, including cyclin E (cycE), E2f1, and Drosophila inhibitor of apoptosis protein 1 (Diap1) (6).
The c-Jun N-terminal kinase (JNK) pathway plays a crucial role in regulating a wide range of cellular activities including proliferation, differentiation, migration, and cell death during tumor progression (7). JNK signaling is also required for tumor growth triggered by loss of cell polarity and oncogenic Ras cooperation in Drosophila (8, 9), and for progenitor cell proliferation and Ras-induced tumorigenesis in mammals (10, 11).
Although both Hpo and JNK signaling have been implicated in cell proliferation and tissue growth, it has remained elusive how the two pathways are coordinated in vivo. Recently some contradictory findings were reported in Drosophila. On one hand, JNK signaling induces Yki activation during compensatory cell proliferation and neoplastic tumor growth (12–14); on the other hand, JNK suppressed Yki elevation in scrib mutant cells during growth regulation (15, 16). However, it remains unknown whether JNK signaling is regulated by the Hpo–Yki pathway and contributes to its growth function.
Here we show that impaired Hpo signaling activates the Rho1–JNK pathway in Drosophila. Compromised Rho1–JNK signaling suppresses Yki-induced cell proliferation and tissue overgrowth in the developing wing, whereas activated Rho1–JNK signaling promotes proliferation and growth when cell death is blocked. Sd is necessary and sufficient for Yki-induced JNK activation. We have further identified rho1 as a direct transcriptional target of the Yki/Sd complex. Our data in Drosophila suggest a possible interaction in growth control between Hpo–Yki and Rho–JNK signaling in mammals.
Results
Impaired Hpo Signaling Up-Regulates JNK Activity.
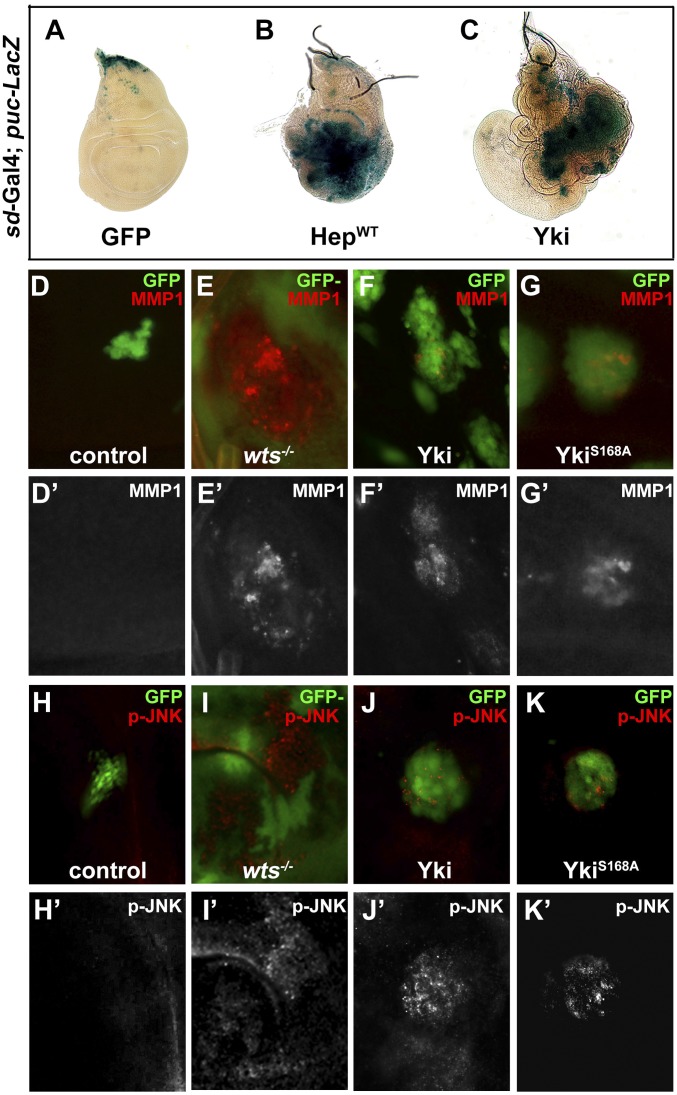
To investigate whether Hpo signaling could modulate JNK activity, we checked the expression of puc, a transcriptional target and a readout of JNK activity in vivo (17). Compared with the control (Fig. 1A), inactivation of Hpo signaling by sd–Gal4-driven Yki expression resulted in up-regulated puc–LacZ expression in the overgrown wing pouch (Fig. 1C). As a positive control, puc–LacZ expression was strongly elevated by expressing the JNK kinase hemipterous (hep) (Fig. 1B). To probe whether JNK is activated by Yki in an autonomous and/or a spatial manner, we examined puc transcription in Flp-out clones expressing a hyperactive form of Yki (YkiS168A). JNK activation was observed throughout the wing disk autonomously (Fig. S1), although occasionally cells adjacent to Yki overexpression clones were also found to have increased puc activity (Fig. S1, white arrow), which is likely caused by the supercompetitive activity of Yki clones (18).
Fig. 1.
Yki activates JNK signaling in Drosophila. Third instar wing discs are shown. Ectopic Yki activates JNK target puc expression. puc-LacZ expression is shown in sd > GFP control (A), sd>HepWT (B), and sd > Yki (C) wing discs. Compared with control clones (D and H), impaired Hippo signaling activates MMP1 expression and JNK phosphorylation in wts mutant clones (E and I), or clones expressing Yki (F and J) or YkiS168A (G and K). Clones were labeled by the presence (D, F, G, H, J, and K) or absence (E and I) of GFP expression. See also Figs. S1 and S2.
Next, to further verify the result, we checked the expression of matrix metalloprotease1 (MMP1), another direct transcriptional target of the JNK pathway (19). MMP1 was normally detected in trachea and a small region of notum in wild-type wing discs (Fig. S2A) (20). We generated loss-of-function and gain-of-function clones in wing discs and found that loss of the wts gene and forced expression of Yki or YkiS168A all resulted in strong MMP1 activation in the clones (Fig. 1 D–G). In addition, both MMP1 and puc–LacZ expression were up-regulated by ectopic Yki or YkiS168A driven by ptc–Gal4 (Figs. S2 B and C and S3F). Finally, we examined JNK activation directly using an antibody specific to the phosphorylated JNK (p-JNK). Elevated p-JNK staining was detected in wts mutant clones as well as cells expressing Yki or YkiS168A (Fig. 1 H–K and Fig. S2E). Together, these data indicate that impaired Hpo signaling results in JNK activation in vivo.
JNK Is Required for Yki-Induced Overgrowth.
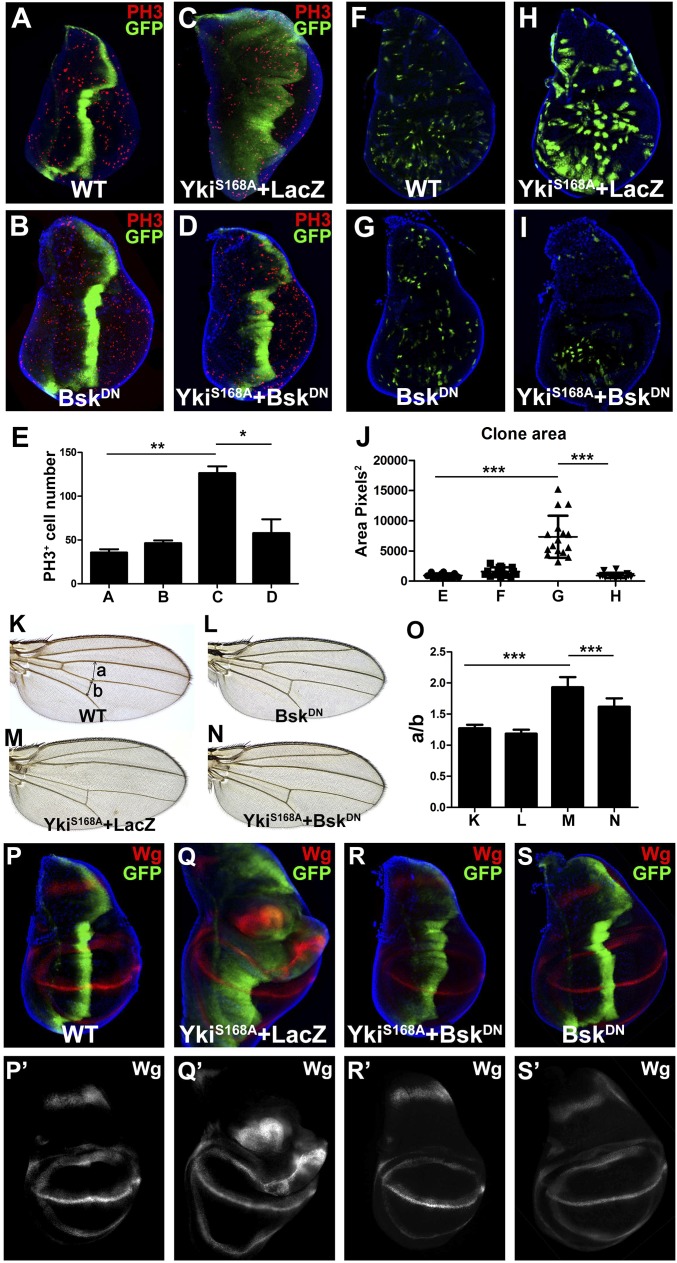
The Hpo pathway plays a key role in organ size control through the regulation of cell number (21). Consistently, immunostaining with an antibody against phospho-histone 3 (PH3), a specific marker for mitotic cells, indicated an increase of PH3+ cells in ptc > YkiS168A wing discs (Fig. 2 C and E). Blocking JNK signaling by expressing a dominant negative form of basket (BskDN, bsk encodes the Drosophila JNK) dramatically suppressed such increase of PH3+ cells and the expansion of the GFP+ region (Fig. 2 D and E). Consistently, reducing JNK activity also strongly impeded loss of wts-induced overgrowth phenotype (Fig. S2 F and G). In addition, clone overgrowth in wing discs resulting from expression of YkiS168A was also suppressed by BskDN (Fig. 2 F–J). Furthermore, expression of YkiS168A along the anterior-posterior (A-P) compartment boundary driven by ptc–Gal4 resulted in an expansion of the intervein region between L3 and L4 in adult wings, which was also suppressed by expression of BskDN (Fig. 2 K–O). However, expression of BskDN by itself had no effect on growth (Fig. 2 B, G, and L) and did not affect YkiS168A-triggered cell death, as revealed by immunostaining for active caspase 3 (Casp3) (Fig. S3 A–C). The small amount of Casp3 activation observed in ptc > YkiS168A discs (Fig. S3B) is consistent with a previous report (18). Together, these results suggest that JNK signaling is required for Yki-induced cell proliferation and organ growth.
Fig. 2.
JNK is required for Yki-induced overgrowth and Wg expression. (A–J) Yki induces JNK-dependent overgrowth in wing discs. Third instar wing discs of ptc > GFP (A), ptc> BskDN (B), ptc > YkiS168A + LacZ (C), or ptc > YkiS168A + BskDN (D) were stained for PH3 and DNA (DAPI). (E) Average PH3+ cells within ptc expression domain in A–D are shown. **P < 0.01, *P < 0.05 (mean + SD, n ≥ 5). (F–I) Third instar wing discs bearing flip-out clones expressing GFP without (F) or with BskDN (G), YkiS168A + LacZ (H), or YkiS168A + BskDN (I). (J) Statistical analyses of clone sizes in F–I. ***P < 0.001 (mean ± SD, n = 15). (K–N) Blocking JNK activity suppressed Yki-induced expansion of the intervein region between L3 and L4 in adult wings. Light micrographs of adult wings from ptc > GFP (K), ptc> BskDN (L), ptc > YkiS168A, tub-Gal80ts + LacZ (M), or ptc > YkiS168A, tub-Gal80ts + BskDN (N) flies. (O) Average ratio of a/b in K–N. a, distance between L3 and L4; b, length of posterior cross-vein between L4 and L5. Positions of a and b are shown in K. ***P < 0.001 (mean + SD, n = 10). (P–S) JNK is required for Yki-induced ectopic Wg expression. Third instar wing discs of ptc > GFP (P), ptc > YkiS168A + LacZ (Q), ptc > YkiS168A + BskDN (R), or ptc> BskDN (S) were stained with DNA (DAPI) and Wg. See also Figs. S3–S5.
Next, to investigate the molecular mechanism by which JNK mediates Yki-induced overgrowth, we analyzed the expression of Yki target genes, including wingless (wg), Expanded (ex), Diap1, dmyc, bantam (ban), and CycE, that are required for growth control (18, 22–24). Blocking JNK signaling fully suppressed Yki-induced up-regulation of Wg (Fig. 2 P–S), but not that of diap1–LacZ, ex–LacZ, dmyc–LacZ, and ban–LacZ (Fig. S4 A–F and I–N). Furthermore, consistent with Wg’s role in regulating Yki-triggered growth, we found Yki-induced proliferation and growth phenotype were significantly blocked by knocking down wg (Fig. S4 G and H). We noted that Yki failed to induce CycE expression in the developing wing (Fig. S4 O and P), in accordance with a previous study that the Hippo pathway regulates CycE in a tissue-specific manner (4). As a positive control, CycE was detected when ectopically expressed in wing discs (Fig. S4Q). As expected, elevated expression of puc and MMP1 induced by YkiS168A were both suppressed by BskDN (Fig. S3 H and I). Together, these data suggest that wg is not a direct transcriptional target of Yki, but rather, regulated by the Hpo–Yki pathway indirectly through JNK signaling. Consistent with this explanation, Wg was reported to act downstream of JNK signaling to promote compensatory cell proliferation (25).
Moreover, to find out whether Yki induces JNK-mediated growth in a tissue-specific manner, we checked genetic interactions between Hpo–Yki and JNK signaling in the developing eye. First, we found expression of Yki or a wts RNAi under GMR promoter induces overgrowth in the adult eye, which cannot be suppressed by coexpression of BskDN (Fig. S5 A–E). Second, the overgrowth phenotype of eye disk clones expressing YkiS168A cannot be suppressed by blocking JNK signaling (Fig. S5 F and G). Thus, the role of JNK in regulating loss of Hpo signal-induced growth is highly context dependent.
Yki Regulates JNK Signaling Through Rho1.
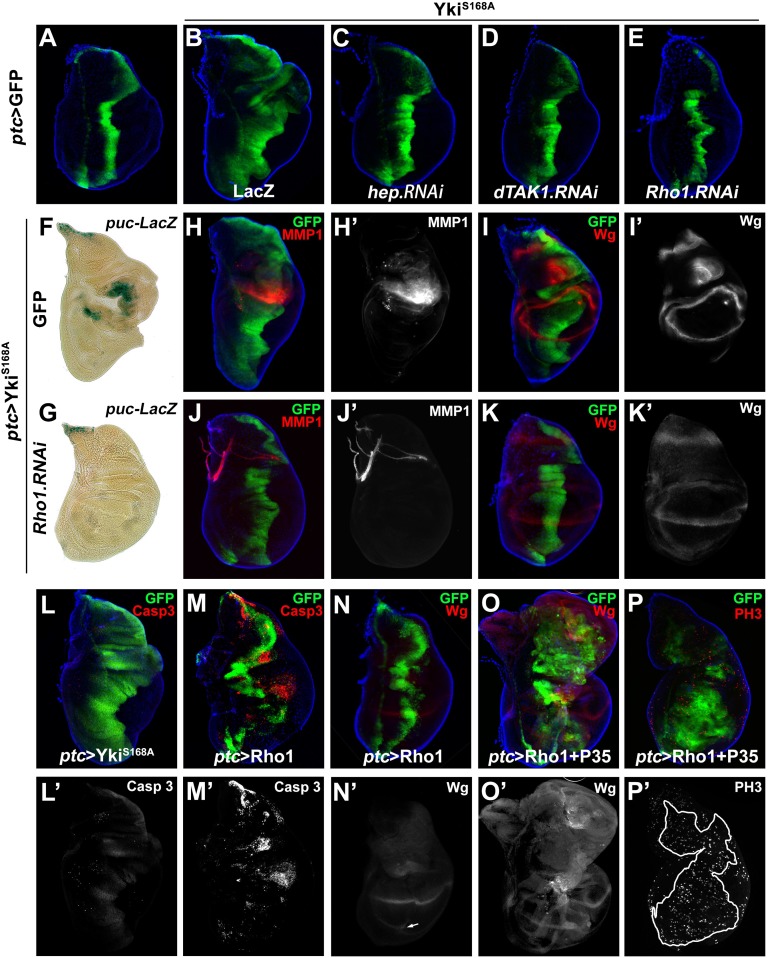
To investigate the molecular mechanism underlying Yki-induced JNK activation, we examined genetic epistasis between Yki and upstream components of JNK signaling. Compromised JNK signaling by knocking down hep or JNKK kinase dTAK1 significantly suppressed ptc > YkiS168A-induced disk overgrowth (Fig. 3 A–D), indicating Yki functions upstream of dTAK1 and Hep. The Rho GTPase family members were known to regulate JNK signaling upstream of dTAK1 (26, 27), and in particular, Rho1 has been shown to induce JNK-mediated apoptosis and compensatory proliferation (28, 29). Consistently, Rho1 expression driven by ptc–Gal4 resulted in elevated puc expression (Fig. S6 D and E). Knocking down Rho1 expression by two independent RNAi significantly blocked Yki-triggered overgrowth (Fig. 3E and Fig. S6C), as well as Yki-induced up-regulation of puc–LacZ, MMP1, and Wg (Fig. 3 F–K). Taken together, these results suggest that Rho1 mediates Yki-induced JNK activation and overgrowth.
Fig. 3.
Rho1 is required for Yki-induced overgrowth and JNK activation. (A–E) Third instar wing discs of control (A) or expressing YkiS168A + LacZ (B), YkiS168A + hep RNAi (C), YkiS168A + dTAK1 RNAi (D), and YkiS168A + Rho1 RNAi (E) are shown. (F–K) Third instar wing discs stained with X-Gal (F and G), MMP1 (H and J), or Wg (I and K). Yki-induced expansion of GFP+ region (H and I), JNK target gene expression (F and H), and Wg expression (I) were all suppressed by knocking down Rho1 (G, J, and K). (L and M) Third instar wing disk stained with cleaved caspase 3. Strong apoptosis was induced by ptc > Rho1 (M), but not ptc > YkiS168A (L). (N–P) Blocking Rho1-triggered apoptosis by P35-stimulated Wg production (O) and cell proliferation (P). See also Figs. S6 and S7.
To examine whether increased Rho1 activity is able to drive cell proliferation and tissue growth, we expressed Rho1 in the wing discs under ptc promoter. In agreement with a previous study (28), Rho1 expression induced strong cell death (Fig. 3M) and mild Wg expression (Fig. 3N) without expansion of the GFP+ region (Fig. 3 M and N). Intriguingly, when cell death was blocked by P35, Rho1 was able to phenocopy YkiS168A-induced overgrowth (Fig. 3 L and O) and proliferation (Fig. 3P), coupled with Wg induction (Fig. 3O). Interestingly, we also observed increased Yki nuclear accumulation in Rho1 + p35 expression cells (Fig. S7D), an indication of Yki activation (4), suggesting a positive feedback loop exits between Yki and Rho1 in growth control. In agreement with this view, we found reducing JNK or Yki activity both significantly impeded Rho1-induced growth (Fig. S7 A–C). Hence, the above data indicate Yki promotes tissue growth by concerted action of downstream signaling pathways of both Rho1–JNK–mediated Wg expression, cell proliferation, and apoptosis and dIAP1-mediated inhibition of apoptosis.
Sd Is Essential for Yki-Induced JNK Activation.
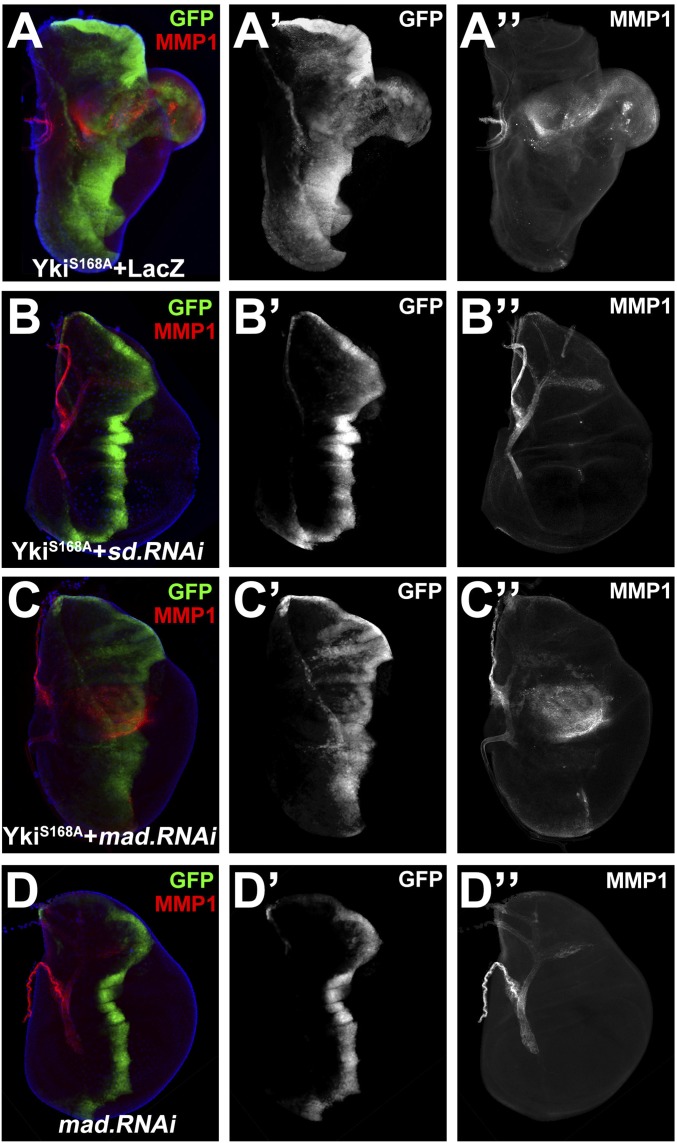
Sd is the best-characterized DNA-binding partner of Yki, and loss of sd fully suppresses Yki-induced tissue overgrowth and elevated target gene transcription (24). Consistently, we found YkiS168A-induced overgrowth and MMP1 expression were dramatically suppressed by knocking down sd (Fig. 4 A and B), whereas ectopic expression of a constitutively active form of Sd (SdGA) (30) was sufficient to induce MMP1 expression (Fig. S8A) and puc transcription (Fig. S8B). Apart from Sd, Mad was recently reported as another transcription factor that cooperates with Yki to regulate growth (31). Consistently, we found loss of mad partially suppressed Yki-induced growth (Fig. 4C′ and Fig. S9); however, the induction of MMP1 remained unaffected (Fig. 4C′′). Collectively, these data indicate that Yki acts through Sd to promote JNK-mediated growth.
Fig. 4.
Yki induces Sd-dependent JNK activation. Knocking down sd suppressed Yki-induced JNK activation. Third instar wing discs were stained for MMP1. YkiS168A-induced MMP1 up-regulation (A) is suppressed by expression of sd RNAi (B), but not that of mad RNAi (C and D). Genotype: ptc > YkiS168A + LacZ (A), ptc > YkiS168A + sd RNAi (B), ptc > YkiS168A + mad RNAi (C), and ptc> mad RNAi (D). See also Figs. S8 and S9.
Yki Directly Induces Rho1 Expression.
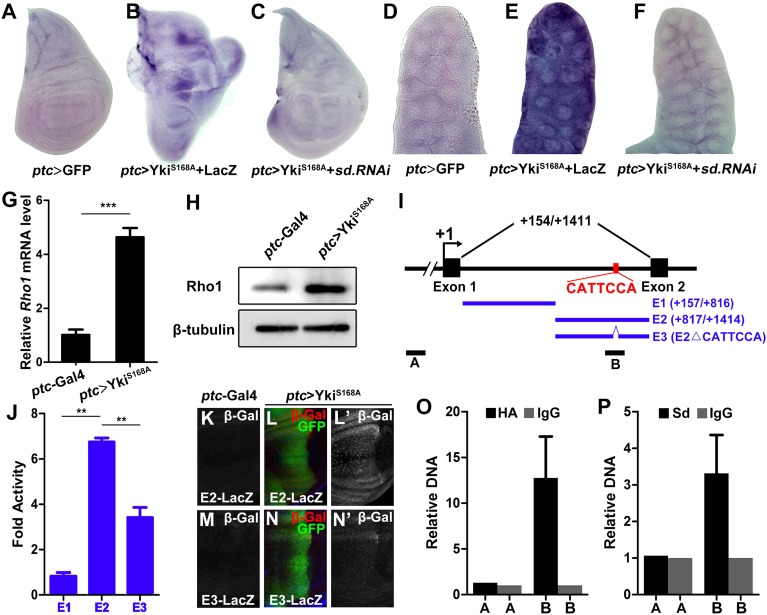
To further explore the molecular mechanism by which the Hippo–Yki pathway modulates Rho1–JNK signaling, we examined whether Yki is able to induce Rho1 transcription. To this end, we first checked Rho1 expression by quantitative real-time PCR (qRT-PCR) and Western blot. Expression of YkiS168A driven by ptc–Gal4 resulted in a significant elevation of Rho1 mRNA (Fig. 5G) and protein (Fig. 5H) in wing discs. This result was confirmed by in situ hybridization, in which Rho1 transcription was significantly up-regulated in wing discs (Fig. 5 A and B) and salivary glands (Fig. 5 D and E). Knocking down sd fully suppressed Yki-induced Rho1 transcription (Fig. 5 C and F), indicating Yki acts through Sd to induce Rho1 transcription.
Fig. 5.
Yki–Sd directly induces Rho1 transcription. (A–F) Knocking down sd suppresses Yki-induced Rho1 transcription. In situ hybridization to Rho1 mRNA of third instar wing discs (A–C) or salivary gland (D–F) expressing GFP (A and D), YkiS168A + LacZ (B and E), YkiS168A + sd RNAi (C and F) driven by ptc–Gal4. (G and H) Ectopic Yki up-regulates Rho1 mRNA (G) and protein (H) levels, as shown by quantitative PCR and Western blot, respectively. (n = 200, mean + SD, ***P < 0.001) (I) Scheme of the Rho1 locus showing the first intron. The Sd binding motif (CATTCCA) is indicated. E1, E2, and E3 were used to drive luciferase expression; A and B represent the control and target region of ChIP assays, respectively. (J) Luciferase assay in Drosophila S2 cells. (n = 3, mean + SD, **P < 0.01) (K–N) The Sd binding motif is required for Yki-induced target gene expression in vivo. Compared with the controls (K and M), ectopic expression of YkiS168A along the A-P compartment boundary results in dramatic up-regulation of E2–LacZ (L), but not E3–LacZ (N). (O and P) Drosophila S2 cells transfected with Sd and Yki–HA were used for quantification of ChIP-PCRs. (n = 3, mean + SD).
To investigate whether the Yki–Sd complex activates Rho1 transcription directly, we examined the Rho1 cis-regulatory region and identified a putative Sd binding motif (CATTCCA) (24, 30) in the first intron (Fig. 5I). To verify this motif as a true Sd binding site, we generated two firefly luciferase reporters containing either the first half (E1) or the second half (E2) of the intron, with the putative Sd binding motif located in the middle of E2 (Fig. 5I), and performed transient dual luciferase assay in S2 cells. Consistent with this prediction, we found E2, but not E1, was able to significantly induce luciferase expression upon Yki transfection (Fig. 5J). Moreover, deletion of the putative Sd binding motif (E3, Fig. 5I) abrogated the increase in luciferase expression (Fig. 5J). To validate the in vitro results, we generated LacZ reporter driven by E2 or E3. Whereas E2–LacZ was significantly induced in the wing disk upon Yki expression (Fig. 5 K and L), E3–LacZ was barely responsive to Yki (Fig. 5 M and N). Collectively, these data suggest that the CATTCCA motif is a true recognition site required for the Yki–Sd complex to activate Rho1 transcription.
Finally, to determine whether the Yki–Sd complex could directly bind to the putative Sd binding motif, we performed chromatin immunoprecipitation (ChIP) experiments in S2 cells cotransfected with HA-tagged Yki (HA–Yki) and Sd, followed by qPCR to generate DNA fragments that match either region A located upstream of the Rho1 transcription start site or region B that harbors the Sd binding motif (Fig. 5I). We found that DNA amplified from chromatin precipitated with antibodies against HA (Fig. 5O) or Sd (Fig. 5P) were appreciably enriched in region B, but not in region A. Whereas neither region was enriched with control IgG antiserum (Fig. 5 O and P). Together, the above results demonstrate that Yki–Sd directly regulates Rho1 transcription by targeting to the Sd binding motif located in the first intron of Rho1.
Discussion
Recent studies have revealed a complex interaction network between Hippo and other key signaling pathways, including TGF-β/SMAD and Wnt/β-catenin pathways (32), whereas its communication with JNK signaling remains elusive (12, 14, 15). Here we provide genetic evidences that impaired Hippo signaling promotes overgrowth through Rho1–JNK signaling in Drosophila. First, loss of Hippo signaling induces JNK activation and its target gene expression. Second, Yki-induced overgrowth is suppressed by blocking Rho1–JNK signaling. Third, ectopic Rho1 expression phenocopies Yki-triggered overgrowth and proliferation when cell death is compromised.
Yki/YAP’s ability in promoting tissue growth depends on transcription factors, including Sd/TEADs and SMADs (33). Consistent with this notion, we found Sd, but not Mad, is essential for Yki-induced JNK activation, whereas ectopic Sd expression is sufficient to activate JNK signaling by itself. We further implicated the Rho1 GTPase as the critical factor that bridges the interaction between Hippo and JNK signaling. Rho1 not only mediates Yki-induced JNK activation and overgrowth, but also serves as a direct transcriptional target of Yki/Sd complex. Intriguingly, we also found that Rho1 activation promotes nuclear translocation of Yki in wing discs, and reducing Yki activity significantly impeded Rho1 induced growth (Fig. S7), implying the existence of a potential positive feedback loop to amplify Yki-induced overgrowth and to help maintain signaling in a steady state. Consistent with our observation, recent studies reported that GPCRs could activate YAP/TAZ through RhoA in mammals (34, 35), whereas elevated JNK signaling in Drosophila could stimulate Yki nuclear translocation during regeneration and tissue growth (12–14). Thus, our results provide the other side of the story about a novel cross-talk between Hippo and JNK signaling.
Intriguingly, we found that ectopic Yki expression driven by ptc–Gal4 induced MMP1 activation (Fig. S2 B′–C′), puc–LacZ expression (Fig. 3F), rho1 transcription (Fig. 5B), and Yki target gene transcription (Fig. S4) predominantly in the proximal region of wing disk, but not that of the dorsal/ventral boundary. This is consistent with a recently published paper showing that tension in the center region of Drosophila wing disk is lower than that in the periphery, which correlates with lower Yki activity (36). It is also worth noting that despite the requirement of JNK signaling in Yki-induced wing overgrowth, JNK was not activated strictly in an autonomous manner upon Yki overexpression (Fig. S1). This could be caused by supercompetitive activity of Yki expression clones (18), or, alternatively, through a propagation of JNK signal into neighboring cells (37), which would be very interesting to study further.
Apart from its role in growth control, the Hippo pathway also regulates tumor invasion and metastasis (2). Similarly, JNK signaling plays a major role in modulating metastasis in both flies and mammals (38, 39). Rho1 was also reported to cooperate with oncogenic Ras to induce large invasive tumors (40). Hence, it is likely that Rho1 also acts as the molecular link between Yki and JNK signaling in modulating metastasis as well.
Materials and Methods
Drosophila Genetics and Stocks.
All stocks were reared on standard Drosophila media at 25 °C unless otherwise indicated. For experiments involving tub-Gal80ts, flies were raised at 18 °C to restrict Gal4 activity for 5 d, then shifted to 29 °C for 2 d to inactivate Gal80ts. The following stocks were used for this study: sd-Gal4, ptc-Gal4, UAS-GFP, UAS-p35, UAS-Rho1 (no. 7334), UAS-YkiS168A, UAS-Rho1 RNAi (nos. 27727 and 9910), UAS-LacZ (no. 3956), dmyc-LacZ (no. 11981), ban-LacZ (no. 10154), and tub-Gal80ts were obtained from the Bloomington Stock Center; UAS-sd RNAi (no. 101497), UAS-wg RNAi (no. 13352), and UAS-yki RNAi (no. 40497) were obtained from the Vienna Drosophila RNAi Center; UAS-SdGA (30), puc-LacZ, UAS-BskDN, UAS-hep RNAi, UAS-dTAK1 RNAi (41), ex-LacZ, diap1-LacZ, UAS-Yki (24), UAS-mad RNAi (42), and wtsX1 (43) were previously described. Clonal analysis is described in SI Materials and Methods.
Immunostaining and in Situ Hybridization.
Wing discs of third instar larvae were fixed in 4% (wt/vol) paraformaldehyde and stained as described previously (24), using mouse anti-MMP1 (1:200; Developmental Studies Hybridoma Bank, DSHB), mouse anti–β-Gal (1:1,000; DSHB), mouse anti-Wg (1:300; DSHB), rabbit antiphospho-JNK (1:200; Calbiochem), rabbit anti-PH3 [1:100; cell signaling technology (CST)], rabbit antiactive caspase 3 (1: 400; CST), Rat anti-CycE (1:200; gift from Helena Richardson, Peter MacCallum Cancer Centre, Melbourne), rabbit anti-Yki (1:1,000; gift from Duojia Pan, Johns Hopkins University School of Medicine, Baltimore). Secondary antibodies were anti-rabbit Alexa (1:1,000; CST) and anti-mouse Cy3 (1:1,000; Jackson Immuno Research). In situ hybridization was performed on wing discs and salivary glands as previously described (44). Antisense probes for full-length Rho1 were generated with digoxigenin-labeled deoxyribonucleotide triphosphates (Roche) using a pCS2–Rho1 construst and synthesized by T7 polymerase.
X-Gal Staining.
Eye and wing discs were dissected from third instar larvae in PBT (1× PBS pH 7.0, 0.1% Triton X-100) and stained for β-galactosidase activity as previously described (45).
Cell Culture, Transfection, Western Blot Analysis, Luciferase Reporter Assay, qRT-PCR, and ChIP Assay.
S2 cells were cultured in Insectagro DS2 (Corning) supplemented with 10% (vol/vol) FBS (HyClone), 50 units/mL of penicillin and 50 μg/mL of streptomycin. Transfection was performed using Effectene transfection reagent (Qiagen) according to the manufacturer’s instructions. qRT-PCR, Western blot analysis, luciferase reporter assay, and ChIP assay are described in SI Materials and Methods.
Statistical Analysis.
Quantification of the data is presented in bar graphs created with GraphPad Prism 5. Data represent mean values ± SD. We used one-way ANOVA with corrected Bonferroni multiple comparison test to calculate statistical significance in all our experiments.
Supplementary Material
Acknowledgments
We thank Dr. Yunyun Jin for communication of unpublished data and Drs. Duojia Pan, Tian Xu, Lei Zhang, and Helena Richardson and the Bloomington Stock Center, the Vienna Drosophila RNAi Center, and the Developmental Studies Hybridoma Bank for fly stocks and reagents. This research was supported by the National Basic Research Program of China (973 Program) (2010CB944901 and 2011CB943903), National Natural Science Foundation of China (31071294, 31171413, and 31371490), the PhD Programs Foundation of Ministry of Education of China (20120072110023), and the Shanghai Committee of Science and Technology (09DZ2260100 and 14JC1406000) (to L.X.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1415020112/-/DCSupplemental.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13(4):246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 3.Staley BK, Irvine KD. Hippo signaling in Drosophila: Recent advances and insights. Dev Dyn. 2012;241(1):3–15. doi: 10.1002/dvdy.22723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell. 2005;122(3):421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Zhang L, Yue T, Jiang J. Hippo signaling pathway and organ size control. Fly (Austin) 2009;3(1):68–73. doi: 10.4161/fly.3.1.7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13(8):877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weston CR, Davis RJ. The JNK signal transduction pathway. Curr Opin Cell Biol. 2007;19(2):142–149. doi: 10.1016/j.ceb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Ma X, et al. Bendless modulates JNK-mediated cell death and migration in Drosophila. Cell Death Differ. 2014;21(3):407–415. doi: 10.1038/cdd.2013.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma X, et al. dUev1a modulates TNF-JNK mediated tumor progression and cell death in Drosophila. Dev Biol. 2013;380(2):211–221. doi: 10.1016/j.ydbio.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 10.Cellurale C, et al. Requirement of c-Jun NH(2)-terminal kinase for Ras-initiated tumor formation. Mol Cell Biol. 2011;31(7):1565–1576. doi: 10.1128/MCB.01122-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sancho R, et al. JNK signalling modulates intestinal homeostasis and tumourigenesis in mice. EMBO J. 2009;28(13):1843–1854. doi: 10.1038/emboj.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun G, Irvine KD. Regulation of Hippo signaling by Jun kinase signaling during compensatory cell proliferation and regeneration, and in neoplastic tumors. Dev Biol. 2011;350(1):139–151. doi: 10.1016/j.ydbio.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohsawa S, et al. Mitochondrial defect drives non-autonomous tumour progression through Hippo signalling in Drosophila. Nature. 2012;490(7421):547–551. doi: 10.1038/nature11452. [DOI] [PubMed] [Google Scholar]
- 14.Sun G, Irvine KD. Ajuba family proteins link JNK to Hippo signaling. Sci Signal. 2013;6(292):ra81. doi: 10.1126/scisignal.2004324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen CL, Schroeder MC, Kango-Singh M, Tao C, Halder G. Tumor suppression by cell competition through regulation of the Hippo pathway. Proc Natl Acad Sci USA. 2012;109(2):484–489. doi: 10.1073/pnas.1113882109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doggett K, Grusche FA, Richardson HE, Brumby AM. Loss of the Drosophila cell polarity regulator Scribbled promotes epithelial tissue overgrowth and cooperation with oncogenic Ras-Raf through impaired Hippo pathway signaling. BMC Dev Biol. 2011;11:57. doi: 10.1186/1471-213X-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agnès F, Suzanne M, Noselli S. The Drosophila JNK pathway controls the morphogenesis of imaginal discs during metamorphosis. Development. 1999;126(23):5453–5462. doi: 10.1242/dev.126.23.5453. [DOI] [PubMed] [Google Scholar]
- 18.Ziosi M, et al. dMyc functions downstream of Yorkie to promote the supercompetitive behavior of hippo pathway mutant cells. PLoS Genet. 2010;6(9):e1001140. doi: 10.1371/journal.pgen.1001140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uhlirova M, Bohmann D. JNK- and Fos-regulated Mmp1 expression cooperates with Ras to induce invasive tumors in Drosophila. EMBO J. 2006;25(22):5294–5304. doi: 10.1038/sj.emboj.7601401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Page-McCaw A, Serano J, Santé JM, Rubin GM. Drosophila matrix metalloproteinases are required for tissue remodeling, but not embryonic development. Dev Cell. 2003;4(1):95–106. doi: 10.1016/s1534-5807(02)00400-8. [DOI] [PubMed] [Google Scholar]
- 21.Tumaneng K, Russell RC, Guan KL. Organ size control by Hippo and TOR pathways. Curr Biol. 2012;22(9):R368–R379. doi: 10.1016/j.cub.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho E, et al. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38(10):1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- 23.Thompson BJ, Cohen SM. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126(4):767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 24.Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14(3):388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 25.Ryoo HD, Gorenc T, Steller H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev Cell. 2004;7(4):491–501. doi: 10.1016/j.devcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 26.Coso OA, et al. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81(7):1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 27.Minden A, Lin A, Claret FX, Abo A, Karin M. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81(7):1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 28.Neisch AL, Speck O, Stronach B, Fehon RG. Rho1 regulates apoptosis via activation of the JNK signaling pathway at the plasma membrane. J Cell Biol. 2010;189(2):311–323. doi: 10.1083/jcb.200912010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warner SJ, Yashiro H, Longmore GD. The Cdc42/Par6/aPKC polarity complex regulates apoptosis-induced compensatory proliferation in epithelia. Curr Biol. 2010;20(8):677–686. doi: 10.1016/j.cub.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, et al. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14(3):377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh H, Irvine KD. Cooperative regulation of growth by Yorkie and Mad through bantam. Dev Cell. 2011;20(1):109–122. doi: 10.1016/j.devcel.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mauviel A, Nallet-Staub F, Varelas X. Integrating developmental signals: A Hippo in the (path)way. Oncogene. 2012;31(14):1743–1756. doi: 10.1038/onc.2011.363. [DOI] [PubMed] [Google Scholar]
- 33.Hong W, Guan KL. The YAP and TAZ transcription co-activators: Key downstream effectors of the mammalian Hippo pathway. Semin Cell Dev Biol. 2012;23(7):785–793. doi: 10.1016/j.semcdb.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu FX, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150(4):780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mo JS, Yu FX, Gong R, Brown JH, Guan KL. Regulation of the Hippo-YAP pathway by protease-activated receptors (PARs) Genes Dev. 2012;26(19):2138–2143. doi: 10.1101/gad.197582.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rauskolb C, Sun S, Sun G, Pan Y, Irvine KD. Cytoskeletal tension inhibits Hippo signaling through an Ajuba-Warts complex. Cell. 2014;158(1):143–156. doi: 10.1016/j.cell.2014.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu M, Pastor-Pareja JC, Xu T. Interaction between Ras(V12) and scribbled clones induces tumour growth and invasion. Nature. 2010;463(7280):545–548. doi: 10.1038/nature08702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rudrapatna VA, Cagan RL, Das TK. Drosophila cancer models. Dev Dyn. 2012;241(1):107–118. doi: 10.1002/dvdy.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9(8):537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 40.Brumby AM, et al. Identification of novel Ras-cooperating oncogenes in Drosophila melanogaster: A RhoGEF/Rho-family/JNK pathway is a central driver of tumorigenesis. Genetics. 2011;188(1):105–125. doi: 10.1534/genetics.111.127910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue L, et al. Tumor suppressor CYLD regulates JNK-induced cell death in Drosophila. Dev Cell. 2007;13(3):446–454. doi: 10.1016/j.devcel.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 42.Boulanger A, Farge M, Ramanoudjame C, Wharton K, Dura JM. Drosophila motor neuron retraction during metamorphosis is mediated by inputs from TGF-β/BMP signaling and orphan nuclear receptors. PLoS ONE. 2012;7(7):e40255. doi: 10.1371/journal.pone.0040255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: The Drosophila lats gene encodes a putative protein kinase. Development. 1995;121(4):1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- 44.O’Neill JW, Bier E. Double-label in situ hybridization using biotin and digoxigenin-tagged RNA probes. Biotechniques. 1994;17(5):870. , 874–875. [PubMed] [Google Scholar]
- 45.Xue L, Noll M. Drosophila female sexual behavior induced by sterile males showing copulation complementation. Proc Natl Acad Sci USA. 2000;97(7):3272–3275. doi: 10.1073/pnas.060018897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.